Abstract
Background
Type 1 diabetes mellitus (T1DM) is one of the most common pediatric diseases and its incidence is rising in many countries. Recently, it has been shown that metabolites other than glucose play an important role in insulin deficiency and the development of diabetes. The aim of our study was to look for discriminating variation in the concentrations of small-molecule metabolites in the plasma of T1DM children as compared to non-diabetic matched controls using proton nuclear magnetic resonance (1H-NMR)-based metabolomics.
Methods
A cross-sectional study was set-up to examine the metabolic profile in fasting plasma samples from seven children with poorly controlled T1DM and seven non-diabetic controls aged 8–18 years, and matched for gender, age and BMI-SDS. The obtained plasma 1H-NMR spectra were rationally divided into 110 integration regions, representing the metabolic phenotype. These integration regions reflect the relative metabolite concentrations and were used as statistical variables to construct (train) a classification model in discriminating between T1DM patients and controls.
Results
The total amount of variation explained by the model between the groups is 81.0% [R2Y(cum)] and within the groups is 75.8% [R2X(cum)]. The predictive ability of the model [Q2(cum)] obtained by cross-validation is 50.7%, indicating that the discrimination between the groups on the basis of the metabolic phenotype is valid. Besides the expected higher concentration of glucose, the relative concentrations of lipids (triglycerides, phospholipids and cholinated phospholipids) are clearly lower in the plasma of T1DM patients as compared to controls. Also the concentrations of the amino acids serine, tryptophan and cysteine are slightly decreased.
Conclusions
The present study demonstrates that metabolic profiling of plasma by 1H-NMR spectroscopy allows to discriminate between T1DM patients and controls. The metabolites that significantly differ between both groups might point to disturbances in biochemical pathways including (1) choline deficiency, (2) increased gluconeogenesis, and (3) glomerular hyperfiltration. Although the sample size of this study is still somewhat limited and a validation should be performed, the proof of principle looks promising and justifies a deeper investigation of the diagnostic possibilities of 1H-NMR metabolomics in follow-up studies.
Trial registration NCT03014908. Registered 06/01/2017. Retrospectively registered
Electronic supplementary material
The online version of this article (doi:10.1186/s13098-017-0246-9) contains supplementary material, which is available to authorized users.
Keywords: 1H-NMR spectroscopy, Metabolomics, Type 1 diabetes, Pediatrics
Background
Type 1 diabetes mellitus (T1DM) is one of the most common pediatric diseases and its incidence is rising in many countries [1]. T1DM is a chronic metabolic disorder that results from a lack of pancreatic β-cell insulin production by autoimmune mechanisms [2]. Insulin is a key hormone to maintain metabolic homeostasis, regulating carbohydrate, lipid and protein metabolism, and insulin deficiency in T1DM subsequently induces a variety of metabolic derangements [3, 4]. To identify novel pathways or early biomarkers indicative of metabolic alterations that are involved in the development of diabetes, metabolomics is an increasingly used tool [5]. Metabolomics research on pediatric study populations is still in its infancy. Up to now, only a few researchers investigated the plasma metabolic fingerprint of T1DM in children, using mass spectrometry as analytical tool [6–8]. One study showed that children who later progressed to T1DM had reduced serum levels of succinic acid and phosphatidylcholine at birth, pointing towards a dysregulated metabolism preceding β-cell autoimmunity and overt T1DM [6]. In addition, methionine deficits in early childhood may lead to an increased risk to develop T1DM later in life [7]. When comparing the plasma metabolic profile of T1DM and healthy children, differences were observed in lipid metabolism (non-esterified fatty acids, lysophospholipids and other fatty acid-derivatives), and some markers of differential activity of the gut microbiota (bile acids, p-cresol sulfate) [8]. However, the use of nuclear magnetic resonance (NMR)-based metabolomics to obtain a deeper knowledge of the plasma metabolic profile of T1DM has not been fully explored in the pediatric population. Proton (1H)-NMR spectroscopy has proven to be a robust and reproducible technique with very limited sample preparation (no extractions) [9], that can detect and quantify a wide variety of metabolites simultaneously, providing information regarding the biochemical pathways involved [10]. The objective of the current study was to investigate metabolic variations in the plasma of T1DM children and adolescents as compared to plasma of non-diabetic matched controls using 1H-NMR spectroscopy combined with multivariate statistics.
