Abstract
Glucuronoarabinoxylan, xyloglucan, and galactomannan are noncellulosic polysaccharides found in plant cell walls. All consist of β-linked glycan backbones substituted with sugar side chains. Although considerable progress has been made in characterizing the structure of these polysaccharides, little is known about the biosynthetic enzymes that produce them. Cellulose synthase-like (Csl) genes are hypothesized to encode Golgi-localized β-glycan synthases that polymerize the backbones of noncellulosic polysaccharides. To investigate this hypothesis, we used heterologous expression in Drosophila Schneider 2 (S2) cells to systematically analyze the functions of the gene products of a group of Csl genes from Arabidopsis and rice (Oryza sativa L.), including members from five Csl gene families (CslA, CslC, CslD, CslE, and CslH). Our analyses indicate that several members of the CslA gene family encode β-mannan synthases. Recombinant CslA proteins produce β-linked mannan polymers when supplied GDP-mannose. The same proteins can produce β-linked glucomannan heteropolymers when supplied both GDP-mannose and GDP-glucose. One CslA protein also produced β-linked glucan polymers when supplied GDP-glucose alone. Heterologous expression studies of additional candidate glycan synthases in insect cells or other systems may help identify other noncellulosic polysaccharide biosynthetic enzymes.
Keywords: Drosophila Schneider 2 cells, heterologous expression, plant cell wall biosynthesis
Plant cells are enveloped by walls consisting of a complex matrix of cellulose, noncellulosic polysaccharides, proteins, and lignin (1, 2). Plant cell walls determine cell shape and size, provide strength and flexibility, and defend plants against pathogens (1, 3). Cell walls also constitute a major source of renewable biomass. Furthermore, the health benefits attributed to the consumption of certain cell wall polysaccharides are well documented (4–6).
Plant cell wall polysaccharides are produced in two subcellular locations. Callose and cellulose, unsubstituted β-linked polymers of glucose, are synthesized at the plasma membrane by callose synthase and cellulose synthase (CesA) proteins, respectively. The CesA genes encode polytopic membrane proteins belonging to the glycosyltransferase family 2 (1, 7). The remaining noncellulosic matrix polysaccharides of the plant cell wall are produced within the Golgi by glycan synthases and glycosyltransferases (3). Despite predictions that hundreds of enzymes may be required to produce the diverse network of glycosidic linkages within this polysaccharide matrix (8), very few genes encoding enzymes involved in the biosynthesis of noncellulosic polysaccharides of the cell wall have been identified and characterized.
Various approaches to establish the identities of enzymes mediating the biosynthesis of the noncellulosic polysaccharide matrix have been pursued. Efforts to identify such enzymes by using biochemical purification strategies have been successful only for Arabidopsis xyloglucan fucosyltransferase (9) and a galactomannan galactosyltransferase from fenugreek (10), both of which are Golgi membrane-localized and contain a single transmembrane domain. No backbone glycan synthases have yet been identified in this manner, perhaps because these proteins are not abundant or labile during purification procedures (1). Mutant screens and reverse genetic strategies have also led to the identification of several hemicellulose biosynthetic enzymes, including Arabidopsis xyloglucan galactosyltransferase and fucosyltransferase (11, 12), and a pectin glucuronyltransferase from tobacco (13). Genetic redundancy and plasticity of cell wall architecture have undoubtedly complicated such efforts (14). Transcriptional profiling of developing cotton fibers undergoing rapid cellulose deposition resulted in the identification of the first plant CesA gene (15). A similar approach was used to identify a candidate β-mannan synthase (ManS) gene from developing guar (Cyamopsis tetragonoloba) seeds undergoing deposition of galactomannan, a seed storage polysaccharide (16). The transcriptional profiling approach is most effective when applied to synchronous and relatively homogenous tissues that devote most cellular resources to a process of interest. Identifying noncellulosic polysaccharide-producing plant tissues that satisfy these criteria has proven difficult.
Heterologous expression is a powerful method to determine the enzymatic activity of gene products. This approach requires functional enzyme assays along with expression systems capable of producing enzymatically active protein. A few cell wall biosynthetic enzymes exhibit activity when expressed in heterologous systems (9, 10, 12, 16). Plants contain a large number of genes encoding cellulose synthase-like (Csl) proteins sharing sequence similarity with the CesA proteins. The backbones of noncellulosic polysaccharides, including glucuronoarabinoxylan, xyloglucan, galactoglucomannan, mixed-linked β-glucan, and the β-galactan side chains of arabinogalactan proteins, may be biosynthesized by Golgi-resident Csl proteins (14, 17). Indeed, Dhugga et al. (16) recently provided compelling evidence in support of this hypothesis by using transcriptional profiling to identify a CslA candidate gene and demonstrating that it imparted ManS activity when expressed in soybean somatic embryos. Of all Csl gene products, the function of only guar ManS is known. By using heterologous expression, we performed a systematic investigation of the function of a group of Csl genes expressed in Drosophila Schneider 2 (S2) cells. Our studies led us to conclude that at least three Arabidopsis CslA genes encode ManS enzymes. In addition, our results demonstrate the feasibility of using this heterologous expression strategy to identify additional glycan synthase gene products that produce the backbones of other noncellulosic polysaccharides.
