Abstract
Intramembrane proteolysis is a new paradigm in biology that controls signaling events throughout evolution. Hydrolysis of peptide bonds is thought to occur within the normally hydrophobic membrane environment, but insights into this unusual activity have been lacking because of difficulty in recapitulating activity in vitro. We have reconstituted intramembrane proteolysis with a pure recombinant substrate and rhomboid proteins in both detergent micelles and artificial membrane environments. Rhomboid proteins from diverse organisms including two model bacteria, a pathogen, an extremophile, and an animal were robustly active in pure form, proving that rhomboids are a new class of enzymes and do not require cofactors to catalyze intramembrane proteolysis. Rhomboid proteins directly recognized their substrates in vitro by the top of the substrate transmembrane domain, displaying specificity apparently reciprocal to that of γ-secretase, the only other activity known to cleave type-I transmembrane domains. Rhomboid proteases represent a different evolutionary path to a serine protease mechanism and exhibited an inhibitor profile unlike other serine proteases. Intriguingly, activity was dramatically modulated by different membrane phospholipid environments, suggesting a mechanism for regulating these proteases. This analysis promises to help reveal the biochemical mechanisms and biological roles of this most widely conserved membrane protein family.
Keywords: cell signaling, presenilin, signal peptide peptidase, site-2 protease, regulated intramembrane proteolysis
Intramembrane proteases are a group of multipass membrane proteins that are thought to catalyze the cleavage of transmembrane domains (TMDs) within the lipid bilayer (1). This activity liberates functional domains that are otherwise inert when anchored to the membrane and thus serves as a regulatory mechanism for rapidly controlling many diverse signaling processes. Intramembrane proteolysis is conserved throughout evolution, suggesting that it may have evolved early, and has subsequently been adapted for controlling many biological processes (2).
Three classes of intramembrane proteases have been identified through the study of different biological processes. Site-2 proteases (S2Ps) are a class of membrane-embedded metalloproteases that were first identified as regulators of cholesterol and fatty acid biosynthesis in metazoans through the regulated liberation of sterol-regulatory element-binding proteins (3). Cleavage of ATF-6 by S2P also triggers the unfolded protein response in metazoa (4), and this process appears to be conserved in bacteria (5, 6). Bacterial S2P homologs also regulate developmental processes, including sporulation in Bacillus subtilis (7) and sexual behavior in Enterococcus (8).
Analysis of Alzheimer's disease focused attention on γ-secretase, an aspartyl intramembrane protease that generates Aβ, a major component of senile plaques in the brain. Subsequent analysis revealed that γ-secretase has broad specificity and cleaves many type-I membrane proteins in the cell (9). The catalytic component of γ-secretase is thought to be presenilin, a protein with eight membrane-spanning domains (10). A related aspartyl protease, signal peptide peptidase, was discovered to be responsible for cleaving signal peptides once they are liberated from precursor proteins (11). Some of these released fragments are bioactive peptides that regulate processes such as immune surveillance (12).
Study of cell signaling during development led to the identification of rhomboid as a key regulator of EGF receptor signaling (13). Rhomboid is responsible for cleaving Spitz, the main ligand of the Drosophila EGF receptor pathway, and in this way functions to initiate signaling. Rhomboid was proposed to function directly as a serine protease that uses a catalytic triad composed of a serine, histidine, and asparagine contributed by different TMDs (14). The active site is thus embedded some shallow distance into the membrane from the extracellular side, presumably to facilitate factor release to the outside of the cell (2).
Rhomboids are nearly ubiquitous (15), but their biological functions are only beginning to be studied. Although little is known about rhomboid function in vertebrates, its signaling function appears to be conserved in the human pathogen Providencia stuartii, where the rhomboid homolog AarA is required for sending an unidentified quorum-sensing signal (16). AarA, like many rhomboids from bacteria and eukaryotes, can cleave Spitz despite significant sequence divergence (17) and even use the same mechanism to achieve substrate specificity (18). In fact, Drosophila rhomboid-1 and AarA can functionally substitute for each other when expressed in Providencia and Drosophila, respectively (19). This process, to our knowledge, is the first signaling mechanism known to be conserved between prokaryotes and eukaryotes.
Despite their central roles in many biological and pathological processes, very little is known about how intramembrane proteases function mechanistically. Rhomboid intramembrane proteolysis has been studied only in cells (14). Although cell-free assays exist for γ-secretase (20) and signal peptide peptidase proteolysis (11), a major shortcoming has been the inability to study intramembrane proteolysis biochemically with individual components. The best understood intramembrane protease is γ-secretase, the catalytic component of which is thought to be presenilin (10). Accordingly, transition-state inhibitors bind directly to presenilin (21, 22), but the proteolytic activity absolutely requires a complex of four membrane proteins (23–25). Recent purification of γ-secretase has further underscored the importance of maintaining the whole complex for catalysis (26). Thus, although there is evidence that S2P, presenilin, signal peptide peptidase, and rhomboid are themselves proteases, biochemical reconstitution of intramembrane proteolysis has proven difficult with these proteins.
