Abstract
We use the pattern of intron conservation in 684 groups of orthologs from seven fully sequenced eukaryotic genomes to provide maximum likelihood estimates of the number of introns present in the same orthologs in various eukaryotic ancestors. We find: (i) intron density in the plant–animal ancestor was high, perhaps two-thirds that of humans and three times that of Drosophila; and (ii) intron density in the ancestral bilateran was also high, equaling that of humans and four times that of Drosophila. We further find that modern introns are generally very old, with two-thirds of modern bilateran introns dating to the ancestral bilateran and two-fifths of modern plant, animal, and fungus introns dating to the plant–animal ancestor. Intron losses outnumber gains over a large range of eukaryotic lineages. These results show that early eukaryotic gene structures were very complex, and that simplification, not embellishment, has dominated subsequent evolution.
Although the discovery of spliceosomal introns in deep branching eukaryotes (1–5) has pushed back the origin of the first introns to the earliest stages of eukaryotic evolution, there still remains considerable debate about the number of introns present in early eukaryotes and the general contours of intron evolution (6–20). According to four alternate theories, either (i) introns were numerous even before the divergence of eukaryotes and prokaryotes (8, 9, 20–22); (ii) most introns invaded the nuclear genome early in eukaryotic evolution, perhaps as converted class II introns from endosymbionts (23); (iii) an explosion of intron number occurred at the beginning of metazoan evolution (24); or (iv) most introns are recently inserted (7, 17, 25, 26).
On the debate between the theories depends not only our understanding of the evolution of genome complexity but also the viability of several fundamental theories about the history of life. If introns are primordial, they could have facilitated the formation of the first genes (8, 9, 20–22). If they were already numerous by early eukaryotic evolution, they could have facilitated a possibly dramatic increase in transcript fidelity by enabling nonsense-mediated mRNA decay (27). If instead they were sparse until a relatively recent invasion in early metazoans, this invasion may have triggered the transition to multicellularity, enabling the intron-mediated creation of some of the multitude of multidomain proteins necessary for extracellular communication (24). If their numbers have grown recently, this could be due to increasing genome complexity as a pathological response to reduced population size (10, 11, 15). Yet, despite the importance of the question and 25 years of research on the topic, very little is certain. Some introns appear to be recently gained, others very old, but their relative numbers and the general history of intron–exon gene structures remain obscure.
It has long been known that at least some introns predate the plant–animal split (e.g., ref. 28). More recent efforts have sought introns in eukaryotes thought to be extremely early-branching. The finding of an intron in Giardia lambia (1), reinforced by the relatively high density of introns in the genes of Plasmodium (2, 3), pushed back the origin of introns further still. Components of the spliceosome, the machinery that extracts intronic sequences from RNA transcripts, have been found in other deeply branching eukaryotes (4, 5). Thus, at least some introns appear to be very old. However, introns with very narrow phylogenetic distributions suggest that some are much more recently gained (29–33).
More recent studies have begun to address questions of the relative numbers of old and recently gained introns. Comparative analyses have shown that intron turnover is very slow in vertebrates (34, 35), with only a few loss/gain events between humans and fish and almost none between humans and rodents. Comparison of the mosquito and fly genomes suggested that at least 50% of introns in each of these genomes, and perhaps as many as 70%, have been in place since their split (12, 36, 37). On a deeper timescale, Fedorov et al. (38) showed that 7–15% of modern-day introns are shared between at least two major eukaryotic kingdoms. Using a different approach, Mourier and Jeffares (6) showed that intron-poor genomes have more 5′ introns and attributed this to a primacy of intron loss, suggesting that ancestral genomes might have been more intron-rich.
Most recently, Rogozin et al. (12) compiled a collection of intron–exon structures from 684 sets of orthologous genes. Each set of orthologs consists of one gene from each of eight fully sequenced eukaryotic genomes: Plasmodium falciparum, Arabidopsis thaliana, Schizosaccharomyces pombe, Saccharomyces cerevisiae, Caenorhabditis elegans, Homo sapiens, Anopheles gambiae, and Drosophila melanogaster. For each gene, the intron positions are marked, allowing detection of an impressive catalog of very old introns. In their data set, one-fourth of Arabidopsis introns are shared with humans, and one-third of Plasmodium introns are shared with another species, suggesting many old introns. Rogozin et al. (12) used a parsimony approach to reconstruct the history of the 684 genes and found a varied picture, with some lineages experiencing massive intron gain, others massive loss, and some a balance between the two.
However, because of the high rates of apparent intron loss along some lineages, parsimony has serious shortcomings. If an intron is present in the ancestor of two species or groups of species but has since been lost in one, this intron will be incorrectly inferred to have been gained in the other lineage. Given that some lineages studied appear to have lost up to 85% of their introns, this failure to account for such a possibility leads to a large systematic bias toward intron insertion in terminal lineages at the expense of intron number in ancestors.