Materials and Methods
Reagents and Enzymes. Radiolabeled nucleotide sugar substrates were obtained from the following sources: GDP-[14C]mannose (10.7 GBq/mmol) and UDP-[14C]galactose (11 GBq/mmol) from Amersham Pharmacia; UDP-[14C]glucose (11.3 GBq/mmol) from ICN; UDP-[14C]xylose (9.8 GBq/mmol) from Perkin–Elmer; GDP-[14C]glucose (11.1 GBq/mmol) from American Radiolabeled Chemicals. Nonradioactive UDP-xylose was obtained from Carbosource Services. Purified endo-1,4-β-mannanase from Bacillus sp., endo-1,4-β-glucanase (cellulase) from Trichoderma sp., endo-1,3-β-glucanase (laminarinase) from Trichoderma sp., endo-1,3(4)-β-glucanase (lichenase) from Bacillus subtilis, and carob galactomannan were from Megazyme International. Thermolysin protease, Sepharose CL-6B, Coomassie brilliant blue R250, [14C]dextran molecular weight standards, and all other nonradioactive UDP- and GDP-sugar substrates were from Sigma. Oligonucleotide primers were synthesized by Integrated DNA Technologies. Gateway vectors, competent Escherichia coli cells, platinum pfx DNA polymerase, LR clonase, S2 cells, vectors, and culture media were from Invitrogen. Horseradish peroxidase-conjugated mouse monoclonal antibodies against the T7 epitope tag were obtained from Novagen. Complete-mini protease inhibitor tablets lacking EDTA were from Roche Diagnostics. The BCA protein assay kit and the Super Signal Pico West chemiluminescence system were from Pierce. Immobilon-P poly(vinylidene difluoride) membrane was from Millipore.
Construction and Expression of Tagged Csl Transgenes. Putative full-length cDNA clones of Arabidopsis and rice (Oryza sativa L.) Csl sequences were obtained from the RIKEN Bioresource Center (18), the Rice Genome Resource Center (19), or the Arizona Genomics Institute. Clones analyzed in this study were AtCslA1 (pda12888), AtCslA 2 (pda05367), AtCslA7 (pda02771), AtCslA9 (pda06746), OsCslA7 (OsJNEb15n15), AtCslC4 (pda04773), AtCslD3 (pda02081), AtCslE1 (pda05727), and OsCslH1 (204759).
A uniform strategy was used to add the sequence encoding the T7 epitope tag directly upstream of and in frame with the start codon. Each ORF was PCR-amplified by using a forward primer encoding the T7 epitope tag plus gene-specific sequence beginning at the start codon and a reverse primer including restriction sites and gene-specific sequence beginning at the stop codon. Primer sequences can be found in Table 2, which is published as supporting information on the PNAS web site. PCR was performed by using Platinum pfx polymerase under the manufacturer's recommended conditions. PCR products were purified as described (20) and ligated with the pENTR-d-TOPO vector before introduction into E. coli cells (21). A positive clone of each construct was selected and fully sequenced to verify the absence of PCR-induced mutations. Tagged Csl ORFs were recombined into the pMT-DEST48 vector for inducible expression in S2 cells.
S2 cells were cotransfected with the pCoBlast vector and the pMT-DEST48 vector containing a tagged Csl or untagged GFP transgene. After transfection, stably transformed lines of S2 cells were cultured with blasticidin selection under the manufacturer's suggested conditions. Copper sulfate was added to a concentration of 0.5 mM to induce protein expression by transformed lines. Twenty-four hours after the addition of copper sulfate, cells were harvested by centrifugation (5 min at 500 × g), and used to prepare microsomal membranes or stored at -80°C.
Preparation of Microsomal Membranes from Drosophila S2 Cells. Unless otherwise noted, all procedures were conducted at 4°C. Cell pellets from 25-ml cultures were disrupted by sonication in 3 ml of extraction buffer consisting of 0.4 M sucrose, 10 mM MgCl2, and a mixture of protease inhibitors in 50 mM Hepes-KOH, pH 7.5. Intact cells and debris were collected by centrifugation for 5 min at 1,000 × g. The resulting supernatant was centrifuged for 30 min at 100,000 × g in an RP100 AT4 rotor (Sorvall) to collect microsomal membranes. Membranes were resuspended in 500 μl of extraction buffer by using a glass homogenizer. Protein content was measured by using the enhanced BCA assay with BSA as standard.
Immunoblot Analysis. Before being loaded on 10% SDS polyacrylamide gels, protein samples (40 μg) in standard SDS/PAGE sample buffer were incubated for 15 min at 42°C (boiling resulted in aggregation of the Csl proteins; data not shown). Proteins were transferred to poly(vinylidene difluoride) membranes by using standard procedures (22). Membranes were blocked with milk proteins and probed with peroxidase-conjugated antibodies against the T7 epitope tag, and the signal was detected by using chemiluminescence. After detection, membranes were stained with Coomassie brilliant blue R250 to verify uniform loading and transfer.
