Abstract
The field of gene editing is undergoing unprecedented growth. The first ex vivo human clinical trial in China started in 2016, more than 1000 US patents have been filed, and there is exponential growth in publications. The ability to edit genes with high fidelity is promising for the development of new treatments for a range of diseases, particularly inherited conditions, infectious diseases, and cancers. For cancer, a major issue is the identification of driver mutations and oncogenes to target for therapeutic effect, and this requires the development of robust models with which to prove their efficacy. The challenge is that there is rarely a single critical gene. However, virally driven cancers, in which cells are addicted to the expression of a single viral oncogene in some cases, may serve as model systems for CRISPR/Cas therapies, as they did for RNAi. These models and systems offer an excellent opportunity to test both preclinical models and clinical conditions to examine the effectiveness of gene editing, and here we review the options and offer a way forward.
Keywords: CRISPR/Cas9, virally-driven cancers, cervical cancer, oncogene-addiction
Main Text
The Concept of Oncogene Addiction as the Basis for Targeted Anti-cancer Therapies
Oncogene addiction may be defined as the unconditional dependence of cancer cells on the expression of a single gene, or multiple genes, in order to survive and grow. The notion of “oncogene addiction” was first described by Bernard Weinstein in 2000,1, 2 based on the observation that despite the involvement of multiple genetic and epigenetic abnormalities in the process of cancer development, a single gene inactivation may halt cancer growth and survival.
Several models were proposed to explain the biological mechanisms of oncogene addiction.3 The first hypothesis was genetic streamlining, which is based on the well-known notion among early evolutionists that useless organs and structures are lost over time.4 This hypothesis postulates that in the presence of selective pressure of the tumorigenic microenvironment, molecular pathways that are deemed non-essential to cell viability would be inactivated.3 In other words, this change renders cell viability entirely reliant on dominant pathways, which if lost, cellular fitness would collapse. On the other hand, the “oncogenic shock” theory hypothesized that oncoproteins possess pro-survival and pro-apoptotic signals.5 In normal cells, this signaling duality allows for rapid switching between biological outputs, which is crucial to counteract oncogenic insults with pro-mitogenic signals. In tumor cells, the survival output dominates over the pro-apoptotic signals. Upon disruption of an oncogene product, the pro-survival signals are believed to last for a shorter duration compared to pro-apoptotic signals, thus resulting in an apoptotic response.6 The synthetic lethality model was put forward to elucidate the molecular events leading to this phenomenon.7 This model states that for any two or more genes to be synthetically lethal, the loss of function of either gene is compatible with cell viability, but the loss of function of both or all genes is lethal to cellular fitness.7 In the context of cancer, the latter model is not limited to gene-gene interactions; it also involves the combination of other epigenetic or pharmacological perturbations that result in a lethal phenotypic outcome.8
More recently, the oncogene amnesia model was proposed to explain tumorigenesis.9 This model accommodates the role of host mechanisms in cancer development. It assumes that for a certain oncogene(s) to be able to signal the malignant transformation of normal cells, physiological barriers (or the safety switches that prompt cell-cycle arrest as a response to genomic stress or instability) must be shut off by other oncogenes or genetic events. Therefore, the cell defense checkpoints are oblivious to genomic insult, or are in a state of cellular “amnesia,” when an oncogene(s) signals malignant transformation.9 In the event of oncogene inactivation, some of these safety switches are restored; therefore, the cell would become aware of such a genomic insult. This cellular “awakening” activates the relevant molecular pathways to promote cell-cycle arrest by apoptosis or senescence.10
Importantly, the notion of oncogene addiction has provided the conceptual framework to characterize and develop cancer-specific therapies. Given that each cancer is different in terms of the molecular events involved in its development, and if these oncogenes are the long sought-after Achilles’ heel of cancer, exploiting this concept may lay the first stone toward developing more targeted therapies. This seems to be particularly relevant to virally driven cancers, where the driver genes are absolutely known.
