SUMMARY
The ecology of forest soils is an important field of research due to the role of forests as carbon sinks. Consequently, a significant amount of information has been accumulated concerning their ecology, especially for temperate and boreal forests. Although most studies have focused on fungi, forest soil bacteria also play important roles in this environment. In forest soils, bacteria inhabit multiple habitats with specific properties, including bulk soil, rhizosphere, litter, and deadwood habitats, where their communities are shaped by nutrient availability and biotic interactions. Bacteria contribute to a range of essential soil processes involved in the cycling of carbon, nitrogen, and phosphorus. They take part in the decomposition of dead plant biomass and are highly important for the decomposition of dead fungal mycelia. In rhizospheres of forest trees, bacteria interact with plant roots and mycorrhizal fungi as commensalists or mycorrhiza helpers. Bacteria also mediate multiple critical steps in the nitrogen cycle, including N fixation. Bacterial communities in forest soils respond to the effects of global change, such as climate warming, increased levels of carbon dioxide, or anthropogenic nitrogen deposition. This response, however, often reflects the specificities of each studied forest ecosystem, and it is still impossible to fully incorporate bacteria into predictive models. The understanding of bacterial ecology in forest soils has advanced dramatically in recent years, but it is still incomplete. The exact extent of the contribution of bacteria to forest ecosystem processes will be recognized only in the future, when the activities of all soil community members are studied simultaneously.
KEYWORDS: bacteria, decomposition, ecosystem processes, forest ecology, global change, litter, nutrient cycling, soil
INTRODUCTION
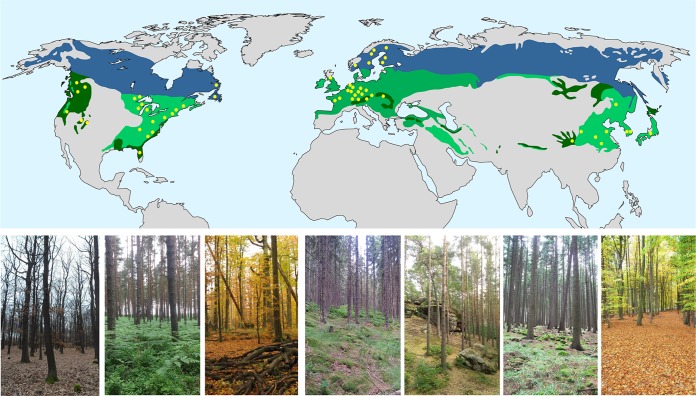
Forests represent one of the largest and most important ecosystems on Earth, covering more than 40 million km2 and representing 30% of the total global land area (1). Forest ecosystems are found in most of Earth's biomes and harbor a large proportion of the global diversity. For example, temperate and boreal forests occupy most of the land surface of the Northern Hemisphere and contain 46% of all trees on Earth—0.66 and 0.74 trillion, respectively (2). Due to their global extension, the processes occurring in these two biomes are of global importance, and an understanding of the composition and function of their microbiomes is thus essential (Fig. 1).
FIG 1.
Distribution of temperate and boreal forests in the Northern Hemisphere. Natural areas of deciduous and mixed temperate forest (light green) and evergreen coniferous forest (dark green) are shown in green, and natural areas occupied by boreal forests are shown in blue. The regions in which microbial communities inhabiting soil have been studied are shown as dots. The bottom images show forest ecosystems dominated by Quercus, Pinus, Fagus, and Picea trees.
Forests typically represent important carbon (C) sinks with large amounts of recalcitrant organic matter in their soils, especially the temperate forest soils, which receive tons of litter per hectare yearly (3). The presence of large trees distinguishes forests from grasslands, wetlands, and agricultural areas in multiple ways (4). Trees represent a multitude of habitats, such as the phyllosphere or rhizosphere, but they also substantially affect the remaining parts of the ecosystem. This is mainly because, as the dominant primary producers, they supply the bulk of the C that enters the ecosystem, and while some of this C is in the form of simple organic molecules, a significant fraction, such as the complex biomass of wood, litter, or roots, is composed of recalcitrant biopolymers (4). Trees also largely contribute to the spatial heterogeneity of forest ecosystems by multiple means, including the penetration of soils by various guilds of roots, generation of patches of litter and ground vegetation, and changes of the morphology of the terrain during uprooting or the production of deadwood (4, 5).
Temperate forests extend approximately from latitudes 25°N to 50°N, gradually changing into boreal forests further north (6). The southern limit of boreal forests is understood as the latitude at which conifers are competitively excluded by temperate tree species with higher rates of photosynthesis. There is, however, no distinct boundary between the two biomes but rather a broad transition zone that includes a mix of coniferous and deciduous tree species (7). Temperate forests are characterized by temperature ranges between −30 and 30°C, with hot summers and cold winters and with 750 to 1,300 mm of precipitation per year. Boreal forests—or mountainous forests in the temperate zone—face lower temperatures, with cold winters lasting more than 6 months and average summer temperatures of around 10°C. Despite the lower precipitation levels (300 to 900 mm of rain per year), boreal forests are typically moist because of the reduced evaporation at low temperatures. In addition, other factors, such as elevation, substrate, drainage, physical soil properties, and nutrient availability, are also responsible for the distribution of forest types on landscape scales.
Temperate forests are composed of a large variety of deciduous trees, while coniferous trees dominate temperate forests at higher altitudes but also occur frequently in plantation forests (8, 9). Boreal forests exhibit a reduced tree diversity with a larger proportion of coniferous trees (10).
Although temperate and boreal forests fulfill several important ecosystem services, their role as large C sinks has gained specific interest in recent decades, especially in the context of global change. Recent approximations indicate that the current C stock stored in forests worldwide is approximately 861 Pg, of which 44% is in soil, 42% in above- and belowground biomass, 8% in deadwood, and 5% in litter (11). Of the global stock of C stored by forests, boreal forests contain 32% and temperate forests another 14%, indicating that their importance for global C sequestration is equivalent to that of tropical forests (11). The large amount of stored C has the potential to influence the feedback between the climate and the global C cycle (12). In this sense, global change, including both climate change and other changes linked to human activities, has been recognized as a major threat to forests (13). Over the millennia, human overexploitation has been the most important risk for forests, and the C balance of forests is largely affected by human activities, including deforestation, the management of production forest, reforestation, afforestation, and others (11, 14). Other disturbances that are associated with climate change and atmospheric drivers are also affecting the C balance of forests. In this sense, the effects of increasing temperature, persistence of droughts, recurrence of fires, or expansion of native and invasive forest pests together with the increasing loads of nitrogen (N) and carbon dioxide affect forest nutrient cycles to a level where forests may potentially become net sources rather than sinks of CO2 (12, 13, 15). Therefore, a better understanding of the role of forests in C fluxes that are largely dependent on the activity of bacteria and fungi has been highlighted as an essential prerequisite for projecting future predictions of the health of our planet (16).
While plants are the key drivers of C uptake from the atmosphere in forests, forest microorganisms contribute greatly to the C balance in these ecosystems. They play an important role as decomposers, symbionts, or pathogens, influencing the C turnover and retention and the availability of other nutrients (17–19). Microbial communities are vital in mediating the biogeochemical cycles, and an understanding of their role in ecosystem processes is essential for the prediction of the forest response to future environmental conditions (8, 17, 20).
Fungi are the most well-studied microbes in temperate and boreal forest soils that harbor abundant and diverse communities of saprotrophic and mycorrhizal fungal taxa (12, 21, 22). Fungi are considered the main decomposers in forest soils because of their ability to produce a wide range of extracellular enzymes that allow them to efficiently degrade the recalcitrant fraction of dead plant biomass (23–25). Moreover, mycorrhizal fungi play a pivotal role in the mobilization and sequestration of N and P in the forest soil and are also responsible for significant soil transport of C (26–29). It is not surprising that most of the research on forest soil ecology so far has focused on fungi (18, 19). This traditional emphasis on fungi, however, is slowly shifting with an increasing appreciation of the role of bacteria as the other major component of forest ecosystems (19).
Bacteria represent another important, though less explored, integral part of the microbial community in forest soils. For example, recent findings indicate that bacteria commonly harbor genes encoding plant cell wall-degrading enzymes (30) and contribute significantly to the decomposition of organic matter (31–35). In addition, bacteria are the major natural agents responsible for N fixation in forest ecosystems (36) and for other ecosystem processes, such as mineral weathering leading to the release of inorganic nutrients (37). The roles of bacteria and fungi, however, should not be viewed as separate. The high abundance of fungal biomass in forest soils has multiple consequences for bacteria, including the creation of specific niches in the soil patches colonized by mycorrhizal fungi (i.e., the mycorrhizosphere) and soil mycelial mats (38, 39), provision of nutrients via organic matter decomposition (40), and an increase in soil connectivity by fungal mycelia that allow certain bacteria to move across the environment (41). Conversely, the functioning of mycorrhiza is modulated by mycorrhiza helper bacteria (MHB) (42).
