Abstract
Efficient detoxification and clearance of cholesterol metabolites such as oxysterols, bile alcohols, and bile acids are critical for survival because they can promote liver and cardiovascular disease. We report here that loss of the nuclear xenobiotic receptor PXR (pregnane X receptor), a regulator of enterohepatic drug metabolism and clearance, results in an unexpected acute lethality associated with signs of severe hepatorenal failure when mice are fed with a diet that elicits accumulation of cholesterol and its metabolites. Induction of a distinct drug clearance program by a high-affinity ligand for the related nuclear receptor, the constitutive androstane receptor, does not overcome the lethality, indicating the unique requirement of PXR for detoxification. We propose that the PXR signaling pathway protects the body from toxic dietary cholesterol metabolites, and, by extension, PXR ligands may ameliorate human diseases such as cholestatic liver diseases and the associating acute renal failure.
Cholesterol metabolism occurs principally in the liver, where excess cholesterol is converted into two primary bile acids, cholic acid (CA) and chenodeoxycholic acid (or β-muricholic acid in rodents), by reaction cascades that involve at least 17 enzymes (1). Bile acids subsequently are excreted through the bile ducts and into the intestine, where they act as detergents that facilitate absorption of cholesterol, fats, and other lipophilic nutrients. In the intestine, primary bile acids can be further metabolized by resident bacteria into secondary bile acids, such as deoxycholic acid and lithocholic acid (LCA). Bile acids and their precursors, when accumulated in excess, elicit harmful effects. For example, genetic defects in cholesterol catabolism can result in accumulation of toxic intermediate metabolites, such as oxysterols, bile alcohols, or bile acids, leading to liver disease, premature cardiovascular disease, or neurological disorders, which cannot be corrected by bile acid replacement therapy (1, 2). In progressive familial intrahepatic cholestasis, a genetic defect in a bile acid transporter leads to retention of hepatic bile acids, biliary cirrhosis, and early death (2, 3). These findings demonstrate the need for tight regulation of cholesterol and bile acid metabolism.
The nuclear receptors comprise a large family of ligand-activated transcription factors that control development, reproduction, energy homeostasis, and other biological processes (4). In particular, the pregnane X receptor (PXR) (NR1I2; also known as SXR or PAR in humans) is expressed primarily in the liver and the intestine and is defined as a xenobiotic receptor that serves to defend against potentially toxic foreign chemicals (5, 6). Numerous structurally unrelated drugs and environmental contaminants can bind and activate PXR, resulting in the transcriptional induction of a battery of genes encoding phase I and II metabolic enzymes as well as membrane-bound transporters, which together enhance the detoxification and clearance of foreign chemicals (7–9). Thus, ligand-induced activation of PXR or transgenic expression of a constitutively active form of PXR in the liver confers resistance against multiple toxins (10–12). Mice lacking PXR (pxr-/- mice) appear normal when maintained under standard laboratory conditions, but ligand-induced toxin resistance is lost, suggesting that PXR is dispensable in the absence of chemical insults (10) and may function only as a xenobiotic receptor.
However, recent studies indicate that PXR also can be activated by endogenous cholesterol metabolites, implicating its involvement in the clearance of potentially toxic intermediates. In cell-based assays, high concentrations of secondary bile acids such as deoxycholic acid and LCA activate both mouse and human PXR (11, 12). In the livers of animals treated with CA or LCA, hepatic expression of PXR target genes, cyp3a11, a major drug-metabolizing cytochrome P450, and organic anion transporter 2 (oatp2; also known as slc21a5), are increased (11–13). Mouse, but not human, PXR also can be activated by two bile acid precursors, 5β-cholestane-3α,7α,12α-triol and 5β-cholestane-3α,7α,12α,25-tetrol, compounds that accumulate in CYP27-deficient mice (14–16). These two bile alcohols accumulate at even higher levels in a human disorder where loss of the CYP27 enzyme results in a condition called cerebrotendious xanthomatosis (17). CYP27-deficient mice do not develop cerebrotendious xanthomatosis, perhaps because of sustained induction of hepatic PXR target genes and bile alcohol detoxification, which does not appear to occur in human cerebrotendious xanthomatosis patients (14–17). In addition to bile acids and bile alcohols, 24(S),25-epoxycholesterol also has been shown to activate PXR in mouse hepatocytes (18). This oxysterol affects hepatic cholesterol synthesis and metabolism through the nuclear oxysterol receptor LXR and the sterol regulatory element binding proteins. Together, these studies clearly indicated that PXR can be activated by endogenous cholesterol metabolites, but the significance of such responses remains unclear.
To address whether PXR has a physiologically significant role in the clearance of cholesterol or its metabolites, we challenged mice lacking PXR with a diet rich in CA, a cholesterol metabolite and a signaling molecule known to block cholesterol catabolism. Unexpectedly, addition of cholesterol to this diet resulted in an acute lethality associated with signs of hepatorenal failure. This unusual phenotype was not rescued by ligand-dependent activation of the related nuclear receptor constitutive androstane receptor (CAR; NR1I4) and the resulting induction of a distinct drug clearance pathway. These results reveal an essential and unique role of PXR in protection from the toxicity of cholesterol and its metabolites.
