Abstract
Salinicoccus sp. BAB 3246 is a halophilic bacterium isolated from a marine water sample collected from the coastal region of Gujarat, India, from a surface water stream. Based on 16sRNA sequencing, the organism was identified as Salinicoccus sp. BAB 3246 (Genebank ID: KF889285). The present work was performed to determine the whole genome sequence of the organism using Ion Torrent PGM platform followed by assembly using the CLC genomics workbench and genome annotation using RAST, BASys and MaGe. The complete genome sequence was 713,204 bp identified by with second largest size for Salinicoccus sp. reported in the NCBI genome database. A total of 652 degradative pathways were identified by KEGG map analysis. Comparative genomic analysis revealed Salinicoccus sp. BAB 3246 as most highly related to Salinicoccus halodurans H3B36. Data mining identified stress response genes and operator pathway for degradation of various environmental pollutants. Annotation data and analysis indicate potential use in pollution control in industrial influent and saline environment.
| Specifications | |
|---|---|
| Organism/cell line/tissue | Salinicoccus sp. BAB 3246 |
| Sex | Not applicable |
| Sequencer or array type | Ion Torrent PGM platform |
| Data format | Fasta complete genome |
| Experimental factors | Marine water sample |
| Experimental features | Shotgun whole genome sequencing followed by genome annotation using RAST, BASys and MaGe. |
| Sample source location | Gujarat, India (21.672439 N 72.275925 E) |
| Data submission |
BioProject: PRJNA342322 RAST: genome ID 1437774.4 - Salinicoccus sp. BAB-3246 |
1. Direct link to deposited data
BioProject: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA342322.
2. Introduction
The genus Salinicoccus, belonging to family Staphylococcaceae was first proposed by Ventosa et al., (1990) and is defined as moderately halophilic, aerobic, Gram-positive, non-motile, non-sporulating, and heterotrophic cocci [1]. The genomic DNA G + C content of the species in this genus lies within the range of 46–51 mol%. Most species in genus Salinicoccus including Salinicoccus albus, Salinicoccus carnicancri, Salinicoccus roseu, Salinicoccus halodurans, Salinicoccus luteus have been found in salty environments, such as fermented foods, solar salterns, salt mines, salt lakes, and saline soils [1], [2], [3], [4], [5], [6], [7]. Alongside, genus Salinicoccus is also reported for production of Amylase, Protease, Gelatinase like enzymes in hyper saline environments [8].
The members of the Salinicoccus genus are abundant in the marine environments suggesting that they play important roles in marine ecosystems, such as the degradation of aromatic compounds and the biogeochemical cycles of carbon and sulfur [5]. S. roseus has been reported to exhibit high salinity and high lactate resistance [9]. Salinicocci have much importance in biotechnology applications such as serine-metabolism strategies to adapt to lactate stress [10]. In order to understand the genetic variability and industrial applications of those genes, genome sequencing and annotation of strain Salinicoccus sp. BAB 3246 was executed. The prime interest was to identify presence of distinctive enzymes for potential industrial applications.
