Abstract
Understanding the mechanisms of HIV-1 drug resistance is critical for developing more effective antiretroviral agents and therapies. Based on our previously described dynamic copy-choice mechanism for retroviral recombination and our observations that nucleoside reverse transcriptase inhibitors (NRTIs) increase the frequency of reverse transcriptase template switching, we propose that an equilibrium exists between (i) NRTI incorporation, NRTI excision, and resumption of DNA synthesis and (ii) degradation of the RNA template by RNase H activity, leading to dissociation of the template-primer and abrogation of HIV-1 replication. As predicted by this model, mutations in the RNase H domain that reduced the rate of RNA degradation conferred high-level resistance to 3′-azido-3′-deoxythymidine and 2,3-didehydro-2,3-dideoxythymidine by as much as 180- and 10-fold, respectively, by increasing the time available for excision of incorporated NRTIs from terminated primers. These results provide insights into the mechanism by which NRTIs inhibit HIV-1 replication and imply that mutations in RNase H could significantly contribute to drug resistance either alone or in combination with NRTI-resistance mutations in reverse transcriptase.
Keywords: drug resistance, recombination, NRTI, reverse transcriptase, dynamic copy choice
The inevitable emergence of drug-resistant HIV-1 variants in response to all antiretroviral therapies presents a major obstacle to reducing morbidity and mortality associated with AIDS (1, 2). A greater understanding of the molecular mechanisms driving evolution of drug resistance is critical for developing more effective antiretroviral agents, delaying the onset of drug resistance, and successfully managing antiviral therapy for individual patients. Nucleoside reverse transcriptase inhibitors (NRTIs) that lead to termination of DNA synthesis during HIV-1 replication comprise a major class of clinically available antiretroviral drugs (3). Antiviral resistance to several NRTIs such as 3′-azido-3′-deoxythymidine (AZT) and 2,3-didehydro-2,3-dideoxythymidine (d4T) arises through a nucleotide excision or primer-unblocking mechanism involving hydrolytic removal of the chain-terminating nucleoside analog from the end of the primer (4, 5). Mutations that confer resistance to thymidine analogs (TAM) increase the rate of nucleotide excision (6, 7). Elucidating the nucleotide excision mechanism has significantly enhanced our understanding of the mechanisms that contribute to NRTI resistance; despite this advance, several aspects of the molecular mechanisms underlying the evolution of drug resistance in vivo are not well understood.
Based on studies designed to elucidate the role of HIV-1 RNase H in viral recombination, we now propose a previously uncharacterized mechanism for NRTI-mediated abrogation of HIV-1 replication. We also propose that mutations reducing RNase H activity could enhance resistance to NRTIs alone or in combination with other well characterized NRTI-resistance mutations that increase the rate of NRTI excision from a blocked primer. To date, the role of RNase H in NRTI-resistance has not been thoroughly evaluated in patients, cell-based assays, or biochemical studies. The results presented here provide insights into the critical role of RNase H in NRTI resistance and NRTI-mediated abrogation of HIV-1 replication.
Materials and Methods
Plasmids and Mutagenesis. The HIV-1-based vector pNLuc expresses the firefly luciferase reporter gene and all HIV-1 proteins except for Nef and Env (8). The pCMVΔR8.2 helper construct expresses all HIV-1 proteins, except for Env, under the control of the human cytomegalovirus (hCMV) immediate early promoter and lacks the packaging signal (Ψ) and adjacent sequences (9). pHCMV-G expresses the G glycoprotein of vesicular stomatitis virus (VSV-G) (10). pNLuc-based constructs containing mutations in reverse transcriptase (RT) were generated by subcloning of an MscI restriction fragment from pCMVΔR8.2 containing the desired mutations, which were generated by site-directed mutagenesis (QuikChange XL Site-Directed Mutagenesis Kit, Stratagene). The presence of the desired mutations and the absence of undesired mutations were verified by sequencing. The sequences of the mutagenic primers are available on request.
Cells, Transfection, and Virus Production. 293T cells (American Type Culture Collection,) and the 293T-based cell line GN-HIV-GFFP (11), containing an HIV-1 provirus that expresses overlapping fragments of GFP (GF and FP) and the hygromycin-resistance gene, were maintained in the presence of DMEM (Cellgro), 10% FCS (HyClone), penicillin (50 units/ml; GIBCO), and streptomycin (50 μg/ml; GIBCO). Hygromycin (Calbiochem) was used at a final concentration of 270 μg/ml.
To produce pNLuc-based viruses, 293T cells were plated at a density of 5 × 106 cells per 100-mm-diameter dish and transfected with pNLuc (or mutant pNLuc) and pHCMV-G DNA by using the MBS Mammalian Transfection kit (Stratagene). Cell supernatants were harvested 24 h after transfection and filtered by using 0.45-μm-pore-size membranes (Nalgene). pNLuc virus was concentrated 20-fold by centrifugation at 25,000 rpm (Surespin, Sorvall) for 90 min at 4°C and stored at -80°C. To produce HIV-GFFP viruses, GN-HIV-GFFP cells were plated at a density of 5 × 106 cells per 100-mm-diameter dish and transfected with pCMVΔR8.2 and pHCMV-G DNA, and the virus produced from the transfected cells was harvested as described above.