Human Papillomavirus and Cervical Cancer: An Example of Oncogene Addiction
The case for a novel therapeutic for cervical cancer is clear. Cervical cancer ranked as the fourth most prevalent cause of cancer deaths worldwide among women in 2012, with 265,000 deaths and another 527,000 new cases.11 The 5-year survival rate has not changed for 20 years, so current treatment strategies have plateaued in efficacy.12 Vaccines cannot treat already-established cancer nor do they provide protection against all types of human papillomavirus (HPV) infection.11, 13, 14 Moreover, current strategies (radiotherapy, chemotherapy, and surgery) suffer from serious drawbacks, are harsh on normal cells, and have not shown adequate selectivity in targeting the malignant tissue of the cervix.12, 15, 16, 17, 18
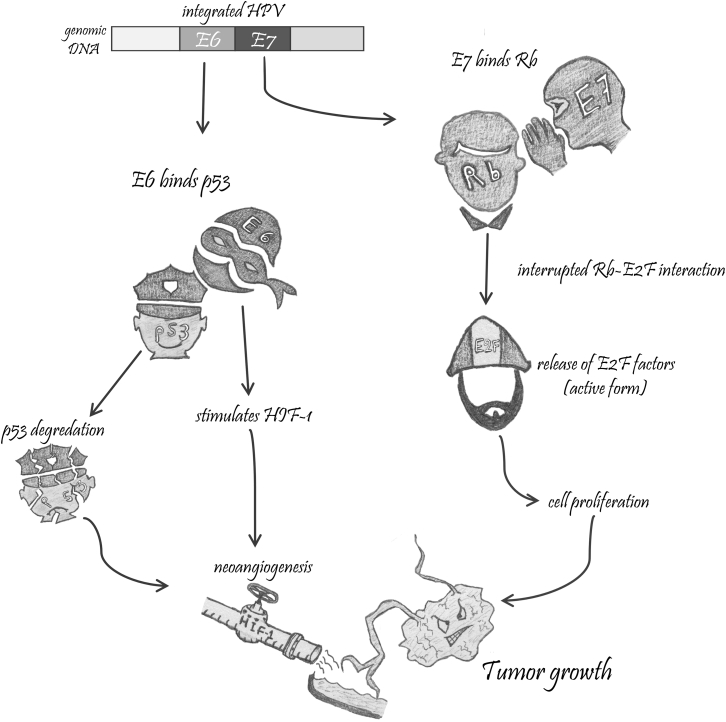
For high-risk HPV types (those that cause cancer), the ongoing expression of E6 and E7 (and to a lesser extent, E5) oncoproteins is required and the loss of this expression results in senescence or apoptosis of these cells.19 It was concluded that the repression of either E6 or E7 viral proteins results in cell-cycle arrest and cell death. Further investigation of the molecular pathways associated with E6 and E7 gene silencing revealed that inactivation of E7 gene expression is associated with re-establishment of the retinoblastoma protein (pRb) pathway, which triggers cell senescence. On the other hand, repression of E6 protein reactivates the p53 pathway, leading to both apoptosis and senescence.19 The course of events involved in the carcinogenesis of the cervix is consistent with the oncogenic amnesia theory. Consistent with this is the long period of time required between the initial HPV infection and the development of cancer, implicating the involvement of multiple epigenetic and genetic alterations. Such changes were associated with the process of overriding checkpoint mechanisms, which are crucial for cell mortality and genomic integrity (Figure 1).9
Figure 1.
Carcinogenesis of the Cervix
HPV-infected cervical cells express viral oncoproteins, E6 and E7. E6 oncoprotein (the militia man) binds the tumor suppressor protein, p53 (the police officer), resulting in its degradation. It also stimulates the hypoxia-inducible factor 1 (HIF-1) transcription factor, an innate hypoxic stress response in normal cells that binds to p53 to induce apoptosis. Because p53 is largely degraded, HIF-1 stimulates the formation of new blood vessels (the supply pipeline), which in turn provide the growing tumor cells with a blood supply. On the other hand, the E7 oncoprotein (the militia leader) binds the “pocket domains” of pRb protein (the corrupt politician), which interrupts the interaction between pRb protein and E2F family transcription factors (the worker), resulting in their release in their transcriptionally active form and leading to cell-cycle activation and a malignant transformation.
Genome Editing
Targeted genome editing, by utilizing programmable endonucleases, has rapidly evolved during the last 5 years. The most well-known system is the CRISPR-associated endonuclease (Cas9), which was first described after the observation that the prokaryotic adaptive immune system can also be exploited as a programmable platform to generate precise insertions or deletions (indels) in living cells.20 Briefly, the best characterized CRISPR system (type II) encodes for Cas protein (Cas9), which induces double-stranded breaks (DSBs) at specific sites of the target DNA. Cas9 uses dual RNAs, or a guide RNA (gRNA), for site-specific DNA cleavage. These gRNAs are made of CRISPR RNA (crRNA), which binds to the target site through Watson-Crick base pairing, and trans-activating crRNA (tracrRNA), which is important for the recognition of and engagement with the Cas9 protein.21 After cleavage, the DSBs are repaired by either the error-prone non-homologous end joining (NHEJ) pathway or the high-fidelity homology-directed repair (HDR) pathway. The latter requires an exogenous DNA repair construct with homology arms flanking the insertion site to introduce a defined modification to the target locus.22
The first description of the CRISPR/Cas9 system was an exciting time for genome-engineering technology. CRISPR/Cas9 is easy to design, highly specific, well suited for high-throughput use, and has the ability to do multiplex gene editing simultaneously. It holds enormous potential for many applications, such as systemic interrogation of genetic elements and functions.23 Basically, CRISPR technology can be harnessed to generate loss-of-function or gain-of-function mutations in tumor suppressor genes, oncogenes, or other modulators of gene expression.24 Upon cleavage of a target DNA by Cas9, and in the absence of a DNA repair template, the genomic DNA is repaired by the NHEJ pathway, hence introducing random indels and resulting in frameshift mutations and gene knockout. Similarly, multiple target sites can be cleaved to generate multiple DSBs and hence mediate larger deletions in the genome.25 The latter editing strategies may be ideal when a loss-of-function type of editing is intended, such as inactivating certain genes or gene deletions. Alternatively, genome editing through the HDR pathway can be leveraged to create a gene knock in, which is particularly effective for making small edits (e.g., “fixing” existing mutations or introducing point mutations or unique restriction sites) or larger edits (e.g., introducing fluorescent proteins or selection cassettes).23
How CRISPR Machinery Can Be Harnessed against Cervical Cancer
