Abstract
The human cytomegalovirus UL99-coded pp28 is a myristoylated phosphoprotein located in the virion tegument domain, which resides between the capsid and envelope. A previous study has demonstrated that BADsubUL99, a pp28-deficient mutant virus, fails to assemble enveloped virus particles. Capsids, coated with tegument proteins, accumulate in the cytoplasm of mutant virus-infected cells. This phenotype indicates that pp28 is required for the acquisition of an envelope; it presumably acts by directing tegument-associated capsids to bud through an intracellular membrane derived from the cell's secretory apparatus that has been modified to contain viral transmembrane glycoproteins. Here we demonstrate that BADsubUL99 can spread from cell to cell, even though highly sensitive assays fail to detect infectious virus progeny in cultures of infected fibroblasts. We propose that, in the absence of pp28, tegument-coated capsids might nevertheless bud through cellular membranes, including the plasma membrane. If this suggestion is correct, the enveloped particle could potentially infect an adjacent cell to mediate the cell-to-cell spread that is observed. This mode of spread might also occur after infection with wild-type virus, and it could facilitate immune evasion, assuming that the resulting particles do not have a normal complement of virus-coded envelope glycoproteins.
Keywords: mutant cytomegalovirus, spread from cell to cell, virus assembly
Human cytomegalovirus (HCMV) is the prototypical member of the β-herpes virus family (1). Epidemiological studies have demonstrated that HCMV infection is widespread in the human population. Infections in healthy children and adults are generally asymptomatic, but HCMV is a leading viral cause of birth defects and a major cause of morbidity and mortality in immunocompromised individuals (2).
The HCMV particle, like the virions of all herpes viruses, contains a double-stranded DNA genome that is enclosed in a protein shell, termed the capsid. The capsid is surrounded by a layer of virus-coded proteins, termed the tegument, and this protein layer is surrounded by a membrane, termed the envelope, that contains a set of virus-coded transmembrane glycoproteins (1). Herpes virus particles also contain virus-coded RNAs (3–6).
The generally accepted model for assembly of herpes virions is complex (reviewed in refs. 7–10). Within the nucleus, viral DNA is packaged into capsids, and several tegument proteins that accumulate within the nucleus associate with the capsid. The capsid–protein complex buds through the inner nuclear membrane acquiring an envelope, which fuses with the outer nuclear membrane, releasing the de-enveloped capsid and associated tegument proteins into the cytoplasm. The majority of the tegument proteins accumulate in the cytoplasm, localizing substantially to a membranous juxtanuclear domain that partially coincides with markers for the trans-Golgi compartment. The capsid, together with tegument proteins derived from both the nucleus and cytoplasm, next buds through a membrane derived from the secretory system located in the cytoplasmic juxtanuclear domain to produce a mature, enveloped virion.
The pp28 tegument protein (11–13), which is encoded by the HCMV UL99 ORF, is a 190-aa myristoylated (14) phosphoprotein (15) that accumulates in the cytoplasm of the infected cell within the juxtanuclear packaging region, together with other tegument proteins and envelope glycoproteins (16). HCMV mutants that are not able to produce the pp28 protein can be propagated in complementing cells that express the viral protein, but they are profoundly defective for replication in normal fibroblasts (10, 17). It was not possible to detect by plaque assay the production of infectious progeny at any time tested after infection of fibroblasts with a point mutant or substitution mutant unable to produce pp28. The mutant viruses generated cytoplasmic, DNA-containing, tegument-associated capsids, but the capsids failed to acquire an envelope. We proposed that pp28 interacts with other tegument constituents through protein– protein interactions and with the virion envelope through its myristic acid moiety, bridging between the tegument and the envelope (10). This view fits well with the failure of the tegument-coated capsids produced by pp28-deficient viruses to associate with an envelope.
Here we demonstrate that a pp28-deficient virus can spread from cell to cell, even though no infectious progeny can be detected in cultures of normal fibroblasts. We propose a model for cell-to-cell spread that bypasses the normal route for acquisition of a virion envelope.
Methods
Cells and Viruses. Primary human foreskin fibroblasts (passages 4–15), referred to in this report as normal fibroblasts, were cultured in medium containing 10% FCS. Complementing cells expressing pp28 were prepared by infection of fibroblasts with a recombinant retrovirus, Retropp28WT, as described in refs. 10 and 18. The pp28-expressing retrovirus grew to relatively low titers, so it was necessary to perform eight sequential infections with an undiluted preparation of Retropp28WT to generate HFFpp28-8x cells. Fibroblasts expressing GFP were produced by one round of infection with a high titer stock of a GFP-expressing retrovirus, RetroGFP.