Materials and Methods
Animals. Generation of pxr-/- animals was performed as described in ref. 10. Male mice backcrossed to the isogenic mouse strain C57B/6J (The Jackson Laboratory) for at least seven or eight times were used in all of the experiments described. Similar results also were obtained with female mice. Wild-type (WT) controls were pxr+/+ littermates from heterozygote crosses or C57B/6J mice purchased from The Jackson Laboratory. Mice were housed in a pathogen-free animal facility under a standard 12-h light/12-h dark cycle with access to chow and water ad libitum. A standard chow (no. 8604, Harlan Teklad, Madison, WI) or chows supplemented with 1.25% cholesterol and/or 0.5% CA (Harlan) were used. For ligand treatments, mice were treated by a single i.p. injection with vehicle (corn oil), 3 mg/kg 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP; Calbio-chem), or 40 mg/kg pregnane-16α-carbonitrile (Sigma). For histology, tissues were fixed in 10% formalin, dehydrated, embedded in paraffin, and sectioned for hematoxylin/eosin staining.
Analytical Procedures. Mice were fasted for 4 h before blood was drawn by heart puncture. Enzymatic assay kits were purchased from Thermo Electron (Milford, MA) except for the total bile acid assay kit from Trinity Biotech (Bray, Ireland). Bile acid composition was determined by liquid chromatography-mass spectrometry analysis as described in ref. 19. All enzymatic assays described were repeated by using independent animal groups.
Oligo-DNA Microarray Gene Expression Analysis. Total tissue RNA were prepared with TRIzol (Invitrogen) according to the manufacturer's instructions from the livers and kidneys of two pairs of age-matched male littermates that had been on the dietary challenge for 7 days. Poly(A) RNA were purified and labeled for hybridization to the Affymetrix Gene Chip Mouse Expression Set 430 2.0. This protocol resulted in four measurements for each analyzed gene for each tissue, i.e., two for the control samples and two for the knockout samples. Probe hybridization to the Affymetrix chips and data scanning were performed by the Salk microarray core facility. Criteria for filtering differentially expressed genes included evaluation of down-regulated targets that were statistically decreased as assigned by the microsuite software and were similarly decreased in four possible pairwise comparisons between two control and two knockout samples as filtered by the bullfrog analysis software (20).
Nothern Blot and Real-Time Quantitative PCR (qPCR). Standard Affymetrix protocols were used for Northern blot analysis using total tissue RNA. We used 32P-labeled cDNA as a probe. The sequences of oligo DNAs used to generate cDNA by RT-PCR are available on request. For qPCR, 1 μg of total RNA treated with DNase I (Ambion) was reverse-transcribed by using Superscript II reverse transcriptase (Invitrogen), and the resulting cDNA subsequently was treated with RNase. Combined cDNA was diluted to 1:16, 1:40, 1:100, 1:400, and 1:1,000 to generate a standard curve, and samples were diluted to 1:40. Amplifications were performed in the ABI Prism 7900HT (Applied Biosystems) in triplicate in 384-well plates by using SYBR Green Master Mix (Applied Biosystems). The sequences of the forward and reverse primers used were described in ref. 8. The sequence detection system software (Applied Biosystems) was used to analyze data, and the absolute values of RNA levels were generated by normalizing the copy number of each target gene to the u36b4 RNA standard.
Statistical Analysis. The number of mice for each group used in experiments is indicated in the figure legends. Values were presented as mean ± SD. A two-tailed Student t test was used to calculate P values.
Results
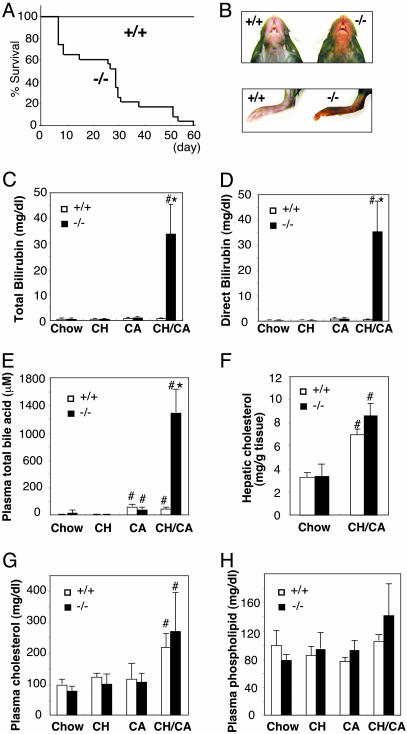
Feeding rodents with a diet rich in both cholesterol and CA is known to increase levels of hepatic cholesterol and catabolic intermediates because of the ability of CA to both increase the intestinal absorption and decrease hepatic cholesterol catabolism (21–23). To address whether PXR plays a role in the clearance of cholesterol metabolites, we challenged pxr-/- and control WT animals with a diet supplemented with 1.25% cholesterol and 0.5% CA (CH/CA diet). Unexpectedly, feeding of the CH/CA diet resulted in ≈40% lethality in pxr-/- animals by day 10 and 100% lethality by day 60 (Fig. 1A). Similar death rates of pxr-/- mice were observed when the concentrations of cholesterol and/or CA in the diet were increased to 2.5% and 1%, respectively (data not shown). In marked contrast, no lethality was observed for WT animals on the CH/CA diet or animals on a diet supplemented with only 1.25% cholesterol (cholesterol diet) or 0.5% CA (CA diet) (Fig. 1 A and data not shown). For unclear reasons, the onset of the CH/CA-induced death of pxr-/- animals varied significantly (5–60 days), and pxr-/- animals remained indistinguishable from control WT animals as judged by various plasma chemistries until they become lethargic (Fig. 1 A and data not shown). Once the lethargic phenotype, which is characterized by hypothermia, jaundice, progressive weight loss, and morbidity, was observed, they typically died within 3 days. Based on these observations, CH/CA-fed pxr-/- animals displaying lethargy between the 7th and 10th day of feeding were killed and used for the subsequent characterizations of the phenotype, unless stated otherwise. In each case, WT mice on the CH/CA diet or mice on the cholesterol or the CA diet were killed at the same time for comparison.
