Highlights
-
•
Fructose-1,6-bisphosphatase (class II) is an essential enzyme and attractive drug target in M. tuberculosis using structure-based drug design.
-
•
T84A mutagenesis in the active site of MtFBPase shows no enzyme activity and no change in binding affinity.
-
•
T84S mutagenesis in the active site of MtFBPase results in reduced enzyme activity (lower Vmax), but retains lithium sensitive.
-
•
The optimal position of the catalytic OH- from the essential Thr84 has likely been subtly altered, resulting in lower enzymatic efficiency.
Keywords: glpX; Mycobacterium tuberculosis; Fructose-1,6-bisphosphatase; Lithium sensitive enzyme; Gluconeogenesis; Site directed mutagenesis
Abstract
The glpX gene encodes for the Class II fructose-1,6-bisphosphatase enzyme in Mycobacterium tuberculosis (Mt), an essential enzyme for pathogenesis. We have performed site directed mutagenesis to introduce two mutations at residue Thr84, T84A and T84S, to explore the binding affinity of the substrate and the catalytic mechanism. The T84A mutant fully abolishes enzyme activity while retaining substrate binding affinity. In contrast, the T84S mutant retains some activity having a 10 times reduction in Vmax and exhibited similar sensitivity to lithium when compared to the wildtype. Homology modeling using the Escherichia coli enzyme structure suggests that the replacement of the critical nucleophile OH− in the Thr84 residue of the wildtype of MtFBPase by Ser84 results in subtle alterations of the position and orientation that reduce the catalytic efficiency. This mutant could be used to trap reaction intermediates, through crystallographic methods, facilitating the design of potent inhibitors via structure-based drug design.
1. Introduction
Infections as a result of Mycobacterium tuberculosis (Mt) continues to be one of the leading causes of death in the world and the emergence of multidrug resistant and extensively drug resistant strains have prolonged and complicated the treatment of the disease. Consequently, new drugs that target pathways or mechanisms not previously exploited by the current regimen of tuberculosis drug treatment are needed, especially for strains already resistant to the first line tuberculosis (TB) drugs.
The gluconeogenesis pathway is essential for many organisms, allowing the synthesis of glucose from organic precursor compounds. It has previously been shown that through the essential phosphoenolpyruvate carboxykinase enzyme in Mt, linking the tricarboxylic acid (TCA) cycle and gluconeogenesis, Mt relies on the products of gluconeogenesis for survival [1]. We have previously shown via knockout inactivation that the glpX gene is essential to the growth and virulence of Mt in vivo [2]. The glpX gene (Rv1099c) of Mt encodes a Class II fructose 1,6-bisphosphatase (FBPaseII) which cleaves a phosphate group from fructose-1,6-bisphosphate (F16BP) through hydrolysis and forms fructose-6-phosphate (F6P) in the rate limiting step of gluconeogenesis. Fructose-6-phosphate remains a key precursor sugar in other biosynthetic pathways, including the generation of mycobacterial cell wall glycolipids [3]. The glpX gene is a promising target for drug development specific to M. tuberculosis as it is the main FBPase in Mt and does not have a human homolog [3]. While a second enzyme with FBPase activity has been identified in Mt, the activity of this enzyme is much lower [4].
Thr84 in MtFBPaseII protein (Thr90 in Escherichia coli) is a conserved residue in the active site as shown in other FBPases (Fig. 1) and is essential to the FBPase activity as determined by an alanine mutation in FBPaseII from E. coli (EcFBPaseII) [5]. It lies within a conserved region for other FBPasesII in what is known as the lithium-sensitive phosphatase motif, in which the leaving phosphate is coordinated by two calcium ions, their waters and the hydroxyl group from threonine [5]. To confirm the catalytic essentiality of this residue in MtFBPaseII and to validate the proposed catalytic mechanism, site directed mutagenesis of this residue to both alanine and serine has been performed. Enzymatic characterization, lithium inhibition, and substrate binding have been also assessed to provide further insight into the properties of the mutant enzymes. A plausible three-dimensional interpretation of the minor changes in the active site of the T84S mutant enzyme is proposed to account for the reduced activity.
Fig. 1.
