Abstract
Changes in the substrate specificities of factors that irreversibly modify the histone components of chromatin are expected to have a profound effect on gene expression through epigenetics. Ezh2 is a histone-lysine methyltransferase with activity dependent on its association with other components of the Polycomb Repressive Complexes 2 and 3 (PRC2/3). Ezh2 levels are increasingly elevated during prostate cancer progression. Other PRC2/3 components also are elevated in cancer cells. Overexpression of Ezh2 in tissue culture promotes formation of a previously undescribed PRC complex, PRC4, that contains the NAD+-dependent histone deacetylase SirT1 and isoform 2 of the PRC component Eed. Eed2 is expressed in cancer and undifferentiated embryonic stem (ES) cells but is undetectable in normal and differentiated ES cells. The distinct PRCs exhibit differential histone substrate specificities. These findings suggest that formation of a transformation-specific PRC complex may have a major role in resetting patterns of gene expression by regulating chromatin structure.
Keywords: methylation, prostate cancer, Ezh2, histone H1
The N-terminal domains of histone polypeptides are subject to multiple posttranslational modifications. These modifications are believed to influence the transitions between the open and compacted chromatin states. One such modification, lysine (K) methylation, plays an important role in epigenetic inheritance of chromatin states (1, 2). The enzymes catalyzing lysine methylation at specific histone residues have been characterized (3). Methylation at K27 of histone H3 is a repressive “mark” catalyzed by the SET-domain containing Enhancer of Zeste protein-2, Ezh2 (4–6). Ezh2 on its own lacks enzymatic activity, which is conferred on its association with specific polypeptides present in the Polycomb Repressive Complexes 2 and 3 (PRC2/3) (7, 8). PRC2/3 complexes contain the Suppressor of Zeste 12 [Su(z)12 or its human homologue, JJAZ1], the Extra Sex Combs protein(s) (or its mammalian homologue Eed), and the histone-binding proteins RbAp46 and RbAp48 (9). The human Eed protein exists in four different isoforms that arise from alternate translation initiation sites from the same mRNA (8, 10), generating different Ezh2-containing complexes (7, 8). The largest form (Eed1) is predominantly present in the Polycomb Repressive Complex 2 (PRC2), whereas the two shortest forms (Eed3 and Eed4) are present in the PRC3 complex (8). Neither PRC2 nor PRC3 contain the Eed isoform 2 (Eed2). The PRC3 complex exclusively targets methylation of histone H3-K27, and the activity is repressed in the presence of histone H1. PRC2 also methylates histone H3-K27 but in the presence of histone H1, PRC2 methylates both H3-K27 and H1-K26 (8).
In this study, we report the characterization of PRC4, a complex containing the Eed isoform 2 (Eed2), which appears to be present only in undifferentiated pluripotent cells as well as in cells that have lost their “normal” regulation. PRC4 contains the NAD+-dependent histone deacetylase SirT1, which we found, together with other subunits of the PRC2/3 complexes, to be overexpressed in breast, colon, and prostate cancers. Importantly, previous studies have indicated that EzH2 is overexpressed in late stages of prostate cancer (11). We found that overexpression of EzH2 results in the accumulation of the PRC4 complex.
Materials and Methods
Histone Lysine Methyltransferase (HKMT) Assays. HKMT assays were performed essentially as described in ref. 12.
Conventional Purification of PRC4. Approximately 10 g of solubilized HeLa nuclear pellet was loaded onto a 1-liter DE-52 column in buffer BC (50 mM Tris, pH 7.9/0.2 mM EDTA/20% glycerol/0.2 mM PMSF/10 mM 2-mercaptoethanol) containing 100 mM ammonium sulfate. After extensive washing of the column with buffer BC, proteins were eluted with an 8-liter linear gradient of ammonium sulfate from 100 to 500 mM in buffer BC. Fractions containing H1KMT-1 activity were pooled, dialyzed against buffer B containing 50 mM KCl, and loaded onto a 100-ml phosphocellulose column. The column was washed with 10 vol by using buffer C and proteins eluted with a 1-liter linear gradient of KCl from 100 mM to 1 M in buffer B. Fractions containing H1KMT-1 activity were pooled, dialyzed against buffer BC containing 50 mM KCl, and loaded onto a 35-ml DEAE-5PW column. The column was washed as described above, and proteins were eluted with a 300-ml linear gradient of KCl from 50 to 700 mM in buffer BC. Fractions containing H1KMT-1 were pooled, precipitated with 65% saturated ammonium sulfate, resuspended in 200 μl of buffer BC containing 500 mM KCl, and fractionated on a 24-ml Superose 6 column in the same buffer.