Fig. 1.
Acute death of CH/CA-fed pxr-/- mice is associated with severe conjugated bilirubinemia. (A) Survival of WT (+/+; n = 20) and pxr-/- (-/-; n = 23) mice on the CH/CA diet. (B) Ventral view of the head (Upper) and hind limb (Lower) of representative WT (+/+) and lethargic pxr-/- (-/-) mice fed the CH/CA diet for 6 days. Note that the skin color of the pxr-/- mouse is significantly darker compared with the WT mouse, indicating the presence of severe jaundice. (C–H) Total plasma bilirubin (C), plasma direct (conjugated) bilirubin (D), plasma total bile acid (E), hepatic cholesterol (F), plasma total cholesterol (G), and plasma phospholipid (H) concentrations in WT (+/+) and pxr-/- (-/-) mice fed the indicated diets for 7–10 days. All values are expressed as mean ± SD, n = 5. *, significant differences compared with WT on the same diet; #, significant differences compared with mice of the same genotype on the chow diet (P < 0.01).
Initial examination of CH/CA-fed lethargic pxr-/- mice suggested hyperbilirubinemia (i.e., high plasma levels of bilirubin) because the skin and tissues were overtly jaundiced (Fig. 1B). Indeed, the plasma total bilirubin levels were on average 54-fold higher in lethargic CH/CA-fed pxr-/- mice compared with control WT mice (Fig. 1C). Bilirubin is an oxidative end product of heme catabolism and in the liver is conjugated by an isoform of uridine diphosphate glucuronosyltransferase (UGT1A1) before excretion into the biliary system (24). We found that almost 100% of the bilirubin in the plasma of lethargic pxr-/- animals (and ≈60% of the total bilirubin in control groups) was present in conjugated form, indicating that hepatic conjugation is fully functional even in the absence of PXR (Fig. 1D). In addition to bilirubin, the levels of plasma bile acids were 18-fold greater in CH/CA-fed pxr-/- animals compared with CH/CA-fed WT mice (Fig. 1E). Analysis of individual bile acids in plasma by liquid chromatography-mass spectrometry revealed markedly elevated levels of two primary bile acids, CA and β-muricholic acid, but not a toxic secondary bile acid, LCA, in pxr-/- mice with severe jaundice (Table 1, which is published as supporting information on the PNAS web site). The levels of hepatic and plasma cholesterol and plasma phospholipids were not significantly different between WT mice and pxr-/- mice, although increased hepatic and plasma cholesterol levels were observed in all of the CH/CA-fed mice as expected (Fig. 1 F–H).
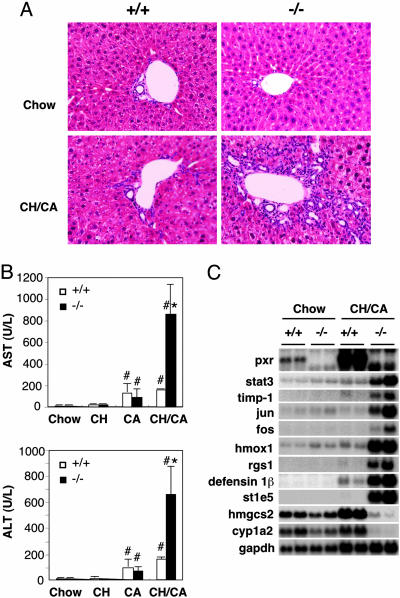
Despite the increases in conjugated bilirubin and bile acids, two markers for hepatobiliary defects, the overall structure of the liver and bile duct morphology were indistinguishable between pxr-/- and WT control mice with or without dietary challenge (Fig. 2A). There was no evidence of bile duct degeneration or proliferation, significant necrosis, hepatic fibrosis, or any typical characteristics of cholestatic liver disease on hematoxylin/eosin (Fig. 2 A) or Masson's Trichrome staining (data not shown). In addition, no significant hepatocyte apoptosis was observed by the TUNEL assay (data not shown). However, in the CH/CA-fed pxr-/- livers, infiltration of mononuclear cells was evidently present both in the periportal and in the midzonal areas of the hepatic lobes, suggesting severe hepatitis with hepatocellular injury. In contrast, in livers of CH/CA-fed WT animals, the inflammatory infiltrate was significantly less pronounced and observed only in the hepatic periportal area (Fig. 2 A). Consistent with the histological signs of hepatitis, two clinically used markers for hepatocellular injury, plasma aspartate transaminase and alanine transaminase, were dramatically increased in pxr-/- animals (5.5- and 4.2-fold, respectively), demonstrating enhanced liver injury in the CH/CA-fed pxr-/- animals. DNA microarray analysis using Affymetrix Gene Chips detected increased expression of genes known to be induced by acute inflammation such as stat-3, tissue inhibitor of metalloproteinase 1 (timp1), jun, fos, heme oxidase-1 (hmox1), regulator of G protein signaling 1 (rgs1), and defensin β1 in CH/CA-fed pxr-/- animals displaying severe jaundice (Fig. 2C). Expression of cyp1a2, a cytochrome P450 enzyme known to be repressed by inflammation, was decreased in the CH/CA-fed pxr-/- animals. In addition, we observed a disregulation of insulin-regulated genes, such as mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase (hmgcs2) and estrogen sulfotransferase (st1e5).