Amino acid sequence alignment of relevant FBPase (Class II) enzymes. Sequence comparison of FBPaseII from Escherichia coli, Mycobacterium tuberculosis, Synechocystis sp. PCC680, and Francisella tularensis were made with Clustal Omega (Sievers et al., and Goujon et al., 2010). Asterisks (*) below the sequence indicate a fully conserved residue across the chosen species. Conserved residues have also been highlighted. Colons (:) below the sequence indicate conservation between residues with strongly similar properties and a period (.) indicates conservation between groups of weakly similar properties. Numbers above the sequence indicate the numbered position of the residue, with the last digit of the number indicating the residue below it. The sequence also indicates the key aspartic acid residue that stabilizes the threonine, as well as the lithium sensitive motif.
2. Materials and methods
Materials were purchased from Fisher Scientific, Waltham, MA. Primers for T84S were purchased from IDT, Coralville, IA. Sequence alignment was prepared with Clustal Omega [6]. All graphs were prepared with Prism software, version 7.0b.
2.1. Site directed mutagenesis
The pET15b plasmid containing the Wt-MtFPBase was previously described [3]. Forward primers and their reverse complement were designed to replace the 84th residue position of MtFBPaseII from threonine to alanine (T84A) (5′-CGTCGACCCCATTGACGGCGCAACGCTGATGAGCAAGGCATGAC-3′) and threonine to serine (T84S) (5′-TGCCGTCGACCCCATTGACGGCAGCACGCTGATGAGCAAG-3′). In the mutations, the codon ACC for Thr84 was replaced by GCA for alanine, and AGC for serine. Site directed mutagenesis was carried out using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). The T84A mutant was amplified by PCR with an initial 2-min incubation at 95 °C, followed by 18 cycles of 95 °C for 20 s, 60 °C for 10 s, and 68 °C for 3.5 min and finally 68 °C for 5 min. The T84S mutant was prepared by PCR amplification with an initial 2 min incubation at 95 °C, 30 cycles of 95 °C for 20 s, 55 °C for 30 s, 65 °C for 3.5 min, and finally 65 °C for 5 min. XL10-Gold Ultracompetent Cells were transformed with the PCR products. The cells were plated on LB agar containing tetracycline (12.5 μg/mL), chloramphenicol (34 μg/mL), and ampicillin (100 μg/mL) and kept at 37 °C overnight. Plasmids were extracted and transformed into BL21 (DE3) competent cells for protein expression and purification. Mutagenesis was confirmed by sequencing.
2.2. Bacterial cell culture
Bacterial strains were grown on LB plates with ampicillin (0.1 mg/mL). A single colony was selected to inoculate 100 mL LB broth and incubated overnight on a shaker incubator at 37 °C after which it was sub-cultured 1:100 in a 2 L flask with 400 mL of LB broth with ampicillin and grown to an OD600 of 0.5. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was then added at the final concentration of 0.5 mM, and the culture was incubated at 25 °C on the shaker for 6 additional hours. Cells were harvested by centrifuging and stored at −20 °C until purification.
2.3. Protein purification
Buffer used for purification contained 50 mM Tris, 300 mM NaCl at pH 8.0. Cells were thawed on ice and resuspended in buffer with DNAse, lysozyme (0.5 mg/mL), and EDTA-free protease inhibitor. Lysozyme, DNAse, Tris, and NaCl were obtained from Sigma-Aldrich, St. Louis, MO. Cells were lysed by sonication for 20 min and the lysate centrifuged at 15,500 RCF for 45 min at 4 °C. The supernatant was vacuum filtered through a 0.45 μM PES membrane and run through a Ni-NTA column equilibrated with 10 mM imidazole in buffer. The column was washed with 20 mM imidazole in buffer until the A280 was zero. The protein was eluted with 250 mM imidazole and then concentrated using Amicon Concentrators with 30 kDa molecular weight cut off and exchanged into a buffer containing 20 mM Tricine, 50 mM KCl, 0.1 mM DTT, 1 mM MgCl2, and 15% glycerol using a Zeba spin desalting column. Both the wildtype enzyme and the mutant enzymes were exchanged and purified using different columns to avoid contamination. Protein purification was verified using SDS-PAGE and is expected to migrate to 36 kDa. Protein concentration was determined using a nanodrop with the absorbance at 280 nm, with the calculated extinction coefficient of 17420 M−1cm−1.