It is only recently that advances in analytical methods have allowed us to assess the role of bacteria in the complex forest ecosystem and to address the important questions that previously remained unanswered, such as the following. Which habitats do bacteria inhabit, and what are the major drivers of their abundance and diversity? What is their role in nutrient cycling and the C balance? How do they interact with other forest organisms? How do they respond to climate change, and how does this response affect ecosystem processes? The aim of this review is to answer these questions for temperate and boreal forests to summarize the present knowledge about the role of bacteria in ecosystem processes and about their response to global change, as well as to motivate further research in the bacterial ecology of forests.
BACTERIAL COMMUNITIES IN FOREST ECOSYSTEMS
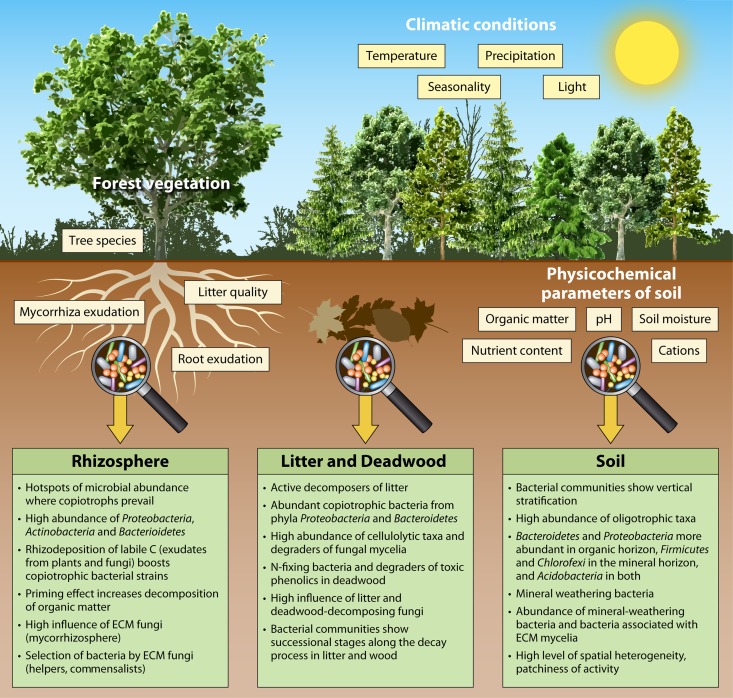
Forest ecosystems provide a broad range of habitats for bacteria, including soil and plant tissues and surfaces, streams, and rocks, among others, but bacteria seem to be especially abundant on the forest floor, in soil and litter (4). Five phyla, Acidobacteria, Actinobacteria, Proteobacteria, Bacteroidetes, and Firmicutes, appear to be abundant in most soils (43). In addition to pH, which seems to be the most important driver of the bacterial community composition in soils, organic matter content, nutrient availability, climate conditions, and biotic interactions (especially the effect of vegetation) affect the composition of bacterial communities (43–47). The spatial variation of these parameters is responsible for the presence of hot spots of microbial activity with increased abundance and activity in the soil, such as in and on plant debris, including litter and deadwood, or on and around plant roots (48–50). Each of these niches has specific properties and, consequently, a specific bacterial community (Fig. 2).
FIG 2.
Drivers of bacterial community composition in forest soils and features of bacterial ecology in the rhizosphere, litter/deadwood, and soil compartments of the forest floor.
Bacterial Communities on Plant Litter and Deadwood
Due to the activity of primary producers, dead plant biomass—litter and deadwood—represents the most important C sources for forest soil microbes. The estimated 1011 tons of fallen leaf litter that accumulates yearly on the forest floor surface and its transformation are of great importance for the cycling of C and other nutrients (51–53). The litter habitat is dominated by a diverse community of fungi that have traditionally been considered to be key players in litter decomposition. In addition to fungi, recent studies have demonstrated an active role of bacteria in litter transformation. In the litter of coniferous forests, bacteria incorporated relatively more cellulose-derived C than that incorporated by fungi (31). Cellulose-C was accumulated mainly by Betaproteobacteria, Bacteroidetes, and Acidobacteria. Similar results were found in different pine forest soils in North America, where members of the Proteobacteria (Burkholderiales, Caulobacteriales, Rhizobiales, and Xanthomonadales), Bacteroidetes (Sphingobacteriales), and subdivision 1 Acidobacteria accumulated the most cellulose-derived C (32). Recently, studies of deciduous forests reported that at least 10% of litter bacteria are able to decompose cellulose. Many of these bacteria belong to the Proteobacteria, Actinobacteria, Bacteroidetes, and Acidobacteria, but members of other phyla are also active (33, 54). Considering the frequency of cellulolytic genes in bacterial genomes (30), bacterial involvement in decomposition seems to be a relatively common trait.
While the acidic soils of coniferous forests harbor mainly Proteobacteria, Acidobacteria, and Actinobacteria (8, 55), in temperate deciduous forests, litter bacterial communities seem to be especially enriched with Proteobacteria and Bacteroidetes (45, 56, 57). Indeed, significant differences in the chemical structure of litter and root exudates among tree species influence soil bacterial communities through changes in substrate chemistry (45, 58, 59). Litter quality, which includes the quantity of nutrients, tissue structure, and the C/N ratio, varies between tree species and results in varying litter decomposability (60–62). Higher litter quality is typically associated with faster litter turnover and the liberation of exchangeable basic cations, such as Ca2+ and Mg2+, helping to maintain low levels of soil acidification (63–65). When trees typical of temperate and boreal forests were ranked in order of increasing acidification ability of their litter, conifers appeared to be the most acidifying, followed by beech, oak, and birch. Maple, hornbeam, ash, and lime comprised the group with the least acidifying forest trees (59). These effects of litter quality on soil nutrient status and pH indirectly drive the bacterial community composition of forest soils and its activity (66–68).
The production of litter is not constant throughout the year; it is limited to a short autumnal period in deciduous forests. These and other aspects of seasonality in the temperate and boreal zones have been identified as key factors affecting the C input into the soil environment and its decomposition by soil bacteria. Cold and dark winters and warm summers with longer photoperiods affect the development and production of foliage and, consequently, microbial community structure and activity in the litter and soil as well as in the foliage (69). Temperature differences affect decomposition rates by accelerating or decelerating enzymatic processes (55, 70). On a shorter time scale, selected weather events, such as precipitation or snowmelt, contribute to the mobilization of nutrients that are washed into the forest soil, and they represent periods of high microbial activity, especially of fast-growing r-strategists (50).
During decomposition, the chemical composition of litter changes with the gradual removal of cellulose, hemicellulose, and lignin (58, 71). In contrast to fungi, bacteria inhabiting the phyllosphere of living leaves do not seem to be largely involved in litter decomposition and are quickly replaced by other taxa (72). Proteobacteria and Bacteroidetes are considered to be copiotrophs that preferentially consume the labile pool of organic C (73) and are thus typical for freshly fallen litter. The fresh litter is also characterized by the quantitative dominance of fungi. The biomass of bacteria increases gradually during decomposition (58, 74), as does their diversity (57, 72). During decomposition, the abundance of mycophagous bacteria coincides with the peak of fungal biomass in the fresh litter. In this stage of decomposition, potentially mycolytic bacteria in Quercus litter represent as much as 40% of the total community (72), highlighting the importance of the fungal biomass as a nutrient source. Bacterial taxa that associate with decomposing fungal mycelia represent a distinct subset of the bacterial community, including members of the genera Pedobacter and Chitinophaga (Bacteroidetes) and Pseudomonas, Variovorax, Ewingella, and Stenotrophomonas (Proteobacteria) (75). With ongoing litter decomposition, the share of cellulolytic bacteria gradually increases (72).
Importantly, a large proportion of plant inputs into forest soils are derived from organs other than leaves, including fine roots, seeds, and twigs (76). While it has been shown that this type of litter represents an important source of soil organic matter (SOM) C (52, 77), bacterial communities associated with this substrate remain unexplored.
