Abstract
The mechanism by which bacteria divide is not well understood. Cell division is mediated by filaments of FtsZ and FtsA (FtsAZ) that recruit septal peptidoglycan synthesizing enzymes to the division site. To understand how these components coordinate to divide cells, we visualized their movements relative to the dynamics of cell wall synthesis during cytokinesis. We found that the division septum was built at discrete sites that moved around the division plane. FtsAZ filaments treadmilled circumferentially around the division ring, driving the motions of the peptidoglycan synthesizing enzymes. The FtsZ treadmilling rate controlled both the rate of peptidoglycan synthesis and cell division. Thus, FtsZ treadmilling guides the progressive insertion of new cell wall, building increasingly smaller concentric rings of peptidoglycan to divide the cell.
Main Text
In most bacteria, cell division involves the inward synthesis of peptidoglycan (PG), creating a septum that cleaves the cell in two. The location of the septal PG synthases is regulated by filaments of the tubulin homolog FtsZ, which associate with the cytoplasmic side of the membrane via the actin-like FtsA and other factors. FtsZ forms membrane-associated filaments with FtsA (FtsAZ) (1, 2). Together, they form a dynamic structure, the Z ring, which encircles the cell at the future division site (3) and recruits PG synthases and other proteins involved in cytokinesis (4). Once the division machinery is mature, the Z ring constricts, while the associated synthases build the septum that partitions the cell in two.
We do not have a clear understanding of how the components of cell division interact in space and time to carry out cytokinesis, as we have been unable to observe the dynamics of each component relative to each other or to the structure they build: the organization and dynamics of FtsZ filaments within the Z ring remain ill-defined, it is not known how FtsAZ filaments control the activity of PG synthases, and the dynamics of septal PG synthesis have never been directly observed. To gain insight into how these components work together to divide bacteria, we visualized the dynamics of septal PG synthesis in relation to the movements of FtsAZ filaments and the septal PG synthase Pbp2B in the Gram-positive Bacillus subtilis.
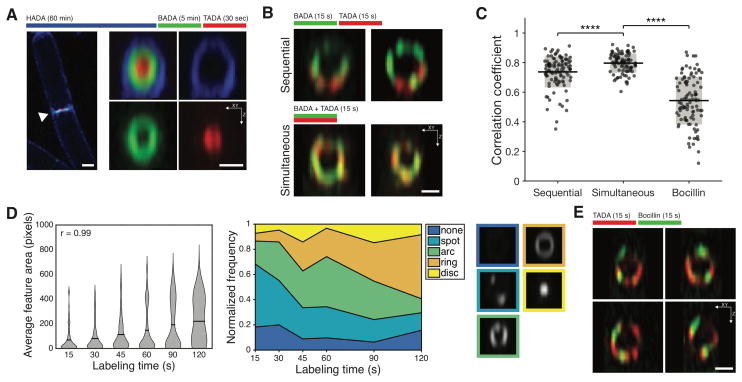
To assess the dynamics of septal PG synthesis, we sequentially pulse-labeled growing cells with different colors of fluorescent D-amino acids (FDAAs) (Table S1), which are incorporated into PG (5) by the D,D-transpeptidation activity of Penicillin Binding Proteins (PBPs) (6). 3D-structured illumination microscopy (3D-SIM) showed that sequential three color FDAA pulse labeling resulted in bullseye patterns at the division plane (Fig. 1A), demonstrating the septum is progressively synthesized inward from the cell surface. Short, sequential pulses of two FDAA colors resulted in discrete spots or arcs distributed around the septum, with the colors more offset compared to cells pulsed simultaneously (Fig. 1B–C, S1A–B). Thus, PG synthesis occurs at discrete sites that move around the division plane.
Figure 1. Septal PG synthesis occurs at discrete, mobile sites.
A Sequential FDAA labeling of division septa shows outside-in synthesis. (Right) 90° rotations of septa.
B Sites of PG synthesis move around the septum.
C Correlation coefficient of overlap between colors in B and E. Line: mean, box: SD.
D FDAA labeling proceeds from puncta to complete rings. (Left) Area of FDAA features increases with pulse length. Lines: mean, r: correlation coefficient of the means. (Center) Blinded classification of FDAA features at various pulse lengths. (Right) representative images of septal PG structures.
E Sites of PG synthesis are offset from synthetic enzymes. Colored bars indicate time course of FDAA labeling. All images taken with 3D-SIM. Scale bars: 0.5 μm.
We next observed how discrete sites of PG synthesis develop into a complete division septum by labeling cells with FDAAs using increasing pulse durations. Both the total amount of labeling and the area of labeled regions increased with pulse duration (Fig. 1D, left; Fig. S2A–C). Following short pulses, septa contained discrete spots or arcs (Fig. S1C). As pulse duration increased, these arcs elongated, gradually transitioning into complete rings at longer pulses (Fig. 1D, right). As expected, PG synthesis inhibitors reduced FDAA incorporation (Fig. S2D). To explore the location of the PG synthases relative to newly incorporated PG, we followed short FDAA pulses with Bocillin, which labels active PBPs (7) while inactivating them. The Bocillin signal was offset from the newly-synthesized PG (Fig. 1C, 1E), suggesting the PG synthases also move around the division plane.
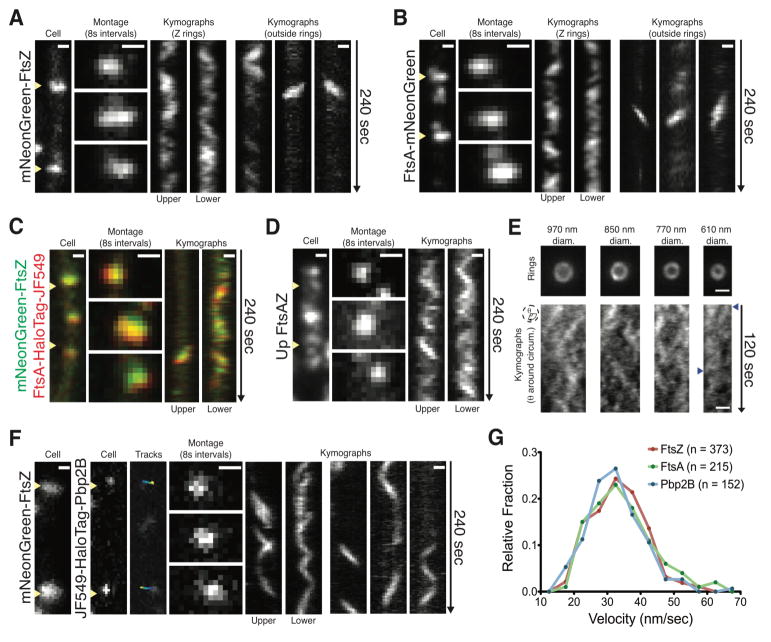
We next examined the motions of the division-specific PG synthases and their associated cytoskeletal polymers. Total internal reflection fluorescence microscopy (TIRFM) of a functional mNeonGreen-FtsZ fusion expressed from the native locus (Fig. S3A–E, Tables S2–3) revealed directional movements within newly assembled Z rings (Fig. 2A, Movie S1). Furthermore, in almost every cell, we observed small mNeonGreen-FtsZ filaments outside the Z ring moving directionally around the cell at the same rate as within Z rings, similar to oscillations previously observed in E. coli (8). A functional FtsA-mNeonGreen fusion showed motions identical to FtsZ (Fig. 2B, Movie S1), and two-color imaging confirmed FtsA and FtsZ colocalized and moved together (Fig. 2C) (9). Overexpression of a second ftsAZ operon (FtsA, mNeonGreen-FtsZ) resulted in many more directionally moving filaments outside the Z ring without affecting fitness (Fig. 2D, S3F). To resolve FtsZ motion in dense, actively constricting Z rings, we vertically immobilized bacteria in agarose microholes, orienting the division plane parallel to the objective (Fig. S4). This revealed multiple FtsZ filaments moving in both directions around the constriction site over a wide range of ring diameters (600–1000 nm) (Fig. 2E, S4E, Movie S2). The movement of multiple FtsZ filaments around Z ring may explain the heterogeneous structures and complex “patch” dynamics observed via super resolution microcopy (10–12), as well as the fast turnover of FtsZ subunits (13).
Figure 2. FtsAZ and Pbp2B move directionally around the division site.
A mNeonGreen-FtsZ (bAB185) shows directional motion inside (left) and outside (right) the Z ring.
B FtsA-mNeonGreen (bAB167) shows directional motion inside (left) and outside (right) the ring.
C mNeonGreen-FtsZ and FtsA-HaloTag-JF549 (bAB229) colocalize and move together.
D FtsAZ overexpression (100 μM IPTG in bAB221) creates increased FtsZ filaments showing directional motion outside the Z ring.
E Vertically immobilized cells (SH41) show multiple, independent mNeonGreen-FtsZ filaments moving in both directions around the division site. Cropped rings and radial kymographs in early (left), mid (center), and late (right) divisional stages. Blue arrows indicate directional FtsZ tracks.
F Single molecules of Pbp2B (bGS31, 15 min incubation of 50 pM JF549) move directionally around the division site. Blue to yellow indicates trajectory time.
G Velocity distributions of FtsA filaments, FtsZ filaments, and single Pbp2B molecules. Kymographs drawn at yellow arrows. Scale bars = 0.5 μm
We next asked if the division-associated transpeptidase Pbp2B moves with FtsAZ. At native expression levels, mNeonGreen-Pbp2B moved directionally along the Z ring; this became more apparent with reduced expression (Fig. S5A–B). To observe the motions of single Pbp2B molecules, we labeled HaloTag-Pbp2B expressed from the native locus with low concentrations of HaloLigand-JF549 (14). TIRFM revealed two types of Pbp2B motion: 1) directional motion around the cell width, always localized to Z rings, and 2) diffusion on the membrane, not localized to Z rings (Fig. 2F, S5C–E, Movie S3). We did not observe diffusive Pbp2B motion along Z rings at any acquisition rate. In some cases, we observed multiple Pbp2B molecules moving directionally within the same ring, sometimes in opposite directions indicating that the Z ring contains multiple, independent synthetic sites. FtsZ, FtsA, and Pbp2B all moved at similar velocities (Fig. 2G, S5D–E), suggesting that their motions are associated.
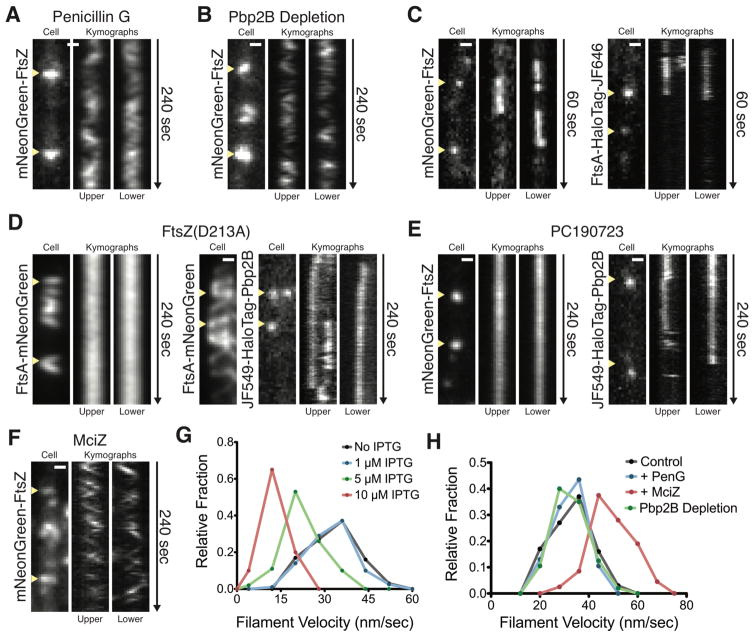
We investigated the mechanism driving FtsAZ/Pbp2B motion, first testing if, similar to MreB (15), Pbp2B inactivation would halt FtsAZ motion. However, FtsAZ motion was unaffected by multiple PG synthesis inhibitors (Fig. 3A, 3H, Movie S4) or depletion of Pbp2B (Fig. 3B, 3H, Movie S5). We next tested if directional FtsAZ motion arose from filament treadmilling, as observed in vitro (16). Consistent with treadmilling, sparse labeling of FtsZ or FtsA in cells demonstrated that single molecules of both proteins were immobile within moving filaments (17) (Fig. 3C, S5F–J Movie S6). Because treadmilling requires nucleotide hydrolysis, we assayed FtsA motion as we modulated the GTPase activity of FtsZ. Exogenous expression of FtsZ(D213A), a mutant shown to have greatly reduced GTPase activity in E. coli (18), gradually reduced FtsAZ velocity, stopping motion at high inductions (Fig. 3D, 3G, Movie S7A). Likewise, addition of PC190723, an inhibitor of FtsZ GTP hydrolysis (19) halted FtsZ movement (Fig. 3E, Movie S8A). Conversely, addition of MciZ, a Z ring antagonist that, at low levels, increases FtsZ GTPase activity (20), increased FtsZ velocity (Fig. 3F, 3H, Movie S7B). We next tested whether FtsZ treadmilling dynamics affected Pbp2B movement. PC190723 or overexpression of FtsZ(D213A) caused Pbp2B molecules to become immobile while remaining FtsZ colocalized (Fig. 3D–E, Movie S8B). Pbp2B velocity scaled with FtsZ treadmilling velocity under various perturbations (Fig. S5K). Thus, FtsZ treadmilling is required for the directional motions of both FtsAZ filaments and septal PG synthases.
Figure 3. Directional FtsAZ motion is driven by treadmilling, independent of cell wall synthesis, and required for Pbp2B motion.
A FtsZ continues to move directionally after treatment with Penicillin G. bAB185 imaged 5 minutes after addition of 3 μl of 10 mg/ml penicillin G to an agarose pad.
B FtsZ continues to move directionally after Pbp2B depletion. Before imaging, bGS31 was grown without IPTG until no divisions were observed (3 hours).
C Single molecules of FtsZ and FtsA are immobile within the division site. Single molecules were obtained by growing bAB219 with no IPTG (left) or bAB229 with 15 min of 250 pM JF646 (right).
D Overexpression of GTPase-deficient FtsZ stops FtsZ and Pbp2B motion. bAB217 (left) and bGS90 (right) were imaged after induction of FtsZ(D213A) (100 μM IPTG, 1 hour).
E Directional motion of FtsZ and Pbp2B is stopped by PC190723. bAB185 (left) and bGS31 with 15 min incubation of 50 pM JF549 (right) 5 minutes after addition of 10 μM PC190723.
F FtsZ velocity increases following exposure to MciZ. bAB185 was imaged following addition of 1 μM MciZ in a microfluidic device.
G Velocity distributions of FtsZ filaments (bAB217) at different levels of FtsZ(D213A) induction.
H Velocity distributions of FtsZ filaments under different perturbations.
