Abstract
Wound healing is a highly complex biological process composed of three overlapping phases: inflammation, proliferation, and remodeling. Impairments at any one or more of these stages can lead to compromised healing. MicroRNAs (miRs) are non-coding RNAs that act as post-transcriptional regulators of multiple proteins and associated pathways. Thus, identification of the appropriate miR involved in the different phases of wound healing could reveal an effective third-generation genetic therapy in chronic wound care. Several miRs have been shown to be upregulated or downregulated during the wound healing process. This article examines the biological processes involved in wound healing, the miR involved at each stage, and how expression levels are modulated in the chronic wound environment. Key miRs are highlighted as possible therapeutic targets, either through underexpression or overexpression, and the healing benefits are interrogated. These are prime miR candidates that could be considered as a gene therapy option for patients suffering from chronic wounds. The success of miR as a gene therapy, however, is reliant on the development of an appropriate delivery system that must be designed to overcome both extracellular and intracellular barriers.
Keywords: microRNA, wound healing, gene therapy, chronic wounds
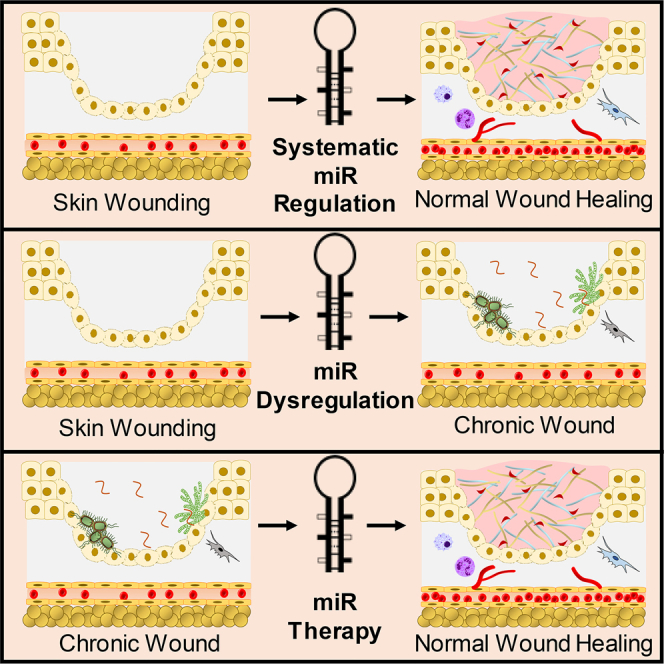
Graphical Abstract
Main Text
Introduction to Wound Healing
The largest organ of the human body is the skin; it accounts for approximately 15% of total body weight in adults.1, 2 The skin acts as a protective barrier against the environment and helps to prevent dehydration. The skin has three distinct layers: (1) epidermis, composed mainly of keratinocyte cells; (2) dermis, which is a collagen-rich tissue containing blood and lymph vessels, fibroblasts, macrophages, and neutrophils; and (3) hypodermis, which is constructed mainly of adipose tissue.3 The development of these layers is a highly coordinated process involving complex gene regulatory networks acting in a temporally resolved integrated way.
A wound is defined as all manner of tissue damage resulting in the disorder of the original tissue structure and homeostasis.4 The mechanisms involved in the repair of wounds involve some of the most complex biological processes.5 The wound healing process in the skin involves three partially overlapping phases: (1) blood clotting and inflammation, (2) new tissue formation, and (3) tissue remodeling (the longest phase).6, 7 These processes result in the wound tissue being repaired within a relatively short period. The scar tissue, however, has reduced tensile strength compared to that of healthy skin tissue,7 demonstrating approximately 70% of the tensile strength of healthy tissue.8
Chronic Wounds
Chronic wounds are the result of multifactorial components within the wound healing process becoming compromised. They can be defined as a wound that is not continuously progressing toward healing9 or not healing in a methodical and timely fashion.8 Pathologically, one of the clearest indicators of a chronic wound is their failure to re-epithelialize.10 The aging population are among those most burdened with chronic wounds, and with low keratinocyte cell turnover, this is a factor affecting their ability to re-epithelialize.11 In general, chronic wounds exhibit reduced mitogenic activity compared to acute wounds, which has been demonstrated not only in keratinocytes but also in skin fibroblasts.12 It has been shown that fibroblast cells isolated from Venus leg ulcers have reduced cell migration properties in comparison to cells isolated from regular skin.12
Overall, factors affecting healing ability are categorized as either local and/or systemic influences. Local refers to causes that directly affect the characteristics of the wound site itself and systemic factors related to the general overall health or disease state of the individual, which affects healing ability. As such, the following sections detail diabetes and infection as examples of a systemic and local factor, respectively, which can influence a wound to become chronically impaired. Differences in microRNA (miR) expression will also be explored within the sections miR and Wound Healing to miR Expression in Chronic Wounds.
Diabetes
Diabetes is a systemic factor that can be linked to wound healing and affects approximately 3.2 million people in the UK, which accounts for 6% of the population.13 This population is at risk of developing chronic non-healing diabetic foot ulcers. The increase in serum glucose has a significant influence on wound healing due to destructive effects showcased by hyperglycemia on the physiology of the affected cells.14 For example, sorbitol, a by-product of glucose metabolism, can accumulate within tissues, resulting in renal and vascular complications.15 Hypoxic regions are prevalent in diabetic wounds and are characterized by an absence of endothelial cells, resulting in reduced angiogenesis and elongation of the inflammatory phase. Diabetic mice models have shown decreased restoration of vasculature in diabetic wounds, with hindered endothelial progenitor cell mobilization and homing and reduced levels of VEGF, compared to a non-diabetic controls in vivo.16
Non-enzymatic glycosylation associated with hyperglycemia also inhibits the function of structural and enzymatic peptides.16 Diabetic animal models of wound healing show decreased rates of granulation. Likewise, in humans, the findings were comparable, showing slow wound maturation and a lower count of fibroblast cells compared to non-diabetic wound sites.14
Several cellular functions are affected in diabetic wounds, such as defective T cell immunity, leukocyte chemotaxis, phagocytosis, bactericidal capacity, and dysfunctions of fibroblasts and epidermal cells. Taken together, such defects contribute to inadequate clearance of infection and delayed healing.17
Obesity is intrinsically linked to diabetes (type 2) and impaired wound healing. By 2050 in the UK, obesity is projected to affect 60% of adult men, 50% of adult women, and 25% of children.18 Obese patients have been reported to experience wound complications more frequently in relation to post-surgery healing, such as higher rates of infection.19 This has been linked to ischemia and hypo-perfusion in the subcutaneous adipose tissue. Skin folds also harbor microorganisms that thrive in moist areas and contribute to infection and tissue breakdown.6
Infection is an example of a local factor that is linked to impaired wound healing. Virtually all open wounds experience colonization of microorganisms, which typically has no clinical consequence because no infection is evident. However, infection within a wound site can influence the healing process by prolonging the inflammatory phase. This is due to incomplete clearance of the wound site. Therefore, there is prolonged elevation of cytokines (e.g., interleukin-1 [IL-1] and tumor necrosis factor alpha [TNF-α]), which are pro-inflammatory.20 If the bacterial count within the wound is more than 105 colony-forming units per gram (CFUs/g) of tissue, the wound will not heal, irrespective of the treatment regime (e.g., skin graft placement and primary sutures).21 Furthermore, no healing will occur if the β-hemolytic strain of Streptococcus is present at the wound site, even at less than 105 CFUs/g.22
Bacterial biofilms are a huge hurdle to overcome in the healing process for chronic wounds. A biofilm is an intricate community of microorganisms, characterized by massive cell densities that are accompanied by an extracellular polymer matrix, made mainly of polysaccharides and proteins.23 This matrix is multifunctional, acting both as a physical protector from biological and pharmaceutical antimicrobials. Further to this, it facilitates the adhesion of bacteria cells to surfaces, particularly foreign bodies.