We have reconstituted intramembrane proteolysis in vitro with five pure rhomboid proteins that were produced recombinantly in Escherichia coli. These rhomboids have limited sequence identity with each other (17), being derived from very diverse organisms, but share biochemical characteristics including activity and substrate specificity (17, 18). Rhomboid activity in vitro recapitulated all of the known properties of this form of intramembrane proteolysis and further revealed unexpected biochemical characteristics of this nearly ubiquitous family of proteases.
Materials and Methods
GST-Rhomboid Expression in Bacteria. Rhomboid genes (14, 17, 27) were cloned into the pGEX-6P-1 for inducible expression as N-terminal GST fusions in E. coli strain C43(DE3). Cultures were grown in LB plus 100 μg/ml ampicillin, shaken at 37°C to an OD600 of 0.5–0.8, induced with 50 μM IPTG, and grown for 12–16 h at 22°C. Coexpression of pET12a-GFP-Spitz-Flag with GST-rhomboids was accomplished by growing cultures in LB plus ampicillin and kanamycin followed by coinduction with 0.25 mM IPTG.
Rhomboid Purification. Cells from 750-ml cultures were lysed in a French press at 1,000–1,100 psi, insoluble material was removed by centrifugation at 10,000 × g for 20 min, and bacterial membranes were isolated from supernatants by ultracentrifugation at 200,000 × g for 1 h. The membranes were washed in PBS and pelleted again. GST-rhomboids were solubilized from purified membranes by rocking at 4°C for 1 h in 1% dodecyl-β-d-maltoside (DDM) (or other detergents). The solutions were centrifuged at 200,000 × g for 1 h, and the resulting supernatant was defined as solubilized GST-rhomboid.
GST-rhomboids were purified by incubating with ≈2 ml of glutathione-Sepharose for 2 h at room temperature. The resin was washed with ≈10-column volumes of PBS plus 0.25% DDM, and the bound rhomboid was eluted by PreScission (Amersham Pharmacia Biotech) protease cleavage according to the manufacturer's instructions. The eluted rhomboid was passed again through ≈0.5 ml of glutathione-Sepharose to remove unbound GST-PreScission.
C100Spi-Flag Production. C100Spi-Flag was generated by replacing the first seven residues of the APP TMD in the pET21a-C100Flag vector with those from Spitz (ASIASGA) using QuikChange mutagenesis (Stratagene) and verified by sequencing. Substrates were produced as described in ref. 28. Briefly, a 500-ml culture of BL21(DE3) harboring the pET21a-C100-Flag plasmid was grown to an OD600 of 1.0 and induced with 1 mM IPTG for 2–3 h at 37°C. Cells were lysed in PBS plus 1% Triton X-100 buffer (containing a protease inhibitor mixture) in a French press. Insoluble material was removed by centrifugation, and the supernatant was incubated with 2 ml of anti-Flag agarose for 2 h. The resin was collected and washed with 10-column volumes of PBS plus 1% Triton X-100, and C100Spi-Flag was eluted with acidic glycine buffer (28).
In Vitro Cleavage Assays. Activity assays were performed with 1–100 ng of pure rhomboid in 50 mM Tris, pH 7.5/150 mM NaCl/0.25% DDM for 2 h at 37°C (or 25°C for YqgP and DmRho4). Cleavage was initiated by adding 0.4 μl of C100Spi-Flag substrate (≈750 ng) to 20-μl reactions and stopped with 7 μl of 4× SDS sample buffer and heating for 10 min at 65°C. Five microliters was resolved by SDS/PAGE and revealed by Western blot analysis by using anti-Flag antibodies and enhanced chemiluminescence (Amersham Pharmacia Biotech). γ-Secretase solubilized from HeLa membranes with 1% 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO) was assayed as described in ref. 28.
Protease inhibitors were purchased from Calbiochem and prepared before use. Phospholipids from natural sources (Avanti Polar Lipids) were prepared fresh by removing the chloroform solvent with a rotary evaporator and high-pressure vacuum. The resulting “lipid films” were hydrated in 50 mM Tris, pH 7.5/150 mM NaCl/0.25% DDM to a final concentration of 10 mg/ml and sonicated to produce small unilamellar vesicles. Rhomboid proteins were reconstituted into membranes by rapid dilution of the detergent to ≈0.05% in the presence of lipids. Under these conditions, most of rhomboid was reconstituted into membranes as quantified by ultracentrifugation.
Results
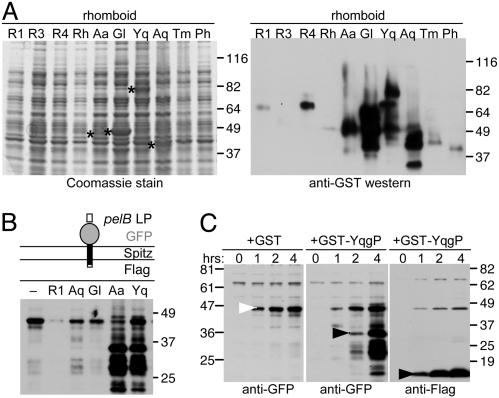
Several Rhomboids Are Active When Expressed in Bacteria. As a first step in reconstituting rhomboid catalysis in vitro, we tested whether rhomboids expressed in E. coli were active. We analyzed 10 different rhomboids as inducible N-terminal fusion proteins with GST, because the rhomboid N terminus is cytoplasmic and activity is often hindered by C-terminal fusions (29). Analysis of whole-cell extracts revealed that several rhomboids can be expressed to high levels in bacteria (Fig. 1A).
