Abstract
To elucidate cellular machinery on a global scale, we performed a multiple comparison of the recently available protein–protein interaction networks of Caenorhabditis elegans, Drosophila melanogaster, and Saccharomyces cerevisiae. This comparison integrated protein interaction and sequence information to reveal 71 network regions that were conserved across all three species and many exclusive to the metazoans. We used this conservation, and found statistically significant support for 4,645 previously undescribed protein functions and 2,609 previously undescribed protein interactions. We tested 60 interaction predictions for yeast by two-hybrid analysis, confirming approximately half of these. Significantly, many of the predicted functions and interactions would not have been identified from sequence similarity alone, demonstrating that network comparisons provide essential biological information beyond what is gleaned from the genome.
Keywords: comparative analysis, multiple alignment, protein network, yeast two-hybrid
A major challenge of postgenomic biology is to understand the complex networks of interacting genes, proteins, and small molecules that give rise to biological form and function. Advances in whole-genome approaches are now enabling us to characterize these networks systematically, by using procedures such as the two-hybrid assay (1) and protein coimmunoprecipitation (2) to screen for protein–protein interactions. To date, these technologies have generated large interaction networks for bacteria (3), yeast (4–7), and, recently, fruit fly (8) and nematode worm (9).
The large amount of protein interaction data now available presents opportunities and challenges in understanding evolution and function. Such challenges involve assigning functional roles to interactions (10), separating true protein–protein interactions from false positives (11), and, ultimately, organizing large-scale interaction data into models of cellular signaling and regulatory machinery. As is often the case in biology, an approach based on evolutionary cross-species comparisons provides a valuable framework for addressing these challenges. However, although methods for comparing DNA and protein sequences have been a mainstay of bioinformatics over the past 30 years, development of similar tools at other levels of biological information, including protein interactions (12–14), metabolic networks (15–17), or gene expression data (18–20), is just beginning.
Recently, we devised a method called pathblast (13) for comparing the protein interaction networks of two species. Just as blast performs rapid pairwise alignment of protein sequences (21), pathblast is based on efficient alignment of two protein networks to identify conserved network regions. Here, we extend this approach to present a computational framework for alignment and comparison of more than two protein networks. We apply this multiple network alignment strategy to compare the recently available protein networks for worm, fly, and yeast, and show that although any single network contains false-positive interactions, embedded beneath this noise are a repertoire of protein interaction complexes and pathways conserved across all three species.
Methods
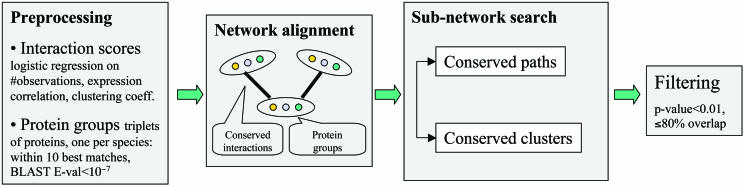
We developed a general framework for comparison and analysis of multiple protein networks. Full details are provided in Supporting Text, Figs. 5–11, and Tables 3–6, which are published as supporting information on the PNAS web site. Briefly, this process integrates interactions with sequence information to generate a network alignment graph. Each node in the graph consists of a group of sequence-similar proteins, one from each species; each link between a pair of nodes represents conserved protein interactions between the corresponding protein groups (Fig. 1). A search over the network alignment is performed to identify two types of conserved subnetwork structures: short linear paths of interacting proteins, which model signal transduction pathways, and dense clusters of interactions, which model protein complexes.
Fig. 1.
Schematic of the multiple network comparison pipeline. Raw data are preprocessed to estimate the reliability of the available protein interactions and identify groups of sequence-similar proteins. A protein group contains one protein from each species and requires that each protein has a significant sequence match to at least one other protein in the group (blast E value < 10–7; considering the 10 best matches only). Next, protein networks are combined to produce a network alignment that connects protein similarity groups whenever the two proteins within each species directly interact or are connected by a common network neighbor. Conserved paths and clusters identified within the network alignment are compared to those computed from randomized data, and those at a significance level of P < 0.01 are retained. A final filtering step removes paths and clusters with >80% overlap.