The wild-type virus used in this study, BADwt, was derived from an infectious bacterial artificial chromosome (BAC) clone of the AD169 strain of HCMV (19). BADsubUL99 is a pp28-deficient mutant virus (10) that was constructed by using the infectious BAC from which BADwt is derived. BADsubUL99 lacks the majority of the UL99 ORF; the sequence spanning from 144392 to 144963 (numbering according to ref. 20) was replaced by a kanamycin resistance gene. Stocks of BADwt were prepared by infection of normal fibroblasts, and stocks of BADsubUL99 were generated by infection of HFFpp28-8x cells at a multiplicity of 0.01 plaque-forming units (pfu) per cell. After full cytopathic effect was observed, the cells were harvested in their medium and disrupted by sonication, and the supernatants obtained after pelleting the debris served as virus stocks. Wild-type and mutant virus titers were determined by plaque assay on HFFpp28-8x cells. In some experiments, cells were treated with citrate buffer (40 mM Na citrate/10 mM KCl/135 mM NaCl, pH 3.0) for 1 min at room temperature to inactivate residual extracellular virus (21), and infected cultures were maintained in medium containing 3% neutralizing Ig to HCMV (CytoGam, MedImmune, Gaithersburg, MD), which was replaced every 24 h.
Detection of Viral Proteins and DNA in Infected Cells. For protein immunofluorescence, normal fibroblasts grown on glass coverslips were infected with BADwt or BADsubUL99 or seeded with infected fibroblasts. At various times after infection, cells were washed twice with PBS at 37°C, fixed for 20 min at room temperature with 2% paraformaldehyde in PBS, and subsequently washed twice with PBS. Fixed cells were permeabilized by treatment for 15 min at room temperature with PBS containing 0.1% Triton X-100, washed twice with PBS, and blocked by incubation at room temperature for 30 min with PBS containing 2% BSA and 0.05% Tween 20. Cells were incubated at room temperature for 1 h with primary antibody diluted in PBS containing 0.05% Tween 20, washed three times with buffer, incubated for 1 h with fluorochrome-conjugated anti-mouse or anti-rabbit secondary antibody, and washed again three times. Coverslips were mounted in SlowFade solution (Molecular Probes). Primary antibodies used in these studies were UL83-encoded pp65 rabbit polyclonal antibody (a gift of M. Schrader, University of Marburg, Marburg, Germany) and UL123-encoded IE1 (1B12; ref. 10) murine monoclonal antibody.
HCMV DNA was detected by fluorescence in situ hybridization (6, 22). Fibroblasts were grown on sterile glass cover slips in six-well plates to confluence and then infected with BADwt or BADsubUL99 at a multiplicity of 0.02 pfu per cell. Cells were fixed in 4% paraformaldehyde in PBS for 15 min, washed with PBS, and permeabilized for 15 min in 0.2% Triton X-100 in PBS. After equilibration in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), cells were treated with DNase-free RNase A (100 mg/ml) in 2× SSC, dehydrated in an ethanol series (70%, 80%, 90%, and 100% ethanol for 5 min each), air-dried, and incubated overnight at 37°C with the hybridization mixture. Digoxigenin-labeled probes corresponding to the complete HCMV genome were generated by nick translation (23) by using DIG-Nick Translation Mix (Roche) prepared according to the manufacturer's protocol. Digoxigenin-labeled probe (≈50 ng), 15 μg of salmon sperm DNA, and 15 μg of yeast tRNA per coverslip were ethanol-precipitated, resuspended in 20 μl of hybridization buffer containing 50% formamide in 2× SSC, 10% dextran sulfate, and 0.1% SDS and heated at 94°C for 4 min to denature the probe DNA. After hybridization, specimens were washed at 37°C with 55% formamide in 2× SSC, pH 7.0 (four times for 5 min each), 2× SSC (two times for 5 min each), and 0.2× SSC (two times for 5 min each) (6). Hybridized probes were detected by using the Fluorescent Antibody Enhancer Set for DIG Detection (Roche) according to the manufacturer's protocol. The third antibody was supplemented with 1 ng/ml DAPI (Molecular Probes) to counterstain the chromosomal DNA. After the last wash, cells were dehydrated in an ethanol series, air-dried, and mounted with SlowFade solution. Fluorescent images were captured on a Zeiss LSM510 confocal microscope.
Results
We were surprised to see the formation of small groups of cells expressing viral antigens after infection of normal fibroblast monolayer cultures with the pp28-deficient mutant, BADsubUL99. Furthermore, evidence of virus replication, i.e., cell rounding, was seen for most cells within cultures of normal fibroblasts at 21 days after infection at a multiplicity of 0.01 pfu per cell with BADsubUL99. This observation would imply that progeny virus was produced in the infected cultures to mediate spread. However, we previously were unable to detect the production of infectious progeny by the mutant in normal fibroblasts (10).