Fig. 1.
Several rhomboids can be expressed at high levels as active proteases in E. coli.(A) Ten prototypic rhomboids from various organisms were screened for expression as inducible GST fusions (overexpressed rhomboids are indicated by asterisks in Left) and by anti-GST Western blot analysis. The rhomboids tested were Drosophila Rho-1 (R1), Rho-3 (R3), Rho-4 (R4), and human RHBDL-2 (Rh); bacterial rhomboids AarA from P. stuartii (Aa), GlpG from E. coli (Gl), and YqgP from B. subtilis (Yq); and rhomboids from thermophilic microbes A. aeolicus (Aq), Thermotoga maritima (Tm), and Pyrococcus horikoshii (Ph). (B) Several GST-rhomboids were active in bacteria. The substrate, depicted with its ectodomain up and two horizontal lines denoting the membrane bilayer, was detected by anti-GFP Western blot analysis of total bacterial protein 4 hours after coinduction. Cleavage products were detected with YqgP (Yq) and AarA (Aa) (further degradation is likely the result of ectodomain release into the protease-rich periplasm). (C) Time course of substrate cleavage by GST-YqgP or GST alone as a control (in hours after coinduction). The uncleaved substrate is indicated by a white arrow, and the cleaved products (N-terminal anti-GFP and C-terminal anti-Flag) are highlighted by black arrows.
Cleavage of Spitz in bacteria upon coexpression of GST-rhomboids suggested that at least several rhomboids were active in bacteria. The substrate designed for this purpose was a chimeric protein composed of the bacterial pelB leader peptide (to target the protein to the membrane), GFP as the extracellular ectodomain, the juxtamembrane–transmembrane–cytosolic residues (123–230) of Spitz, and a C-terminal flag epitope (Fig. 1B). Coexpression of this substrate with B. subtilis YqgP and Providencia AarA resulted in exceptionally strong substrate cleavage in E. coli (Fig. 1 B and C).
Substrate Design. To study rhomboid catalysis in vitro, it was essential to include more substrate than protease in the cleavage reaction to avoid spurious results. Although the Spitz chimera was cleaved efficiently by rhomboids in bacteria, this protein itself was poorly expressed and thus not suitable to serve as a recombinant substrate in vitro. Instead, we adapted C100-Flag, a recombinant C-terminal fragment of APP that expresses to very high levels and has been used as a substrate for γ-secretase (20). Cleavage was achieved by replacing the first seven residues of the APP TMD with those from Spitz, which we have shown to be necessary and sufficient to convert proteins into substrates for rhomboids (18). Cleavage in a mammalian cell-based transfection assay confirmed that C100Spi-Flag, but not C100-Flag, was an efficient substrate for all rhomboids tested (data not shown).
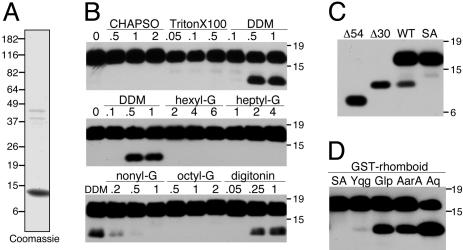
Rhomboid Catalyzes Intramembrane Proteolysis in Detergent Micelles. To develop an in vitro assay for rhomboid intramembrane proteolysis, GST-rhomboids were solubilized from purified bacterial membranes with eight different detergents at three concentrations each. The rhomboid YqgP from B. subtilis, which shares activity and specificity characteristics with other Spitz-cleaving rhomboids (17, 18), was first chosen for characterization because it was expressed at high levels and was active in bacteria. Solubilization trials focused on DDM, 3-[(3-cholamidopropyl)-dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO), Triton X-100, digitonin, and the alkyl glucoside series hexyl-β-d-glucopyranoside, heptyl-β-d-glucopyranoside, octyl-β-d-glucopyranoside, and nonyl-β-d-glucopyranoside.
Solubilized GST-YqgP was incubated with C100Spi-Flag that had been purified by using Flag affinity chromatography (Fig. 2A), and the cleavage products were revealed by anti-Flag Western blot analysis. Under these conditions, a cleaved product was detected with GST-YqgP solubilized in DDM and digitonin and, to a lesser extent, with nonyl-β-d-glucopyranoside, suggesting that nonionic glucoside detergents with long alkyl chains were best for solubilizing active rhomboid (Fig. 2B). The cleavage product resulted from intramembrane proteolysis by GST-YqgP because it was absolutely dependent on the presence of the active serine in GST-YqgP (Fig. 2C). Moreover, comparing the size of the cleaved product to engineered C100Spi-Flag deletions as standards revealed that cleavage resulted from intramembrane proteolysis in the top portion of the TMD. This site is expected for only rhomboid cleavage.
Fig. 2.