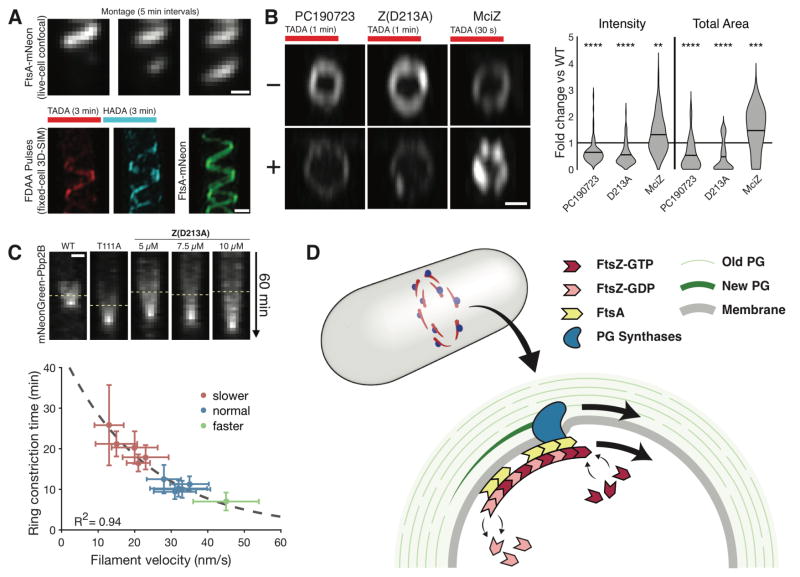
Given the mobile nature of septal PG synthesis, we reasoned that the directional movements of FtsAZ/Pbp2B around the division plane could be coupled to septal PG synthesis. To test this, we labeled cells with FDAAs as we altered FtsZ dynamics. Overexpression of FtsZ(D213A) created long, slowly-growing FtsA-mNeonGreen spirals which incorporated FDAA along their entire length (Fig. 4A, Movie S9). Likewise, long PC190723 treatments resulted in fragmented patches of both FtsZ and FDAA incorporation (Fig. S6A), indicating Pbp2B activity is constrained by FtsAZ location. However, these strong inhibitions of FtsZ dynamics required much longer pulses to achieve FDAA labeling, suggesting that FtsZ treadmilling limits PG synthesis. To test this, we altered FtsAZ velocity as we pulse-labeled cells with FDAAs. This revealed that both the total amount and total area of PG synthesis within the ring are modulated by FtsAZ velocity: conditions that slowed dynamics decreased both the total amount and area of FDAA labeling (Fig. 4B, S6B–C). Conversely, increasing FtsAZ velocity (with MciZ) increased both the total amount and area of labeling. Thus, in B. subtilis, both the amount and spatial distribution of septal PG synthesis are directly coupled to, and limited by, the rate of FtsZ filament treadmilling.
Figure 4. Cytokinesis is controlled by directional motion of FtsAZ filaments.
A FtsZ(D213A) overexpression (1 mM IPTG in bAB217) produces slowly growing FtsA spirals with spiral FDAA insertion. (Top) Montage of growing FtsA spiral acquired with spinning-disk confocal. (Bottom) Sequential FDAA labeling and FtsA localization imaged with 3DSIM in a fixed cell.
B Altering FtsZ velocity changes the amount and total area of FDAA incorporation. (left) PY79 with 1 μg/ml PC190723, 10 min, (middle) bAB217 with 20 μM IPTG for 1 hour, (right) AH93 with 50 mM xylose, 5 min. Following treatment, cells were incubated with TADA for time indicated, fixed, then imaged with 3D-SIM. (Far right) Total intensity and area of septal FDAA incorporation. Lines: mean.
C Cytokinesis scales with FtsZ treadmilling velocity. For each condition, pairs of strains were used to measure 1) FtsZ velocity (using mNeonGreen-FtsZ) and 2) septation rates (using mNeonGreen-Pbp2B). (Top) Kymographs of constricting Pbp2B rings in different FtsZ backgrounds (line marks start of constriction). (Bottom) Plot of constriction time vs. FtsZ velocity in different conditions. Treadmilling velocity and septation rates were acquired in identical conditions, save for measurements with MciZ (see SOM). Error bars: SD of the mean.
D Model for treadmilling-coupled cell division. (Top) The Z ring contains multiple FtsAZ filaments that treadmill around the division plane, pulling associated PG synthases. (Bottom) FtsZ treadmilling both regulates and distributes the activity of the PG synthases, building sequentially smaller uniform arcs of PG to divide the cell.
Because FtsAZ dynamics control the rate of septal synthesis, we asked whether the rate of cytokinesis depended on FtsAZ treadmilling. We modulated treadmilling velocity by 1) introducing mutations affecting GTP hydrolysis into FtsZ at the native locus, 2) titrating exogenous FtsZ(D213A), 3) expressing MciZ, and 4) other perturbations (Table S4). This revealed the cytokinesis rate scaled with FtsZ treadmilling: division was slower when velocity was decreased, and faster when velocity was increased (Fig. 4C, S7A–C, Movie S10). Even under the strongest perturbations, these decreased rates of cytokinesis did not alter the rate of cell elongation (Fig. S7D–F). Thus, in B. subtilis, FtsAZ treadmilling is both coupled to, and limiting for, septal PG synthesis and cell constriction.
Our results indicate that cell division occurs by the action of discrete enzyme-filament complexes that, driven by FtsZ treadmilling, move around the division plane, building new PG during their transit (Fig. 4D). FtsZ treadmilling creates long range order from the local activity of the PG synthases, linking circumferential enzyme motion to the insertion of cell wall. This tight coupling may yield uniform insertion of new material around the division plane, building the septum inward in progressively smaller concentric rings. Cell division slightly differs in E. coli, where FtsZ treadmilling also distributes PG synthesis around the ring, perhaps because the rate of PG synthesis is limiting relative to FtsZ treadmilling (10). This difference may arise from different levels of cell wall precursors between the two organisms, or that E. coli must also couple PG synthesis to outer membrane insertion.
The coupling between FtsZ treadmilling and PG synthesis can unify previously conflicting models of cell division. FtsZ filaments have been proposed to generate force to bend membranes (2, 22), and to scaffold PG synthesis (10, 23). If FtsZ filaments deform membranes, coupling their movement to PG synthesis would allow each deformation to be reinforced via synthesis of PG (24). Thus, multiple sites of local deformation and coupled reinforcing synthesis moving around the division site would iteratively build the invaginating septum.
Materials and Methods
Culture growth
All B. subtilis strains were prepared for experimentation as described below, unless otherwise noted. Strains were streaked from −80°C freezer stocks onto LB agar plates, and grown overnight at 37°C. Single colonies were transferred to liquid cultures in CH medium or PHMM Media - used to grow cells that were not chained (equal parts of CH and S750, with final concentrations of 1 mM glutamate, 0.5% glucose and 100 mM MgCl2). Cells were placed on a roller drum for agitation and grown at 37°C. After cultures reached OD600 < 0.5, serial dilutions were grown for one more round until OD600 ~ 0.5. Alternatively, cultures were grown overnight at 25°C and then these starter cultures were diluted into the same media and grown at 37°C. bGS28 and bGS31 were grown with 50–100 μM IPTG to induce Pbp2B expression.
Western blots
Wild type (PY79), mNeonGreen-FtsZ (bAB185), FtsA-mNeonGreen (bAB167) and mNeonGreen-Pbp2B (ME7) were grown in 3 mL LB at 37°C until they reached OD600 ~ 0.5, then diluted 10-fold to OD600 ~ 0.05 and grown again until OD600 ~ 0.5. Cultures were centrifuged at 13,000 rpm for 2 minutes, washed once with lysis buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 10 mM EDTA, 1 mM PMSF) and pellets stored at −80°C overnight. Pellets were resuspended in 50 μl of lysis buffer with 10 μg/ml DNAse I, 100 μg/ml RNAse A and 10 mg/ml of lysozyme and incubated at 37°C for 20 minutes. Small aliquots were used to determine total protein mass (Pierce BCA Protein Assay Kit). 10, 25 and 50 μg of the lysates were then separated by SDS PAGE in 10% Bis-Tris gels, and blotted to 0.2 μm nitrocellulose membranes (Bio-Rad). The One-Hour Western Chemiluminescence Detection System kit (GenScript) was used coupled with primary antibiodies against FtsZ (anti-rabbit), FtsA (anti-mice) and Pbp2B (anti-rabbit). Antibodies were a gift from Jeff Errington, Frederico Gueiros Filho and Richard Daniel, respectively. Images were taken by a CCD camera coupled to the Chemilmager 5500 System (Alpha Innotech).
Fluorescent D-amino acids synthesis
HADA (7-hydroxycoumarin-3-carboxylic acid-D-alanine), BADA (BODIPY FL-D-alanine) and TADA (TAMRA-D-alanine) were synthesized as reported previously (5, 25). 7-hydroxycoumarin-3-carboxylic acid (for HADA, 1.45 mmole) or BODIPY FL (for BADA, 1.45 mmole) was added to a 100 ml, flame-dried round flask containing 14.5 ml anhydrous DMF and a stir bar. After adding 1.45 mmole carbonyldiimidazole to the flask, the reaction was stirred at room temperature (RT) for 2 hours, following by directly adding 1.45 mmole Nα-Boc-D-2,3-diaminopropionic acid. The reaction was stirred overnight under argon atmosphere. The solvent was removed in vacuo and the product was extracted with EtOAc and 0.33N HCl/H2O, and treated with trifluoroacetic acid and dichloromethane in 1:1 ratio for 30 min with stirring at RT. The final product was purified via reverse-phase HPLC with 10–90% MeCN/H2O gradient. For TADA synthesis, Nα-Boc-D-2,3-diaminopropionic acid (1.45 mmole), 5-(and 6-) carboxytetramethylrhodamine succinimidyl ester (1.45 mmole), and diisopropylethylamine (200 μl) were added in one portion to a 100 ml round flask containing 14.5 ml anhydrous DMF. The reaction was stirred under argon overnight at RT. The product was de-protected and purified as described above.
Fluorescent D-amino acids labeling
FDAA stock solutions were prepared in DMSO (Sigma-Aldrich ReagentPlus >99.5%) at a concentration of 100 mM and stored at −20°C before use. The labeling conditions, including incubation interval and color order, are indicated in each FDAA figure and summarized in Table S1.
FDAA-I
For the sequential labeling in Fig. 1A, exponential phase cells (PY79) at OD600 ~ 0.5 were diluted with PHMM containing 1 mM HADA to OD600 ~ 0.1, and then grown for 1 hour at 37°C. Cells were washed twice with fresh PHMM (37°C), and centrifuged between washes (6,000 g for 1 minute at room temperature). For the second labeling, the cell pellets were resuspended in pre-warmed PHMM containing 1 mM BADA for 5 minutes. After this, cells were washed twice, and labeled a third time by resuspending in pre-warmed PHMM containing 2 mM TADA for 30 seconds. Cells were then immediately treated with 70% ice-cold ethanol, and incubated on ice for 1 hour. Ethanol-fixed cells were collected via centrifugation (10,000 g for 5 minutes at 4°C), washed twice with 4°C 1 × Phosphate Buffered Saline (PBS, pH 7.4), resuspended in PBS, and stored on ice before imaging.
FDAA-II
For the short-pulse labeling in Fig. 1B, 1E and S1A, cells (PY79) were first labeled with HADA (0.5mM, 60–90 minutes) and washed as described in FDAA-I. Cells were then transferred to a syringe filter (Acrodisc, 0.2 μm HT Tuffryn, 25 mm), and pre-warmed, fresh PHMM media (10 ml) was added to wash out excess dye. The cells were then treated sequentially with PHMM-FDAA media (2 mM BADA or TADA in 1 ml media for 15 sec), PHMM media (for washing), and PHMM-FDAA/Bocillin media (2 mM FDAA or 50 μg/ml Bocillin in 1 ml media for 15 sec) to label the cells. After washing with 10 ml PBS, cells were collected by back-sucking fixation reagent through the filter into the syringe. For Fig. 1B and S1A, 70% ethanol was used for fixation as described in FDAA-I. For Fig. 1E, to preserve the Bocillin signal, cells were fixed in 4% para-formaldehyde for 30 minutes at room temperature. Cells were collected by centrifugation (10,000 g for 5 minutes at 4°C), washed, and stored in PBS on ice before imaging.
FDAA-III
For time-course labeling in Fig. 1D, cells (PY79) were first labeled with HADA (1 mM, 60 minutes) as described above. The collected cells were resuspended in pre-warmed PHMM containing TADA (2 mM in 0.3 ml) and incubated for 15–120 seconds in a 37°C water bath. The cells were then fixed with 70% ethanol as described above. Labeling in Fig. S1B–C was performed similarly, using the FDAAs in a different order (see Table S1).
FDAA-IV
For FDAA labeling with antibiotic treatments in Fig. S2D, cells (PY79) were first labeled with HADA (1 mM, 60 minutes) as described above. Cell pellets were then resuspended in pre-warmed PHMM containing antibiotics (4 μg/ml Vancomycin or 50 μg/ml Penicillin G) and incubated for 3 minutes in a 37°C water bath. Next, TADA stock solution was added directly to the cells to a final concentration of 2 mM. After a 2 minute incubation, cells were fixed with 70% ethanol and collected as described above.
FDAA-V
For FDAA labeling with Pbp2B depletions in Fig. S2D, exponential phase cells (bGS3) were diluted with PHMM containing 1 mM HADA to OD600 ~ 0.1, and grown for 3 hours at 37°C without IPTG to deplete Pbp2B. Following two washing steps with fresh PHMM (37°C), cell pellets were resuspended in pre-warmed PHMM containing 2 mM TADA and incubated for 2 minutes. The cells were fixed with 4% para-formaldehyde as described above.
FDAA-VI
For FDAA labeling with FtsZ(D213A) expression in Fig. 4A, exponential phase cells (bAB217) were grown in PHMM containing 100 μM IPTG for 60 min, treated with TADA (2mM, 3 min), washed twice with pre-warmed PHMM, and then labeled with HADA (2 mM, 3 min). The cells were washed and then fixed with 4% para-formaldehyde as described above. For Fig. 4B, exponential phase cells (bAB217) were diluted with PHMM containing 1 mM HADA to OD600 ~ 0.1. After 30 minutes of growth at 37°C, IPTG was added to the culture to a final concentration of 20 μM, and incubated for another hour. Following two washing steps with fresh PHMM (37°C) containing IPTG, cells were resuspended in fresh PHMM containing 2 mM TADA and IPTG, then incubated for 1 minute. The cells were then fixed with 70% ethanol and washed.
FDAA-VII
For FDAA labeling with PC190723 treatment in Fig. 4B, exponential phase cells (PY79) were diluted with PHMM containing 1 mM HADA to OD600 ~ 0.05. After a 90-minute growth at 37°C, PC190723 (10 mg/ml, DMSO stock) was added to the culture to a final concentration of 1 μg/ml, and incubated for another 10 min. Following two washing steps with fresh PHMM (37°C) containing PC190723, cells were resuspended in fresh PHMM containing 2 mM TADA and PC190723, then incubated for 1 minute. The cells were then fixed with 70% ethanol as described above. Labeling in Fig. S6A was performed similarly, using 10 μg/ml PC190723 and 2 min of TADA labeling.