Methods
Subjects and characteristics
Children with poorly controlled T1DM (n = 7) and non-diabetic controls (n = 7) were recruited at the Department of Pediatrics of the Jessa Hospital Hasselt (Belgium) between June 2012 and November 2013. Inclusion criteria were: (1) aged between 8 and 18; (2) normal-weight according to the International Obesity Task Force (IOTF) BMI criteria [11]; and (3) fasted for at least 8 h. All subjects were matched for gender (four males and three females in both groups), age (12.0 ± 3.0 and 13.5 ± 2.7 years, respectively), and BMI-SDS (0.03 ± 0.62 and 0.05 ± 0.55, respectively). Subject characteristics are presented in Table 1. T1DM patients were diagnosed according to international consensus guidelines [12]. T1DM patients had diabetes for 6.8 ± 3.8 years, and were treated with exogenous insulin (mean insulin dose per kg: 0.83 ± 0.22/kg). T1DM patients show high fasting plasma glucose levels (mean: 187 ± 82 mg/dl) and hemoglobin A1c (HbA1c) concentrations above 6.5% (mean: 9.7 ± 2.8%) (Additional file 1: Table S1). None of the subjects was using lipid-lowering drugs or other medication. The study was conducted in accordance with the ethical rules of the Helsinki Declaration and Good Clinical Practice. The study protocol was approved by the medical-ethical committees of the Jessa Hospital and Hasselt University (12.27/ped12.02). Informed and written consent was obtained from all participants and their parents or legal guardian.
Table 1.
Subject characteristics
| Subject alias | Age | Gender | Tanner stage | Height (in cm) | Weight (in kg) | BMI | BMI-SDS |
|---|---|---|---|---|---|---|---|
| T1DM patients | |||||||
| 1 | 17.2 | 1 | 5 | 167.0 | 54.0 | 19.4 | −0.38 |
| 2 | 15.0 | 0 | 5 | 159.0 | 58.0 | 22.9 | 1.01 |
| 3 | 11.4 | 1 | 2 | 152.0 | 37.0 | 16.0 | −0.52 |
| 4 | 10.0 | 0 | 1 | 148.0 | 34.5 | 15.8 | −0.31 |
| 5 | 8.7 | 0 | 1 | 140.0 | 33.2 | 16.9 | 0.59 |
| 6 | 11.8 | 1 | 2 | 149.0 | 40.5 | 18.2 | 0.37 |
| 7 | 10.1 | 1 | 1 | 139.5 | 30.0 | 15.4 | −0.53 |
| Controls | |||||||
| 1 | 15.0 | 1 | 5 | 172.8 | 58.9 | 19.7 | 0.23 |
| 2 | 9.9 | 1 | 1 | 139.0 | 30.3 | 15.7 | −0.38 |
| 3 | 11.6 | 1 | 1 | 147.0 | 42.2 | 19.5 | 0.92 |
| 4 | 12.6 | 0 | 2 | 158.7 | 40.9 | 16.2 | −0.81 |
| 5 | 12.5 | 0 | 3 | 151.9 | 41.0 | 17.8 | −0.11 |
| 6 | 17.8 | 0 | 5 | 168.4 | 60.1 | 21.2 | 0.14 |
| 7 | 15.2 | 1 | 4 | 179.1 | 64.1 | 20.0 | 0.32 |
No significant differences were found between T1DM and control group for age, gender, Tanner stage (pubertal status), height, weight, BMI and BMI-SDS
T1DM type 1 diabetes, BMI body mass index, BMI-SDS body mass index standard deviation score
Biochemical measurements
Fasting venous blood of T1DM patients was collected in 2-ml fluoride-oxalate tubes for biochemical analysis at the Clinical Laboratory of Jessa Hospital. Plasma glucose was measured by the glucose oxidase method using a Synchron LX20 analyzer (Beckman Coulter, Brea, CA, USA) and HbA1c was measured using ion exchange chromatography (Menarini HA-8160 HbA1c auto-analyzer, Menarini Diagnostics, Belgium).
Sample collection, preparation and 1H-NMR analysis
Fasting venous blood was collected in 6-ml lithium heparin tubes and stored at 4 °C within 10 min. Within 30 min, samples were centrifuged at 1600g for 15 min and plasma aliquots of 500 µl were transferred into cryovials and stored at −80 °C [13]. Detailed protocols regarding sample preparation and 1H-NMR analysis have been previously described elsewhere [14]. Plasma 1H-NMR spectra were rationally divided into 110 integration regions defined on the basis of spiking experiments with known metabolites [15]. These integration regions reflect the relative metabolite concentrations—i.e. the metabolic phenotype—and were used as statistical variables to construct (train) a classification model in discriminating between T1DM patients and controls.