By using a maximum likelihood analysis to incorporate rates of intron loss, we provide estimates for intron densities in very deep eukaryotic ancestors. The results are striking. First, the last common ancestor of worms, insects, and chordates is estimated to have had the same number of introns (3,321 in the 684 studied genes) as modern humans (3,345). This contradicts the common assumption (supported by the earlier authors' parsimony analysis) that the large number of introns in humans relative to well studied invertebrates is due to massive gain in the lineage leading to humans. Instead, the data support an apparent equilibration in the past perhaps 600 million years of the evolutionary history of humans in which intron gains have been matched by intron losses. The much smaller numbers of introns in worms (1,468 in the studied genes) and insects (675 in mosquito, 723 in flies) appear to be secondarily derived, due to massive loss along those lineages over the same time period.
The second striking result is that the last common ancestor of all crown group eukaryotes (plants, animals, and fungi, but not deeply branching protists) is estimated to have harbored ≈2,000 introns in the studied genes, a number exceeding that of all studied species except Arabidopsis (2,933) and humans. This suggests that intron-rich gene structures such as those found in modern vertebrates date to the earliest epochs of eukaryotic evolution, and that genomes with smaller numbers of introns have experienced a net genomic streamlining since. Indeed, the dominance of intron loss over intron gain appears quite general. Of 10 studied branches, 6 show a significant decrease in the total number of introns, whereas only 2 show a significant increase. Thus, although intron gain and loss may both be fairly common, overall losses appear to outweigh gains.
Finally, we estimate the number of introns from each modern genome present at each ancestral node represented in the data set. We find that a large number of modern introns date back very far. Roughly two-thirds of animal introns studied date back to the bilateran ancestor, and two-fifths of plant, fungi, and human ancestors predate the plant–animal ancestor. Thus, introns are not the recent insertions envisioned by some but instead the remnants of very deep molecular processes.
Our results suggest that early eukaryotes were far more intron-rich than previously appreciated, and that eukaryotic evolution has been dominated by intron loss, with only a very few studied lineages showing an increase in intron number. Introns are generally very old, with large fractions of introns dating back to very deep eukaryotic ancestors. These results contradict fundamental assumptions about the evolution of genome complexity and demand a rethinking of early eukaryotic evolution.
Materials and Methods
The Data Set and Programs. Amino acid-level sequence alignments and corresponding intron positions with presence–absence matrices for each position at which an intron is present in the conserved regions of 684 clusters of orthologous genes, previously used by Rogozin et al. (12), were downloaded from the National Center for Biotechnology Information web site (ftp://ftp.ncbi.nlm.nih.gov./pub/koonin/intron_evolution). Introns at the exact same position (between the exact corresponding pair of nucleotides in the alignment) were considered homologous and not due to independent multiple insertions. Following the original authors' finding of no evidence for intron movement (“drift” or “sliding”) in this data set, only introns present at the exact same position were considered homologous (see ref. 12 for details). S. cerevisiae was excluded due to its very small number of introns. Computer programs were written in Perl programming language to conduct the analyses described.
Estimating Numbers of Introns in Ancestors. Consider a node X, which represents the last common ancestor of two descendant groups 1 and 2 with a group of outgroup species (nondescendants of X) that we collectively call group 3. It will be possible to determine that an intron was present at X if it is present in modern species from at least two of the three groups, because the path connecting a pair of species from different groups necessarily passes through X. Other introns present in ancestor X have been lost in one or both descendant groups and/or are absent in the outgroups and thus are present in only one or even zero of the three modern groups. These will not be known by modern phylogenetic distribution to have been present in ancestor X. If we call the probabilities that an intron present in X is present in some species in 1, in 2, and in 3 o1, o2, and o3, respectively, the probabilities of an intron present at X having various modern phylogenetic distributions are simply:
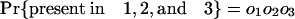 |
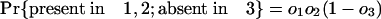 |
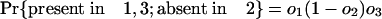 |
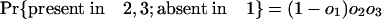 |
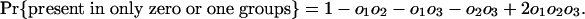 |
Thus, the probability of the observed pattern of intron conservation among groups given values for o1, o2, o3, and NX (the total introns present in ancestor X) is:
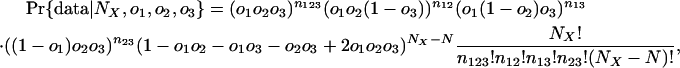 |
where n123 is the number of introns present in all three groups, n12 the number present in only groups 1 and 2, and so forth, and N = n123 + n12 + n13 + n23. The likelihood of a set of parameters is then:
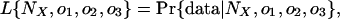 |
which has its maximum likelihood estimator at
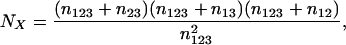 |
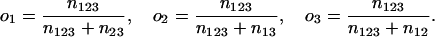 |
To derive confidence intervals for NX, we used the profile-likelihood method, which treats all parameters other than one as nuisance parameters and maximizes over them. Thus,
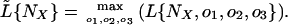 |
We note that, although these o values may be complicated to interpret biologically (particularly o3, which will involve parameters of both intron gain and loss in a potentially large number of lineages), our concern is not in the values of these variables themselves but in what they tell us about the more concrete value NX. There presumably is some real value o for each group, which is the fraction of introns at X found in some member of the group, and it is this value that concerns us, not its more intricate decomposition into rates of loss and gain along different individual lineages. Use of these summary variables rather than the more concrete individual probabilities for each terminal branch and internode does not affect the relative likelihoods of different values of NX (proven in the Appendix).