FDAA-VIII
For FDAA labeling with MciZ expression in Fig. 4B, exponential phase cells (strain AH93) were diluted with PHMM containing 1 mM HADA to OD600 ~ 0.05. After a 90-minute incubation at 37°C, 50 mM xylose was added to the culture, and the cells were incubated for 5 minutes. Following two washing steps with fresh PHMM (37°C) containing xylose, cells were resuspended in fresh PHMM containing 2 mM TADA and 50 mM xylose, then incubated for 30 seconds. The cells were fixed with 70% ethanol as described above.
HaloTag labeling with Halo-TMR and JF- dyes
For the Fig. 2D, full labeling of FtsA-HaloTag(SW) (see strain construction for details) was obtained by incubating strain bAB229 with 500 nM HaloTag-JF549 ligand for 15 min. For Fig. 2F and 3E, Pbp2B single molecules were obtained by incubating bGS31 (HaloTag-pbp2B) with 40–100 pM of HaloTag-JF549 ligand for 15 min; cells were washed once in fresh media to remove free dye before imaging. For Fig. 3C, FtsA single molecules were obtained by incubating bAB213 (ftsA-HaloTag(SW)) with 250 pM of HaloTag-JF646 ligand for 15 min. HaloTag ligands were a gift of Luke Lavis (14).
Depletions in liquid culture
For the Pbp2B depletions shown in Fig. 3B and Movie S5, bGS31 cells (ftsZ::mNeonGreen-ftsZ, erm-Phyperspank-HaloTag-pbp2B) were grown in liquid culture with 10 μM IPTG until OD600 ~ 0.5, the inducer was washed out, and then cells were shifted to media lacking IPTG. Before imaging mNeonGreen-FtsZ, cells were observed under microscope every 30 minutes until no divisions were observed to ensure Pbp2B depletion.
Expression of ftsAZ constructs
For all experiments in which the expression of a second copy of tagged FtsA and/or FtsZ was required, fusions were made in a way to maintain the operon structure to avoid perturbations on the FtsA/FtsZ ratio (26, 27). As shown in Fig. S3F, increased levels of either FtsA or FtsZ can impair cell division, while high levels of FtsAZ did not present an observable phenotype.
Perturbations in CellASIC microfluidic system
To observe the effects of MciZ (Fig. 3F, Movie S7B) and PC190723 (Fig. 3E, Movie S8A) on FtsAZ motion, we used the microfluidic CellASIC system from EMD Millipore with B04A plates. Cells in optimal growth conditions (as described in the culture growth section) were loaded into the device and equilibrated for 1 hour in CH media at a flow of 3 PSI. Afterward, based on the observation that exogenous MciZ is capable of penetrating cells and inhibiting Z ring assembly (28), cells were flowed with 1 μM of synthetic MciZ peptide or 10 μM of PC190723 at 5 PSI and imaged by TIRF microscopy. Pulses and washes were performed as needed and described in each figure legend. Synthetic MciZ peptide and PC190723 drug were purchased from PSL Peptide Specialty Laboratories GmbH and EMD Millipore, respectively.
Antibiotic treatments
For the effect of different antibiotics on FtsAZ motion (Fig. 3A and Movie S4), cells were grown in liquid as described above and placed under agarose pads (500 μl of media with 2% agarose). After FtsAZ motion was confirmed by TIRF microscopy, 3 μl of the antibiotic was added on top of the pad, followed by 5 minutes of incubation. Time lapses were then acquired from different fields of view. Antibiotic concentrations were as follows: 10 mg/μl Ampicillin (blocks transpeptidation); 10 mg/μl Penicillin G (blocks transpeptidation); 50 mg/μl Vancomycin (blocks transpeptidation and transglycosylation); 100 mg/μl Fosfomycin (blocks PG precursor synthesis); 100 mg/μl Cephalexin (blocks transpeptidation). To confirm the effect of each specific treatment, we conducted separate experiments where we incubated liquid cultures with each antibiotic (at a final concentration of 10 μg/μl) for 5 minutes, followed by imaging.
Correlation between FtsAZ motion and cytokinesis rates
For the effects of different genetic backgrounds and overexpression of a number of components on FtsAZ motion and cytokinesis rates (shown in Fig. 4C, Table S4, Movie S10), cells were grown in liquid as described above and placed under agarose pads. Where inducible promoters were used, the appropriate IPTG concentration (0, 1, 5, 7.5, 10 and 100 μM) was mixed into to the pad. The exception to this protocol was the experiments with MciZ-expressing cells. Because very low concentrations of MciZ can quickly block cell division (20, 29), MciZ was only induced after cells were under the agarose pad, by addition of 5 μl of a 3M xylose solution on top of the 500 μL pad (as with the antibiotic perturbations described above). Each condition was imaged on a spinning disk confocal (for Pbp2B ring constriction) and on a TIRF microscope (for FtsAZ filament dynamics). The strains, conditions, and measurements for each experiment are shown in Table S4. mNeonGreen-Pbp2B was used as a reporter of cytokinesis, and the constriction times were measured by manual kymographs. These measurements were then validated by automated analysis. Manual tracking: For each condition, 100 random rings were cropped, and kymographs were drawn along the ring axis using Fiji. From these kymographs we determined when ring constriction initiated by using the time point where the slope of the edge of the signal switched from being vertical to angled (top dotted line). The disappearance of Pbp2B was used to indicate the end constriction period (assayed by the last bright pixel in the kymograph). The difference in these two time points was taken as the constriction interval for each ring. Automated analysis: To independently validate these manual measurements, we applied an automated analysis of the cytokinesis time. We exploited the fact that mNeonGreen-Pbp2B rings get brighter up until they begin constriction to set a threshold for each image based on the average of intensity of 100 constricting rings. We then applied this threshold to entire fields of cells. We then tracked these thresholded rings with the TrackMate plugin to determine each ring’s lifetime (30). We set a lifetime cutoff between 5–30 minutes, to exclude rings that that were already at late stages of constriction, or anomalous constrictions.
Microholes nanofabrication
Microholes are patterned through electron-beam lithography as described previously (31) and etched through a reactive-ion etching (32). Briefly, a silicon wafer was spin-coated with resist NEB-22 (Sumitomo Chemical). Predesigned features were patterned through Leica EBPG 5000+ with a beam step size of 20 nm. The exposed resist was removed by solvent Microposit MF-322 solution and the exposed wafer surface was etched using an AMS Bosch Etcher. The remaining resist was removed using oxygen plasma.
Culture growth for vertical microhole immobilization
B. subtilis strains to be imaged by vertical microhole immobilization were streaked from −80°C freezer stocks onto LB agar plates, grown at 30°C in Time Lapse Medium (TLM) (33) overnight, re-diluted in pre-warmed Chemically Defined Medium (CDM) (33) and grown at 30°C to OD600 ~ 0.4 before being immobilized in agarose microholes (see below).
Imaging – slide preparation
Coverslips were sonicated in 2% Hellmanex (Hellma GmbH&Co, Milllheim, FRG) for 30 minutes, followed by multiple washes with deionized water, then sonicated with 1 M KOH for 30 minutes, followed by multiple washes with water. Coverslips were washed twice with 100% ethanol, and then sonicated for 30 minutes in 100% ethanol, followed by one more wash in 100% ethanol. They were stored in ethanol and dried before use.
Imaging – Pbp2B and FtsZ ring constriction rates
Images were collected on a Nikon TI spinning disk confocal microscope with Yokogawa CSU-10 spinning disk and a Hamamatsu ImagEM (EM-CCD) camera (effective pixel size 110 nm), and Nikon 100X NA 1.45 TIRF objective. Illumination was accomplished using fiber coupled Spectral lasers, using a 488 laser for imaging of mNeonGreen, and a 494 nm laser for imaging of FM5-95. For the experiments following ring constriction, time lapses were acquired with 1 minute intervals for 2 hours with exposure times of 50–150 milliseconds.
Imaging – FtsAZ and Pbp2B dynamics by TIRFm
Images were collected on a Nikon TI microscope equipped with a 6.5 μm pixel CMOS camera and an EMCCD camera, together with a Nikon 60X NA 1.40 and a Nikon 100X NA 1.45 objective set. Exposure times varied between 0.1 and 3 seconds, as indicated. Illumination was accomplished using a fiber coupled Agilent launch with for 405 nm, 488 nm (imaging of mNeonGreen and msfGFP fluorescent proteins), 561 nm (imaging of JF549 dye) and 647 nm (imaging of JF646 dye).
Imaging – 3D-SIM of FDAA labeling
3D-SIM images were collected on DeltaVision OMX system (Applied Precision Inc, Issaquah, USA) equipped with 1.4 NA Olympus 100X oil objective, laser source (405, 488, 561 and 642 nm) and four photometrics Cascade II EM-CCD cameras. 24X50 mm coverslips (# 1.5) were used as imaging support for the inverted system. Cell samples were loaded on the coverslips, followed by laying an 8X8 mm wide, 2 mm thick PBS-agar pad on top of the cells. The coverslip-pad combination was placed onto the OMX microscope in a sample stage plate with the pad facing upwards. 3D-SIM imaging was controlled with the DV-OMX software. FDAA signal was captured by using: 1) for HADA, 405 nm laser with 419–465 nm emission filter; 2) for BADA, 488 nm laser with 500–550 nm filter; and 3) for TADA, 561 nm laser with 609–654 nm filter. The z-axis scanning depth ranged from 2.4 to 3 μm. The immersion oil was optimized for each experiment with a range from 1.514 to 1.518.
Imaging – agarose microhole sample preparation
Agarose microholes were created by pouring molten 6% agarose onto a silicon micropillar array (Fig. S4). Patterned agarose was transferred into a Geneframe (Thermo Scientific AB-0577) mounted on a glass slide. The regions either side of the microholes were cut away, leaving a thin strip of agarose in the center of the Geneframe, to ensure sufficient oxygen. Cells at OD600~0.4 were concentrated 100× by centrifugation. 4 μL of sample was then loaded onto the agarose pad and a cover slip was placed on top.
Imaging – FtsZ dynamics in agarose microholes
FtsZ dynamics were imaged by near-TIRF illumination on a Nikon N-STORM microscope equipped with an NA 1.49 Nikon 100× TIRF objective, laser source (488 nm), additional 2.5× magnification optics and a 16 μm pixel Andor iXon DU897 EMCCD, giving a final pixel size at the image plane of 64 nm. Exposure times were 1 s and images were taken at 1 s intervals. The temperature at the sample was 30.0°C.
Image processing
All image processing unless otherwise specified was performed in Fiji (34). Images used for particle tracking were unaltered, except for trimming two pixels from the edges of some videos to remove edge artifacts detected by the tracking software. Images were cropped, contrast adjusted, rotated and scaled (no interpolation) for figures. SIM images were reconstructed using softWoRx and the quality was confirmed by SIMcheck (35). 3D projections were obtained with the Volume Viewer function of softWoRx with 25 stacks. The image of the 13th stack (90°-rotated) was used for data analysis.
Data analysis – kymographs
Kymographs were generated by the Fiji kymograph plugin with a line width of 3 (for FtsA and FtsZ) or 5 (for Pbp2B). For kymograph-based velocity measurements, velocities were quantitated by measuring the length of fluorescent trajectories in pixels and converting to nm and sec, respectively. For constriction times, kymographs of each condition were plotted and the intervals converted from the length of line segments in pixels: see “Correlation between FtsAZ motion and cytokinesis rates” above for details.
Data analysis – particle tracking
For particle tracking, tracks were generated using the TrackMate plugin in Fiji (30). Particles were detected with the Laplacian of Gaussians (LoG) detector, with a 400 nm blob diameter. Tracks were generated using the Linear motion LAP Tracker, with a 100 nm search radius and no frame gaps allowed. Tracks were then exported into MATLAB for further processing. For the characterization of FtsZ and A single molecules, we included all tracks containing 5 or more frames, since frames 4 and under cannot be fit by the velocity calculating algorithm; no additional filtering was done. For the characterization of FtsZ, FtsA and Pbp2B velocity, we applied additional filters to select for tracks with quantifiable directional motion. Using a custom script, tracks were filtered to select for directionally-moving tracks and exclude background, erroneous assignments, and, in the case of Pbp2B, rapidly diffusing molecules outside of the Z ring. Tracks shorter than 10 seconds or longer than 25 seconds were excluded, as were tracks whose start to endpoint displacement was less than 50 nm. For each track, the velocity, the diffusion coefficient, and the scaling exponent α were calculated as described previously (15). Briefly, mean squared displacement vs time delay (MSD vs t) was calculated for each track. Linear fits were made to log(MSD(t)) vs log(t); α is the slope of this fit. To ensure the quality of the tracks, only tracks with r2 > 0.95 for this linear fit were included in the data set. Velocity and diffusion coefficient were then calculated by fitting MSD(t) = 4D(t) + (Vt)2 using nonlinear least-squares fitting (15).
Data analysis – photobleaching traces
Images for photobleaching analysis were collected with 200 ms exposures, acquiring continuously (5 frames/second) to ensure sufficient time resolution. Representative particles were chosen manually. Using Fiji, intensity traces were generated from a 5×5 pixel region that was centered on the particle. To reduce noise, intensity traces were then filtered using a median intensity filter with a 4 second time window. The filtered trajectories were normalized to their minimum and maximum values: this was necessary because, particularly in TIRF, illumination intensity is non-uniform across the field of view. For plotting, intensity traces were aligned such that time t=0 corresponds to the frame in which the particle appears.
Data analysis – vertical microhole imaging
Images were corrected for translational drift using the Fiji StackReg plugin. Kymographs were automatically calculated along the circumference of individual Z rings using a custom MATLAB script. The best fitting circle for each Z ring was identified by least squares fitting, and the intensity around the circumference of the ring calculated with sub-pixel precision by bicubic interpolation. FtsZ velocities were calculated as per the kymographs above. Mean filament speed in Fig. S4E measured by Gaussian fitting.
Data analysis – plots
Plots were generated using Prism and MATLAB. Violin plots were generated in MATLAB using the violin.m function (36). Horizontal error bars were added to Fig. 4C using the ploterr.m function (37). In plots, ns and * is used to indicate the result of an unpaired two-sample t-test: ns p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Data analysis – 3D-SIM
To confirm that 3D-SIM images could be used for intensity quantitation, we demonstrated that intensity of Tetraspeck beads scaled linearly with exposure time after reconstruction (Fig. S2A). For quantitative analyses, septal PG images were collected by cropping the division site of the cells (~1.5 × 0.5 μm) using softWoRx software (Applied Precision, Issaquah, WA). Long-pulse labeling of FDAA (usually HADA) was used to identify the division sites and cell division state. Only the cells having a clear division site but not yet a complete septum were collected and analyzed. To quantify total intensity, we summed the intensity of 3D projections of Z rings. To quantify area labeled, we counted the number of pixels in the image that were above a set intensity threshold. Because the choice of threshold was arbitrary, where this metric was used, we demonstrated that the observed trends were independent of threshold choice (Fig. S2B). For total area labeled, we summed all the labeled pixels in the image; this represents total amount of FDAA labeling. For average feature area, we counted the number of pixels in each discrete labeled region of FDAA; this indicates the amount of FDAA labeling per spot. For the time course data in Fig. 1D and S2B–C, images from the longer labeling times (60, 90, and 120 seconds) were acquired with half the exposure time of the shorter labeling times to avoid saturating intensities. To compare these time points, we corrected the intensity for the later time points by multiplying their values by 2; we confirm that intensity scales linearly with exposure time in Fig. S2A. Violin plots were generated in MATLAB using the violin.m function (36). To compute the correlation between images, we first background subtracted each image in Fiji (rolling ball radius = 10). The correlation coefficient was then computed in MATLAB.