Rhomboids are active in detergent micelles. (A) Purified C100Spi-Flag substrate was analyzed by SDS/PAGE. Note the appearance of aggregated species ≈40 kDa, which is typical for this hydrophobic protein. (B) GST-YqgP was solubilized from bacterial membranes in detergents at or severalfold above their critical micelle concentration (% wt/vol above each lane) and subsequently tested for C100Spi-Flag cleavage in vitro.(C) A cleaved product was detected with WT but not with a serine-to-alanine mutant (SA) of GST-YqgP. This product was larger than deletion 54, which left residues 54–100 (corresponding to a cleavage at the bottom of the TMD), but about the size of deletion 30, which left residues 30–100 (corresponding to a cleavage near the top of the TMD). (D) In addition to GST-YqgP, robust rhomboid activity was recovered in vitro with GST-AarA, GST-GlpG, and GST-AqRho (Aq) solubilized in 1% DDM.
Using the approach developed for GST-YqgP, we were also able to detect strong in vitro activity with all GST-rhomboids that expressed well in bacteria, including GlpG from E. coli, AarA from P. stuartii, and AqRho from the extreme thermophile Aquifex aeolicus, when solubilized from membranes with 1% DDM (Fig. 2D). Taken together, these observations indicate that diverse rhomboids catalyze intramembrane proteolysis in vitro and could serve as valuable prototypes for studying the rhomboid protease family.
Requirements for Rhomboid Activity in Vitro. Development of an in vitro activity assay allowed us to examine the requirements for rhomboid catalysis. Rhomboid proteolysis in vitro was robust and did not depend on specific assay conditions (Fig. 3). Intramembrane proteolysis did not require ATP and was not enhanced when 5 mM ATP was added. Different rhomboids exhibited higher activity at specific temperatures, and these optima correlated with their physiological temperature ranges; YqgP, which comes from a soil bacterium, was more active at 25°C than at 37°C, whereas GlpG from E. coli was much more active at 37°C. Activity of most rhomboids was not affected by salt concentrations from 0 to 250 mM or at a pH of 6–8.5. The condition that had the most impact on catalysis was the type and concentration of detergent and membrane lipids in the cleavage reaction. For most rhomboids, these effects were reciprocal: higher concentrations of DDM reduced intramembrane proteolysis, whereas addition of exogenous phospholipids stimulated activity (Fig. 3).
Fig. 3.
Requirements for rhomboid activity in vitro. Rhomboids GST-YqgP and GST-GlpG solubilized in 1% DDM from bacterial membranes were tested for C100Spi-Flag cleavage under various conditions. All reactions were carried out at 37°C in 0.25% DDM unless otherwise noted. ATP was added at 5 mM, DTT was added at 1 mM, and phospholipids were added at ≈0.1% (PL).
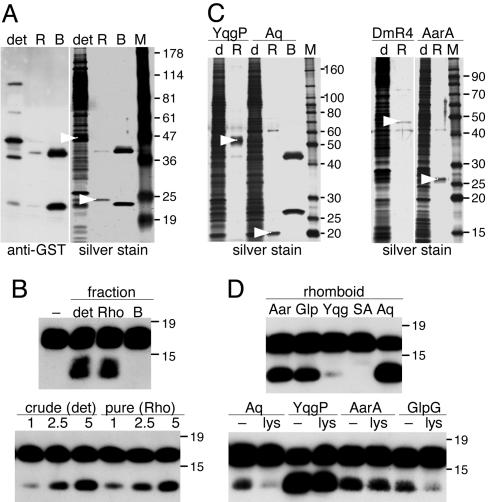
Pure Rhomboid Proteins Catalyze Intramembrane Proteolysis. A key unresolved issue is whether rhomboids are themselves proteases or whether they require auxiliary factors to achieve intramembrane proteolysis. To address this biochemically, we purified GST-rhomboids using glutathione-Sepharose affinity chromatography, removing the GST tag by on-column cleavage (Fig. 4 A and C). Importantly, the conditions used to solubilize and purify rhomboids are harsh and have been observed to dissociate γ-secretase components during their purification (30). Thus, we expected that, if rhomboid required other cofactors for proteolysis, the conditions used for its purification would result in their dissociation. All pure fractions retained very robust proteolytic activity, and even Drosophila Rho-4, which expressed at low levels, was active alone in pure form but required addition of lipids (see Fig. 6A). The amount of activity in crude and pure fractions directly correlated only with the amount of rhomboid protein present and not other factors (Fig. 4B). Furthermore, addition of E. coli proteins to the pure rhomboid fractions did not enhance activity, suggesting that a limiting cofactor was not being supplied by the host cells (Fig. 4D). These results indicate that rhomboid proteins themselves catalyze intramembrane proteolysis without requiring cofactors.
Fig. 4.
Purified rhomboids catalyze intramembrane proteolysis. (A) GST-GlpG was purified by glutathione-Sepharose affinity chromatography and released by on-column cleavage with GST-PreScission protease. The solubilized membrane fraction (det), eluted rhomboid (R), and material remaining on the resin after elution (B) were analyzed. The eluted rhomboid (arrowheads) was pure, with a small amount of unbound GST-PreScission as a contaminant (also in C). The material remaining on the resin was GST (lower band) and GST-PreScission (upper band). (B) Solubilized GST-GlpG and pure GlpG catalyzed robust intramembrane proteolysis, whereas the material bound to the resin after GlpG elution (including GST-PreScission) had no activity. Standardizing the amount of GlpG in the solubilized and pure fractions (Lower) resulted in the same amount of proteolytic activity (numbers represent the relative amounts of enzyme). (C) YqgP, AarA, AqRho, and Drosophila Rho-4 were purified and analyzed under the same conditions (rhomboids are highlighted with an arrowhead). (D) All of the purified rhomboids retained robust proteolytic activity in pure form (low activity of YqgP was due to delipidation). GST-YqgP, with its active-site serine mutated to alanine, was used as a control (SA). Importantly, addition of DDM-solubilized cell lysates to purified rhomboids (Lower) did not enhance proteolytic activity.