As previously explained, cervical cancer cells rely entirely on the continuous expression of the HPV E6 and E7 proteins to survive, which renders these proteins an ideal therapeutic target. Past attempts were made to inhibit the expression or translation of these oncoproteins.26 One of the early publications in this area used phosphorothioate oligonucleotides complementary to the regions flanking the start codons of HPV-16 E6 and E7 genes and thus resulted in growth inhibition of cervical cancer cell lines CaSki and SiHa.27 Several biological systems have since been exploited to interfere with the gene expression of E6 and E7 genes or their molecular interactions, such as using HPV E7 antagonist peptides,28 re-introducing the wild-type (WT) p53 into cervical cancer cell lines via a recombinant adenovirus vector,29 interrupting the binding between E6 oncoprotein and E6AP and E6BP via zinc-ejecting inhibitors,30 targeting E6 mRNA by anti-HPV-16 E6 ribozymes,31 or inhibiting the long control region (LCR) of HPV-18 by interrupting the binding of transcription factor AP1, using heparinoid GAG-hed.32 More recently, RNAi has been explored to silence the expression of E6/E7 genes via short-interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs).33, 34, 35
After the CRISPR/Cas9 revolution, researchers aimed to exploit this technology to target specific sites in the E6 and E7 genes. In HPV-16 and HPV-18 cervical cancer cell lines, targeting E6 and E7 genes with the WT Cas9 restored p53 and pRb cellular levels respectively, resulting in cell death and apoptosis.36, 37, 38 The same significant effect was also observed when HPV-16 E6 and E7 genes were knocked out in nude mice, leading to inhibition of tumor growth.39 The latter study also demonstrated that the E6 and E7 genes can be knocked out by targeting their common early promoter region, p97, which is located just upstream of E6.39 Given that both E6 and E7 genes are short, choosing unique target sequences within these genes can be challenging; thus, the notion of targeting other regulatory regions might serve as an alternative strategy. In fact, HPV gene expression is largely regulated at the post-transcriptional level, including RNA processing, nuclear export, mRNA stability, and translation, in addition to several early and late promoters controlling the expression at the transcriptional level.40 Therefore, interrupting these regulatory networks may be a promising approach. Apart from targeting the early p97 promoter for HPV-16 E6 and E7 genes (or the p105 promoter for HPV-18), CRISPR-based therapies could be designed to target, for example, the region extending from the end of the L1 open reading frame (ORF) to the late 3′ UTR, the region that encodes for key RNA stability regulators that preclude the expression of the viral capsid proteins.41 Because these capsid proteins are highly immunogenic, restoring their expression would trigger a robust immune response and thus lead to the clearance of the virus.40 Besides the early and late promoters, several other promoters have been described in various HPV genomes, such as a late promoter located at the beginning of the E4 ORF, which regulates the expression of L1 capsid protein,42 and one located at the E5 ORF that encodes for L2 as a first ORF.42 However, exploiting these regulatory pathways to enhance virus clearance or interrupt gene expression remains hindered by our limited understanding of the molecular interactions involving multiple genomic regions.
The Therapeutic Potential of CRISPR/Cas9 Systems in Other Virally Driven Cancers
Designing an effective cancer therapy is challenging because the drivers of tumorigenesis and the tumor microenvironment are complex and constantly changing.43 However, some cancer types are driven by well-characterized viral infections that if targeted may lead to a viable cancer treatment. One example is the role of Epstein-Barr virus (EBV) in the development of Burkitt’s lymphoma and nasopharyngeal carcinoma.44 EBV is a double-stranded DNA virus with 85 genes that infects lymphocytes and epithelial cells during the initial replication stage.45 When latent, EBV expresses few of its proteins that play a role in the malignant transformation, such as EBV nuclear antigen 1 (EBNA-1), EBNA-2, and latent membrane protein 1 (LMP-1) and LMP-2.46 These genes are involved in the activation of multiple molecular pathways that promote cell transformation, invasion, and cancer metastasis. Despite our understanding of its role in malignancy, current anti-viral treatments for EBV have largely failed to clear latent EBV infection. The approved anti-viral drugs for EBV target DNA polymerase, which is abundant during the active stage but not in the latent (non-replicating) stage of the infection. Therefore, attempts were made to utilize CRISPR/Cas9 systems to target EBV in the latent stage. When human cells derived from Burkitt’s lymphoma with Cas9/gRNAs against the EBNA-1,47, 48 LMP-1, or EBNA-3C genes were targeted,48 a significant reduction in cell proliferation was observed. This resulted in a decrease in viral load48 and almost a complete clearance of the latent EBV infection.47 Another research group also used CRISPR/Cas9 to generate a targeted deletion of 558 bp in the promoter region of the BamHI A rightward transcript (BART), which encodes for viral microRNAs (miRNAs), in latently infected nasopharyngeal carcinoma C666-1 cells.49 The results confirmed the loss of miRNA expression, indicating that this approach could be utilized as a therapy against latent EBV infection.
In addition, persistent viral infections also seem to play a role in the development of hepatocellular carcinoma.50 Similarly to EBV, the currently approved drugs against hepatitis B virus (HBV) have failed to fully eliminate viral infection from the body.51 This is because of the highly stable HBV covalently closed circular (ccc) DNA, a plasmid-like episome in the nucleus of the infected hepatocyte.52 Genome-editing platforms, including CRISPR/Cas9 systems, were utilized to target and disrupt HBV genome expression with favorable effects (reviewed in Lin et al.51 and White et al.53).