Enzyme Assays. A standard assay buffer consisting of 50 mM Hepes-KOH (pH 7.5), containing 2.5 mM DTT, 2.5 mM MgCl2, 5 mM MnCl2, and 6% glycerol was used for all in vitro glycan synthase reactions. Typical assays were conducted at room temperature for 15–30 min in a total volume of 40 μl containing 100–200 μg of protein from S2 microsomal fractions. Nucleotide sugar concentrations used for glycan synthase activity assays were selected based on assay results using microsomal membrane preparations from pea or corn plants (data not shown). The concentrations selected for the various assays were as follows (concentration of radiolabeled substrate in parentheses): xylan synthase, 10 μM UDP-xylose (1.9 μM); ManS, 25 μM GDP-mannose (2.1 μM); Glc*ManS, 12.5 μM GDP-mannose, 12.5 μM GDP-glucose (1.4 μM); GlcMan*S, 12.5 μM GDP-mannose (2.1 μM), 12.5 μM GDP-glucose; UDP-GlcS, 50 μM UDP-glucose (3.3 μM); GDP-GlcS, 25 μM GDP-glucose (1.4 μM); galactan synthase, 50 μM UDP-galactose (2.1 μM). Reactions were terminated by adding 1 ml of 70% ethanol containing 2 mM EDTA and 100 μg of carob galactomannan as carrier. Products were precipitated at -20°C and pelleted at 13,000 × g; pellets were washed four times with 70% ethanol containing 2 mM EDTA to remove unincorporated nucleotide sugar substrate. Washed pellets were resuspended in water and subjected to scintillation counting or further characterization. Reactions were scaled up 5- to 10-fold to make products for characterization.
Characterization of Radiolabeled Polysaccharides. To determine the identity of the labeled monosaccharides present in polysaccharides produced in vitro, aliquots of each product were hydrolyzed in 2 M trifluoroacetic acid at 120°C for 1 h. Product hydrolysates were dried completely by warming under a nitrogen stream and dissolved in water. Monosaccharides were separated by using high pH anion exchange chromatography through a Dionex CarboPac PA10 column by using 3 mM sodium hydroxide as the eluant. This eluant provides satisfactory resolution between a mixture of arabinose, fucose, galactose, glucose, mannose, and xylose standards (data not shown). One-milliliter fractions were collected, and their radioactivity was measured with a scintillation counter. To identify labeled monosaccharides, the elution time of a radioactive monosaccharide was compared with those of known standards added to the samples. For each product, the radioactive monosaccharide eluted as a single peak, the location of which corresponded with the peak of a known standard sugar detected by pulsed amperometry; no epimerization of the labeled sugars was observed.
To characterize the structures of in vitro assay products, aliquots of large-scale reactions were treated with buffer alone or incubated with various hydrolytic enzymes. The following enzymes were used to characterize enzyme products: endo-β-mannanase, cellulase, laminarinase, lichenase, and thermolysin. Digestion reactions were carried out for 90 min at optimal temperature and pH for the various enzymes. Typical reactions were conducted in 150-μl total volume containing 0.1–1 milliunits of hydrolytic enzyme. Reactions were terminated by adding 1.2 ml of 70% ethanol containing 2 mM EDTA and 100 μg of carob galactomannan as carrier. Products were precipitated at -20°C and sedimented at 13,000 × g. The supernatant fraction was carefully withdrawn and both the supernatant and pellet fractions were subjected to scintillation counting.
The sizes of in vitro assay products were estimated by using gel permeation chromatography on a Sepharose CL-6B column (1.5 cm × 100 cm) in 200 mM sodium hydroxide. Aliquots of large-scale products were dissolved in 200 mM sodium hydroxide at 65°C for 1 h; these were then mixed with a blue dextran standard (2.0 × 106 Da) and loaded on the column. The radioactivity present in aliquots of 2.5 ml-column fractions was measured by scintillation counting.
All of the results presented here are from representative experiments that were performed two or more times with similar results.
Results
Heterologous Expression of Cellulose Synthase-Like Proteins. Sequences related to CesA genes may encode enzymes that produce the backbones of noncellulosic polysaccharides of the plant cell wall (14, 17). To test this hypothesis, we selected Csl genes from various Csl subfamilies from Arabidopsis and rice for heterologous expression studies. cDNA clones encoding AtCslA9, AtCslC4, AtCslD3, AtCslE1, and OsCslH1 were obtained from stock centers (18, 19), and a short oligonucleotide sequence encoding the T7 epitope tag was added by PCR immediately upstream of, and in frame with the start codon of, each Csl gene. S2 cells were transfected with vectors containing the tagged Csl transgenes under the control of the inducible metallothionein promoter. This expression system was selected because S2 cells are of eukaryotic origin and more easily cultured in vitro than the mammalian cells, e.g., COS cells used in earlier work (9). Furthermore, these cells do not contain endogenous plant cell wall biosynthetic genes, so we expected little interference by Drosophila proteins in our cell wall biosynthetic enzyme activity assays.
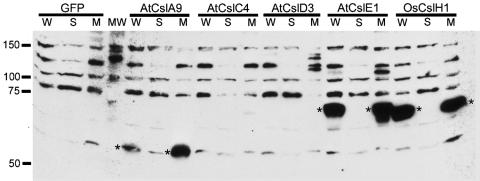
We fractionated lysates of induced S2 cells transformed with constructs encoding amino-terminal T7-tagged Csl fusion proteins or untagged GFP as a control by differential centrifugation and analyzed the presence of recombinant proteins by immunoblot analysis. The Csl proteins are predicted to contain multiple transmembrane domains, and were therefore expected to be present in the membrane fractions. After fractionation, tagged Csl proteins were detected exclusively in microsomal protein samples of S2 cells expressing AtCslA9, AtCslE1, and OsCslH1, but appeared to be absent from corresponding fractions of AtCslC4, AtCslD3, and GFP control samples and from soluble fractions of all samples (Fig. 1). The tagged Csl fusion proteins displayed slightly anomalous migration patterns on protein gels, migrating somewhat more rapidly than expected based on their predicted molecular mass. This phenomenon has also been observed with guar ManS produced in soybean somatic embryos (16).