Antiviral Drugs. AZT, d4T, and 2′,3′-dideoxyinosine (ddI) were obtained from Sigma-Aldrich; 2′,3′-dideoxy-3′-thiacytidine (3TC) was obtained from Moravek Biochemicals; and nonnucleoside reverse transcriptase inhibitor (NNRTI) efavirenz (EFV) was obtained from the NIH AIDS Research & Reference Reagent Program.
Single-Replication-Cycle Drug-Susceptibility Assay. 293T cells were plated at a density of 1 × 104 cells per well of a 24-well plate and used as targets of infection. Concentrated pNLuc-based virus stock was diluted 10- to 1,000-fold and used to infect the 293T cells. The 293T target cells were incubated with medium containing serial dilutions of a drug or lacking any drugs for 24 h before infection, during infection and after infection. Replication was monitored by measuring luciferase activity in infected target cells ≈48 h after infection, using the firefly luciferase assay system with reporter lysis buffer (Promega). Luciferase activity in cell lysates was determined by using a 96-well luminometer (LUMIstar Galaxy, BMG LABTECH). Data were plotted as the percent inhibition of luciferase activity versus log10 drug concentration, and the percent inhibition was calculated as follows: [1 - (luciferase activity in the presence of drug/luciferase activity in the absence of drug] × 100%. Inhibition curves defined by the four-parameter sigmoidal function y = y0 + a/[1 + (x/x0)b], as described in ref. 12, were fit to the data by sigmaplot 8.0 software to calculate the drug concentration required to inhibit virus replication by 50% (IC50).
Determination of ex Vivo RT Template-Switching Frequency. The 293T target cells were plated at a density of 5 × 104 cells per 60-mm-diameter dish. The next day, the culture supernatant was replaced with fresh medium containing different dilutions of drug (including a no-drug control). Serial dilutions of HIV-GFFP virus were used to infect the target cells 24 h later in the presence or absence (negative control) of the drug. The infected cells were subjected to hygromycin selection 24 h after infection in the presence of the drug for an additional 72 h. The medium was replaced with fresh hygromycin-containing medium every 3 or 5 days for 10–14 days. Hygromycin-resistant colonies were quantified to determine viral titers. Approximately 500–2,000 hygromycin-resistant colonies were pooled and expanded. Subsequently, the cells were harvested and resuspended in 0.5–2 ml of PBS supplemented with 1% FCS and analyzed by flow cytometry to determine the percentage of GFP-expressing cells (FACScan, Becton Dickinson).
Results
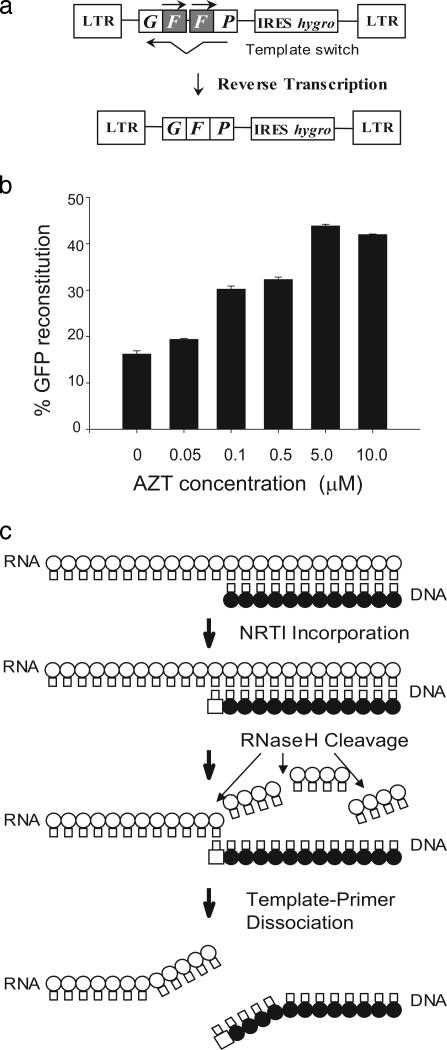
RT Template-Switching Frequency Is Increased in the Presence of NRTIs. Genetic recombination during HIV-1 replication occurs when RT switches templates between copackaged genomic RNAs (13). To determine the effects of NRTIs on RT template switching, we used a previously described HIV-1 vector that contains a direct repeat of the middle (“F”) portion of the green fluorescent protein gene (GFP) (Fig. 1a) (11). The vector cannot express a functional GFP protein; however, if a homologous template switch occurs during reverse transcription within the F repeats, a functional GFP is reconstituted and expressed, providing a sensitive measure of the frequency of RT template-switching events in a single cycle of viral replication. We determined the frequency of GFP reconstitution for wild-type HIV-1 RT in the absence or presence of increasing concentrations of AZT (Fig. 1b). The results indicated that as the AZT concentration increased, the frequency of RT template switching increased from 16% to 40%. We also observed that the frequency of RT template switching increased in the presence of d4T in a dose-dependent manner but was not influenced by the presence of EFV (data not shown).