Mouse Model and Microarray Gene Expression Profiling. Nkx3.1; Pten mutant mice are described in ref. 13. Histological, immunohistochemical, and gross morphological analyses of the prostate at 2–24 months of age were evaluated as described in refs. 13–15. Anti-Ezh2 and anti-Su(z)12 antibodies were affinity purified by using the respective antigen. Anti-Ki67 antibody was purchased from Novo-Castra Laboratories (Newcastle, U.K.), and anti-SirT1 (sir2-α) was purchased from Upstate Biotechnology (Lake Placid, NY). For gene expression profiling, laser-capture microdissection (PixCell IIE, Arcturus, Mountain View, CA) was performed on snap-frozen prostate tissues to isolate epithelial cells from wild-type (WT) prostate, prostatic intraepithelial neoplasia (PIN), or cancer lesions. RNA was prepared by using the PicoPure RNA isolation kit (Arcturus), followed by one round of amplification using the RiboAmp RNA amplification kit (Arcturus). Samples were labeled by using a BioArray High Yield RNA transcript labeling kit (Enzo Life Sciences, Farmingdale, NY) and hybridized to Affymetrix GeneChips. For statistical analyses, initial data acquisition and normalization was performed by using Affymetrix microarray suite 5.0 software. Data filtering and statistical analyses were conducted in three steps. First, the analysis of variance (ANOVA) method was used to compare samples at different stages of cancer by using a P value cutoff of 0.005. The Benjamini and Hochberg procedure was used to control for false discovery rate also by using a P value cutoff of 0.005. Next, we applied expression level detection restrictions, retaining only genes with detection of P < 0.001 in at least one sample. Finally, we performed a two-way hierarchical clustering analysis on the 1,308 remaining genes by using gene-cluster and tree view software. The complete array data will be presented elsewhere (X.O. and C.A.-S., unpublished data).
Results and Discussion
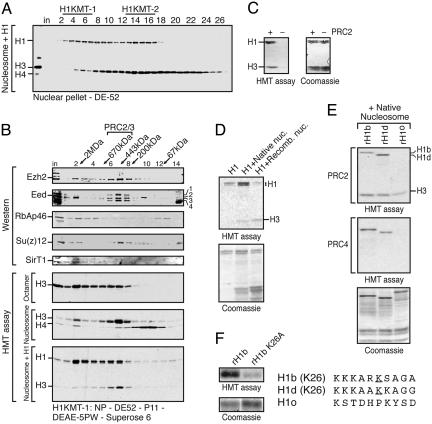
PRC4 I a Polycomb Repressive Complex That Preferentially Methylates Histone H1b. Histone H1 is rich in lysines; therefore, several HK-MTs, like G9a and Set9, can methylate H1 in vitro; however, these enzymes cannot methylate H1 in the context of H1-containing oligonucleosomes (data not shown). When we assayed crude fractions derived from HeLa nuclear pellets by using H1 together with native oligonucleosomes as substrate, we detected two peaks of H1-specific activities that bind to DEAE-cellulose resin (H1KMT-1 and H1KMT-2; Fig. 1A). Partial purification of the H1KMT-2 activity revealed a mixture of the PRC2 and PRC3 complexes characterized in ref. 8, as well as another H1-specific HKMT activity (or activities) not yet characterized (data not shown).
Fig. 1.
Characterization of histone H1 methyltransferase activity and PRC4 substrate specificity. (A) Histone H1 methylation assay of the first purification step, DE-52 column, showing separation of two histone H1-specific activities, H1KMT-1 and H1KMT-2. The assay was performed using H1-containing oligonucleosomes. Positions of H1 and core histones are indicated. Fraction numbers are indicated at the top. The fractions corresponding to H1KMT-1 activity were pooled as indicated. (B) Gel filtration analysis of the PRC4 complex. Fraction numbers and corresponding molecular mass standards are indicated at the top. The first five rows from the top are Western blots of column fractions using the indicated antibodies. The bottom three rows are HKMT assays of column fractions performed with substrates indicated on the left. Positions of core histones and histone H1 are indicated. A trace amount of nucleosomal H4 specific activity was detected in the fractions eluting at ≈1.5 MDa, likely due to a contaminating HMT activity. (C) H1 and native nucleosomes were assayed in the presence or absence of native PRC2. (D) PRC4 histone methyltransferase activity toward H1 in the presence and absence of native or recombinant oligonucleosomes. (E) Comparison of PRC2 and PRC4 methylation of H1 isoforms. Recombinant H1b, H1d, or H1o were purified from Escherichia coli and used as substrate with native oligonucleosomes. (F) PRC4 methylated K26 residue on H1b. Recombinant H1b WT or mutant with a substitution of K26 to A were used as substrates for methylation by PRC4. On the right is a comparison of the amino acid sequences of H1b, H1d, or H1o sequences with the K26 residue underlined.
Partial purification of H1KMT1 revealed that the activity is composed of the PRC2/3 complexes as well as another HKMT activity that eluted from a gel filtration column with an apparent native mass of ≈1.5 MDa (Fig. 1B and see below). Surprisingly, we found that subunits of the PRC2/3 complexes, specifically Ezh2, Su(z)12, Eed, and RbAp46 polypeptides, copurified with the larger molecular mass HKMT activity. This finding was evidenced by Western blot analyses of aliquots of the fractions derived from gel filtration on a Superose 6 column and a DEAE-5PW column (Fig. 1B and data not shown, respectively). Most interestingly, only one form of the four Eed isoforms described in ref. 8, Eed2, coeluted with the larger-molecular-mass H1KMT-1 activity (Fig. 1B). Association of Eed2 with this larger complex was selective because the three other Eed isoforms copurified with the ≈400-kDa HKMT activity corresponding to PRC2/3 on the Superose 6 column (Fig. 1B and data not shown). We designated the larger complex containing Eed2 as PRC4.