BADwt and BADsubUL99 Spread from Cell to Cell in Fibroblasts. We performed a controlled experiment to monitor spread in normal fibroblasts by BADsubUL99 compared with its wild-type parent, BADwt. Neutralizing Ig to HCMV was included in the culture medium (3% by volume) to limit cell-free spread by the wild-type virus. This amount of antibody reduced the infectivity of an HCMV virus stock by a factor of 3–8 × 103 in repeated experiments. By day 5 after infection, the wild-type virus had spread to produce foci of infected cells in the fibroblast monolayer that included 20–100 cells. The mutant virus spread somewhat more slowly, producing foci ranging in size from 10 to 30 cells at this time. Importantly, when cultures were infected at the same input multiplicity with mutant or wild-type virus, approximately the same number of infected-cell foci were produced, demonstrating that BADsubUL99 spread from an initially infected cell as efficiently as did BADwt.
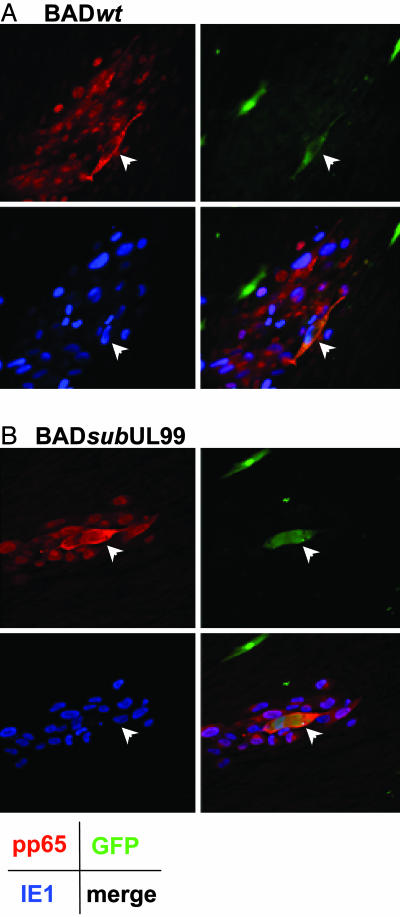
Fig. 1 documents the spread of viral proteins from an infected cell to its neighbors. In this experiment, normal fibroblasts expressing GFP from a recombinant retrovirus were infected at a multiplicity of 0.02 pfu per cell with either BADwt or BADsubUL99. At 24 h after infection, individual cells were isolated, treated sequentially with citrate buffer and neutralizing Ig to HCMV to inactivate residual extracellular virus, and replated onto a monolayer of uninfected normal fibroblasts. The cultures were then maintained in medium containing anti-HCMV Ig to inhibit cell-free spread of virus. Virus spread was monitored by immunofluorescent detection of the immediate-early IE1 and late pp65 proteins. At 5 days after infection of the original cell, the wild-type and mutant viruses produced similar numbers of foci in which cells expressed both IE1 and pp65 (Fig. 1). Two additional late proteins, pp150 and the major capsid protein, were also monitored and found to be expressed after infection with the wild-type or mutant virus (data not shown). As discussed above, the BADsubUL99-generated foci usually contained fewer cells than those produced in BADwt-infected cultures.
Fig. 1.
Cell-to-cell spread of HCMV-coded proteins. Fibroblasts expressing GFP were infected with BADwt (A) or BADsubUL99 (B) at a multiplicity of 0.02 pfu per cell. After 24 h, the culture was treated sequentially with citrate buffer and medium containing 3% anti-HCMV Ig to inactivate residual extracellular virus, and the cells were transferred to cultures of normal fibroblasts lacking GFP in medium containing 3% anti-HCMV Ig. Five days after infection, cells were processed for immunofluorescence by using antibodies specific for the HCMV IE1 protein (blue), HCMV pp65 protein (red), or GFP (green). A Lower Right and B Lower Right display images in which the three colors are merged. (Final magnification: ×250.)
In this experiment, GFP fluorescence identifies the original infected cell that was seeded onto a monolayer of uninfected cells that do not express GFP. The original infected cell that is responsible for the focus of infected cells is marked by an arrow in the images. Fibroblasts become enlarged after infection with HCMV, and, for both wild-type and mutant, the GFP-expressing cell is larger than surrounding cells, consistent with the fact that it was infected for a longer period. In the case of the wild-type virus (Fig. 1 A), the cytoplasmic GFP is limited to the single larger cell that seeded the infected focus. If the virus had spread to adjacent cells by cell–cell fusion, then the GFP would have spread beyond the original cell, and this is clearly not the case. Rare foci showed possible evidence for a two-cell fusion event, as is presented for the mutant virus (Fig. 1B). In this case, it appears that two nuclei share the cytoplasmic GFP fluorescence that marks the founder cell of the focus. However, even if a fusion event occurred involving the founder cell plus one additional cell, the remaining cells in the infected focus do not share the GFP. Further, the viral pp65 protein (Fig. 1, red signal), which is concentrated at a juxtanuclear assembly domain late after HCMV infection (16) but is also found throughout the cytoplasm, clearly delineates individual cells in the foci, rather than fused, multinucleate cells. Failure of the GFP marker to spread between cells also rules out the possibility that the spread of viral antigens results from cell division after infection. Cell division is also precluded as a possible explanation for spread by the fact that HCMV blocks cell cycle progression (24–27).