Fig. 1.
Effect of AZT treatment on RT template switching and model for NRTI-mediated abrogation of HIV-1 replication. (a) Simplified structure of GN-HIV-GFFP provirus, which contains long terminal repeats (LTR) and all cis-acting elements of HIV-1 (not shown). Overlapping fragments (GF and FP) of reporter gene GFP with the directly repeated 250-bp F portion (shaded) and the hygromycin-resistance gene (hygro), which is expressed by using an internal ribosomal entry site (IRES) of encephalomyocarditis virus, are shown. During reverse transcription, the repeated F portion is deleted with a high frequency to reconstitute a functional GFP. (b) Direct-repeat deletion and GFP-reconstitution frequencies of HIV-1 RT in the presence of different AZT concentrations. The percentage of GFP reconstitution represents the proportion of infected 293T target cells that exhibited fluorescence after hygromycin selection. Bar graphs and error bars represent the mean and standard error of the percentage of GFP reconstitution, respectively. (c) A model for NRTI-mediated abrogation of HIV-1 replication. A schematic outline of HIV-1 minus-strand DNA synthesis is presented. Thick arrows indicate the sequence of events postulated to be critical for NRTI-mediated abrogation of HIV-1 replication. These events consist of NRTI incorporation, RNase H cleavage, and template–primer dissociation. Open circles, RNA template; filled circles, growing DNA strand; open squares, incorporated NRTI; thin arrows, RNase H cleavage of the template RNA after it is copied.
A Proposed Mechanism for NRTI-Mediated Abrogation of HIV-1 Replication. Our previous studies have provided strong evidence for a mechanism of RT template switching that we have named dynamic copy choice (11, 14). Mutations in RT or depletion of intracellular deoxynucleoside triphosphate pools, both of which slowed down the rate of DNA synthesis, increased the frequency of RT template switching, whereas mutations that reduced the rate of RNA degradation decreased this frequency. These observations indicated that an equilibrium exists between the rates of DNA synthesis and RNA degradation during reverse transcription and that this equilibrium is an important determinant of RT template switching. Our observations that AZT and d4T increased the frequency of RT template switching, and predictions of the dynamic copy choice model for retroviral recombination, suggested that the presence of these NRTIs altered the steady state between the rate of DNA synthesis and the rate of RNA degradation, resulting in an increase in the frequency of RT template switching. These observations led us to propose the mechanism for NRTI-mediated abrogation of HIV-1 replication shown in Fig. 1c. We hypothesize that NRTI incorporation leads to chain termination and stops DNA synthesis until nucleotide excision occurs and allows synthesis to resume. Degradation of the RNA template by RNase H leads to dissociation of the template and primer strands, termination of reverse transcription, and abrogation of HIV-1 replication. Thus, we postulate that an equilibrium exists between excision of an incorporated NRTI by wild-type RT, resumption of DNA synthesis, and RNase H activity; mutations in the polymerase domain of RT that confer AZT resistance increase the rate of excision of AZT, allowing DNA synthesis and viral replication to be resumed before RNA degradation leads to dissociation of the template and primer.
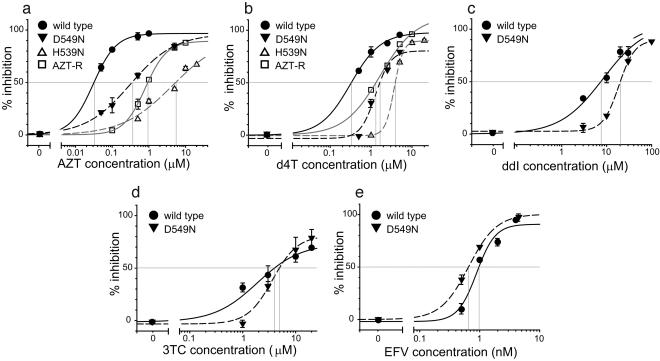
Mutations in RNase H Confer Resistance to AZT and d4T. A prediction of the model for NRTI-mediated abrogation of HIV-1 replication (Fig. 1c) is that mutations reducing the rate of RNA degradation will increase the time available for excision of AZT or d4T from a terminated primer, leading to an increase in drug resistance. To test this prediction, we determined the antiretroviral drug sensitivity of wild-type RT, an RT mutant (AZT-R) containing a well characterized cluster of TAM (D67N, K70R, T215Y, K219Q) (6, 12), and two mutants of RNase H that reduce RNase H activity, namely, D549N (15) and H539N (16, 17) (Fig. 2). The antiretroviral drug sensitivity was determined in a single cycle of replication by using a retroviral vector expressing the luciferase reporter gene.