The search is guided by reliability estimates for each protein interaction (computed based on a method by Bader et al., ref. 22), which are combined into a probabilistic model for scoring candidate subnetworks. Under the model, a log likelihood ratio score is used to compare the fit of a subnetwork to the desired structure (path or cluster) versus its likelihood given that each species' interaction map was randomly constructed. The underlying model assumptions are that (i) in a real subnetwork, each interaction should be present independently with high probability, and (ii) in a random subnetwork, the probability of an interaction between any two proteins depends on their total number of connections in the network.
The search algorithm exhaustively identifies high-scoring subnetwork seeds and expands them in a greedy fashion. The significance of the identified subnetworks is evaluated by comparing their scores to those obtained on randomized data sets, in which each of the interaction networks is shuffled along with the protein similarity relationships between them.
Results
We applied the multiple network alignment framework (Fig. 1) to perform a three-way alignment of the protein–protein interaction networks of Caenorhabditis elegans, Drosophila melanogaster, and Saccharomyces cerevisiae. These species span the largest sets of protein interactions in the public databases to-date and, along with mouse, comprise the major model organisms used to study cellular physiology, development, and disease. Protein interaction data were obtained from the Database of Interacting Proteins (23) (February 2004 download) and contained 14,319 interactions among 4,389 proteins in yeast, 3,926 interactions among 2,718 proteins in worm, and 20,720 interactions among 7,038 proteins in fly. Protein sequences obtained from the Saccharomyces Genome Database (24), WormBase (25), and FlyBase (26) were combined with the protein interaction data to generate a network alignment of 9,011 protein similarity groups and 49,688 conserved interactions for the three networks.
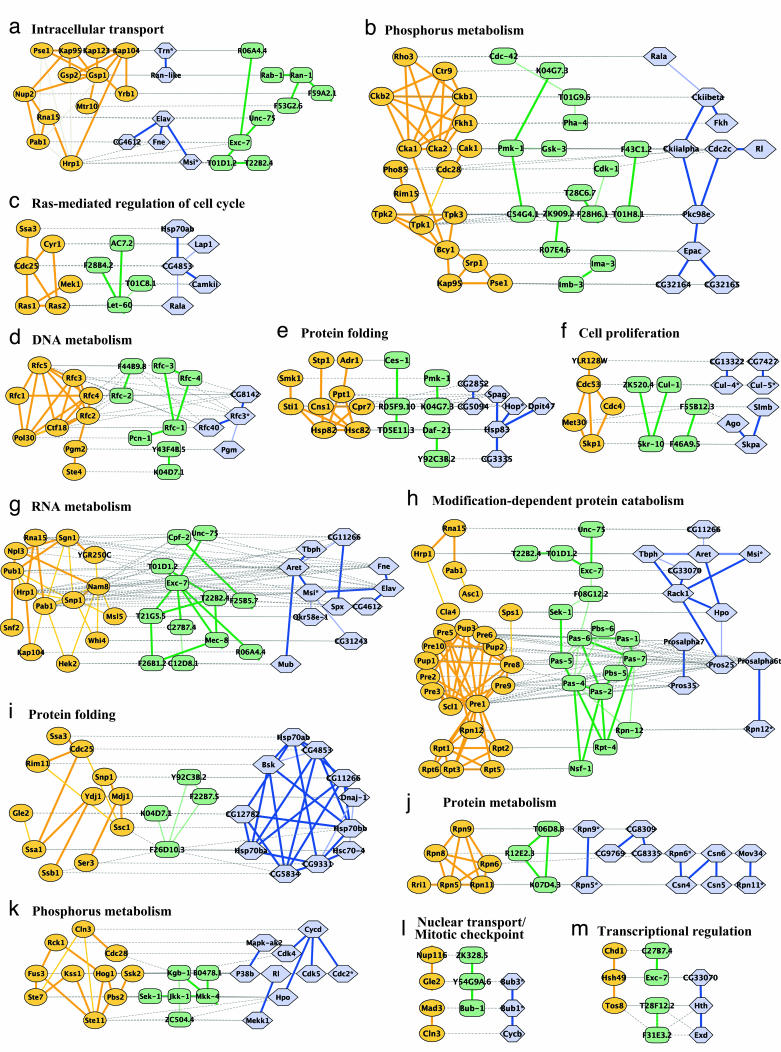
A search over the network alignment identified 183 protein clusters and 240 paths conserved at a significance level of P < 0.01. These covered a total of 649 proteins among yeast, worm, and fly. Representative examples of conserved clusters and paths are shown in Fig. 2. The identified conserved clusters and paths, along with their graphical layouts, are available from the authors upon request.