In addition to litter, coarse deadwood is also an important structural component of forest ecosystems, especially in unmanaged forests. Wood debris represents a rich yet recalcitrant source of organic matter, mainly due to its physical impermeability and high lignin content. These characteristics benefit filamentous, macroscopic fungi (78), which are consequently the dominant decomposers of deadwood. However, bacteria are also important inhabitants of decaying wood, especially during the initial phases of decay (79, 80). The physicochemical properties of wood that are tree specific, such as the density, pH, and water and N content, determine the composition of bacterial communities (80, 81). Proteobacteria, Acidobacteria, Actinobacteria, and Firmicutes belong to the most abundant phyla (79, 80, 82). Because fungal decomposition changes the properties of wood substantially via the removal of easily available C, acidification, and the production of fungal mycelia, bacteria that can thrive at an acidic pH and utilize C from fungal mycelia are positively selected (80, 83, 84). As observed for litter decomposition, the bacterial community in deadwood shows successional development with wood decay. The assembly of the bacterial community appears to be a stochastic process during the initial stage of wood decay, and the bacterial abundance in fresh wood is very low, with <0.2 × 109 16S rRNA copies g−1. During decomposition, the bacterial abundance increases, reaching levels as high as 3 × 109 to 13 × 109 16S rRNA copies g−1 in highly decayed Picea abies wood, with a density of 0.15 g cm−3 (80). While the involvement of wood-associated bacteria in wood biopolymer decomposition versus the use of substrates liberated by fungal decay remains largely unexplored, recent studies support a role for bacteria in other important processes, including the degradation of toxic wood compounds and N fixation. Genera such as Burkholderia, Phenylobacterium, and Methylovirgula, which are abundant in the middle and late stages of wood decomposition, are known to degrade aromatic compounds and to use methanol as a sole carbon source (79, 82). Moreover, the N-fixation ability of bacteria that are abundant during the late stages of wood decomposition (such as the members of the Rhizobiales, which may account for 25% of all bacteria in this phase) offers the possibility of mutualistic interactions with fungi that provide C via wood decomposition (82). Despite these initial studies, little is still known about the community dynamics of wood-inhabiting bacteria or their interactions with wood-decaying fungi (81). Considering that multiple bacteria are able to utilize cellulose and other recalcitrant plant polymers and that the fungal biomass is highly abundant in deadwood, the involvement of bacteria in the decomposition of lignocellulose and fungal mycelia appears to be highly probable.
Bacteria in Forest Soils
Forest soils are among the most diverse microbial habitats on Earth, in which bacteria are the most abundant group of microorganisms (4, 85). Forest soils are characterized by a sharp vertical stratification resulting from the decomposition of litter-derived organic matter and the weathering of the mineral matrix. The decreasing content and quality of organic matter with soil depth are accompanied by decreases in microbial biomass, respiration, and activity of extracellular enzymes. For example, in a Quercus petraea forest soil, organic matter decreased 10-fold, bacterial biomass 8-fold, and enzyme activity 5-fold to 20-fold across the top 5 cm of the soil profile (86). The temperate forest soil profile typically comprises the organic horizon, representing a mixture of processed, plant-derived organic matter and soil components, and the mineral soil horizon, with a lower content of organic matter, originating from both the decomposition of organic matter and exudation from abundant tree roots. The bacterial communities are horizon specific, although they display a high level of taxon overlap (56). For example, the organic and mineral horizons in a temperate Quercus forest are dominated by Acidobacteria (accounting for 40 to 50% of all sequences), together with members of the Actinobacteria, Proteobacteria, and Bacteroidetes (56). Bacterial communities in coniferous forest soils also differ among horizons. Although Acidobacteria, Proteobacteria, and Actinobacteria are most abundant in organic as well as mineral horizons, the organic horizon is richer in Proteobacteria and Bacteroidetes (8, 87), which have been proposed to preferentially utilize easily accessible carbon substrates (73, 88). In contrast, communities in the mineral soil harbor a larger proportion of Firmicutes and Chlorofexi organisms that are more adapted to the use of recalcitrant carbon substrates and inorganic nutrients.
The high abundance of Acidobacteria, Actinobacteria, and Proteobacteria across forest soils (87, 89–92) appears to indicate their functional importance. These three phyla also dominated the active fraction of a P. abies soil, comprising >80 to 90% of rRNA molecules, and were responsible for the bulk of bacterial transcription (8, 55).
The soil bacterial community composition has been shown to display biogeographical patterns on a continental scale that are predictable but differ from the well-studied plant and animal community patterns (44). The first comprehensive study indicated that the soil pH is the most important driver of the bacterial community composition, and this observation was confirmed in multiple studies that focused specifically on forest soils (43, 93–97). Whereas Acidobacteria and Alphaproteobacteria are abundant in acidic soils (8, 96, 98, 99), the abundances of Bacteroidetes and Actinobacteria increase with a rising pH (43, 94, 100).
The preference of bacterial taxa for niches with certain nutrient contents and organic matter quality is indicative of their ecological strategy. The abundance of Acidobacteria was initially negatively correlated with the C availability in soils, corroborating the idea that Acidobacteria are slow-growing oligotrophs that have adapted to resource limitations (73, 101, 102). However, the present results show that members of the Acidobacteria inhabit soils across a wide range of C contents, representing, on average, 20% of all bacteria; their abundance may exceed 60% in acidic forest soils where both the content of organic matter and the allocation of C by tree roots are high (43, 103). Acidobacteria show a high level of metabolic versatility that allows them to decompose complex C substrates derived from the recalcitrant SOM pool (69, 101). Recently, subdivision 1 Acidobacteria from acidic coniferous forest soil were demonstrated to utilize C from cellulose and to produce high titers of extracellular enzymes (31, 104), indicating their involvement in decomposition in these soils. Unfortunately, due to their limited culturability, little is known about the ecophysiology of other subdivisions of Acidobacteria and the Planctomycetes, except that they are common in forest soils (105, 106). Verrucomicrobia, which are also abundant in forest soils (56, 107, 108), show an increased abundance with soil depth (109), but it is unclear whether this is due to their preference for the oligotrophic environment or to the occupation of specific microniches. Members of the phylum Bacteroidetes, such as the genus Mucilaginibacter, which are common in forest soils, have been shown to be potent decomposers of cellulose and other biopolymers (31, 33, 104).
Importantly, some soil bacteria are involved in the weathering of minerals, a process of great importance in nutrient-poor soils, where minerals represent an important pool of inorganic nutrients (37, 110). Members of the Betaproteobacteria, such as the genera Burkholderia and Collimonas, have been recognized as efficient mineral-weathering bacteria. These taxa are common in both deciduous and coniferous forest soils (31, 56, 106, 108, 110–112).
For linking of the presence of bacteria or their activity to soil properties, it is important that soil is a complex of microniches with heterogeneous physicochemical properties on various scales. Because bacteria inhabit small niches, the properties of their immediate environment rather than the mean soil properties affect the local bacterial community. This spatial heterogeneity has been shown to result in the heterogeneity of bacterial communities on small scales of <1 cm (113). Furthermore, local dispersal limitations can also remarkably influence the bacterial community composition (93, 114). Considering the high level of spatial variation of forest C stocks on the same scale (115), the occurrence of individual taxa in forest soil may actually be highly variable on a small scale and may differ among activity hot spots, such as the rhizosphere and the bulk soil (49, 50). The transient nature of some physicochemical factors, such as soil moisture, can strongly influence the physical connectivity of the soil matrix, affecting bacterial and nutrient dispersion and, as a consequence, rates of key soil processes (116–118). Our ability to define and identify activity hot spots and to analyze their properties, as well as the abundance, composition, and function of their bacterial inhabitants, will be critical in future analyses to increase our mechanistic understanding of the roles of bacteria in biogeochemical processes.
Rhizosphere Bacterial Communities
In temperate and boreal forests, most C enters the soil food web via the roots in the form of labile C compounds, such as sugars, amino acids, and organic acids (119, 120), with approximately one-third of the plant net primary production (NPP) allocated to roots and soil (121). Indeed, root exudation, water and nutrient uptake by roots, decay, respiration, and physicochemical changes in soil are important factors influencing the composition and function of the microbial community in the rhizosphere, with biogeochemical consequences for the entire soils (46, 122).
The roots and rhizospheres of trees as well as ground vegetation associate with fungi to form mycorrhiza. While the ground vegetation mostly forms a symbiotic relationship with arbuscular mycorrhizal (AM) or ericoid mycorrhizal (ErM) fungi (29), more than 90% of forest trees in temperate and boreal zones participate in ectomycorrhizal (ECM) symbiosis, and only a minority of trees associate with AM (123). Thus, ECM is the quantitatively most prevalent and explored mycorrhizal type in forests (124). ECM fungi form mantels around tree root tips, and their mycelia extend into surrounding soils that are used to provide mineral nutrients to their hosts (62, 124, 125). Fungal associations of AM with trees are much less common and are confined to a few tree species (38). Due to the large absorptive area and exudation of labile compounds by ECM hyphae, the mycorrhizosphere and mycosphere represent important niches with features that differ from the nonmycorrhizal rhizosphere and bulk soil, hosting different bacterial communities (62, 126, 127). The exudation of labile C by tree roots or mycorrhizal hyphae enhances the availability of C in the rhizosphere and, consequently, the microbial abundance and activity of extracellular enzymes in comparison to those in the bulk soil (128–131). The input of labile C compounds into the rhizosphere selects for bacterial strains with rapid growth and low-affinity substrate enzymes (r-strategists) that are enriched in comparison to the bulk soil profile (73, 132). The quantity and quality of exudates select for specific microbial communities and the expression of specific genes and may prime the decomposition of recalcitrant organic matter (133, 134). Considering the seasonal changes in NPP, seasonality is a major driver of microbial community composition and functioning in the rhizosphere (135).