2.4. Protein identity confirmation
Protein identity was confirmed via mass spectrometry by the Research Resouce Center at University of Illinois at Chicago. Samples were centrifuged, then injected into a 6550 iFunnel QTOF MS coupled to a 1290 UPLC system. Intact proteins were resolved on a Poroshell 300SB-C3 Analytical Column 5.0 μm on a linear gradient. Mass spectrometry data was acquired via Agilent Jet Stream − Electrospray Ionization at 1 spectra/s from 100 to 3000 m/z. All mass spectrometry equipment was from Agilent Technologies (Santa Clara, CA).
2.5. Enzymatic activity
To measure enzyme activity, a malachite green assay was used as previously described with minor modifications [3]. Protein of each the wildtype and mutants were added in equal concentrations to the substrate fructose-1,6-bisphosphate with a final concentration of 15 μM. A negative control containing only the reaction buffer was also tested. Reaction buffer contained 50 mM KCl, 20 mM Tricine at pH 8.0, 8 mM MgCl2, and 15 nM MtFBPaseII. The malachite green solution was added after 6 min and then incubated at room temperature for 10 min. Absorbance levels were read at 630 nm.
A coupled assay was also used to compute the specific activity of the enzyme. The procedure was modified slightly from previous methods by using 0.5 mM NADP+, yeast glucose-6-phosphate dehydrogenase (2.0 units/mL), yeast phosphoglucoisomerase (2.5 units/mL), 15 nM wildtype (Wt) MtFBPaseII and 15 μM F16BP [3]. Enzymes were purchased from Sigma-Aldrich. Lithium sensitivity testing was performed with 30 nM enzyme.
2.6. Surface plasmon resonance
For surface plasmon resonance (SPR) experiments, proteins were thawed from frozen sample in tricine buffer. Protein was exchanged three times into phosphate buffered saline, pH 7.4, with 15% glycerol with a Zeba spin desalting column. Protein was immobilized to a CM5 chip by diluting concentrated protein (A280 = 2.2) with sodium acetate pH 4.0 by factor of 10/200 to give a final concentration of 5 μM with a Biacore T200 at the University of Illinois at Chicago’s Research Resources Center. EDC and NHS were used to prepare the immobilization surface and ethanolamine was used to fill any unbound sites. Final response after immobilization was about 15,000 RFUs. The protein was then allowed to equilibrate at 30 μL/min for at least 2 h in 20 mM Tricine buffer with 50 mM KCl, 8 mM MgCl2, and 0.5 mM TCEP. F16BP was titrated in duplicate with final concentrations of 0.3–100 μM. Data was analyzed with Biacore T200 analysis software version 3.0 (GE Lifesciences, Pittsburgh, PA), fitting the binding and dissociation curves separately. Values are reported as an average of duplicate runs.
3. Results
3.1. Confirmation of mutagenesis by sequencing
Sequencing of the PCR product revealed two mutations for T84A site directed mutagenesis. The first mutation was the intended mutation for threonine to alanine at residue 84. There was also a silent alanine mutation of GCC to GCT found at residue 77. Sequencing of the T84S mutant showed a 100% identity for the threonine to serine mutation with no extra mutations in the gene.