Data analysis – blinded manual classification
All images to be classified were combined into a single data set. First, the user was given instructions on how to use the software and shown sample images that typified each category. Next, the user classified 20 images in a practice round, using a custom MATLAB GUI; these responses were not recorded. The user was then asked to sort each image in the data set into one of the categories, with the option to skip images they felt could not be classified. All images in the combined data set were shuffled and presented to the user in a random order, with no information about which experimental condition they came from.
Data analysis – correlation between filament speed and cytokinesis
FtsZ filament average velocities and constriction times were obtained from kymographs as described above (see “Correlation between FtsAZ motion and cytokinesis rates” and “Data analysis – kymographs”). Filament velocities versus constriction times were plotted using MATLAB. An exponential fit to the data was generated in MATLAB.
Data analysis – single cell growth rates
Wild type (PY79), FtsZ(D213A) (10 μM IPTG, strain bAB273) and FtsZ(T111A) (strain bAB270) cells were grown in LB at 37°C, placed onto LB+2% agarose pads and imaged with phase contrast for 90 minutes at 2 minute intervals. Individual cell growth was analyzed by a custom MATLAB program. First, the area of the cells was measured based on the segmentation of phase contrast images. Cells were then tracked over time. The growth rate of each cell is calculated by the slope of the linear fit of the natural logarithm of total areas. For each linear fit of the total area of cells, the R squared threshold is set to 0.95.
Sequences of Fluorescent Proteins, Tags and Linkers
msfGFP
ATGCGAAAAGGGGAAGAATTGTTTACAGGCGTAGTACCGATATTAGTTGAGCTGGA TGGAGATGTTAACGGACACAAGTTTTCCGTACGCGGAGAGGGAGAAGGCGATGCCA CCAATGGCAAACTCACCCTTAAATTTATTTGTACGACAGGCAAGCTACCAGTGCCGT GGCCTACATTGGTCACTACACTCACGTATGGTGTGCAATGCTTTGCAAGATATCCCG ACCACATGAAGCAACATGACTTTTTCAAATCAGCCATGCCTGAAGGATATGTCCAAG AAAGGACTATTAGCTTTAAGGACGACGGCACCTACAAAACACGCGCTGAAGTCAAA TTCGAAGGTGATACGTTAGTTAATCGTATCGAACTGAAAGGTATAGACTTTAAAGAA GATGGAAATATCCTGGGGCATAAGTTGGAGTACAATTTCAACAGCCATAACGTATA CATCACAGCTGACAAGCAGAAAAATGGCATTAAAGCAAATTTCAAAATCAGACATA ACGTTGAAGATGGGTCAGTTCAGCTTGCGGATCATTATCAACAGAACACACCGATTG GCGATGGTCCGGTGTTACTACCTGATAACCATTATCTGTCGACTCAGAGTAAACTGT CTAAAGATCCAAATGAAAAACGGGACCACATGGTCCTTTTAGAGTTTGTGACGGCA GCTGGGATTACGCATGGAATGGATGAACTTTACAAATAA
mNeonGreen
ATGGTTTCGAAAGGAGAGGAGGATAATATGGCTAGCCTCCCAGCGACCCACGAACT GCATATTTTTGGCAGCATTAATGGCGTTGACTTTGATATGGTGGGGCAGGGAACAGG GAACCCTAACGATGGCTATGAGGAGCTCAATCTCAAGAGTACAAAAGGAGATTTGC AATTTTCACCTTGGATCCTGGTTCCGCATATTGGCTACGGCTTTCATCAATACTTGCC TTATCCGGACGGCATGTCCCCGTTCCAAGCTGCGATGGTGGATGGTTCTGGGTACCA GGTGCACCGTACTATGCAGTTTGAGGACGGTGCCTCACTGACGGTCAACTATAGATA TACTTATGAAGGCTCACACATTAAGGGTGAGGCCCAAGTTAAAGGAACAGGGTTTC CTGCGGATGGACCGGTAATGACAAACAGTTTAACCGCTGCGGACTGGTGTCGCTCG AAAAAAACATACCCAAACGATAAAACGATCATCTCGACCTTCAAATGGAGCTATAC TACGGGCAACGGCAAACGCTATCGTTCCACAGCACGCACGACTTATACGTTTGCTAA ACCGATGGCCGCAAACTACCTCAAAAATCAACCTATGTACGTGTTCAGAAAAACCG AGTTAAAACATTCAAAAACGGAACTTAATTTTAAAGAGTGGCAAAAGGCGTTTACA GACGTGATGGGTATGGATGAACTCTATAAGTGA
HaloTag
ATGGCAGAAATCGGTACTGGCTTTCCATTCGACCCGCATTATGTGGAAGTCCTGGGC GAGAGAATGCATTACGTTGACGTGGGTCCGAGAGATGGAACTCCGGTCCTTTTTCTG CACGGGAATCCTACAAGCTCTTATGTTTGGCGCAATATCATCCCTCATGTAGCTCCG ACGCATCGCTGTATTGCGCCGGACCTGATTGGTATGGGAAAATCTGATAAACCAGAC CTGGGTTACTTTTTCGATGATCATGTGCGTTTCATGGATGCCTTCATTGAGGCATTAG GGCTTGAAGAAGTCGTCTTGGTGATTCATGATTGGGGCTCAGCTCTGGGATTTCACT GGGCTAAAAGAAATCCTGAACGCGTAAAAGGCATCGCGTTTATGGAGTTCATTCGTC CAATTCCGACTTGGGATGAATGGCCTGAGTTCGCGAGAGAAACATTTCAAGCATTTC GCACGACCGATGTAGGCCGGAAGTTAATCATCGATCAGAATGTCTTTATCGAAGGG ACATTGCCGATGGGAGTCGTTCGTCCGTTAACAGAAGTCGAAATGGATCACTATAGA GAACCTTTTCTTAATCCTGTGGACAGAGAGCCGCTGTGGCGGTTTCCGAACGAACTG CCGATTGCAGGCGAGCCTGCTAACATTGTAGCGCTGGTTGAAGAGTATATGGATTGG CTTCATCAGTCTCCAGTTCCGAAGTTATTGTTTTGGGGTACGCCTGGCGTGCTTATTC CACCGGCCGAAGCGGCACGTTTGGCAAAAAGCCTGCCAAATTGCAAAGCCGTTGAC ATTGGCCCTGGACTTAACTTGCTTCAAGAGGATAACCCGGACTTAATCGGGAGCGAA ATTGCCCGGTGGCTTTCTACCTTAGAAATCAGCGGCTAG
15aa Linker A
CTCGAGGGATCTGGCCAGGGACCGGGCTCAGGCCAAGGAAGCGGC
15aa Linker B
CTTGAGGGTAGCGGACAAGGTCCTGGATCTGGTCAAGGCAGTGGG
Strain Construction
ME7 [pbp2B::mNeonGreen-15aa-pbp2B] was created by transforming PY79 with a Gibson assembly consisting of 4 fragments: 1) PCR with primers oME32 and oME22 and PY79 genomic DNA template (containing the region upstream of the pbp2B gene); 2) PCR with primers oZB46 and oZB47 and bMK93 genomic DNA template (containing the construct of first 30 bp mNeonGreen-cat cassette-PxylA promoter-mazF gene-Pveg promoter); 3) PCR with primers oZB33 and oZB34 and the synthetic mNeonGreen gene as template (ordered from IDT, see fluorophore sequences section); 4) PCR with primers oME20 and oME21 and PY79 genomic DNA template (containing the pbp2B gene). Transformations were plated on LB-Cm plates with 2% glucose and incubated at 37°C. Transformants were grown in LB with 2% glucose until OD600 ~ 0.5, plated on LB plates with 2% xylose, and incubated at 37°C. Colonies were then patched onto LB, LB-Cm and LB-Xylose plates and candidates that were sensitive to Cm but resistant to xylose were stocked and confirmed by PCR and sequencing.
bGS3 [pbp2B::erm-Phyperspank-mNeonGreen-15aa-pbp2B] was created by transforming PY79 with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oAB81 and oAB82 and PY79 genomic DNA template (containing the region upstream of the pbp2B gene); 2) PCR with primers oJM29 and oMD232 and bMD352 genomic DNA template (containing the construct erm-Phyperskpank); 3) PCR with primers oAB137 and oAB87 and ME7 genomic DNA template (containing the mNeonGreen-15aa-pbp2B fusion). Transformations were plated on LB-MLS plates with 100 μM IPTG and colonies were patched onto LB and LB+IPTG plates. Candidates that presented resistance to MLS and IPTG dependence were confirmed by PCR and sequencing.
bGS28 [pbp2B::erm-Phyperspank-HaloTag-15aa-pbp2B] was created by transforming PY79 with a Gibson assembly consisting of 4 fragments: 1) PCR with primers oAB81 and oAB82 and PY79 genomic DNA template (containing the region upstream of the pbp2B gene); 2) PCR with primers oJM29 and oMD232 and bMD352 genomic DNA template (containing the construct erm-Phyperskpank); 3) PCR with primers oGS7 and oAB14 and the synthetic HaloTag gene as template (ordered from DNA 2.0, see fluorophore sequences section); 4) PCR with primers oME20 and oAB87 and PY79 genomic DNA template (containing the pbp2B gene). Transformations were plated on LB-MLS plates with 100 μM IPTG and colonies were patched onto LB and LB+IPTG plates. Candidates that presented resistance to MLS and IPTG dependence were confirmed by PCR and sequencing.
bGS60 [pbp2B::erm-Phyperspank-HaloTag-15aa-pbp2B, ftsZΩftsZ(T111A) (tet)] was created by transforming bGS28 with genomic DNA from AB62 (a gift from Frederico Gueiros Filho).
bGS90 [pbp2B::erm-Phyperspank-HaloTag-15aa-pbp2B, amyE::erm-Phyperspank-ftsAZ(D213A)] was created by transforming bGS28 with genomic DNA from bAB215.
bGS31 [pbp2B::erm-Phyperspank-HaloTag-15aa-pbp2B, ftsZ::mNeonGreen-15aa-ftsZ] was created by transforming bAB185 with genomic DNA from bGS28.
bAB94 [amyE::erm-Phyperspank-ftsA] was created by transforming PY79 with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oMD191 and oMD232 and bMD352 genomic DNA template (containing the region upstream of the amyE gene, the erm cassette and the Phyperspank promoter); 2) PCR with primers oAB78 and oAB93 and PY79 genomic DNA template (containing the ftsA gene); 3) PCR with primers oMD196 and oMD197 and PY79 genomic DNA template (containing the region downstream of amyE).
bAB96 [amyE::erm-Phyperspank-ftsZ] was created by transforming PY79 with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oMD191 and oMD232 and bMD352 genomic DNA template (containing the region upstream of the amyE gene, the erm cassette and the Phyperspank promoter); 2) PCR with primers oAB95 and oAB94 and PY79 genomic DNA template (containing the ftsA gene); 3) PCR with primers oMD196 and oMD197 and PY79 genomic DNA template (containing the region downstream of amyE).
bAB98 [amyE::erm-Phyperspank-ftsAZ] was created by transforming PY79 with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oMD191 and oMD232 and bMD352 genomic DNA template (containing the region upstream of the amyE gene, the erm cassette and the Phyperspank promoter); 2) PCR with primers oAB78 and oAB94 and PY79 genomic DNA template (containing the ftsAZ operon); 3) PCR with primers oMD196 and oMD197 and PY79 genomic DNA template (containing the region downstream of amyE).
bAB146 [amyE::erm-Phyperspank-mNeonGreen-15aa-pbp2B] was created by transforming PY79 with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oMD191 and oMD232 and bMD352 genomic DNA template (containing the upstream region to amyE gene, the erm cassette and the Phyperspank promoter); 2) PCR with primers oAB137 and oAB134 and ME7 genomic DNA template (containing the mNeonGreen-15aa-pbp2B construct); 3) PCR with primers oMD196 and oMD197 and PY79 genomic DNA template (containing the region downstream to amyE).
bAB167 [ftsA::ftsA-mNeonGreen(SW)] was created by inserting mNeonGreen between helices 6 and 7 of FtsA, the equivalent of a sandwich fusion (SW) that proved to be functional for MreB-mCherry (38). PY79 cells were transformed with a Gibson assembly consisting of 7 fragments: 1) PCR with primers oAB74 and oAB75 and template PY79 genomic DNA (containing the region upstream of the ftsAZ promoters); 2) PCR with primers oJM28 and oJM29 and template pWX467a (containing the erm cassette); 3) PCR with primers oAB76 and oAB33 and template PY79 genomic DNA (containing the first 801 bp of the ftsA gene); 4) PCR with primers oAB135 and oAB136 and the synthetic mNeonGreen gene as template (ordered from IDT, see fluorophore sequences section); 5) PCR with primers oAB48 and oAB28 and template PY79 genomic DNA (containing the last 522 bp of the ftsA gene and the ftsZ region); 6) PCR with primers oJM28 and oJM29 and template pWX465a (containing the cat cassette); 7) PCR with primers oAB29 and oAB30 and template PY79 genomic DNA (containing the region downstream of ftsAZ terminators). After proper integration was confirmed by PCR and sequencing, the resistance cassettes were looped-out using pDR244 (containing Cre recombinase, a gift from David Rudner), and incubating transformed cells overnight at 30°C. Colonies were streaked onto LB, LB-MLS, LB-Cm and LB-Spec plates and incubated overnight at 45°C. After confirming that only LB plates presented colonies, the loop-outs were confirmed by PCR and sequencing.
bAB185 [ftsZ::mNeonGreen-15aa-ftsZ] was created by transforming PY79 with a Gibson assembly consisting of 7 fragments: 1) PCR with primers oAB74 and oAB75 and template PY79 genomic DNA (containing the region upstream of the ftsAZ promoters); 2) PCR with primers oJM28 and oJM29 and template pWX467a (containing the erm cassette); 3) PCR with primers oAB76 and oAB139 and template PY79 genomic DNA (containing the region upstream of the ftsZ gene); 4) PCR with primers oZB33 and oZB34 and the synthetic mNeonGreen gene as template (ordered from IDT, see fluorophore sequences section); 5) PCR with primers oAB140 and oAB28 and template PY79 genomic DNA (containing the ftsZ gene region); 6) PCR with primers oJM28 and oJM29 and template pWX465a (containing the cat cassette); 7) PCR with primers oAB29 and oAB30 and template PY79 genomic DNA (containing the region downstream of the ftsAZ terminators). After proper integration was confirmed by PCR and sequencing, the resistance cassettes were looped out by transforming the plasmid pDR244 (containing Cre recombinase and a temperature sensitive origin of replication, a gift from David Rudner), and incubating transformed cells overnight at 30°C. Colonies were streaked onto LB, LB-MLS, LB-Cm and LB-Spec plates and incubated overnight at 45°C to remove the plasmid. After confirming that only LB plates presented colonies, the loop-outs were confirmed by PCR and sequencing.
