Abstract
We report the genome sequence of Bifidobacterium animalis subspecies lactis BL3, which has preventive properties on acute colitis and colon cancer. The genome of BL3, which was isolated from Korean faeces, consisted of a 1 944 323 bp size single chromosome, and its G+C content was 60.5%. Genome comparison against the closest Bifidobacterium animalis strain revealed that BL3 had particularly different regions of four areas encoding flavin-nucleotide-binding protein, transposase, multidrug ABC transporter and ATP binding protein.
Keywords: Bifidobacterium, genome sequence, IBD, PacBio, probiotics
Introduction
Inflammatory bowel disease (IBD) is a group of chronic inflammatory disorders occurring in the digestive tract [1]. Ulcerative colitis and Crohn disease, as principal types of IBD, can affect the entire gastrointestinal tract [2]. While ulcerative colitis affects the large intestine and rectum with continues inflammation, Crohn disease causes inflammation of the lining of overall digestive tract and can even spread deep into tissue [3], [4], [5].
IBD symptoms include abdominal pain, vomiting, diarrhoea, rectal bleeding and weight loss [6], [7]. Currently there are no drugs for the treatment of IBD and only few therapeutic options for modulating intestinal inflammation; sustained IBD can be an increased risk factor for colorectal cancer [2]. IBD a complex disease caused by various factors such as environment, genetics, immunologic responses and inflammation [8]. However, recent studies have paid attention to gut microbiota and have suggested that alterations of the intestinal microbiome may contribute to inflammation and the progression of IBD [9], [10]. Therefore, modulation of intestinal flora could be a therapy for IBD treatment.
Previously we isolated Bifidobacterium animalis subspecies lactis BL3 strain from Korean faeces, which showed a preventive effect on acute colitis and colitis-associated colon cancer by inhibiting NF-κB activity [11]. In order to gain better insight into the preventive effects of probiotic Bifidobacterium on IBD, we analysed the genome sequence of B. animalis subspecies lactis BL3. Currently only seven genomes of B. animalis subspecies lactis strains are available, so the genetic information of this species is still insufficient. Therefore, in this study we analysed the whole genome sequence of B. animalis subspecies lactis BL3 to elucidate and understand preventive effect of probiotics on IBD and related disorders. Further characterization of genomic contents in probiotic Bifidobacterium such as B. animalis subspecies lactis BL3 will be needed to develop health-promoting probiotics.
Materials and methods
Bacteria strains and DNA preparation
Previously isolated B. animalis subspecies lactis BL3 from Korean faeces was cultivated on BL medium at 37°C for 18 hours in an anaerobic condition. Genomic DNA was extracted from the cultured bacterium with a QIAamp DNA Mini Kit (Qiagen, Germantown, MD, USA). The purity, quality and quantity of extracted DNA was examined by a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher, Waltham, MA, USA) and Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA) respectively.
Genome sequencing, assembly and annotation
The whole genome of B. animalis subspecies lactis was sequenced by the PacBio RS II platform. A 20 kb DNA library, constructed according to the manufacturer's instructions, was sequenced by single-molecule real-time sequencing technology with P6 DNA polymerase and C4 chemistry. A total of 1038 high-quality sequences (182 595 subreads) were obtained from the sequencing. The sequences were assembled using HGAP 3.0, and annotation was carried out with National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline [12] through the NCBI Genome submission portal (GenomeSubmit, http://ncbi.nlm.nih.gov). DNAPlotter [13] was used to draw the chromosome topology of this genome. Functional classification of the coding genes was performed using BLASTP search against the Clusters of Orthologous Groups (COGs) database [14], [15]. The CRISPR region was detected using CRISPRFinder [16]. The genomic similarity between two genome sequences was estimated by the computation of orthologous average nucleotide identity (OrthoANI) using the Orthologous Average Nucleotide Identity Tool [17]. Pan-genomic study using Panseq [18] was performed to investigate the genomic conservation and find novel sections in the sequenced genome.