3. Experimental design, materials and methods
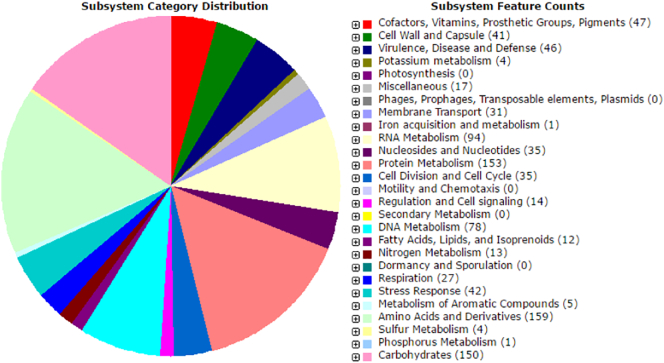
The halophilic organism was isolated from marine water collected from surface streams of coastal region near Bhavnagar, Gujarat, India (latitude, longitude: 21.67 N, 72.27E). The isolation was performed by providing 15% Sodium Chloride containing Medium. The identification of Salinococcus sp. BAB 3246 was validated by 16 s rRNA sequencing and submitted to Genebank (accession no: KF889285.1). Furthermore, the DNA was extracted using Hi-Media Kit for Genomic DNA isolation Kit. The genome sequencing was performed using Ion Torrent PGM generating 15,26,815 sequencing reads. Initially all reads were subjected to preprocessing and conversion of BAM to fasta file format using Galaxy NGS: BamTools, online server using default parameters provided by the developer [11]. The genome data were assembled using CLC Genomic Workbench 5. The final whole genome assembly size was reported is 7,13,204 bp. The genome annotation was performed using RAST (Rapid Annotation using Subsystem Technology) [12], BASys (a web server for automated bacterial genome annotation) [13] and MaGe (Microscope Genome Annotation) [14]. The RAST analysis revealed total 1691 coding sequences (Table 1). A total of 1009 subsystems were identified, including Stress Response (42), Sulfur Metabolism (4), Potassium metabolism (4) and Iron metabolism (1). However, the highest numbers of subsystems were observed for Amino Acids and Derivatives (159), Protein Metabolism (153) and Carbohydrate synthesis (150) (Fig. 1). KEGG pathway analysis was performing using seed viewer system of RAST. The KEGG map analysis revealed 652 pathways associated with only degradation of metabolites (Table 2).
Table 1.
Summary of RAST annotation.
| Genome | Salinicoccus sp. BAB 3246 |
|---|---|
| Size (bp) | 7,13,204 |
| G + C content | 49.1 |
| Number of coding sequences | 1691 |
| Number of features | 1762 |
| Number of subsystems | 1009 |
| Number of RNAs | 71 |
| Number of contigs | 1 |
Fig. 1.
Subsystem category distribution.
Table 2.
KEGG map analysis for degradation pathway.
| No | Name of derivative | KEGG map | Salinicoccus sp. BAB-3246 |
|---|---|---|---|
| 1 | 1,1,1-Trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) degradation | Tyrosine metabolism | 4 |
| 2 | 1,2-Dichloroethane degradation | 1,2-Dichloroethane degradation | 1 |
| Glyoxylate and dicarboxylate metabolism | 9 | ||
| 3 | 1,4-Dichlorobenzene degradation | Benzoate degradation via hydroxylation | 2 |
| Glycolysis/gluconeogenesis | 17 | ||
| Glyoxylate and dicarboxylate metabolism | 9 | ||
| Pyruvate metabolism | 14 | ||
| 4 | 1- and 2-Methylnaphthalene degradation | 1- and 2-Methylnaphthalene degradation | 2 |
| Naphthalene and anthracene degradation | 1 | ||
| 5 | 2,4-Dichlorobenzoate degradation | Benzoate degradation via hydroxylation | 2 |
| Naphthalene and anthracene degradation | 1 | ||
| 6 | 3-Chloroacrylic acid degradation | 3-Chloroacrylic acid degradation | 1 |
| Pyruvate metabolism | 14 | ||
| 7 | Atrazine degradation | Atrazine degradation | 1 |