Fig. 2.
Single-replication-cycle drug-susceptibility assays. Phenotypic drug-susceptibility testing for AZT (a), d4T (b), ddI (c), 3TC (d), and EFV (e) was performed with pNLuc-based viruses. The genotypes of the viruses were wild type (filled circles), AZT-resistant mutant (AZT-R) with four TAM (D67N, K70R, T215Y, and K219Q) (open squares), RNase H mutant D549N (inverted filled triangles), and RNase H mutant H539N (open triangles). Intersections of vertical lines with the drug concentration axis show the IC50 value for each curve. Representative graphs are shown.
Results obtained in the presence of AZT showed that the D549N mutation increased the AZT concentration needed to inhibit viral replication by 50% (IC50) ≈12-fold, similar to the increase observed for the AZT-R virus containing the TAM (≈23-fold) (Fig. 2a and Table 1). The D549N mutation resulted in a 5- to 10-fold reduction in viral titers in a single-replication-cycle assay (data not shown). Even more dramatically, the H539N mutation in RNase H, which had a greater defect in viral replication than the D549N mutation (100-fold decrease), increased the AZT IC50 180-fold relative to wild-type RT and 9-fold relative to the TAM.
Table 1. Effects of mutations in HIV-1 RNase H on sensitivity to RT inhibitors.
| AZT
|
d4T
|
ddI
|
3TC
|
EFV
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 ± SE, μM | Fold inc. | IC50 ± SE, μM | Fold inc. | IC50 ± SE, μM | Fold inc. | IC50 ± SE, μM | Fold inc. | IC50 ± SE, μM | Fold inc. | |
| Wild type | 0.04 ± 0.004* | - | 0.45 ± 0.04* | - | 10.99 ± 1.94† | - | 5.23 ± 0.98† | - | 1.4 ± 0.33† | - |
| AZT-R | 0.99 ± 0.36‡ | 23 | 1.08 ± 0.17‡ | 2.4 | 10.87 ± 2.7†§ | None | >20† | >3.8 | 1.2 ± 0.16†§ | None |
| D549N | 0.52 ± 0.15† | 12 | 1.06 ± 0.24‡ | 2.4 | 20.78 ± 2.76‡ | 1.9 | 8.85 ± 2.9†§ | None | 0.99 ± 0.3†§ | None |
| H539N | 7.96 ± 1.69† | 185 | 4.51 ± 0.22† | 10 | ND | ND | ND | |||
| AZT-R+D549N | ≈50‡¶ | ≈1,250 | 5.62 ± 1.55‡ | 12.5 | 9.94∥ | None | >20∥ | >3.8 | 0.89∥ | None |
| WT+E478Q mixed virions | 0.22 ± 0.07† | 5.5 | 1.6 ± 0.6† | 3.6 | ND | ND | ND | |||
IC50 means and standard errors were calculated by using sigmaplot 8.0 as described in Methods. Fold increases (Fold inc.) were calculated only for statistically significant differences in IC50 (P < 0.05, relative to wild type) by dividing the mutant virus IC50 mean by the wild-type virus IC50 mean for each drug. All infections were performed in duplicate, and the results were averaged. ND, not determined; -, not applicable.
IC50 mean calculated based on measurements of 9-15 separate infections
IC50 mean calculated based on measurements of two to four separate infections
IC50 mean calculated based on measurements of five to seven separate infections
IC50 is not statistically different from the wild type (P > 0.03)
For four of the five experiments, the highest drug concentration tested (10 μM) inhibited viral replication only by 10-30%; in one experiment, 50 μM AZT inhibited viral replication by 52%
IC50 mean calculated based on measurements of one infection
To determine whether the D549N and H539N mutations conferred resistance to another NRTI, we measured the sensitivity of the RNase H mutants to d4T (Fig. 2b and Table 1). Similar to the results obtained with AZT, we observed that the d4T IC50 was substantially increased relative to wild-type RT or the AZT-R mutant containing the TAM. The D549N mutation and the TAM increased the d4T IC50 2.4-fold, whereas the H539N mutation increased the d4T IC50 10-fold relative to wild-type RT and 4-fold relative to the TAM. Dideoxyadenosine (ddA), the active metabolite derived from ddI, is excised from blocked primers less efficiently than AZT or d4T (7, 18); consistent with the predictions of the model outlined in Fig. 1c, the D549N mutation in RNase H decreased sensitivity to ddI by a small (<2-fold) but statistically significant amount (Fig. 2c and Table 1; P < 0.05). These observations indicated that, in the absence of TAM, the mutations in the RNase H domain of HIV-1 RT conferred a high level of resistance to those NRTIs that could be efficiently removed from a blocked primer by the nucleotide excision mechanisms (e.g., AZT and d4T).