3.2. Protein purification and mass spectrometry
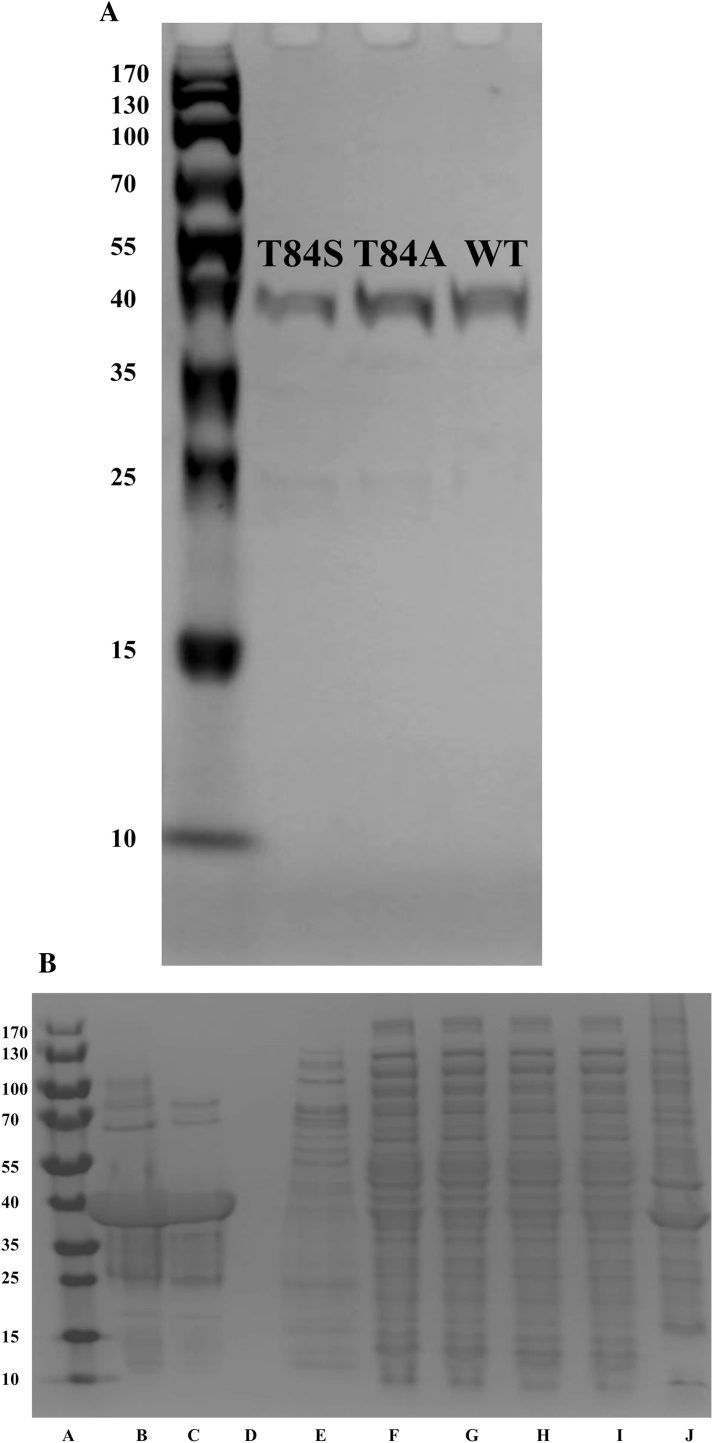
Purification of enzymes for wildtype and mutants was performed as previously described [3] (Fig. 2). The overloaded protein in Fig. 2 B shows some impurities that cannot be detected at lower concentrations (Fig. 2A). Both mutants followed the same procedure for purification with similar results, only the gel for T84A is presented. The yield for T84S was 0.1 mg protein per gram of cell. A typical purification of wildtype MtFPBaseII is 0.7 mg/g of cells. Mass spectrometry molecular mass determination indicated 36584.1 Da for T84A and 36598.7 Da for T84S. Expected masses of T84A and T84S are 36583.37 and 36599.37 Da respectively.
Fig. 2.
Purification SDS-PAGE gels of MtFBPase. Equal amounts of protein from both mutants and the wildtype were loaded on an SDS-PAGE gel (A). The purification process of T84A was documented (B). In order from left to right, the lanes are: A) Ladder (EZ Run Prestained REC protein ladder), B) T84A in exchange buffer, C) T84A eluted in buffer with 250 mM imidazole, D) second wash with 20 mM imidazole, E) first wash with 20 mM imidazole, F) second flow through sample of filtrate through Ni-NTA column, G) first flow through sample of filtrate through Ni-NTA column, H) supernatant after running through 0.45 μm filter, I) supernatant from the lysate after centrifugation and J) lysate after running on sonicator.