Despite the importance of the associated ecological processes, bacterial communities inhabiting rhizospheres in forests have been explored much less than those from grasslands or agricultural systems. Recent studies from nonforest environments, such as grasslands (136, 137), croplands (138–140), and other ecosystems (134, 141–143), concluded that the rhizosphere contains a certain subset of the bulk soil microbiome, which is enriched in members of the Proteobacteria, Actinobacteria, and Bacteroidetes (136, 140, 142, 143). The dominance of Alphaproteobacteria, Betaproteobacteria, Actinobacteria, and Bacteroidetes was also observed in the rhizospheres of beeches in a mountainous forest (144). These observations suggest the enrichment of copiotrophic bacterial taxa (73, 138, 145).
The microbiomes surrounding the ECM roots and hyphae also differ from those in bulk soil (106, 127, 136). An enrichment of Proteobacteria (e.g., Burkholderia, Rhizobium, and Pseudomonas) and Actinobacteria (such as Streptomyces) in the mycorrhizosphere was reported based on culture-dependent studies (41, 124, 146–148). A recent molecular study of Pinus sylvestris mycorrhizospheres indicated that the community composition is much more complex and includes both copiotrophic and oligotrophic bacteria (149).
Various roles, ranging from mycorrhiza helpers to mycophages, have been assigned to the ECM-associated bacteria (124). The mycorrhiza helper bacteria (MHB) isolated from the mycorrhizospheres of different ECM symbionts either enhance the formation of mycorrhiza or support the previously established symbiosis (126, 150, 151). In the mycorrhizosphere, plants, ECM fungi, and bacteria form tripartite associations in which each player critically affects the metabolism of the others. It has been proposed that ECM fungi select for bacterial communities through the exudation of low-molecular-weight compounds, such as organic acids and amino acids (116, 152). Different ECM fungi in a similar environment can host very similar bacterial communities that are suited for this niche (146). MHB influence both ECM fungi and their plant host. For example, MHB from the genera Pseudomonas and Streptomyces promote mycorrhiza formation by modifying the gene expression of the ECM, leading to accelerated mycelial growth and stimulating the formation of new lateral roots (92, 150, 153). Rhizosphere bacteria also provide additional benefits to plants. For example, Streptomyces AcH 505 is an antagonist of plant pathogens and can induce defense mechanisms in P. abies and Quercus robur (125, 154, 155). Other very-well-known bacteria promoting plant vigor are endophytes, such as the members of the genus Rhizobium, which form nodules on the roots of Alnus plants, fix gaseous N, and provide it to their host.
BACTERIAL INVOLVEMENT IN ECOSYSTEM PROCESSES
Role of Bacteria in the Carbon Cycle
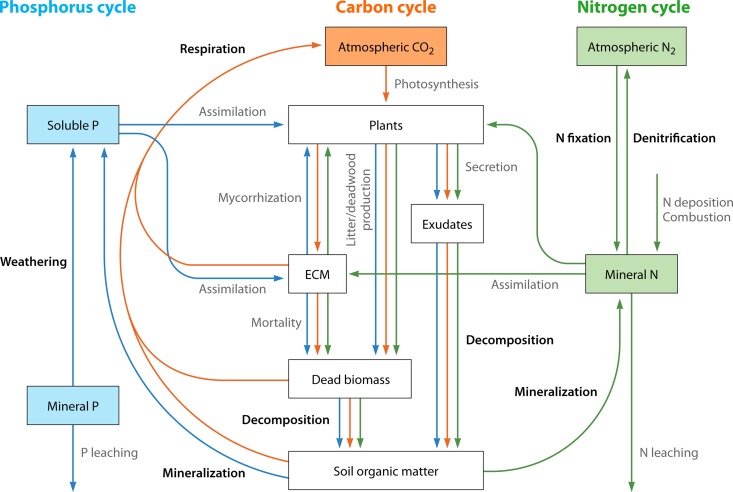
Forest soils store two-thirds of the terrestrial C (156). The flux of C is initiated by the fixation of atmospheric CO2 by photosynthesis and mediated by the allocation of recalcitrant and simple organic compounds into the soil (Fig. 3). When plant debris and simple organic compounds are decomposed and used by microorganisms to build their biomass, some C is returned to the atmosphere via respiration. Because root exudates can readily be assimilated by soil microorganisms, decomposition of the dead plant biomass has been highlighted as the key process regulating C flow in soil systems that influences the ratio between C mineralization and immobilization (156).
FIG 3.
Schematic view of the coupled biogeochemical cycles of carbon, nitrogen, and phosphorus in forest ecosystems. Colored arrows show the transfer of elements (C in orange, N in green, and P in blue) between ecosystem compartments. Ecological processes with the active involvement of bacteria are highlighted in bold.
Recent studies indicated that bacteria play a more important role in the transformation of the dead plant biomass than was previously assumed and significantly contribute to decomposition processes in litter and soil (31–33). The plant biomass is composed largely of lignocellulose, a highly organized and interlinked mix of different polymers that contains various amounts of cellulose, hemicelluloses, and lignin (157). Because of the presence of hemicelluloses in a recalcitrant complex with lignin, cellulose represents the most accessible biopolymer, and its degradation is thus a key step in the C cycle. The most important enzymes involved in this process are endocellulases, exocellulases, and β-glucosidases. Together with these enzymes, a diverse set of hemicellulases, such as endoxylanases, xylosidases, xyloglucanases, endomannanases, mannosidases, fucosidases, arabinosidases, pectinases, and ligninolytic enzymes, are necessary for the hydrolysis of other polymers that are present in the plant biomass (158). The genes encoding cellulases are present in 24% of all sequenced bacterial genomes (159), and glycosyl hydrolases that degrade other plant structural biopolymers are also common (30). Recently, López-Mondéjar et al. (33) isolated cellulolytic bacteria belonging to a variety of phyla from a deciduous forest topsoil, and they demonstrated, for the first time, the presence of cellulolytic members among some abundant soil genera, e.g., Mucilaginibacter, Luteibacter, and Pedobacter. Other studies highlight the important ecological role of strains of the phyla Acidobacteria and Actinobacteria as degraders of plant biomass polysaccharides in the acidic soils of temperate forests (31, 104, 160).
Genomes of several forest soil bacteria encode proteins involved in decomposition of dead plant biomass (33, 161–163). In addition to promotion by hydrolytic enzymes, efficient hydrolysis is often promoted by the presence of carbohydrate-binding modules (CBM) that are part of either the enzymes or bacterial cell surfaces (157). Genes coding for proteins that are involved in signal transduction, nutrient binding, and transport are often found in the same operons as hydrolase genes. Some of these proteins, such as the TonB-dependent receptors, have been established as plant carbohydrate scavengers (164). Other auxiliary proteins, such as the substrate-binding proteins (SBPs) from ATP-binding cassette (ABC) transporters, have been shown to be capable of binding a variety of plant cell wall soluble and insoluble saccharides, including microcrystalline or amorphous cellulose, xyloglucan, xylan, and mannan (165). The rich suite of these auxiliary proteins expressed by a Paenibacillus sp. strain from a temperate forest soil suggests their important involvement in polysaccharide decomposition (162). A suite of carbohydrate-binding proteins, such as type IV pili and an atypical oxidative cellulase, in a cellulolytic Luteibacter species from temperate forest soil demonstrates the variability among the cellulolytic systems of bacteria, which remain far from being completely described (33, 166, 167). Since plant biomass represents a renewable and abundant resource for the production of environmentally friendly chemicals and biofuels (168), forest topsoils represent a rich potential source of bacterial strains with biotechnological relevance (169, 170).
Soil bacteria are also able to contribute to the breakdown of phenolic compounds, including lignin, although their efficiency is typically much lower than that of fungi (34, 160). Ligninolytic bacteria can be found among the Proteobacteria (genera such as Sphingomonas, Burkholderia, Enterobacter, Ochrobacterium, and Pseudomonas, among others), Firmicutes (Bacillus and Paenibacillus), and Actinobacteria (Rhodococcus, Mycobacterium, Microbacterium, and Streptomyces) (35, 171), and there is evidence that potentially ligninolytic taxa are common in soils. For example, abundant bacterial laccase genes have been found in temperate hardwood forests (172), and bacterial taxa possessing laccases, such as Burkholderia, Bradyrhizobium, and Azospirillum, are highly active in forest soils (173). However, even if the enzymology of bacterial lignin degradation is poorly understood compared to that for fungi, bacteria may use ligninolytic peroxidases and laccases to reduce the toxicity of phenolic compounds rather than to degrade lignin.