Statistical analysis
Multivariate statistics was performed using SIMCA-P+ (Version 13.0, Umetrics, Sweden). After mean-centering and Pareto scaling of the variables, unsupervised principal component analysis (PCA) was performed in order to look for clustering and possible confounders within the dataset, and to identify possible outliers by a Hotelling’s T2 range test and a distance to model plot. In a next step, orthogonal partial least squares discriminant analysis (OPLS-DA) was used to build (train) a model (statistical classifier) to discriminate between T1DM patients and controls [16]. The validity of the established model was evaluated on one hand by the total amount of variation between and within the groups explained by the model [denoted as R2Y(cum) and R2X(cum), respectively] and on the other hand by the predictive ability of the model as determined by a sevenfold cross-validation [denoted as Q2(cum)]. To be classified as a variable that strongly contributes to the group discrimination, three selection criteria have to be fulfilled: (1) significantly different in univariate statistics (a student t test corrected for multiple testing by the Benjamini–Hochberg method), (2) an OPLS-DA absolute value of p(corr), i.e. the loading scaled as a correlation coefficient, exceeding 0.6 and (3) an OPLS-DA variable importance for the projection (VIP) value exceeding 0.5 [16].
Results
Multivariate OPLS-DA statistics was used to train a classification model (classifier) in discriminating between T1DM patients and controls based on data input from their metabolic profile or phenotype. A PCA analysis was conducted first to look for clustering and possible confounders. Figure 1 shows that the subjects were clustered in a way that allowed T1DM patients to be clearly differentiated from controls and no outliers were detected. Moreover, staining the PCA score plots for gender, age and BMI-SDS clearly shows that none of these factors have a confounding effect on the discrimination between T1DM patients and controls, as was expected for matched groups (data not shown).
Fig. 1.
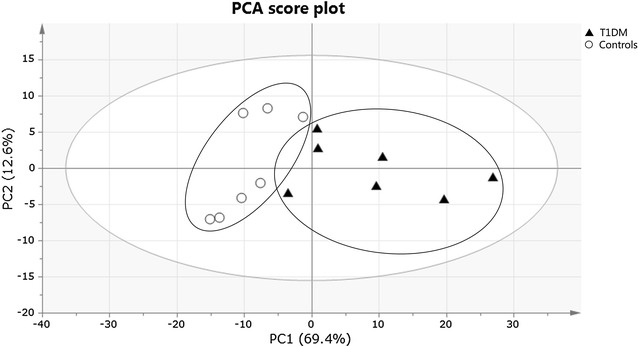
PCA score plot obtained for T1DM patients (filled triangle) and healthy controls (circle). Each participant is represented by its metabolic profile and visualized as a single symbol of which the location is determined by the contributions of the 110 variables in the 1H-NMR spectrum. The PCA score plot shows the first principal component (PC1: 69.4%), explaining the largest variance within the dataset, versus the second principal component (PC2: 12.6%) that explains the second largest variance
In a next step, OPLS-DA was used to build a model (statistical classifier) to differentiate between T1DM patients and controls (Fig. 2a). The total amount of variation between the groups that can be explained by the model is 81.0% [R2Y(cum)] while this within the groups is 75.8% [R2X(cum)]. The predictive ability of the model, obtained by cross-validation, is quite high with a Q2(cum) of 50.7%, indicating that the discrimination between the groups on the basis of the metabolic phenotype is valid. The OPLS-DA S-line plot shown in Fig. 2b visualizes the covariance [left y-axis; p(ctr)] and correlation coefficient [right y-axis; abs(p(corr)] between the variables and the classification score in the model (see caption of Fig. 2b for more information). Strongly discriminating variables combine a clear covariance [p(ctr)] with a high absolute value for the correlation coefficient [abs(p(corr)] (Table 2).
Fig. 2.