3.3. Enzymatic activity
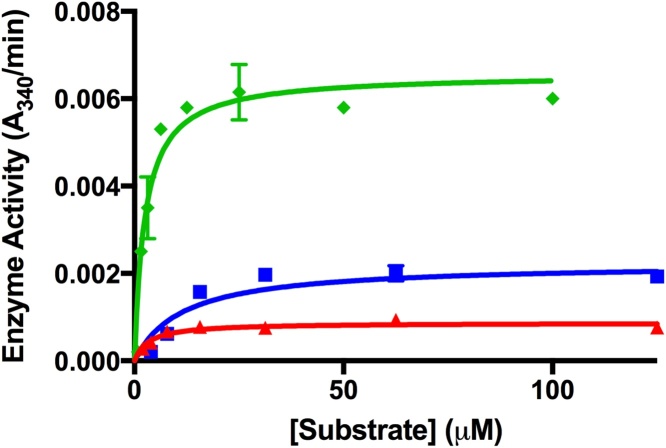
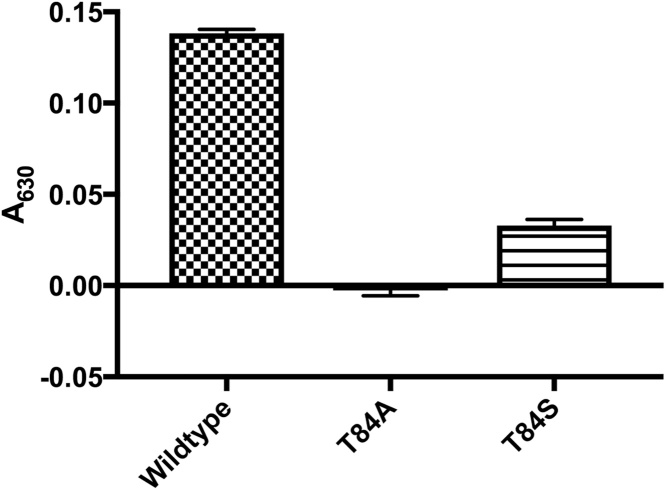
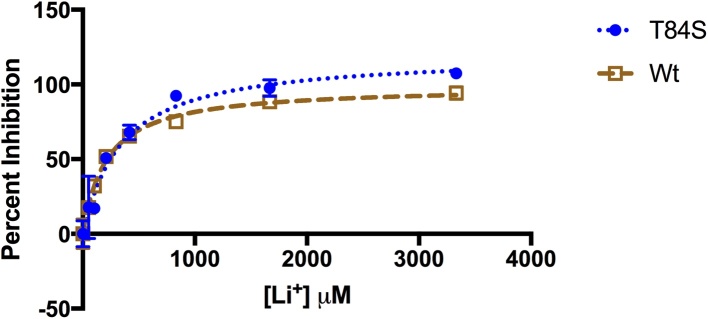
T84A showed no enzyme activity, with an absorbance matching that of the negative control (Fig. 3). In contrast, the T84S mutant retained some activity. Km values of 3.7 μM for Wt-MtFPBaseII and 5.7 μM for T84S were determined from the fitted curve (Fig. 4). Vmax values were 0.37 and 3.5 units/mg for the serine mutant and wildtype, respectively (Table 1). This equates to 20% and 11% of the activity of the serine mutant in the malachite green assay and in the coupled assay, respectively, as compared to the wildtype. In addition, we observed a reduced sensitivity of the enzyme to lithium in the T84S mutant when compared to the wildtype enzyme (Fig. 5).
Fig. 3.
Enzyme activity vs. substrate concentration. The substrate affinity for T84S (blue squares at 50 nM and red triangles at 30 nM) and wildtype (green diamonds at 15 nM) enzymes were calculated by varying the substrate concentration. Data were fitted using Prism software Michaelis-Menten equation to find Km and its error. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Enzyme activity. Malachite green A630 values are the average of 8 trials for each protein. Buffer was used as the blank. No FBPaseII activity was detected in the T84A mutant. T84S mutant retained about 20% activity as compared to the wildtype. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Wildtype and mutant enzyme kinetic parameters.
| Sample | IC50 Li+ (μM) | ka (M−1s−1) |
kd (s−1) |
KD (μM) | Km (μM) |
Vmax (μmol min−1 mg−1) |
Kcat (s−1) |
Kcat/Km (s−1 mM−1) |
|---|---|---|---|---|---|---|---|---|
| Wt | 250 ± 40 | 112 | 0.0214 | 190 | 3.7 ± 0.1 | 3.5 ± 2.4 | 2.1 ± 1.5 | 570 |
| T84A | nd | 141 | 0.0208 | 147 | nd | nd | nd | nd |
| T84S | 280 ± 70 | 121 | 0.0172 | 142 | 5.7 ± 1.2 | 0.37 ± 0.26 | 0.22 ± 0.16 | 39 |
Fig. 5.