Fig. 6.
Effect of different membrane phospholipid environments on rhomboid intramembrane proteolysis in vitro.(A) Activity of purified rhomboids in DDM micelles (DDM) was compared to activity in various phospholipid environments: lipid extracts from E. coli (coli), brain (Br), heart (H) and liver (Liv) and defined phospholipids cardiolipin (CL), PE, phosphatidylcholine (PC), sphingomyelin (SM), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylglycerol (PG) were used at their optimal concentration (0.1%). Note that ly/PE is a 1:1 mixture of lysoPE and PE, which readily form bilayers, unlike PE alone. (B) Various lipid concentrations were tested for their effect on rhomboid intramembrane proteolysis; the effects of E. coli and brain extracts on purified YqgP are shown as an example. (C) Unlike for rhomboids, a liver lipid extract and phosphatidylcholine had the strongest stimulatory effect on γ-secretase ε cleavage of C100Flag.
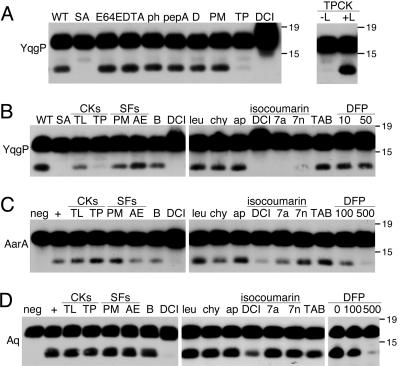
Rhomboid Is Insensitive to Most Protease Inhibitors. Rhomboid is the first serine protease of its kind, and it represents a completely different evolutionary path to a serine protease mechanism. A direct in vitro activity assay allowed us to analyze rhomboid sensitivity to different classes of protease inhibitors, many of which could not be tested previously because of inability to penetrate cells or cytotoxicity. Consistent with being a serine protease, rhomboid activity was insensitive to broad-spectrum metalloprotease (EDTA, o-phenanthroline), cysteine protease (E64), and aspartyl protease (pepstatin A, DAPT) inhibitors (Fig. 5A).
Fig. 5.
Sensitivity of rhomboid activity in vitro to protease inhibitors. (A) Cleavage of C100Spi-Flag by GST-YqgP was insensitive to the cysteine protease inhibitor E64 (20 μM), metalloprotease inhibitors EDTA (5 mM) and o-phenanthroline (1 mM; ph), and 10 μM aspartyl protease inhibitors pepstatin A and DAPT. YqgP activity was inhibited by 100 μM TPCK (TP) and DCI but was insensitive to 1 mM PMSF. Note that, although TPCK inhibited YqgP in DDM micelles, it became completely ineffective in the presence of phospholipids (Right, +L). (B–D) Rhomboids YqgP, AarA, and AqRho were tested for sensitivity to mechanistically diverse serine protease inhibitors. Chloromethylketones (CKs) Nα-p-tosyl-l-lysine chloromethyl ketone (TL) and TPCK (TP) were used at 100 μM, sulfonyl fluorides (SFs) PMSF (PM) and 4-(2-aminoethyl)benzenesulfonyl fluoride (AE) and benzamidine (B) were tested at 1 mM, DCI, leupeptin (leu) and chymostatin (chy) were tested at 100 μM, and aprotinin (ap) was tested at 2 μg/ml. DFP, diisopropylfluorophosphonate (in μM). TAB indicates the Roche complete protease inhibitor tablet. The relative efficacy of isocoumarins DCI, 7-amino-4-chloro-3-methoxyethoxy-isocoumarin (7a), and 7-nitro-4-chloro-3-methoxyethoxy-isocoumarin (7n) was compared at 25 μM. Rhomboids were tested at reduced concentrations and by preincubating with inhibitors to ensure that even weak inhibition could be detected.
Dichloroisocoumarin (DCI) was the only serine protease inhibitor that potently inhibited intramembrane proteolysis by all rhomboids tested (Fig. 5). Importantly, this inhibition was specific because it occurred at or below 100 μM and because other isocoumarin derivatives (31) had different potencies against rhomboids, with the 7-nitro-4-chloro-3-methoxyethoxy-isocoumarin being less potent than either the 7-amino-4-chloro-3-methoxyethoxyisocoumarin or DCI. Cleavage by YqgP was also sensitive to N-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK), but only in detergent micelles (Fig. 5A), and was specific in that Nα-p-tosyl-l-lysine chloromethyl ketone, a chloromethylketone similar to TPCK but with a lysine instead of a phenylalanine, did not inhibit YqgP. TPCK and DCI are the only two compounds known to inhibit Spitz cleavage in cell culture (14).
