Abstract
Background
With the alarming rise in antibiotic resistance in African countries, the need for a surveillance system in the region has become pressing. The rapid expansion of data networks makes it possible to set up healthcare applications that can be both cost-efficient and effective. Large data sets are available for assessment of current antibiotic resistance among Ethiopian patients. Based on the data-presentation, a practical approach is proposed on how diagnostic laboratories can participate remedial action against antibiotic resistance in Ethiopia.
Methods
In Addis Ababa (Ethiopia), raw data comprising bacterial species name, specimen type and antibiograms covering the period January 2014 to May 2015 was accessed from the laboratory information management system. Using R code, the data was read and fitted into data-frames and analyzed to assess antibiotic resistance in the Ethiopian patient population.
Results
Susceptibility to an antibiotic was tested with 14.983 cultures of 54 different bacterial species or subgroups, isolated from 16 types of specimen. Half of the cultures (n = 6444) showed resistance to an antibiotic. Resistance against penicillin was highest with, on average, 91.1% of 79 bacterial cultures showing resistance. Very high resistance rates were also observed for ampicillin, whereas resistance was lowest with cefoxitin.
Conclusions
Extraction and analysis of raw-data from the laboratory database is relatively simple and can provide valuable insight into the relationships between type of sample and drug-resistance in countries where such data is still scarce. With the largest number of antibiotic resistance tests described for Ethiopia, a tool is proposed for consistent data collection with specified core variables. Trends in antibiotic resistance can be revealed and treatment failures avoided when used as an easy accessible reference application for healthcare providers.
Keywords: Antibiogram, Antibiotic, Ethiopia, R, Resistance, Surveillance
Background
Irrational use of antibiotics has been perceived to be a major problem worldwide and in particular in the African continent [1–3]. To improve rational drug use in Ethiopia, national standard treatment guidelines were issued to assist health care workers in their treatment of infectious diseases [4]. Treatment guidelines must be continuously revised as susceptibility to antibiotic drugs is constantly threatened due to an empirical approach to treatment and high self-medication of humans and animals without a medical prescription [5, 6]. Enforcing a strict adherence policy in the healthcare sector to reduce the development and spread of drug-resistant bacterial strains goes hand in hand with nationwide antimicrobial surveillance. Routine clinical diagnostic laboratories can contribute to the national surveillance network by sharing routine antibiograms from clinical samples [7, 8]. At the International Clinical Laboratories, routine diagnostic analyses are performed on clinical samples collected at patient service centers across the country and the results are reported back automatically via a virtual private network. The laboratory seeks to extend its services by promoting a healthy life-style to the general public and setting up network applications allowing clinicians to prescribe using mobile networks. Extracting raw data from the laboratory information system and then entering it into a customized data-frame requires some knowledge of programming. In this study, the open source R computing language was used to visualize the prevalence of antibiotic resistance in routine clinical samples in Ethiopia on the basis of raw data extracted from the laboratory information management system, which is the first step towards automated real-time presentation of antibiograms.
Methods
Data and sites
Retrospective data from January 2014 to May 2015 was exported from the Polytech laboratory information management system (LIMS) (Comp Pro Med Inc., Santa Rosa, USA) at International Clinical Laboratories, Addis Ababa, Ethiopia. Parameters in the raw data included bacterial species name, type of specimen and antibiotic sensitivity of the bacterial cultures. The data was obtained from routine analysis of clinical specimens collected from individual patients visiting the patient service site, and from clinics and hospitals. All specimens were received and analyzed at the central laboratory in Addis Ababa, Ethiopia.
Type of specimens
All types of specimen were included in the data query. From patient service sites outside Addis Ababa, only urine specimens were considered feasible for transportation and bacterial analysis at the central laboratory. Specimens that did not match the set categories in the LIMS were renamed as routine samples.