Current Treatments
Gauze has been historically popular for wound dressings. However, the use of gauze promotes desiccation of the wound base, which is not advantageous for efficient healing.24 Furthermore, dry gauze often adheres to the surface of the wound, which can be painful for the patient upon removal. Such gauze dressings are susceptible to complete saturation of wound fluid and so can be ineffective in the protection against bacterial invasion.24, 25 Films are alternatives to gauze dressings and can be utilized as multifunctional adhesive dressings, as in they can be used as primary dressings directly on the wound or alternatively as a secondary dressing to secure other primary dressings to the wound. Film membranes are thin and semi-permeable, enabling the exchange of oxygen and water vapor to the wound in addition to preventing liquid and microorganism contaminants.
Topical antiseptic and antibiotic treatments can be employed to reduce the bioburden of the wound. Limitations associated with topical antiseptics and antibiotics include toxicity to human tissue that can lead to prolonged inflammation and microbial resistance, in addition to the necessity for many reapplications to achieve the desired effects.26 Acetic acid is an example of a topical treatment typically administered at concentrations of between 0.25% and 1.0% (v/v). It is effective against most gram-positive and gram-negative organisms. Acetic acid is an inexpensive product; however, it can cause cytotoxicity in vitro and has limited activity against biofilms. The latter is a particular problem associated with chronic wounds.27, 28
The management and treatment of chronic wounds is influenced by many multifactorial elements; however, the current modes of treatments and dressing are largely inadequate for chronic wounds. Therefore, miR replacement or inhibitory therapy is an alternative mode of treatment because miR has the potential to affect an array of downstream gene targets.
miR
miRs are instrumental in the regulation of gene expression through the promotion of mRNA degradation and inhibition of mRNA translation. Intronic miRs are encoded within a gene transcript precursor, sharing the same promoter with the encoded gene transcripts.29 Unidentified promoters transcribe intergenic miR, located in the non-coding regions. Finally, polycistronic miRs derived from primary transcripts contain a diverse range of hairpins, giving rise to different miRs.30
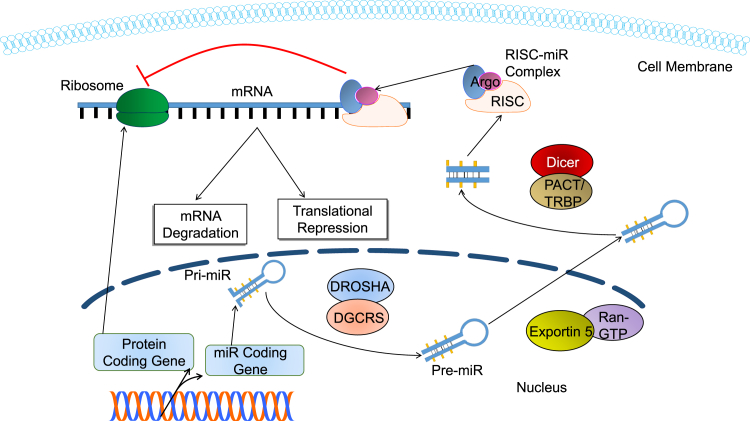
The process of miR biogenesis (Figure 1) is a highly complex process. In mammalian cells, miRs are transcribed by RNA polymerase II.31 Primary miR (pri-miR) is cleaved by the RNase III intra-nuclear enzyme (i.e., Drosha) into many miR precursors (pre-miRs).32 Drosha and DGCR8/Pasha establish the microprocessor complex, which cuts the pri-miR at the ssRNA/dsRNA junction found at the base of the pri-miR hairpins and results in pre-miR production.31 The resulting pre-miRs are ∼65 nt in length, with characteristic stem-loop hairpin secondary structures.33 The pre-miR is bound by Exportin 5 in the presence of the co-factor Ran-guanine triphosphatase (Ran-GTP) and is transported to the cell cytoplasm. Once the pre-miR has moved to the cytoplasm, a second RNase III enzyme (i.e., Dicer) processes the pre-miR with the help of several co-factors (e.g., double-stranded RNA-activated protein kinase [PACT] and TAR RNA-binding protein [TRBP]).34 Processing by Dicer leads to a mature miR strand and an opposing strand that degrades. Thus, the miR is incorporated into the RNA-induced silencing complex (RISC).35
Figure 1.
Biogenesis of miR
Biogenesis of miR and the resulting post-transcriptional gene silencing mechanisms. Pri-miR is transcribed within the nucleus and then cleaved by the DROSHA complex to form pre-miR. Exportin 5 transports this pre-miR into the cytoplasm of the cell. There it is further sliced to form mature miR. This miR then binds with the RISC to form a RISC-miR complex, which acts as a translational repressor and degrades mRNA.
miR Function
miR modulates gene expression through association of an Argonaute protein, which loads the miR into the RISC at the 3′ end.35 The incorporated miR guides the RISC to its specific mRNA target through base-pair complementarity, which results in the disruption of translation.
Depending on the grade of complementarity between the target mRNA and the miR, two different mechanisms of RISC-mediated gene regulation may ensue. In the case of almost complete complementarity, the RISC cleaves the target mRNA, preventing translation.33 In the case of imperfect complementarity, translation is suppressed by miR decapping and/or deadenylation (Figure 1).31
miR and Wound Healing
Several studies have demonstrated that miR expression is upregulated or downregulated during the overlapping phases of wound healing. Table 1 details key miRs that have been found to be instrumental during the different phases of wound healing. Therefore, provided the optimal miR can be identified, new therapies could be developed.