When treatment requires the full elimination of the virus, CRISPR/Cas9 systems seem ideal to achieve that goal. This can be applicable in many other virally driven cancers such as Merkel cell carcinoma, caused by Merkel cell polyomavirus,54 or Kaposi’s sarcoma, caused by human herpes virus 8.55 One could utilize the newer CRISPR/Cas effector C2c2 system for RNA viruses such as hepatitis C, which causes liver cancer,56 or human T lymphotrophic virus type I, which causes adult T cell leukemia/lymphoma57; this system is capable of editing RNA instead of DNA.
Apart from cancers driven by viral infection, the first human trial is still ongoing to utilize this technology to knock out programed cell death protein (PD-1) in T cells in patients with metastatic non-small cell carcinoma of the lung.58 The targeted PD-1 protein helps to halt the immune response; thus, its knockout should boost the immune response against cancer cells. While the latter trial is ex vivo, which allows for the selection of the edited cells and careful analysis of the product before it is infused back into patients, it should provide insight on the feasibility and the safety of gene editing in humans.58 More recently, a more ambitious human trial has been approved in the United States to use the CRISPR/Cas9 system to insert an extra gene to enable T cells to recognize and target several types of cancers, together with disabling the PD-1 gene and two other genes.59
Challenges to CRISPR-Based Therapeutics
To design an effective therapy with the potential for its clinical translation, the treatment needs to be efficacious against the disease of interest and deliverable to the target of interest, with tolerable or no serious adverse effects. To date, CRISPR/Cas9-based therapies have been associated with certain major limitations, such as the targeting specificity and the possibility of off-target binding of Cas9 protein,60 on-target editing efficiency,61 and the induction of the host immune response against the delivery vectors.62
On-Target Specificity
Previous studies revealed that not every nucleotide in the gRNA is needed to pair with the target DNA in order to edit genes, suggesting the possibility of off-target binding.20 Off-target mutations and single-nucleotide variants (SNVs) were also reported in sites with poor sequence homology to the gRNAs, and these mutations were not in the predicted regions as per the in silico modeling.63 This issue raises many safety concerns regarding potential CRISPR/Cas9-based therapeutics, and whether it might be associated with undesired mutations or disruption of normal gene function. Therefore, off-target effects need to be minimized to nearly zero or, alternatively, they must be precisely profiled.64 Targeting specificity is broadly determined by factors related to the gRNA characteristics, the Cas9 characteristics, the abundance of gRNA-Cas9 complexes, and the uniqueness of the chosen target sites.65 Several modifications were introduced to enhance the targeting specificity,66 such as truncating the gRNAs to shorter than 20 nt (increased specificity by more than 5,000-fold),67 introducing point mutations at the active sites of Cas9 nucleases to modify its cutting capacity and force it to use two-enzyme binding to create the DSB,68 or using catalytically inactive Cas9-fused FokI nucleases or FokI-dCas9.69 The added requirement of two-enzyme binding to generate DSBs has significantly increased the targeting specificity, but at the expense of on-target editing efficiency.66 Other modifications also involved manipulating the electropositive charge of the HNH/RucC groove of Cas9 endonuclease as a means to destabilize off-target binding between non-target DNA and Cas9 and reduce off-target editing (eSpCas9 variant).70 Disrupting the interaction between Cas9 and the phosphate backbone of the target DNA by introducing four mutations in the engineered Cas9 (SpCas9-HF1 variant) has also been described.71 Finally, newer Cas9 endonucleases such as SaCas9 were shown to be more specific.72 Others have shown that the delivery of purified Cas9 protein complexed with gRNA, rather than as plasmid DNA, improved on-target editing efficiency and reduced long-term accumulation of off-target mutations.73, 74
Since Cas9 endonucleases are capable of introducing mutations anywhere in the genome, regardless of the degree of sequence similarity to the on-target sites,75 therapies with such a nonspecific editing potential might pose a serious risk of activating alternative molecular pathways and altering the intrinsic tumor microenvironment. This may contribute to the emergence of treatment resistance. Indeed, several oncogene-targeted therapies previously showed the development of resistance to treatment through several mechanisms.76 For example, the acquisition of second-site mutations in the binding site of the tyrosine kinase inhibitor, imatinib, as a mechanism to develop resistance to therapy and reinstate the oncogene function in patients diagnosed with chronic myeloid leukemia.77 Moreover, mutations of the alternate components of oncogene-induced signaling pathways might be another theme for emerging resistance to oncogene-targeted therapies. For instance, the development of resistance to vemurafenib, a BRAF enzyme inhibitor approved for the treatment of late-stage melanoma, occurs by the acquisition of activating mutations in other components of mitogen-activated protein kinase (MAPK) oncogenic pathways, such as NRAS,78 or by loss-of-function mutations in the negative regulator NF1.79 Therefore, such possibilities should not be overlooked with CRISPR/Cas9-based treatments, especially in the case of cervical cancer. In spite of all the exciting preclinical results, none of the previously discussed studies that utilized the CRISPR/Cas9 system to treat cervical cancer assessed the specificity of the treatment. Thus, it is not possible to know whether it had any off-target toxicity. Therefore, genome-wide off-target detection by unbiased deep sequencing-based methods is imperative to ensure the stringency of on-target effects with no or precisely defined off-target edits.66
Knock-In Editing Efficiency