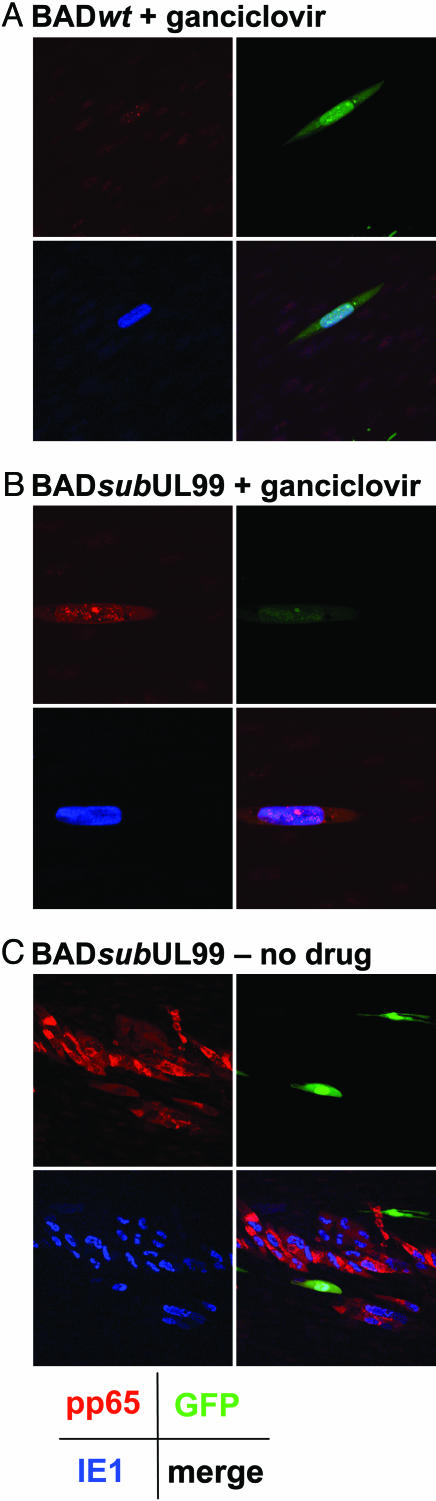
It remained possible that only viral proteins were moving from cell to cell, without spread of the viral genome. To investigate this possibility, we tested whether ganciclovir, which blocks HCMV DNA replication, can block the spread (Fig. 2). Again, GFP-expressing fibroblasts were infected with HCMV and subsequently seeded into cultures of uninfected fibroblasts that did not express GFP. Ten days after infection of the original cell, BADsubUL99 had produced large colonies of infected cells (Fig. 2C). However, in the presence of the drug, both BADwt (Fig. 2 A) and BADsubUL99 (Fig. 2B) failed to spread from the initial GFP-positive cell, showing that virus DNA replication is required for the formation of foci of cells expressing viral antigens.
Fig. 2.
Ganciclovir blocks cell-to-cell spread of viral proteins. Fibroblasts expressing GFP were infected with BADwt (A) or BADsubUL99 (B and C) at a multiplicity of 0.02 pfu per cell. After 24 h, the culture was treated sequentially with citrate buffer and medium containing 3% anti-HCMV Ig to inactivate residual extracellular virus, and the cells were transferred to cultures of normal fibroblasts lacking GFP in medium containing 3% anti-HCMV Ig and 100 mM ganciclovir (A and B) or in medium with no drug (C). Ten days after infection, cells were processed for immunofluorescence by using antibodies specific for the HCMV IE1 protein (blue), HCMV pp65 protein (red), or GFP (green). A Lower Right, B Lower Right, and C Lower Right display images in which the three colors are merged. (Final magnification: ×500 in A and B and ×250 in C.)