Fig. 2.
Representative conserved network regions. Shown are conserved clusters (a–k) and paths (l and m) identified within the networks of yeast, worm, and fly. Each region contains one or more overlapping clusters or paths (see Fig. 3). Proteins from yeast (orange ovals), worm (green rectangles), or fly (blue hexagons) are connected by direct (thick line) or indirect (connection via a common network neighbor; thin line) protein interactions. Horizontal dotted gray links indicate cross-species sequence similarity between proteins (similar proteins are typically placed on the same row of the alignment). Automated layout of network alignments was performed by using a specialized plug-in to the cytoscape software (34) as described in Supporting Text.
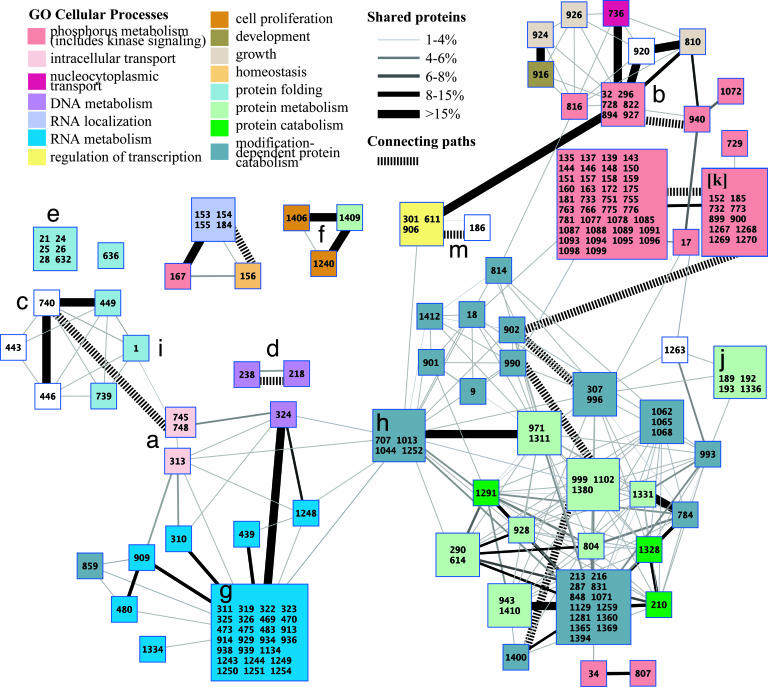
Fig. 3 shows a global map of all clusters and paths conserved among the yeast, worm, and fly protein networks. The map shows evidence of modular structure, groups of conserved clusters overlap to define 71 distinct network regions, most enriched for one or more well defined biological functions. The largest numbers of conserved clusters were involved in protein degradation (green boxes at lower right), RNA polyadenylation and splicing (blue boxes at lower left), and protein phosphorylation and signal transduction (red boxes at upper right). Other significant conserved clusters were involved in DNA synthesis, nuclear-cytoplasmic transport, and protein folding. The map also reveals conserved links between different biological processes, for instance linking kinase signaling (red) to protein catabolism (green; lower right) or to regulation of transcription (yellow; upper middle).
Fig. 3.
Modular structure of conserved clusters among yeast, worm, and fly. Multiple network alignment revealed 183 conserved clusters, organized into 71 network regions represented by colored squares. Regions group together clusters that share >15% overlap with at least one other cluster in the group and are all enriched for the same GO cellular process (P < 0.05 with the enriched processes indicated by color). Cluster ID numbers are given within each square; numbers are not sequential because of filtering. Solid links indicate overlaps between different regions, where thickness is proportional to the percentage of shared proteins (intersection/union). Hashed links indicate conserved paths that connect clusters together. Labels a–k and m mark the network regions exemplified in Fig. 2.