To date, several hundred publications have demonstrated the high efficiency of Cas9 endonucleases to achieve gene knockouts.80, 81 However, inserting a precise modification via the HDR repair pathway seems less efficient in most cases.82 Unlike the NHEJ, HDR repair occurs only during the S and G2 phases of the cell cycle83 and concurrently with the NHEJ84 and thus is less frequent than the NHEJ repair mechanism.82 The latter observation raises concerns about the risk of generating error-prone random indels, which may introduce unwanted mutations or contribute to the emergence of treatment resistance. In addition to its low frequency, the HDR repair pathway is also limited by the possibility of the continuous re-editing of the target sites; thus, the Cas9 endonucleases may continue re-cutting the edited loci until they are sufficiently modified by NHEJ to prevent further editing.81 To increase HDR editing efficiency, Maruyama et al.82 targeted DNA ligase IV, a key enzyme in the NHEJ repair pathway, and demonstrated that inhibiting this enzyme resulted in an up to 19-fold increase in the frequency of HDR repair. The gene silencing of other key NHEJ pathway proteins, such as KU70 and KU80 in addition to DNA ligase IV, by shRNAs has shown a significant increase in the frequency of HDR repair, which was also correlated with NHEJ inhibition.85 Apart from inhibiting the NHEJ pathway, modifying the repair template seems to be another strategy to promote HDR on the expense of NHEJ pathway. Renaud et al.86 tested different lengths of homology arms flanking the cleavage site and chemically modified the 3′ and 5′ ends of the repair oligonucleotides. The latter study reported that phosphorothioate-modified oligonucleotides with around 100 nt of the total homology region strongly improved the homologous recombination compared to the conventional phosphodiester oligonucleotides with shorter or longer homology regions.86 Moreover, Richardson et al.87 mapped the Cas9-DNA interaction and described the asymmetrical dissociation of the Cas9-DNA complex, releasing the 3′ end of the non-target DNA strand first; thus, designing a donor DNA template complementary to the non-target strand enhanced HDR efficiency by up to 60%. To prevent re-editing of already HDR-edited loci, Paquet et al.81 proposed introducing a Cas9-blocking mutation in the protospacer adjacent motifs (PAMs) or gRNA target sequence, which CRISPR/Cas9 requires for targeting. The findings from this study showed a significant increase in HDR efficiency by 2- to 10-fold when silent mutations were introduced to either PAMs or gRNA target sequences in various cell lines.81 The combination of NHEJ pathway inhibitors together with modifying the donor template may also be considered as a strategy to achieve precise, HDR-mediated edits.
Conclusions and Lessons from Previous Oncogene-Targeted Therapies
The utilization of CRISPR machinery as a versatile genome-editing platform represents a promising opportunity to design effective and highly efficient oncogene-targeted therapies against virally driven cancers. Despite their high editing efficiency, CRISPR/Cas9 systems remain limited by several challenges that need to be addressed. Therefore, a fierce debate has been triggered about whether this technology is sufficiently mature to be used in humans.88 These concerns are justified, given our limited understanding of how Cas9 endonucleases are actually editing the genome, the high frequency of random edits generated, and the possibility of accumulation of mutations. This dilemma is not unique to CRISPR/Cas9 systems; one study investigated the fate of HPV-16 E7 protein after silencing with siRNA and reported that the truncated 5′ end of the targeted protein is still actively expressed after treatment.89 This observation raises a critical question about the events that follow the editing and whether the generated knockouts (or knock ins) would be associated with any unexpected outcome. Therefore, it is imperative to roadmap how the designed treatment would affect the target of interest, the proteins expressed from that target site, the other known genes that interact with the target sites, the mechanism of effect (or cell death if intended), and the specificity of targeting by using unbiased sequencing methods.66
As previously discussed, many oncogene-targeted therapeutics have suffered from the emergence of treatment resistance.76 Despite the promising initial response, chronic exposure to such therapies often gives way to relapse. Since it seems fairly common, the emergence of resistance to future CRISPR-based cancer therapies should be anticipated. Several strategies were proposed to overcome the acquired resistance to therapy.76 Although targeting either E6 or E7 oncogenes should hypothetically cure cancer, the single-oncogene targeting strategy might be associated with resistance to therapy as a result of the acquisition of mutations in the target site. Therefore, targeting multiple oncogenes and molecular pathways simultaneously should be the recommended strategy to achieve a sustained clinical response. Moreover, tumor heterogeneity should be thoroughly characterized. While cervical cancer is almost always triggered by the expression of E6 and E7 oncoproteins, previous evidence reported the high percentage of intra- and inter-tumor heterogeneity in gene expression profiles among patients with cervical cancer.90 Further profiling of cervical cancer showed that it is not only metabolically heterogeneous, but this metabolic heterogeneity could also predict the response to the treatment.91 Although carcinogenesis of the cervix is initiated by the integration and expression of E6 and E7 oncoproteins, most infected women develop transient dysplasia that spontaneously regresses to normal. Therefore, it is safe to assume that cervical cancer development is multi-factorial, with the involvement of “other” genetic elements in addition to E6 and E7 oncoproteins. Therefore, in-depth characterization and profiling of these genetic alterations with the genetic definition of the patients who are most likely to respond, together with targeting multiple signaling pathways by employing the more specific Cas9 variants/modified gRNAs, should enable the design of more personalized, and yet durable, targeted therapies.