Table 1.
miR Involved in the Normal Wound Healing Process
| miR | Targets | Reference | |
|---|---|---|---|
| Pro-inflammatory | miR-155 | SOCS1, SHIP1, IL-12 | 34, 65 |
| miR-140 | PDGF | 66 | |
| Anti-inflammatory | miR-16 | COX2 | 67 |
| miR-105 | TLR2 | 68 | |
| miR-21 | PDCD4, PTEN | 34, 69 | |
| miR-125b | TNF-α | 70, 71 | |
| miR-223 | Mef2c | 72 | |
| miR-203 | TNF-α, IL-24 | 73 | |
| miR-146a,b | TRAF6, IRAK1, STAT1, TNF-α, COX2 | 34, 71, 74 | |
| Proliferation | miR-21 | TIMP3, TIAM1 | 43 |
| miR-31 | EMP-1 | 49, 50 | |
| miR-155 | KGF, FGF-7 | 75 | |
| miR-99 | IGF1R, mTOR, AKT1 | 76 | |
| miR-198 | DIAPH1, PLAU, LAMC2 | 77 | |
| miR-184 | AKT | 38, 78, 79, 80 | |
| miR-205 | SHIP2, Rho-ROCK1 | 38 | |
| miR-203 | RAN, RAPH1 | 81 | |
| miR-210 | E2F3, ISCU1, ISCU2 | 82 | |
| miR-483-3p | MK2, MK167, YAP1 | 83 | |
| Pro-angiogenic | miR-17-5p | TIMP1 | 34 |
| miR-17-92 | TSP-1, CTGF | 51, 84, 85 | |
| miR-31 | FIH-1, Spred1 | 86, 87 | |
| miR-126 | Spred1, PIK2R2 | 79, 88, 89 | |
| miR-130a | GAX, HOXA5 | 90 | |
| miR-210 | EFNA3 | 91, 92 | |
| miR-296 | HGS | 93 | |
| miR-378 | Fus-1, Sufu | 94 | |
| miR-424 | CUL2 | 47 | |
| Anti-angiogenic | miR-92a | Integrin-α5 | 34, 95 |
| miR-17 | JAK1 | 96 | |
| miR-15b | VEGF | 97 | |
| miR-16 | VEGF | 97 | |
| miR-20a | MKK3,VEGF | 97, 98 | |
| miR-20b | HIF-1α, VEGF | 97, 99 | |
| miR-221 | c-kit | 100, 101 | |
| miR-222 | c-kit | 93, 94 | |
| miR-320 | IGF-1 | 102, 103 | |
| miR-503 | CCNE1,cdc25A | 104 | |
| Remodeling | miR-29a | TAB-1, collagen I and II | 39, 105 |
| miR-29b | SMADs, β-catenin | 106 | |
| miR-29c | SMADs, β-catenin | 107 | |
| miR192/215 | E-cadherin, SIP1 | 41, 108 |
miR and the Inflammatory Phase
Inflammatory response and resolve is paramount to successful and rapid wound healing. Due to the tight regulation of pro-inflammatory and anti-inflammatory signaling, disruption of miR biogenesis may result in an imbalance of these signals adversely affecting the healing cascade. Evolving studies demonstrate that miR-21, miR-146a/b, and miR-155 play key roles in regulating the inflammatory process. miR-146 and miR-155 are promoted by TNF-α and IL-1β, and miR-146 has been shown to silence interleukin-1 receptor-associated kinase 1 (IRAK) and cyclooxygenase-2 (COX2), whereas miR-155 silences Src homology 2 domain-containing inositol 5-phosphatase (SHIP1), suppressor of cytokine signaling 1 (SOCS1), and IL-12.36, 37
miR and the Proliferation Phase
Proliferation of keratinocyte cells is critical for re-epithelialization of the wound, ergo miR regulation is vital. For this process to be efficient and progress rapidly, an abundant oxygen and nutrient supply is required and angiogenesis is key. Keratinocytes have been shown to migrate faster upon silencing of SHIP2 and the enhanced activation of the serine-threonine protein kinase AKT signaling pathway.38 miR-205 has been shown to suppress SHIP2, a negative regulator of the AKT pathway and enhance cofilin expression by a reduction in the regulation of phosphor-cofilin, which increases cell motility.38 Another important miR is miR-21, which has been shown to be an inhibitor of phosphatase and tensin homolog PTEN. PTEN inhibits the AKT pathway, thereby activating many cell survival and proliferative pathways.
miR and the Remodeling Phase
An important aspect of this phase of wound healing is the deposition of collagen. It has been observed that miR-29a has a direct influence on the expression of collagen at a post-transitional level.39 Fetal skin from mammals can heal without scarring. However, during advanced gestation, the transition is to a scarring phenotype.40 miR-192, miR-29b, and miR-29c are highly induced in the later phase of this process. Transforming growth factor β (TGF-β) (anti-fibrotic) and proteins such as SMADs that are involved in the pathways for scar-free healing are targets of miR-29b/c. miR-192 also enhances collagen 1-α-2 expression through the targeting of SMAD-interacting protein 1 (SIP1).34, 41
miR Expression in Chronic Wounds
It has also been observed that many miRs expressed during the normal wound healing process are dysregulated during chronic wound healing.34 A recent study investigating the expression of miRs in rodents induced with diabetes showed that 18 miRs were upregulated and 65 miRs were downregulated compared to control wound healing groups (i.e., non-diabetic rodents). Therefore, many different miRs could be used as a therapeutic in the healing of chronic wounds.42 On the basis of the amalgamation of these observations, the following sections explore several specific miRs and show how they could be exploited for advanced wound healing applications.
miR-21
Murine studies have demonstrated that miR-21 plays an important role in keratinocyte migration and re-epithelialization during wound healing. miR-21 has been found to be upregulated during wound healing, coincident with TGF-β temporal expression. Consistently, knockdown of endogenous miR-21 using a specific antagomir dramatically delayed re-epithelialization, possibly due to the reduced keratinocyte migration. Likewise, Yang et al.43 found that through the utilization of wound scratch healing assays, transfection with miR-21 significantly increased keratinocyte cell migration by 62% compared to scrambled controls. TIMP3 and TIAM1 are regulated by miR-21 in vitro and in vivo. TIMP3 inhibits the activation of the EDK pathway with downstream activation of ERK1/2, thus promoting proliferation. Therefore, it can be postulated that miR-21 promotes keratinocyte migration and boosts re-epithelialization during skin wound healing.