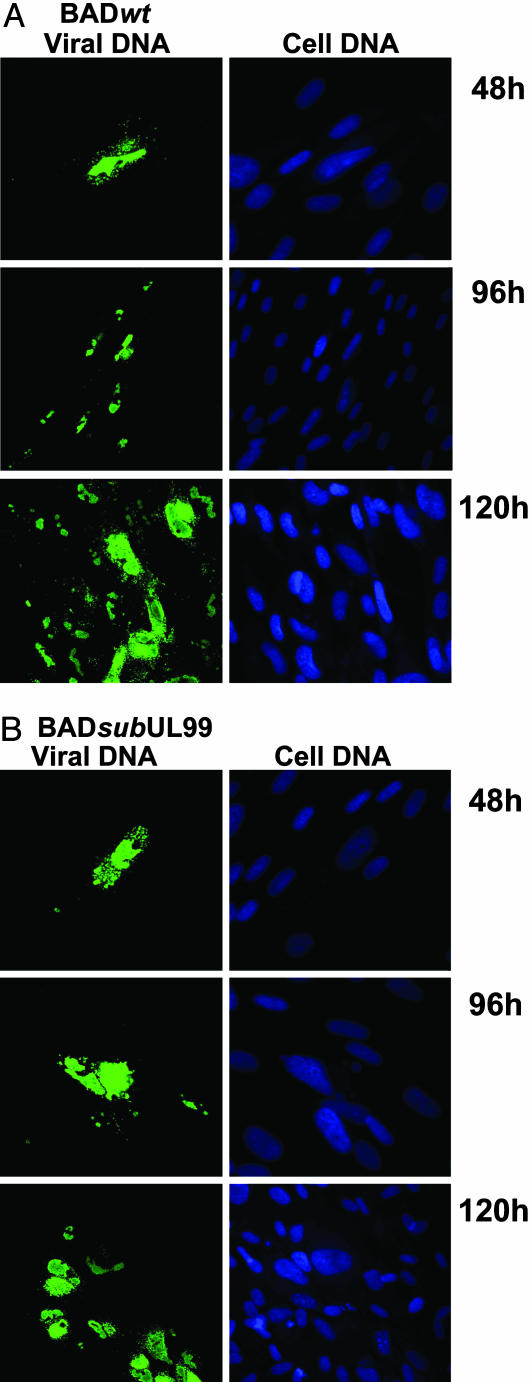
To demonstrate directly that viral DNA spread together with viral antigens, we performed a fluorescent in situ hybridization analysis (Fig. 3). Normal fibroblasts were infected at a multiplicity of 0.02 pfu per cell with the wild-type or mutant virus. Twenty-four hours later, the cultures were treated with citrate buffer to inactivate residual extracellular virus and then fed with medium containing anti-HCMV Ig to inhibit spread by extracellular virus. At 2, 4, and 5 days after infection, samples were processed for analysis. At 2 days after infection, viral DNA was evident in single cells, and foci of HCMV DNA-containing cells were evident at 4 and 5 days after infection.
Fig. 3.
Cell-to-cell spread of HCMV DNA. Normal fibroblasts were infected with BADwt (A) or BADsubUL99 (B), and 24 h later the cultures were treated with citrate buffer to inactivate residual extracellular virus. After maintenance in medium containing 3% anti-HCMV antibody until 48, 96, or 120 h after infection, cells were processed for fluorescent in situ hybridization by using an HCMV-specific probe DNA. Viral DNA (green) and total cellular DNA (blue) were visualized by confocal microscopy. (Final magnification: ×500.)
Viral DNA and proteins spread from cell to cell after infection of normal fibroblasts with a pp28-deficient virus.
BADsubUL99 Fails to Produce Detectable Progeny in Normal Fibroblasts. We have reported previously that pp28-deficient mutants, including BADsubUL99, fail to produce detectable infectious progeny in normal fibroblasts (10). In our earlier analysis, we monitored virus production by plaque assay on pp28-expressing fibroblasts (HFFpp28-8x cells), where the constitutively expressed viral protein could complement the growth defect of the mutant virus. However, BADsubUL99 replicated to relatively low titers on the pp28-expressing cells, suggesting that pp28-expressing fibroblasts did not efficiently complement the mutant. Consequently, it was possible that infectious virus was produced in BADsubUL99-infected cultures, but we failed to detect it by plaque assay on HFFpp28-8x cells. Therefore, we used more sensitive assays to search for the production of infectious progeny by BADsubUL99 in normal fibroblasts.
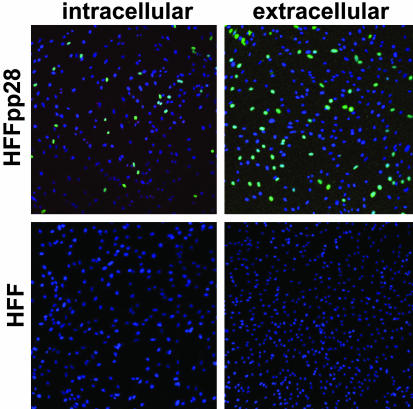
First, we assayed for the ability of the mutant virus to generate single infected cells expressing the most abundant immediate-early protein, IE1 (Fig. 4). BADsubUL99 expresses its gene products normally and exhibits a growth defect at the level of assembly in normal fibroblasts. Therefore, infectious mutant particles should readily express IE1 protein. Both intracellular and extracellular BADsubUL99 particles could be detected by this assay at 10 days after infection of complementing HFFpp28-8x cells (Fig. 4 Upper). In contrast, no IE1-expressing cells could be detected at 10 days after infection of normal fibroblasts (Fig. 4 Lower). Indeed, in two independent experiments, BADsubUL99-infected normal fibroblast cultures that contained >103 infected foci failed to produce a single IE1-positive cell.