Author Contributions
L.J. contributed to the conception and design of this review, performed the literature search and acquisition of data, drafted the review, and approved the final version of this manuscript. N.A.J.M. contributed to the conception and design of this review, revised the initial draft critically for important intellectual content, and approved the final version of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
L.J. is funded by Griffith University. N.A.J.M. has received funding from the Cancer Council of Queensland and the National Health and Medical Research Council of Australia.
References
- 1.Weinstein I.B. Disorders in cell circuitry during multistage carcinogenesis: the role of homeostasis. Carcinogenesis. 2000;21:857–864. doi: 10.1093/carcin/21.5.857. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein I.B., Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. discussion 3080. [DOI] [PubMed] [Google Scholar]
- 3.Torti D., Trusolino L. Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: promises and perils. EMBO Mol. Med. 2011;3:623–636. doi: 10.1002/emmm.201100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamb A. Consequences of nonadaptive alterations in cancer. Mol. Biol. Cell. 2003;14:2201–2205. doi: 10.1091/mbc.E02-11-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S.V., Fischbach M.A., Haber D.A., Settleman J. “Oncogenic shock”: explaining oncogene addiction through differential signal attenuation. Clin. Cancer Res. 2006;12:4392s–4395s. doi: 10.1158/1078-0432.CCR-06-0096. [DOI] [PubMed] [Google Scholar]
- 6.Brunet C.L., Gunby R.H., Benson R.S., Hickman J.A., Watson A.J., Brady G. Commitment to cell death measured by loss of clonogenicity is separable from the appearance of apoptotic markers. Cell Death Differ. 1998;5:107–115. doi: 10.1038/sj.cdd.4400334. [DOI] [PubMed] [Google Scholar]
- 7.Kaelin W.G., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 8.Fece de la Cruz F., Gapp B.V., Nijman S.M. Synthetic lethal vulnerabilities of cancer. Annu. Rev. Pharmacol. Toxicol. 2015;55:513–531. doi: 10.1146/annurev-pharmtox-010814-124511. [DOI] [PubMed] [Google Scholar]
- 9.Felsher D.W. Oncogene addiction versus oncogene amnesia: perhaps more than just a bad habit? Cancer Res. 2008;68:3081–3086. doi: 10.1158/0008-5472.CAN-07-5832. discussion 3086. [DOI] [PubMed] [Google Scholar]
- 10.Lowe S.W., Cepero E., Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 11.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 12.Dueñas-Gonzalez A., Cetina L., Coronel J., Cano C., Dolores R. New pharmacotherapy options for cervical cancer. Expert Opin. Pharmacother. 2014;15:51–60. doi: 10.1517/14656566.2014.855198. [DOI] [PubMed] [Google Scholar]
- 13.Gabrielli B., Bokhari F., Ranall M.V., Oo Z.Y., Stevenson A.J., Wang W., Murrell M., Shaikh M., Fallaha S., Clarke D. Aurora A is critical for survival in HPV-transformed cervical cancer. Mol. Cancer Ther. 2015;14:2753–2761. doi: 10.1158/1535-7163.MCT-15-0506. [DOI] [PubMed] [Google Scholar]
- 14.Tabrizi S.N., Brotherton J.M., Kaldor J.M., Skinner S.R., Liu B., Bateson D., McNamee K., Garefalakis M., Phillips S., Cummins E. Assessment of herd immunity and cross-protection after a human papillomavirus vaccination programme in Australia: a repeat cross-sectional study. Lancet Infect. Dis. 2014;14:958–966. doi: 10.1016/S1473-3099(14)70841-2. [DOI] [PubMed] [Google Scholar]
- 15.Covens A., Rosen B., Murphy J., Laframboise S., DePetrillo A.D., Lickrish G., Colgan T., Chapman W., Shaw P. Changes in the demographics and perioperative care of stage IA(2)/IB(1) cervical cancer over the past 16 years. Gynecol. Oncol. 2001;81:133–137. doi: 10.1006/gyno.2001.6158. [DOI] [PubMed] [Google Scholar]
- 16.Rose P.G. Chemoradiotherapy for cervical cancer. Eur. J. Cancer. 2002;38:270–278. doi: 10.1016/s0959-8049(01)00352-5. [DOI] [PubMed] [Google Scholar]
- 17.Reade C.J., Eiriksson L.R., Covens A. Surgery for early stage cervical cancer: how radical should it be? Gynecol. Oncol. 2013;131:222–230. doi: 10.1016/j.ygyno.2013.07.078. [DOI] [PubMed] [Google Scholar]
- 18.Alexander-Sefre F., Chee N., Spencer C., Menon U., Shepherd J.H. Surgical morbidity associated with radical trachelectomy and radical hysterectomy. Gynecol. Oncol. 2006;101:450–454. doi: 10.1016/j.ygyno.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 19.DeFilippis R.A., Goodwin E.C., Wu L., DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 2003;77:1551–1563. doi: 10.1128/JVI.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sontheimer E.J., Barrangou R. The bacterial origins of the CRISPR genome-editing revolution. Hum. Gene Ther. 2015;26:413–424. doi: 10.1089/hum.2015.091. [DOI] [PubMed] [Google Scholar]
- 22.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez-Rivera F.J., Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung H.S., Rajasekaran N., Ju W., Shin Y.K. Human papillomavirus: current and future RNAi therapeutic strategies for cervical cancer. J. Clin. Med. 2015;4:1126–1155. doi: 10.3390/jcm4051126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan T.M., Ting R.C. In vitro and in vivo inhibition of human papillomavirus type 16 E6 and E7 genes. Cancer Res. 1995;55:4599–4605. [PubMed] [Google Scholar]