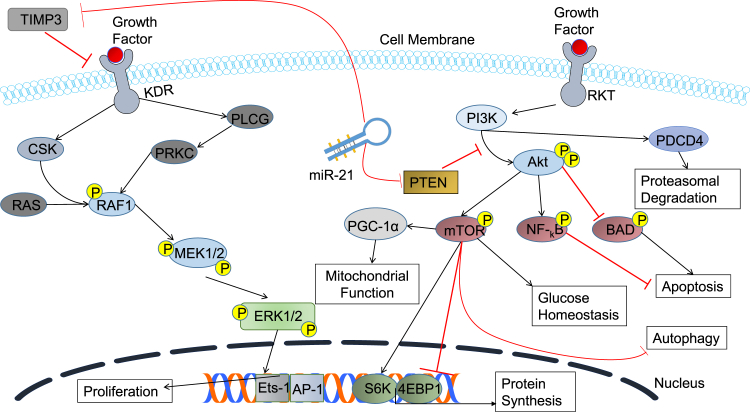
miR-21 has also been found to be a key regulator of vascular smooth muscle cell proliferation and apoptosis via BCL2 activation and PTEN suppression. It also regulates MMP-2 via the PTEN pathway as a result of myocardial infarction.44 Inhibition of the PTEN pathway also activates many downstream targets, such as mTOR, which regulates protein synthesis, glucose homeostasis, autophagy, and proliferation (Figure 2).45 This suggests that introduction of miR-21 within the diabetic chronic wound site could have potential as a therapeutic by increasing the extent of cell migration.46
Figure 2.
Therapeutic Wound Healing Targets of miR-21
Schematic diagram of miR-21 targets TIMP3 and PTEN. By inhibiting PTEN, miR-21 promotes the activation of many cell survival pathways. The Akt pathway functionality increases; thus, the mTOR pathway, for example, is heightened, which regulates protein synthesis, mitochondrial function, and glucose homeostasis. Nf-kB inhibits apoptosis and Akt inhibits BAD, which is a pro-apoptotic pathway.
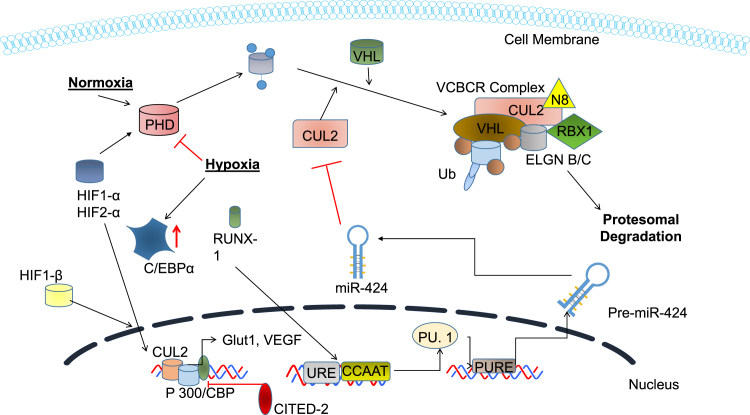
miR-424
In normal wound healing, miR-424 is upregulated in response to hypoxic conditions (Figure 3).47 It works through the stabilization of transcription factor HIF-α. In normoxic conditions, oxygen targets HIF for degradation by post-translational hydroxylation at specific prolyl residues domains (PHD) within these subunits. This increases the affinity for the ubiquitin ligases for proteolytic destruction by the ubiquitin/proteasome pathway. The oxygen-dependent hydroxylation process, however, is suppressed during hypoxia, leading to stabilization of HIF-1α and subsequent attachment to its constitutive partner HIF-1β to induce transactivation. Stabilization of HIF has been classically known to induce the transcription of coding genes (e.g., vascular endothelial growth factor [VEGF], erythropoietin, and nitric oxide synthase-2).48
Figure 3.
Therapeutic Wound Healing Targets of miR-424
Schematic diagram of miR-424 targeting CUL2. By targeting CUL2, the levels on HIF-1 increase within the cell and its proteosomal degradation is inhibited. Elevated HIF-1 increases the transcription of VEGF and Glut1.
With regards to miR-424, this results in transactivation genes that govern angiogenesis and metabolic pathways. miR-424 cleaves to the CUL2 3′ UTR and inhibits CUL2 expression. A drop in CUL2 levels leads to the destabilization of VCBCR U3-ligase complex, which leads to the stabilization and nuclear translocation of HIF-1α in endothelial cells. In vitro wound scratch assays found that miR-424 significantly increased cell migration, with endothelial cells transfected with miR-424, decreasing the wound width by 40% compared to a 16% decrease in the control group. Furthermore, angiogenesis tubular formation assays with endothelial cells transfected with miR-424 also showed increased tube establishment.47 This miR (mu-miR-322), which is the rodent homolog of miR-424, has been demonstrated to downregulate in a diabetic rodent model.42 However, in rodent models with ischemia, it has been shown that this miR is significantly upregulated.47 If there are prolonged periods of hypoxia, the natural response of cells is the initiation of apoptosis. Angiogenesis is crucial for rapid wound healing because it provides nutrients and oxygen to the wound site. With angiogenesis being compromised in patients with diabetes, it is crucial that it is stimulated to progress healing. Furthermore, fibroblasts that are exposed to prolonged periods of hypoxia may not participate in the formation of extracellular matrix (ECM), which again causes delays in healing. miR-424 is, therefore, a promoter of angiogenesis, validated by in vitro and in vivo rodent models.
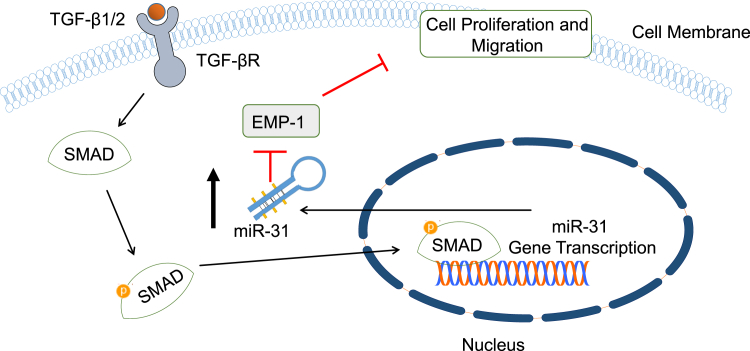
miR-31
Human wound healing studies have demonstrated that miR-31 upregulation occurs in keratinocytes at the wound edge during proliferation. It has also been shown that overexpression of miR-31 increases proliferation, leading to an increased rate in re-epithelialization. Given that a low rate of re-epithelialization is typical of chronic wounds, miR-31 may be able to aid in the rapid closure of the wound.49
Both TGF-β1 and TGF-β2 have been found to be upregulated in wounds and promote keratinocyte migration. TGF-β1 and TGF-β2 receptor binding results in the activation of downstream SMAD proteins, which in turn induce the expression of TGF-β target genes. Epithelial membrane protein-1 (EMP-1) has been identified as a suppresser of keratinocyte cell proliferation and migration (Figure 4). miR-31 expression is regulated by TGF-β2 and is a direct target of EMP-1. Silencing of EMP-1 expression results in a decreased level of miR-31 in keratinocytes; this suggests that a high level of EMP-1 during inflammation may trigger miR-31 expression.49
Figure 4.