In addition to plant biomass, fungal mycelia represent an important pool of organic matter in forest litter and soil (174). Forest ecosystems are dominated by ectomycorrhizal fungi (ECM), with a biomass of up to 600 kg ha−1 (147, 151), and the annual production of fungal mycelia in a spruce forest ranges from 100 to 300 kg ha−1 (77). Fungal mycelia represent a large C and N pool that typically contains chitin and other polysaccharides, such as glucans and glucomannans, as well as phenolics, such as melanin (75, 175). Fungal biomass represents a more readily decomposable substrate than lignocellulose, and bacteria have been reported to be more important contributors than fungi to its decomposition (75, 176). The bacteria associated with fungal biomass decomposition represent a specific subset of the litter and soil communities, dominated by Pseudomonas, Ewingella, Pedobacter, Variovorax, Stenotrophomonas, and Chitinophaga, genera that are known to produce chitinolytic enzymes (75). Chitinolytic enzymes, which are represented by chitinases and N-acetylglucosaminidases, appear to be widespread in bacterial genomes, especially Actinobacteria (30, 177). The members of this phylum often contain several chitinase genes as well as a diverse array of corresponding CBMs, and they often also possess β-1,3-glucanases, thus encompassing a complex set of enzymes that potentially target fungal cell walls (178). Chitinase production by soil bacteria is a complex adaptation that combines a nutritional strategy with potential involvement in the antagonistic interactions with fungi (178, 179). Although the bacterial biomass in forest soils has a size similar to that of fungi (55), the process of bacterial cell wall polymer decomposition is far less understood. The involvement of Planctomycetes, Verrucomicrobia, Armatinomonadetes, or candidate division OD1 in the degradation of bacterial exopolysaccharides produced in soil was recently demonstrated (180).
Finally, a large fraction of C in forest ecosystems is allocated belowground during the vegetation period via tree roots, in the form of root exudates (181, 182). Up to 40% of newly photosynthesized C can be allocated to the soil via roots, where it is rapidly respired or incorporated into the microbial biomass (183, 184). The rhizosphere, including the root-associated mycelia of mycorrhizal fungi, is thus probably the most important C allocation hot spot in forest soils and is strongly influenced by plant activity (185). Because ECM fungi cover most of the fine roots and root tips, the largest fraction of this C is allocated to them (186). The fraction of assimilates that is exuded directly from roots into the rhizosphere is estimated to range from 1 to 5% and is composed mainly of carbohydrates, amino acids, and organic acids (120, 187). The rhizodeposition of C by plants exhibits severalfold seasonal differences that correspond to the intensity of photosynthesis throughout the year (182). While direct evidence of a net C flux from plants to rhizosphere microbiota has been shown for many crops, little is known about the structure and function of bacterial and fungal communities that are actively involved in the assimilation of root exudates in forest ecosystems (188). Although decomposer fungi can find suitable conditions in the nonmycorrhizal rhizospheres of trees, this niche has traditionally been considered bacterium dominated because their rapid growth favors their utilization of root exudates (185). Recent studies indicated that soil bacteria indeed use root exudates in forest soils (189), and the seasonal production of root exudates leads to an increase in bacterial biomass as well as in the share of the rhizosphere- and mycorrhizosphere-specific bacteria in mineral soil in summer (56, 70). In the same way as that of tree roots, hyphae of ectomycorrhizal fungi exude low-molecular-weight organic compounds that support diverse bacterial communities (78, 190). The rhizosphere of roots colonized by mycorrhizal fungi is specific in that a portion of the plant-derived C is released into the soil through the ECM mycelium (155). As a consequence, ECM fungi constitute a nutrient hot spot for other microorganisms, because the fungal symbionts have a very large surface area and receive a direct supply of photosynthetic C (149). Soil bacteria have been found to grow abundantly and rapidly at the expense of the exudates from P. sylvestris roots colonized by Piloderma, supporting a role for the bacteria in the metabolism of the organic acids and other compounds produced by the fungus (152). The observation that these bacteria are able to utilize fungal sugars more readily than plant sugars supports the existence of a bacterial guild that is specialized for hyphal exudates (191).
Methane represents a gaseous form of organic C that can be oxidized by certain forest soil bacteria under aerobic conditions (192). These methanotrophic bacteria are the main consumers of atmospheric methane as well as methane generated by methanogenic archaea in waterlogged anaerobic horizons of certain soils (193–196). Due to the presence of methanotrophic bacteria, forest soils (especially those of the boreal forests) may represent a sink of atmospheric methane. Although methanotrophs are phylogenetically located in two bacterial phyla, most of the characterized isolates belong to the Alphaproteobacteria and Gammaproteobacteria (197). The discovery of new acidophilic methanotrophs related to Verrucomicrobia recently revealed a higher level of diversity of methanotrophic bacteria in as yet unexplored acidic environments (198). In addition to using organic C sources, certain soil bacteria are able to fix CO2. These autotrophic taxa were described for the genus Bradyrhizobium (Betaproteobacteria), whose members are abundant inhabitants of forest soils (199, 200), and a recent transcriptomic study confirmed the expression of the components of a photosynthetic apparatus in the Cyanobacteria and Proteobacteria. The input of C into forest soils through fixation is, however, apparently minimal (55).
Role of Bacteria in Nitrogen and Phosphorus Cycles
Nitrogen is the most common limiting nutrient in soils, entering the ecosystem largely through fixation (Fig. 3). This process is dominated by bacteria that are estimated to be responsible for more than 95% of the N input in unmanaged environments (36, 201). The presence of the nifH gene of the Alphaproteobacteria (Bradyrhizobium, Azospirillum, Hyphomicrobium, and Gluconacetobacter) and Deltaproteobacteria (Geobacter spp.) was observed in different temperate forest soils, demonstrating the ubiquity of some N-fixing bacteria, not only as symbiotic but also as free-living taxa (200). The observed diversity of N-fixing bacteria may rapidly change if there is an exogenous input of N (36). Particularly in temperate forest soils that are strongly affected by anthropogenic N deposition, N input is a good predictor of N fixation activity (202).
Nitrification and denitrification lead to the loss of N from soils through NO, N2O, and N2 gas emissions as well as through the leaching of NO3 if its concentration exceeds the ecosystem retention capacity (203, 204). Among these, nitrous oxide is a particularly powerful greenhouse gas with a potentially warming effect that is almost 300 times larger than that of CO2 (205). Nitrification is the biological oxidation of ammonia to nitrate via a multienzymatic process. The amoA gene, encoding ammonia oxidase, which performs the first step of nitrification, is present in ammonia-oxidizing bacteria (AOB) as well as ammonia-oxidizing archaea (AOA), but AOB rather than AOA seem to dominate in soils (206, 207). The reaction catalyzed by this enzyme is often limiting in temperate and boreal forest soils (208). All known AOB belong to the Betaproteobacteria and Gammaproteobacteria, and they are mostly represented by the genera Nitrosomonas, Nitrosococcus, and Nitrosospira. Nitrosomonas and Nitrosospira are further subdivided into clusters with different responses to environmental drivers, such as acidity, pH, and ammonia availability, and due to that, changes in soil N availability can result in dramatic shifts in the composition of the AOB community (206, 208, 209). Nitrosospira seems to be the most abundant AOB in acidic forest soils with low NH4+ contents (206, 210, 211). The amoA gene content, however, appears to vary across forest soils (212). Nitrification rates seem to be correlated with the AOB abundance in soil (209, 213). AOB are also involved in N losses from soil by producing N2O during denitrification (214). Denitrification is essential for the global N cycle, maintaining the reduction of nitrate or nitrite to gaseous nitrogen compounds, such as N2 and nitrous oxide. The denitrification pathway is known as dissimilatory nitrate reduction because nitrate or nitrogen oxides are the final electron acceptors, allowing bacteria to produce energy at the expense of reduced electron donors in the absence of oxygen. Denitrifying bacteria are not always able to perform all the steps of the dissimilatory pathway, and incomplete denitrification is the major source of N2O emissions from soils (215). Denitrification can be addressed by studying marker genes involved in the individual enzymatic steps, such as the reduction of nitrate (narG), nitrite (nirK/nirS), nitric oxide (norB), and nitrous oxide (nosZ) (205). Denitrifying bacteria are abundant and widespread in forest soils, and denitrification genes have been found in bacterial strains belonging to the Acidobacteria, Proteobacteria, and Firmicutes, as well as in other bacterial phyla (216–218). A significant correlation was described among gene abundances, denitrification rates, and NO and N2O fluxes, confirming the importance of their carriers for the environmental processes that occur in forest soils (69, 203, 219). In addition to mineral N forms, bacteria may obtain N from a range of organic compounds present in forest soils, namely, the chitin present in the polysaccharides of fungal mycelia and the amino acids and proteins in dead organic matter. The utilization of these compounds is expected to be subject to intensive competition among ectomycorrhizal fungi, saprotrophic fungi, and bacteria (146).