Results and discussion
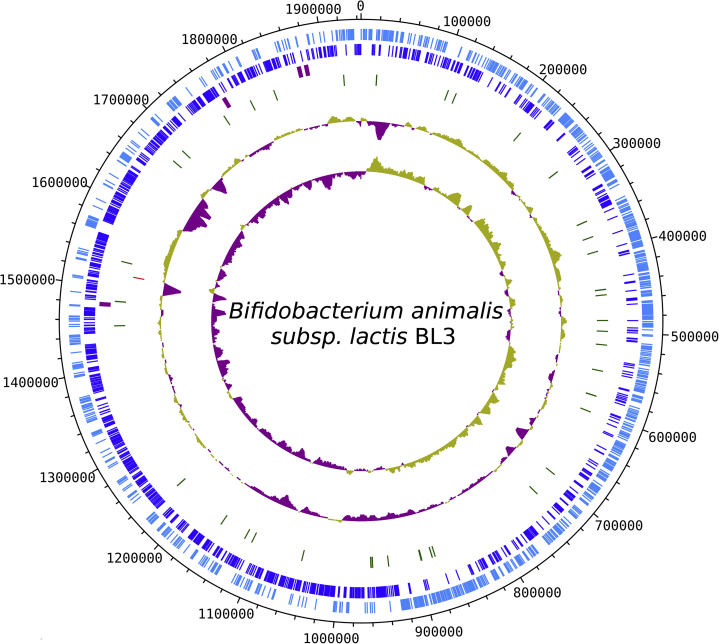
The high depth of single-molecule real-time sequencing enabled to produce a finished-level complete genome sequence in the analysis of B. animalis subspecies lactis BL3. No ambiguous base and no circular gaps were found in the genome sequence of BL3. The genome of BL3 consisted of a 1 944 323 bp size single chromosome, and its G+C content was 60.5%. In total 1613 genes were identified by Prokaryotic Genome Annotation Pipeline (PGAAP 3.3) with both the best-placed reference protein set and an ab initio prediction program, GeneMarkS+. The number of protein-coding gene was 1532, and 14 pseudo genes were discovered. Twelve rRNA genes (four 5S, four 16S and four 23S), 52 tRNA genes and three noncoding genes were also found. A 1,618 nt long CRISPR array was found in this genome at approximately the 1 517 114 to 1 518 732 position. Twenty-two spacers were detected in the array, and the DR sequence was [C/T]CCTCAATGAAGCTCCGAAGCCGAGACTTCGGAGAT. Genomic features of B. animalis subspecies lactis BL3 are presented in Fig. 1.
Fig. 1.
Circular map of genomic features of Bifidobacterium animalis subspecies lactis BL3 plotting seven tracks. Track 1 (light blue; outset): forward-stranded coding CDS. Track 2 (blue): reverse-strand coding CDS. Track 3 (light purple): rRNA including 5S, 16S and 23S. Track 4 (green): tRNA. Track 5 (red): CRISPR array. Track 6 (light green and purple): GC content. Track 7 (light green and purple): GC skew.
We found that the closest strain was Bifidobacterium animalis subspecies lactis DSM 10140 (accession no. NC_012815) from the BLAST search against the 16S targeted loci nucleotide database; the sequence identity was 100%. The genomic features of DSM 10140 were similar to the BL3 genome (Table 1). Moreover, genome-to-genome similarity was estimated as 99.98% via OrthoANI computation. Genomic differences between DSM 10140 and BL3 were found at four regions: 881 501∼882 000, 1 305 501∼1306 000, 1 309 501∼1310 000 and 1 820 501∼1821 000. The first region encodes two tRNA genes and a coding gene (locus_tag: BGL50_03705), which produces flavin–nucleotide binding protein. The second region has no annotation. The third one encodes a coding gene (locus_tag: BGL50_05610), which is annotated as ‘transposase.’ The last region encodes two coding genes (locus_tag: BGL50_07615 and BGL50_07620), which produce ‘multidrug ABC transporter’ and ‘ATP-binding protein.’
Table 1.