Fig. 1.
Immunoblot analysis of epitope-tagged proteins from Arabidopsis and rice Csl genes produced in S2 cells. Samples of whole cell lysates (W), microsomal membranes (M), or the soluble (S) fraction from transgenic S2 cells producing T7-tagged Csl proteins or untagged GFP were subjected to immunoblot analysis. The positions and sizes (in kDa) of molecular mass standards (MW) are indicated to the left of the immunoblot. Because the antibodies used to detect the T7 tag also crossreact with several endogenous proteins present in S2 cell samples, T7-tagged Csl proteins are marked with asterisks. Some variability between samples was observed with regard to the intensity of interaction of the anti-T7 tag antibodies with the endogenous S2 proteins; however, the interaction of the antibodies with the tagged Csl proteins was consistently strong.
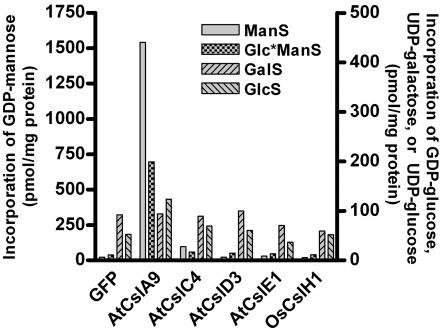
To determine whether any of the recombinant Csl proteins displayed glycan synthase activity, we conducted enzyme activity assays using microsomal fractions from transformed S2 cells and the radiolabeled nucleotide sugar donors UDP-[14C]galactose, UDP-[14C]glucose, UDP-[14C]xylose, GDP-[14C]mannose, and GDP-[14C]glucose. Control reactions with corn microsomal membranes or pea Golgi samples showed high activity for each assay (data not shown). In contrast, all S2 samples other than the AtCslA9 samples displayed a very low level of incorporation, similar to GFP controls, with every substrate assayed. Microsomes from S2 cells expressing AtCslA9 incorporated GDP-mannose and GDP-glucose into ethanol-insoluble polymers at levels greatly exceeding controls (Fig. 2).
Fig. 2.
Glycan synthase activity assays of Csl proteins from Arabidopsis and rice produced in S2 cells. Microsomal membrane fractions of transgenic S2 cells producing T7-tagged Csl proteins or untagged GFP were assayed for various β-glycan synthase activities. GDP-[14C]glucose was used for the Glc*ManS assay, and UDP-[14C]glucose was used for the GlcS assay. Xylan synthase assays were also conducted, and, because incorporation was very low (<3 pmol per mg of protein) in all samples, the data are not shown.
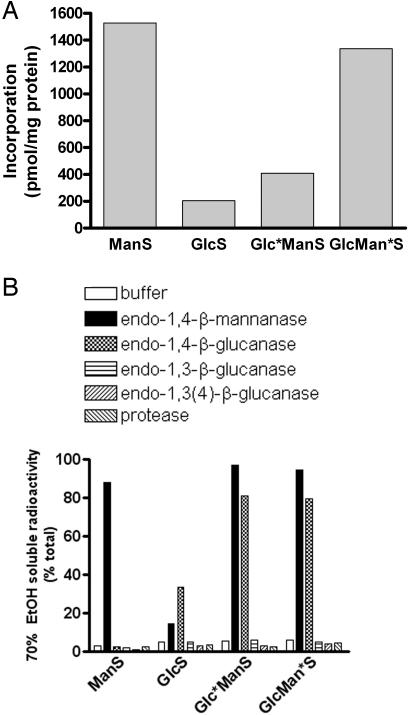
Characterization of Recombinant AtCslA9 and Its Products. It has been reported that plant membrane preparations containing ManS activity can catalyze various reactions depending on the sugar nucleotide supplied, producing a β-mannan when supplied GDP-mannose, a β-glucan when supplied GDP-glucose, or a β-glucomannan when supplied a combination of these substrates (23–25). Crude particulate preparations were used for these experiments, so it was not possible to determine whether these different activities resulted from a single enzyme. Because recombinant AtCslA9 displayed activity with both GDP-mannose and a mixture of GDP-glucose and GDP-mannose, we further characterized the enzyme and its products. Recombinant AtCslA9 possessed ManS, β-glucan synthase (GlcS), and glucomannan synthase (GlcManS) activities, producing ethanol-insoluble products when assayed with GDP-mannose, GDP-glucose, or a combination of these substrates in vitro (Fig. 3A).
Fig. 3.
Glucomannan synthase activity assays and analysis of in vitro products of AtCslA9 produced in S2 cells. (A) Incorporation of GDP-mannose and GDP-glucose into 70% ethanol-insoluble products by microsomal membrane fractions of S2 cells producing AtCslA9. The Glc*ManS product was labeled with GDP-[14C]glucose in the presence of nonradioactive GDP-mannose, and the GlcMan*S product was labeled with GDP-[14C]mannose in the presence of nonradioactive GDP-glucose. (B) In vitro assay products of recombinant AtCslA9 were treated with various hydrolytic enzymes, or buffer alone. The percentage of total radioactivity present in the 70% ethanol-soluble fraction after each treatment is shown.