Fig. 4.
BADsubUL99 fails to make detectable infectious progeny. Normal fibroblasts (HFF) and fibroblasts expressing pp28 protein (HFFpp28) were infected with BADsubUL99 at a multiplicity of 0.01 pfu per cell. At 10 days after infection, the supernatant was removed from the cultures for assay of extracellular virus and cells were lysed in a small volume of medium by sonication for assay of intracellular virus. Fibroblasts were infected with the preparations and assayed 24 h later for expression of HCMV IE1 (green) by immunofluorescence. Nuclei are blue in the images. (Final magnification: ×100.)
We used transwell culture vessels as a second approach to search for the production of infectious BADsubUL99. These culture vessels contain two chambers separated by a membrane with 0.45-μm pores; cells cannot pass through the membrane, but the medium and HCMV particles readily communicate between chambers. Normal fibroblasts were plated in the upper chamber, and pp28-expressing, HFFpp28-8x cells were plated in the bottom chamber. When the normal fibroblasts in three separate vessels were infected by BADwt at a multiplicity of 0.01 pfu per cell, the virus spread in each vessel to the lower chamber and killed the HFFpp28-8x cells by day 25 after infection. In contrast, when the normal fibroblasts in three vessels were infected at the same multiplicity with BADsubUL99, they developed the morphology of infected cells, but there was no evidence for spread of the infection to the lower chamber containing HFFpp28-8x cells in any of the vessels after 45 days, when the experiment was terminated.
Two highly sensitive assays failed to detect the production of infectious virus after infection of normal fibroblasts by BADsubUL99.
Discussion
Earlier work demonstrated that, after infection of normal fibroblasts, pp28-deficient HCMV mutants failed to generate infectious progeny that can be detected by plaque assay on fibroblasts constitutively expressing the viral protein (10). Here we tested for infectious BADsubUL99 progeny using two more sensitive assays. No progeny able to express IE1 protein was detected after infection of normal fibroblasts (Fig. 4). The defect in pp28-deficient viruses occurs at the end of the replication cycle, and the assay for IE1 does not require the mutant virus to advance beyond the immediate-early phase of the cycle to score as an infectious particle. Consequently, the failure to detect even a single IE1-positive cell, when BADsubUL99-infected cultures were assayed in their entirety, argues strongly that no infectious particles were generated in normal fibroblasts. In a second assay, mutant virus failed to spread across a membrane from infected normal fibroblasts to pp28-expressing fibroblasts in a transwell culture vessel. This assay reinforces the conclusion that BADsubUL99 does not produce infectious extracellular progeny, and it demonstrates that, in contrast to the wild-type virus, cell–cell contact is required for spread of the mutant.
Despite our inability to detect infectious virus, we readily observed spread of BADsubUL99 in monolayers of normal fibroblasts. Immediate-early and late proteins (Fig. 1) and viral DNA (Fig. 3) were identified in the cells comprising the infected foci, and no spread occurred when viral DNA replication was blocked by addition of ganciclovir to infected cultures (Fig. 2). The pp28-deficient mutant produced about the same number of infected cell foci as did its parent, indicating that the mutant was as likely as the wild-type virus to spread.
Cell-to-cell spread in herpes virus-infected cultures is generally thought to involve the directed release of mature virus particles into the space between adjacent cells with the subsequent adsorption and entry into a neighboring cell (reviewed in ref. 28). For cells in close apposition, such as endothelial cells with tight junctions, the exposure of the spreading virus to the external environment can be short and privileged in that the virus is not readily susceptible to neutralizing antibodies in the extracellular environment. In the case of α-herpes viruses, specific viral glycoproteins have been shown to facilitate cell-to-cell spread (reviewed in refs. 28 and 29).
This study detects a different type of spread, the movement of HCMV to adjacent cells in the absence of an essential assembly protein. It is conceivable that tegument-coated capsids move to adjacent cells through intracellular channels. There is precedent for such movement in plant viruses, which encode movement proteins that facilitate cell-to-cell spread of subviral complexes through intercellular bridges termed plasmodesmata. For example, tobacco mosaic virus replication complexes spread between cells through plasmodesmata (30). However, we are not aware of intercellular connections that could potentially support such spread in fibroblasts. Furthermore, if such connections were induced by infection, one might expect that they would facilitate the spread of GFP as well as virus. Spread of this marker protein was not evident in experiments in which the initially infected cell contained GFP but the surrounding cells did not (Fig. 1).