bAB199 [ftsZ::mNeonGreen-15aa-ftsZ, amyE::erm-Phyperspank-ftsA] was created by transforming bAB185 with genomic DNA from bAB94.
bAB209 [ftsZ::mNeonGreen-15aa-ftsZ, amyE::PXylA-mciZ (cat)] was created by transforming bAB185 with genomic DNA from AH93.
bAB213 [ftsAZ::erm-ftsA-HaloTag(SW)-ftsZ-cat] was created by inserting HaloTag between helices 6 and 7 of FtsA, the equivalent of a sandwich fusion (SW) that proved to be functional for MreB-mCherry (38). PY79 cells were transformed with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oAB74 and oAB33 and template bAB163 genomic DNA (containing the region beginning upstream of the ftsAZ promoters and including the first 801 bp of the ftsA gene, erm cassette included); 2) PCR with primers oAB21 and oAB44 and the synthetic HaloTag gene as template (ordered from DNA 2.0, see fluorophore sequences section); 3) PCR with primers oAB48 and oAB30 and template bAB167 genomic DNA (containing the region from the last 522 bp of the ftsA gene to downstream of the ftsAZ terminators, cat cassette included). Proper integration was confirmed by PCR and sequencing.
bAB215 [amyE::erm-Phyperspank-ftsAZ(D213A)] was created by transforming PY79 with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oMD191 and oMD232 and bMD352 genomic DNA template (containing the region upstream of the amyE gene, the erm cassette and the Phyperspank promoter); 2) PCR with primers oAB78 and oAB124 and bAB98 genomic DNA template (containing the first 2047 bp of the ftsAZ operon, inserting the mutation GCT at bp 638 of the ftsZ gene); 3) PCR with primers oMD123 and oMD197 and bAB98 genomic DNA template (containing the last 529 bp of the ftsZ gene, inserting the mutation GCT at the 638 bp position of the ftsZ gene, and the region downstream of amyE).
bAB217 [amyE::erm-Phyperspank-ftsA-mNeonGreen(SW)-ftsZ(D213A)] was created by transforming PY79 with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oMD191 and oMD232 and bMD352 genomic DNA template (containing the region upstream of the amyE gene, the erm cassette and the Phyperspank promoter); 2) PCR with primers oAB78 and oAB124 and bAB163 genomic DNA template (containing the first 2845 bp of the ftsA-mNeonGreen( SW)-ftsZ operon, inserting the mutation GCT at bp 638 of the ftsZ gene); 3) PCR with primers oAB123 and oMD197 and bAB98 genomic DNA template (containing the last 529 bp of the ftsZ gene, inserting the mutation GCT at the 638 bp position of the ftsZ gene, and the region downstream of amyE).
bAB219 [amyE::erm-Phyperspank-ftsA-mNeonGreen-15aa-ftsZ] was created by transforming PY79 with a Gibson assembly consisting of 3 fragments: 1) PCR with primers oMD191 and oMD232 and bMD352 genomic DNA template (containing the region upstream of the amyE gene, the erm cassette and the Phyperspank promoter); 2) PCR with primers oAB78 and oAB94 and bAB185 genomic DNA template (containing the ftsA-mNeonGreen-15aa-ftsZ operon); 3) PCR with primers oMD196 and oMD197 and PY79 genomic DNA template (containing the region downstream of amyE).
bAB221 [ftsZ::mNeonGreen-15aa-ftsZ, amyE::erm-Phyperspank-ftsA-mNeonGreen-15aa-ftsZ] was created by transforming bAB185 with genomic DNA from bAB219.
bAB229 [ftsAZ::erm-ftsA-HaloTag(SW)-mNeonGreen-15aa-ftsZ-cat] was created by transforming PY79 with a Gibson assembly consisting of 2 fragments: 1) PCR with primers oAB74 and oAB44 and template bAB213 genomic DNA (containing the region from upstream of the ftsAZ promoters to the end of the HaloTag gene, erm cassette included); 2) PCR with primers oAB48 and oAB30 and template bAB181 genomic DNA (containing the region from the last 522 bp of the ftsA gene to downstream of the ftsAZ terminators, cat cassette included). Proper integration was confirmed by PCR and sequencing.
bAB248 [ftsZ::mNeonGreen-15aa-ftsZ, amyE::Phyperspank-minCD (spc)] was created by transforming bAB185 with genomic DNA from JB60 (a gift from Frederico Gueiros Filho).
bAB270 [pbp2B::mNeonGreen-15aa-pbp2B, ftsZΩftsZ(T111A) (tet)] was created by transforming ME7 with genomic DNA from AB62 (a gift from Frederico Gueiros Filho).
bAB271 [pbp2B::mNeonGreen-15aa-pbp2B, ftsZΩftsZ(G106S) (tet)] was created by transforming ME7 with genomic DNA from Z-84 (a gift from Frederico Gueiros Filho).
bAB272 [pbp2B::mNeonGreen-15aa-pbp2B, amyE::erm-Phyperspank-ftsAZ] was created by transforming ME7 with genomic DNA from bAB98.
bAB273 [pbp2B::mNeonGreen-15aa-pbp2B, amyE::erm-Phyperspank-ftsAZ(D213A)] was created by transforming ME7 with genomic DNA from bAB215.
bAB274 [pbp2B::mNeonGreen-15aa-pbp2B, amyE::PXylA-mciZ (cat)] was created by transforming ME7 with genomic DNA from AH93.
bAB281 [amyE::erm-Phyperspank-ftsA-mNeonGreen-15aa-ftsZ, ftsZΩftsZ(T111A) (tet)] was created by transforming bAB219 with genomic DNA from AB62 (a gift from Frederico Gueiros Filho).
bAB282 [amyE::erm-Phyperspank-ftsA-mNeonGreen-15aa-ftsZ, ftsZΩftsZ(G106S) (tet)] was created by transforming bAB219 with genomic DNA from Z-G106S (a gift from Frederico Gueiros Filho).
bAB285 [pbp2B::mNeonGreen-15aa-pbp2B, amyE::Phyperspank-minCD (spc)] was created by transforming ME7 with genomic DNA from JB60 (a gift from Frederico Gueiros Filho).
bAB310 [pbp2B::mNeonGreen-15aa-pbp2B, amyE::erm-Phyperspank-ftsA] was created by transforming ME7 with genomic DNA from bAB94.
SH013 [trpC2, ftsZ:: ftsZ-GFP::cat] was created by transforming B. subtilis 168CA [trpC2] with genomic DNA from PL642 (a gift from Petra Anne Levin).
SH033 [trpC2, ftsZ:: ftsZ-GFP::cat, Δhag::aph(Kan)] was created by transforming SH013 with genomic DNA from PB5250 (obtained from BGSC catalogue number 1A842).
SH41 [ftsZ::mNeonGreen-15aa-ftsZ, Δhag::aph (Kan)] was created by transforming bAB185 with genomic DNA from PB5250 (obtained from BGSC catalogue number 1A842).
Supplementary Material
Figure S1: The FDAA labeling pattern is independent of the FDAA type or the sequence in which they are used
A Sequential short-pulse labeling of FDAAs with reversed color sequence compared to Fig. 1B. This demonstrates the pattern of FDAA sequential labeling is independent of the sequence of FDAA labeling.
B Labeling cells with FDAAs for one 45-second pulse yields the same label intensity as labeling for three sequential 15-second pulses. This indicates that FDAA pulses of different durations are comparable, and that repeated pulse labeling with FDAAs is non-perturbing. ns: the comparison is not statistically significant by t-test.
C FDAA sequential labeling using different orders of dyes (indicated in diagrams). Discrete spots of FDAA labeling were seen in short-pulse labeling regardless of what FDAA color was used for the pulse. Images were background subtracted in Fiji (rolling ball radius = 10).
Colored bars indicate the time course of FDAA labeling in each experiment. All images were taken with 3D-SIM. Scale bars = 0.5 μm.
Figure S2: Quantitation of FDAA labeling from 3D-SIM data
A The fluorescence intensity measured from 3D-SIM imaging varies linearly with exposure times both at high (left) and low (right) laser powers, validating intensity-based quantitation. Plot shows the maximum intensity of TetraSpeck beads measured at varying exposure times for three different fields of view. Black lines are linear fits to each field of view.
B The area of FDAA-labeled features at the septum increases with increased labeling time (as in Fig. 1D) independent of the threshold cutoff used to binarize images. Labeled area is measured as the number of pixels in each region above a chosen intensity threshold. Fig. 1D shows that for one such threshold (45 units) FDAA labeling time and average labeled area are positively correlated. This graph shows the correlation coefficients for the complete range of possible threshold choices, indicating that this correlation holds regardless of threshold choice, except at the extreme values.
C The total amount of FDAA labeling at the septum increases with labeling time. Total amount of labeling is calculated as the sum of the fluorescence intensity in each image; distribution shown as violin plots, black lines: means of the distributions. r: correlation coefficient of means.
D Disruption of cell wall synthesis causes decreased FDAA labeling at the septum. To decrease cell wall synthesis, cells were treated with 4 μg/ml Vancomycin for 3 min (Van), 50 μg/ml Penicillin G for 3 min (PenG), or Pbp2B was depleted for 3 hours. Following perturbations, cells were labeled with 2 mM TADA for 2 minutes. (Left) representative TADA images for each condition. Representative images shown are those with the median intensity of their distribution. (Right) total FDAA intensity for each condition. Black lines indicate the mean.
Figure S3: Viability and fitness of native fusions
Native mNeonGreen fusions of Pbp2B (strain ME7), FtsA (strain bAB167), and FtsZ (strain bAB185) have similar fitness and protein levels compared to wild type cells (strain PY79).
A Growth curves of native fusion strains. Cells were grown in triplicate in LB until O.D.600 < 1.0, and then diluted to O.D. 600 = 0.1. Aliquots were taken every hour and O.D.600 was measured in a plate reader (Synergy HT plate reader, Bio-Tek).
B Cell length distributions. Cells were grown until mid-exponential phase in LB, concentrated and incubated for 1 minute with 1 μg/ml of FM5-95 dye, and imaged in a confocal spinning disk microscope. Cell length was measured using 250 cells for each strain.
C Cell viability from spot dilutions. Cells were grown until O.D.600 ~ 1.0 in LB, 5 μl of each culture was spotted from serial dilutions on LB plates, and incubated for 12 hours.
D Relative protein levels of native fusions compared to wild type assayed by Western blotting. Different lysate dilutions of each strain carrying its respective native fusion were blotted side by side with PY79 control. Cells were incubated with specific polyclonal antibodies for FtsZ, FtsA and Pbp2B, respectively. Bands were probed and visualized using a chemiluminescence system.
E Relative quantification of native mNeonGreen fusions. Mean values were obtained from the average of 3 independent experiments, and normalized to the measurements of the respective untagged band from PY79. FtsA-mNeonGreen:FtsA and mNeonGreen-Pbp2B:Pbp2B ratios show similar protein levels (0.98±0.11 and 0.99±0.07, respectively). For the mNeonGreen-FtsZ fusion, considerably lower mNeonGreen-FtsZ:FtsZ ratios were observed (0.71±0.08). However, a proteolysis product from mNeonGreen-FtsZ construct was observed in all blots, and the sum of the intensities of mNeonGreen-FtsZ and its proteolysis product gave a balanced ratio (1.07±0.09).
F Increased FtsAZ levels have no measurable effect on fitness or cell division, indicating that while overexpression of either FtsA or FtsZ alone may be toxic, balanced expression of both causes no observable cellular defects. Membrane staining of cells overexpressing a second copy of ftsZ (strain bAB96, bottom left panel), ftsA (bAB94, top right panel) or ftsAZ (bAB98, bottom right panel). The control (top left panel) is a representative image of bAB96 cells without FtsZ overexpression. Cells were grown in LB until mid-exponential phase, diluted into fresh medium with 100 μM IPTG (or 0 μM IPTG as a control for each culture) and grown for more 2 hours prior imaging. Images were acquired on a spinning-disk confocal microscope after cells were incubated with 1 μg/ml of FM5-95 dye for 30 seconds at room temperature. Because the overexpression of either FtsZ or FtsA alone affected cell length and width, most of our experiments using inducible constructs were performed by expressing both FtsA and FtsZ together, which had no such effects. Growth rates of cells overexpressing FtsAZ are shown in S7A (brown curve), with no observable change from wildtype.
Figure S4: Imaging FtsZ in microholes
A Scanning electron microscopy images of silicon micropillars nanofabricated through electron-beam lithography and reactive-ion etching.
B Schematic of the stamping process to produce agarose pads containing microholes.
C Schematic of B. subtilis in microholes. FtsZ filaments are shown in green.
D (i) Brightfield images of microholes without bacteria. (ii) Widefield epifluorescence images of B. subtilis (SH33) inoculated in microholes. Green indicates GFP-FtsZ, red indicates FM4-64-labeled cell membrane.
E Quantification of mNeonGreen-FtsZ filament dynamics in actively constricting vertically immobilized cells. FtsZ filaments move in both directions with equal probability. Vectorial velocities (i) and speeds (ii) of mNeonGreen-FtsZ filaments (strain SH41). Mean filament speed was 42 nm ± 23 nm/sec (mean ± S.D., N = 110). 25% of filaments were immobile (speed of ~ 0 nm/s).
F A FtsZ-GFP fusion has similar dynamics compared to mNeonGreen-FtsZ. Vertically immobilized cells (strain SH33) display multiple FtsZ-GFP filaments with independent directional motion around the division site. (top) Cropped rings in early (left), mid (center), and late (right) divisional stages. (bottom). Blue arrow indicates start of a directional FtsZ track. Scale bars = 0.5 μm.
Figure S5: Divisome imaging and particle tracking
A Pbp2B-mNeonGreen expressed at native levels (ME7) shows Pbp2B motion at the Z ring, imaged by TIRF microscopy. Pbp2B can be seen throughout the cell, but localizes preferentially to the Z ring. Indicated kymograph (right) was drawn at the arrowhead. Scale bars: 1 μm (cell), 0.5 μm (kymograph).
B Pbp2B-mNeonGreen expressed at low levels (bGS3) shows Pbp2B motion at the Z ring as imaged by TIRF microscopy. Cells were grown in 2 μM IPTG to induce low levels of Pbp2B expression. This decreased the overall signal density, allowing dynamics to be resolved more clearly. Kymographs are drawn at arrowheads. Scale bars: 1 μm (cell), 0.5 μm (kymograph).