Mutation in RNase H Does Not Confer Resistance to 3TC or EFV. The NRTI 3TC is not excised efficiently from blocked primers (7). We therefore predicted that RNase H mutations would not increase resistance to 3TC. Consistent with this prediction, we did not observe a significant increase in resistance to 3TC by the presence of the D549N mutation in RNase H (Fig. 2d and Table 1; P > 0.2).
The proposed mechanism outlined in Fig. 1c for NRTI-mediated abrogation of HIV-1 replication predicts that mutations in RNase H will not influence sensitivity to NNRTIs, which are not chain terminators and inhibit HIV-1 RT by a different mechanism (19). To test this prediction, we determined the sensitivity of wild-type RT and the D549N RNase H mutant to EFV (Fig. 2e and Table 1; P > 0.4). The results were consistent with the predictions of the model and indicated that the D549N mutation had no influence on the sensitivity to EFV.
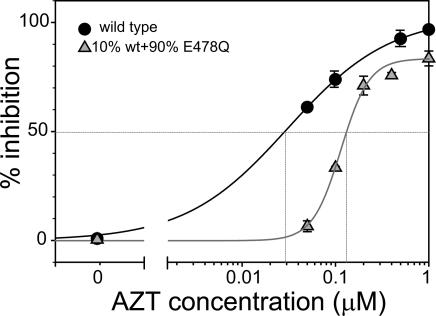
Reducing RNase H Activity by Phenotypic Mixing Confers Resistance to AZT. To determine whether the D549N and H539N mutations in RNase H increase resistance to NRTIs by reducing the overall RNase H activity or by altering the structure of RT, we generated phenotypically mixed virions with reduced RNase H activity as described in ref. 20 and determined their sensitivity to AZT (Fig. 3 and Table 1). 293T cells were cotransfected at a 1:9 ratio with DNAs encoding wild-type RT and RNase H mutant E478Q, which is almost completely defective in RNase H activity (21). HIV-1 RT is a p66:p51 heterodimer wherein the p66 monomer contains catalytically active polymerase and RNase H domains and the p51 subunit contains only a catalytically inactive polymerase domain (22). Approximately 90% of the RT heterodimers in the phenotypically mixed virions were expected to contain a p66 monomer in which the RNase H is inactivated by the E478Q mutation, and ≈10% of heterodimers were expected to contain a p66 monomer with a functional RNase H domain. Thus, the phenotypically mixed virions are expected to contain ≈10% of the RNase H activity present in the wild-type virions. We determined the sensitivity to AZT of these phenotypically mixed virions containing reduced RNase H activity, which exhibited a 10-fold reduced infectivity relative to wild type. Reducing RNase H activity by phenotypic mixing increased the AZT IC50 ≈5.5-fold, indicating that overall reduction in RNase H activity was sufficient to confer resistance to AZT.
Fig. 3.
AZT-susceptibility profile for phenotypically mixed RNase-H-defective HIV-1 virions. RNase-H-defective HIV-1 virions were generated by cotransfection of wild-type pNLuc DNA and RNase-H-defective mutant pNLuc-E478Q DNA at a 1:9 ratio. Intersections of vertical lines with the drug concentration axis show the IC50 value for each curve. Representative graphs are shown.
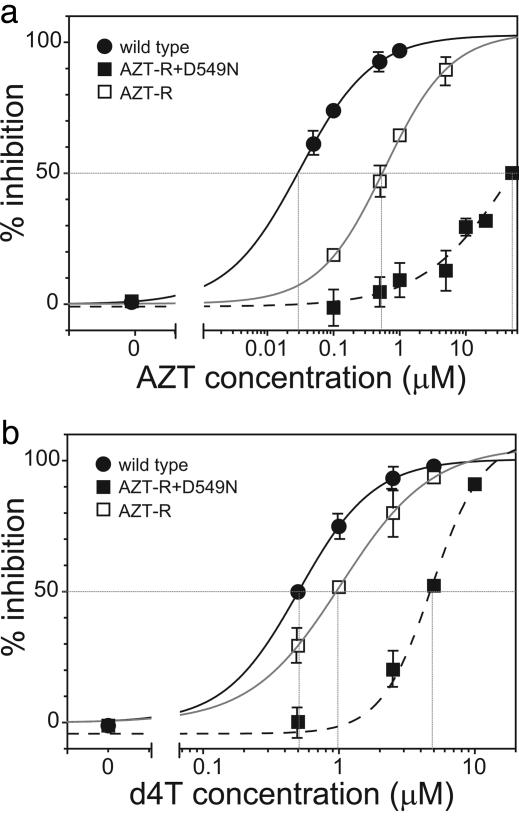
Mutations in RNase H and TAM Increase Resistance to NRTIs in a Synergistic Manner. To determine whether mutations in RNase H influenced the sensitivity to NRTIs in the presence of TAM, we constructed mutants that contained the four TAM present in the AZT-R mutant (D67N, K70R, T215Y, and K219Q) as well as the D549N substitution in RNase H. We then compared the sensitivity to AZT and d4T of wild-type RT, the AZT-R mutant, and the mutant containing the TAM and D549N (AZT-R+D549N) (Fig. 4 and Table 1). The results showed that the AZT-R+D549N mutant was substantially more resistant to AZT and d4T than wild-type RT or the RT containing the TAM alone. For the AZT-R+D549N mutant, the AZT IC50 was ≈50 μM, revealing a 1,250-fold reduced susceptibility to AZT compared with the wild-type virus. The increase in the AZT IC50 exhibited by the AZT-R+D549N mutant (1,250-fold) was also greater than the product of the AZT IC50 values for the AZT-R mutant (23-fold) and the D549N mutant (12-fold), indicating that the TAM and the D549N mutation synergistically increased the resistance to AZT.