Inhibition of MtFBPaseII by Li+. Activity of Wt and T84S mutant were assessed for lithium sensitivity. IC50 curves for Wt-MtFBPaseII and T84S-MtFBPaseII, both at 30 nM enzyme concentration, show that the serine mutant and wildtype protein have similar sensitivity to lithium. Lithium concentration was tested from 0 to 3333 μM. Since threonine is part of a conserved lithium sensitive motif, the effect of mutagenesis of this residue on activity is important.
3.4. Surface plasmon resonance
SPR experiments indicated a KD value of 190 μM for Wt-MtFBPaseII, 147 μM for T84A-FBPaseII and 142 μM for T84A-FBPaseII (Table 1), confirming that the binding constants for the substrate (F16BP) is approximately the same for the wildtype and T84 mutant enzymes.
4. Discussion
Wt-MtFBPaseII was found to have a Vmax of 3.5 units/mg specific activity while previously it was found to have 1.6 units/mg [3]. We also observed a Km of 3.7 μM as compared to 44 μM [3]. These inconsistencies in the kinetic parameters of the enzyme could be explained by a few modifications that we made in the purification procedures compared to the previously reported method. We changed the purification buffer to Tris pH 8.0 and buffer exchange was performed with a desalting column rather than excessive dilution and concentration by centrifugation. The Vmax of the enzyme was found to be higher, the Km for F16BP was found to be about 10 times lower and the catalytic efficiency much higher 570 compared to 22.7 s−1 mM−1 as reported before [3]. This purification protocol leads to a more active enzyme possibly due to a better buffering system and less stress from centrifugal forces.
The catalytic and affinity parameters of the mutant enzymes are presented in Table 1. The mutant T84A is essentially inactive demonstrating the importance of the threonine hydroxyl for the catalytic mechanism as previously suggested [5]. In contrast, the T84S has reduced activity with catalytic parameters of Kcat = 0.22 ± 0.16 s−1, Km = 5.7 ± 1.2 μM, Vmax = 0.37 ± 0.26 μmol min−1 mg−1. This suggests that the overall activity has been decreased by approximately ten-fold, while keeping a similar binding affinity for the substrate. Most likely there are no major conformational changes in the active site of the T84S mutant, explaining its partial activity.
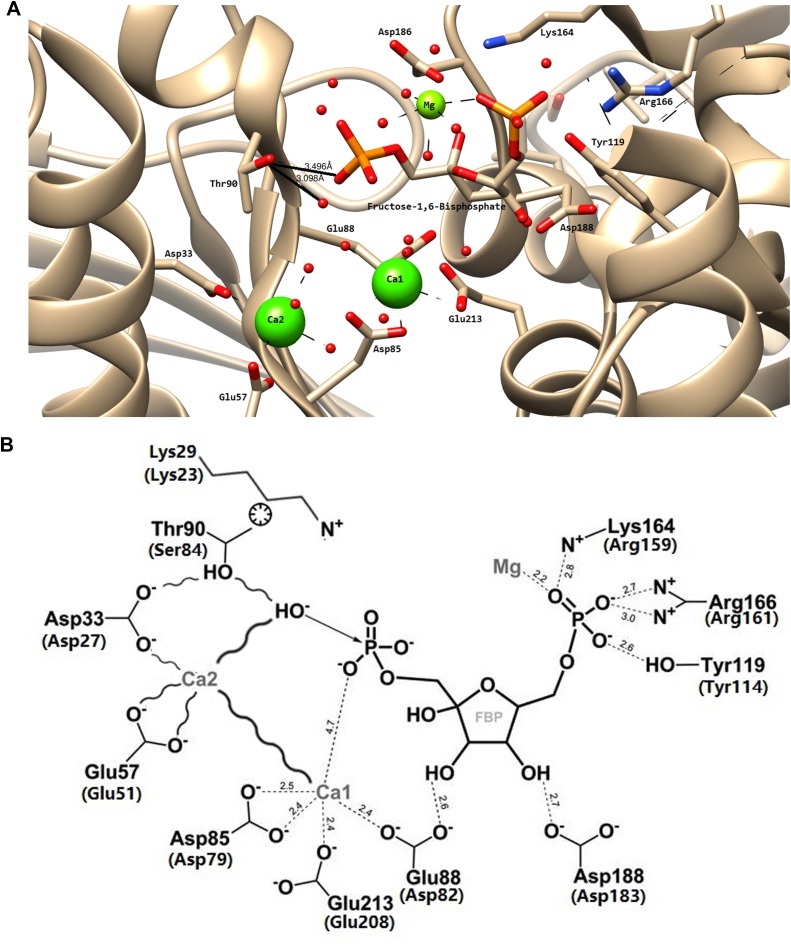
The interpretation of these results in terms of the three-dimensional structure of the homologous E. coli enzyme (PDB 3D1R) suggests that the replacement of the critical nucleophile OH− in the Thr84 residue of the wildtype of MtFBPase by the hydroxyl of Ser84 results in minor alterations of the position and orientation of this group, reducing the catalytic efficiency of the mutant enzyme (Fig. 6b). Thr90 in the E. coli enzyme has been proposed to be critical for the activation of the OH− hydroxyl of a neighboring water molecule. In turn, this water molecule nucleophile would attack the leaving phosphate of the F16BP. The changes in the position and orientation of the equivalent serine hydroxyl in the T84S mutant would result in a cascading effect on Asp27 and Glu51 mediated by the Mg2+ ions (Ca2+ positions in the figure), also affecting the position of the activating water (Fig. 6b).