C Particle tracking shows directional motion of FtsZ filaments (left), FtsA filaments (center), and Pbp2B single molecules (right). mNeonGreen-FtsZ (bAB185), FtsA-mNeonGreen (bAB167), and Halo-Pbp2B (bGS31) labeled with 40 μM JF-549 were imaged by TIRF microscopy. Particles were identified and tracked using TrackMate and analyzed using custom MATLAB code. Tracks are overlaid on a frame from each time lapse, and are colored blue to yellow along their duration. Scale bar: 1 μm.
D Particle tracking shows that FtsA, FtsZ, and Pbp2B all move at the same speed. Particle tracks were filtered to select for directional motion, and velocity was calculated from fits of particle tracks to MSD(t)=4D(t)+(Vt)2 (15). The small difference in the means of these distributions compared to the velocity distributions in Fig. 2G (velocity measurement by kymograph) is likely due to differences in the two methods of measuring velocity. However, the velocities of FtsA, FtsZ and Pbp2B agree with each other when assayed using two independent velocity measurements, reinforcing the conclusion that they move at the same speed.
E Distributions of the scaling coefficient α for particle tracks of FtsA, FtsZ, and Pbp2B indicates that they move directionally. α was calculated as described previously (15). This distribution of α values is consistent with directed motion (1 < α < 2).
F Distributions of the velocity of unfiltered FtsZ and A particle tracks calculated from fits of (MSD vs t) to MSD(t)=4D(t)+(Vt)2 for both single molecules (solid lines) and filaments (dashed lines). FtsA and FtsZ filament trajectories show a mobile population (visible as a second peak in the 10–20 nm/s region), whereas the motions of single molecules show a negligible value for directional motion (~ 10−8 nm/s).
G Detailed velocity distributions of FtsZ and FtsA single molecules and filaments. Left: Distributions of all velocities less than 10−7 nm/s, representing the non-moving population of particles. Right: Distributions of all velocities greater than 10−7 nm/s, representing the mobile population. Single molecules of FtsA and FtsZ have more non-moving particles and very few moving particles compared to filaments, indicating that, while filaments of FtsZ and FtsA move, single molecules do not.
H Distributions of the diffusion coefficients for unfiltered FtsZ and FtsA particle tracks calculated from fits of (MSD vs t) to MSD(t)=4D(t)+(Vt)2. Single molecules of mNeonGreen-FtsZ (bAB219) and Halo-FtsA (bAB213) labeled with 100 pM JF-549 were imaged by TIRF microscopy. Particles were tracked and diffusion coefficients calculated as described above. The very low value of the diffusion coefficient is consistent with immobile particles, and similar to that of immobile class A PBPs measured in E. coli and B. subtilis (39, 40).
For panels F–H, n = 2,695 (FtsZ single molecules), 10,358 (FtsA single molecules), 4,718 (FtsZ filaments), 7,164 (FtsA filaments).
I Single-step photobleaching of FtsZ and FtsA single molecules. Intensity traces are aligned such that the frame in which the molecule appears is time = 0. Faded, thin lines show the raw data; the bold lines are data which has been filtered to reduce noise. The fluorescence signal drops to baseline in a single time step, indicating that these are single molecules.
J Simultaneous imaging of directionally-moving FtsZ filaments and immobile FtsA single molecules. Dual-color TIRF imaging of native levels of mNeonGreen-FtsZ (green) and single molecules of FtsA-HaloTag-JF549 (red). Images were taken at 2 second intervals with 250 ms exposure times.
K Pbp2B velocity correlates with FtsAZ filament velocity. JF549-HaloTag-Pbp2B single molecules were imaged under 5 different conditions, their velocities measured by kymographs and plotted against the FtsAZ filament velocity in that same condition. Pbp2B velocities (mean ± S.D.): wild type (strain bGS31, n = 152): 32.6 ± 9.1 nm/sec; FtsA overexpression (strain bAB310 with 20 μM IPTG, n = 87): 22.0 ± 5.8 nm/sec; FtsZ(D213A) (strain bGS90 with 100 μM IPTG): no motion detected; FtsZ(T111A) (strain bGS60, n = 108): 20.7 ± 5.8 nm/s; PC190723 treatment (strain bGS31 with 10 μM PC190723): no motion detected.
Figure S6: FDAA labeling with FtsAZ velocity perturbations
A Treatment with high concentrations of PC190723 causes PG synthesis to localize to small patches along the cell, mirroring the localization of FtsZ following similar treatment. Cells were treated with 10 μg/ml PC190723 for 10 minutes, and labeled with TADA as shown. A cell not treated with PC190723 that was labeled equivalently with FDAAs is shown for comparison. Scale bar: 1 μm.
B The trends observed in Fig. 4B are independent of threshold used for analysis. For each perturbation (PC190723, FtsZ(D213A), MciZ), data plotted is the mean FDAA-labeled area with perturbation minus the mean FDAA-labeled area without perturbation. Negative values of the curves indicate that the perturbation decreases labeled area, while positive values indicate that the perturbation increases labeled area. These measurements are consistent across all possible thresholds for labeling except extreme values, indicating that the trends reported in Fig. 4B are independent of threshold choice.
C Blinded classification of FDAA labeling under various FtsZ perturbations. This demonstrates that perturbations that decrease FtsZ velocity also cause more heterogeneous labeling (more spots and arcs, fewer complete rings), whereas perturbations that increase FtsZ velocity yield a more uniform distribution of FDAA signal. + indicates sample with perturbation, – indicates unperturbed sample for comparison. Note that the labeling and imaging conditions varied for the three experiments shown here, explaining the variation between the three unperturbed samples. See Table S1 for details.
Figure S7: Perturbation of FtsAZ dynamics affects cytokinesis time, but not growth rate
A Violin distributions of (left) mNeonGreen-Pbp2B constriction times (measured from kymographs) and (right) FtsAZ filament velocities under different conditions. Cells were grown and imaged as described in methods.
B Comparison between constriction times of Pbp2B rings measured by kymograph (Kymo) and automated tracking (Track) analysis. Scatter plot of constriction time distributions was obtained as described in methods; statistics are shown in Table S4.
C Distribution of constriction times obtained from cells under slow growth conditions (S750-glucose at 25°C). Perturbations that changed FtsAZ filament motion and constriction times in rich media at 37°C had the same effect in minimal media at 25°C, either slowing down (FtsZ(T111A) - strain bAB270, FtsZ(D213A) - strain bAB273 with 10 μM IPTG) or accelerating (MciZ overexpression - strain bAB274 with 5 μL of 3 M xylose on top of the pad) the rate of cytokinesis.
D Bulk growth curves of cells under all FtsZ perturbations that significantly changed constriction rates. Cells were grown in LB until OD600 < 1.0, and then diluted to OD600 = 0.1 and split into 8 replicates. Growth curves were taken in a plate reader (Epoch-2 plate reader, Biotek) with continuous shaking at 37°C.
E Linear segments from growth curves in D (40 to 110 minutes). Curves were converted into a ln(OD600) vs. time (min) plot, and doubling times were then calculated using linear regression. Doubling times (mean ± S.D.): WT (strain PY79): 21.3 ± 2.6 min; FtsZ(T111A) (strain bAB270): 20.7 ± 2.7 min; FtsZ(D213A) (5 μM IPTG, strain bAB273): 20.2 ± 1.45 min; FtsZ(D213A) (10 μM IPTG, strain bAB273): 19.4 ± 2.1 min; FtsA overexpression (100 μM IPTG, strain bAB199): 21.0 ± 2.1 min; and MciZ overexpression (60 mM xylose, strain bAB274): 20.4 ± 1.9 min.
F Representative growth rate curves extracted from single cell time lapses. Doubling times (mean ± S.D.): wild type (strain PY79): 24.8 ± 1.4 min (n = 14); FtsZ(T111A) (strain bAB270): 22.3 ± 2.1 min (n = 29); FtsZ(D213A) (10 μM IPTG, strain bAB273, 10 μM IPTG): 24.3 ± 2.2 min (n = 18).
Table S1: FDAA experimental conditions
Table S2: Strains and oligonucleotides used in this study Strain
Table S3: Live-cell imaging conditions
Table S4: Measurements of FtsAZ filament velocities and Pbp2B ring constriction periods with different genetic perturbations, growth conditions and measurement methods
Movie S1: FtsA and FtsZ filaments move directionally together inside and outside of the division site. Cells were imaged using TIRF microscopy at 3 second intervals. Representative 5-minute acquisitions, displayed at 15 frames per second (45× actual speed). Scale bar: 2 μm.
Column 1: mNeonGreen-FtsZ (bAB185) showing filament motion.
Column 2: FtsA-mNeonGreen(SW) (bAB167) showing filament motion.
Column 3: FtsA-HaloTag(SW)-JF549 (red) and mNeonGreen-FtsZ (green) showing filament motion. bAB229 cells were incubated with 500 nM of HaloTag-JF549 ligand for 15 minutes.
Column 4: mNeonGreen-FtsZ (bAB221) showing filament motion with FtsAZ overexpressed. Cells were grown with 100 μM IPTG for 1 hour.
mNeonGreen-FtsZ (strain SH41) dynamics in vertically immobilized cells with 600–1000 nm radii. Cells were imaged by epifluorescence at 1 second intervals. 2-minute acquisitions displayed at 15 frames per second (15× actual speed). Scale bar: 1 μm.
A. JF549-HaloTag-Pbp2B (bGS31) shows Pbp2B single molecules by TIRF microscopy. Cells were incubated with 50 pM of HaloTag-JF549 ligand for 15 minutes, washed, and prepared as indicated in methods. Cells were imaged at 1 second intervals using TIRF microscopy. Images of mNeonGreen-FtsZ were used to mark the position of the division machinery (first 60 frames of the movie) before imaging of JF549. Arrowheads indicate Z rings that colocalize with directionally moving Pbp2B. 4-minute acquisition displayed at 30 frames per second (30× actual speed).
B Overexpression of exogenous mNeonGreen-Pbp2B (100μM IPTG, bAB146) shows diffusion of Pbp2B molecules along the cell by TIRF microscopy. Cells were imaged at 100 millisecond intervals. 10-second acquisition displayed at 30 frame per second (3× actual speed). Scale bar: 2 μm.
FtsA-mNeonGreen(SW) (bAB167) showing filament motion during perturbations to cell wall synthesis. Antibiotics were added to the top of the pad 5 minutes before imaging. Images shown as follows: no antibiotic control (top left); 50 mg/μl Vancomycin (top right); 10 mg/μl Ampicillin (center left); 10 mg/μl Penicillin G (center right); 100 mg/μl Cephalexin (bottom left) and 100 mg/μl Fosfomycin (bottom right). Cells were imaged at 1 second intervals with TIRF microscopy. Representative 5-minute acquisitions, displayed at 30 frames per second (30× actual speed). Scale bar: 2 μm.
A mNeonGreen-FtsZ (bAB185) shows absence of Z ring constriction following depletion of Pbp2B. Cells were grown to mid-exponential phase with 10 μM IPTG, inducer was washed out from medium and cells were imaged after 1 hour of growth without IPTG. Images were acquired at 30 second intervals for 1.5 hours using epifluorescence illumination. Movie displayed at 30 frames per second (900× actual speed).
B mNeonGreen-FtsZ (bAB185) continues to show FtsZ filament motion following depletion of Pbp2B. Cells were imaged with 2 hours of Pbp2B depletion (after no Z ring constriction was detected). Cells were imaged at 1 second intervals using TIRF microscopy. Representative 5-minute acquisitions, displayed at 30 frames per second (30× actual speed). Scale bars: 2 μm.
A Low expression of mNeonGreen-FtsZ (bAB219) shows that single molecules of FtsZ are immobile at division sites. Cells were imaged at 500 millisecond intervals for 2 minutes by TIRF microscopy. Movie displayed at 30 frames per second (15× actual speed).
B Single molecules of FtsA-HaloTag(SW)-JF646 (bAB213) are immobile within division sites. Cells were imaged at 200 millisecond intervals for 1 minute using TIRF microscopy. Representative videos are displayed at 30 frames per second (6× actual speed). Scale bars: 2 μm.
A Inducible FtsA-mNeonGreen(SW)-FtsZ(D213A) (bAB217) titration showing motion of FtsA filaments in the presence of different induction levels of the GTPase-defective FtsZ(D213A). Cells were grown until mid-exponential phase, diluted and incubated 1 hour with 1 μM (top left), 5 μM (top right), 10 μM (bottom left) and 100 μM IPTG (bottom right). Cells were imaged at 3 second intervals for 5 minutes with TIRF microscopy. Movie displayed at 15 frames per second (45× actual speed).
B mNeonGreen-FtsZ (bAB185) showing motion of FtsZ filaments after the addition of MciZ peptide by perfusion in a CellAsic microfluidic chamber. Cells were imaged in CH medium (top left), then 10 minutes (top right) and 20 minutes (bottom left) after the addition of 1 μM MciZ. Sequentially, MciZ was washed out from cells for 10 minutes and cells were imaged again (bottom right). Cells were imaged at 3 second intervals for 5 minutes with TIRF microscopy. The exogenous addition (or expression) of MciZ causes two phases of effects on FtsZ dynamics: 1) Initially (between 0 and 10 minutes after exogenous addition or induction of MciZ), small FtsZ filaments move around the cell at a faster rate. During this period, all Z rings constricted faster than usual. At later time points (15–20 minutes), as the MciZ incubation time (or expression) increased, all FtsZ filaments and Z rings in the cell disappeared and did not return. Movie displayed at 15 frames per second (45× actual speed). Scale bars: 2 μm.
A mNeonGreen-FtsZ (bAB185) showing motion of FtsZ filaments during treatment with PC190723, added by perfusion in a CellAsic microfluidic chamber. Cells were imaged at 3 second intervals in CH medium for 10 minutes, PC190723 (10 μM) was flowed over the cells, and cells were imaged for 10 minutes (top right). After this, PC190723 was washed out, and cells were imaged for 20 minutes. Movie displayed at 30 frames per second (90× actual speed).
B JF549-HaloTag-Pbp2B (bGS31) showing Pbp2B single molecules by TIRF microscopy during PC190723 treatment. Cells were incubated with 50 pM of HaloTag-JF549 ligand for 15 minutes prior to imaging. Cells were imaged 18 minutes after the addition of 3 μl of 10 mg/ml PC190723 on top of a 500 μl 2% agarose pad. Snapshot images of mNeonGreen-FtsZ were used to mark the position of the division machinery (first 60 frames of the movie, white arrowheads). Cells were imaged at 1 second intervals for 4 minutes with TIRF microscopy. Movie displayed at 30 frames per second (30× actual speed). Scale bars: 2 μm.