Bacterial species identification
Preliminary identification was based on number of colonies, types of colonies, morphological appearance and gram staining after growth on both selective and non-selective media. Further bacterial characterization involved biochemical testing and specific growth characteristics under different conditions. All identification tests were carried out according to validated standard operation procedures [9, 10]. After appropriate incubation and based on the site from which the specimen was obtained, the bacterial species were classified as pathogenic or contaminant bacteria. The group of bacteria classified as Streptococcus spp. includes all streptococcal species with the exception of the beta-hemolytic streptococcal species, which are categorized as a separate group.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the Kirby Bauer disk diffusion technique. Standard discs were used to detect and measure induced inhibition for specified antibiotic concentrations, placed on Mueller–Hinton agar supplemented with 5% sheep blood seeded with 0.5 McFarland of bacteria. The plates were incubated overnight under specified conditions (e.g. temperature and atmosphere depending on bacteria species and type of specimen). After incubation was complete, the zone inhibition diameter, in mm, was measured. The zones were interpreted as susceptible, intermediate or resistant [11].
Quality control
Commercial pre-plated and quality control-passed media were used with 15 different ATCC strains and blank incubation controls to check the transportation and storage conditions of the pre-plated media. The quality control was based on retesting of retained strains and correlation between results of different characteristics of a strain. New batches of ATCC strains for media were checked and compared with previously used ATCC strains and gram stained control slides were used to check the quality of the staining reagents.
Data analysis
Scripts were written in R (version 3.2.2) to read and fit the raw data into data frames. Test results from Mycobacterium cultures were deleted from the data set. The remaining data was summarized and plotted. Anaerobic and aerobic blood cultures were merged in one group. Drug resistance was calculated by dividing the number of resistant cultures by the sum of resistant and sensitive cultures, disregarding the intermediate sensitivity results. The R scripts and raw data are accessible in the Github repository [12].
Results
Drug susceptibility was tested on 14.983 cultures excluding Mycobacterium spp. over the period from January 2014 until May 2015. The total numbers of resistant, intermediate and sensitive cultures were 7440, 1099 and 6444, respectively. Specimens were obtained from urine (n = 11.034), wounds (n = 1346), blood (n = 672), body fluid (n = 466), pediatric blood (n = 411), ear (n = 322), pus (n = 154), stool (n = 136), sputum (n = 134), cerebral spinal fluid (CSF) (n = 94), routine samples (n = 78), eye (n = 45), throat (n = 62), nasal swab (n = 11), nasal discharge (n = 10) and urethral discharge (n = 8) (Table 1).
Table 1.
Eighteen months of retrospective bacterial analyses showing the most prevalent bacterial species for each sample
| Source | Bacterial species most frequently isolated | Frequency of bacterial species | Percentage of bacterial species (%) |
|---|---|---|---|
| Blood | Coagulase-negative Staphylococcus species | 214 | 31.8 |
| Blood-pediatric | Coagulase-negative Staphylococcus species | 157 | 38.2 |
| Body fluid | Escherichia coli | 183 | 39.3 |
| Cerebral spinal fluid | Staphylococcus aureus | 23 | 24.5 |
| Ear culture | Staphylococcus aureus | 79 | 24.5 |
| Eye culture | Streptococcus pneumoniae | 26 | 57.8 |
| Nasal discharge | Group G Streptococcus | 10 | 100.0 |
| Nasal swab | Coagulase-negative Staphylococcus species | 11 | 100.0 |
| Pus | Escherichia coli | 73 | 47.4 |
| Routine | Escherichia coli | 36 | 46.2 |
| Sputum | Klebsiella oxytoca | 49 | 36.6 |
| Stool | Salmonella group non paratyphi A/B | 43 | 31.6 |
| Throat | Streptococcus pyogenes (Group A) | 22 | 35.5 |
| Urethral discharge | Escherichia coli | 8 | 100.0 |
| Urine | Escherichia coli | 7140 | 64.7 |
| Wound | Staphylococcus aureus | 565 | 36.0 |
In total, 54 different bacterial species or subgroups were identified (Appendix 1). To visualize resistance patterns, data was further tailored by merging bacterial species into genus groups. Antibiogram groups containing few cultures and bacterial cultures with intermediate resistance levels were considered not representative of the general antibiotic drug resistance (ADR) profiles and therefore omitted from further analyses. As a result, 1836 cultures were filtered out when the sum of resistant plus sensitive cultures was less than 7 within a group of bacterial species and for a given antibiotic. The remaining 13.147 antibiograms contained 927 intermediate cultures, which were subsequently excluded from the calculation of antibiotic resistance rates. The bacterial antibiotic resistance rate was calculated for each group of bacteria for the 21 different antibiotics (Fig. 1).