An example may help to elucidate the method. Consider the dipteran ancestor. We know that an intron present in D. melanogaster and A. gambiae was present in the dipteran ancestor. In addition, we know that an intron present in D. melanogaster or A. gambiae as well as some nondipteran (C. elegans, H. sapiens, or a nonanimal) was present in the ancestral dipteran. Presumably, for other ancestral dipteran introns, intron loss has erased the phylogenetic record of the presence in an ancestor.
The 451 introns present in A. gambiae and a nondipteran are known to be present in the dipteran ancestor without respect to presence in D. melanogaster and are thus an unbiased set for estimating rates of intron retention in the D. melanogaster lineage (that is, the o value for the D. melanogaster lineage). Of these, 295 are present in D. melanogaster, thus ≈65% of ancestral dipteran introns are retained in D. melanogaster (o1 ≈ 0.65). Similarly, 295/489 introns found in D. melanogaster and a nondipteran are retained in A. gambiae, thus ≈60% of ancestral dipteran introns are retained in A. gambiae (o2 ≈ 0.60). Finally, 382 introns are present in both dipteran species and thus known to be present in the dipteran ancestor. Of these, 295 are also found in a nondipteran, thus ≈77% of ancestral dipteran introns are also found in a nondipteran (o3 ≈ 0.77).
The probability that a true ancestral dipteran intron will be present in D. melanogaster, A. gambiae, and a nondipteran is o1o2o3 ≈ 0.30. The probability that it will be present only in D. melanogaster and A. gambiae is o1o2(1–o3) ≈ 0.09, in D. melanogaster and a nondipteran but not A. gambiae is o1(1–o2)o3 ≈ 0.16, and in A. gambiae and a nondipteran but not D. melanogaster is (1–o1)o2o3 ≈ 0.20. In these and only these cases, the intron will be known to be ancestral. Thus the overall probability of correct identification of an ancestral bilateran ancestor is o1o2o3 + o1o2(1–o3) + o1(1–o2)o3 + (1–o1)o2o3 = o1o2 + o1o3 + o2o3 – 2o1o2o3 ≈ 0.75. Thus, the 732 known ancestral introns suggest some 732/0.75 ≈ 968 total ancestral dipteran introns.
The Number of Introns in a Modern Species Present in an Ancestor. We can also ask how many introns present in a particular species A in group 1 were also present in ancestor X. Here we define oA as the probability that an intron present in ancestor X has been retained in A and ignore other species in group 1. An intron present in X has the following probabilities of various phylogenetic distributions with respect to A, 2, and 3:
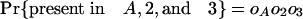 |
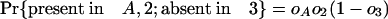 |
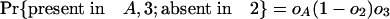 |
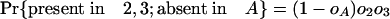 |
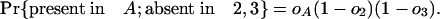 |
And the total probability of all these possibilities is
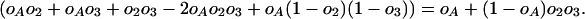 |
If NXA is the total number of introns in A that were present in ancestor X, the probability of seeing the data given values for oA, o2, o3, and NXA simplifies to:
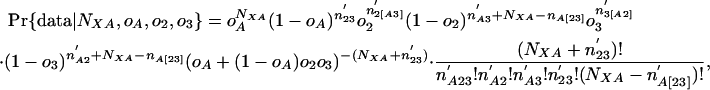 |
where the primes simply indicate that the rest of the species in group 1 are not considered (e.g., if group 1 comprised species A, B, and C, n′A2 would be the number of introns present in A as well as some species in 2, absent in all group 3 species, and possibly but not necessarily present in B and/or C); and brackets indicate that an intron must be present in at least one species inside the brackets (e.g., n′A[23] = n′A23 + n′A2 + n′A3). The likelihood of a given value of NXA is then just the maximum of Pr{data| NXA, oA, o2, o3} over all o values, which has its maximum likelihood estimator at
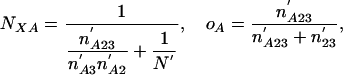 |
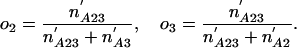 |
Results
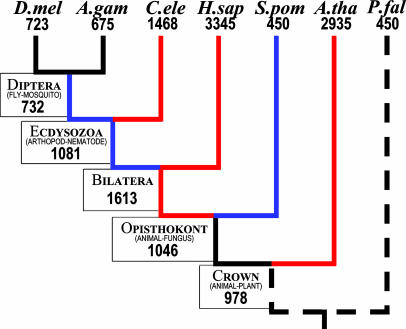
Ancestral Intron Densities. A previous study reconstructed ancestral intron numbers by a Dollo parsimony analysis of the current data (12). Such a parsimony reconstruction is given in Fig. 1 for comparison with our results.