Other serine protease inhibitors were ineffective against rhomboid activity, including sulfonyl fluorides PMSF and 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), even when they were preincubated with the enzyme at saturating concentrations. Inhibition could be forced with organophosphonates (diisopropylfluorophosphonate) only by preincubating the enzymes with concentrations approaching 1 mM (Fig. 5 C and D), but these high concentrations also had effects on γ-secretase activity (data not shown). Rhomboid activity was also insensitive to other commonly used peptidic inhibitors such as aprotinin, leupeptin, and chymostatin and even a complete protease inhibitor mixture (Roche). Therefore, rhomboid activity is insensitive to most serine protease inhibitor classes, unlike other serine proteases.
Membrane Lipids Modulate Rhomboid Catalysis in Vitro. Rhomboids are unlike all other known serine proteases, being multispanning membrane proteins with their active site embedded within the membrane bilayer. Interestingly, different membrane lipids had markedly different effects on rhomboid activity. Biological membranes are composed of hundreds of different lipids; therefore, we first tested the effect of total lipid extracts from four different cell/tissue types: E. coli, brain, liver, and heart. YqgP, AarA, and Rho-4 had the strongest dependence on lipids for activity. Whereas membrane lipids from liver and heart had mild effects on YqgP activity, lipids from brain and E. coli stimulated activity by as much as 100-fold (Fig. 6). Similarly, lipids from brain strongly stimulated Drosophila Rho-4. Conversely, these same lipids reduced activity of GlpG, with the native E. coli lipid extract almost completely inhibiting GlpG activity. AqRho and γ-secretase were only mildly affected by these lipids, whereas AarA was strongly stimulated by all lipids tested. These effects were specific to rhomboid catalysis and not due to other differences, like effects on the substrate, because the same lipid extracts had differential effects on different rhomboids and on γ-secretase activity.
As a first attempt to define the basis of specificity in the dramatic stimulation of rhomboid activity, we compared the effects of defined membrane lipids. Phosphatidylethanolamine (PE) constitutes ≈70% of E. coli membranes, but it had mild effects on catalysis by prokaryotic rhomboids. YqgP catalysis was stimulated by phosphatidylglycerol and phosphatidylserine, but the highest stimulation occurred with choline-containing lipids phosphatidylcholine and sphingomyelin, although these lipids are rare in bacterial membranes. In contrast, AarA was strongly stimulated by all lipids and, in particular, cardiolipin, whereas cardiolipin inhibited cleavage by other rhomboids. Drosophila Rho-4 was strongly stimulated only by PE and phosphatidylinositol. Conversely, AqRho activity was inhibited by anionic lipids, whereas GlpG was inhibited by most lipids. These observations indicate that different lipid environments can have both stimulatory and inhibitory effects on rhomboid activity and raise the intriguing possibility that rhomboid proteolysis may be regulated by different membrane environments.
Substrate Specificity Is Determined by Rhomboid Directly. Rhomboid proteases initiate cell signaling events by activating ligands and thus must be specific for their targets to achieve their biological roles. The first seven residues of the Spitz TMD are necessary and sufficient for cleavage by rhomboids from diverse organisms, and the identity of the residues suggested that rhomboids require a destabilized TMD helix (18). However, these experiments were performed in cells, and indirect effects could not be excluded.
To determine whether rhomboids themselves directly recognize their substrates, we examined the ability of rhomboids and γ-secretase as a control to cleave C100-Flag versus C100Spi-Flag. These two substrates were ≈95% identical, the sole difference being that the first seven residues of the APP TMD were replaced with those from Spitz in C100Spi-Flag. All pure rhomboids cleaved C100Spi-Flag very efficiently but could not cleave C100-Flag (Fig. 7). Conversely, γ-secretase could cleave C100-Flag but was dramatically less active against C100Spi-Flag. Thus, because the specificity for substrates was directly recapitulated by all purified rhomboids in vitro, rhomboids themselves are directly responsible for recognizing their substrates.
Fig. 7.
Purified rhomboid proteases display substrate specificity in vitro. Cleavage of C100Spi-Flag versus C100-Flag was assessed with purified rhomboid proteases, GST-YqgP-SA as a negative control, and γ-secretase (γ). The APP substrates are depicted in white, with the seven TMD residues of Spitz in black. Note that YqgP and DmRho-4 cleavage was tested in the presence of a brain lipid extract because these purified rhomboids displayed little activity without lipids.
Discussion
Intramembrane proteolysis is a key regulatory mechanism that controls many biological processes and is conserved throughout evolution (1, 2). At the heart of this paradigm are a class of membrane proteins that are thought to acts as proteases, but how they function has remained uncertain because activity could not been recapitulated in vitro with defined components.
We have reconstituted intramembrane proteolysis in vitro with a pure substrate and five pure rhomboids, establishing that rhomboids themselves are a new class of enzymes and, in contrast to γ-secretase (24), act alone. Therefore, intramembrane proteolysis in general does not require large membrane complexes. Together with our analysis of five different pure rhomboids, the recent analysis of the E. coli S2P homolog RseP in pure form (32) further indicates that the requirement for cofactors for intramembrane proteolysis may be the exception rather than the rule. Importantly, we found that Drosophila Rho-4 was also active in pure form, establishing for the first time, to our knowledge, that a eukaryotic intramembrane protease also does not require cofactors for activity.