Fig. 1.
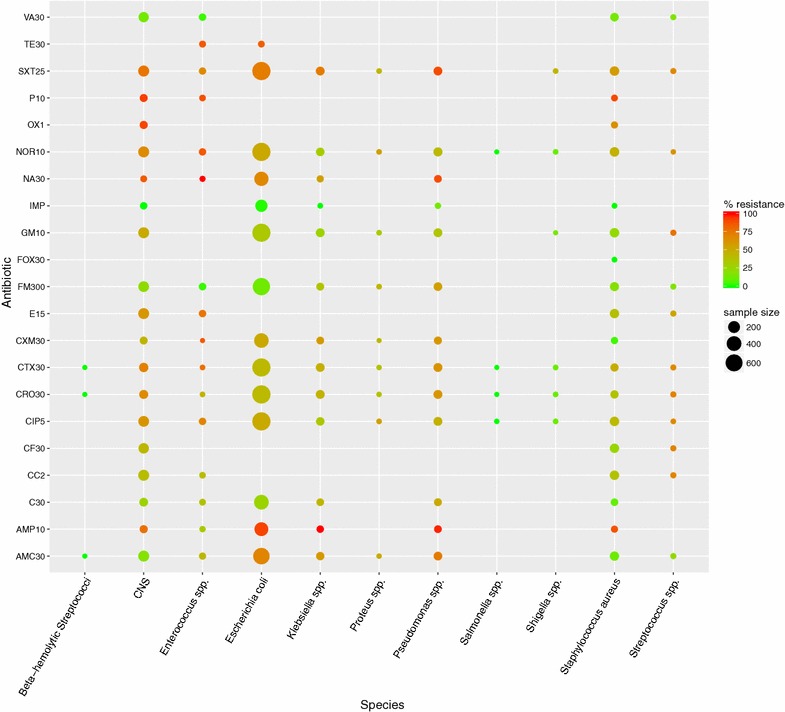
Percentages of resistance of bacterial species (n = 12.220) against antibiotics represented by color and number of tested samples represented by dot-size (January 2014–May 2015). CNS coagulase negative staphylococci, SAMC30 amoxicillin/clavulanic acid, AMP10 ampicillin, C30 chloramphenicol, CC2 clindamycin, CF30 cephalophin, CIP5 ciprofloxacin, CRO30 ceftriaxone, CTX30 cefotaxime, CXM30 cefuroxime, E15 erythromycin, FM300 nitrofurantoin, FOX30 cefoxitin, GM10 gentamicin, IMP imipenem, NA30 nalidixic acid, NOR10 norfloxacin, OX1 oxacillin, P10 penicillin, SXT25 sulfamethoxazole, TE30 tetracycline, VA30 vancomycin
The highest resistance rates were observed for penicillin (P10) with on average 91.1% of 79 bacterial cultures showing resistance, with coagulase negative Staphylococcus (CNS) (92.9%; n = 42), Staphylococcus aureus (90.0%; n = 20) and Enterococcus spp. (88.2%; n = 17). High percentages of resistant cultures were also observed for ampicillin (AMP10) with the highest four Klebsiella spp. (100%; n = 30), CNS (90.2%; n = 51), Pseudomonas spp. (93.3%; n = 60) and Escherichia coli (91.5%; n = 331). The lowest resistance rates were observed with cefoxitin in S. aureus cultures (FOX30) (0%; n = 10). For imipenem (IMP), no or low resistance rates were observed in cultures with CNS (0%; n = 34), Klebsiella spp. (0%; n = 10), S. aureus (0%; n = 10), E. coli (1.3%, n = 224) and Pseudomonas spp. (13.3%; n = 15). Resistance to vancomycin (VA30) was observed with Enterococcus spp. (3.3%; n = 30), CNS (13.4%; n = 119), S. aureus (14.0%; n = 57) and Streptococcus spp. (16.7%; n = 12). A table showing percentages of resistance rates and numbers of cultures from this study is presented in Appendix 2.