EMP-1 as a Therapeutic Wound Healing Target of miR-31
Schematic diagram of miR-31 targeting EMP-1. EMP-1 inhibits migration and proliferation of cells; thus, its inhibition will result in an increase of these functions.
Another target of miR-31 is factor-inhibiting HIF-1 (FIH-1) (Figure 5), which is an asparaginyl B-hydroxylase enzyme. It has been shown that FIH-1 regulates HIF-1 and subsequently VEGF, thus having a direct effect on the rate of angiogenesis. Upregulation of miR-31 would therefore increase the levels of VEGF through the silencing of FIH-1.50
Figure 5.
FIH-1 as a Therapeutic Wound Healing Target of miR-31
Schematic diagram of miR-31 targeting FIH-1. Thus, increasing the levels of HIF-1 within the cells. This leads to an increase in the levels of VEGF, thus increasing angiogenesis.
Furthermore, it has been demonstrated that miR-31 is downregulated in wounds within the diabetic rodent model.42 Taken together, all the evidence indicates that delivery of miR-31 is a candidate therapy for the inflammatory and proliferative phases of wound healing.
miR-221 and miR-222
Infection is another key consideration in the wound healing process, and nitric oxide (NO) is a potent antibacterial agent. miR-221 and miR-222 have been found to reduce the expression of endothelial NO, which is essential for many cellular functions.51 Many studies have demonstrated that NO levels are lower in diabetic wounds.52, 53 In vivo studies using eNOS knock-out mice have demonstrated delayed closure of wounds54 and a reduced rate of angiogenesis.55 Thus, the silencing of miR-221 and miR-222 could serve as a potential therapeutic to target infection in chronic wound healing.
Modes of Upregulating/Downregulating miR
For the introduction of miR into cells, many types of cargo could be utilized, for example, plasmid DNA coding for specific miRs to upregulate expression or antagomirs for silencing of expression.56, 57 For this, numerous genetic delivery systems are currently utilized in research, with viral vectors being the most efficient.58 However, there are apprehensions surrounding viral vectors as genetic carriers due to mutagenesis, toxicity, and its limited capacity for genetic cargo.59 Examples of non-viral options include polymers of cationic nature, liposomes, and peptides, which exhibit the ability to not only package genetic cargo, but also deliver it to the nucleus of cells.60 Liposomes are composed of a membrane composed of lipids, in which nucleic acid can be encapsulated within. Liposomes can take three forms: anionic, neutral, and, cationic, with cationic being the most frequently utilized for nucleic acid delivery due to its efficacy in interacting with cell membranes.61 There are many commercial options of cationic liposomes, such as Lipofectamine RNAi-MAX (Invitrogen)62 or SiPORT (Invitrogen).63 In vitro assays looking at the efficiency of liposomes at transfecting non-small-cell lung cancer cells with miR-29b found that there was a 5-fold increase in their expression compared to non-treated cells, validating their power as an miR delivery system.64 For wound healing, the ideal non-viral delivery vector must be non-immunogenic and non-toxic so as not to compromise the already delicate wound tissue.
Conclusions
Utilization of miR presents an attractive proposition for the development of therapeutic targets that could act on various pathways associated with chronically impaired wound healing. With varying levels of many miRs during the different phases of wound healing, there are several possible targets that could be employed.
Examples of prime miR candidates for wound healing include miR-21 and miR-31. miR-21, a target for the proliferation and inflammatory phase, could stop inflammation by targeting PDCD4 and promote proliferation and cell survival by activation of the mTOR pathway. miR-31 targets FIH-1; this increases the levels of VEGF intracellularly, which is a key promoter of angiogenesis. A significant advantage of using miRs such as these is that they can be detected in blood serum. With dose and response correlation being a prevalent issue within gene therapy options, detection of miRs as biomarkers in serum will provide translational information with regards to the successful delivery of the therapeutic.
However, the rate-limiting factor in the implementation of an miR therapy is a delivery system. This delivery system must be designed to overcome extracellular and intercellular barriers to ensure the successful delivery of the therapeutic cargo to the desired targets without evoking an immune response. Because it is of principal importance to maintain an optimum healing environment, it would be prudent to incorporate a gene therapy delivery system as part of a dressing instrument. Adopting such a system could confer multi-functionality with the dressing, ensuring a moist and protected micro-environment in addition to the efficacious delivery of the miR gene therapy. It can be rationalized, therefore, that miR holds the potential to become a momentous third-generation nucleic acid therapeutic for the treatment and management of chronic wounds, provided the optimal targeting and delivery system can be designed.
Contributor Information
Nicholas Dunne, Email: nicholas.dunne@dcu.ie.
Helen O. McCarthy, Email: h.mccarthy@qub.ac.uk.
References
- 1.Tobin D.J. Biochemistry of human skin--our brain on the outside. Chem. Soc. Rev. 2006;35:52–67. doi: 10.1039/b505793k. [DOI] [PubMed] [Google Scholar]
- 2.Kanitakis J. Anatomy, histology and immunohistochemistry of normal human skin. Eur. J. Dermatol. 2002;12:390–399. [PubMed] [Google Scholar]
- 3.Templeton N., editor. Gene and Cell Therapy. CRC Press; 2015. [Google Scholar]
- 4.Gurtner G.C., Werner S., Barrandon Y., Longaker M.T. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 5.Gould L., Abadir P., Brem H., Carter M., Conner-Kerr T., Davidson J., DiPietro L., Falanga V., Fife C., Gardner S. Chronic wound repair and healing in older adults: current status and future research. J. Am. Geriatr. Soc. 2015;63:427–438. doi: 10.1111/jgs.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo S., Dipietro L.A. Factors affecting wound healing. J. Dent. Res. 2010;89:219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werner S., Antsiferova M. Encyclopedia of Cell Biology. Elsevier; 2016. Wound healing: an orchestrated process of cell cycle, adhesion, and signaling; pp. 216–222. [Google Scholar]
- 8.Orgill D., Blanco C. Elsevier; 2009. Biomaterials for Treating Skin Loss. [Google Scholar]
- 9.Falanga V. The chronic wound: impaired healing and solutions in the context of wound bed preparation. Blood Cells Mol. Dis. 2004;32:88–94. doi: 10.1016/j.bcmd.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Adair H.M. Epidermal repair in chronic venous ulcers. Br. J. Surg. 1977;64:800–804. doi: 10.1002/bjs.1800641113. [DOI] [PubMed] [Google Scholar]
- 11.Grove G.L., Kligman A.M. Age-associated changes in human epidermal cell renewal. J. Gerontol. 1983;38:137–142. doi: 10.1093/geronj/38.2.137. [DOI] [PubMed] [Google Scholar]
- 12.Raffetto J.D., Mendez M.V., Marien B.J., Byers H.R., Phillips T.J., Park H.Y., Menzoian J.O. Changes in cellular motility and cytoskeletal actin in fibroblasts from patients with chronic venous insufficiency and in neonatal fibroblasts in the presence of chronic wound fluid. J. Vasc. Surg. 2001;33:1233–1241. doi: 10.1067/mva.2001.113297. [DOI] [PubMed] [Google Scholar]
- 13.Diabetes UK. (2014). Diabetes: facts and stats. https://www.diabetes.org.uk/Documents/AboutUs/Statistics/Diabetes-key-stats-guidelines-April2014.pdf.