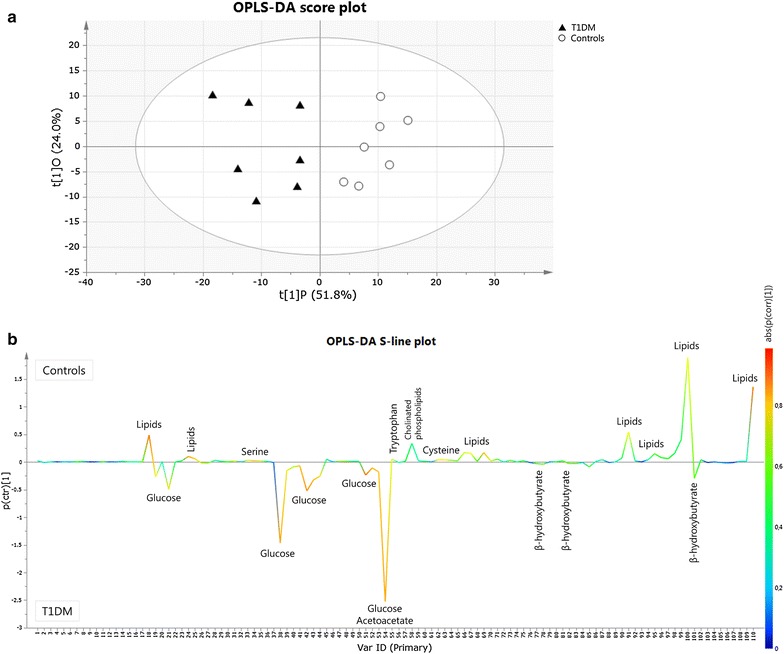
OPLS-DA score plot (a) and S-line plot (b) obtained for T1DM patients (filled triangle) and healthy controls (circle). Each participant is represented by its metabolic profile and visualized as a single symbol of which the location is determined by the contributions of the 110 variables in the 1H-NMR spectrum. The OPLS-DA score plot shows the first predictive component (t[1]P: 51.8%), explaining the variation between the groups, versus the first orthogonal component (t[1]O: 24.0%) that explains the variation within the groups. The OPLS-DA S-line plot visualizes differences between T1DM patients (negative) and controls (positive). The left y-axis represents p(ctr)[1], the covariance between a variable and the classification score. It indicates if an increase or decrease of a variable is correlated to the classification score. The magnitude of the covariance is however difficult to interpret since covariance is scale dependent. This means that a high value for the covariance does not necessary imply a strong correlation, as the covariance is also influenced by the intensity of the signal with respect to the noise level. Therefore this measure will likely indicate variables with large signal intensities. The right y-axis shows p(corr)[1], the correlation coefficient between a variable and the classification score (i.e. the normalized covariance). It gives a linear indication of the strength of the correlation. As the correlation is independent of the intensity of the variable, it will be a better measure for the reliability of the variable in the classification process. In b, the red color stands for the highest absolute value of the correlation coefficient. Strongly discriminating variables have a large intensity and large reliability
Table 2.
Plasma variables that significantly differ between T1DM and control subjects by multivariate statistics
| VAR | Spectral range (ppm) | Assigned metabolite | Model based on all 110 VARs (including glucose related VARs) | Model based on 95 VARs (excluding glucose related VARs) | ||
|---|---|---|---|---|---|---|
| p(corr)[1] | VIP ± cvSE | p(corr)[1] | VIP ± cvSE | |||
| 18 | 5.4300–5.2752 | –HC=CH– in fatty acid chain | 0.890 | 2.07 ± 0.71 | 0.787 | 2.77 ± 0.82 |
| 24 | 4.4100–4.3159 | C1 H and C3 H in glycerol backbone of triglycerides + OCH 2 of choline in sphingomyelin/phosphatidylcholine | 0.856 | 0.89 ± 0.47 | 0.740 | 1.19 ± 0.63 |
| 34 | 4.0310–4.0136 | l-Serine | 0.809 | 0.48 ± 0.30 | 0.743 | 0.65 ± 0.40 |
| 35 | 4.0136–4.0010 | l-Serine | 0.715 | 0.43 ± 0.26 | 0.669 | 0.57 ± 0.38 |
| 38 | 3.9590–3.8330 | d-Glucose | −0.837 | 3.56 ± 0.24 | ||
| 41 | 3.7956–3.7820 | d-Glucose | −0.815 | 0.83 ± 0.24 | ||
| 42 | 3.7820–3.7550 | d-Glucose | −0.861 | 2.19 ± 0.12 | ||
| 43 | 3.7550–3.7390 | d-Glucose | −0.825 | 1.66 ± 0.14 | ||
| 44 | 3.7390–3.7141 | d-Glucose | −0.782 | 1.42 ± 0.19 | ||
| 51 | 3.5914–3.5649 | d-Glucose | −0.901 | 1.44 ± 0.27 | ||
| 52 | 3.5649–3.5510 | d-Glucose | −0.828 | 1.04 ± 0.19 | ||
| 54 | 3.5360–3.3980 | d-Glucose | −0.836 | 4.63 ± 0.54 | ||
| 55 | 3.3980–3.3765 | l-Tryptophan | 0.721 | 0.81 ± 0.28 | 0.870 | 1.08 ± 0.35 |
| 63 | 3.1090–3.0860 | l-Cysteine | 0.754 | 0.57 ± 0.37 | 0.701 | 0.77 ± 0.54 |
| 91 | 2.1230–1.9720 | –CH 2–CH=CH– in fatty acid chain | 0.752 | 2.17 ± 0.44 | 0.734 | 2.91 ± 0.65 |
| 100 | 1.3450–1.2458 | CH3–(CH 2)n– in fatty acid chain | 0.747 | 4.04 ± 1.52 | 0.728 | 5.41 ± 1.75 |
| 110 | 0.9660–0.8000 | CH 3–(CH2)n– in fatty acid chain | 0.898 | 3.14 ± 0.73 | 0.735 | 4.21 ± 1.03 |
cvSE standard error of cross-validation, p(corr)[1] correlation scaled loading, VAR variable, VIP variable influence on projection