Discussion
This study describes the results from almost fifteen thousand antibiograms analyzed with open-source software; it is the largest number of ADR test results to have been described in Ethiopia. As is the case throughout the rest of Africa, data on antibiotic resistance levels in Ethiopia is minimal and mostly derived from literature. Estimates of ADR percentages are rendered ambiguous as a result of quality assurance issues, small sample sizes and biases related to hospital-based studies with isolates from specific patient groups [6, 13, 14]. Although the interpretation of the data presented in this study must be taken with caution, it does provide a general overview of the current situation, showing similarities with ADR described in other recent studies.
One of the study limitations is the fact that the query in the laboratory database converged one time-period and was restricted to sample source, bacteria species and antibiotic susceptibility result. With the available LIMS query method, it was impossible to distinguish whether multiple specimens originated from one patient. Samples obtained from hospitals might have included patients hospitalized >48 h, which makes it impossible in this study to distinguish between hospital acquired infections and resistance patterns at community level. In addition, because information on antibiotic use prior to the bacterial culture was not available it is not possible to distinguish between patients who had been treated with antibiotics and those who were not. The tradition of empirical antibiotic treatment of presumed infectious illnesses in Ethiopia is such that it can be expected that patients will seek medical advice after treatment failure, which explains the high ADR levels described in this study.
The majority of antibiograms in this study were derived from urine samples in which E. coli was most often determined. More than 80% of the cultures showed resistance to ampicillin and tetracycline, while resistance rates to chloramphenicol, nitrofurantoin and imipenem were less than 27%. Overall, the resistance patterns were similar to the results described by Abejew et al. for the Dessie regional health research laboratory [15] in which also high rates was described for ampicillin and tetracycline while susceptible to nitrofurantoin. High rates of multi-drug resistant extended-spectrum beta-lactamase (ESBL) producing enterobacteriaceae uropathogens were described in Gondor hospital [16]. The results of the ESBL tests could not be entered in the LIMS during the study timeframe and were directly reported to physicians. Examination of these reports from the study time also suggests high prevalence of ESBL. After the recent installation of the BD Phoenix 100 instrument at International Clinical Laboratories (ICL) and with the data now being stored in the LIMS, suspected and confirmed ESBL-producing microorganisms are now encountered almost daily (private communication with ICL microbiologist).
The second most numerous specimens received at the laboratory were obtained from wounds, with most infections due to S. aureus. These bacteria demonstrated less than 21% resistance to most antibiotics, with the exception of penicillin (n = 20; resistance rate = 90.0%), ampicillin (n = 24; resistance rate = 87.5%) and oxacillin (n = 25; resistance rate = 64.0%). Presence of methicillin-resistant S. aureus (MRSA) could not be proven in this study. Other studies indicate high rates of MRSA in the community and amongst health-care workers, however, reliance on phenotypic tests usually provides an overestimation [17–19]. There are no recent estimates of the prevalence of MRSA in the Ethiopian community, therefore, making molecular confirmation methods more accessible in Ethiopia to be crucial for future national surveillance programs.
A comparison of the data obtained with the recommendations in the Ethiopian standard treatment guidelines [4] clearly shows that a high treatment failure rate can be expected when drugs are administered according to national protocols. There are extensive differences in the rates of resistance observed for different bacterial species within one type of specimen. It is therefore important to back up treatment consultations with bacterial species identification accompanied by antibiograms.