Fig. 6.
a) The active site of EcFBPaseII in the refined structure of the D61A mutant. The active site of EcFBPaseII (PDB code 3D1R) is displayed with F16BP in the active site. Distances from the catalytic T90 (T84 in MtFBPaseII) to the neighboring phosphate in the substrate and surrounding water molecules are displayed together with the most prominent residues near the active site of the E. coli. Ca2+ are shown at the positions of the catalytic Mg2+ ions. b) A schematic representation of the effects of the replacement of Thr84 by serine in the active site of MtFBPase. (Adapted from Brown et al. [5]Fig. 1). This figure corresponds to a two-dimensional representation of Fig. 6a. Residues listed in parentheses indicate corresponding residues in Mycobacterium tuberculosis T84S mutant. Undetermined distances due to the mutation are denoted by a curvy line. The proposed cause for the stability and orientation of the critical threonine residue in the active is indicated by a circle included between the sides chains of T90 and K29, suggesting hydrophobic interactions.
Previous research has shown the important contribution of Thr90 to the activity of EcFBPaseII and its place within the lithium-sensitive phosphatase motif, 85DPIDGT90, as the key in its enzymatic function [5]. This motif is conserved in Mt, and it has been shown MtFBPaseII, as predicted, also can be inhibited by lithium [3]. In Fig. 1, the Li+ sensitive region is indicated, and the threonine (Thr84 in MtFBPaseII) is conserved among Mt, Mycobacterium smegmatis, Escherichia coli, Francisella tularensis, and Corynebacterium glutanicum.
The motif relies on an aspartic acid residue to activate the threonine, which in turn activates a catalytic water molecule (Fig. 6a and 6b) for a nucleophilic attack [5], [7], which is shown to be conserved in the selected species in Fig. 1 (Asp27 in Mt and Asp33 in Ec). Two mechanisms for the involvement of the bivalent metal ion have been proposed. A three metal model [8] and a two metal model [7]. The more probable mechanism is the two metal model in Class II FBPases where the bivalent metal ions stabilize the transition state charge, leading to the release of the inorganic phosphate [7]. In the case of MtFBPase, Mg2+ and Mn2+ are the only two ions that resulted in any significant FBPase activity [5]. The same lithium-sensitive motif is present in other phosphatase enzymes, such as inositol monophosphatases, inositol polyphosphate 1-phosphatases and 3′-phosphoadenosine-5′-phosphatases and the introduction of lithium ions has also inhibited the phosphatase activity of these enzymes [9]. The aforementioned aspartic acid residue has also been conserved across many species exhibiting this motif, including all of the organisms listed above (residue 27 in Mt). Mutagenesis of this residue in EcFBPaseII shows reduced FBPase activity [5] and may show similar activity reduction in MtFBPase.