The plot shows that the concentrations of lipids (triglycerides, phospholipids and cholinated phospholipids) are clearly decreased in the plasma of T1DM patients as compared to controls, whereas serine, tryptophan and cysteine concentrations seem to be decreased slightly. The glucose levels on the other hand are clearly increased in the plasma of T1DM patients. These changes were also observed as significant by a univariate t test with post hoc Benjamini–Hochberg correction (Table 3). The plot further shows that ketone levels (i.e. acetoacetate and β-hydroxybutyrate) are slightly elevated in T1DM patients. In order to look if T1DM patients and controls can be differentiated by a model constructed without the 15 variables related to the strong glucose signals in the 1H-NMR spectra, the variables representing glucose were removed from the metabolic profile prior to the OPLS-DA model building (construction of the model with only 95 variables). The results are also presented in Table 2 and confirm that the relative concentrations of lipids (triglycerides, phospholipids and cholinated phospholipids) and some amino acids (serine, tryptophan and cysteine) are reduced in the plasma of T1DM patients.
Table 3.
Plasma variables that significantly differ between T1DM and control subjects by univariate statistics
| VAR | Spectral range (ppm) | Assigned metabolite | Relative concentration | Increased/decreased in T1DM | p value* | %-change | |
|---|---|---|---|---|---|---|---|
| T1DM | Controls | ||||||
| 18 | 5.4300–5.2752 | –HC=CH– in fatty acid chain | 21.95 ± 3.63 | 30.95 ± 4.00 | ↓ | 0.001 | −29.1 |
| 24 | 4.4100–4.3159 | C1 H and C3 H in glycerol backbone of triglycerides + OCH 2 of choline in sphingomyelin/phosphatidylcholine | 3.28 ± 1.16 | 5.09 ± 1.16 | ↓ | 0.005 | −35.6 |
| 34 | 4.0310–4.0136 | l-Serine | 0.70 ± 0.26 | 1.21 ± 0.22 | ↓ | 0.003 | −42.1 |
| 35 | 4.0136–4.0010 | l-Serine | 0.84 ± 0.28 | 1.29 ± 0.23 | ↓ | 0.008 | −34.9 |
| 38 | 3.9590–3.8330 | d-Glucose | 91.99 ± 16.9 | 64.44 ± 6.58 | ↑ | 0.004 | 42.8 |
| 41 | 3.7956–3.7820 | d-Glucose | 7.62 ± 0.60 | 6.18 ± 0.60 | ↑ | <0.001 | 23.3 |
| 42 | 3.7820–3.7550 | d-Glucose | 30.58 ± 5.63 | 20.56 ± 1.95 | ↑ | 0.003 | 48.7 |
| 43 | 3.7550–3.7390 | d-Glucose | 17.92 ± 3.83 | 11.85 ± 1.35 | ↑ | 0.005 | 51.2 |
| 44 | 3.7390–3.7141 | d-Glucose | 17.76 ± 3.24 | 13.05 ± 1.49 | ↑ | 0.007 | 36.1 |
| 51 | 3.5914–3.5649 | d-Glucose | 14.45 ± 2.19 | 10.20 ± 1.06 | ↑ | 0.002 | 41.7 |
| 52 | 3.5649–3.5510 | d-Glucose | 5.84 ± 1.10 | 3.65 ± 0.50 | ↑ | 0.002 | 60.0 |
| 54 | 3.5360–3.3980 | d-Glucose | 116.37 ± 30.40 | 69.27 ± 8.54 | ↑ | 0.005 | 68.0 |
| 55 | 3.3980–3.3765 | l-Tryptophan | 0.70 ± 0.32 | 1.94 ± 0.41 | ↓ | <0.001 | −63.9 |
| 63 | 3.1090–3.0860 | l-Cysteine | 2.28 ± 0.47 | 3.05 ± 0.39 | ↓ | 0.006 | −25.2 |
| 91 | 2.1230–1.9720 | –CH 2–CH=CH– in fatty acid chain | 53.13 ± 7.19 | 64.03 ± 3.76 | ↓ | 0.007 | −17.0 |
| 100 | 1.3450–1.2458 | CH3–(CH 2)n– in fatty acid chain | 117.69 ± 17.62 | 155.40 ± 22.01 | ↓ | 0.004 | −24.3 |
| 110 | 0.9660–0.8000 | CH 3–(CH2)n– in fatty acid chain | 89.10 ± 14.79 | 111.75 ± 7.83 | ↓ | 0.006 | −20.3 |
* Benjamini–Hochberg adjusted p value, calculated using the independent samples t test. %-change is the increase (+) or decrease (−) of the mean in the T1DM group with respect to the control group
VAR variable
Discussion