Resistance to antibiotics poses serious threats not only in the developing world, but internationally [20]. National and international surveillance initiatives are on-going to identify and publicize trends in resistance rates and, in some instances, have resulted in rapid changes to national treatment guidelines [21, 22]. The crucial ingredient lacking in Ethiopian health policy is the availability of reliable data. The Ethiopia Food, Medicine, Healthcare Administration and Control Authority (EFMHACA) is currently establishing a five strategic plan to decrease antibiotic resistance. This strategy is based on awareness, evidence-based information, infection prevention, optimized use of antimicrobials in human and animal health and strengthening national partnerships. Microbiology laboratories are the first line of identification of antibiotic resistance among the general public. Besides delivering awareness to its clients, the laboratories play a key role in providing evidence-based data for antimicrobial surveillance. Reliable trends are directly correlated with reliable data input. The World Health Organisation published a manual on how to aggregate and validate national ADR files [23]. Additional guidance on how to structure raw data into correct data frames and how to visualize the data into clarifying figures could facilitate and encourage microbiology laboratories to assist in data collection. With currently available information technology, it takes relatively little effort to set up the application with real-time data feed as an additional service from diagnostic laboratories towards all levels: diagnostic laboratories providing the additional service of a real-time data feed that reaches all levels ranging from the clinicians prescribing antibiotics to national surveillance programs.
Conclusions
The extraction and analysis of raw-data from the laboratory database provides valuable insights into the relationships between type of sample and drug-resistance in countries where such data is still scarce. When used as an easy accessible reference application for healthcare providers, computer-based surveillance can reveal trends in antibiotic resistance levels and thus prevent treatment failures.
Authors’ contributions
RTH: writing script, data analysis and drafting the manuscript; MT: laboratory work and drafting manuscript and Methods; WTH: writing script and data analysis. MN: supervision and drafting the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Mr. Kebede Fufa and the staff of the microbiology section for their input on microbiology diagnostics, and the International Clinical Laboratories for making the data accessible. Thanks also to Miss. Asnakech Alemu who was so kind as to elucidate us on the EFMHACA strategy plan. We extend our sincere appreciation to Sara Chauvin for reviewing the manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset supporting the conclusions of this article is available in the Github repository, https://github.com/grebbel/antibiogram.git.
Ethics approval and consent to participate
The research is based on retrospective analysis of data extracted from the laboratory management system. The data was analyzed anonymously and results cannot be traced to individual patients. Ethical approval is therefore not applicable. Raw data in connection with a paper is publically accessible.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ADR
antibiotic drug resistance
- ATCC
American Type Culture Collection
- CNS
coagulase negative Staphylococcus
- CSF
cerebral spinal fluid
- ESBL
extended-spectrum beta-lactamase
- EFMHACA
Ethiopia Food, Medicine, Healthcare Administration and Control Authority
- ICL
International Clinical Laboratories
- LIMS
laboratory information management system
- MRSA
methicillin-resistant S. aureus
Appendix 1
See Table 2.
Table 2.