Biochemical characterization of PRC4 revealed that, in the absence of H1, PRC4 methylates histone H3 both in the form of octamers and oligonucleosomes (Fig. 1B). When PRC4 was assayed by using H1 containing oligonucleosomes as a substrate, methylation of histone H3 was greatly reduced, and methylation of histone H1 became predominant (Fig. 1B). This observation suggested that the histone substrate specificity of this complex was similar to that of PRC2, although it was not identical (see below).
In our purification assay, we used a mixture of native histone H1 and native oligonucleosomes as substrate. The oligonucleosomes appeared free of any contaminating HKMT activities. Whereas H1 and H3 are methylated in the presence of native PRC2, there is no detectable signal with substrate alone (Fig. 1C). We next characterized the substrate specificity of PRC4 in more detail. As shown in Fig. 1D, PRC4 can methylate native histone H1 to some extent (lane 1). Similar to PRC2, its activity toward histone H1 is markedly stimulated by the addition of native nucleosomes that incorporate H1 readily (lane 2 and data not shown). PRC4 was less active on native histone H1 in the presence of recombinant nucleosomes (Fig. 1D, compare lane 1 with 3), suggesting that posttranslational modifications of core histones might be required for optimal PRC4 activity. We noticed that, unlike PRC2, PRC4 preferentially methylates the slower-migrating histone H1 species (8). The observed difference in mobility of this H1 species was not due to phosphorylation because phosphatase treatment was ineffectual (data not shown). Further studies uncovered that PRC4 methylated preferentially the histone H1b isoform, whereas PRC2 methylated preferentially the histone H1d isoform (Fig. 1E). PRC2 and PRC4 did not exhibit methylation activity toward the H1o isoform that lacks K26 (Fig. 1E). Consistently, PRC4 failed to methylate a mutant form of H1b in which K26 was substituted to alanine (Fig. 1F). Therefore, these analyses indicated that PRC4 prefers to methylate histone H1b as compared with H1d, the preferred substrate for PRC2. Moreover, unlike PRC2, PRC4 loses its ability to methylate nucleosomal histone H3 in the presence of histone H1 (Fig. 1E).
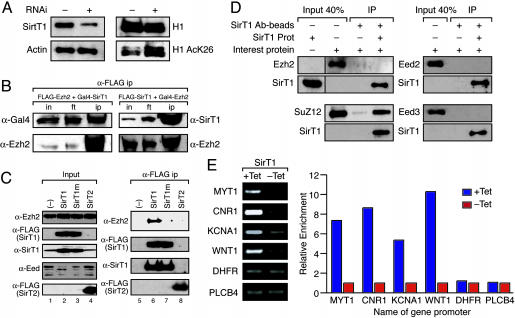
Sirt1 Deacetylates H1-K26 and Interacts with the PRC4 Complex. Our previous studies demonstrated that SirT1 is a NAD+-dependent histone deacetylase that interacts with histone H1 and deacetylates histone H1-K26 (16), a residue conserved among most of the histone H1 isoforms. Most importantly, H1-K26 is the residue that is methylated by PRC2 (8) and by PRC4 (this study). We speculated that SirT1-mediated deacetylation of H1-K26 might be required for PRC2- or PRC4-mediated methylation of H1-K26, much like deacetylation by HDAC1 facilitates H3-K9 methylation (17). By using RNA interference, we gauged the ability of SirT1 to deacetylate H1-K26 in vivo (Fig. 2A). After transfection of dsRNA directed toward SirT1 in HeLa cells, we analyzed the levels of SirT1, total histone H1, and H1 acetylated at K26. Whereas decreasing the levels of SirT1 did not substantially affect the total amount of H1, we observed a striking increase in the levels of acetylated H1-K26, suggesting that SirT1 deacetylates this residue in vivo (Fig. 2A).
Fig. 2.
Interaction between SirT1 and PRC components. (A) RNA interference (RNAi) experiments using SMARTpool of dsRNA for SirT1 (Dharmacon, Lafayette, CO) as described in ref. 16. HeLa cells were transfected with or without the SirT1 SMARTpool and analyzed by Western blot for the presence of SirT1, actin, H1, and H1-AcK26. (B) We transfected 293 cells with expression vectors as indicated. Extracts from transfected cells were immunoprecipitated with anti-FLAG antibodies. Aliquots of the immunoprecipitation input (in), flowthrough (ft), and eluate (ip) were analyzed by Western blot using the indicated antibodies. (C) Analysis of specificity of SirT1-Ezh2 interaction. Anti-FLAG immunoprecipitation was performed with extracts from 293 cells transfected with expression vectors encoding untagged Ezh2 and FLAG-tagged SirT proteins as follows: WT SirT1 (SirT1), active site mutant SirT1 (SirT1m), or WT SirT2 protein (SirT2). Inputs for immunoprecipitation (Input, Left) and anti-FLAG immunoprecipitates (α-FLAG ip, Right) were analyzed by Western blot using the indicated antibodies. (D) Recombinant Ezh2, Su(z)12, EED (30–535), or EED (95–535) were incubated separately with or without recombinant SirT1 protein and immunoprecipitated by using SirT1-specific antibody (2G10) coupled to beads. Inputs and elutions were analyzed by Western blot using the indicated antibodies. (E) Chromatin immunoprecipitation experiments by using an antibody to Gal4 were performed as described in ref. 16 in 293f cells expressing Gal4Sirt1 in the presence(+Tet) or absence (–Tet) of tetracycline. The precipitated chromatin was analyzed with PCR by using primers specific to the promoters targeted by PRC2/3 (MYT1, CNR1, KCNA1, and WNT1) and to the negative control promoters (DHFR and PLCB4) as indicated. The relative enrichment of the signal in the absence or presence of tetracycline (which causes the induction of Gal4-SirT1) is shown.