To determine the identity of the labeled sugar incorporated into the products of AtCslA9, we subjected acid hydrolysates of assay products to high pH anion exchange chromatography. Our analyses revealed no detectable conversion of the sugars (i.e., epimerization of GDP-mannose or GDP-glucose) by the S2 microsomes expressing AtCslA9 (data not shown). We treated each of the products of the ManS, GlcS, and GlcManS reactions with a series of hydrolytic enzymes, which revealed unique structural features of each product (Fig. 3B). Simple incubation in aqueous buffer did not alter the ethanol insolubility of enzyme products. Digestion of the ManS product with endo-β-mannanase released 70% ethanol-soluble fragments, whereas treatment of this product with protease, cellulase, lichenase, or laminarinase did not. In contrast, the GlcS product was partially digested by cellulase, but not susceptible to digestion by endo-β-mannanase or any of the other enzymes (the GlcS product was fully digested when higher levels of cellulase were added; data not shown). The products of GlcManS reactions were susceptible to digestion by endo-β-mannanase and cellulase, but not to any of the other enzymes. These results indicate that recombinant AtCslA9 produces at least three types of products in vitro: a β-mannan when supplied GDP-mannose, a β-glucan when supplied GDP-glucose, or a β-linked mixed polymer containing glucose and mannose residues when both substrates are present. The AtCslA9 protein may thus be considered a β-glucomannan synthase.
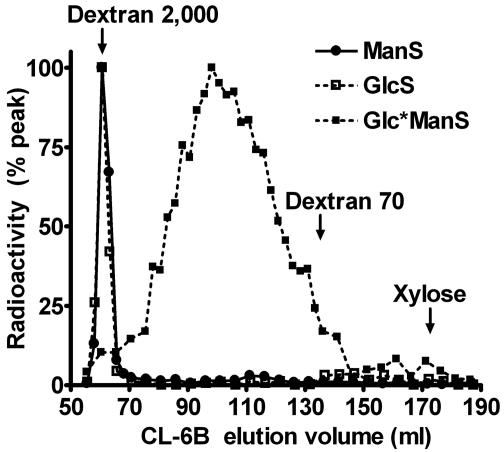
In vitro assay products of AtCslA9 were further characterized by using gel permeation chromatography on a Sepharose CL-6B column (Fig. 4). The ManS and GlcS products coeluted with a dextran blue standard (average molecular weight of 2 × 106 Da) in the column void, indicating that these products have a very high molecular weight. The products of GlcManS reactions eluted over a molecular weight range of ≈6.4 × 104 to 5.6 × 105 Da, with a peak around 1.3 × 105 Da. These data suggest that the ManS and GlcS products either have a much higher degree of polymerization (exceeding 6,000 sugar units) or they more readily self-associate when compared to the GlcManS products (≈800 sugar units). The large size of these polymers is in agreement with previous size estimates of products synthesized in vitro (16, 26–28).
Fig. 4.
Estimating the size of in vitro products of recombinant AtCslA9 with gel permeation chromatography. A Sepharose CL-6B column was used to fractionate the products of recombinant AtCslA9. The polysaccharide fractionation range of this column is 1.0 × 104 to 1.0 × 106 Da. The locations of elution peaks of dextran 2,000, dextran 70, and xylose standards are indicated. Because the fractionation profiles of GlcManS products labeled with GDP-mannose and GDP-glucose were similar, the profile of the GlcMan*S product has been omitted from the graph.
Expression and Characterization of AtCslA2 and AtCslA7. The Csl gene superfamily of Arabidopsis contains 30 members in six subfamilies, the largest being the CslA group with 9 members (14). The CslA9 gene of Arabidopsis is most closely related in sequence to guar ManS, but other members of the Arabidopsis CslA family may also encode ManS enzymes. Four additional CslA genes were selected from Arabidopsis and rice for further analysis: AtCslA1, AtCslA2, AtCslA7, and OsCslA7. Transgenes encoding these enzymes with amino-terminal T7 tags were introduced into S2 cells and assayed for protein expression and enzyme activity. Neither AtCslA1 nor OsCslA7 were detected on immunoblots of microsomal membrane fractions of S2 cells (data not shown). However, AtCslA2 and AtCslA7 were detected in S2 microsomes (Fig. 5). Microsomal membranes from transgenic S2 cells producing AtCslA2 and AtCslA7 incorporated GDP-mannose into a β-mannan or glucomannan at levels greatly exceeding the GFP control (Table 1). Microsomes from S2 cells producing AtCslA2 also incorporated GDP-glucose into a glucomannan; however, this activity was not detected in AtCslA7 samples, possibly because of a lower level of synthesis of this recombinant protein by the S2 cells. The in vitro products of ManS reactions with recombinant AtCslA2 and AtCslA7 as well as a GlcManS product of AtCslA2 exhibited properties similar to those produced by AtCslA9 in terms of size and susceptibility to the action of various hydrolytic enzymes (data not shown). These observations indicate that multiple members of the Arabidopsis CslA gene family encode β-mannan synthases, and that these enzymes may differ somewhat in their substrate specificity and kinetic properties.