Fig. 4.
Drug-susceptibility profile for HIV-1 RT mutants containing a combination of TAM and RNase H mutations. Phenotypic drug-susceptibility testing for AZT (a) and d4T (b) was performed with pNLuc-based viruses. The genotypes of the viruses were wild type (filled circles), AZT-resistant mutant (AZT-R) with four TAM (D67N, K70R, T215Y, and K219Q) (open squares), and mutant containing a combination of four TAM and RNase H mutation D549N (AZT-R+549N) (filled squares). Intersections of vertical lines with the drug concentration axis show the IC50 value for each curve. Representative graphs are shown.
Similarly, the increase in the d4T IC50 exhibited by the AZT-R+D549N mutant (12.5-fold) was greater than the product of the increase in the d4T IC50 values for the AZT-R mutant (2.4-fold) and the D549N mutant (2.4-fold), indicating that the TAM and the D549N mutation also increased the resistance to d4T in a synergistic manner.
Discussion
These studies strongly support a mechanism we propose here in which the interplay between NRTI incorporation, nucleotide excision, and RNase H activity is critical in NRTI-mediated abrogation of HIV-1 replication as well as in NRTI resistance. It is generally believed that NRTI incorporation during viral DNA synthesis leads to chain termination followed by abrogation of viral replication (23). The results of our studies provide a more complete picture of the events that lead to termination of viral DNA synthesis upon NRTI incorporation, taking into account the effects of the nucleotide excision activity and the dynamic steady state between polymerase and RNase H activities. The effects of the RNase H mutations on increasing NRTI resistance parallel the efficiency with which the NRTIs are excised from a blocked primer (AZT > d4T > ddI ≈ 3TC) (7). The results of these studies also imply that incorporation of some NRTIs will terminate viral replication only during RNA-dependent DNA synthesis because the time available for nucleotide excision is limited by the degradation of the template by RNase H. In contrast, viral replication will not be significantly inhibited during DNA-dependent DNA synthesis because the time available for nucleotide excision is not limited by the RNase H activity.
The observation that mutations in RNase H did not increase resistance to EFV suggests that, in contrast to AZT and d4T, inhibition of reverse transcription by EFV is not influenced by reductions in RNase H activity. EFV and other NNRTIs inhibit reverse transcription by binding to RT at the specific site, and their inhibition is not influenced by the presence of mutations that enhance nucleotide excision. Similar to the model outlined in Fig. 1, it is conceivable that inhibition of DNA synthesis by NNRTI–RT interactions depends on RNase H degradation of the template RNA, leading to a dissociation of the primer–template complex. This model suggests that the affinity of the NNRTI to the RT could determine the rate with which the NNRTI is released from the RT and DNA synthesis is reinitiated. One possibility is that EFV binds to RT in an irreversible manner, and slowing down the rate of RNA degradation does not lead to reinitiation of DNA synthesis before the template–primer complex is dissociated. It is possible that RNase H might influence resistance to other NNRTIs that bind to RT with a lower affinity.
It has been shown that AZT monophosphate inhibits RNase H activity (24). One hypothesis is that the mutations in RNase H that increase AZT resistance alter the inhibition of RNase H by AZT monophosphate. Two observations argue against this hypothesis. First, phenotypically mixed virions containing lower levels of wild type RNase H exhibit AZT resistance, indicating that a reduction in RNase H activity, rather than the specific mutations, is correlated with AZT resistance. Second, the same RNase H mutations also increased resistance to d4T.
These studies have important implications for genotypic and phenotypic analysis of HIV-1 for clinical drug resistance. In nearly all of the sequencing analyses of HIV-1 genomes for drug resistance performed by commercial genotyping services, PCR products that encompass the protease and ≈350 aa of RT are analyzed, which does not include the RNase H domain (12, 25, 26); for example, the GenBank sequence database contains >100,000 HIV-1 RT sequences but only ≈1,000 RNase H sequences. It is important to include the RNase H domain in future genotypic analyses to determine the potential contribution of RNase H domain to NRTI resistance. Similarly, current vectors used to generate recombinant virus for phenotypic analyses of drug resistance do not include the RNase H domain and could result in underestimation of drug resistance.