Fig. 1.
A Dollo parsimony reconstruction of the data, for comparison with our results. D.mel, D. melanogaster; A.gam, A. gambiae; C.ele, C. elegans, H.sap, H. sapiens; S.pom, S. pombe; A.tha, A. thaliana; P.fal, P. falciparum.
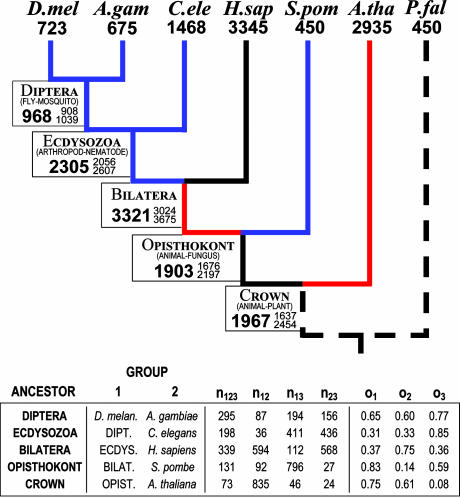
We used maximum likelihood to estimate the number of introns present in the conserved regions of 684 genes at each ancestral node. The results are given in Fig. 2. Two results are particularly surprising. First, the ancestral bilateran equaled humans in intron density (3,321 vs. 3,345 in the 684 genes), thus differences between vertebrates and less intron-dense animals are due to massive overall intron loss, not gain, since the Cambrian explosion. Second, the crown group (plant–animal) ancestor harbored ≈2,000 introns in the genes studied, not only dwarfing moderately intron-dense modern genomes [flies (723), S. pombe (450)], but also approaching extremely intron-dense ones [humans (3,345), Arabidopsis (2,933)]. The dominance of loss over gain is quite general. In Fig. 1, six lineages (blue) show pronounced net loss, whereas only two (red) show net gain. Overall, four of six species have suffered net intron loss since the crown group ancestor, four of five since the animal–fungi ancestor. These observations stand the prevalent view of eukaryotic evolution on its head: instead of introns invading intron-poor ancestral genomes to yield higher modern densities, intron-rich ancestral genomes have been stripped of their introns to yield relatively low modern densities.
Fig. 2.
Estimates of the numbers of introns in 684 genes for various eukaryotic ancestors. Numbers of introns for modern species are known, numbers for ancestors estimated. Large numbers give maximum likelihood estimates, small numbers confidence intervals (2 units of log-likelihood score). Groups 1 and 2 are the two descendant groups for each ancestor. Group 3 is then all species not in groups 1 or 2. o1–3 columns give the maximum likelihood values. See Fig. 1 legend for abbreviations.
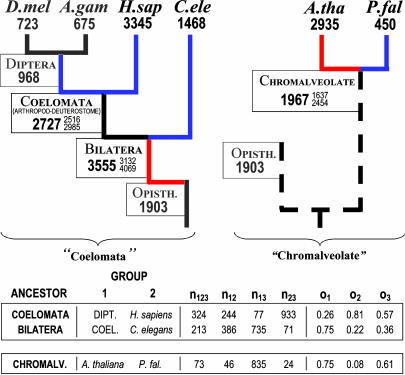
Contention over two phylogenetic issues complicates matters: (i) Diptera may be more closely related to humans [Coelomata, (39, 40)], not C. elegans [Ecdysozoa, (41)], as we have assumed, and (ii) A. thaliana may be more closely related to P. falciparum [Chromalveolate (42, 43)], not opisthokonts, as we have assumed. We also estimated ancestral densities assuming these alternatives. The results are given in Fig. 3. (Fortunately, the intron densities of ancestors in one part of the tree do not depend on the phylogeny in other parts, e.g., the postulated coelomata ancestor will have the same estimate regardless of whether the phylogeny is basal apicomplexan or chromalveolate.) Under these assumptions, the basic results are unchanged. Assuming Coelomata, the ancestral bilateran still appears very intron-rich, with C. elegans and Diptera having lost large numbers of introns since that time. Assuming the Chromalveolate phylogeny, the very deep plant–apicomplexan ancestor had a high intron density. Thus, the finding of very old, very intron-rich ancestors is not an artifact of any incorrect phylogenetic assumptions.
Fig. 3.
Estimates for the numbers of introns present in various eukaryotic ancestors, assuming alternative phylogenies. See Fig. 1 legend for abbreviations.
If complex gene structures are extremely old, how old are individual modern introns themselves? Among human introns, 104 are present in Plasmodium and an additional 722 in Arabidopsis. However, the Arabidopsis lineage shows ≈38% loss (39/104 shared human–Plasmodium introns are absent in Arabidopsis), so the 722 human introns retained in Arabidopsis represent only ≈62% of some 1,155 = 722/62% total ancestral plant–animal introns retained in humans but absent in P. falciparum. Along with the 104 introns shared with Plasmodium, ≈1,259/3,345 (38%) of modern human introns predate the plant–animal divergence. We used the pattern of intron conservation to estimate the fraction of introns in descendant species that were present in each ancestor. As Table 1 shows, among descendant species, an estimated three-quarters of introns were present in the ancestral bilateran, and two-fifths predate the opisthokont and crown ancestors. Thus, most introns are extremely old, not recently acquired.