To validate our results, we compared these conserved clusters to known complexes in yeast as annotated by the Munich Information Center for Protein Sequences (MIPS) (27). We only considered MIPS complexes that were manually annotated independently from the Database of Interacting Proteins interaction data (i.e., excluding complexes in MIPS category 550 that are based on high-throughput experiments). Overall, the network alignment contained 486 annotated yeast proteins spanning 57 categories at level 3 of the MIPS hierarchy. We defined a cluster to be pure if it contained three or more annotated proteins and at least half of these shared the same annotation. Ninety-four percent of the conserved clusters were pure, indicating the high specificity of our approach, compared to a lower percentage of 83% when applying a noncomparative variant of our method to data from yeast only (i.e., applying the same methodology to search for high-scoring clusters within the yeast network only).
We further checked whether the conserved clusters were biased by spurious interactions, resulting from “sticky” proteins that lead to positive two-hybrid tests without interaction. Of 39 proteins with >50 network neighbors, only 10 were included in conserved clusters. These 10 proteins were involved in 60 intracluster interactions, 85% of which were supported by coimmunoprecipitation experiments. This finding indicates that the clusters were not biased because of artifacts of the yeast two-hybrid assays.
Three-Way Versus Two-Way Network Alignments. In addition to the three-way comparison, we also performed all possible pairwise network alignments: yeast/worm, yeast/fly, and worm/fly. This process identified 220 significant conserved clusters for yeast/worm, 835 for yeast/fly, and 132 for worm/fly. Several examples of these are shown in Fig. 9. Global overviews of the pairwise conserved clusters (similar to Fig. 3) are provided in Figs. 6–8.
Analysis of the proteins shared among the different pairwise and three-way network comparisons led to two general findings. First, the density and number of conserved clusters found in the yeast/fly comparison were considerably greater than for the other comparisons, because of the large amounts of interaction data for these species relative to worm (see Table 6 and Fig. 11). Second, the worm/fly conserved clusters were largely distinct from the clusters arising from the other analyses. For example, only 29% of the proteins in the worm/fly clusters were assigned to conserved clusters in the three-way analysis (135 of 462). This observation is consistent with the closer taxonomic relationship of worm and fly compared to yeast and the particular selection of protein “baits” for the C. elegans protein-protein interaction screen: roughly one-quarter were specifically chosen to be metazoan specific, and almost two-thirds had no clear yeast ortholog (9).
Prediction of Protein Functions. Conserved subnetworks that contain many proteins of the same known function suggest that their remaining proteins also have that function. Based on this concept, we predicted protein functions whenever the set of proteins in a conserved cluster or path (combined over all species) was significantly enriched for a particular Gene Ontology (GO) (28) annotation (P < 0.01) and at least half of the annotated proteins in the cluster or path had that annotation. When these criteria were met, all remaining proteins in the subnetwork were predicted to have the enriched GO annotation (see Supporting Text).
This process resulted in 4,669 predictions of previously undescribed GO Biological Process annotations spanning 1,442 distinct proteins in yeast, worm, and fly; and 3,221 predictions of GO Molecular Function annotations spanning 1,120 proteins. We estimated the specificity of these predictions by using cross validation, in which one hides part of the data, uses the rest of the data for prediction, and tests the prediction success by using the held-out data (see Supporting Text). As shown in Table 1, depending on the species, 58–63% of our predictions of GO Processes agreed with the known annotations (see also Tables 3 and 4). This analysis outperformed a sequence-based method of annotating proteins based on the known functions of their best sequence matches, for which the accuracy ranged between 37% and 53% (see Supporting Text). The complete list of protein function predictions is provided in Table 7, which is published as supporting information on the PNAS web site.
Table 1. Cross-validation results for protein cellular process prediction.
| Species | No. correct | No. of predictions | Success rate, % |
|---|---|---|---|
| Yeast | 114 | 198 | 58 |
| Worm | 57 | 95 | 60 |
| Fly | 115 | 184 | 63 |
For each species, the number of correct predictions, the total number of predictions, and the success rate in 10-fold cross-validation are listed.