| Folate biosynthesis | 5 | ||
| 8 | Benzoate degradation via CoA ligation | Benzoate degradation via CoA ligation | 4 |
| Benzoate degradation via hydroxylation | 2 | ||
| Butanoate metabolism | 9 | ||
| Ethylbenzene degradation | 1 | ||
| Phenylalanine metabolism | 1 | ||
| Pyruvate metabolism | 14 | ||
| 9 | Benzoate degradation via hydroxylation | Benzoate degradation via CoA ligation | 4 |
| Benzoate degradation via hydroxylation | 2 | ||
| Caprolactam degradation | 2 | ||
| Glycolysis/gluconeogenesis | 17 | ||
| Naphthalene and anthracene degradation | 1 | ||
| Phenylalanine metabolism | 1 | ||
| Pyruvate metabolism | 14 | ||
| Tryptophan metabolism | 9 | ||
| Tyrosine metabolism | 4 | ||
| 10 | Biphenyl degradation | Benzoate degradation via CoA ligation | 4 |
| Benzoate degradation via hydroxylation | 2 | ||
| Glycolysis/gluconeogenesis | 17 | ||
| Pyruvate metabolism | 14 | ||
| 11 | Bisphenol A degradation | Benzoate degradation via hydroxylation | 2 |
| 12 | Caprolactam degradation | Benzoate degradation via hydroxylation | 2 |
| Caprolactam degradation | 2 | ||
| 13 | Carbazole degradation | Benzoate degradation via CoA ligation | 4 |
| Benzoate degradation via hydroxylation | 2 | ||
| Glycolysis/gluconeogenesis | 17 | ||
| Pyruvate metabolism | 14 | ||
| Tryptophan metabolism | 9 | ||
| 14 | Ethylbenzene degradation | Benzoate degradation via CoA ligation | 4 |
| Ethylbenzene degradation | 1 | ||
| Glycolysis/gluconeogenesis | 17 | ||
| Propanoate metabolism | 6 | ||
| Pyruvate metabolism | 14 | ||
| 15 | Fluorene degradation | Benzoate degradation via hydroxylation | 2 |
| Glycolysis/gluconeogenesis | 17 | ||
| Pyruvate metabolism | 14 | ||
| 16 | Fluorobenzoate degradation | Benzoate degradation via hydroxylation | 2 |
| 17 | Geraniol degradation | Geraniol degradation | 3 |
| Valine, leucine and isoleucine degradation | 9 | ||
| 18 | Limonene and pinene degradation | Limonene and pinene degradation | 3 |
| 19 | Lysine degradation | Biotin metabolism | 1 |
| Citrate cycle (TCA cycle) | 14 | ||
| Lysine biosynthesis | 5 | ||
| Lysine degradation | 6 | ||
| 20 | Naphthalene and anthracene degradation | Benzoate degradation via hydroxylation | 2 |
| Naphthalene and anthracene degradation | 1 | ||
| Pyruvate metabolism | 14 | ||
| Tryptophan metabolism | 9 | ||
| Tyrosine metabolism | 4 | ||
| 21 | Other glycan degradation | Glycosphingolipid biosynthesis - ganglio series | 1 |
| 22 | Styrene degradation | Citrate cycle (TCA cycle) | 14 |
| Ethylbenzene degradation | 1 | ||
| Glycolysis/gluconeogenesis | 17 | ||
| Propanoate metabolism | 6 | ||
| Pyruvate metabolism | 14 | ||
| 23 | Synthesis and degradation of ketone bodies | Butanoate metabolism | 9 |
| Fatty acid metabolism | 5 | ||
| Glycolysis/gluconeogenesis | 17 | ||
| Pyruvate metabolism | 14 | ||
| 24 | Tetrachloroethene degradation | Glyoxylate and dicarboxylate metabolism | 9 |
| Pyruvate metabolism | 14 | ||
| 25 | Toluene and xylene degradation | Benzoate degradation via CoA ligation | 4 |
| Benzoate degradation via hydroxylation | 2 | ||