Mutations in RNase H that might confer resistance to NRTIs are not known. The results of these studies suggest that any mutations that substantially reduce RNase H activity will confer NRTI resistance; if this prediction is correct, then a large number of mutations in the RNase H domain might be associated with the NRTI-resistance phenotype. Interestingly, a survey of the HIV-1 RNase H sequences that are available in GenBank for the presence of mutations in the RNase H domain revealed one sequence containing the D549N substitution that was associated with TAM in a drug-experienced patient (27) (GenBank accession no. AAK12154.1); additionally, we found 32 other substitutions in the RNase H primer grip motif [N474 (2), Q475 (6), K476 (21), Y501 (2), and I505 (1)] that might reduce RNase H activity and contribute to NRTI resistance. The presence of the D549N substitution and other RNase H mutations in HIV-1 genomes isolated from patients suggests that RNase H mutations contribute to NRTI resistance in vivo. The effect of the D549N or other RNase H mutations on NRTI resistance may be different in clinical isolates or in other HIV-1 subtypes. It is also important to point out that the D549N and H539N mutations analyzed in this study might not be frequently selected in vivo in response to therapy; selection of drug-resistant variants will depend on the balance between the level of resistance to the antiviral drug and the effect of the mutation on the viral fitness.
The observation that AZT increases the frequency of RT template switching indicates that AZT therapy could enhance the frequency of recombination and thereby increase variation in HIV-1 populations in infected individuals. Previous studies have shown that AZT treatment can increase the mutation rates of oncoretroviruses (28) and HIV-1 (29). Increasing variation in HIV-1 could potentially enhance the rate of viral evolution and facilitate escape from the host immunological responses or development of drug resistance. The increased frequency of RT template switching within a short homologous repeat (250 bp) also suggests that cycles of AZT incorporation/nucleotide excision are likely to be carried out frequently by wild-type HIV-1 RT during the course of viral DNA synthesis. The frequencies with which NRTIs are incorporated and the subset of incorporation events in which nucleotide excision fails to occur are unknown.
Another implication of these studies is that inhibitors of HIV-1 RNase H that are currently being identified as antiretroviral agents (30–35) will enhance resistance to some NRTIs. It will be important to determine whether the anti-RNase H inhibitors under development will antagonize the activity of some NRTIs so that effective antiretroviral combination therapies can be devised.
In summary, our studies indicate that an equilibrium between nucleotide excision and RNase H degradation plays an important role in NRTI-mediated inhibition of HIV-1 replication, as well as NRTI drug resistance, and point to a need for more comprehensive investigations of the in vivo role of RNase H in NRTI resistance.
Acknowledgments
We especially thank Wei-Shau Hu for helpful discussions and critical reading of the manuscripts and Anne Arthur for expert editorial help. We thank Vitaly Boyko for help with GenBank database analysis and David Thomas for critical reading of the manuscript.
Abbreviations: 3TC, 2′,3′-dideoxy-3′-thiacytidine; AZT, 3′-azido-3′-deoxythymidine; ddI, 2′,3′-dideoxyinosine; d4T, 2,3-didehydro-2,3-dideoxythymidine; EFV, efavirenz; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-NRTI; RT, reverse transcriptase; TAM, thymidine analogs.
References
- 1.Havlir, D. V. (2002) Proc. Natl. Acad. Sci. USA 99, 4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shafer, R. W., Winters, M. A., Palmer, S. & Merigan, T. C. (1998) Ann. Intern. Med. 128, 906-911. [DOI] [PubMed] [Google Scholar]
- 3.Goldschmidt, V. & Marquet, R. (2004) Intern. J. Biochem. Cell Biol. 36, 1687-1705. [DOI] [PubMed] [Google Scholar]
- 4.Arion, D., Kaushik, N., McCormick, S., Borkow, G. & Parniak, M. A. (1998) Biochemistry 37, 15908-15917. [DOI] [PubMed] [Google Scholar]
- 5.Meyer, P. R., Matsuura, S. E., Mian, A. M., So, A. G. & Scott, W. A. (1999) Mol. Cell 4, 35-43. [DOI] [PubMed] [Google Scholar]