Table 1. Estimated fraction of introns in modern taxa present in various ancestors.
| Fraction present in ancestor, %
|
|||||
|---|---|---|---|---|---|
| Modern genome | Crown | Opisthokont | Bilatera | Ecdysozoa | Diptera |
| D. melanogaster | 35 (29-48) | 37 (32-49) | 70 (67-74) | 75 (71-79) | 87 (85-91) |
| A. gambiae | 40 (30-50) | 35 (31-41) | 69 (66-72) | 73 (70-77) | 87 (84-90) |
| C. elegans | 34 (25-50) | 31 (26-38) | 52 (49-55) | 51 (49-54) | - |
| H. sapiens | 38 (33-44) | 41 (37-45) | 75 (70-79) | - | - |
| S. pombe | 48 (40-62) | 60 (57-63) | - | - | - |
| A. thaliana | 41 (37-46) | - | - | - | - |
| Average | 39 | 41 | 67 | 66 | 87 |
Discussion
These results push back the origin of very intron-dense genome structures over a billion years to the plant–animal split. Indeed, ancestors at the divergences between major eukaryotic kingdoms as well as the ancestral bilateran appear to have harbored nearly as many introns as the most intron-dense modern organisms. This is a sharp repudiation of the common assumption that intron-riddled gene structures arose only recently.
In addition, our analysis shows that the majority of modern introns are themselves very old. Two-thirds of bilateran introns were present in the bilateran ancestor; 40% of opisthokont introns were present in the ophisthokont ancestor; and 40% of plant, animal, and fungal introns were present in the plant–animal ancestor. This is quite different from what is commonly assumed and surprising in light of relatively fast rates of intron turnover observed in nematodes and flies (44–47).
Frequent Episodes of Massive Intron Loss. Our results show a general trend toward net intron loss in eukaryotes from intron-rich ancestors to the moderate or intron-sparse genes observed in many modern species. There were at least six episodes of massive intron loss in the analyzed lineages. Genomes have experienced a significant net reduction in intron number in fungi, nematodes, arthopods, and possibly in apicomplexans (if, as some think, they are not an outgroup to plants and animals). Furthermore, the evolutionary positions of many species not analyzed here imply several other episodes of genome streamlining. S. cerevisiae has experienced net intron loss since its divergence from S. pombe. Encephalitozoon cuniculi, with only ≈0.005 introns per gene, must have lost large numbers of ancestral fungal introns. The large number of Plasmodium introns shared with plants/animals implies that the tiny numbers of introns in Cryptosporidium parvum and trypanosomes are due to additional episodes of massive loss. Finally, depending on the evolutionary position of Euglenozoa, the lineage leading to Leishmania may have also experienced a tremendous reduction in intron number.
Our results suggest that the bias toward intron loss previously found in nematodes and in mammals is a general trend in eukaryotic evolution (32, 33, 35, 47). This is in agreement with earlier findings that intron-sparse genomes tend to have their introns concentrated near the 5′ end of the gene (6, 19), as would be expected if these genomes have lost most of their ancestral introns through gene conversion by reverse-transcription products of spliced mRNAs. However, it is in tension with other reports of an excess of intron gains in families of paralogous genes (14, 16). However, an increase in the rate of intron turnover between paralogs relative to orthologs has been found in Plasmodium (48), and Babenko et al. (14) specifically observed an increase in the rate of gain relative to loss after gene duplication, thus the difference between studies could reflect the data sets used: orthologs here, paralogs in other studies.
Implications for Models of Intron Evolution. Two of several pictures of intron evolution are compatible with our results. If either introns were numerous before the divergence of prokaryotes and eukaryotes, or introns arrived en masse as converted type II introns from early intracellular endosymbionts, introns would be expected to be numerous in early eukaryotic evolution (8, 9, 20–23, 28). On the other hand, these results are clearly not amenable to the notion that introns are mostly recently inserted or appeared in large numbers first in early metazoans (24–26, 29–31).
Lynch (7, 11) has recently suggested that introns are slightly deleterious elements that arose as a pathological response to diminishing population size. On this model, selection against introns is roughly constant, with the greater efficiency of selection in large populations prohibiting intron establishment. This model sees a scenario in which introns were sparse in the genomes of ancient unicellular organisms with large population size and have become intron-dense only recently with the decrease in population size associated with multicellularity and organismal complexity. However, to explain the observed recurrent independent episodes of massive intron loss, such a model would have to invoke small population sizes at the plant–animal and animal–fungus splits and multiple independent subsequent increases in population size, an unexpected and highly nonparsimonious explanation. It seems much more likely that different selection or mutation regimes for introns along different lineages are driving the observed instances of gene streamlining.