- 14.Broughton G., 2nd, Janis J.E., Attinger C.E. Wound healing: an overview. Plast. Reconstr. Surg. 2006;117(Suppl):1e-S–32e-S. doi: 10.1097/01.prs.0000222562.60260.f9. [DOI] [PubMed] [Google Scholar]
- 15.He Z., King G.L. Microvascular complications of diabetes. Endocrinol Metab. Clin. North Am. 2004;33:215–238. doi: 10.1016/j.ecl.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Brem H., Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J. Clin. Invest. 2007;117:1219–1222. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gary Sibbald R., Woo K.Y. The biology of chronic foot ulcers in persons with diabetes. Diabetes Metab. Res. Rev. 2008;24(Suppl 1):S25–S30. doi: 10.1002/dmrr.847. [DOI] [PubMed] [Google Scholar]
- 18.Swanton, K. (2008). Healthy weight, healthy lives: a toolkit for developing local strategies. London Department of Health, http://webarchive.nationalarchives.gov.uk/20130107105354/http:/www.dh.gov.uk/prod_consum_dh/groups/dh_digitalassets/documents/digitalasset/dh_088967.pdf.
- 19.Momeni A., Heier M., Bannasch H., Stark G.B. Complications in abdominoplasty: a risk factor analysis. J. Plast. Reconstr. Aesthet. Surg. 2009;62:1250–1254. doi: 10.1016/j.bjps.2008.03.043. [DOI] [PubMed] [Google Scholar]
- 20.Fedyk E.R., Jones D., Critchley H.O.D., Phipps R.P., Blieden T.M., Springer T.A. Expression of stromal-derived factor-1 is decreased by IL-1 and TNF and in dermal wound healing. J. Immunol. 2001;166:5749–5754. doi: 10.4049/jimmunol.166.9.5749. [DOI] [PubMed] [Google Scholar]
- 21.Robson M.C. Wound infection. A failure of wound healing caused by an imbalance of bacteria. Surg. Clin. North Am. 1997;77:637–650. doi: 10.1016/s0039-6109(05)70572-7. [DOI] [PubMed] [Google Scholar]
- 22.Bowler P.G., Duerden B.I., Armstrong D.G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001;14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schierle C.F., De la Garza M., Mustoe T.A., Galiano R.D. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen. 2009;17:354–359. doi: 10.1111/j.1524-475X.2009.00489.x. [DOI] [PubMed] [Google Scholar]
- 24.Fonder M.A., Lazarus G.S., Cowan D.A., Aronson-Cook B., Kohli A.R., Mamelak A.J. Treating the chronic wound: a practical approach to the care of nonhealing wounds and wound care dressings. J. Am. Acad. Dermatol. 2008;58:185–206. doi: 10.1016/j.jaad.2007.08.048. [DOI] [PubMed] [Google Scholar]
- 25.Baranoski S. Choosing a wound dressing, part 1. Nursing. 2008;38:60–61. doi: 10.1097/01.NURSE.0000305919.47233.61. [DOI] [PubMed] [Google Scholar]
- 26.Atiyeh B.S., Dibo S.A., Hayek S.N. Wound cleansing, topical antiseptics and wound healing. Int. Wound J. 2009;6:420–430. doi: 10.1111/j.1742-481X.2009.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjarnsholt T., Alhede M., Jensen P.Ø., Nielsen A.K., Johansen H.K., Homøe P., Høiby N., Givskov M., Kirketerp-Møller K. Antibiofilm properties of acetic acid. Adv. Wound Care (New Rochelle) 2015;4:363–372. doi: 10.1089/wound.2014.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipsky B.A., Hoey C. Topical antimicrobial therapy for treating chronic wounds. Clin. Infect. Dis. 2009;49:1541–1549. doi: 10.1086/644732. [DOI] [PubMed] [Google Scholar]
- 29.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 30.Sand M., Gambichler T., Sand D., Skrygan M., Altmeyer P., Bechara F.G. MicroRNAs and the skin: tiny players in the body’s largest organ. J. Dermatol. Sci. 2009;53:169–175. doi: 10.1016/j.jdermsci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 32.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 33.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee J., Chan Y.C., Sen C.K. MicroRNAs in skin and wound healing. Physiol. Genomics. 2011;43:543–556. doi: 10.1152/physiolgenomics.00157.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chendrimada T.P., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 36.Roy S., Sen C.K. MiRNA in innate immune responses: novel players in wound inflammation. Physiol. Genomics. 2011;43:557–565. doi: 10.1152/physiolgenomics.00160.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu N., Zhang D., Chen S., Liu X., Lin L., Huang X., Guo Z., Liu J., Wang Y., Yuan W. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis. 2011;215:286–293. doi: 10.1016/j.atherosclerosis.2010.12.024. [DOI] [PubMed] [Google Scholar]
- 38.Yu J., Ryan D.G., Getsios S., Oliveira-Fernandes M., Fatima A., Lavker R.M. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. USA. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maurer B., Stanczyk J., Jüngel A., Akhmetshina A., Trenkmann M., Brock M., Kowal-Bielecka O., Gay R.E., Michel B.A., Distler J.H. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62:1733–1743. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 40.Beanes S.R., Dang C., Soo C., Ting K. Skin repair and scar formation: the central role of TGF-beta. Expert Rev. Mol. Med. 2003;5:1–22. doi: 10.1017/S1462399403005817. [DOI] [PubMed] [Google Scholar]
- 41.Kato M., Zhang J., Wang M., Lanting L., Yuan H., Rossi J.J., Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y.-F., Ding M., Liu D.-W., Liu Y., Mao Y.-G., Peng Y. MicroRNA profiling in cutaneous wounds of diabetic rats. Genet. Mol. Res. 2015;14:9614–9625. doi: 10.4238/2015.August.14.24. [DOI] [PubMed] [Google Scholar]
- 43.Yang X., Wang J., Guo S.L., Fan K.J., Li J., Wang Y.L., Teng Y., Yang X. miR-21 promotes keratinocyte migration and re-epithelialization during wound healing. Int. J. Biol. Sci. 2011;7:685–690. doi: 10.7150/ijbs.7.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banerjee J., Sen C.K. MicroRNAs in skin and wound healing. Methods Mol. Biol. 2013;936:343–356. doi: 10.1007/978-1-62703-083-0_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarbassov D.D., Ali S.M., Sabatini D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Madhyastha R., Madhyastha H., Nakajima Y., Omura S., Maruyama M. MicroRNA signature in diabetic wound healing: promotive role of miR-21 in fibroblast migration. Int. Wound J. 2012;9:355–361. doi: 10.1111/j.1742-481X.2011.00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh G., Subramanian I.V., Adhikari N., Zhang X., Joshi H.P., Basi D., Chandrashekhar Y.S., Hall J.L., Roy S., Zeng Y. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J. Clin. Invest. 2010;120:4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang D., Shi Z., Li M., Mi J. Hypoxia-induced miR-424 decreases tumor sensitivity to chemotherapy by inhibiting apoptosis. Cell Death Dis. 2014;5:e1301. doi: 10.1038/cddis.2014.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D., Li X., Wang A., Meisgen F., Pivarcsi A., Sonkoly E., Ståhle M., Landén N.X. MicroRNA-31 promotes skin wound healing by enhancing keratinocyte proliferation and migration. J. Invest. Dermatol. 2015;135:1676–1685. doi: 10.1038/jid.2015.48. [DOI] [PubMed] [Google Scholar]