- 6.Meyer, P. R., Matsuura, S. E., Tolun, A. A., Pfeifer, I., So, A. G., Mellors, J. W. & Scott, W. A. (2002) Antimicrob. Agents Chemother. 46, 1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naeger, L. K., Margot, N. A. & Miller, M. D. (2002) Antimicrob. Agents Chemother. 46, 2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiernan, R. E., Ono, A., Englund, G. & Freed, E. O. (1998) J. Virol. 72, 4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naldini, L., Blomer, U., Gage, F. H., Trono, D. & Verma, I. M. (1996) Proc. Natl. Acad. Sci. USA 93, 11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yee, J. K., Miyanohara, A., Laporte, P., Bouic, K., Burns, J. C. & Friedmann, T. (1994) Proc. Natl. Acad. Sci. USA 91, 9564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikolenko, G. N., Svarovskaia, E. S., Delviks, K. A. & Pathak, V. K. (2004) J. Virol. 78, 8761-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petropoulos, C. J., Parkin, N. T., Limoli, K. L., Lie, Y. S., Wrin, T., Huang, W., Tian, H., Smith, D., Winslow, G. A., Capon, D. J. & Whitcomb, J. M. (2000) Antimicrob. Agents Chemother. 44, 920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, W. S. & Temin, H. M. (1990) Proc. Natl. Acad. Sci. USA 87, 1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svarovskaia, E. S., Delviks, K. A., Hwang, C. K. & Pathak, V. K. (2000) J. Virol. 74, 7171-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristofaro, J. V., Rausch, J. W., Le Grice, S. F. J. & DeStefano, J. J. (2002) Biochemistry 41, 10968-10975. [DOI] [PubMed] [Google Scholar]
- 16.Tisdale, M., Schulze, T., Larder, B. A. & Moelling, K. (1991) J. Gen. Virol. 72, 59-66. [DOI] [PubMed] [Google Scholar]
- 17.Volkmann, S., Wohrl, B. M., Tisdale, M. & Moelling, K. (1993) J. Biol. Chem. 268, 2674-2683. [PubMed] [Google Scholar]
- 18.Meyer, P. R., Smith, A. J., Matsuura, S. E. & Scott, W. A. (2004) J. Biol. Chem. 279, 45389-45398. [DOI] [PubMed] [Google Scholar]
- 19.Shafer, R. W. (2002) Clin. Microbiol. Rev. 15, 247-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julias, J. G., Ferris, A. L., Boyer, P. L. & Hughes, S. H. (2001) J. Virol. 75, 6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schatz, O., Cromme, F. V., Gruningerleitch, F. & Le Grice, S. F. J. (1989) FEBS Lett. 257, 311-314. [DOI] [PubMed] [Google Scholar]
- 22.Huang, H., Chopra, R., Verdine, G. L. & Harrison, S. C. (1998) Science 282, 1669-1675. [DOI] [PubMed] [Google Scholar]
- 23.Menendez-Arias, L. (2002) Trends Pharmacol. Sci. 23, 381-388. [DOI] [PubMed] [Google Scholar]
- 24.Tan, C. K., Civil, R., Mian, A. M., So, A. G. & Downey, K. M. (1991) Biochemistry 30, 4831-4835. [DOI] [PubMed] [Google Scholar]
- 25.Shafer, R. W., Hertogs, K., Zolopa, A. R., Warford, A., Bloor, S., Betts, B. J., Merigan, T. C., Harrigan, R. & Larder, B. A. (2001) J. Clin. Microbiol. 39, 1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tural, C., Ruiz, L., Holtzer, C., Schapiro, J., Viciana, P., Gonzalez, J., Domingo, P., Boucher, C., Rey-Joly, C. & Clotet, B. (2002) AIDS 16, 209-218. [DOI] [PubMed] [Google Scholar]
- 27.Lukashov, V. V., Huismans, R., Jebbink, M. F., Danner, S. A., De Boer, R. J. & Goudsmit, J. (2001) AIDS Res. Human Retroviruses 17, 807-818. [DOI] [PubMed] [Google Scholar]
- 28.Julias, J. G., Kim, T., Arnold, G. & Pathak, V. K. (1997) J. Virol. 71, 4254-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansky, L. M. & Bernard, L. C. (2000) J. Virol. 74, 9532-9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loya, S. S. & Hizi, A. (1993) J. Biol. Chem. 268, 9323-9328. [PubMed] [Google Scholar]
- 31.Min, B. S., Miyashiro, H. & Hattori, M. (2002) Phytother. Res. 16, S57-S62. [DOI] [PubMed] [Google Scholar]
- 32.Gabbara, S., Davis, W. R., Hupe, L., Hupe, D. & Peliska, J. A. (1999) Biochemistry 38, 13070-13076. [DOI] [PubMed] [Google Scholar]
- 33.Borkow, G., Fletcher, R. S., Barnard, J., Arion, D., Motakis, D., Dmitrienko, G. I. & Parniak, M. A. (1997) Biochemistry 36, 3179-3185. [DOI] [PubMed] [Google Scholar]
- 34.Shaw-Reid, C. A., Munshi, V., Graham, P., Wolfe, A., Witmer, M., Danzeisen, R., Olsen, D. B., Carroll, S. S., Embrey, M., Wai, J. S., et al. (2003) J. Biol. Chem. 278, 2777-2780. [DOI] [PubMed] [Google Scholar]
- 35.Parniak, M. A., Min, K. L., Budihas, S. R., Le Grice, S. F. & Beutler, J. A. (2003) Anal. Biochem. 322, 33-39. [DOI] [PubMed] [Google Scholar]