Prediction of Protein Interactions. We also used the multiple network alignment to predict protein–protein physical interactions. We predicted an interaction between a pair of proteins based on (i) evidence that proteins with similar sequences interact within other species (directly or by a common network neighbor) and, optionally, (ii) cooccurrence of these proteins in the same conserved cluster or path. The accuracy of these predictions was evaluated by using 5-fold cross validation, as described in Supporting Text. In cross validation, strategy i achieved 77–84% specificity and 23–50% sensitivity, depending on the species for which the predictions were made (see Tables 2 and 5). These results were highly significant for the three species. Combining both strategies resulted in eliminating virtually all false positive predictions (specificity, >99%), while greatly reducing the number of true positives, yielding sensitivities of 10% and lower (see Table 2). Given the elevated specificity of the combined strategies, we were able to predict 176 previously undescribed interactions for yeast, 1,139 for worm, and 1,294 for fly with high confidence. Thus, although protein interactions have been used previously to predict interactions among the orthologous proteins of other species (9, 29), screening these against conserved paths and clusters markedly improves the specificity of prediction. The complete list of predicted protein interactions is provided in Table 8, which is published as supporting information on the PNAS web site.
Table 2. Cross-validation results for protein interaction prediction.
| Species | Sensitivity, % | Specificity, % | P value | Strategy |
|---|---|---|---|---|
| Yeast | 50 | 77 | 1.1e-25 | i |
| Worm | 43 | 82 | 1e-13 | i |
| Fly | 23 | 84 | 5.3e-5 | i |
| Yeast | 9 | 99 | 1.2e-6 | i + ii |
| Worm | 10 | 100 | 6e-4 | i + ii |
| Fly | 0.4 | 100 | 0.5 | i + ii |
For each species, the specificity and sensitivity of the predictions in 5-fold cross-validation, the significance of the results, and the prediction strategy (see text) are listed.
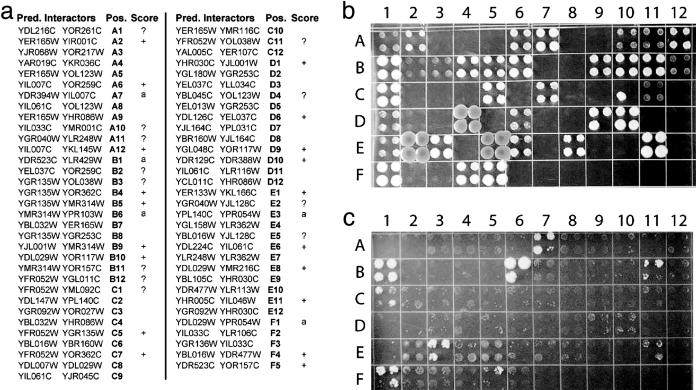
To further evaluate the utility of protein interaction prediction based on network conservation, we tested experimentally 65 of the interactions that were predicted for yeast by using the combined strategies i and ii above (Fig. 4a). The tests were performed by using two-hybrid assays (1, 4), which are based on a reporter gene that is transcriptionally activated if the two tested proteins (bait and prey) can physically interact (see Supporting Text and Fig. 4b). Five of the tests involved baits that induced reporter activity in the absence of any prey (Fig. 4c). Of the remaining 60 putative interactions, 31 tested positive (more conservatively, 19 of 48, see Fig. 4), yielding an overall success rate in the range of 40–52%.
Fig. 4.
Verification of predicted interactions by two-hybrid testing. (a) Sixty-five pairs of yeast proteins were tested for physical interaction based on their cooccurrence within the same conserved cluster and the presence of orthologous interactions in worm and fly. Each protein pair is listed along with its position on the agar plates shown in b and c and the outcome of the two-hybrid test. (b) Raw test results are shown, with each protein pair tested in quadruplicate to ensure reproducibility. Protein 1 vs. 2 of each pair was used as prey vs. bait, respectively. (c) This negative control reveals activating baits, which can lead to positive tests without interaction. Protein 2 of each pair was used as bait, and an empty pOAD vector was used as prey. Activating baits are denoted by “a” in the list of predictions shown in a. Positive tests with weak signal (e.g., A1) and control colonies with marginal activation are denoted by “?” in a; colonies D4, E2, and E5 show evidence of possible contamination and are also marked by a “?”. Discarding the activating baits, 31 of 60 predictions tested positive overall. A more conservative tally, disregarding all results marked by a “?,” yields 19 of 48 positive predictions.