Fig. 2.
The liver phenotype of CH/CA-fed WT and pxr-/- mice. (A) Gross morphology of livers from chow- or CH/CA-fed WT (+/+, Left) and pxr-/- (-/-, Right) mice. Liver sections were prepared (n = 5 per group) for histology and stained with hematoxylin/eosin. Note the absence of abnormality in chow-fed WT (Left Upper) and pxr-/- (Right Upper) mice and the presence of inflammatory infiltration in both CH/CA-fed WT (Left Lower) and pxr-/- (Right Lower) mice. (B) Plasma activities of aspartate transaminase (AST; Upper) and alanine transaminase (ALT; Lower). Animals used in this study were the same as in Fig. 1. All values are expressed as mean ± SD, n = 5. *, significant differences compared with WT on the same diet; #, significant differences compared with mice of the same genotype on the chow diet (P < 0.05). (C) Northern blot analysis of total RNA isolated from livers of chow- or CH/CA-fed WT (+/+) and pxr-/- (-/-) mice.
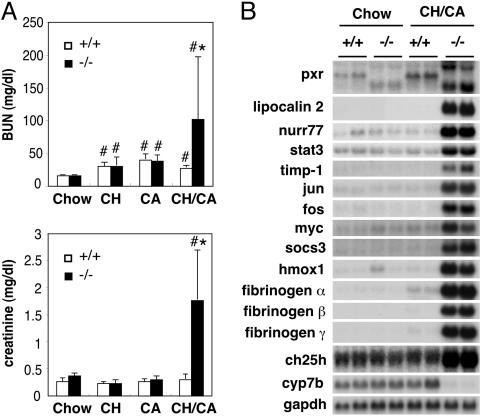
Because functional renal failure is a common cause of death among human patients with advanced liver disease (25), we analyzed renal function in the CH/CA-fed animals. Plasma concentrations for blood urea nitrogen and creatinine, two markers of kidney dysfunction, were found to be 3.7- and 5.7-fold higher, respectively, for lethargic CH/CA-fed pxr-/- animals compared with WT animals on the same diet (Fig. 3A), indicating that acute renal failure was at least a partial factor in the observed morbidity. Although histological examination of the kidneys revealed no significant signs of tissue injury or inflammation (data not shown), an Affymetrix oligonucleotide array analysis identified alterations in the expression of genes known to be induced by acute inflammation, such as stat-3, lipocalin 2, hmox-1, timp-1, nuclear receptor nurr77, jun, fos, myc, suppressor of cytokine signaling-3 (socs3), and fibrinogen isoforms, suggesting the presence of acute inflammation in the kidney (Fig. 3B). We also observed dramatic changes in the expression of genes involved in cholesterol catabolism, such as cholesterol 25-hydroxylase (ch25h) and cyp7b, reflecting disrupted cholesterol homeostasis in the CH/CA-fed pxr-/- animals. We conclude that the CH/CA diet induces severe functional disruption and inflammation in both the liver and the kidney in the absence of PXR.
Fig. 3.
The kidney phenotype of CH/CA-fed WT and pxr-/- mice. (A) Plasma levels of blood urea nitrogen (BUN; Upper) and creatinine (Lower). Animals used in this study were the same as in Fig. 1. All values are expressed as mean ± SD, n = 5. *, significant differences compared with WT on the same diet; #, significant differences compared with mice of the same genotype on the chow diet (P < 0.05). (B) Northern blot analysis of total RNA isolated from kidney of chow- or CH/CA-fed WT (+/+) and pxr-/- (-/-) mice.