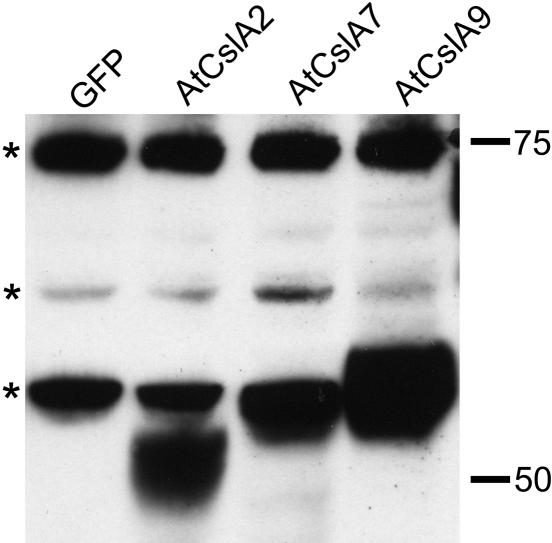
Fig. 5.
Heterologous expression of members of the Arabidopsis CslA gene family in S2 cells. Microsomal membranes from S2 cells expressing T7-tagged CslA proteins or GFP were subjected to immunoblot analysis. Several endogenous S2 proteins crossreact with the T7-tag antibodies, including one with an apparent molecular mass just above 50 kDa that comigrates with the T7-tagged AtCslA7 and AtCslA9 proteins and displays a slightly larger molecular mass than the T7-tagged AtCslA2 protein. The positions of the endogenous crossreactive Drosophila proteins are marked to the left of the immunoblot with asterisks; the positions and sizes (in kDa) of protein standards are indicated to the right.
Table 1. Comparison of the enzymatic activites of various recombinant CslA proteins produced in S2 cells.
| Sample
|
||||
|---|---|---|---|---|
| Assay | GFP | AtCslA2 | AtCslA7 | AtCslA9 |
| ManS | 37 | 2,428 | 486 | 2,651 |
| GlcS | 9 | 12 | 10 | 215 |
| Glc*ManS | 8 | 99 | 18 | 338 |
| GlcMan*S | 22 | 636 | 124 | 1,369 |
Asterisks designate the labeled nucleotide sugar substrate used in GlcManS reactions. All activities are expressed as pmol per mg of protein.
Discussion
Plants contain an extensive family of Csl genes sharing significant sequence similarity with CesA genes that encode the catalytic subunits of cellulose synthase. The Csl genes are hypothesized to encode processive β-glycosyltransferases involved in the biosynthesis of noncellulosic polysaccharides (14, 17). Demonstrating conclusive links between Csl genes and β-glycan synthase activities has proven difficult, and before this study, the biochemical function of no Arabidopsis Csl protein was known. We used a heterologous expression strategy to express a small group of Csl genes in S2 cells and assayed for β-glycan synthase activities. Our analyses demonstrate that the Arabidopsis CslA2, CslA7, and CslA9 genes all encode ManS enzymes. In vitro activity assays of these enzymes indicate that a single CslA gene product is capable of incorporating GDP-mannose or GDP-glucose into large β-linked homo- or heteropolymers, depending on the presence or absence of the other nucleotide sugar in the assay mixture. The mannan, glucan, and glucomannan products differ from one another in terms of their sizes and susceptibility to various polysaccharide hydrolases. Whether additional CslA genes, especially those from divergent species, encode proteins with ManS activity is a question that requires investigation. However, based on our analyses, it seems reasonable to postulate that all CslA genes from diverse groups of plants encode enzymes with ManS activity.
The observation that several Csl gene products exhibited ManS activity when expressed in S2 cells indicates that this system may be suitable to examine the activity of other glycan synthases. However, we have not yet been able to measure glycan synthase activity associated with a protein from any Csl family outside of the CslA group. There are several potential explanations for the lack of activity we observed when producing some of the Csl transgenes in S2 cells. The simplest explanation for this lack of glycan synthase activity is that AtCslA1, OsCslA7, AtCslC4, and AtCslD3 were not detectably produced by S2 cells. It is not clear why these Csl proteins were poorly synthesized in S2 cells, and yet both AtCslC4 and AtCslD3 were detectable in transgenic tobacco BY-2 cells (data not shown).
Lack of expression does not explain the absence of glycan synthase activity of the AtCslE1 and OsCslH1 gene products. Both of these proteins were detected at levels exceeding any of the AtCslA proteins. The observed lack of activity may be due to the absence of factor(s) crucial for enzyme activity such as additional members of a protein complex or a suitable acceptor molecule to which sugar residues may be transferred. For example, the β-glucan synthase that produces the xyloglucan backbone is thought to lose activity in the absence of an associated xylosyltransferase (29), and multiple isoforms of CesA proteins are required for cellulose biosynthesis (30). Of course, there are many other possible explanations ranging from interference with enzyme activity by the epitope tag to the possibility that the AtCslE1 and OsCslH1 gene products do not use the substrates tested to catalyze biosynthetic reactions.
Polysaccharides with β-mannan and β-glucomannan backbones are abundant constituents of the wood of gymnosperms (31, 32) and also serve as carbohydrate reserves in a variety of plant species (28, 31, 33–35). Several groups have biochemically characterized glucomannan synthase activities from a variety of plant species (25, 31–34). Because crude plant membrane preparations containing these enzyme activities were used for previous studies, researchers were unable to determine conclusively whether incorporation of GDP-mannose and GDP-glucose into glucomannan polymers was mediated by the same enzyme. Our data demonstrate that the expression of a single CslA gene is sufficient to impart ManS, GlcS, and GlcManS activities to S2 cells in vitro. Furthermore, because no acceptor compound was added to in vitro assays of S2 cells expressing CslA genes, these enzymes may not need acceptors or may be able to use an acceptor present in Drosophila cells. The acceptor requirements of CslA gene products require further investigation.