The Problem with Parsimony. Our results stand in contrast with those of Rogozin et al. (12), although using exactly the same data. They analyzed the phylogenetic patterns of the introns in the data set using Dollo parsimony. Such a reconstruction is given in Fig. 1. To understand parsimony's shortcomings here, consider an intron gained before the divergence of humans from worms and insects. Because ≈63% of introns present at this divergence are absent in all studied invertebrates and the rate of intron loss in humans is small (≈25%), there is an ≈45% (= 63% × 75%) chance that the intron will be found only in humans. However, in this most likely case of loss in worms and insects and maintenance in humans, parsimony will incorrectly infer that the intron has been gained in the lineage leading to humans. As the rate of loss in S. pombe is ≈86%, there is a 35% (= 40% × 86%) chance that an intron originating as deeply as the fungi–animal split will show up only in humans, leading to an inference that the intron originated at least a half billion years later than it actually did.
Comparison of Figs. 1 and 2 shows general differences in the estimates of ancestral intron number. At each node, our maximum likelihood estimate is much larger than the parsimony estimate. There are vast discrepancies in the picture of net intron loss or gain through evolution. The lineage leading to C. elegans, which appeared on a parsimony analysis to have relative stasis in intron number, is shown by our analysis to have reduced its intron number by 40%. The lineage leading to humans is even more striking, with parsimony suggesting a doubling in the number of introns, whereas our method shows complete intron number stasis. Relative to parsimony, our analysis tends to move intron insertions from the terminal branches to internodes by estimating the number of ancestral introns whose loss in some lineage(s) has led to a deceptively parochial modern phylogenetic distribution.
Parallel Insertions. We have assumed here that introns found at the exact same site in orthologs are in fact homologous, descendants of an intron present in the ancestor. However, if intron insertion occurs at only a limited number of sites, some such intron correspondences could be due to independent insertions at the same site along different lineages (16, 17, 49, 50). If such “parallel insertions” are common, our estimates will be biased. However, other results suggest that this is not the case.
The largest number of intron correspondences among species in the data set is between humans and Arabidopsis. Absence of these introns in S. pombe, C. elegans, and Diptera is interpreted as loss along those lines, with rates of loss estimated from these cases yielding the large ancestral intron densities estimated. Alternatively, some fraction of the Arabidopsis–human intron correspondences could be due to more recent parallel insertion. These introns would be absent in invertebrates and appear incorrectly to have been lost there, leading us to overestimate intron loss rates and in turn ancestral intron number. A recent paper by Bányai and Patthy (18) looks at the fate of introns implicated in the formation of their resident genes and thus known to be present ancestrally by non-phylogenetic criteria. These introns show the same pattern inferred here, dominated by intron retention in humans and loss in C. elegans and D. melanogaster (Table 2), suggesting the pattern of ancestral retention and loss inferred here, not parallel insertion, is responsible for the multitude of human–Arabidopsis correspondences.
Table 2. Retention of introns in the present study and the Bányai—Patthy study.
| Fraction introns retained, %
|
|||
|---|---|---|---|
| C. elegans | D. melanogaster | H. sapiens | |
| Bányai—Patthy study | 24.9 | 19.4 | 77.7 |
| Present study | 22.8 | 18.5 | 75.2 |
For the present study, introns known to be present in the bilateran ancestor due to the presence in both animals and nonanimals are included. For the Bányai—Patthy study, introns known to be present at the time of gene formation are considered.
Conclusion
These results contradict the assumption that genome complexity has increased through evolution. Instead, species have repeatedly abandoned complex gene structures for simpler ones, questioning the purpose and value of intricate gene structures. These results suggest a reconsideration of the genomics of eukaryotic emergence.
Acknowledgments
We thank Alex Platt for invaluable advice through all stages of this project.
Appendix
The o values we use constitute simplifications we must justify. The value o1 is not a simple biological quantity for a multispecies group but instead a function of the probabilities of retention along many independent branches. o3 may be an even more complex quantity incorporating rates of both loss and gain along multiple branches. Thus, we must show that using o values rather than these individual quantities does not influence the relative likelihoods of different values of NX.
For any given collection of one of more group 1 species, which we can call α, we can define the probability that an intron present in ancestor X will be present in exactly that set of species among all species in group 1 as Pr{α|present at X}. We can also define node Y as the node “in the direction of” group 1 that shares an internode with X and r as the probability that an intron present in X is also present in Y. That is, if the species of group 1 are descendants of X (either all or none will be descendants), Y is the last common ancestor of group 1, and r is the probability that an intron is retained along the X-Y internode; if the species of group 1 are not descendants of X, Y is the most recent ancestor of X and some member of group 1, and r is the probability that an intron present at X was not inserted along the X-Y internode.