Discussion
Comparison to Existing Methods. We have developed pairwise network alignment algorithms that were used to detect linear paths (13) or dense clusters (14) that are conserved between yeast and the bacteria Helicobacter pylori. The multiple network alignment scheme that we have presented here is an extension of our earlier approaches to handle more than two species. Additional advantages of the current approach over the previous approaches are: (i) it is a unified method to detect both paths and clusters, which generalizes to other network structures; (ii) this approach incorporates a refined probabilistic model for protein interaction data; and (iii) it includes an automatic system for laying out and visualizing the resulting conserved subnetworks.
A related method that uses cross-species data for predicting protein interactions is the interolog approach (12, 18): a pair of proteins in one species is predicted to interact if their best sequence matches in another species were reported to interact. In comparison, our proposed scheme can associate proteins that are not necessarily each other's best sequence match. This advantage confers increased flexibility in detecting conserved function by allowing for paralogous family expansion and contraction or gene loss. Because conservation is evaluated in the context of a protein interaction subnetwork and not independently for each interaction, the high specificity of the resulting predictions can be maintained (see below).
Best blast Hits May Not Imply Functional Conservation. Frequently, the network alignment associates sequence-similar proteins between species even though they are not each other's best sequence match. For instance, the conserved network region in Fig. 2h suggests that the worm protein exc-7 plays the same functional role as yeast Pab1 and fly CG33070 (blast E-value ≈ 10–42) based on the conserved interactions with Asc1/F08G12.2/Rack1 (yeast/worm/fly), Rna15/Unc-75 (yeast/worm), and T01D1.2/Tbph (worm/fly). However, CG33070 is only the fifth best blast match in fly overall (the best being CG3151 at E value ≈ 10–70).
Overall, of the 679 protein triples aligned at the same position within a three-way conserved cluster, only 177 contained at least one pair of best sequence matches; of the 129 additional triples in conserved paths, only 31 contained best sequence matches. Clearly, in some cases, the best matches are not present within conserved clusters because of missing interactions in the protein networks of one or more species. However, it is unlikely that true interactions with the best-matching proteins would be missed repeatedly across multiple proteins in a cluster and across multiple species. These observations suggest that protein network comparisons provide essential information about function conservation.
Functional Links Within Conserved Networks. Conserved network regions enriched for several functions point to cellular processes that may work together in a coordinated fashion. Because of the appreciable error rates inherent in measurements of protein–protein interactions, an interaction in a single species linking two previously unrelated processes would typically be ignored as a false positive. However, an observation that two or three networks reinforce this interaction is considerably more compelling, especially when the interaction is embedded in a densely connected conserved network region. For example, Fig. 2h links protein degradation to the process of poly(A) RNA elongation. Although these two processes are not connected in this region of the yeast network, several protein interactions link them in the networks of worm and fly (e.g., Pros25-Rack1-Msi or Pros25-Rack1-Tbph). These findings are consistent with previously documented association of proteasomes with mRNA-binding proteins, although the exact nature of this association has been controversial (30, 31). A related functional link between the proteasome and nucleic acid synthesis was detected in our earlier network comparison of yeast and the bacteria H. pylori (13).
As another example, Fig. 9l shows a worm/fly conserved cluster for which ≈40% of the proteins have no significant yeast ortholog (blast E value > 0.01). The cluster links functions such as proteolysis (Pros25, Pros28.1, Pas-1–7), actin binding (Cher,W04D2.1), ion transport (CG32810, C40A11.7, C52B11.2), and axon guidance (Fra). Taken together, these functions suggest a role for this cluster in growth cone formation during axon guidance. Guidance of axons to their synaptic targets is an initial step in the development of the central nervous system (32) and is mediated by special compartments called growth cones at the tips of the extending neurites. Formation of growth cones is induced by elevated levels of Ca2+ ions, which trigger local proteolysis and restructuring of the actin cytoskeleton (33). Thus, as implicated by our findings, axon guidance requires synergy between proteolysis, actin binding, and ion transport within an intricate network of protein interactions.