- 28.Guo C.P., Liu K.W., Luo H.B., Chen H.B., Zheng Y., Sun S.N., Zhang Q., Huang L. Potent anti-tumor effect generated by a novel human papillomavirus (HPV) antagonist peptide reactivating the pRb/E2F pathway. PLoS ONE. 2011;6:e17734. doi: 10.1371/journal.pone.0017734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamada K., Zhang W.W., Alemany R., Wolf J., Roth J.A., Mitchell M.F. Growth inhibition of human cervical cancer cells with the recombinant adenovirus p53 in vitro. Gynecol. Oncol. 1996;60:373–379. doi: 10.1006/gyno.1996.0057. [DOI] [PubMed] [Google Scholar]
- 30.Beerheide W., Bernard H.U., Tan Y.J., Ganesan A., Rice W.G., Ting A.E. Potential drugs against cervical cancer: zinc-ejecting inhibitors of the human papillomavirus type 16 E6 oncoprotein. J. Natl. Cancer Inst. 1999;91:1211–1220. doi: 10.1093/jnci/91.14.1211. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y.F., Rao Z.G., Zhang J.R. Effects of anti-HPV16 E6-ribozyme on the proliferation and apoptosis of human cervical cancer cell line CaSKi. Di Yi Jun Yi Da Xue Xue Bao. 2002;22:496–498. [PubMed] [Google Scholar]
- 32.Villanueva R., Morales-Peza N., Castelán-Sánchez I., García-Villa E., Tapia R., Cid-Arregui A., García-Carrancá A., López-Bayghen E., Gariglio P. Heparin (GAG-hed) inhibits LCR activity of human papillomavirus type 18 by decreasing AP1 binding. BMC Cancer. 2006;6:218. doi: 10.1186/1471-2407-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu W., Putral L., Hengst K., Minto K., Saunders N.A., Leggatt G., McMillan N.A. Inhibition of cervical cancer cell growth in vitro and in vivo with lentiviral-vector delivered short hairpin RNA targeting human papillomavirus E6 and E7 oncogenes. Cancer Gene Ther. 2006;13:1023–1032. doi: 10.1038/sj.cgt.7700971. [DOI] [PubMed] [Google Scholar]
- 34.Nishida H., Matsumoto Y., Kawana K., Christie R.J., Naito M., Kim B.S., Toh K., Min H.S., Yi Y., Matsumoto Y. Systemic delivery of siRNA by actively targeted polyion complex micelles for silencing the E6 and E7 human papillomavirus oncogenes. J. Control. Release. 2016;231:29–37. doi: 10.1016/j.jconrel.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Chapoy-Villanueva H., Martinez-Carlin I., Lopez-Berestein G., Chavez-Reyes A. Therapeutic silencing of HPV 16 E7 by systemic administration of siRNA-neutral DOPC nanoliposome in a murine cervical cancer model with obesity. J. BUON. 2015;20:1471–1479. [PubMed] [Google Scholar]
- 36.Hu Z., Yu L., Zhu D., Ding W., Wang X., Zhang C., Wang L., Jiang X., Shen H., He D. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. BioMed Res. Int. 2014;2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu L., Wang X., Zhu D., Ding W., Wang L., Zhang C., Jiang X., Shen H., Liao S., Ma D. Disruption of human papillomavirus 16 E6 gene by clustered regularly interspaced short palindromic repeat/Cas system in human cervical cancer cells. Onco Targets Ther. 2014;8:37–44. doi: 10.2147/OTT.S64092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Kennedy E.M., Kornepati A.V.R., Goldstein M., Bogerd H.P., Poling B.C., Whisnant A.W., Kastan M.B., Cullen B.R. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Graham S.V. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010;5:1493–1506. doi: 10.2217/fmb.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy I.M., Haddow J.K., Clements J.B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J. Virol. 1991;65:2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milligan S.G., Veerapraditsin T., Ahamet B., Mole S., Graham S.V. Analysis of novel human papillomavirus type 16 late mRNAs in differentiated W12 cervical epithelial cells. Virology. 2007;360:172–181. doi: 10.1016/j.virol.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagano J.S. Epstein-Barr virus: the first human tumor virus and its role in cancer. Proc. Assoc. Am. Physicians. 1999;111:573–580. doi: 10.1046/j.1525-1381.1999.t01-1-99220.x. [DOI] [PubMed] [Google Scholar]
- 45.Baer R., Bankier A.T., Biggin M.D., Deininger P.L., Farrell P.J., Gibson T.J., Hatfull G., Hudson G.S., Satchwell S.C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 46.Rowe M., Lear A.L., Croom-Carter D., Davies A.H., Rickinson A.B. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J. Virol. 1992;66:122–131. doi: 10.1128/jvi.66.1.122-131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Diemen F.R., Kruse E.M., Hooykaas M.J., Bruggeling C.E., Schürch A.C., van Ham P.M., Imhof S.M., Nijhuis M., Wiertz E.J., Lebbink R.J. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Quake S.R. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc. Natl. Acad. Sci. USA. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuen K.S., Chan C.P., Wong N.H., Ho C.H., Ho T.H., Lei T., Deng W., Tsao S.W., Chen H., Kok K.H., Jin D.Y. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J. Gen. Virol. 2015;96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 50.Block T.M., Mehta A.S., Fimmel C.J., Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- 51.Lin G., Zhang K., Li J. Application of CRISPR/Cas9 technology to HBV. Int. J. Mol. Sci. 2015;16:26077–26086. doi: 10.3390/ijms161125950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shlomai A., Rice C.M. Virology. Getting rid of a persistent troublemaker to cure hepatitis. Science. 2014;343:1212–1213. doi: 10.1126/science.1252186. [DOI] [PubMed] [Google Scholar]