| Glycerolipid metabolism | 3 | ||
| Glycolysis/gluconeogenesis | 17 | ||
| Pyruvate metabolism | 14 | ||
| 26 | Trinitrotoluene degradation | Trinitrotoluene degradation | 1 |
| 27 | Valine, leucine and isoleucine degradation | Biosynthesis of type II polyketide backbone | 1 |
| Citrate cycle (TCA cycle) | 14 | ||
| Propanoate metabolism | 6 | ||
| Pyrimidine metabolism | 17 | ||
| Valine, leucine and isoleucine biosynthesis | 12 | ||
| Valine, leucine and isoleucine degradation | 9 | ||
| 28 | Gamma-Hexachlorocyclohexane degradation | Benzoate degradation via hydroxylation | 2 |
| Citrate cycle (TCA cycle) | 14 | ||
| Glyoxylate and dicarboxylate metabolism | 9 | ||
| Naphthalene and anthracene degradation | 1 |
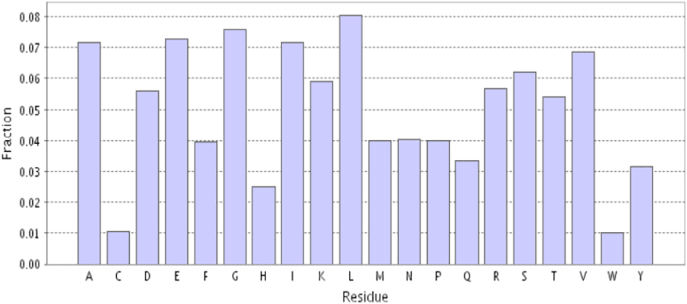
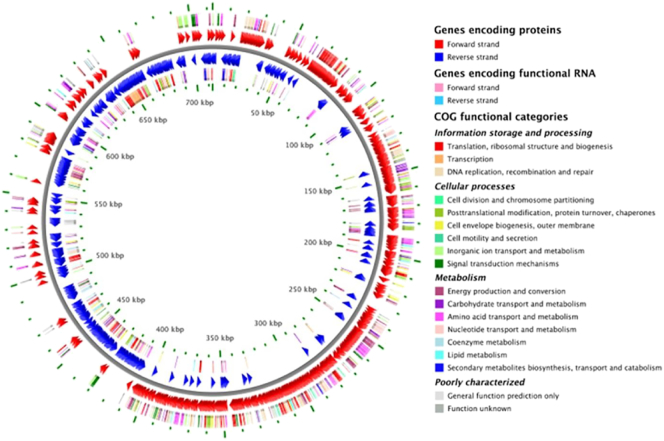
The genome annotation using BASys annotate 955 genes amongst total 2330 genes reported in and automated mode. The amino acid composition was also examined using BASys (Fig. 2). The highest amino acid residue content was predicted for Leucine followed by Glycine, Glutamic acid and Alanine. Annotated data were displayed in the form of circular DNA as a genome browser map for easy representation of genome data (Fig. 3). The genome annotation using Microscope Genome Annotation identified 1772 Genomic Objects (without artifacts): CDS, 1326; fCDS, 358; misc_RNA, 16; rRNA, 12; tRNA, 60.
Fig. 2.
Amino acid composition of Salinicoccus sp. BAB 3246.
Fig. 3.
Genome browser map for Salinicoccussp. BAB 3246.
4. Quantitative comparison of coding sequences, rna and subsystem
The comparison of genome size for six different strains available in NCBI genome database revealed that, S. halodurans strain had the largest genome size of 2,778,379 bp followed by 873,136 bp, 713,204 bp, 679,606 bp, 461,933 bp and 342,819 bp respectively for S. carnicancri Crm, Salinicoccus sp. BAB 3246, S. luteus DSM 17002, S. roseus and S. albus DSM 19776 strain. A maximum of 2839 coding sequences was reported for S. halodurans followed by 1691, 863, 668, 449 and 334 respectively for Salinicoccus sp. BAB 3246, S. carnicancri Crm, S. luteus DSM 17002, S. roseus and S. albus DSM 19776 strain (Table 3).
Table 3.
Quantitative comparison of coding sequence, RNA and subsystem.
| Genome | Size (bp) | G + C content | Coding sequences | Features | RNAs | Subsystems | BioProject |
|---|---|---|---|---|---|---|---|
| Salinicoccus sp. BAB_3246 | 713,204 | 49.1 | 1691 | 1762 | 71 | 202 | PRJNA342322 |
| Salinicoccus roseus | 461,933 | 49.9 | 449 | 459 | 10 | 80 | PRJNA272357 |
| Salinicoccus carnicancri Crm | 873,136 | 47.6 | 863 | 909 | 46 | 138 | PRJNA175941 |
| Salinicoccus albus DSM 19776 | 342,819 | 45.2 | 334 | 334 | 0 | 77 | PRJNA185242 |
| Salinicoccus luteus DSM 17002 | 679,606 | 49.7 | 668 | 669 | 1 | 114 | PRJNA235106 |
| Salinicoccus halodurans | 2,778,379 | 44.5 | 2839 | 2912 | 73 | 388 | PRJNA282445 |
5. Nucleotide sequence accession number
The complete sequence of Salinicoccus sp. BAB 3246 genome can be accessed under the NCBI BioProject: PRJNA342322.