To expand the studies, we investigated whether there is a physical interaction between Ezh2-containing complexes and SirT1. This hypothesis is supported by studies in Drosophila demonstrating that both proteins are involved in position effect variegation (18, 19). Additionally, it was reported recently that SirT1 and Drosophila Ezh2 appear to be components of a high-molecular-weight complex in larvae (20). Supporting our speculation, we observed coelution of SirT1 specifically with the PRC4 complex (Fig. 1B). Moreover, cotransfection of FLAG-tagged versions of Ezh2 and SirT1 resulted in coimmunoprecipitation of both proteins, regardless of which protein was tagged (Fig. 2B). Importantly, FLAG-SirT1, but not FLAG-SirT2, coimmunoprecipitated Ezh2 (Fig. 2C, lanes 6 and 8, respectively), hence, indicating that the interaction between the Ezh2 complex and SirT1 was specific. A SirT1 protein harboring a mutation that abolished catalytic activity was unable to coimmunoprecipitate Ezh2, suggesting that the NAD+-dependent activity of SirT1 is required for interaction (Fig. 2C, lane 7). Immunoprecipitation experiments using purified individual subunits of the PRC2/3/4 complexes expressed from baculovirus-infected Sf9 cells and mixed with purified SirT1 demonstrated a specific and direct interaction between SirT1 and Su(z)12 (Fig. 2D).
Of note, we found that overexpression of WT SirT1 in 293f cells gave rise to a change in the relative abundance of the Eed isoforms present in nuclear extracts (Fig. 2C, lane 2). Specifically, the amounts of Eed isoforms 1, 3, and 4 were reduced. In contrast, the levels of the second largest Eed protein (Eed2) were significantly increased. This altered pattern of Eed isoform expression required SirT1 enzymatic activity. The results obtained from cells overexpressing the catalytically inactive form of SirT1 were indistinguishable from those obtained with untransfected cells (Fig. 2B, compare lanes 1 and 3). Moreover, this finding was specific for WT SirT1, because overexpression of WT SirT2 was ineffectual (Fig. 2B, compare lanes 1 and 4). The underlying mechanism for this change in EED isoform distribution upon SirT1 overexpression is still elusive.
To analyze the association of SirT1 with the PRC2/3-associated genes in vivo, we performed chromatin immunoprecipitation experiments. Our previous chromatin immunoprecipitation analyses led to the identification of specific genes that contained the common PRC2/3 complex components at their promoters and also contained methyl H3-K27 indicative of PRC2 substrate specificity (21). By using a tetracycline-inducible system to regulate SirT1 expression (16), we found that SirT1 was present at these same promoters upon its overexpression (Fig. 2E). Importantly, the association of SirT1 with these genes was specific, because two genes whose expression was independent of PRC2/3 components (21) did not show SirT1 at their promoters (Fig. 2E). We concluded that SirT1 has the potential to associate with the PRC2/3 complexes through its interaction with the Su(z)12 subunit. Yet, biochemical experiments demonstrated that SirT1 association is specific to PRC4. SirT1 failed to interact with the PRC2/3 complexes [either native (Fig. 1B) or reconstituted from baculovirus-infected Sf9 cells (data not shown)]. The possibility exists that the epitope of Su(z)12 necessary for interaction with SirT1 is masked in the case of PRC2 and PRC3. These results, collectively, established that the genes that are regulated by the PRC2/3 complexes also can be regulated by the PRC4 complex.
Gene expression would be expected to be impacted differently depending on the composition of the PRCs, given that PRC2, -3, and -4 exhibit differential substrate specificities. Thus, we next sought the cellular state(s)/condition(s) that may exhibit such a change in PRC composition.
Expression of PRC Components Are Regulated During Differentiation and During Cancer. Previous studies demonstrated overexpression of Ezh2 during advanced stages of prostate (11) and breast (22) cancer in human cells. Yet, given that Ezh2 methyltransferase activity requires its association with other PRC subunits (9), overexpression of Ezh2 during prostate (and breast) cancer progression may result in the disruption of the integrity of the PRC complexes or may favor the formation of one or more of these complexes.