Type 1 diabetes mellitus is a serious health concern worldwide that is usually diagnosed in children and young adults [1]. T1DM is a metabolic disorder, and in recent decades it has been shown that metabolites other than glucose play an important role in insulin deficiency and the development of diabetes [4, 5]. Metabolomics, the study of small-molecule metabolites, has developed into an important tool in diabetes research [5]. In this study, we investigated metabolic variations in T1DM children and adolescents using NMR-spectroscopy-based metabolomics to gain inside into biochemical pathways that are altered in early stages of T1DM. Besides the expected higher concentration of glucose, we found lower relative concentrations for lipids (triglycerides, phospholipids and cholinated phospholipids) and the amino acids serine, tryptophan and cysteine in plasma of T1DM children and adolescents as compared to non-diabetic controls.
Our findings of relatively lower levels of lipids in the plasma of T1DM as compared to controls are in agreement with other metabolomics studies [3, 4, 6]. In a prospective Finnish study, it was found that children who developed T1DM have reduced serum levels of phosphatidylcholine at birth, next to lower levels of multiple triglycerides and phospholipids throughout the follow-up [6]. In addition, it has been demonstrated that children developing T1DM before 4 years of age have low cord-blood levels of phospholipids, mainly phosphatidylcholines [17]. It is suggested that T1DM progressors are choline-deficient at birth, and that choline deficiency is the key mechanism leading to lower serum triglyceride levels and their increased accumulation in the liver [6]. Choline metabolism also depends on the gut microbial composition [18], making the latter an attractive target for early prevention and treatment of T1DM [19]. In addition, low levels of phosphatidylcholine may play a role in oxidative damage affecting the pancreatic β-cell insulin production, because phosphatidylcholines are thought to have anti-inflammatory properties [20]. According to an NMR-based metabolomics study in adults, lower levels of triglycerides in T1DM patients may also be attributable to treatment with insulin [3], which is known to have an anti-lipolytic action [21]. We further observed lower plasma levels of serine, tryptophan and cysteine in T1DM as compared to controls. In current literature, only a limited number of papers can be found regarding the relationship between these amino acids and T1DM. A study in diabetic db−/db− mice suggested that a strongly decreased concentration of the gluconeogenic amino acids serine, alanine and glycine, resulted in increased gluconeogenesis [22]. In addition, a study in rats suggested that tryptophan suppresses the elevation of blood glucose and lessens the burden associated with insulin secretion from β-cells [23]. Finally, reduced plasma levels of cysteine in T1DM patients can be explained by glomerular hyperfiltration, resulting in an increased renal clearance of cysteine [24].
Although the sample size of this study is still somewhat limited, the experiments were carried out according to a strictly controlled protocol. This pilot study demonstrates the proof of principle that metabolic phenotyping of T1DM in children by proton-NMR spectroscopy allows to differentiate between T1DM patients and controls and therefore justifies the start-up of larger studies.
Because NMR metabolomics can be used to search for subtle changes in the plasma of children prone to develop T1DM, it might become an important tool for the early diagnosis and prognosis of T1DM in children. Hence, restoring or improving the plasma metabolic profile, e.g. by re-establishing lipid and amino acid availability or by modulating gut microbial composition, might prevent β-cell destruction and delay T1DM progression in children and adolescents.