Acknowledgments
We/authors are thankful to Department of Life Sciences and The Virtual Institute of Bioinformatics, HNGU, Patan, India to provide sequencing and computational facility.
References
- 1.Ventosa A., Márquez M.C., Ruiz-Berraquero F., Kocur M. Salinicoccus roseus gen. nov., sp. nov., a new moderately halophilic gram-positive coccus. Syst. Appl. Microbiol. 1990;13:29–33. [Google Scholar]
- 2.França L., Rainey F.A., Nobre M.F., da Costa M.S. Salinicoccus salsiraiae sp. nov.: a new moderately halophilic gram-positive bacterium isolated from salted skate. Extremophiles. 2006;10:531-6. doi: 10.1007/s00792-006-0532-1. [DOI] [PubMed] [Google Scholar]
- 3.Aslam Z., Lim J.H., Im W.-T., Yasir M., Chung Y.R., Lee S.-T. Salinicoccus jeotgali sp. nov., isolated from jeotgal, a traditional Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 2007;57:633-8. doi: 10.1099/ijs.0.64586-0. [DOI] [PubMed] [Google Scholar]
- 4.Gao M., Wang L., Chen S., Zhou Y., Liu H. Salinicoccus kekensis sp. nov., a novel alkaliphile and moderate halophile isolated from Keke Salt Lake in Qinghai, China. Antonie Van Leeuwenhoek. 2010;98:351-7. doi: 10.1007/s10482-010-9449-x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y.Q., Yu L.Y., Liu H.Y., Zhang Y.Q., Xu L.H., Li W.J. Salinicoccus luteus sp. nov., isolated from a desert soil. Int. J. Syst. Evol. Microbiol. 2007;57:1901–1905. doi: 10.1099/ijs.0.64967-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y.G., Cui X.L., Wang Y.X., Zhang Y.Q., Li Q.Y., Liu Z.X., Wen M.L., Peng Q., Li W.J. Salinicoccus albus sp. nov., a halophilic bacterium from a salt mine. Int. J. Syst. Evol. Microbiol. 2009;59:874–879. doi: 10.1099/ijs.0.003251-0. [DOI] [PubMed] [Google Scholar]
- 7.Jung M.J., Kim M.S., Roh S.W., Shin K.S., Bae J.W. Salinicoccus carnicancri sp. nov., a halophilic bacterium isolated from a Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 2010;60:653–658. doi: 10.1099/ijs.0.012047-0. [DOI] [PubMed] [Google Scholar]
- 8.de Lourdes Moreno M., Pérez D., García M.T., Mellado E. Halophilic bacteria as a source of novel hydrolytic enzymes. Lifestyles (Basel, Switzerland) 2013;3:38–51. doi: 10.3390/life3010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Wang L., Yang H., Cai Y., Sun L., Xue Y., Yu B., Ma Y. Comparative proteomic insights into the lactate responses of halophilic Salinicoccus roseus W12. Sci Rep. 2015;5:13776. doi: 10.1038/srep13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ventosa A. Bergey's Man. Syst. Archaea Bact. John Wiley & Sons, Ltd; Chichester, UK: 2015. Salinicoccus; pp. 1–7. [Google Scholar]
- 11.Barnett D.W., Garrison E.K., Quinlan A.R., Str̈mberg M.P., Marth G.T. Bamtools: a C ++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aziz R.K., Bartels D., Best A. a, DeJongh M., Disz T., Edwards R. a, Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R. a, McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genet. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Domselaar G.H., Stothard P., Shrivastava S., Cruz J.A., Guo A.C., Dong X., Lu P., Szafron D., Greiner R., Wishart D.S. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33:455–459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallenet D., Belda E., Calteau A., Cruveiller S., Engelen S., Lajus A., Le Févre F., Longin C., Mornico D., Roche D., Rouy Z., Salvignol G., Scarpelli C., Smith A.A.T., Weiman M., Médigue C. MicroScope - an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013;41:636–647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]