Then Pr{α|present at X} = Pr{α|present at Y}r, and our simplified quantity o1 is just:
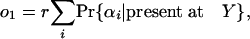 |
and the probability that an intron present at X is absent in all species in group 1 is
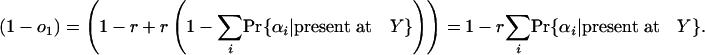 |
Let ni2 be the number of introns present in 2 as well as the group of species denoted by αi; ni3 be the number present in 3 and αi; and ni23 be the number present in 2, 3, and αi. The probability of seeing the data given values for NX, o2, o3, r, and a value of Pr{αi|present at Y} for each αi (hereafter simply “Pr{αi|Y} for all i”) is then:
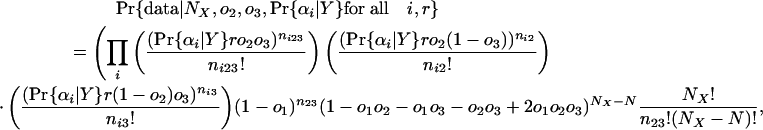 |
and we can define the likelihood of a given set of parameters as
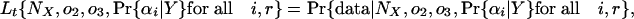 |
and the profile likelihood for a given value of NX for the complete model is:
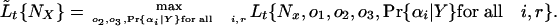 |
We use the t subscript to distinguish the likelihood for a total set of parameters from the nonsubscripted likelihood values for a set of simplified o value parameters. The use of the simplifying variable o1 will not affect the relative likelihood of different values of NX (that is, the ratio of likelihoods for two NX values will be equal under the complete and simplified models) if kL̃{NX} = L̃t{NX} for some value k that does not depend on NX, which will be the case if
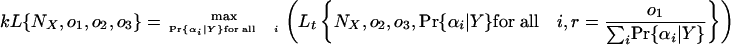 |
(that is, if the likelihood for the simplified model is equal to the maximum likelihood for the complete model for the same NX, o2, o3, and any set of Pr{αi|Y} times a constant). This yields
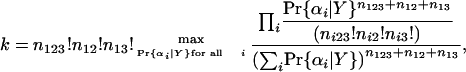 |
which is indeed independent of NX, o1, o2, and o3. Thus, the use of the simplifying variable o1 does not influence the relative likelihoods of different values of NX.
References
- 1.Nixon, J. E., Wang, A., Morrison, H. G., McArthur, A. G., Sogin, M. L., Loftus, B. J. & Samuelson, J. (2002) Proc. Natl. Acad. Sci. USA 99, 3701–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner, M. J., Shallom, S. J., Carlton, J. M., Salzberg, S. L., Nene, V., Shoaibi, A., Ciecko, A., Lynn, J., Rizzo, M., Weaver, B., et al. (2002) Nature 419, 531–534. [DOI] [PubMed] [Google Scholar]
- 3.Hall, N., Pain, A., Berriman, M., Churcher, C., Harris, B., Harris, D., Mungall, K., Bowman, S., Atkin, R., Baker, S., et al. (2002) Nature 419, 527–531. [DOI] [PubMed] [Google Scholar]
- 4.Fast, N. M. & Doolittle, W. F. (1999) Mol. Biochem. Parasitol. 99, 275–278. [DOI] [PubMed] [Google Scholar]
- 5.Fast, N. M., Roger, A. J., Richardson, C. A. & Doolittle, W. F. (1998) Nucleic Acids Res. 26, 3202–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mourier, T. & Jeffares, D. C. (2003) Science 300, 1393. [DOI] [PubMed] [Google Scholar]
- 7.Lynch, M. (2002) Proc. Natl. Acad. Sci. USA 99, 6118–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Souza, S. J. (2003) Genetica 118, 117–121. [PubMed] [Google Scholar]
- 9.Roy, S. W. (2003) Genetica 118, 251–266. [PubMed] [Google Scholar]
- 10.Lynch, M. & Richardson, A. O. (2002) Curr. Opin. Genet. Dev. 12, 701–710. [DOI] [PubMed] [Google Scholar]
- 11.Lynch, M. & Conery, J. S. (2003) Science 302, 1401–1404. [DOI] [PubMed] [Google Scholar]
- 12.Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G. & Koonin, E. V. (2003) Curr. Biol. 13, 1512–1517. [DOI] [PubMed] [Google Scholar]
- 13.Sverdlov, A. V., Rogozin, I. B., Babenko, V. N. & Koonin, E. V. (2003) Curr. Biol. 13, 2170–2174. [DOI] [PubMed] [Google Scholar]
- 14.Babenko, V. N., Rogozin, I. B., Mekhedov, S. L. & Koonin, E. V. (2004) Nucleic Acids. Res. 32, 3724–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin, E. V. (2004) Cell Cycle 3, 280–285. [PubMed] [Google Scholar]
- 16.Qiu, W. G., Schisler, N. & Stoltzfus, A. (2004) Mol. Biol. Evol. 21, 1252–1263. [DOI] [PubMed] [Google Scholar]
- 17.Sadusky, T., Newman, A. J. & Dibb, N. J. (2004) Curr. Biol. 14, 505–509. [DOI] [PubMed] [Google Scholar]