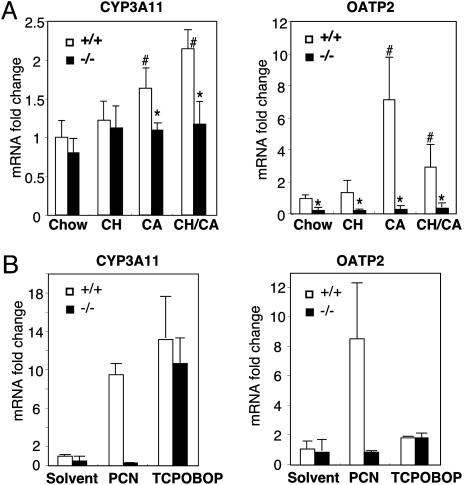
To examine whether feeding of the CH/CA diet generates ligands for PXR, we examined the expression of known PXR target genes in WT and pxr-/- animals on the CH/CA diet in comparison with animals on control diets. To avoid indirect influences of the lethal phenotype on gene expression, RNA was isolated from animals that had not yet developed severe jaundice (i.e., bilirubin level of <5 mg/dl). As shown in Fig. 4A, two known PXR target genes, cyp3a11 and oatp2, were found to be induced by CA or CH/CA feeding in a PXR-dependent manner in the liver. We did not observe PXR-dependent changes in the hepatic expression of other known PXR target genes, such as cyp2b10, ugt1a1, std, gsta1, and gstm1, or genes implicated in primary bile formation, such as cyp7a, cyp7b, cyp8b, oatp1, fic1, bsep, mdr1, mdr3, mrp2/cmoat, bcrp, abcg5, abcg8, and ntcp (data not shown). Indeed, oatp2 and cyp3a11 are the only genes that are differentially expressed between CH/CA-fed WT and pxr-/- animals before development of jaundice according to an Affymetrix oligonucleotide array analysis (data not shown). In addition to the liver, PXR is expressed at high levels in the intestine (26) and at a lower level in the kidney (Fig. 3B). However, we did not detect significant PXR-dependent changes in the expression of putative PXR target genes in these two tissues (data not shown). Thus, only a subset of hepatic PXR target genes including cyp3a11 and oatp2 were differentially expressed in CH/CA-fed WT and pxr-/- animals.
Fig. 4.
Regulatory target genes for PXR in CH/CA-fed mice. (A) Real-time qPCR analysis for cyp3a11 and oatp2 gene expression in the liver. RNA samples were isolated from livers of animals used in Fig. 1, except that RNA samples for CH/CA-fed pxr-/- mice were collected from animals that had not yet developed jaundice as described in the text. Values for qPCR analysis were normalized by the level of u36b4 gene encoding a ribosomal protein subunit, are expressed as fold change over chow-fed WT control, and represent the mean ± SD, n = 4. *, significant differences compared with WT on the same diet; #, significant differences compared with mice of the same genotype on the chow diet (P < 0.05). (B)WT(+/+) and pxr-/- (-/-) mice were subjected to i.p. injection of pregnane-16α-carbonitrile (PCN) (40 mg/kg), TCPOBOP (3 mg/kg), or solvent. After 24 h, total RNA was isolated from the liver and subjected to qPCR analysis for cyp3a11 and oatp2 mRNA expression. Values for qPCR analysis were normalized by the level of u36b4 gene, are expressed as fold change over chow-fed WT control, and represent the mean ± SD, n = 3.
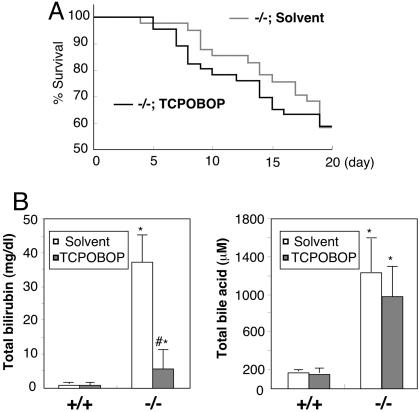
PXR and the related nuclear receptor CAR regulate overlapping sets of genes that function in the hepatic clearance of toxic compounds by binding to common cis-acting sites in the promoter regions (8, 27). For example, ligand-induced activation of CAR or PXR both result in induction of cyp3a11, cyp2b, and the bile acid sulfotransferase std genes in the mouse liver and confer a common resistance to toxicants, such as tribromoethanol, zoxazolamine, and LCA (11, 12, 28–31). These two receptors also regulate distinct target genes. For example, in the liver, oatp2 can be induced only by a PXR-specific ligand pregnane-16α-carbonitrile, but not by a CAR-specific ligand TCPOBOP (Fig. 4B) (8). To determine whether activation of CAR rescues pxr-/- animals from the CH/CA diet-induced toxicity, we pretreated pxr-/- animals with TCPOBOP, a potent and stable CAR agonist, at a dose known to chronically activate CAR for >12 weeks (32, 33). Animals commenced on the CH/CA diet the next day. We observed no increased survival in the CH/CA diet-fed pxr-/- animals pretreated with TCPOBOP (Fig. 5A). Notably, however, the lethality in TCPOBOP-pretreated pxr-/- animals was not associated with severe jaundice (Fig. 5B). As expected, TCPOBOP-pretreated CH/CA-fed animals expressed high levels of cyp2b and cyp3a in the liver despite the lack of PXR (data not shown). Other signs of toxicity, such as plasma bile acids and blood urea nitrogen, were found to be equivalently high in both TCPOBOP-treated pxr-/- and mock-treated animals (Fig. 5B and data not shown). We propose that PXR protects the body from the toxicity of the CH/CA diet, presumably through the induction of a subset of CAR-independent genes. The results also confirm the previously proposed role of CAR in the clearance of bilirubin (34, 35).
Fig. 5.
The effects of CAR activation in CH/CA-fed pxr-/- mice. (A) Survival of pxr-/- mice pretreated with TCPOBOP or solvent before the feeding of the CH/CA diet for 20 days. No significant difference in the rate of death was observed. (B) Total bilirubin (Left) and total bile acid (Right) concentrations in plasma from WT (+/+) and lethargic pxr-/- (-/-) mice pretreated with TCPOBOP or solvent on the CH/CA diet for 7–10 days. All values are expressed as mean ± SD, n = 5. *, significant differences compared with WT on the same treatment; #, significant differences compared with mice of the same genotype without TCPOBOP pretreatment (P < 0.005).