Identified bacteria with frequency in and percentage of the total number of isolates
| Bacteria isolated | Frequency | Percentage (%) |
|---|---|---|
| Acinetobacter species | 12 | 0.08 |
| Actinomyces viscosus | 9 | 0.06 |
| Bacteroides fragilis | 2 | 0.01 |
| Beta hemolytic non-group A | 7 | 0.05 |
| Citrobacter braakii | 1844 | 12.31 |
| Citrobacter farmeri | 16 | 0.11 |
| Citrobacter freundii | 8 | 0.05 |
| Citrobacter koseri | 8 | 0.05 |
| Coagulase-negative Staphylococcus species | 16 | 0.11 |
| Enterobacter cloacae | 59 | 0.39 |
| Enterobacter species | 15 | 0.10 |
| Enterococcus faecalis | 362 | 2.42 |
| Enterococcus species | 12 | 0.08 |
| Escherichia coli | 8057 | 53.77 |
| Group A Streptococcus | 33 | 0.22 |
| Group B Streptococcus | 85 | 0.57 |
| Group C Streptococcus | 26 | 0.17 |
| Group F Streptococcus | 9 | 0.06 |
| Group G Streptococcus | 10 | 0.07 |
| Haemophilus influenzae biotype I | 11 | 0.07 |
| Haemophilus influenzae | 5 | 0.03 |
| Klebsiella ornithinolytica | 36 | 0.24 |
| Klebsiella oxytoca | 446 | 2.98 |
| Klebsiella pneumoniae | 279 | 1.86 |
| Klebsiella species | 54 | 0.36 |
| Klebsiella terrigena | 24 | 0.16 |
| Moraxella lacunata | 8 | 0.05 |
| Morganella morganii | 8 | 0.05 |
| Pantoea species 4 (Erwina species) | 8 | 0.05 |
| Proteus mirabilis | 20 | 0.13 |
| Proteus penneri | 8 | 0.05 |
| Proteus species | 50 | 0.33 |
| Proteus vulgaris | 32 | 0.21 |
| Pseudomonas aeruginosa | 154 | 1.03 |
| Pseudomonas species | 771 | 5.15 |
| Salmonella group | 18 | 0.12 |
| Salmonella paratyphi A | 8 | 0.05 |
| Salmonella paratyphi B | 11 | 0.07 |
| Salmonella species | 41 | 0.27 |
| Serratia species | 13 | 0.09 |
| Shigella boydii | 19 | 0.13 |
| Shigella dysenteriae | 8 | 0.05 |
| Shigella group A1 | 9 | 0.06 |
| Shigella sonnei | 16 | 0.11 |
| Shigella species | 22 | 0.15 |
| Staphylococcus aureus | 1554 | 10.37 |
| Staphylococcus lugdunensis | 294 | 1.96 |
| Staphylococcus saprophyticus | 80 | 0.53 |
| Streptococcus agalactiae (group B) | 11 | 0.07 |
| Streptococcus anginosus/milleri | 21 | 0.14 |
| Streptococcus pneumoniae | 63 | 0.42 |
| Streptococcus pyogenes (group A) | 76 | 0.51 |
| Streptococcus species | 210 | 1.40 |
| Viridans streptococci | 5 | 0.03 |
| Total | 14.983 | 100.00 |
Appendix 2
See Table 3.
Table 3.
Percentage resistance and number of bacterial cultures (in brackets) per antibiotic after merging bacterial species and filtering out antibiogram groups containing less than 7 observations
| β-Hemolytic Streptococci | CNS | Enterococcus spp. | Escherichia coli | Klebsiella spp. | Proteus spp. | Pseudomonas spp. | Salmonella spp. | Shigella spp. | Staphylococcus aureus | Streptococcus spp. | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VA30 | 3.4 (119) | 3.3 (30) | 3.5 (57) | 8.3 (12) | |||||||
| TE30 | 4.8 (21) | 5.3 (19) | |||||||||
| SXT25 | 2.6 (156) | 3.7 (27) | 0.7 (740) | 4.6 (65) | 11.1 (9) | 4.6 (65) | 11.1 (9) | 3.4 (87) | 8.3 (12) | ||
| P10 | 4.8 (42) | 5.9 (17) | 5.0 (20) | ||||||||