The ability to manipulate the conditions in vitro revealed that proteolytic activity is strongly affected by specific membrane lipid environments. This fact has not been appreciated previously, but it is likely to be important in considering the function and regulation of these enzymes in cells. For example, many mammalian rhomboids are present on the cell surface (33), which is enriched in different phospholipid species compared to internal organelles (34). Regional heterogeneity in membrane lipid composition has also been observed in the plasma membranes of both metazoan and microbial cells. Thus, rhomboids could become activated only when they reach the appropriate organelle or membrane domain by virtue of the altered lipid environment, although this hypothesis awaits further study.
The assay that we have developed is likely to be a powerful tool for further deciphering the mechanistic function of rhomboid proteases in detail, because it uses defined components; pure rhomboids and a pure recombinant substrate were assayed in detergent micelles or reconstituted into defined membrane lipids. Moreover, we used 20–1,000 times more substrate than rhomboid protein in these assays, and extended incubation times could drive cleavage to completion. Rhomboids thus truly act catalytically in our assays, making this system well suited to a precise enzymological analysis of rhomboid catalysis. Moreover, proteolytic activity and substrate specificity in vitro were also conserved in the rhomboid from Aquifex, which had not previously shown activity against Spitz in other assays (17). Therefore, our in vitro analysis now indicates that the biochemical properties of rhomboids may be more conserved than first anticipated.
An additional advantage of this system is that γ-secretase can be studied in parallel, which should reveal the similarities and differences between intramembrane proteases. Our current analysis has revealed that rhomboids alone can discriminate between highly similar (i.e., 95% identical) proteins in vitro, and this specificity is determined by the N-terminal-most residues of the substrate TMD. Surprisingly, these residues hindered cleavage of APP by γ-secretase, the only other activity known to catalyze intramembrane cleavage of type-I membrane proteins. Because γ-secretase is thought to be able to cleave any type-I TMD (35), this, to our knowledge, is the first demonstration that it may be more dependent on a helical TMD for cleavage than was previously thought and further suggests that its specificity is reciprocal to that of rhomboid proteases.
Although rhomboids are serine proteases, it is striking that most canonical serine protease inhibitors proved ineffective against their activity. Although the paucity of prototypes will make developing rhomboid inhibitors challenging, this observation is intriguing mechanistically because rhomboids followed a different evolutionary path to a serine protease mechanism. In particular, inhibitors that directly attack the active-site serine were ineffective at inhibiting rhomboid (isocoumarins and chloromethylketones inhibit proteases in whole or in part by acylating the histidine). This disparity might suggest that the catalytic triad in rhomboid is not preformed without substrate binding; a conformation change that has been proposed to result in substrate internalization for γ-secretase (28) might also induce formation of the catalytic triad in rhomboid proteases. Although a structural analysis will be required to address this issue directly and to prove whether rhomboids do indeed use a classical serine protease mechanism, this approach should be feasible with the rhomboid proteases analyzed, because they express to high levels in bacteria and can be purified in active form without the need for other cofactors. Considering the widespread nature of these key regulatory enzymes and their expected roles in pathogenic events, understanding rhomboids promises to have important implications for understanding development and disease.
Acknowledgments
We are grateful to Rosanna Baker for helpful comments on the manuscript. 7-nitro-4-chloro-3-methoxyethoxy-isocoumarin and 7-amino-4-chloro-3-methoxyethoxy-isocoumarin were a kind gift from Dr. Frédéric Bihel (Harvard Medical School and Brigham and Women's Hospital). This work was supported by a grant from the International Human Frontier Science Program Organization (to S.U.) and National Institutes of Health Grant AG17574 (to M.S.W.).
Author contributions: S.U. designed research, performed research, analyzed data, and wrote the paper; and S.U. and M.S.W. contributed new reagents/analytic tools.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TMD, transmembrane domain; PE, phosphatidylethanolamine; S2P, site-2 protease; DDM, dodecyl-β-d-maltoside; DCI, dichloroisocoumarin; TPCK, N-p-tosyl-l-phenylalanine chloromethyl ketone.