Genome summary of Bifidobacterium animalis subspecies lactis BL3 and DSM 10140
| Characteristic | B. animalis subsp. lactis BL3 | B. animalis subsp. lactis DSM 10140 |
|---|---|---|
| Genome | ||
| Accession | CP017098 | NC_012815 |
| Sequence | 1 chromosome | 1 chromosome |
| Total size | 1 944 323 | 1938 483 |
| G-C content | 60.48 | 60.48 |
| Assembly level | Complete genome | Complete genome |
| Annotation | ||
| Genes | 1613 | 1610 |
| Coding genes | 1532 | 1534 |
| Pseudo genes | 14 | 12 |
| RNAs | 67 | 64 |
| rRNAs (5S, 16S, 23S) | 4, 4, 4 | 4, 4, 4 |
| tRNAs | 52 | 51 |
| ncRNAs | 3 | 1 |
| CRISPRs | 1 | 1 |
The genomic and comparative genomic analyses of probiotic microorganisms will provide valuable information on the detailed functional properties and genotype-level safety of probiotics. The genetic study on the current probiotic microorganism will increase our knowledge of their biologic mechanisms against preventing human diseases and may lead to genome-based biotechnologic applications in the human healthcare and food industries utilizing Bifidobacterium strains.
Nucleotide sequence accession number
The assembled and annotated genome of B. animalis subspecies lactis BL3 has been deposited at DNA Data Bank of Japan, European Molecular Biology Laboratory, and GenBank under accession number CP017098. The version described here is the first version, CP017098. The genome sequence data are available in FASTA, annotated GenBank flat file and ASN.1 formats. This strain has also been deposited in the Korea Collection for Type Culture (KCTC 11904BP).
Acknowledgements
This research was supported by Main Research Program of the Korea Research Food Institute funded by the Ministry of Science, ICT and Future Planning (E0170602-01).
Contributor Information
S. Lim, Email: shlim@cellbiotech.com.
Y.-D. Nam, Email: youngdo98@kfri.re.kr.
Conflict of Interest
None declared.
References
- 1.Baumgart D.C., Sandborn W.J. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 2.Kaser A., Zeissig S., Blumberg R.S. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lockhart-Mummery H.E., Morson B.C. Crohn’s disease (regional enteritis) of the large intestine and its distinction from ulcerative colitis. Gut. 1960;1:87–105. doi: 10.1136/gut.1.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho J.H., Brant S.R. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140:1704–1712. doi: 10.1053/j.gastro.2011.02.046. e1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ford A.C., Moayyedi P., Hanauer S.B. Ulcerative colitis. BMJ. 2013;346:f432. doi: 10.1136/bmj.f432. [DOI] [PubMed] [Google Scholar]
- 6.Hanauer S.B., Sandborn W. Management of Crohn’s disease in adults. Am J Gastroenterol. 2001;96:635–643. doi: 10.1111/j.1572-0241.2001.3671_c.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu S., Ren J., Zhao Y., Han G., Hong Z., Yan D. Nonthyroidal illness syndrome: is it far away from Crohn’s disease? J Clin Gastroenterol. 2013;47:153–159. doi: 10.1097/MCG.0b013e318254ea8a. [DOI] [PubMed] [Google Scholar]
- 8.Baumgart D.C., Carding S.R. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 9.Mukhopadhya I., Hansen R., El-Omar E.M., Hold G.L. IBD—what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 10.Aroniadis O.C., Brandt L.J. Fecal microbiota transplantation: past, present and future. Curr Opin Gastroenterol. 2013;29:79–84. doi: 10.1097/MOG.0b013e32835a4b3e. [DOI] [PubMed] [Google Scholar]
- 11.Kim S.W., Kim H.M., Yang K.M., Kim S.A., Kim S.K., An M.J. Bifidobacterium lactis inhibits NF-κB in intestinal epithelial cells and prevents acute colitis and colitis-associated colon cancer in mice. Inflamm Bowel Dis. 2010;16:1514–1525. doi: 10.1002/ibd.21262. [DOI] [PubMed] [Google Scholar]
- 12.Tatusova T., Dicuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke G.P., Beiko R.G., Ragan M.A., Charlebois R.L. Inferring genome trees by using a filter to eliminate phylogenetically discordant sequences and a distance matrix based on mean normalized BLASTP scores. J Bacteriol. 2002;184:2072–2080. doi: 10.1128/JB.184.8.2072-2080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grissa I., Vergnaud G., Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee I., Kim Y.O., Park S.C., Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 18.Laing C., Buchanan C., Taboada E.N., Zhang Y., Kropinski A., Villegas A. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics. 2010;11:1. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]