- 50.Mahon P.C., Hirota K., Semenza G.L. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suárez Y., Fernández-Hernando C., Pober J.S., Sessa W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 52.Witte M.B., Thornton F.J., Tantry U., Barbul A. L-Arginine supplementation enhances diabetic wound healing: involvement of the nitric oxide synthase and arginase pathways. Metabolism. 2002;51:1269–1273. doi: 10.1053/meta.2002.35185. [DOI] [PubMed] [Google Scholar]
- 53.Schäffer M.R., Tantry U., Efron P.A., Ahrendt G.M., Thornton F.J., Barbul A. Diabetes-impaired healing and reduced wound nitric oxide synthesis: a possible pathophysiologic correlation. Surgery. 1997;121:513–519. doi: 10.1016/s0039-6060(97)90105-7. [DOI] [PubMed] [Google Scholar]
- 54.Witte M.B., Barbul A. Role of nitric oxide in wound repair. Am. J. Surg. 2002;183:406–412. doi: 10.1016/s0002-9610(02)00815-2. [DOI] [PubMed] [Google Scholar]
- 55.Lee P.C., Salyapongse A.N., Bragdon G.A., Shears L.L., 2nd, Watkins S.C., Edington H.D., Billiar T.R. Impaired wound healing and angiogenesis in eNOS-deficient mice. Am. J. Physiol. 1999;277:H1600–H1608. doi: 10.1152/ajpheart.1999.277.4.H1600. [DOI] [PubMed] [Google Scholar]
- 56.Jin H.Y., Gonzalez-Martin A., Miletic A.V., Lai M., Knight S., Sabouri-Ghomi M., Head S.R., Macauley M.S., Rickert R.C., Xiao C. Transfection of microRNA mimics should be used with caution. Front. Genet. 2015;6:340. doi: 10.3389/fgene.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haftmann C., Riedel R., Porstner M., Wittmann J., Chang H.D., Radbruch A., Mashreghi M.F. Direct uptake of Antagomirs and efficient knockdown of miRNA in primary B and T lymphocytes. J. Immunol. Methods. 2015;426:128–133. doi: 10.1016/j.jim.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morille M., Passirani C., Vonarbourg A., Clavreul A., Benoit J.P. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29:3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 59.Wilson J.M. Lessons learned from the gene therapy trial for ornithine transcarbamylase deficiency. Mol. Genet. Metab. 2009;96:151–157. doi: 10.1016/j.ymgme.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 60.Tros de Ilarduya C., Sun Y., Düzgüneş N. Gene delivery by lipoplexes and polyplexes. Eur. J. Pharm. Sci. 2010;40:159–170. doi: 10.1016/j.ejps.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 61.Yang N. An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig. 2015;5:179–181. doi: 10.4103/2230-973X.167646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hara E.S., Ono M., Eguchi T., Kubota S., Pham H.T., Sonoyama W., Tajima S., Takigawa M., Calderwood S.K., Kuboki T. miRNA-720 controls stem cell phenotype, proliferation and differentiation of human dental pulp cells. PLoS One. 2013;8:e83545. doi: 10.1371/journal.pone.0083545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sluijter J.P.G., van Mil A., van Vliet P., Metz C.H., Liu J., Doevendans P.A., Goumans M.J. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler. Thromb. Vasc. Biol. 2010;30:859–868. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- 64.Wu Y., Crawford M., Mao Y., Lee R.J., Davis I.C., Elton T.S., Lee L.J., Nana-Sinkam S.P. Therapeutic delivery of microRNA-29b by cationic lipoplexes for lung cancer. Mol. Ther. Nucleic Acids. 2013;2:e84. doi: 10.1038/mtna.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu L.-F., Thai T.H., Calado D.P., Chaudhry A., Kubo M., Tanaka K., Loeb G.B., Lee H., Yoshimura A., Rajewsky K. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eberhart J.K., He X., Swartz M.E., Yan Y.L., Song H., Boling T.C., Kunerth A.K., Walker M.B., Kimmel C.B., Postlethwait J.H. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat. Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shanmugam N., Reddy M.A., Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J. Biol. Chem. 2008;283:36221–36233. doi: 10.1074/jbc.M806322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benakanakere M.R., Li Q., Eskan M.A., Singh A.V., Zhao J., Galicia J.C., Stathopoulou P., Knudsen T.B., Kinane D.F. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J. Biol. Chem. 2009;284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheedy F.J., Palsson-McDermott E., Hennessy E.J., Martin C., O’Leary J.J., Ruan Q., Johnson D.S., Chen Y., O’Neill L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 70.Xu N., Brodin P., Wei T., Meisgen F., Eidsmo L., Nagy N., Kemeny L., Ståhle M., Sonkoly E., Pivarcsi A. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J. Invest. Dermatol. 2011;131:1521–1529. doi: 10.1038/jid.2011.55. [DOI] [PubMed] [Google Scholar]
- 71.Sonkoly E., Ståhle M., Pivarcsi A. MicroRNAs: novel regulators in skin inflammation. Clin. Exp. Dermatol. 2008;33:312–315. doi: 10.1111/j.1365-2230.2008.02804.x. [DOI] [PubMed] [Google Scholar]
- 72.Johnnidis J.B., Harris M.H., Wheeler R.T., Stehling-Sun S., Lam M.H., Kirak O., Brummelkamp T.R., Fleming M.D., Camargo F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 73.Primo M.N., Bak R.O., Schibler B., Mikkelsen J.G. Regulation of pro-inflammatory cytokines TNFα and IL24 by microRNA-203 in primary keratinocytes. Cytokine. 2012;60:741–748. doi: 10.1016/j.cyto.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 74.Park H., Huang X., Lu C., Cairo M.S., Zhou X. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J. Biol. Chem. 2015;290:2831–2841. doi: 10.1074/jbc.M114.591420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pottier N., Maurin T., Chevalier B., Puisségur M.P., Lebrigand K., Robbe-Sermesant K., Bertero T., Lino Cardenas C.L., Courcot E., Rios G. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS One. 2009;4:e6718. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin Y., Tymen S.D., Chen D., Fang Z.J., Zhao Y., Dragas D., Dai Y., Marucha P.T., Zhou X. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS ONE. 2013;8:e64434. doi: 10.1371/journal.pone.0064434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sundaram G.M., Common J.E.A., Gopal F.E., Srikanta S., Lakshman K., Lunny D.P., Lim T.C., Tanavde V., Lane E.B., Sampath P. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature. 2013;495:103–106. doi: 10.1038/nature11890. [DOI] [PubMed] [Google Scholar]