References
- 1.Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. (2000) Cell 100, 391–398. [DOI] [PubMed] [Google Scholar]
- 2.Urban, S. & Freeman, M. (2002) Curr. Opin. Genet. Dev. 12, 512–518. [DOI] [PubMed] [Google Scholar]
- 3.Rawson, R. B., Zelenski, N. G., Nijhawan, D., Ye, J., Sakai, J., Hasan, M. T., Chang, T. Y., Brown, M. S. & Goldstein, J. L. (1997) Mol. Cell 1, 47–57. [DOI] [PubMed] [Google Scholar]
- 4.Ye, J., Rawson, R. B., Komuro, R., Chen, X., Dave, U. P., Prywes, R., Brown, M. S. & Goldstein, J. L. (2000) Mol. Cell 6, 1355–1364. [DOI] [PubMed] [Google Scholar]
- 5.Alba, B. M., Leeds, J. A., Onufryk, C., Lu, C. Z. & Gross, C. A. (2002) Genes Dev. 16, 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehara, K., Ito, K. & Akiyama, Y. (2002) Genes Dev. 16, 2147–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudner, D. Z., Fawcett, P. & Losick, R. (1999) Proc. Natl. Acad. Sci. USA 96, 14765–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An, F. Y., Sulavik, M. C. & Clewell, D. B. (1999) J. Bacteriol. 181, 5915–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Struhl, G. & Adachi, A. (2000) Mol. Cell 6, 625–636. [DOI] [PubMed] [Google Scholar]
- 10.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T. & Selkoe, D. J. (1999) Nature 398, 513–517. [DOI] [PubMed] [Google Scholar]
- 11.Weihofen, A., Binns, K., Lemberg, M. K., Ashman, K. & Martoglio, B. (2002) Science 296, 2215–2218. [DOI] [PubMed] [Google Scholar]
- 12.Lemberg, M. K., Bland, F. A., Weihofen, A., Braud, V. M. & Martoglio, B. (2001) J. Immunol. 167, 6441–6446. [DOI] [PubMed] [Google Scholar]
- 13.Bier, E., Jan, L. Y. & Jan, Y. N. (1990) Genes Dev. 4, 190–203. [DOI] [PubMed] [Google Scholar]
- 14.Urban, S., Lee, J. R. & Freeman, M. (2001) Cell 107, 173–182. [DOI] [PubMed] [Google Scholar]
- 15.Koonin, E. V., Makarova, K. S., Rogozin, I. B., Davidovic, L., Letellier, M. C. & Pellegrini, L. (2003) Genome Biol. 4, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rather, P. N., Ding, X., Baca-DeLancey, R. R. & Siddiqui, S. (1999) J. Bacteriol. 181, 7185–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urban, S., Schlieper, D. & Freeman, M. (2002) Curr. Biol. 12, 1507–1512. [DOI] [PubMed] [Google Scholar]
- 18.Urban, S. & Freeman, M. (2003) Mol. Cell 11, 1425–1434. [DOI] [PubMed] [Google Scholar]
- 19.Gallio, M., Sturgill, G., Rather, P. & Kylsten, P. (2002) Proc. Natl. Acad. Sci. USA 99, 12208–12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y. M., Lai, M. T., Xu, M., Huang, Q., DiMuzio-Mower, J., Sardana, M. K., Shi, X. P., Yin, K. C., Shafer, J. A. & Gardell, S. J. (2000) Proc. Natl. Acad. Sci. USA 97, 6138–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esler, W. P., Kimberly, W. T., Ostaszewski, B. L., Diehl, T. S., Moore, C. L., Tsai, J. Y., Rahmati, T., Xia, W., Selkoe, D. J. & Wolfe, M. S. (2000) Nat. Cell Biol. 2, 428–434. [DOI] [PubMed] [Google Scholar]
- 22.Li, Y. M., Xu, M., Lai, M. T., Huang, Q., Castro, J. L., DiMuzio-Mower, J., Harrison, T., Lellis, C., Nadin, A., Neduvelil, J. G., et al. (2000) Nature 405, 689–694. [DOI] [PubMed] [Google Scholar]
- 23.Edbauer, D., Winkler, E., Regula, J. T., Pesold, B., Steiner, H. & Haass, C. (2003) Nat. Cell Biol. 5, 486–488. [DOI] [PubMed] [Google Scholar]
- 24.Kimberly, W. T., LaVoie, M. J., Ostaszewski, B. L., Ye, W., Wolfe, M. S. & Selkoe, D. J. (2003) Proc. Natl. Acad. Sci. USA 100, 6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takasugi, N., Tomita, T., Hayashi, I., Tsuruoka, M., Niimura, M., Takahashi, Y., Thinakaran, G. & Iwatsubo, T. (2003) Nature 422, 438–441. [DOI] [PubMed] [Google Scholar]
- 26.Fraering, P. C., Ye, W., Strub, J. M., Dolios, G., LaVoie, M. J., Ostaszewski, B. L., Van Dorsselaer, A., Wang, R., Selkoe, D. J. & Wolfe, M. S. (2004) Biochemistry 43, 9774–9789. [DOI] [PubMed] [Google Scholar]
- 27.Urban, S., Lee, J. R. & Freeman, M. (2002) EMBO J. 21, 4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esler, W. P., Kimberly, W. T., Ostaszewski, B. L., Ye, W., Diehl, T. S., Selkoe, D. J. & Wolfe, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 2720–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J. R., Urban, S., Garvey, C. F. & Freeman, M. (2001) Cell 107, 161–171. [DOI] [PubMed] [Google Scholar]
- 30.Fraering, P. C., LaVoie, M. J., Ye, W., Ostaszewski, B. L., Kimberly, W. T., Selkoe, D. J. & Wolfe, M. S. (2004) Biochemistry 43, 323–333. [DOI] [PubMed] [Google Scholar]
- 31.Bihel, F., Quelever, G., Lelouard, H., Petit, A., Alves da Costa, C., Pourquie, O., Checler, F., Thellend, A., Pierre, P. & Kraus, J. L. (2003) Bioorg. Med. Chem. 11, 3141–3152. [DOI] [PubMed] [Google Scholar]
- 32.Akiyama, Y., Kanehara, K. & Ito, K. (2004) EMBO J. 23, 4434–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohi, O., Urban, S. & Freeman, M. (2004) Curr. Biol. 14, 236–241. [DOI] [PubMed] [Google Scholar]
- 34.Munro, S. (2003) Cell 115, 377–388. [DOI] [PubMed] [Google Scholar]
- 35.Kopan, R. & Ilagan, M. X. (2004) Nat. Rev. Mol. Cell Biol. 5, 499–504. [DOI] [PubMed] [Google Scholar]