The functions of mannan polysaccharides in Arabidopsis are unclear. Aside from a recent immunological study (36), the localization and functions of β-mannan polysaccharides have received relatively little attention in Arabidopsis, presumably because they are present at a lower level than many of the other noncellulosic polysaccharides (37). Nonetheless, mannans and/or glucomannans are present in Arabidopsis and appear to be most abundant in cells with thickened walls, e.g., xylem cells (36). Recently, transglycosylase enzymes that modify the architecture of mannan polysaccharides in plant cell walls have also been discovered (38). It seems paradoxical that although mannan and glucomannan polysaccharides appear to be relatively minor components of Arabidopsis cell walls, CslA genes encoding ManS enzymes are among the most highly expressed of the Csl genes (39).
Although it seems likely that these polysaccharides are present as a structural component of Arabidopsis cell walls, mannans may serve additional roles in these and other plants. For instance, oligosaccharides derived from galactoglucomannan have been implicated as signaling molecules that inhibit auxin-stimulated elongation in pea and spruce segments (40). In light of our observations that the AtCslA7 and AtCslA9 genes encode enzymes with ManS activity, it is interesting to consider the phenotypes of Arabidopsis mutant plants containing insertions in these genes. Analyses of Arabidopsis mutant plants (SGT4425) containing a transposon insertion in exon seven of the CslA7 gene indicate that disruption of this gene results in defective pollen tube growth and disruption of embryonic development in homozygous seeds. Transcripts of the AtCslA7 gene were detected in a variety of tissues; thus, it is not clear why the phenotype is most severe in germinating pollen tubes and developing embryos (41). Mutants of RAT4 containing a T-DNA insertion in the 3′ untranslated region of the AtCslA9 gene display resistance to Agrobacterium tumefaciens transformation, apparently caused by decreased binding of bacterial cells to roots. AtCslA9 promotor-GUS fusions indicated that this gene is expressed in a variety of Arabidopsis tissues, including lateral roots and the elongation zone, where the root is most susceptible to Agrobacterium transformation (42). In both mutant studies, the authors suggested that the mutant phenotypes resulted from alterations in polysaccharide content; however, in neither case was such a defect observed.
Microarray analysis of expression patterns of Arabidopsis CslA genes indicates that, although many of these genes are expressed in multiple tissues, transcripts of others appear to be quite tissue specific (39). Thus, alterations in polysaccharide content or structure in these mutants may be quite subtle or possibly specific to particular cell types. These mutant analyses and our biochemical studies of Arabidopsis CslAs suggest that β-mannan and/or glucomannan polysaccharides may serve important and previously unknown roles in plants. Studies are underway to investigate these possibilities.
Supplementary Material
Acknowledgments
We thank Ms. Linda Danhof for tireless dedication and skillful handling of S2 cell cultures and protein expression; all of the members of the Keegstra laboratory and the Cell Wall Group at Michigan State University and the University of California, Riverside, including Drs. Teruko Konishi, David Cavalier, Jonathan Walton, and John Froehlich for helpful discussions and technical advice; and Ms. Karen Bird for editorial assistance. A.H.L. is grateful to have the support of a postdoctoral fellowship from the American Heart Association. Research in the laboratory of K.K. is supported in part by funds from the U.S. Department of Energy (Energy Bioscience Program) and the National Science Foundation (Plant Genome Program).
Author contributions: A.H.L. and K.K. designed research; A.H.L. performed research; A.H.L. and C.G.W. contributed new reagents/analytic tools; A.H.L. analyzed data; A.H.L. wrote the paper; and K.K. provided supervision.
Abbreviations: CesA, cellulose synthase; Csl, cellulose synthase-like; GlcS, β-glucan synthase; GlcManS, glucomannan synthase; ManS, β-mannan synthase; S2, Drosophila Schneider 2.
See Commentary on page 1815.
References
- 1.Dhugga, K. S. (2001) Curr. Opin. Plant Biol. 4, 488-493. [DOI] [PubMed] [Google Scholar]
- 2.Doblin, M. S., Vergara, C. E., Read, S., Newbigin, E. & Bacic, A. (2003) in The Plant Cell Wall, ed. Rose, J. K. C. (Blackwell, Oxford), Vol. 8, pp. 183-222. [Google Scholar]
- 3.Keegstra, K. & Raikhel, N. V. (2001) Curr. Opin. Plant Biol. 4, 219-224. [DOI] [PubMed] [Google Scholar]
- 4.Wolk, A., Manson, J. E., Stampfer, M. J., Colditz, G. A., Hu, F. B., Speizer, F. E., Hennekens, C. H. & Willett, W. C. (1999) J. Am. Med. Assoc. 281, 1998-2004. [DOI] [PubMed] [Google Scholar]