FtsA-mNeonGreen(SW)-FtsZ(D213A) (bAB217) showing the growth of FtsA spirals by spinning-disk confocal microscopy. Cells were grown without inducer, placed under a 2% agarose CH pad containing 1 mM IPTG, and imaged at 30 second intervals for 2 hours. To make the transition from ring to spirals visible, histogram bleach correction was applied to all time lapses in Fiji to normalize signal between frames. Representative videos are displayed at 30 frames per second (900× actual speed). Scale bar: 2 μm.
mNeonGreen-Pbp2B rings within cells of different genetic backgrounds were followed over time by confocal microscopy. Cells were imaged at 1 minute intervals for 2 hours. 6 examples out of 12 different conditions are shown as follows: wild type (top left), FtsAZ overexpression (top right), MciZ overexpression (center left), native FtsZ(T111A) mutant (center right), FtsZ(D213A) mutant expressed with 5 μM IPTG (bottom left), FtsZ(D213A) mutant expressed with 10 μM IPTG (bottom right). Representative 30-minute videos are displayed at 8 frames per second (480× actual speed). Scale bar: 2 μm.
Acknowledgments
We thank F. Gueiros-Filho, R. Losick, M. Erb, and P. Levin for strains; J Xiao for discussions; L. Lavis for JF dyes; B. Murphy and E. Pasciak for FDAA synthesis help; F. Gueiros-Filho, R. Daniel and J. Errington for antibodies; R. Losick, B. LaSarre, and D. Kearns for comments. This work was supported by NIH grants GM113172 to MSV and YVB; GM51986 to YVB; DP2AI117923-01 to ECG; a Newcastle University Research Fellowship and Royal Society Research Grant RG150475 to SH; a Science Without Borders Research Fellowship to AWB-F; an ERC Advanced Grant SynDiv 669598 to CD, and an NSF GRFP (DGE1144152) to GRS. SIM was performed in the Indiana University Light Microscopy Imaging Center supported by S10RR028697-01
Footnotes
Movies S1–S10 available at http://garnerlab.fas.harvard.edu/FtsZ/
Code available at https://bitbucket.org/garnerlab/bisson-2016
Author Contributions
This work represents the combined research directions of 3 groups (EG, YB/MV, SH/CD). FDAA labeling and 3D-SIM by YH and EK. Strains constructed by AB, GS, and CJ. Confocal and TIRF imaging by AB and GS. Microholes designed by SH and CD, fabricated by FW, and imaged by SH and CJ. Programming by GS, SH and YS. All authors designed experiments and wrote the paper.
References
- 1.Chen Y, Erickson HP. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J Biol Chem. 2005;280:22549–22554. doi: 10.1074/jbc.M500895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szwedziak P, Wang Q, Bharat TAM, Tsim M, Lowe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuru E, et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lupoli TJ, et al. Transpeptidase-Mediated Incorporation of d-Amino Acids into Bacterial Peptidoglycan. J Am Chem Soc. 2011;133:10748–10751. doi: 10.1021/ja2040656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberhardt C, Kuerschner L, Weiss DS. Probing the catalytic activity of a cell division-specific transpeptidase in vivo with beta-lactams. J Bacteriol. 2003;185:3726–3734. doi: 10.1128/JB.185.13.3726-3734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thanedar S, Margolin W. FtsZ Exhibits Rapid Movement and Oscillation Waves in Helix-like Patterns in Escherichia coli. Current Biology. 2004;14:1167–1173. doi: 10.1016/j.cub.2004.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowlett VW, Margolin W. 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys J. 2014;107:L17–20. doi: 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coltharp C, Buss J, Plumer TM, Xiao J. Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci USA. 2016;113:E1044–E1053. doi: 10.1073/pnas.1514296113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holden SJ, et al. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci USA. 2014;111:4566–4571. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strauss MP, et al. 3D-SIM Super Resolution Microscopy Reveals a Bead-Like Arrangement for FtsZ and the Division Machinery: Implications for Triggering Cytokinesis. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm JB, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Meth. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu L, Yu J. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys J. 2008;95:2009–2016. doi: 10.1529/biophysj.108.128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redick SD, Stricker J, Briscoe G, Erickson HP. Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J Bacteriol. 2005;187:2727–2736. doi: 10.1128/JB.187.8.2727-2736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haydon DJ, et al. An Inhibitor of FtsZ with Potent and Selective Anti-Staphylococcal Activity. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 20.Bisson-Filho AW, et al. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc Natl Acad Sci USA. 2015;112:E2130–E2138. doi: 10.1073/pnas.1414242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X, et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell-wall synthesis. 2016:077610. doi: 10.1126/science.aak9995. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA. 2013;110:11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egan AJF, Vollmer W. The stoichiometric divisome: a hypothesis. Front Microbiol. 2015;6:0–6. doi: 10.3389/fmicb.2015.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nature protocols. 2015;10:33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang HC, Gayda RC. High-level expression of the FtsA protein inhibits cell septation in Escherichia coli K-12. Journal of bacteriology. 1990;172:4736–4740. doi: 10.1128/jb.172.8.4736-4740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dewar SJ, Begg KJ, Donachie WD. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araujo-Bazan L, Ruiz-Avila LB, Andreu D, Huecas S, Andreu JM. Cytological Profile of Antibacterial FtsZ Inhibitors and Synthetic Peptide MciZ. Frontiers in microbiology. 2016;7:1558. doi: 10.3389/fmicb.2016.01558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handler AA, Lim JE, Losick R. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol Microbiol. 2008;68:588–599. doi: 10.1111/j.1365-2958.2008.06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nick Perry J-YT, Schindelin Johannes. TrackMate. 2012 [Google Scholar]
- 31.Wu F, van Schie BGC, Keymer JE, Dekker C. Symmetry and scale orient Min protein patterns in shaped bacterial sculptures. Nature nanotechnology. 2015;10:719–726. doi: 10.1038/nnano.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannik J, et al. Robustness and accuracy of cell division in Escherichia coli in diverse cell shapes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6957–6962. doi: 10.1073/pnas.1120854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jong IG, Beilharz K, Kuipers OP, Veening J-W. Live Cell Imaging of Bacillus subtilis and Streptococcus pneumoniae using Automated Time-lapse Microscopy. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindelin J, et al. Fiji: an open source platform for biological image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ball G, et al. SIMcheck: a Toolbox for Successful Super-resolution Structured Illumination Microscopy. Scientific reports. 2015;5:15915. doi: 10.1038/srep15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman H. Simple violin plot using matlab default kernel density estimation. 2015 [Google Scholar]
- 37.Zoergiebel F. PLOTERR general error bar plot. 2008 [Google Scholar]
- 38.Bendezu FO, Hale CA, Bernhardt TG, de Boer PAJ. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. The EMBO journal. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee TK, Meng K, Shi H, Huang KC. Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nature communications. 2016;7:13170. doi: 10.1038/ncomms13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho H, et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nature microbiology. 2016:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blasios V, et al. Genetic and Biochemical Characterization of the MinC-FtsZ Interaction in Bacillus subtilis. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0060690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunst F, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 44.Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senesi S, et al. Surface-associated flagellum formation and swarming differentiation in Bacillus subtilis are controlled by the ifm locus. Journal of bacteriology. 2004;186:1158–1164. doi: 10.1128/JB.186.4.1158-1164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: The FDAA labeling pattern is independent of the FDAA type or the sequence in which they are used
A Sequential short-pulse labeling of FDAAs with reversed color sequence compared to Fig. 1B. This demonstrates the pattern of FDAA sequential labeling is independent of the sequence of FDAA labeling.
B Labeling cells with FDAAs for one 45-second pulse yields the same label intensity as labeling for three sequential 15-second pulses. This indicates that FDAA pulses of different durations are comparable, and that repeated pulse labeling with FDAAs is non-perturbing. ns: the comparison is not statistically significant by t-test.
C FDAA sequential labeling using different orders of dyes (indicated in diagrams). Discrete spots of FDAA labeling were seen in short-pulse labeling regardless of what FDAA color was used for the pulse. Images were background subtracted in Fiji (rolling ball radius = 10).
Colored bars indicate the time course of FDAA labeling in each experiment. All images were taken with 3D-SIM. Scale bars = 0.5 μm.
Figure S2: Quantitation of FDAA labeling from 3D-SIM data
A The fluorescence intensity measured from 3D-SIM imaging varies linearly with exposure times both at high (left) and low (right) laser powers, validating intensity-based quantitation. Plot shows the maximum intensity of TetraSpeck beads measured at varying exposure times for three different fields of view. Black lines are linear fits to each field of view.
B The area of FDAA-labeled features at the septum increases with increased labeling time (as in Fig. 1D) independent of the threshold cutoff used to binarize images. Labeled area is measured as the number of pixels in each region above a chosen intensity threshold. Fig. 1D shows that for one such threshold (45 units) FDAA labeling time and average labeled area are positively correlated. This graph shows the correlation coefficients for the complete range of possible threshold choices, indicating that this correlation holds regardless of threshold choice, except at the extreme values.
C The total amount of FDAA labeling at the septum increases with labeling time. Total amount of labeling is calculated as the sum of the fluorescence intensity in each image; distribution shown as violin plots, black lines: means of the distributions. r: correlation coefficient of means.
D Disruption of cell wall synthesis causes decreased FDAA labeling at the septum. To decrease cell wall synthesis, cells were treated with 4 μg/ml Vancomycin for 3 min (Van), 50 μg/ml Penicillin G for 3 min (PenG), or Pbp2B was depleted for 3 hours. Following perturbations, cells were labeled with 2 mM TADA for 2 minutes. (Left) representative TADA images for each condition. Representative images shown are those with the median intensity of their distribution. (Right) total FDAA intensity for each condition. Black lines indicate the mean.
Figure S3: Viability and fitness of native fusions
Native mNeonGreen fusions of Pbp2B (strain ME7), FtsA (strain bAB167), and FtsZ (strain bAB185) have similar fitness and protein levels compared to wild type cells (strain PY79).
A Growth curves of native fusion strains. Cells were grown in triplicate in LB until O.D.600 < 1.0, and then diluted to O.D. 600 = 0.1. Aliquots were taken every hour and O.D.600 was measured in a plate reader (Synergy HT plate reader, Bio-Tek).
B Cell length distributions. Cells were grown until mid-exponential phase in LB, concentrated and incubated for 1 minute with 1 μg/ml of FM5-95 dye, and imaged in a confocal spinning disk microscope. Cell length was measured using 250 cells for each strain.
C Cell viability from spot dilutions. Cells were grown until O.D.600 ~ 1.0 in LB, 5 μl of each culture was spotted from serial dilutions on LB plates, and incubated for 12 hours.
D Relative protein levels of native fusions compared to wild type assayed by Western blotting. Different lysate dilutions of each strain carrying its respective native fusion were blotted side by side with PY79 control. Cells were incubated with specific polyclonal antibodies for FtsZ, FtsA and Pbp2B, respectively. Bands were probed and visualized using a chemiluminescence system.
E Relative quantification of native mNeonGreen fusions. Mean values were obtained from the average of 3 independent experiments, and normalized to the measurements of the respective untagged band from PY79. FtsA-mNeonGreen:FtsA and mNeonGreen-Pbp2B:Pbp2B ratios show similar protein levels (0.98±0.11 and 0.99±0.07, respectively). For the mNeonGreen-FtsZ fusion, considerably lower mNeonGreen-FtsZ:FtsZ ratios were observed (0.71±0.08). However, a proteolysis product from mNeonGreen-FtsZ construct was observed in all blots, and the sum of the intensities of mNeonGreen-FtsZ and its proteolysis product gave a balanced ratio (1.07±0.09).
F Increased FtsAZ levels have no measurable effect on fitness or cell division, indicating that while overexpression of either FtsA or FtsZ alone may be toxic, balanced expression of both causes no observable cellular defects. Membrane staining of cells overexpressing a second copy of ftsZ (strain bAB96, bottom left panel), ftsA (bAB94, top right panel) or ftsAZ (bAB98, bottom right panel). The control (top left panel) is a representative image of bAB96 cells without FtsZ overexpression. Cells were grown in LB until mid-exponential phase, diluted into fresh medium with 100 μM IPTG (or 0 μM IPTG as a control for each culture) and grown for more 2 hours prior imaging. Images were acquired on a spinning-disk confocal microscope after cells were incubated with 1 μg/ml of FM5-95 dye for 30 seconds at room temperature. Because the overexpression of either FtsZ or FtsA alone affected cell length and width, most of our experiments using inducible constructs were performed by expressing both FtsA and FtsZ together, which had no such effects. Growth rates of cells overexpressing FtsAZ are shown in S7A (brown curve), with no observable change from wildtype.
Figure S4: Imaging FtsZ in microholes
A Scanning electron microscopy images of silicon micropillars nanofabricated through electron-beam lithography and reactive-ion etching.
B Schematic of the stamping process to produce agarose pads containing microholes.
C Schematic of B. subtilis in microholes. FtsZ filaments are shown in green.
D (i) Brightfield images of microholes without bacteria. (ii) Widefield epifluorescence images of B. subtilis (SH33) inoculated in microholes. Green indicates GFP-FtsZ, red indicates FM4-64-labeled cell membrane.
E Quantification of mNeonGreen-FtsZ filament dynamics in actively constricting vertically immobilized cells. FtsZ filaments move in both directions with equal probability. Vectorial velocities (i) and speeds (ii) of mNeonGreen-FtsZ filaments (strain SH41). Mean filament speed was 42 nm ± 23 nm/sec (mean ± S.D., N = 110). 25% of filaments were immobile (speed of ~ 0 nm/s).
F A FtsZ-GFP fusion has similar dynamics compared to mNeonGreen-FtsZ. Vertically immobilized cells (strain SH33) display multiple FtsZ-GFP filaments with independent directional motion around the division site. (top) Cropped rings in early (left), mid (center), and late (right) divisional stages. (bottom). Blue arrow indicates start of a directional FtsZ track. Scale bars = 0.5 μm.
Figure S5: Divisome imaging and particle tracking
A Pbp2B-mNeonGreen expressed at native levels (ME7) shows Pbp2B motion at the Z ring, imaged by TIRF microscopy. Pbp2B can be seen throughout the cell, but localizes preferentially to the Z ring. Indicated kymograph (right) was drawn at the arrowhead. Scale bars: 1 μm (cell), 0.5 μm (kymograph).
B Pbp2B-mNeonGreen expressed at low levels (bGS3) shows Pbp2B motion at the Z ring as imaged by TIRF microscopy. Cells were grown in 2 μM IPTG to induce low levels of Pbp2B expression. This decreased the overall signal density, allowing dynamics to be resolved more clearly. Kymographs are drawn at arrowheads. Scale bars: 1 μm (cell), 0.5 μm (kymograph).
C Particle tracking shows directional motion of FtsZ filaments (left), FtsA filaments (center), and Pbp2B single molecules (right). mNeonGreen-FtsZ (bAB185), FtsA-mNeonGreen (bAB167), and Halo-Pbp2B (bGS31) labeled with 40 μM JF-549 were imaged by TIRF microscopy. Particles were identified and tracked using TrackMate and analyzed using custom MATLAB code. Tracks are overlaid on a frame from each time lapse, and are colored blue to yellow along their duration. Scale bar: 1 μm.