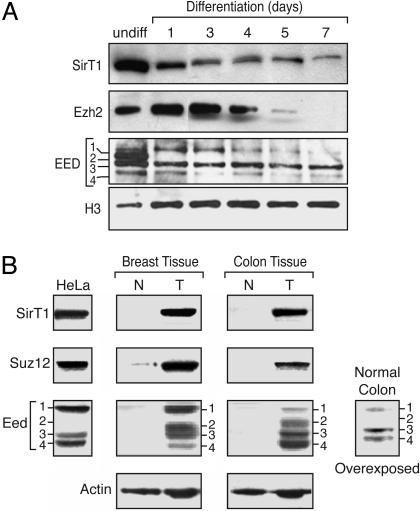
Because transformed cells exhibit partial dedifferentiation, we first compared the levels of several of the PRC components as a function of the state of cellular differentiation. We examined the expression levels of the different Eed proteins that differentially associate with PRC2/3/4, in this case during in vitro differentiation of mouse embryonic stem (ES) cells. Previous studies have shown that the overall levels of Ezh2 and Eed proteins are down-regulated during this process (23). In agreement, we found that the levels of Ezh2 protein gradually declined as cells were induced to differentiate (Fig. 3A). In undifferentiated mouse ES cells, the four Eed isoforms were observed. Strikingly, the Eed protein isoforms showed different patterns of expression during ES cell differentiation. The PRC4-specific Eed2 was down-regulated upon induction to differentiate. The levels of the other Eed isoforms also changed during the differentiation process, each of them with apparently different kinetics and with Eed3 predominantly remaining. This finding is consistent with our analysis using cells derived from normal fresh mouse tissue, in which mainly Eed3 was detectable (data not shown). The levels of SirT1 also decreased during ES cell differentiation (Fig. 3A).
Fig. 3.
Expression of PRC components is modulated as a function of differentiation and cellular transformation. (A) Western blot analysis of nuclear extracts from PGK12.1 ES cells and differentiated cells. Days of differentiation are indicated. The Western blot was probed with antibodies against SirT1, Ezh2, and Eed as well as control antibody directed against histone H3. (B) Western blot analysis of PRC components using HeLa cell nuclear extracts or whole-tissue extracts prepared from normal (N) and tumor (T) tissues obtained from a breast cancer patient and a colon cancer patient. Tissue samples obtained from the Cooperative Human Tissue Network were processed as described in ref. 21. The blots were probed with antibodies to Su(z)12, SirT1, Eed, and actin, which served as a loading control. A long exposure of the same blot containing the normal colon extract probed with Eed antibody is also shown.
Interestingly, analysis of the Eed isoforms in tissue culture cells uncovered great variability. We found that 293f cells expressed all four isoforms (see below), whereas HeLa cells expressed predominantly Eed isoforms 1, 3, and 4 (Fig. 3B). Eed2 is present in HeLa cells, but at much reduced levels compared with the other isoforms, and is detectable only after concentration through purification of the PRC4 complex (see Fig. 1). Importantly, all of the other subunits of the PRC complexes, including SirT1 that is specific to PRC4, were found to be markedly overexpressed in tissue culture cells, relative to their expression levels in normal human and mouse tissues (see below).
To further analyze the expression of the Eed subunits and other PRC4 components such as SirT1 and Su(z)12, we performed Western blot analyses using human colon and breast cells derived from normal and cancerous tissues. As control, we used HeLa cell nuclear extracts that displayed SirT1 and Su(z)12 and, in agreement with our previous observations, Eed isoforms 1, 3, and 4, but not Eed2 (Fig. 3B). We were unable to detect any of the PRC proteins in human cells derived from normal tissues, although actin was as readily detectable in this case as in cancer cells. Cancer cells displayed elevated levels of Su(z)12, as expected (24), but SirT1 and the four isoforms of Eed also were elevated (Fig. 3B). In an attempt to analyze the Eed isoforms present in normal tissue, we increased the exposure time of the Western blots. Under these conditions, Eed isoforms 1, 3, and 4 were now detectable, with Eed3 present at the greatest levels (Fig. 3B). Yet, Eed2 was still undetectable in the case of cells derived from normal tissue, similar to our findings with differentiated ES cells (Fig. 3A). Thus, in accordance with previous studies demonstrating overexpression of Ezh2 in human prostate cancer, there is an overexpression of PRC subunits in cancer cells, including the PRC4-specific SirT1. Remarkably, these cells also displayed an altered pattern of Eed isoform expression with marked elevation of the PRC4-specific Eed2, which was undetectable in cells derived from normal tissue.
Ezh2 Overexpression Leads to PRC4 Formation. That only PRC4 is comprised of Eed2 and SirT1 prompted us to investigate conditions that may promote its formation. We initially detected PRC4 by using conditions favorable for isolating Ezh2-containing complexes that involved overexpression of FLAG-tagged Ezh2 (data not shown). We next tested for the effects of Ezh2 overexpression on PRC4 complex formation.