- 78.Yu J., Peng H., Ruan Q., Fatima A., Getsios S., Lavker R.M. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 2010;24:3950–3959. doi: 10.1096/fj.10-157404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harris T.A., Yamakuchi M., Ferlito M., Mendell J.T., Lowenstein C.J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W., Mouneimne G., Sidani M., Wyckoff J., Chen X., Makris A., Goswami S., Bresnick A.R., Condeelis J.S. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Viticchiè G., Lena A.M., Cianfarani F., Odorisio T., Annicchiarico-Petruzzelli M., Melino G., Candi E. MicroRNA-203 contributes to skin re-epithelialization. Cell Death Dis. 2012;3:e435. doi: 10.1038/cddis.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biswas S., Roy S., Banerjee J., Hussain S.R., Khanna S., Meenakshisundaram G., Kuppusamy P., Friedman A., Sen C.K. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc. Natl. Acad. Sci. USA. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bertero T., Gastaldi C., Bourget-Ponzio I., Imbert V., Loubat A., Selva E., Busca R., Mari B., Hofman P., Barbry P. miR-483-3p controls proliferation in wounded epithelial cells. FASEB J. 2011;25:3092–3105. doi: 10.1096/fj.10-168401. [DOI] [PubMed] [Google Scholar]
- 84.Suárez Y., Fernández-Hernando C., Yu J., Gerber S.A., Harrison K.D., Pober J.S., Iruela-Arispe M.L., Merkenschlager M., Sessa W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dews M., Homayouni A., Yu D., Murphy D., Sevignani C., Wentzel E., Furth E.E., Lee W.M., Enders G.H., Mendell J.T. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu J., Chen C., Liu Q., Liu B., Song C., Zhu S., Wu C., Liu S., Yu H., Yao D. The role of the miR-31/FIH1 pathway in TGF-β-induced liver fibrosis. Clin. Sci. 2015;129:305–317. doi: 10.1042/CS20140012. [DOI] [PubMed] [Google Scholar]
- 87.Edmonds M.D., Boyd K.L., Moyo T., Mitra R., Duszynski R., Arrate M.P., Chen X., Zhao Z., Blackwell T.S., Andl T. MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer. J. Clin. Invest. 2016;126:349–364. doi: 10.1172/JCI82720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang S., Aurora A.B., Johnson B.A., Qi X., McAnally J., Hill J.A., Richardson J.A., Bassel-Duby R., Olson E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuhnert F., Mancuso M.R., Hampton J., Stankunas K., Asano T., Chen C.Z., Kuo C.J. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 90.Chen Y., Gorski D.H. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fasanaro P., D’Alessandra Y., Di Stefano V., Melchionna R., Romani S., Pompilio G., Capogrossi M.C., Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pulkkinen K., Malm T., Turunen M., Koistinaho J., Ylä-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 93.Würdinger T., Tannous B.A., Saydam O., Skog J., Grau S., Soutschek J., Weissleder R., Breakefield X.O., Krichevsky A.M. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee D.Y., Deng Z., Wang C.-H., Yang B.B. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc. Natl. Acad. Sci. USA. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bonauer A., Carmona G., Iwasaki M., Mione M., Koyanagi M., Fischer A., Burchfield J., Fox H., Doebele C., Ohtani K. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 96.Doebele C., Bonauer A., Fischer A., Scholz A., Reiss Y., Urbich C., Hofmann W.K., Zeiher A.M., Dimmeler S. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115:4944–4950. doi: 10.1182/blood-2010-01-264812. [DOI] [PubMed] [Google Scholar]
- 97.Hua Z., Lv Q., Ye W., Wong C.K., Cai G., Gu D., Ji Y., Zhao C., Wang J., Yang B.B. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pin A.-L., Houle F., Guillonneau M., Paquet E.R., Simard M.J., Huot J. miR-20a represses endothelial cell migration by targeting MKK3 and inhibiting p38 MAP kinase activation in response to VEGF. Angiogenesis. 2012;15:593–608. doi: 10.1007/s10456-012-9283-z. [DOI] [PubMed] [Google Scholar]
- 99.Lei Z., Li B., Yang Z., Fang H., Zhang G.M., Feng Z.H., Huang B. Regulation of HIF-1α and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One. 2009;4:e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., Liuzzi F., Lulli V., Morsilli O., Santoro S. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Poliseno L., Tuccoli A., Mariani L., Evangelista M., Citti L., Woods K., Mercatanti A., Hammond S., Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 102.Kuehbacher A., Urbich C., Dimmeler S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol. Sci. 2008;29:12–15. doi: 10.1016/j.tips.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 103.Wang X.H., Qian R.Z., Zhang W., Chen S.F., Jin H.M., Hu R.M. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin. Exp. Pharmacol. Physiol. 2009;36:181–188. doi: 10.1111/j.1440-1681.2008.05057.x. [DOI] [PubMed] [Google Scholar]
- 104.Caporali A., Meloni M., Völlenkle C., Bonci D., Sala-Newby G.B., Addis R., Spinetti G., Losa S., Masson R., Baker A.H. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282–291. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 105.Ciechomska M., O’Reilly S., Suwara M., Bogunia-Kubik K., van Laar J.M. MiR-29a reduces TIMP-1 production by dermal fibroblasts via targeting TGF-β activated kinase 1 binding protein 1, implications for systemic sclerosis. PLoS One. 2014;9:e115596. doi: 10.1371/journal.pone.0115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Z., Hassan M.Q., Jafferji M., Aqeilan R.I., Garzon R., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Rooij E., Sutherland L.B., Thatcher J.E., DiMaio J.M., Naseem R.H., Marshall W.S., Hill J.A., Olson E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang B., Herman-Edelstein M., Koh P., Burns W., Jandeleit-Dahm K., Watson A., Saleem M., Goodall G.J., Twigg S.M., Cooper M.E. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes. 2010;59:1794–1802. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]