Conclusion
The present study demonstrates the proof of principle that metabolic phenotyping of plasma by 1H-NMR spectroscopy allows to discriminate between T1DM patients and controls. T1DM children and adolescents show lower relative plasma concentrations of lipids (triglycerides, phospholipids and cholinated phospholipids), serine, tryptophan and cysteine as compared to non-diabetic controls. NMR-spectroscopy-based metabolomics appears to be a promising tool for the identification of disturbed biochemical pathways related to the development of T1DM. Nevertheless, further identification and validation is needed in order to evaluate the use of NMR metabolomics in the prediction, diagnosis and monitoring of T1DM in children. Therefore, deeper follow-up studies in larger pediatric cohorts are of utmost importance to further explore the potential of metabolomics in the field of pediatric diabetes.
Authors’ contributions
LB, GM and PA conceived and designed the study. LB and GM collected the data. LB carried out the experiments. LB analyzed the data statistically. LB, PA, WG, EL and JPN interpreted the data. LB did literature research. LB generated figures and tables. LB wrote the manuscript with help of GM and PA. GM, WG, EL, JPN and PA revised the paper. LB had full access to all of the data in the study and takes responsibility for the integrity of the data and accuracy of the data analysis. All authors read and approved the final manuscript.
Acknowledgements
We thank all children and adolescents for their participation in this study. This study is part of the ‘Limburg Clinical Research Program (LCRP) UHasselt-ZOL-Jessa’, supported by the foundation Limburg Sterk Merk, province of Limburg, Flemish government, Hasselt University, Ziekenhuis Oost-Limburg and Jessa Hospital. We also thank the Research Foundation Flanders for their support via the MULTIMAR project and G. Reekmans for his assistance in the 1H-NMR analysis of plasma samples.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was conducted in accordance with the ethical rules of the Helsinki Declaration and Good Clinical Practice. The study protocol was approved by the medical-ethical committees of the Jessa Hospital and Hasselt University (12.27/ped12.02).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- 1H-NMR
proton nuclear magnetic resonance
- BMI
body mass index
- BMI-SDS
body mass index standard deviation score
- cvSE
standard error of cross-validation
- HbA1c
hemoglobin A1c
- IOTF
International Obesity Task Force
- OPLS-DA
orthogonal partial least squares discriminant analysis
- PCA
principal component analysis
- p(corr)[1]
correlation scaled loading
- T1DM
type 1 diabetes mellitus
- VAR
variable
- VIP
variable influence on projection
Additional file
Additional file 1: Table S1. Characteristics of T1DM patients.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13098-017-0246-9) contains supplementary material, which is available to authorized users.
Contributor Information
Liene Bervoets, Email: lienebervoets@live.com.
Guy Massa, Email: guy.massa@jessazh.be.
Wanda Guedens, Email: wanda.guedens@uhasselt.be.
Evelyne Louis, Email: evelynelouis@hotmail.be.
Jean-Paul Noben, Email: jeanpaul.noben@uhasselt.be.
Peter Adriaensens, Phone: +3211268396, Email: peter.adriaensens@uhasselt.be.
References
- 1.Patterson C, Guariguata L, Dahlquist G, Soltesz G, Ogle G, Silink M. Diabetes in the young—a global view and worldwide estimates of numbers of children with type 1 diabetes. Diabetes Res Clin Pract. 2014;103:161–175. doi: 10.1016/j.diabres.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383:69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brugnara L, Mallol R, Ribalta J, Vinaixa M, Murillo S, Casserras T, et al. Improving assessment of lipoprotein profile in type 1 diabetes by 1H NMR spectroscopy. PLoS ONE. 2015;10:e0136348. doi: 10.1371/journal.pone.0136348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanza IR, Zhang S, Ward LE, Karakelides H, Raftery D, Nair KS. Quantitative metabolomics by H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS ONE. 2010;5:e10538. doi: 10.1371/journal.pone.0010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bain JR. Targeted metabolomics finds its mark in diabetes research. Diabetes. 