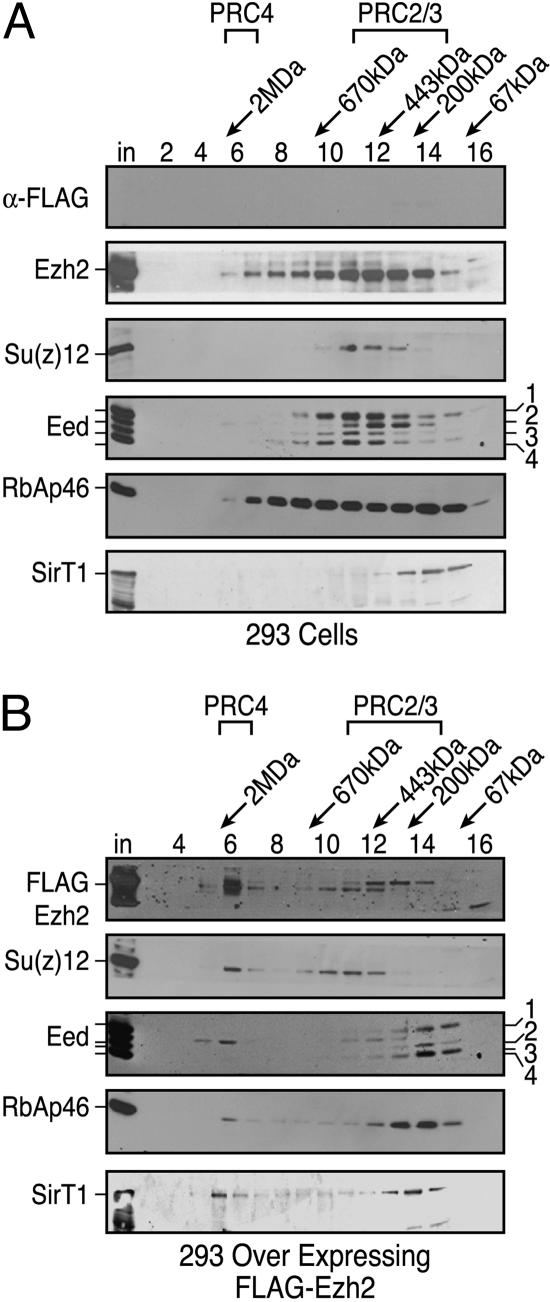
Western blot analysis using monoclonal antibodies against Eed (M26) detected the four isoforms in 293f cells during gel filtration chromatography (Fig. 4A). Forms 1, 3, and 4 were found in the ≈400-kDa region, which coelutes with endogenous Ezh2 and Su(z)12 and likely represents a mixture of the PRC2/PRC3 complexes (8). Within these same fractions, Eed2 did not coelute with Eed forms 1, 3, and 4; rather, the Eed2 elution profile was present in the ≈200 kDa range (Fig. 4A). SirT1 eluted with an apparent mass of ≈350 kDa, in agreement with our previous studies indicating that SirT1 exists in cells as a trimer (16).
Fig. 4.
PRC complexes in 293f cells overexpressing Ezh2. Gel filtration of nuclear extracts from 293f (A) and 293-FLAG-Ezh2 (B) cell lines on Sephacryl-400 columns. Fraction numbers and molecular mass standards are indicated; “in” corresponds to column input. Western blots were probed with antibodies against proteins as indicated. The amount of protein analyzed in A was approximately twice that of B.
However, when 293f cells overexpressed Ezh2, a fraction of Ezh2 was now found in the ≈1.5-MDa complex (Fig. 4B). Most importantly, a redistribution of Eed isoforms also was observed. Specifically, Eed2 now was predominantly associated with the 1.5-MDa complex, i.e., PRC4, and was not the result of aggregation because the other Eed isoforms (1/3/4) comprising the PRC2/3 complexes remained in the ≈400-kDa region (Fig. 4B). A fraction of Su(z)12, and RbAp46 also were found in the 1.5-MDa region (Fig. 4B). Additionally, a fraction of SirT1 then was found to coelute with Ezh2 in the molecular mass range of ≈1.5 MDa, in agreement with our observation that SirT1 copurifies with PRC4 (Fig. 4B).
PRC Components and Target Gene Regulation in a Mouse Model for Human Prostate Cancer. Thus far, our studies demonstrated that the components of the PRC2/3/4 complexes are up-regulated in transformed cells and that up-regulation of Ezh2 results in the formation of PRC4 containing SirT1 and the Eed2 isoform. Up to this point, we used tissue culture cells and human tissue specimens to compare the levels of PRC components at the fixed stages of normalcy or full transformation. To extend these studies to an in vivo model system, we next used a well defined mouse model of prostate cancer to track changes in PRC component profiles over a range of carcinogenesis, during inception and progression of cancer lesions in vivo.
Previous studies established that mice heterozygotic for the tumor suppressor Pten and the prostate-specific homeobox gene, Nkx3.1, represent an ideal model system to study different stages of prostate cancer in an age-dependent manner (13, 15). These Nkx3.1+/–; Pten+/– compound mutant mice develop PIN lesions by 6 months of age and adenocarinoma by 12 months (13, 15). After androgen deprivation by castration, these mice develop metastasis to the lung and lymph nodes (15).