D Particle tracking shows that FtsA, FtsZ, and Pbp2B all move at the same speed. Particle tracks were filtered to select for directional motion, and velocity was calculated from fits of particle tracks to MSD(t)=4D(t)+(Vt)2 (15). The small difference in the means of these distributions compared to the velocity distributions in Fig. 2G (velocity measurement by kymograph) is likely due to differences in the two methods of measuring velocity. However, the velocities of FtsA, FtsZ and Pbp2B agree with each other when assayed using two independent velocity measurements, reinforcing the conclusion that they move at the same speed.
E Distributions of the scaling coefficient α for particle tracks of FtsA, FtsZ, and Pbp2B indicates that they move directionally. α was calculated as described previously (15). This distribution of α values is consistent with directed motion (1 < α < 2).
F Distributions of the velocity of unfiltered FtsZ and A particle tracks calculated from fits of (MSD vs t) to MSD(t)=4D(t)+(Vt)2 for both single molecules (solid lines) and filaments (dashed lines). FtsA and FtsZ filament trajectories show a mobile population (visible as a second peak in the 10–20 nm/s region), whereas the motions of single molecules show a negligible value for directional motion (~ 10−8 nm/s).
G Detailed velocity distributions of FtsZ and FtsA single molecules and filaments. Left: Distributions of all velocities less than 10−7 nm/s, representing the non-moving population of particles. Right: Distributions of all velocities greater than 10−7 nm/s, representing the mobile population. Single molecules of FtsA and FtsZ have more non-moving particles and very few moving particles compared to filaments, indicating that, while filaments of FtsZ and FtsA move, single molecules do not.
H Distributions of the diffusion coefficients for unfiltered FtsZ and FtsA particle tracks calculated from fits of (MSD vs t) to MSD(t)=4D(t)+(Vt)2. Single molecules of mNeonGreen-FtsZ (bAB219) and Halo-FtsA (bAB213) labeled with 100 pM JF-549 were imaged by TIRF microscopy. Particles were tracked and diffusion coefficients calculated as described above. The very low value of the diffusion coefficient is consistent with immobile particles, and similar to that of immobile class A PBPs measured in E. coli and B. subtilis (39, 40).
For panels F–H, n = 2,695 (FtsZ single molecules), 10,358 (FtsA single molecules), 4,718 (FtsZ filaments), 7,164 (FtsA filaments).
I Single-step photobleaching of FtsZ and FtsA single molecules. Intensity traces are aligned such that the frame in which the molecule appears is time = 0. Faded, thin lines show the raw data; the bold lines are data which has been filtered to reduce noise. The fluorescence signal drops to baseline in a single time step, indicating that these are single molecules.
J Simultaneous imaging of directionally-moving FtsZ filaments and immobile FtsA single molecules. Dual-color TIRF imaging of native levels of mNeonGreen-FtsZ (green) and single molecules of FtsA-HaloTag-JF549 (red). Images were taken at 2 second intervals with 250 ms exposure times.
K Pbp2B velocity correlates with FtsAZ filament velocity. JF549-HaloTag-Pbp2B single molecules were imaged under 5 different conditions, their velocities measured by kymographs and plotted against the FtsAZ filament velocity in that same condition. Pbp2B velocities (mean ± S.D.): wild type (strain bGS31, n = 152): 32.6 ± 9.1 nm/sec; FtsA overexpression (strain bAB310 with 20 μM IPTG, n = 87): 22.0 ± 5.8 nm/sec; FtsZ(D213A) (strain bGS90 with 100 μM IPTG): no motion detected; FtsZ(T111A) (strain bGS60, n = 108): 20.7 ± 5.8 nm/s; PC190723 treatment (strain bGS31 with 10 μM PC190723): no motion detected.
Figure S6: FDAA labeling with FtsAZ velocity perturbations
A Treatment with high concentrations of PC190723 causes PG synthesis to localize to small patches along the cell, mirroring the localization of FtsZ following similar treatment. Cells were treated with 10 μg/ml PC190723 for 10 minutes, and labeled with TADA as shown. A cell not treated with PC190723 that was labeled equivalently with FDAAs is shown for comparison. Scale bar: 1 μm.
B The trends observed in Fig. 4B are independent of threshold used for analysis. For each perturbation (PC190723, FtsZ(D213A), MciZ), data plotted is the mean FDAA-labeled area with perturbation minus the mean FDAA-labeled area without perturbation. Negative values of the curves indicate that the perturbation decreases labeled area, while positive values indicate that the perturbation increases labeled area. These measurements are consistent across all possible thresholds for labeling except extreme values, indicating that the trends reported in Fig. 4B are independent of threshold choice.
C Blinded classification of FDAA labeling under various FtsZ perturbations. This demonstrates that perturbations that decrease FtsZ velocity also cause more heterogeneous labeling (more spots and arcs, fewer complete rings), whereas perturbations that increase FtsZ velocity yield a more uniform distribution of FDAA signal. + indicates sample with perturbation, – indicates unperturbed sample for comparison. Note that the labeling and imaging conditions varied for the three experiments shown here, explaining the variation between the three unperturbed samples. See Table S1 for details.
Figure S7: Perturbation of FtsAZ dynamics affects cytokinesis time, but not growth rate
A Violin distributions of (left) mNeonGreen-Pbp2B constriction times (measured from kymographs) and (right) FtsAZ filament velocities under different conditions. Cells were grown and imaged as described in methods.
B Comparison between constriction times of Pbp2B rings measured by kymograph (Kymo) and automated tracking (Track) analysis. Scatter plot of constriction time distributions was obtained as described in methods; statistics are shown in Table S4.
C Distribution of constriction times obtained from cells under slow growth conditions (S750-glucose at 25°C). Perturbations that changed FtsAZ filament motion and constriction times in rich media at 37°C had the same effect in minimal media at 25°C, either slowing down (FtsZ(T111A) - strain bAB270, FtsZ(D213A) - strain bAB273 with 10 μM IPTG) or accelerating (MciZ overexpression - strain bAB274 with 5 μL of 3 M xylose on top of the pad) the rate of cytokinesis.
D Bulk growth curves of cells under all FtsZ perturbations that significantly changed constriction rates. Cells were grown in LB until OD600 < 1.0, and then diluted to OD600 = 0.1 and split into 8 replicates. Growth curves were taken in a plate reader (Epoch-2 plate reader, Biotek) with continuous shaking at 37°C.
E Linear segments from growth curves in D (40 to 110 minutes). Curves were converted into a ln(OD600) vs. time (min) plot, and doubling times were then calculated using linear regression. Doubling times (mean ± S.D.): WT (strain PY79): 21.3 ± 2.6 min; FtsZ(T111A) (strain bAB270): 20.7 ± 2.7 min; FtsZ(D213A) (5 μM IPTG, strain bAB273): 20.2 ± 1.45 min; FtsZ(D213A) (10 μM IPTG, strain bAB273): 19.4 ± 2.1 min; FtsA overexpression (100 μM IPTG, strain bAB199): 21.0 ± 2.1 min; and MciZ overexpression (60 mM xylose, strain bAB274): 20.4 ± 1.9 min.
F Representative growth rate curves extracted from single cell time lapses. Doubling times (mean ± S.D.): wild type (strain PY79): 24.8 ± 1.4 min (n = 14); FtsZ(T111A) (strain bAB270): 22.3 ± 2.1 min (n = 29); FtsZ(D213A) (10 μM IPTG, strain bAB273, 10 μM IPTG): 24.3 ± 2.2 min (n = 18).
Table S1: FDAA experimental conditions
Table S2: Strains and oligonucleotides used in this study Strain
Table S3: Live-cell imaging conditions
Table S4: Measurements of FtsAZ filament velocities and Pbp2B ring constriction periods with different genetic perturbations, growth conditions and measurement methods
Movie S1: FtsA and FtsZ filaments move directionally together inside and outside of the division site. Cells were imaged using TIRF microscopy at 3 second intervals. Representative 5-minute acquisitions, displayed at 15 frames per second (45× actual speed). Scale bar: 2 μm.
Column 1: mNeonGreen-FtsZ (bAB185) showing filament motion.
Column 2: FtsA-mNeonGreen(SW) (bAB167) showing filament motion.
Column 3: FtsA-HaloTag(SW)-JF549 (red) and mNeonGreen-FtsZ (green) showing filament motion. bAB229 cells were incubated with 500 nM of HaloTag-JF549 ligand for 15 minutes.
Column 4: mNeonGreen-FtsZ (bAB221) showing filament motion with FtsAZ overexpressed. Cells were grown with 100 μM IPTG for 1 hour.
mNeonGreen-FtsZ (strain SH41) dynamics in vertically immobilized cells with 600–1000 nm radii. Cells were imaged by epifluorescence at 1 second intervals. 2-minute acquisitions displayed at 15 frames per second (15× actual speed). Scale bar: 1 μm.
A. JF549-HaloTag-Pbp2B (bGS31) shows Pbp2B single molecules by TIRF microscopy. Cells were incubated with 50 pM of HaloTag-JF549 ligand for 15 minutes, washed, and prepared as indicated in methods. Cells were imaged at 1 second intervals using TIRF microscopy. Images of mNeonGreen-FtsZ were used to mark the position of the division machinery (first 60 frames of the movie) before imaging of JF549. Arrowheads indicate Z rings that colocalize with directionally moving Pbp2B. 4-minute acquisition displayed at 30 frames per second (30× actual speed).
B Overexpression of exogenous mNeonGreen-Pbp2B (100μM IPTG, bAB146) shows diffusion of Pbp2B molecules along the cell by TIRF microscopy. Cells were imaged at 100 millisecond intervals. 10-second acquisition displayed at 30 frame per second (3× actual speed). Scale bar: 2 μm.
FtsA-mNeonGreen(SW) (bAB167) showing filament motion during perturbations to cell wall synthesis. Antibiotics were added to the top of the pad 5 minutes before imaging. Images shown as follows: no antibiotic control (top left); 50 mg/μl Vancomycin (top right); 10 mg/μl Ampicillin (center left); 10 mg/μl Penicillin G (center right); 100 mg/μl Cephalexin (bottom left) and 100 mg/μl Fosfomycin (bottom right). Cells were imaged at 1 second intervals with TIRF microscopy. Representative 5-minute acquisitions, displayed at 30 frames per second (30× actual speed). Scale bar: 2 μm.
A mNeonGreen-FtsZ (bAB185) shows absence of Z ring constriction following depletion of Pbp2B. Cells were grown to mid-exponential phase with 10 μM IPTG, inducer was washed out from medium and cells were imaged after 1 hour of growth without IPTG. Images were acquired at 30 second intervals for 1.5 hours using epifluorescence illumination. Movie displayed at 30 frames per second (900× actual speed).
B mNeonGreen-FtsZ (bAB185) continues to show FtsZ filament motion following depletion of Pbp2B. Cells were imaged with 2 hours of Pbp2B depletion (after no Z ring constriction was detected). Cells were imaged at 1 second intervals using TIRF microscopy. Representative 5-minute acquisitions, displayed at 30 frames per second (30× actual speed). Scale bars: 2 μm.
A Low expression of mNeonGreen-FtsZ (bAB219) shows that single molecules of FtsZ are immobile at division sites. Cells were imaged at 500 millisecond intervals for 2 minutes by TIRF microscopy. Movie displayed at 30 frames per second (15× actual speed).
B Single molecules of FtsA-HaloTag(SW)-JF646 (bAB213) are immobile within division sites. Cells were imaged at 200 millisecond intervals for 1 minute using TIRF microscopy. Representative videos are displayed at 30 frames per second (6× actual speed). Scale bars: 2 μm.
A Inducible FtsA-mNeonGreen(SW)-FtsZ(D213A) (bAB217) titration showing motion of FtsA filaments in the presence of different induction levels of the GTPase-defective FtsZ(D213A). Cells were grown until mid-exponential phase, diluted and incubated 1 hour with 1 μM (top left), 5 μM (top right), 10 μM (bottom left) and 100 μM IPTG (bottom right). Cells were imaged at 3 second intervals for 5 minutes with TIRF microscopy. Movie displayed at 15 frames per second (45× actual speed).
B mNeonGreen-FtsZ (bAB185) showing motion of FtsZ filaments after the addition of MciZ peptide by perfusion in a CellAsic microfluidic chamber. Cells were imaged in CH medium (top left), then 10 minutes (top right) and 20 minutes (bottom left) after the addition of 1 μM MciZ. Sequentially, MciZ was washed out from cells for 10 minutes and cells were imaged again (bottom right). Cells were imaged at 3 second intervals for 5 minutes with TIRF microscopy. The exogenous addition (or expression) of MciZ causes two phases of effects on FtsZ dynamics: 1) Initially (between 0 and 10 minutes after exogenous addition or induction of MciZ), small FtsZ filaments move around the cell at a faster rate. During this period, all Z rings constricted faster than usual. At later time points (15–20 minutes), as the MciZ incubation time (or expression) increased, all FtsZ filaments and Z rings in the cell disappeared and did not return. Movie displayed at 15 frames per second (45× actual speed). Scale bars: 2 μm.
A mNeonGreen-FtsZ (bAB185) showing motion of FtsZ filaments during treatment with PC190723, added by perfusion in a CellAsic microfluidic chamber. Cells were imaged at 3 second intervals in CH medium for 10 minutes, PC190723 (10 μM) was flowed over the cells, and cells were imaged for 10 minutes (top right). After this, PC190723 was washed out, and cells were imaged for 20 minutes. Movie displayed at 30 frames per second (90× actual speed).
B JF549-HaloTag-Pbp2B (bGS31) showing Pbp2B single molecules by TIRF microscopy during PC190723 treatment. Cells were incubated with 50 pM of HaloTag-JF549 ligand for 15 minutes prior to imaging. Cells were imaged 18 minutes after the addition of 3 μl of 10 mg/ml PC190723 on top of a 500 μl 2% agarose pad. Snapshot images of mNeonGreen-FtsZ were used to mark the position of the division machinery (first 60 frames of the movie, white arrowheads). Cells were imaged at 1 second intervals for 4 minutes with TIRF microscopy. Movie displayed at 30 frames per second (30× actual speed). Scale bars: 2 μm.
FtsA-mNeonGreen(SW)-FtsZ(D213A) (bAB217) showing the growth of FtsA spirals by spinning-disk confocal microscopy. Cells were grown without inducer, placed under a 2% agarose CH pad containing 1 mM IPTG, and imaged at 30 second intervals for 2 hours. To make the transition from ring to spirals visible, histogram bleach correction was applied to all time lapses in Fiji to normalize signal between frames. Representative videos are displayed at 30 frames per second (900× actual speed). Scale bar: 2 μm.
mNeonGreen-Pbp2B rings within cells of different genetic backgrounds were followed over time by confocal microscopy. Cells were imaged at 1 minute intervals for 2 hours. 6 examples out of 12 different conditions are shown as follows: wild type (top left), FtsAZ overexpression (top right), MciZ overexpression (center left), native FtsZ(T111A) mutant (center right), FtsZ(D213A) mutant expressed with 5 μM IPTG (bottom left), FtsZ(D213A) mutant expressed with 10 μM IPTG (bottom right). Representative 30-minute videos are displayed at 8 frames per second (480× actual speed). Scale bar: 2 μm.