- 53.White M.K., Hu W., Khalili K. The CRISPR/Cas9 genome editing methodology as a weapon against human viruses. Discov. Med. 2015;19:255–262. [PMC free article] [PubMed] [Google Scholar]
- 54.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 56.Alberti A., Chemello L., Benvegnù L. Natural history of hepatitis C. J. Hepatol. 1999;31(Suppl 1):17–24. doi: 10.1016/s0168-8278(99)80369-9. [DOI] [PubMed] [Google Scholar]
- 57.Poiesz B.J., Ruscetti F.W., Gazdar A.F., Bunn P.A., Minna J.D., Gallo R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 59.Reardon S. First CRISPR clinical trial gets green light from US panel. Nature. 2016 Published online June 22, 2016. [Google Scholar]
- 60.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding Y., Li H., Chen L.L., Xie K. Recent advances in genome editing using CRISPR/Cas9. Front. Plant Sci. 2016;7:703. doi: 10.3389/fpls.2016.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng R., Lin G., Li J. Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 2016;283:1218–1231. doi: 10.1111/febs.13586. [DOI] [PubMed] [Google Scholar]
- 63.Schaefer K.A., Wu W.-H., Colgan D.F., Tsang S.H., Bassuk A.G., Mahajan V.B. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat. Methods. 2017;14:547–548. doi: 10.1038/nmeth.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wen W.S., Yuan Z.M., Ma S.J., Xu J., Yuan D.T. CRISPR-Cas9 systems: versatile cancer modelling platforms and promising therapeutic strategies. Int. J. Cancer. 2016;138:1328–1336. doi: 10.1002/ijc.29626. [DOI] [PubMed] [Google Scholar]
- 65.Wu X., Kriz A.J., Sharp P.A. Target specificity of the CRISPR-Cas9 system. Quant. Biol. 2014;2:59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tycko J., Myer V.E., Hsu P.D. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol. Cell. 2016;63:355–370. doi: 10.1016/j.molcel.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X., Skarnes W.C. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 69.Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cho S.W., Lee J., Carroll D., Kim J.-S., Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9–sgRNA ribonucleoproteins. Genetics. 2013;195:1177–1180. doi: 10.1534/genetics.113.155853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Cowan C.A., Talkowski M.E., Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pagliarini R., Shao W., Sellers W.R. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gorre M.E., Mohammed M., Ellwood K., Hsu N., Paquette R., Rao P.N., Sawyers C.L. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 78.Van Allen E.M., Wagle N., Sucker A., Treacy D.J., Johannessen C.M., Goetz E.M., Place C.S., Taylor-Weiner A., Whittaker S., Kryukov G.V., Dermatologic Cooperative Oncology Group of Germany (DeCOG) The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maertens O., Johnson B., Hollstein P., Frederick D.T., Cooper Z.A., Messiaen L., Bronson R.T., McMahon M., Granter S., Flaherty K. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3:338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 82.Maruyama T., Dougan S.K., Truttmann M., Bilate A.M., Ingram J.R., Ploegh H.L. Inhibition of non-homologous end joining increases the efficiency of CRISPR/Cas9-mediated precise [TM: inserted] genome editing. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pierce A.J., Hu P., Han M., Ellis N., Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 86.Renaud J.B., Boix C., Charpentier M., De Cian A., Cochennec J., Duvernois-Berthet E., Perrouault L., Tesson L., Edouard J., Thinard R. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep. 2016;14:2263–2272. doi: 10.1016/j.celrep.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 87.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 88.Li C.X., Qian H.L. A double-edged sword: CRISPR-Cas9 is emerging as a revolutionary technique for genome editing. Mil Med Res. 2015;2:25. doi: 10.1186/s40779-015-0054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singhania R., Pavey S., Payne E., Gu W., Clancy J., Jubair L., Preiss T., Saunders N., McMillan N.A. Short interfering RNA induced generation and translation of stable 5′ mRNA cleavage intermediates. Biochim. Biophys. Acta. 2016;1859:1034–1042. doi: 10.1016/j.bbagrm.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 90.Bachtiary B., Boutros P.C., Pintilie M., Shi W., Bastianutto C., Li J.-H., Schwock J., Zhang W., Penn L.Z., Jurisica I. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin. Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 91.Kidd E.A., Grigsby P.W. Intratumoral metabolic heterogeneity of cervical cancer. Clin. Cancer Res. 2008;14:5236–5241. doi: 10.1158/1078-0432.CCR-07-5252. [DOI] [PubMed] [Google Scholar]