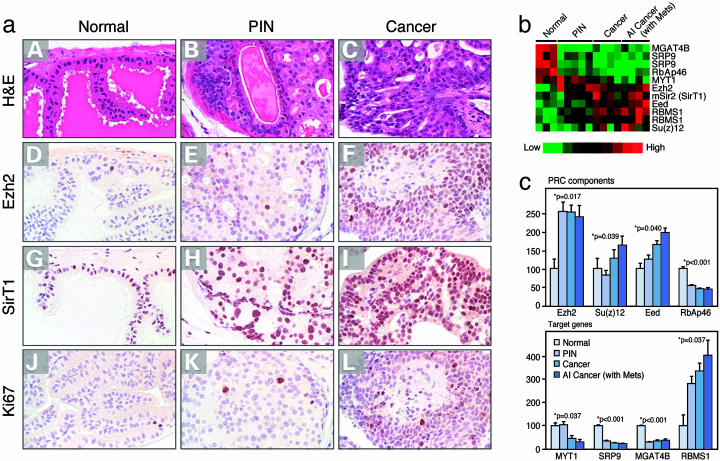
By using this system, we gauged the levels of PRC2/3/4 components using immunohistochemistry performed with highly specific antibodies directed against Ezh2 and SirT1 in normal prostate, PIN, and adenocarcinomas. This analysis revealed low levels of Ezh2 and SirT1 in normal prostate, which was restricted to epithelial cells with some expression of SirT1 to the stroma (Fig. 5a). PIN lesions displayed a moderate elevation in cells expressing Ezh2 and SirT1, and further elevated levels of expression were observed in cancer-derived tissue (Fig. 5a). It has been suggested that elevated Ezh2 levels may be attributable to the extent of cellular proliferation (25). However, in this mouse model, increased levels of Ezh2 and also SirT1 did not correlate with elevated expression of the proliferation marker Ki67 (Fig. 5a).
Fig. 5.
PRC4 analyses in vivo. (a) Immunohistochemical detection of SirT1 and Ezh2 in PIN and cancer lesions. Sections from anterior prostates of WT and Nkx3.1+/–; Pten+/– mice were processed for hematoxylin/eosin (H&E) staining (A–C) or immunostained by using Ezh2 (D–F), SirT1 (G–I), or Ki67 (J–L) antisera. (A, D, G, and J) Sections from a WT Nkx3.1+/+; Pten+/+ mouse at 9 months of age. (B, E, H, and K) Sections from a Nkx3.1+/–; Pten+/– mouse at 12 months of age. (C, F, I, and L) Sections from a Nkx3.1+/–; Pten+/– mouse at 15 months of age. Note that Ezh2 immunostaining is infrequent in normal tissue but is more common in PIN and carcinoma, whereas SirT1 is broadly expressed in normal epithelium and stroma but is up-regulated in epithelial cells in PIN and carcinoma. (b and c) Expression profiling of PRC components and target genes from normal to cancer lesions of prostate from compound mutant mice performed by using RNA obtained by laser-capture microdissection. Tree (b) and quantitative (c) representations of microarray results.
To expand these observations, we performed microarray gene expression profiling by using RNA obtained by laser-capture microdissection of prostatic lesions representative of different stages of cancer. The analyses uncovered that the expression of Ezh2, Su(z)12, and Eed all were elevated relative to normal tissue, but the expression level of the histone-binding protein RbAp46 was down-regulated (Fig. 5 b and c). This finding is not surprising because RbAp46 and RbAp48 are components of multiple complexes that affect chromatin structure (26), and RbAp46 down-regulation might play an important role in breast cancer progression (27).
We also investigated the expression pattern of the PRC-regulated genes that we identified previously (21). The PRC down-regulated genes, MYT1, SRP9, and MGAT4B, exhibited decreased expression in prostate cancer in vivo (Fig. 5c). The PRC up-regulated gene, RBMS1, exhibited elevated expression in prostate cancer in vivo (Fig. 5c).
Therefore, our findings using the mouse model for human prostate cancer established a biological relevance for the increased expression of PRC2/3/4 components and differential expression of PRC-regulated genes. However, it is important to emphasize that it is currently unknown whether overexpression of PRC2/3/4 components during cancer progression is causal or not. Nonetheless, our studies demonstrate that the formation of the PRC4 complex, which contains unique histone methylation specificity, is induced in transformed cells. These studies emphasize that the composition of a complex composing a HKMT can be dynamic with resultant changes in substrate specificity. In the case of PRC4, this divergent composition/substrate specificity aligns with cellular transformation.
Acknowledgments
We thank Dr. A. Otte (Swammerdam Institute for Life Science, Amsterdam) for providing anti-Eed antibodies (M26). We thank Drs. Michael Shen and Lynne Vales for valuable comments on the manuscript, Xiaohui Sun for immunohistochemistry, and Jayshree Rao and Yong Lin for assistance with microarray analyses. This work was supported by National Institutes of Health Grants GM3712O (to D.R.), CA84294 (to C.A.-S.), and CA45290 (to P.F.). D.R. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: N.B., C.A.-S., P.F., and D.R. designed research; A. Kuzmichev, R.M., A.V., T.S.P., M.S., A. Kirmizis, and X.O. performed research; and D.R. wrote the paper.
Abbreviations: HKMT, histone lysine methyltransferase; PIN, prostatic intraepithelial neoplasia; PRC, Polycomb Repressive Complex.