Discussion
Although several cholesterol metabolites, such as bile acids, bile alcohols, and epoxycholesterols, have been shown to activate PXR, there has been little evidence for the physiological or pathological importance of PXR function in their detoxification. The results presented here provide genetic evidence that PXR is essential for survival of animals when cholesterol and bile acids accumulate to high levels. Elevated cholesterol is often associated with cholestatic liver diseases in humans such as primary biliary cirrhosis (36) or arteriohepatic dysplasia (Alagille syndrome) (37). Ligand-mediated activation of PXR in such patients may enhance the hepatic detoxification of toxic metabolites. The antibiotic Rifampicin, a potent agonist for human PXR, has been used to ameliorate the pruritus that commonly accompanies cholestasis (38, 39). Our studies in mice suggest that PXR agonists may have additional benefits for patients with cholestasis.
Feeding of CA together with cholesterol dramatically increased the levels of hepatic cholesterol and oxysterols by facilitating intestinal cholesterol absorption and suppressing hepatic cholesterol breakdown (Fig. 1F) (21–23, 40). However, we do not think that accumulation of cholesterol per se is toxic in pxr-/- animals. Feeding of a cholesterol-rich diet to lxrα-/- animals results in accumulation of hepatic and plasma lowdensity lipoprotein cholesterol (41) and in an increased hepatic expression of PXR target genes (J.S., unpublished data). Mice lacking both pxr and lxrα on a cholesterol-rich diet also accumulate high levels of hepatic cholesterol, but premature death or jaundice was not observed even after 8 weeks of feeding (J.S., unpublished data). Apparently, the mechanism of hepatic cholesterol accumulation differs between CH/CA-fed WT mice and cholesterol-fed lxrα-/- mice; in the former, both major hepatic cholesterol breakdown pathways, the neutral and the acidic pathways, are repressed (42), whereas in the latter, only the neutral pathway is suppressed (41). Perhaps repression of both pathways (together with increased intestinal cholesterol absorption) by CH/CA feeding results in accumulation of metabolic intermediates such as hydroxy- or epoxycholesterols or their derivatives, some of which will impair hepatorenal function unless detoxified by PXR.
In this study, hepatic cyp3a11 and oatp2 are the only putative PXR target genes found to be differentially expressed in the CH/CA-fed WT and pxr-/- animals (Fig. 4A). Of these, cyp3a is unlikely to play a major role in protection of animals from CH/CA diet feeding, because TCPOBOP induced cyp3a expression but did not rescue the lethality of pxr-/- animals. OATP2 is localized in the sinusoidal membranes in the hepatocytes and is proposed to transport bile acids into the hepatocytes (43). Therefore, oatp2 represents the best candidate target gene for mediating CH/CA protection. Although the phenotype of oatp2 deficiency is not known, inactivating mutations in several other transporters are known to cause hepatobiliary diseases. For example, the loss of the MRP2/cMOAT transporter results in conjugated bilirubinemia known as Dubin–Johnson syndrome and the loss of FIC1, BSEP, or MDR3 transporter results in progressive familial intrahepatic cholestasis (2, 3). Genetic dissection of OATP2 function and identification of additional PXR target genes will be required to further understand the mechanism of the PXR-mediated protection.
Although it is not clear what actually triggers the acute death of pxr-/- mice on the CH/CA diet, it is unlikely that high levels of conjugated bilirubin or bile acids are the direct cause, because liver-specific hnf1β-/- mice that also show a high level of plasma-conjugated bilirubin and bile acids typically survive for several months after birth (44). We instead speculate that the renal failure might be the direct cause of the death (Fig. 3A). In fact, renal failure represents a major cause of death for patients with advanced liver diseases. In a large follow-up study of advanced cirrhotic patients, acute renal failure occurred in 40% of the patients over 5 years, with a median survival of 1.7 weeks or a 90% mortality at 10 weeks (25). In addition, Alagille syndrome, characterized by a paucity of bile ducts and cholestasis, is often accompanied by renal and cardiovascular abnormalities (45). If the CH/CA-induced renal failure in pxr-/- animals and renal failure associated with liver diseases are caused by common endogenous toxins, PXR ligands may be beneficial not only to treat pruritus but also to prevent acute renal failure and death in patients with advanced liver diseases.
Supplementary Material
Acknowledgments
We thank Elaine Stevens and Lita Ong for administrative assistance; Yu-Hua Zou for help in DNA microarray analysis; University of California, San Diego, histology shared resources for preparation of histological sections and staining; Nissi Varki for pathological consultation; and Ruth Yu for critical reading of the manuscript. J.S. is supported by Damon Runyon Cancer Research Foundation Postdoctoral Fellowship DRG 1711-02. G.D.B. is supported by National Institutes of Health Public Health Services Grant CA09370-23. The DNA microarray analysis was supported by the National Institutes of Health Nuclear Receptor Signaling Atlas orphan receptor program Grant U19DK62434-01. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by the Howard Hughes Medical Institute.