While the potential source of N—the atmosphere—is theoretically unlimited, this is not the case for phosphorus (P), which is supplied mostly by weathering of minerals; P thus represents the limiting nutrient in many soils (Fig. 3). Bacteria are important mediators of the P cycle because some are able to solubilize mineral P while others may immobilize it in their biomass. Unfortunately, the involvement of bacteria in the P cycle has received far less attention than that in the N and C cycles. The main mechanism responsible for P uptake by bacteria is through the solubilization and uptake of inorganic P, its most abundant source. The ability to solubilize inorganic P has been described for different bacteria and ectomycorrhizal fungi present in the rhizosphere and soil. This ability is related to the release of organic anions, such as citrate, gluconate, oxalate, and succinate, into the soil (220). Phosphatase enzymes release phosphate from organic phosphate esters during organic matter decay (221). In a recent metagenomic analysis of temperate forest soils, it was highlighted that members of the Alphaproteobacteria, Betaproteobacteria, Actinobacteria, and Acidobacteria dominated the processes related to P turnover (222, 223). While genes related to P solubilization are more abundant in P-rich soil, those that play a role in P uptake systems are more abundant in P-limited soil (222).
EFFECTS OF GLOBAL CHANGE ON BACTERIAL COMMUNITIES IN FOREST ECOSYSTEMS
Bacteria strongly contribute to key ecological processes that are important for society in light of the ongoing global change (114). The determination of how relationships between site factors and bacterial communities affect the equilibrium between soil organic matter decomposition and C sequestration in forests is of paramount importance for prediction of the responses of terrestrial ecosystems to climate change (224). Future climatic conditions are predicted to produce tree species migration (225, 226), an increase in extreme events, such as droughts and wildfires, and an increase in vegetation productivity due to longer growth seasons (13, 227, 228). These effects in combination with other phenomena, such as increased N deposition, may cause dramatic shifts in global C fluxes, in particular affecting the boreal and temperate forests of the northern continental regions (229–231). The high diversity of C compounds released into the soils and the complex structures of bacterial and fungal communities, with their overlapping functions, create a very complex system that is difficult to incorporate into predictive models (16). The complex nonlinear interactions between forest and atmosphere have the capacity to constrain or amplify climate change-associated processes, depending on the activity of soil bacteria and fungi (232–234). Recent studies investigating the effects of global change on ecosystem properties offer the first clues to the prediction of future developments (Table 1).
TABLE 1.
Effects of carbon dioxide, temperature, and nitrogen increases on bacterial communities and biogeochemistry in northern forests in manipulative experimentsa
| Manipulation | Habitat | Vegetation | Effects on biogeochemistry | Effects on bacterial populations | Effects on bacterial taxa | Selected reference(s) |
|---|---|---|---|---|---|---|
| [CO2] | Temperate forest | Pinus taeda, Populus tremuloides, Populus alba, Pinus nigra, Acer saccharum, Betula papyrifera | ↑ NPP, ↑ efficiency of C-fixing enzymes, ↑ N sequestration | = bacterial biomass, = AOB biomass, ↑ bacterial diversity | = Proteobacteria (↓ Bradyrhizobium, ↑ Rhodoplanes, ↑ Pseudomonas, ↑ Nitrosospira), ↑ Actinobacteria, ↑ Bacteroidetes, ↓ Acidobacteria (except Gp4 and Gp6) | 207, 235, 236, 241, 271 |
| Temperate forest | Pinus taeda | ↑ dichotomous branching index, ↑ root soil exploration, ↑ exudation (growing season), ↑ NAGase activity, ↑ phenol oxidase, ↑ NH4 turnover | ↑ microbial biomass (growing season) | 120, 121 | ||
| Temperate forest | Populus tremuloides, Betula papyrifera, Acer saccharum, Pinus sylvestris | ↑ SOM decomposition, ↑ decomposition of low-molecular-weight organic acids | = bacterial abundance, ↑ bacterial diversity | ↓ Acidobacteria, ↓ Bacteroidetes, ↑ Burkholderia, ↑ Nitrosospira | 152, 240 | |
| Temperate forest | Fagus sylvatica | = NPP, ↑ SOM decomposition, ↑ genes for decomposition of labile C, = NH4, ↑ NO3 depletion, ↑ exudation | ↓ bacterial diversity | ↑ Pseudomonas, ↑ Actinobacteria, ↑ Planctomycetes, ↑ Firmicutes, ↓ Acidobacteria (except Gp5, Gp6, and Gp7) | 144 | |
| Temperate forest | Fagus sylvatica, Quercus robur, Quercus petraea, Carpinus betulus | = NPP, = NH4, ↑ NO3 accumulation, = C/N ratio for trees | 243 | |||
| Temp | Boreal forest | Abies balsanea, Pinus sylvestris, Picea abies | ↑ soil respiration, ↑ enzyme activities on labile C, = SOM decomposition | = microbial biomass, ≠ microbial community composition | 231, 272 | |
| Temperate forest | Picea asperata | ↑ root exudation rates, ↑ C/N ratio for exudates, ↑ SOM decomposition | 273 | |||
| Temperate forest | Picea rubens, Abies balsamea, Fagus grandifolia, Acer saccharum, Acer rubrum, Betula lenta, Prunus serotine, Quercus rubra | ↑ soil respiration, ↑ SOM decomposition, ↑ Vmax of enzymes | 274, 275 | |||
| Temperate forest | Betula papyrifera, Betula lenta, Acer rubrum, Quercus velutina, Quercus rubra, Fagus grandifolia, Picea abies, Fagus sylvatica, Abies alba | ↑ soil respiration, ↑ extracellular enzyme activity, = C, = N | = bacterial biomass, ≠ bacterial community composition | ↓ Gram-negative bacteria, ↑ Acidobacteria, ↑ Alphaproteobacteria | 16, 246, 276 | |
| Temperate forest | Acer rubrum, Betula papyrifera, Quercus velutina, Acer pensylvanicum | ↑ oxidative activity | ↑ bacterial diversity | ↑ Acidobacteria, ↑ Actinobacteria | 234 | |
| N | Boreal forest | Picea abies | ↑ soil C (high N deposition), ↓ soil respiration (high N deposition) | ↓ bacterial biomass, = amoA gene abundance, ≠ AOB community composition | 263, 267 | |
| Temperate forest | Acer saccharum | ↓ SOM decomposition, ↓ soil respiration, = NPP, ↑ OM in forest floor, ↓ extracellular enzyme activity, ↑ N in soil organic matter, ↑ NO3 leaching | ↓ bacterial diversity (site specific), ↑ carbohydrate metabolism genes, ↑ aromatic metabolism genes, ↑ respiration metabolism genes, ↓ chitin decomposition genes (site specific), ↓ lignocellulose decomposition genes (site specific), ↓ N cycle genes (site specific) | ↓ Bacteroidetes, ↓ Gammaproteobacteria, ↑ Firmicutes, ↑ Actinobacteria, ↓ Acidobacteria, ↓ Verrucomicrobia | 213, 260–262, 264 | |
| Temperate forest | Fagus grandifolia, Acer saccharum, Acer rubrum, Picea rubens | ↓ soil respiration (P. rubens site) | ↓ microbial biomass (P. rubens site) | 257 | ||
| Temperate forest | Pinus resinosa, Quercus velutina, Quercus rubra, Betula lenta, Acer rubrum, Fagus grandifolia | ↓ soil respiration, ↓ SOM decomposition, ↑ C stocks (except at Pinus site), ↑ tree biomass, ↓ extracellular enzyme activity, ↑ lignin recalcitrant C, ↑ NO3, ↑ nitrification, = litter fall (except ↑ at Pinus site), = root biomass (except ↓ at Pinus site), = root productivity | ↓ microbial biomass, ↑ bacterial richness, ↑ specific operational taxonomic units | ↑ Proteobacteria, = Acidobacteria | 257, 266, 268 | |
| Temperate and boreal forests | ↑ aboveground NPP, ↑ belowground C, ↑ tree growth, ↑ litter fall, = litter decomposition, = soil respiration, ↑ C sink | ↓ microbial biomass | 277 |
Arrows indicate increases and decreases of the listed parameters. =, no significant change in the parameter; ≠, significant change in the parameter.
Carbon Dioxide
Increasing CO2 concentrations are predicted to boost NPP in boreal and temperate forests (235); however, their capacity to maintain such high levels of NPP will strongly depend on their ability to overcome nutrient limitations (236). Unfortunately, the degree to which forests will persist as C sinks remains uncertain (237); in particular, the rhizosphere priming effect may be reduced, and the increased NPP may increase soil C storage (238, 239). It is clear that the degree to which rising CO2 levels will be offset by C sequestration in forest ecosystems will strongly depend on the adaptation of both trees and soil microorganisms to the altered environmental conditions.