| OX1 | 6.7 (30) | 4.0 (25) | |||||||||
| NOR10 | 2.6 (151) | 3.6 (28) | 0.7 (726) | 3.5 (57) | 11.1 (9) | 5.4 (74) | 14.3 (7) | 11.1 (9) | 3.4 (89) | 12.5 (8) | |
| NA30 | 5.0 (40) | 7.7 (13) | 0.6 (363) | 4.0 (25) | 5.6 (36) | ||||||
| IMP | 6.7 (30) | 1.3 (224) | 10.0 (10) | 6.7 (15) | 10.0 (10) | ||||||
| GM10 | 2.7 (150) | 0.7 (726) | 4.5 (66) | 11.1 (9) | 4.6 (65) | 14.3 (7) | 3.5 (86) | 7.7 (13) | |||
| FOX30 | 10.0 (10) | ||||||||||
| FM300 | 2.8 (143) | 3.1 (32) | 0.6 (643) | 5.1 (39) | 11.1 (9) | 3.4 (59) | 2.7 (73) | 8.3 (12) | |||
| E15 | 2.7 (149) | 3.4 (29) | 3.7 (81) | 7.7 (13) | |||||||
| CXM30 | 5.0 (40) | 14.3 (7) | 1.0 (388) | 5.6 (36) | 14.3 (7) | 4.7 (43) | 3.8 (26) | ||||
| CTX30 | 14.3 (7) | 2.4 (82) | 11.1 (9) | 0.7 (735) | 4.6 (65) | 12.5 (8) | 5.9 (68) | 14.3 (7) | 11.1 (9) | 6.4 (47) | 8.3 (12) |
| CRO30 | 14.3 (7) | 1.4 (71) | 11.1 (9) | 0.7 (746) | 4.5 (67) | 12.5 (8) | 5.9 (68) | 14.3 (7) | 11.1 (9) | 6.1 (49) | 7.7 (13) |
| CIP5 | 2.8 (143) | 3.4 (29) | 0.7 (739) | 3.6 (55) | 11.1 (9) | 4.5 (66) | 12.5 (8) | 11.1 (9) | 3.5 (86) | 11.1 (9) | |
| CF30 | 3.1 (129) | 3.6 (83) | 7.7 (13) | ||||||||
| CC2 | 2.6 (152) | 5.9 (17) | 3.6 (84) | 7.7 (13) | |||||||
| C30 | 4.8 (62) | 5.3 (19) | 1.0 (400) | 5.9 (34) | 5.0 (40) | 6.3 (32) | |||||
| AMP10 | 6.8 (44) | 6.7 (15) | 0.9 (331) | 6.7 (30) | 5.6 (36) | 4.2 (24) | |||||
| AMC30 | 14.3 (7) | 2.6 (152) | 4.0 (25) | 0.9 (559) | 5.9 (51) | 12.5 (8) | 5.0 (60) | 3.7 (3.7) | 7.7 (13) |
CNS coagulase negative staphylococci, SAMC30 amoxicilline/clavulate acid, AMP10 ampicillin, C30 chloroamphenicol, CC2 clindamycin, CF30 cephalophin, CIP5 ciprofloxacin, CRO30 ceftriaxone, CTX30 cefotaxime, CXM30 cefuroxime, E15 erythromycin, FM300 nitrofurantoin, FOX30 cefoxitin, GM10 gentamicin, IMP imipenem, NA30 nalidixic acid, NOR10 norfloxacin, OX1 oxacillin, P10 penicillin, SXT25 sulfamethoxazole, TE30 tetracycline, VA30 vancomycin
Contributor Information
Robert-Jan ten Hove, Phone: +33 69 33 19 93, Email: robert@theparasitologist.com.
Melaku Tesfaye, Email: melaku@icladdis.com.
Witold Frederik ten Hove, Email: wfth1969@gmail.com.
Mesfin Nigussie, Email: mesfin@icl.com.et.
References
- 1.Essack SY, Desta AT, Abotsi RE, Agoba EE. Antimicrobial resistance in the WHO African region: current status and roadmap for action. J. Public Health. 2016 doi: 10.1093/pubmed/fdw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global action plan on antimicrobial resistance. WHA68.7. Sect. Agenda item 15.1 May 26. 2015. p 1–4. http://apps.who.int/gb/ebwha/pdf_files/WHA68/A68_R7-en.pdf.
- 3.Draft political declaration of the high-level meeting of the General Assembly on antimicrobial resistance. 16-16108 Sep 21. 2016. p 1–5. http://www.un.org/pga/71/wp-content/uploads/sites/40/2016/09/DGACM_GAEAD_ESCAB-AMR-Draft-Political-Declaration-1616108E.pdf.
- 4.Drug Administration and Control Authority of Ethiopia. Standard Treatment guidelines for health centers; 2010.