Validation of Predicted Interactions. Our two-hybrid tests of predicted interactions yielded a success rate in the range of 40–52%. These results are satisfactory for three reasons. First, the performance is clearly significant compared to the chance of identifying protein interactions at random (0.024%, estimated from an earlier two-hybrid screen (4) of 192 baits × 6,000 preys that yielded 281 interacting pairs). Second, two-hybrid analysis is known to miss a substantial portion of true interactions (11); this is particularly likely in our case where protein pairs were checked in only one orientation of bait and prey. For instance, two of the pairs that tested negative (YJR068W-YOR217W and YBL105C-YHR030C) have been shown to interact genetically in synthetic-lethal screens (27), suggesting a possible physical interaction. Third, predicting interactions by using a multiple network alignment approach compares favorably to previous approaches based on conservation of individual protein interactions. For instance, in ref. 12, the interolog approach was applied to a set of 72 reported interactions in yeast, predicting 71 previously undescribed interactions in worm. Seven of the predicted worm interactions tested positive by using a two-hybrid assay (10%), whereas 19 of the previously reported yeast interactions (26%) retested positive. Considering only the worm interactions that were predicted based on the 19 confirmed interactions in yeast, six of these tested positive, upper bounding the prediction accuracy at 31%. In tests of 145 additional predictions, 28 were confirmed, obtaining an overall accuracy of 16%. Similar results were obtained in a subsequent study by Yu et al. (29), where the accuracies of the interolog approach and an extension of it were estimated at 30–31%.
Conclusion
Nearly all comparative genomic studies of multiple species have been based on DNA and protein sequence analysis. Here, we transcend that framework by presenting a comparative study of the protein–protein interaction networks of three model eukaryotes. These comparisons show that many circuits embedded within the protein networks are conserved over evolution, and that these circuits cover a variety of well defined functional categories. Because measurements of protein interactions tend to be noisy and incomplete, it would have been difficult, if not impossible, to find these mechanisms by looking at only a single species. Moreover, many of these similarities and the network connections they imply would not have been suggested by sequence similarity alone, as the proteins involved are frequently not best sequence matches. The multiple network alignment also allows us to ascribe unique functions to many proteins and predict previously unobserved protein–protein interactions. Therefore, comparative network analysis is a powerful approach for elucidating network organization and function.
Supplementary Material
Acknowledgments
We thank Jacob Scott and Shen-Hong Chen for help with the analyses and Steven Brenner, Jens Gramm, and Markus Herrgard for critical reading of the manuscript. This research was supported by grants from the National Science Foundation and National Institutes of Health.
Author contributions: R.S., R.M. Karp, and T.I. designed research; R.S., S.S., R.M. Kelley, T.K., S.M., T.S., and T.I. performed research; R.S., S.S., R.M. Kelley, P.U., R.M. Karp, and T.I. contributed new reagents/analytic tools; R.S., S.S., R.M. Kelley, T.K., S.M., P.U., T.S., and T.I. analyzed data; and R.S., P.U., R.M. Karp, and T.I. wrote the paper.
Abbreviation: GO, gene ontology.
References
- 1.Fields, S. & Song, O. (1989) Nature 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold, R. & Mann, M. (2003) Nature 422, 198–207. [DOI] [PubMed] [Google Scholar]
- 3.Rain, J. C., Selig, L., De Reuse, H., Battaglia, V., Reverdy, C., Simon, S., Lenzen, G., Petel, F., Wojcik, J., Schachter, V., et al. (2001) Nature 409, 211–215. [DOI] [PubMed] [Google Scholar]
- 4.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 5.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. & Sakaki, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 7.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 8.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., et al. (2003) Science 302, 1727–1736. [DOI] [PubMed] [Google Scholar]
- 9.Li, S., Armstrong, C. M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P. O., Han, J. D., Chesneau, A., Hao, T., et al. (2004) Science 303, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samanta, M. P. & Liang, S. (2003) Proc. Natl. Acad. Sci. USA 100, 12579–12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S. G., Fields, S. & Bork, P. (2002) Nature 417, 399–403. [DOI] [PubMed] [Google Scholar]
- 12.Matthews, L. R., Vaglio, P., Reboul, J., Ge, H., Davis, B. P., Garrels, J., Vincent, S. & Vidal, M. (2001) Genome Res. 11, 2120–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelley, B. P., Sharan, R., Karp, R. M., Sittler, T., Root, D. E., Stockwell, B. R. & Ideker, T. (2003) Proc. Natl. Acad. Sci. USA 100, 11394–11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharan, R., Ideker, T., Kelley, B. P., Shamir, R. & Karp, R. M. (2004) in Proceedings of the Eighth Annual International Conference on Research in Computational Molecular Biology–RECOMB, eds. Bourne, P. E. & Gusfield, D. (Am. Soc. Microbiol. Press, Washington, DC), pp. 282–289.