Thus, how do CO2 levels affect bacterial populations in forest topsoils? And is it possible to generalize the observations obtained across ecosystems for different soils with various vegetations? The effects of elevated CO2 levels do not directly affect soil bacterial populations: CO2 concentrations in soil pores are normally higher than those in the atmosphere, and the response of bacteria to small CO2 changes is thus negligible (240). As a result, the observed effects are mostly indirectly driven by altered plant production and root exudation, which are both tree species specific. In addition, changes in organic acid exudation by mycorrhizal fungi as a result of the presence of CO2 will affect the bacterial community composition (152), and this process also depends on the vegetation properties. It is difficult to determine whether the functional responses of bacterial communities to CO2 will be similar across forests or will depend on soil and vegetation characteristics. The expected increase in root exudation should generally favor the development of soil copiotrophic bacteria over specialized oligotrophs. A good example of this response is the decrease in abundance of oligotrophic Acidobacteria in various soils following an increase in CO2 (144, 152, 238, 240). Such a general response is rather exceptional considering that different members of the same phylum often respond differently to the same disturbance (241). The increased abundance of certain bacterial taxa will have functional consequences only if their functional traits do not change in response to the treatment, which may explain the sometimes controversial results obtained for responses to CO2 in different forest soils, despite the fairly consistent response of bacterial abundance after an increase in CO2 (238, 240, 242). For example, while some studies have indicated that an elevated CO2 level does not affect the AOB abundance or nitrifying activity (207), others have described an increase in nitrification or nitrifying taxa (240, 243), even in an unaltered bacterial community. Thus, it is unclear whether the effects of increased CO2 levels on bacterial biomass, richness, community composition, and functionality will be consistent across different forest ecosystems.
Global Warming
The rise in temperature is intimately linked to the increased CO2 concentration in the atmosphere, because the latter largely causes the former. Although both processes appear globally, the extent of the rise in temperature is predicted to be pronounced in the Northern Hemisphere, where temperate and boreal forest ecosystems dominate. The long-term scenarios are, however, not clear because multiple long-term events, such as the depletion of labile C limiting bacterial growth, thermal adaptation of microbial processes, and shifts in microbial communities or in the expression of their metabolic potential, may attenuate positive feedbacks of warming (244). The C balance after warming will largely depend on the efficiency of soil microbes in accessing and using C in response to the altered environmental conditions (12). A very recent work showed that an increase in temperatures can increase tree exudation rates (229). As a consequence, warming can alter soil drivers through exudation, which in turn might produce different responses of soil rhizosphere bacterial communities, affecting SOM decomposition differently in different environments, depending on the tree-specific exudation (229).
After years of experimentally increased warming in boreal and temperate forest soils, bacterial biomass values are usually not significantly different from those of nonwarmed controls, suggesting an attenuation effect (234, 245–248). However, it has been demonstrated that warming affects bacterial diversity and that these changes reflect the magnitude of the temperature change (249). These shifts in bacterial communities may affect the C cycling processes that involve bacteria. Multiple studies have shown that short- and long-term warming can affect the composition of the soil bacterial community toward populations that are more adapted to the use of complex sources of C (234, 250). Members of the Alphaproteobacteria and Acidobacteria display increased abundances after simulated warming, suggesting a change in soil bacterial lifestyle from copiotrophy to oligotrophy, which coincides with the depletion of labile C while maintaining high levels of respiration (16, 234). This finding is contradictory to the aforementioned effect of increased atmospheric CO2, which should boost copiotrophic bacterial populations, highlighting the necessity of studies that combine the manipulation of both variables. However, other studies have documented no shifts in bacterial communities after years of warming (90, 246).
Considering the warming effects on N cycle processes, we may rely only upon a few contradictory works examining that topic. An increase in temperature was observed to accelerate nitrification and denitrification processes, affecting the N cycle in soils (229, 251). The warming effect may be offset by other factors, such as N deposition, which can increase the temperature sensitivity of the enzymes involved in SOM decomposition or suppress the decomposition process, counterbalancing the priming effects (245, 252).
Nitrogen Deposition
Global anthropogenic N inputs are estimated to be 30% to 50% higher than those from natural sources. They have increased 10-fold in the past 150 years and are projected to double during the next century (253, 254). In contrast to global warming and increased levels of CO2, however, their effects are largely local. Since N is a limiting nutrient in most terrestrial ecosystems, including boreal and temperate forests (255), anthropogenic N deposition may strongly influence NPP in these environments by reducing this limitation (256). N deposition also has a range of additional consequences that range from shifts in the soil C/N ratio, soil acidification, and root exudation to changes in the vegetation and microbiota (257). Bacterial soil communities are clearly sensitive to increased levels of N itself as well as to all the associated soil physicochemical changes (213, 258, 259).
There appears to be a general consensus that N deposition increases soil C sequestration due to the decline in SOM decomposition via the reduction of biomass and activity of soil microbial populations in many different soil environments, including temperate and boreal forests (258, 260–264). The most probable reason for C sequestration is the observed reduction of tree root exudation (256), although direct effects of a pH decrease induced by N deposition were proposed as an alternative explanation (265). Although the reduction of biomass following N input was mostly observed in fungi (256, 266), it was recently noted that the biomass of soil bacteria can be reduced as well, by up to 50% (258, 263, 267). Reduction of the bacterial biomass, however, depends on the level of N input. At chronically low levels of N, which are typical of boreal and temperate forest soils, the bacterial biomass may remain stable while the fungal biomass markedly drops. This decrease in the fungal/bacterial biomass ratio due to N addition is consistently found across forest soils, where it ranges from 25% to 70% (256, 257, 263, 266).
N deposition also changes the composition of bacterial communities in forest soils. The abundances of Acidobacteria and Verrucomicrobia decrease, while those of Actinobacteria and Firmicutes increase, following the addition of N to a variety of soils (264). In a report on a temperate forest, the share of Acidobacteria did not respond to N addition, but some groups within the phylum were significantly affected, suggesting that the response of the bacterial community may be complex (268). Also, in other cases, the deposition of N significantly affected relative abundances of core bacterial phyla, but it reduced bacterial diversity and altered the bacterial composition at lower taxonomic levels (258, 261).
A decrease in oligotrophic bacteria is often found concomitantly with a decrease in the activity of enzymes related to cellulose breakdown, again supporting the hypothesis that the addition of N negatively affects microbial SOM decomposition (262, 264). When genes of the N cycle (fixation, nitrification, and denitrification) were assessed in N deposition experiments, the metabolic potential of the bacterial community remained stable or even decreased in terms of gene abundance (213, 267). However, in studies assessing the effects of N addition on N cycle processes in forest soils, it was reported that after N saturation of the environment, bacterial and archaeal nitrification and denitrification gain importance, initiating the cycle, and result in the efflux of N compounds, such as nitrate, N2, or nitrous oxide gas, from the soil ecosystem (150, 269, 270). This phenomenon suggests that the abundances of particular genes, in this case, are not correlated with the rates of ecological processes. In this sense, it was recently proposed that even in forest soil environments where the addition of N highly increases nitrification, it is possible that bacteria with more efficient nitrification pathways are selected and that this leads to higher rates of nitrification despite the reduced abundance of nitrifiers and genes related to N cycle processes (213). Clearly, metatranscriptomic or metaproteomic studies are needed to gain insight into the regulation of the N cycle by N availability.
CONCLUSIONS
Undoubtedly, boreal and temperate forests will be important for the global C balance considering the global change. The role of bacteria in these ecosystems has been understudied, and it will be important to clarify whether these biomes will continue to be a large C sink for anthropogenic emissions. Here we show that despite the important role of fungi in forest soils, bacteria fulfill multiple important ecosystem roles in the forest environment, including organic matter decomposition, regulation of mycorrhizal symbiosis, and involvement in N cycle processes. More complex and comparable studies to assess the variables affected by global change in a wide range of ecosystems will be required to predict the future of these environments. The effects produced by greenhouse gas emissions and anthropogenic deposition of chemical compounds may provide a counterbalancing effect in some ecosystems, while the changes may be dangerous in others due to positive feedback. Although our knowledge is still very limited, we stress that the scientific data collected over the past years and even decades is never contradictory with regard to the need for boreal and temperate forest ecosystem protection by strict policies. A greater decrease in global forest area will be particularly detrimental because it will prevent other climate change mitigation efforts, such as the reduction of greenhouse emissions.