2013;62:349–351. doi: 10.2337/db12-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oresic M, Simell S, Sysi-Aho M, Nanto-Salonen K, Seppanen-Laakso T, Parikka V, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205:2975–2984. doi: 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pflueger M, Seppanen-Laakso T, Suortti T, Hyotylainen T, Achenbach P, Bonifacio E, et al. Age- and islet autoimmunity-associated differences in amino acid and lipid metabolites in children at risk for type 1 diabetes. Diabetes. 2011;60:2740–2747. doi: 10.2337/db10-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balderas C, Ruperez FJ, Ibanez E, Senorans J, Guerrero-Fernandez J, Casado IG, et al. Plasma and urine metabolic fingerprinting of type 1 diabetic children. Electrophoresis. 2013;34:2882–2890. doi: 10.1002/elps.201300062. [DOI] [PubMed] [Google Scholar]
- 9.Dumas ME, Maibaum EC, Teague C, Ueshima H, Zhou B, Lindon JC, et al. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: the INTERMAP Study. Anal Chem. 2006;78:2199–2208. doi: 10.1021/ac0517085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 11.Cole TJ, Lobstein T. Extended international (IOTF) body mass index cut-offs for thinness, overweight and obesity. Pediatr Obes. 2012;7:284–294. doi: 10.1111/j.2047-6310.2012.00064.x. [DOI] [PubMed] [Google Scholar]
- 12.Craig ME, Jefferies C, Dabelea D, Balde N, Seth A, Donaghue KC, ISPAD Clinical Practice Consensus Guidelines ISPAD, et al. Definition, epidemiology, and classification of diabetes in children and adolescents. Pediatr Diabetes. 2014;2014(15):4–17. doi: 10.1111/pedi.12186. [DOI] [PubMed] [Google Scholar]
- 13.Bervoets L, Louis E, Reekmans G, Mesotten L, Thomeer M, Adriaensens P, et al. Influence of preanalytical sampling conditions on the 1H NMR metabolic profile of human blood plasma and introduction of the Standard PREanalytical Code used in biobanking. Metabolomics. 2015;11:1197–1207. doi: 10.1007/s11306-015-0774-y. [DOI] [Google Scholar]
- 14.Louis E, Adriaensens P, Guedens W, Vanhove K, Vandeurzen K, Darquennes K, et al. Metabolic phenotyping of human blood plasma: a powerful tool to discriminate between cancer types? Ann Oncol. 2016;27:178–184. doi: 10.1093/annonc/mdv499. [DOI] [PubMed] [Google Scholar]
- 15.Louis E, Bervoets L, Reekmans G, De Jonge E, Mesotten L, Thomeer M, et al. Phenotyping human blood plasma by 1H-NMR: a robust protocol based on metabolite spiking and its evaluation in breast cancer. Metabolomics. 2015;11:225–236. doi: 10.1007/s11306-014-0690-6. [DOI] [Google Scholar]
- 16.Eriksson L, Byrne T, Johansson E, Trygg J, Wikström C. Multi- and megavariate data analysis: basic principles and applications. 3. Umetrics Academy: Umea; 2013. [Google Scholar]
- 17.La Torre D, Seppanen-Laakso T, Larsson HE, Hyotylainen T, Ivarsson SA, Lernmark A, et al. Decreased cord-blood phospholipids in young age-at-onset type 1 diabetes. Diabetes. 2013;62:3951–3956. doi: 10.2337/db13-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He C, Shan Y, Song W. Targeting gut microbiota as a possible therapy for diabetes. Nutr Res. 2015;35:361–367. doi: 10.1016/j.nutres.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Treede I, Braun A, Sparla R, Kuhnel M, Giese T, Turner JR, et al. Anti-inflammatory effects of phosphatidylcholine. J Biol Chem. 2007;282:27155–27164. doi: 10.1074/jbc.M704408200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verges B. Lipid disorders in type 1 diabetes. Diabetes Metab. 2009;35:353–360. doi: 10.1016/j.diabet.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Altmaier E, Ramsay SL, Graber A, Mewes HW, Weinberger KM, Suhre K. Bioinformatics analysis of targeted metabolomics—uncovering old and new tales of diabetic mice under medication. Endocrinology. 2008;149:3478–3489. doi: 10.1210/en.2007-1747. [DOI] [PubMed] [Google Scholar]
- 23.Inubushi T, Kamemura N, Oda M, Sakurai J, Nakaya Y, Harada N, et al. L-tryptophan suppresses rise in blood glucose and preserves insulin secretion in type-2 diabetes mellitus rats. J Nutr Sci Vitaminol. 2012;58:415–422. doi: 10.3177/jnsv.58.415. [DOI] [PubMed] [Google Scholar]
- 24.Wollesen F, Brattstrom L, Refsum H, Ueland PM, Berglund L, Berne C. Plasma total homocysteine and cysteine in relation to glomerular filtration rate in diabetes mellitus. Kidney Int. 1999;55:1028–1035. doi: 10.1046/j.1523-1755.1999.0550031028.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