The pp28 protein is membrane-associated (14). When it is expressed in the absence of other viral proteins, it targets to the endoplasmic reticulum–Golgi-intermediate compartment (16), and it is localized to a juxtanuclear packaging domain that partially coincides with markers for the trans-Golgi compartment in HCMV-infected cells (14). Myristoylation is required for the proper localization (16, 31) and function of pp28 (10, 17), further arguing that the protein functions in association with membranes. BADsubUL99-infected cells accumulate tegument-associated capsids throughout the cytoplasm (10). Therefore, it is likely that pp28 normally directs tegument-associated capsids to bud across the trans-Golgi membrane, where it resides, to generate mature, enveloped particles. In the absence of pp28, it is conceivable that tegument-coated capsids nevertheless bud, perhaps very inefficiently, through alternative membranes. If the capsids bud through an intracellular membrane, we would expect that occasional enveloped particles would be present in the cytoplasm of BADsubUL99-infected cells, but this was not observed (10). Perhaps budding through intracellular membranes does occur, but at too low a frequency to be seen in our experiments. Alternatively, the capsid might bud across the plasma membrane. The resulting extracellular virion-like particle would have the potential to fuse with the membrane of an adjacent cell, mediating cell-to-cell spread. If such virion-like particles are produced, they fail to score in our assays for infectious particles. They could fail to score because they are generated only very rarely, because they are unstable, or because they do not have a full complement of virus-coded glycoproteins, and they cannot absorb to a cell unless placed in intimate contact with its membrane, the sort of contact that permits cell-to-cell spread that is resistant to neutralizing antibodies.
The presence of capsids throughout the cytoplasm is not unique to cells infected with a pp28-deficient virus. Tegument-coated capsids are dispersed throughout the cytoplasm of fibroblasts infected with wild-type HCMV as well (10, 32). Perhaps a portion of these capsids bud independent of pp28 through the plasma membrane to produce virion-like particles that contribute to cell-to-cell spread of the wild-type virus.
The virion-like particles postulated here could be functionally similar to the virions produced by glycoprotein D-deficient mutants of pseudorabies virus (reviewed in ref. 33). Glycoprotein D is the principal envelope protein of α-herpes viruses that binds cell surface receptors (34). Pseudorabies virions that lack glycoprotein D are not infectious as extracellular particles (35, 36), but they can spread from cell to cell in the nervous system of animals and in cultured cells (37, 38). Furthermore, pseudorabies virus has been proposed to assemble virions within axons that can spread from cell to cell (39). Although the details of assembly might be different, the work with psuedorabies virus provides a precedent for the assembly of herpes virions at a site distant from the normal Golgi component-associated assembly site, as we are proposing here for HCMV.
If this speculative model for an alternative mode of virus spread is correct, might it be advantageous to the virus? If the particle does not contain a normal complement of virion glycoproteins, it could enable the virus to evade neutralizing antibodies as it spreads. The production of alternatively packaged genomes would add to the numerous immune evasion strategies that have been described for HCMV (reviewed in refs. 40 and 41).
Acknowledgments
We thank L. Enquist (Princeton University, Princeton) for critical reading of the manuscript and M. Schrader for a generous gift of antibody. This work was supported by National Institutes of Health Grant CA82396, and M.C.S. was supported by a predoctoral fellowship from the Brazilian government (Conselho Nacional de Desenvolvimento Científíco e Tecnológico-CNPq).
Author contributions: M.C.S. and T.E.S. designed research; M.C.S. and J.S. performed research; and M.C.S. and T.E.S. wrote the paper.
Abbreviations: HCMV, human cytomegalovirus; pfu, plaque-forming unit.