Author contributions: J.S. designed research; J.S., L.W.C., S.C., and C.L. performed research; J.S., M.D., S.C., C.L., and C.-H.L. contributed new reagents/analytic tools; J.S., G.D.B., S.C., C.L., and C.-H.L. analyzed data; and J.S. and R.M.E. wrote the paper.
Abbreviations: CA, cholic acid; CAR, constitutive androstane receptor; CH/CA diet, 1.25% cholesterol/0.5% CA diet; LCA, lithocholic acid; PXR, pregnane X receptor; qPCR, quantitative PCR; TCPOBOP, 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene.
References
- 1.Russell, D. W. (2003) Annu. Rev. Biochem. 72, 137-174. [DOI] [PubMed] [Google Scholar]
- 2.Koopen, N. R., Muller, M., Vonk, R. J., Zimniak, P. & Kuipers, F. (1998) Biochim. Biophys. Acta 1408, 1-17. [DOI] [PubMed] [Google Scholar]
- 3.Elferink, R. O. & Groen, A. K. (2002) Biochim. Biophys. Acta 1586, 129-145. [DOI] [PubMed] [Google Scholar]
- 4.Chawla, A., Repa, J. J., Evans, R. M. & Mangelsdorf, D. J. (2001) Science 294, 1866-1870. [DOI] [PubMed] [Google Scholar]
- 5.Xie, W. & Evans, R. M. (2001) J. Biol. Chem. 276, 37739-37742. [DOI] [PubMed] [Google Scholar]
- 6.Kliewer, S. A., Goodwin, B. & Willson, T. M. (2002) Endocr. Rev. 23, 687-702. [DOI] [PubMed] [Google Scholar]
- 7.Blumberg, B., Sabbagh, W., Jr., Juguilon, H., Bolado, J., Jr., van Meter, C. M., Ong, E. S. & Evans, R. M. (1998) Genes Dev. 12, 3195-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maglich, J. M., Stoltz, C. M., Goodwin, B., Hawkins-Brown, D., Moore, J. T. & Kliewer, S. A. (2002) Mol. Pharmacol. 62, 638-646. [DOI] [PubMed] [Google Scholar]
- 9.Rosenfeld, J. M., Vargas, R., Jr., Xie, W. & Evans, R. M. (2003) Mol. Endocrinol. 17, 1268-1282. [DOI] [PubMed] [Google Scholar]
- 10.Xie, W., Barwick, J. L., Downes, M., Blumberg, B., Simon, C. M., Nelson, M. C., Neuschwander-Tetri, B. A., Brunt, E. M., Guzelian, P. S. & Evans, R. M. (2000) Nature 406, 435-439. [DOI] [PubMed] [Google Scholar]
- 11.Staudinger, J. L., Goodwin, B., Jones, S. A., Hawkins-Brown, D., MacKenzie, K. I., LaTour, A., Liu, Y., Klaassen, C. D., Brown, K. K., Reinhard, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 3369-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie, W., Radominska-Pandya, A., Shi, Y., Simon, C. M., Nelson, M. C., Ong, E. S., Waxman, D. J. & Evans, R. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuetz, E. G., Strom, S., Yasuda, K., Lecureur, V., Assem, M., Brimer, C., Lamba, J., Kim, R. B., Ramachandran, V., Komoroski, B. J., et al. (2001) J. Biol. Chem. 276, 39411-39418. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin, B., Gauthier, K. C., Umetani, M., Watson, M. A., Lochansky, M. I., Collins, J. L., Leitersdorf, E., Mangelsdorf, D. J., Kliewer, S. A. & Repa, J. J. (2003) Proc. Natl. Acad. Sci. USA 100, 223-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dussault, I., Yoo, H. D., Lin, M., Wang, E., Fan, M., Batta, A. K., Salen, G., Erickson, S. K. & Forman, B. M. (2003) Proc. Natl. Acad. Sci. USA 100, 833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda, A., Salen, G., Matsuzaki, Y., Batta, A. K., Xu, G., Leitersdorf, E., Tint, G. S., Erickson, S. K., Tanaka, N. & Shefer, S. (2001) J. Biol. Chem. 276, 34579-34585. [DOI] [PubMed] [Google Scholar]
- 17.Honda, A., Salen, G., Matsuzaki, Y., Batta, A. K., Xu, G., Leitersdorf, E., Tint, G. S., Erickson, S. K., Tanaka, N. & Shefer, S. (2001) J. Lipid Res. 42, 291-300. [PubMed] [Google Scholar]
- 18.Shenoy, S. D., Spencer, T. A., Mercer-Haines, N. A., Alipour, M., Gargano, M. D., Runge-Morris, M. & Kocarek, T. A. (2004) Drug Metab. Dispos. 32, 66-71. [DOI] [PubMed] [Google Scholar]
- 19.Stedman, C., Robertson, G., Coulter, S. & Liddle, C. (2004) J. Biol. Chem. 279, 11336-11343. [DOI] [PubMed] [Google Scholar]
- 20.Zapala, M. A., Lockhart, D. J., Pankratz, D. G., Garcia, A. J. & Barlow, C. (2002) Genome Biol. 3, SOFTWARE0001. [DOI] [PMC free article] [PubMed]
- 21.Tepperman, J., Caldwell, F. T. & Tepperman, H. M. (1964) Am. J. Physiol. 206, 628-634. [DOI] [PubMed] [Google Scholar]