- 5.Liu, S., Manson, J. E., Stampfer, M. J., Rexrode, K. M., Hu, F. B., Rimm, E. B. & Willett, W. C. (2000) J. Am. Med. Assoc. 284, 1534-1540. [DOI] [PubMed] [Google Scholar]
- 6.Greenwald, P., Clifford, C. K. & Milner, J. A. (2001) Eur. J. Cancer 37, 948-965. [DOI] [PubMed] [Google Scholar]
- 7.Coutinho, P. M., Deleury, E., Davies, G. J. & Henrissat, B. (2003) J. Mol. Biol. 328, 307-317. [DOI] [PubMed] [Google Scholar]
- 8.Perrin, R. M., Wilkerson, C. & Keegstra, K. (2001) Plant Mol. Biol. 47, 115-130. [PubMed] [Google Scholar]
- 9.Perrin, R. M., DeRocher, A. E., Bar-Peled, M., Zeng, W., Norambuena, L., Orellana, A., Raikhel, N. V. & Keegstra, K. (1999) Science 284, 1976-1979. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, M. E., Dickson, C. A., Chengappa, S., Sidebottom, C., Gidley, M. J. & Reid, J. S. G. (1999) Plant J. 19, 691-697. [DOI] [PubMed] [Google Scholar]
- 11.Vanzin, G. F., Madson, M., Carpita, N. C., Raikhel, N. V., Keegstra, K. & Reiter, W.-D. (2002) Proc. Natl. Acad. Sci. USA 99, 3340-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G. F., Caplan, J., Shoue, D. A., Carpita, N. C. & Reiter, W.-D. (2003) Plant Cell 15, 1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwai, H., Masaoka, N., Ishii, T. & Satoh, S. (2002) Proc. Natl. Acad. Sci. USA 99, 16319-16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richmond, T. & Somerville, C. R. (2001) Plant Mol. Biol. 47, 131-143. [PubMed] [Google Scholar]
- 15.Pear, J. R., Kawagoe, Y., Schrechengost, W. E., Delmer, D. P. & Stalker, D. M. (1996) Proc. Natl. Acad. Sci. USA 93, 12637-12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhugga, K. S., Barreiro, R., Whitten, B., Stecca, K., Hazebroek, J., Randhawa, G. S., Dolan, M., Kinney, A. J., Tomes, D., Nichols, S., et al. (2004) Science 303, 363-366. [DOI] [PubMed] [Google Scholar]
- 17.Hazen, S. P., Scott-Craig, J. S. & Walton, J. D. (2002) Plant Physiol. 128, 336-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y., et al. (2002) Science 296, 141-145. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi, S., Satoh, K., Nagata, T., Kawagashira, N., Doi, K., Kishimoto, N., Yazaki, J., Ishikawa, M., Yamada, H., Ooka, H., et al. (2003) Science 301, 376-379.12869764 [Google Scholar]
- 20.Liepman, A. H. & Olsen, L. J. (2003) Plant Physiol. 131, 215-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 22.Harlow, E. & Lane, D. (1988) Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 23.Heller, J. S. & Villemez, C. L. (1972) Biochem. J. 129, 645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalessandro, G., Piro, G. & Northcote, D. H. (1988) Planta 175, 60-70. [DOI] [PubMed] [Google Scholar]
- 25.Piro, G., Zuppa, A., Dalessandro, G. & Northcote, D. H. (1993) Planta 190, 206-220. [DOI] [PubMed] [Google Scholar]
- 26.Villemez, C. L. (1974) Arch. Biochem. Biophys. 165, 407-412. [DOI] [PubMed] [Google Scholar]
- 27.Hinman, M. B. & Villemez, C. L. (1975) Plant Physiol. 56, 608-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clermont, S., Said, R. & Percheron, F. (1982) Phytochemistry 21, 1951-1954. [Google Scholar]
- 29.Hayashi, T. & Matsuda, K. (1981) J. Biol. Chem. 256, 11117-11122. [PubMed] [Google Scholar]
- 30.Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K. & Turner, S. R. (2003) Proc. Natl. Acad. Sci. USA 100, 1450-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbein, A. D. (1969) J. Biol. Chem. 244, 1608-1616. [PubMed] [Google Scholar]
- 32.Dalessandro, G., Piro, G. & Northcote, D. H. (1986) Planta 169, 564-574. [DOI] [PubMed] [Google Scholar]
- 33.Franz, G. (1973) Phytochemistry 12, 2369-2373. [Google Scholar]
- 34.Edwards, M. E., Bulpin, P. V., Dea, I. C. M. & Reid, J. S. G. (1989) Planta 178, 41-51. [DOI] [PubMed] [Google Scholar]
- 35.Meier, H. & Reid, J. S. G. (1982) in Encyclopedia of Plant Physiology, eds. Loewus, F. A. & Tanner, W. (Springer, Berlin), Vol. 13A, pp. 418-471. [Google Scholar]
- 36.Handford, M. G., Baldwin, T. C., Goubet, F., Prime, T. A., Miles, J., Yu, X. & Dupree, P. (2003) Planta 218, 27-36. [DOI] [PubMed] [Google Scholar]
- 37.Zablackis, E., Huang, J., Darvill, A. G. & Albersheim, P. (1995) Plant Physiol. 107, 1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroder, R., Wegrzyn, T. F., Bolitho, K. M. & Redgwell, R. J. (2004) Planta 219, 590-600. [DOI] [PubMed] [Google Scholar]
- 39.Hamann, T., Osborne, E., Youngs, H., Misson, J., Nussaume, L. & Somerville, C. (2004) Cellulose 11, 279-286. [Google Scholar]
- 40.Auxtova, O., Liskova, D., Kakoniova, D., Kubackova, M., Karacsonyi, S. & Bilisics, L. (1995) Planta 196, 420-424. [Google Scholar]
- 41.Goubet, F., Misrahi, A., Park, S. K., Zhang, Z., Twell, D. & Dupree, P. (2003) Plant Physiol. 131, 547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, Y., Nam, J., Carpita, N., Matthysse, A. G. & Gelvin, S. B. (2003) Plant Physiol. 133, 1000-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.