- 15.Dandekar, T., Schuster, S., Snel, B., Huynen, M. & Bork, P. (1999) Biochem. J. 343, 115–124. [PMC free article] [PubMed] [Google Scholar]
- 16.Forst, C. V. & Schulten, K. (2001) J. Mol. Evol. 52, 471–489. [DOI] [PubMed] [Google Scholar]
- 17.Ogata, H., Fujibuchi, W., Goto, S. & Kanehisa, M. (2000) Nucleic Acids Res. 28, 4021–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart, J. M., Segal, E., Koller, D. & Kim, S. K. (2003) Science 302, 249–255. [DOI] [PubMed] [Google Scholar]
- 19.Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. (2002) Science 296, 340–343. [DOI] [PubMed] [Google Scholar]
- 20.Bergmann, S., Ihmels, J. & Barkai, N. (2004) PLoS Biol. 2, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader, J. S., Chaudhuri, A., Rothberg, J. M. & Chant, J. (2004) Nat. Biotechnol. 22, 78–85. [DOI] [PubMed] [Google Scholar]
- 23.Xenarios, I., Salwinski, L., Duan, X. J., Higney, P., Kim, S. M. & Eisenberg, D. (2002) Nucleic Acids Res. 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christie, K. R., Weng, S., Balakrishnan, R., Costanzo, M. C., Dolinski, K., Dwight, S. S., Engel, S. R., Feierbach, B., Fisk, D. G., Hirschman, J. E., et al. (2004) Nucleic Acids Res. 32, D311–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, T. W., Chen, N., Cunningham, F., Tello-Ruiz, M., Antoshechkin, I., Bastiani, C., Bieri, T., Blasiar, D., Bradnam, K., Chan, J., et al. (2004) Nucleic Acids Res. 32, D411–D417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flybase Consortium (2003) Nucleic Acids Res. 31, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mewes, H. W., Amid, C., Arnold, R., Frishman, D., Guldener, U., Mannhaupt, G., Munsterkotter, M., Pagel, P., Strack, N., Stumpflen, V., et al. (2004) Nucleic Acids Res. 32, D41–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie, H., Wasserman, A., Levine, Z., Novik, A., Grebinskiy, V., Shoshan, A. & Mintz, L. (2002) Genome Res. 12, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, H., Luscombe, N. M., Lu, H. X., Zhu, X., Xia, Y., Han, J. D., Bertin, N., Chung, S., Vidal, M. & Gerstein, M. (2004) Genome Res. 14, 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid, H. P., Pouch, M. N., Petit, F., Dadet, M. H., Badaoui, S., Boissonnet, G., Buri, J., Norris, V. & Briand, Y. (1995) Mol. Biol. Rep. 21, 43–47. [DOI] [PubMed] [Google Scholar]
- 31.Gautier-Bert, K., Murol, B., Jarrousse, A. S., Ballut, L., Badaoui, S., Petit, F. & Schmid, H. P. (2003) Mol. Biol. Rep. 30, 1–7. [DOI] [PubMed] [Google Scholar]
- 32.Yu, T. W. & Bargmann, C. I. (2001) Nat. Neurosci. 4, Suppl., 1169–1176. [DOI] [PubMed] [Google Scholar]
- 33.Spira, M. E., Oren, R., Dormann, A., Ilouz, N. & Lev, S. (2001) Cell Mol. Neurobiol. 21, 591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B. & Ideker, T. (2003) Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.