References
- 1.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 2.Sims, R. J., III, Nishioka, K. & Reinberg, D. (2003) Trends Genet. 19, 629–639. [DOI] [PubMed] [Google Scholar]
- 3.Lachner, M. & Jenuwein, T. (2002) Curr. Opin. Cell Biol. 14, 286–298. [DOI] [PubMed] [Google Scholar]
- 4.Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R. S. & Zhang, Y. (2002) Science 298, 1039–1043. [DOI] [PubMed] [Google Scholar]
- 5.Czermin, B., Melfi, R., McCabe, D., Seitz, V., Imhof, A. & Pirrotta, V. (2002) Cell 111, 185–196. [DOI] [PubMed] [Google Scholar]
- 6.Muller, J., Hart, C. M., Francis, N. J., Vargas, M. L., Sengupta, A., Wild, B., Miller, E. L., O'Connor, M. B., Kingston, R. E. & Simon, J. A. (2002) Cell 111, 197–208. [DOI] [PubMed] [Google Scholar]
- 7.Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. (2002) Genes Dev. 16, 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuzmichev, A., Jenuwein, T., Tempst, P. & Reinberg, D. (2004) Mol. Cell 14, 183–193. [DOI] [PubMed] [Google Scholar]
- 9.Cao, R. & Zhang, Y. (2004) Mol. Cell 15, 57–67. [DOI] [PubMed] [Google Scholar]
- 10.Denisenko, O., Shnyreva, M., Suzuki, H. & Bomsztyk, K. (1998) Mol. Cell. Biol. 18, 5634–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varambally, S., Dhanasekaran, S. M., Zhou, M., Barrette, T. R., Kumar-Sinha, C., Sanda, M. G., Ghosh, D., Pienta, K. J., Sewalt, R. G., Otte, A. P., et al. (2002) Nature 419, 624–629. [DOI] [PubMed] [Google Scholar]
- 12.Nishioka, K., Chuikov, S., Sarma, K., Erdjument-Bromage, H., Allis, C. D., Tempst, P. & Reinberg, D. (2002) Genes Dev. 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, M. J., Cardiff, R. D., Desai, N., Banach-Petrosky, W. A., Parsons, R., Shen, M. M. & Abate-Shen, C. (2002) Proc. Natl. Acad. Sci. USA 99, 2884–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, M. J., Bhatia-Gaur, R., Banach-Petrosky, W. A., Desai, N., Wang, Y., Hayward, S. W., Cunha, G. R., Cardiff, R. D., Shen, M. M. & Abate-Shen, C. (2002) Cancer Res. 62, 2999–3004. [PubMed] [Google Scholar]
- 15.Abate-Shen, C., Banach-Petrosky, W. A., Sun, X., Economides, K. D., Desai, N., Gregg, J. P., Borowsky, A. D., Cardiff, R. D. & Shen, M. M. (2003) Cancer Res. 63, 3886–3890. [PubMed] [Google Scholar]
- 16.Vaquero, A., Scher, M., Lee, D., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. (2004) Mol. Cell 16, 93–105. [DOI] [PubMed] [Google Scholar]
- 17.Czermin, B., Schotta, G., Hulsmann, B. B., Brehm, A., Becker, P. B., Reuter, G. & Imhof, A. (2001) EMBO Rep. 2, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg, M. I. & Parkhurst, S. M. (2002) Cell 109, 447–458. [DOI] [PubMed] [Google Scholar]
- 19.Laible, G., Wolf, A., Dorn, R., Reuter, G., Nislow, C., Lebersorger, A., Popkin, D., Pillus, L. & Jenuwein, T. (1997) EMBO J. 16, 3219–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuyama, T., Banerjee, R., Breen, T. R. & Harte, P. J. (2004) Curr. Biol. 14, 1812–1821. [DOI] [PubMed] [Google Scholar]
- 21.Kirmizis, A., Bartley, S., Kuzmichev, A., Margueron, R., Reinberg, D., Green, R. & Farnham, P. (2004) Genes Dev. 18, 1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleer, C. G., Cao, Q., Varambally, S., Shen, R., Ota, I., Tomlins, S. A., Ghosh, D., Sewalt, R. G., Otte, A. P., Hayes, D. F., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva, J., Mak, W., Zvetkova, I., Appanah, R., Nesterova, T. B., Webster, Z., Peters, A. H., Jenuwein, T., Otte, A. P. & Brockdorff, N. (2003) Dev. Cell 4, 481–495. [DOI] [PubMed] [Google Scholar]
- 24.Kirmizis, A., Bartley, S. M. & Farnham, P. J. (2003) Mol. Cancer Ther. 2, 113–121. [PubMed] [Google Scholar]
- 25.Bracken, A. P., Pasini, D., Capra, M., Prosperini, E., Colli, E. & Helin, K. (2003) EMBO J. 22, 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loyola, A. & Almouzni, G. (2004) Biochim. Biophys. Acta 1677, 3–11. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, T. F., Yu, S. Q., Deuel, T. F. & Wang, Z. Y. (2003) Anticancer Res. 23, 3735–3740. [PubMed] [Google Scholar]