- 18.Bányai, L. & Patthy, L. (2004) FEBS Lett. 565, 127–132. [DOI] [PubMed] [Google Scholar]
- 19.Sverdlov, A. V., Babenko, V. N., Rogozin, I. B. & Koonin, E. V. (2005) Gene 338, 85–91. [DOI] [PubMed] [Google Scholar]
- 20.De Roos, A. D. G. (2005) Bioinformatics 21, 2–9. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, W. (1978) Nature 271, 501. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert, W. (1987) Cold Spring Harbor Symp. Quant. Biol. 52, 901–905. [DOI] [PubMed] [Google Scholar]
- 23.Cavalier-Smith, T. (1991) Trends Genet. 7, 145–148. [PubMed] [Google Scholar]
- 24.Patthy, L. (1999) Gene 238, 103–114. [DOI] [PubMed] [Google Scholar]
- 25.Logsdon, J. M., Jr. (1998) Curr. Opin. Genet. Dev. 8, 637–648. [DOI] [PubMed] [Google Scholar]
- 26.Logsdon, J. M., Jr. (2004) Proc. Natl. Acad. Sci. USA 101, 11195–11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch, M. & Kewalramani, A. (2003) Mol. Biol. Evol. 20, 563–571. [DOI] [PubMed] [Google Scholar]
- 28.Marchionni, M. & Gilbert, W. (1986) Cell 46, 133–141. [DOI] [PubMed] [Google Scholar]
- 29.Palmer, J. & Logsdon, J.M., Jr. (1991) Curr. Opin. Genet. Dev. 1, 470–477. [DOI] [PubMed] [Google Scholar]
- 30.Dibb, N. J. & Newman, A. J. (1989) EMBO J. 8, 2015–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logsdon, J. M., Jr., Tyshenko, M. G., Dixon, C., Jafari, J. D., Walker, V. K. & Palmer, J. (1995) Proc. Natl. Acad. Sci. USA 92, 8507–8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho, S., Jin, S. W., Cohen, A. & Ellis, R. E. (2004) Genome Res. 14, 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kionthke, K., Gavin, N. P., Raynes, Y., Roehrig, C., Piano, F. & Fitch, D. H. (2004) Proc. Natl. Acad. Sci. USA 101, 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elgar, G. (1996) Hum. Mol. Genet. 5, 1437–1442. [DOI] [PubMed] [Google Scholar]
- 35.Roy, S. W., Fedorov, A. & Gilbert, W. (2003) Proc. Natl. Acad. Sci. USA 100, 7158–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zdobnov, E. M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R. R., Christophides, G. K., Thomasova, D., Holt, R. A., Subramanian, G. M., et al. (2002) Science 298, 149–159. [DOI] [PubMed] [Google Scholar]
- 37.Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., Wincker, P., Clark, A. G., Ribeiro, J. M., Wides, R., et al. (2002) Science 298, 129–149.12364791 [Google Scholar]
- 38.Fedorov, A., Merican, A. F. & Gilbert, W. (2002) Proc. Natl. Acad. Sci. USA 99, 16128–16133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knoll, A. H. & Carroll, S. B. (1999) Science 284, 2129–2137. [DOI] [PubMed] [Google Scholar]
- 40.Wolf, Y. I., Rogozin, I. B. & Koonin, E. V. (2004) Genome Res. 14, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguinaldo, A. M., Turbeville, J. M., Linford, L. S., Rivera, M. C., Garey, J. R., Raff, R. A. & Lake, J. A. (1997) Nature 387, 489–493. [DOI] [PubMed] [Google Scholar]
- 42.Baldauf, S. L., Rogers, A. J., Wenk-Siefert, I. & Doolittle, W. F. (2000) Science 290, 972–977. [DOI] [PubMed] [Google Scholar]
- 43.Cavalier-Smith, T. (1999) Eukar. Microbiol. 46, 347–366. [DOI] [PubMed] [Google Scholar]
- 44.Moriyama, E. N., Petrov, D. A. & Hartl, D. L. (1998) Mol. Biol. Evol. 15, 770–773. [DOI] [PubMed] [Google Scholar]
- 45.Kent, W. J. & Zahler, A. M. (2000) Genome Res. 10, 1115–1125. [DOI] [PubMed] [Google Scholar]
- 46.Robertson, H. M. (1998) Genome Res. 8, 449–463. [DOI] [PubMed] [Google Scholar]
- 47.Robertson, H. M. (2000) Genome Res. 10, 192–203. [DOI] [PubMed] [Google Scholar]
- 48.Castillo-Davis, C. I., Bedford, T. B. & Hartl, D. L. (2004) Mol. Biol. Evol. 21, 1422–1427. [DOI] [PubMed] [Google Scholar]
- 49.Cho, G. & Doolittle, R. F. (1997) J. Mol. Evol. 44, 573–584. [DOI] [PubMed] [Google Scholar]
- 50.Stolzfus, A. (2004) Curr. Biol. 14, R351–R352. [DOI] [PubMed] [Google Scholar]