- 22.Vergnes, L., Phan, J., Strauss, M., Tafuri, S. & Reue, K. (2003) J. Biol. Chem. 278, 42774-42784. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, Z., Li, D., Blanchard, D. E., Lear, S. R., Erickson, S. K. & Spencer, T. A. (2001) J. Lipid Res. 42, 649-658. [PubMed] [Google Scholar]
- 24.Tukey, R. H. & Strassburg, C. P. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 581-616. [DOI] [PubMed] [Google Scholar]
- 25.Gines, A., Escorsell, A., Gines, P., Salo, J., Jimenez, W., Inglada, L., Navasa, M., Claria, J., Rimola, A., Arroyo, V., et al. (1993) Gastroenterology 105, 229-236. [DOI] [PubMed] [Google Scholar]
- 26.Kliewer, S. A., Moore, J. T., Wade, L., Staudinger, J. L., Watson, M. A., Jones, S. A., McKee, D. D., Oliver, B. B., Willson, T. M., Zetterstrom, R. H., et al. (1998) Cell 92, 73-82. [DOI] [PubMed] [Google Scholar]
- 27.Xie, W., Barwick, J. L., Simon, C. M., Pierce, A. M., Safe, S., Blumberg, B., Guzelian, P. S. & Evans, R. M. (2000) Genes Dev. 14, 3014-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, P., Zhang, J., Egan-Hafley, M., Liang, S. & Moore, D. D. (2000) Nature 407, 920-923. [DOI] [PubMed] [Google Scholar]
- 29.Sonoda, J., Xie, W., Rosenfeld, J. M., Barwick, J. L., Guzelian, P. S. & Evans, R. M. (2002) Proc. Natl. Acad. Sci. USA 99, 13801-13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saini, S. P., Sonoda, J., Xu, L., Toma, D., Uppal, H., Mu, Y., Ren, S., Moore, D. D., Evans, R. M. & Xie, W. (2004) Mol. Pharmacol. 65, 292-300. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, J., Huang, W., Qatanani, M., Evans, R. M. & Moore, D. D. (2004) J. Biol. Chem. 279, 49517-49522. [DOI] [PubMed] [Google Scholar]
- 32.Tzameli, I., Pissios, P., Schuetz, E. G. & Moore, D. D. (2000) Mol. Cell. Biol. 20, 2951-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, G., Henderson, C. J., Parker, M. G., White, R., Bars, R. G. & Wolf, C. R. (1993) Biochem. J. 289, 807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, W., Zhang, J., Chua, S. S., Qatanani, M., Han, Y., Granata, R. & Moore, D. D. (2003) Proc. Natl. Acad. Sci. USA 100, 4156-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, W., Zhang, J. & Moore, D. D. (2004) J. Clin. Invest. 113, 137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crippin, J. S., Lindor, K. D., Jorgensen, R., Kottke, B. A., Harrison, J. M., Murtaugh, P. A. & Dickson, E. R. (1992) Hepatology 15, 858-862. [DOI] [PubMed] [Google Scholar]
- 37.Davit-Spraul, A., Pourci, M. L., Atger, V., Cambillau, M., Hadchouel, M., Moatti, N. & Legrand, A. (1996) Gastroenterology 111, 1023-1032. [DOI] [PubMed] [Google Scholar]
- 38.Bachs, L., Pares, A., Elena, M., Piera, C. & Rodes, J. (1992) Gastroenterology 102, 2077-2080. [DOI] [PubMed] [Google Scholar]
- 39.Cancado, E. L., Leitao, R. M., Carrilho, F. J. & Laudanna, A. A. (1998) Am. J. Gastroenterol. 93, 1510-1517. [DOI] [PubMed] [Google Scholar]
- 40.Wang, L., Han, Y., Kim, C. S., Lee, Y. K. & Moore, D. D. (2003) J. Biol. Chem. 278, 44475-44781. [DOI] [PubMed] [Google Scholar]
- 41.Peet, D. J., Turley, S. D., Ma, W., Janowski, B. A., Lobaccaro, J. M., Hammer, R. E. & Mangelsdorf, D. J. (1998) Cell 93, 693-704. [DOI] [PubMed] [Google Scholar]
- 42.Sinal, C. J., Tohkin, M., Miyata, M., Ward, J. M., Lambert, G. & Gonzalez, F. J. (2000) Cell 102, 731-744. [DOI] [PubMed] [Google Scholar]
- 43.Reichel, C., Gao, B., Van Montfoort, J., Cattori, V., Rahner, C., Hagenbuch, B., Stieger, B., Kamisako, T. & Meier, P. J. (1999) Gastroenterology 117, 688-695. [DOI] [PubMed] [Google Scholar]
- 44.Coffinier, C., Gresh, L., Fiette, L., Tronche, F., Schutz, G., Babinet, C., Pontoglio, M., Yaniv, M. & Barra, J. (2002) Development (Cambridge, U.K.) 129, 1829-1838. [DOI] [PubMed] [Google Scholar]
- 45.Emerick, K. M., Rand, E. B., Goldmuntz, E., Krantz, I. D., Spinner, N. B. & Piccoli, D. A. (1999) Hepatology 29, 822-829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.