References
- 1.Mocarski, E. S. & Courcelle, C. T. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. & Straus, S. E. (Lippincott-Raven, Philadelphia), Vol. 2, pp. 2629–2673. [Google Scholar]
- 2.Pass, R. F. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. & Straus, S. E. (Lippincott-Raven, Philadelphia), Vol. 2, pp. 2675–2705. [Google Scholar]
- 3.Bresnahan, W. A. & Shenk, T. (2000) Science 288, 2373–2376. [DOI] [PubMed] [Google Scholar]
- 4.Greijer, A. E., Dekkers, C. A. J. & Middeldorp, J. M. (2000) J. Virol. 74, 9078–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sciortino, M.-T., Suzuki, M., Taddeo, B. & Roizman, B. (2001) J. Virol. 75, 8105–8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terhune, S. S., Schroer, J. & Shenk T. (2004) J. Virol. 78, 10390–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson, W. (1996) Intervirology 39, 389–400. [DOI] [PubMed] [Google Scholar]
- 8.Enquist, L. W., Husak, P. J., Banfield, B. W. & Smith, G. A. (1999) Adv. Virus Res. 51, 237–347. [DOI] [PubMed] [Google Scholar]
- 9.Mettenleiter, T. C. (2002) J. Virol. 76, 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva, M. C., Yu, Q. C., Enquist, L. & Shenk, T. (2003) J. Virol. 77, 10594–10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez, J. & St. Jeor, S. C. (1986) J. Virol. 60, 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landini, M. P., Severi, B., Furlini, G. & de Giorgi, B. (1987) Virus Res. 8, 15–23. [DOI] [PubMed] [Google Scholar]
- 13.Baldick, C. J., Jr., & Shenk, T. (1996) J. Virol. 70, 6097–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez, V., Sztul, E. & Britt, W. J. (2000) J. Virol. 74, 3842–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer, H., Bankier, A. T., Landini, M. P., Brown, C. M., Barrell, B. G., Ruger, B. & Mach, M. (1988) J. Virol. 62, 2243–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez, V., Greis, K. D., Sztul, E. & Britt, W. J. (2000) J. Virol. 74, 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Britt, W. J., Jarvis, M., Seo, J. Y., Drummond, D. & Nelson, J. (2004) J. Virol. 78, 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinsella, T. M. & Nolan, G. P. (1996) Hum. Gene Ther. 7, 1405–1413. [DOI] [PubMed] [Google Scholar]
- 19.Yu, D., Smith, G. A., Enquist, L. W. & Shenk, T. (2002) J. Virol. 76, 2316–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chee, M. S., Bankier, A. T., Beck, S., Bohni, R., Brown, C. M., Cerny, R., Horsnell, T., Hutchison, C. A., III, Kouzarides, T., Martignetti, J. A., et al. (1990) Curr. Top. Microbiol. Immunol. 154, 125–169. [DOI] [PubMed] [Google Scholar]
- 21.Tirabassi, R. S. & Enquist, L. W. (1998) J. Virol. 72, 4571–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichter, P., Tang, C. J., Call, K., Hermanson, G., Evans, G. A., Housman, D. & Ward, D. C. (1990) Science 247, 64–69. [DOI] [PubMed] [Google Scholar]
- 23.Rigby, P.W., Dieckmann, M., Rhodes, C. & Berg, P. (1977) J. Mol. Biol. 113, 237–251. [DOI] [PubMed] [Google Scholar]
- 24.Jault, F. M., Jault, J. M., Ruchti, F., Fortunato, E. A., Clark, C., Corbeil, J., Richman, D. D. & Spector, D. H. (1995) J. Virol. 69, 6697–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bresnahan, W. A., Boldogh, I., Thompson, E. A. & Albrecht, T. (1996) Virology 224, 150–160. [DOI] [PubMed] [Google Scholar]
- 26.Lu, M. & Shenk, T. (1996) J. Virol. 70, 8850–8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dittmer, D. & Mocarski, E. S. (1997) J. Virol. 71, 1629–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, D. C. & Huber, M. T. (2002) J. Virol. 76, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamichi, K., Matsumoto, Y. & Otsuka, H. (2002) Virology 294, 22–30. [DOI] [PubMed] [Google Scholar]
- 30.Kawakami, S., Watanabe, Y. & Beachy, R. N. (2004) Proc. Natl. Acad. Sci. USA 101, 6291–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, T. R. & Lee, S. W. (2004) J. Virol. 78, 1488–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, J. D. & De Harven, E. (1973) J. Virol. 12, 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomishima, M. J., Smith, G. A. & Enquist, L. W. (2001) Traffic 2, 429–436. [DOI] [PubMed] [Google Scholar]
- 34.Shukla, D., Dal Canto, M. C., Rowe, C. L. & Spear, P. G. (2000) J. Virol. 74, 11773–11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauh, I. & Mettenleiter, T. C. (1991) J. Virol. 65, 5348–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peeters, B., de Wind, N., Hooisma, M., Wagenaar, F., Gielkens, A. & Moormann, R. (1992) J. Virol. 66, 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babic, N., Mettenleiter, T. C., Flamand, A. & Ugolini, G. (1993) J. Virol. 67, 4421–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peeters, B., Pol, J., Gielkens, A. & Moormann, R. (1993) J. Virol. 67, 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomishima, M. J. & Enquist, L. W. (2002) J. Virol. 76, 8310–8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loenen, W. A., Bruggeman, C. A. & Wiertz, E. J. (2001) Semin. Immunol. 13, 41–49. [DOI] [PubMed] [Google Scholar]
- 41.Basta, S. & Bennink, J. R. (2003) Viral Immunol. 16, 231–242. [DOI] [PubMed] [Google Scholar]