- 5.Groot MJ, van’t Hooft KE. The hidden effects of dairy farming on public and environmental health in the Netherlands, India, Ethiopia, and Uganda, considering the use of antibiotics and other agro-chemicals. Front Public Health. 2016;4:1–9. doi: 10.3389/fpubh.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zenebe T, Kannan S, Yilma D, Beyene G. Invasive bacterial pathogens and their antibiotic susceptibility patterns in Jimma Specialized Hospital, Jimma, Southwest Ethiopia. Ethiop J Health Sci. 2011;21:1–8. doi: 10.4314/ejhs.v21i1.69038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SS, Yokoe DS, Stelling J, Placzek H, Kulldorff M, Kleinman K, et al. Automated detection of infectious disease outbreaks in hospitals: a retrospective cohort study. PLoS Med. 2010;7:e1000238. doi: 10.1371/journal.pmed.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Brien TF, Stelling J. Integrated multilevel surveillance of the world’s infecting microbes and their resistance to antimicrobial agents. Clin Microbiol Rev. 2011;24:281–295. doi: 10.1128/CMR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isenberg H. Clinical microbiology procedures handbook. American Society for Microbiology; 2004.
- 10.Murray PA. Manual of clinical microbiology. 7th ed; 1999.
- 11.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests; approved standard; 2012.
- 12.ten Hove R-J, ten Hove W, Tesfayem M. Github repository for the antibiogram data set and R scripts. 2016. https://github.com/grebbel/antibiogram.git.
- 13.Shiferaw B, Gelaw B, Assefa A, Assefa Y, Addis Z. Bacterial isolates and their antimicrobial susceptibility pattern among patients with external ocular infections at Borumeda hospital, Northeast Ethiopia. BMC Ophthalmol. 2015;15:103. doi: 10.1186/s12886-015-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, et al. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital, Northern Ethiopia. SpringerPlus. 2015;4:314. doi: 10.1186/s40064-015-1056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abejew AA, Denboba AA, Mekonnen AG. Prevalence and antibiotic resistance pattern of urinary tract bacterial infections in Dessie area, North-East Ethiopia. BMC Res Notes. 2014;7:687. doi: 10.1186/1756-0500-7-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4:12. doi: 10.1186/s13756-015-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falagas ME, Karageorgopoulos DE, Leptidis J, Korbila IP. MRSA in Africa : filling the global map of antimicrobial resistance. PloS ONE. 2013;8:e68024. doi: 10.1371/journal.pone.0068024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibabaw A, Abebe T, Mihret A. Nasal carriage rate of methicillin resistant Staphylococcus aureus among Dessie Referral Hospital Health care workers; Dessie, Northeast Ethiopia. Antimicrob Resist Infect Control. 2013;2:1. doi: 10.1186/2047-2994-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tibebu M, Embiyale W. Community acquired multi drug resistant Staphylococcus aureus in a rural setting of North Western Ethiopia: a tough challenge. Ethiop Med J. 2014;52:147–150. [PubMed] [Google Scholar]
- 20.World Health Organization. The evolving threat of antimicrobial resistance—options for action. 2012. http://www.who.int/patientsafety/implementation/amr/publication/en/.
- 21.Ison CA, Town K, Obi C, Chisholm S, Hughes G, Livermore DM, et al. Decreased susceptibility to cephalosporins among gonococci: data from the Gonococcal Resistance to Antimicrobials Surveillance Programme (GRASP) in England and Wales, 2007–2011. Lancet Infect Dis. 2013;13:762–768. doi: 10.1016/S1473-3099(13)70143-9. [DOI] [PubMed] [Google Scholar]
- 22.Johnson AP. Surveillance of antibiotic resistance. Phil Trans R Soc B. 2015;370:20140080. doi: 10.1098/rstb.2014.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. Antimicrobial resistance. GLASS documents and tools. 2017. http://www.who.int/antimicrobial-resistance/global-action-plan/surveillance/glass-documents/en/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is available in the Github repository, https://github.com/grebbel/antibiogram.git.