We have shown that the replacing the threonine hydroxyl by the corresponding serine hydroxyl in the mutant enzyme results in an enzyme with slower catalytic turnover. The availability of the T84S mutant with reduced catalytic turnover could allow the trapping of interesting reaction intermediates, using crystallographic methods, which could be used as starting points for the design of potent inhibitors of MtFBPase using structure-based drug design methods. Upon further optimization, testing and development, the best compounds could be used as therapeutic agents against tuberculosis.
In this manuscript, mutagenesis of threonine to serine in the active site of Mycobacterium tuberculosis fructose-1,6-bisphosphatase (Class II) retains partial enzyme activity, there is no conflict of interest.
Acknowledgements
Sequencing was performed by the University of Illinois at Chicago’s (UIC) Research Resources Center (RRC) DNA Services Facility. We would like to acknowledge Potts Memorial Foundation Grant number G3541 and Chicago Biomedical Consortium Grant number 084679-00001.
References
- 1.Marrero J., Rhee K.Y., Schnappinger D., Pethe K., Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U. S. A. 2010;107(21):9819–9824. doi: 10.1073/pnas.1000715107. (PubMed PMID: 20439709; PubMed Central PMCID: PMCPMC2906907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutka H.J., Wang Y., Franzblau S.G., Movahedzadeh F. glpx gene in Mycobacterium tuberculosis is required for in vitro gluconeogenic growth and In vivo survival. PLoS One. 2015;10(9):e0138436. doi: 10.1371/journal.pone.0138436. (PubMed PMID: 26397812; PubMed Central PMCID: PMCPMC4580611.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutka H.J., Rukseree K., Wheeler P.R., Franzblau S.G., Movahedzadeh F. glpX gene of Mycobacterium tuberculosis: heterologous expression, purification, and enzymatic characterization of the encoded fructose 1,6-bisphosphatase II. Appl. Biochem. Biotechnol. 2011;164(8):1376–1389. doi: 10.1007/s12010-011-9219-x. (PubMedPMID:21451980) [DOI] [PubMed] [Google Scholar]
- 4.Ganapathy U., Marrero J., Calhoun S., Eoh H., de Carvalho L.P., Rhee K. Two enzymes with redundant fructose bisphosphatase activity sustain gluconeogenesis and virulence in Mycobacterium tuberculosis. Nat. Commun. 2015;6:7912. doi: 10.1038/ncomms8912. (PubMedPMID:26258286;PubMedCentralPMCID:PMCPMC4535450.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown G., Singer A., Lunin V.V., Proudfoot M., Skarina T., Flick R. Structural and biochemical characterization of the type II fructose-1,6-bisphosphatase GlpX from Escherichia coli. J. Biol. Chem. 2009;284(6):3784–3792. doi: 10.1074/jbc.M808186200. (PubMedPMID:19073594;PubMedCentralPMCID:PMCPMC2635049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sievers F., Wilm A., Dineen D., Gibson T., Karplus K., Li W. Fast, scalable, generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7(539) doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel S., Martinez-Ripoll M., Blundell T.L., Albert A. Structural enzymology of Li(+)-sensitive/Mg(2 +)-dependent phosphatases. J. Mol. Biol. 2002;320(5):1087–1094. doi: 10.1016/s0022-2836(02)00564-8. (PubMed PMID: 12126627) [DOI] [PubMed] [Google Scholar]
- 8.Stieglitz K.A., Johnson K.A., Yang H., Roberts M.F., Seaton B.A., Head J.F. Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus The story of a mobile loop. J. Biol. Chem. 2002;277(25):22863–22874. doi: 10.1074/jbc.M201042200. (PubMedPMID:11940584) [DOI] [PubMed] [Google Scholar]
- 9.York J.D., Ponder J.W., Majerus P.W. Definition of a metal-dependent/Li(+)-inhibited phosphomonoesterase protein family based upon a conserved three-dimensional core structure. Proc. Natl. Acad. Sci. U. S. A. 1995;92(11):5149–5153. doi: 10.1073/pnas.92.11.5149. (PubMed PMID: 7761465; PubMed Central PMCID: PMCPMC41866). [DOI] [PMC free article] [PubMed] [Google Scholar]