Considering the importance of the transfer of C from the roots of primary producers to soil, the rhizosphere is likely the most important hot spot of bacterial activity in soils. The available nutrients provided by plants and mycorrhizal fungi lead to the formation of an interactive network of different soil microbes (not only bacteria) that makes the rhizosphere a fascinating and very complex system in which conditions are far from those in individual pores of the bulk soil. The number of drivers to consider, including the specificities of compounds and microbial diversity depending on the type of tree or plant and a large influence of seasonal effects (among others), hampers the accessibility to microbial ecologists of the processes that occur in the rhizosphere. However, the function of this niche very likely offers the key to understanding the biogeochemistry of forest soils. Studies combining the assessment of metabolites present in the rhizosphere at different times, their turnovers and transformations, and the effects and implications of bacterial taxa in terms of fluxes are the next step toward elucidating the effects of forest trees on the rhizospheres and how they relate to the cycling of C and other nutrients.
The large number of studies examining the bacterial communities in forest ecosystems that were published in recent years are shedding light on the high diversity and complex structure of these communities. However, to provide insights into the ecological roles and contributions of individual bacterial taxa to biogeochemical processes, it is necessary to link the community composition to specific functions. Although DNA-based analyses aid in unraveling community dynamics and changes in microbial activity, RNA-based studies offer important information about the active bacterial community and the processes that occur in ecosystems. RNA-based studies of forests confirm how thousands of microbial taxa are transcriptionally active in forest litter and soil, underscoring the involvement of many players in important processes that occur in soil and the importance of taxa with a low abundance of DNA.
ACKNOWLEDGMENTS
This work was supported by the Czech Science Foundation (grant 14-09040P), the Ministry of Education, Youth and Sports of the Czech Republic (grant LD15086), and the Institutional Research Concept of the Institute of Microbiology of the CAS (grant RVO61388971).
We declare that we have no conflicts of interest.
Biographies

Salvador Lladó is a researcher whose main career focus is to relate bacterial identity to functionality related to the C cycle in temperate forest soils. During his Ph.D. research in the Department of Microbiology of the University of Barcelona, he worked on characterizing bacterial populations involved in the decomposition of hydrocarbon pollutants in contaminated soils. After he obtained his Ph.D., in December 2013, he moved to the laboratory of Petr Baldrian in the Laboratory of Environmental Microbiology of the Czech Academy of Sciences, where he is applying DNA and RNA high-throughput sequencing and metabolomics, combined with classical microbiological methods of isolation and physiological characterization, to obtain a better understanding of the roles of the most abundant bacteria in the C and N cycles in forest soils. His broader interest is to be able to relate bacterial community structure and functionalities in soils with the changing C fluxes due to climate change.

Rubén López-Mondéjar studied biology at the University of Murcia (Spain) and carried out his research in the Department of Soil and Water Conservation at the CEBAS-CSIC (Spain), working on the application of molecular methods to improve the use of beneficial microorganisms in agriculture, and he received his Ph.D. in applied biology from the University of Alicante (Spain) in 2011. Afterwards, he become a postdoctoral researcher in the Laboratory of Environmental Microbiology of the CAS (Czech Republic) and worked on the study of the composition and ecology of soil bacteria by using multiomics methods. His current research is focused on exploring the bacterial systems involved in polysaccharide degradation in forest soil and in discovering new microorganisms and enzymes useful for biomass conversion and for the production of environmentally friendly chemicals and biofuels. His broader interest is to unveil the enormous potential of microbial strains as natural resources for the different fields of biotechnology.

Petr Baldrian received his Ph.D. in microbiology at the Charles University in Prague, Czech Republic, based on work describing the effects of metals on wood-rotting fungi. He later changed his topic to environmental microbiology, focusing on enzyme activities and the fungal contribution to decomposition of litter and soil organic matter as well as the characterization of enzymatic systems of microorganisms and their biotechnological potential. He is now Head of the Laboratory of Environmental Microbiology of the Institute of Microbiology of the Czech Academy of Sciences in Prague. Recently, he contributed to the identification of the active fraction of the soil microbiome and the analysis of the relative contributions of bacteria and fungi to ecosystem processes. His work is mostly focused on the functioning of forest soils, the seasonality of microbial processes, and the drivers of community assembly of soil bacteria and fungi.
REFERENCES
- 1.Keenan RJ, Reams GA, Achard F, de Freitas JV, Grainger A, Lindquist E. 2015. Dynamics of global forest area: results from the FAO Global Forest Resources Assessment 2015. For Ecol Manage 352:9–20. doi: 10.1016/j.foreco.2015.06.014. [DOI] [Google Scholar]
- 2.Crowther TW, Glick HB, Covey KR, Bettigole C, Maynard DS, Thomas SM, Smith JR, Hintler G, Duguid MC, Amatulli G, Tuanmu MN, Jetz W, Salas C, Stam C, Piotto D, Tavani R, Green S, Bruce G, Williams SJ, Wiser SK, Huber MO, Hengeveld GM, Nabuurs GJ, Tikhonova E, Borchardt P, Li CF, Powrie LW, Fischer M, Hemp A, Homeier J, Cho P, Vibrans AC, Umunay PM, Piao SL, Rowe CW, Ashton MS, Crane PR, Bradford MA. 2015. Mapping tree density at a global scale. Nature 525:201–205. doi: 10.1038/nature14967. [DOI] [PubMed] [Google Scholar]
- 3.Lebeis SL. 2015. Greater than the sum of their parts: characterizing plant microbiomes at the community-level. Curr Opin Plant Biol 24:82–86. doi: 10.1016/j.pbi.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Hardoim PR, van Overbeek LS, Berg G, Pirttila AM, Compant S, Campisano A, Doring M, Sessitsch A. 2015. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Štursová M, Bárta J, Šantrùčková H, Baldrian P. 2016. Small-scale spatial heterogeneity of ecosystem properties, microbial community composition and microbial activities in a temperate mountain forest soil. FEMS Microbiol Ecol 92:fiw185. doi: 10.1093/femsec/fiw185. [DOI] [PubMed] [Google Scholar]
- 6.Taggart RE, Cross AT. 2009. Global greenhouse to icehouse and back again: the origin and future of the boreal forest biome. Glob Planet Chang 65:115–121. doi: 10.1016/j.gloplacha.2008.10.014. [DOI] [Google Scholar]
- 7.Goldblum D, Rigg LS. 2010. The deciduous forest-boreal forest ecotone. Geogr Compass 4:701–717. doi: 10.1111/j.1749-8198.2010.00342.x. [DOI] [Google Scholar]
- 8.Baldrian P, Kolařík M, Štursová M, Kopecký J, Valaskova V, Větrovský T, Žifčáková L, Šnajdr J, Rídl J, Vlček C, Voříšková J. 2012. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6:248–258. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vesterdal L, Clarke N, Sigurdsson BD, Gundersen P. 2013. Do tree species influence soil carbon stocks in temperate and boreal forests? For Ecol Manage 309:4–18. doi: 10.1016/j.foreco.2013.01.017. [DOI] [Google Scholar]
- 10.Machacova K, Back J, Vanhatalo A, Halmeenmaki E, Kolari P, Mammarella I, Pumpanen J, Acosta M, Urban O, Pihlatie M. 2016. Pinus sylvestris as a missing source of nitrous oxide and methane in boreal forest. Sci Rep 6:23410. doi: 10.1038/srep23410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Y, Birdsey RA, Fang J, Houghton R, Kauppi PE, Kurz WA, Phillips OL, Shvidenko A, Lewis SL, Canadell JG, Ciais P, Jackson RB, Pacala SW, McGuire AD, Piao S, Rautiainen A, Sitch S, Hayes D. 2011. A large and persistent carbon sink in the world's forests. Science 333:988–993. doi: 10.1126/science.1201609. [DOI] [PubMed] [Google Scholar]
- 12.Allison SD, Treseder KK. 2011. Climate change feedbacks to microbial decomposition in boreal soils. Fungal Ecol 4:362–374. doi: 10.1016/j.funeco.2011.01.003. [DOI] [Google Scholar]
- 13.Gauthier S, Bernier P, Kuuluvainen T, Shvidenko AZ, Schepaschenko DG. 2015. Boreal forest health and global change. Science 349:819–822. doi: 10.1126/science.aaa9092. [DOI] [PubMed] [Google Scholar]
- 14.Alkama R, Cescatti A. 2016. Biophysical climate impacts of recent changes in global forest cover. Science 351:600–604. doi: 10.1126/science.aac8083. [DOI] [PubMed] [Google Scholar]
- 15.Millar CI, Stephenson NL. 2015. Temperate forest health in an era of emerging megadisturbance. Science 349:823–826. doi: 10.1126/science.aaa9933. [DOI] [PubMed] [Google Scholar]
- 16.DeAngelis KM, Pold G, Topcuoglu BD, van Diepen LT, Varney RM, Blanchard JL, Melillo J, Frey SD. 2015. Long-term forest soil warming alters microbial communities in temperate forest soils. Front Microbiol 6:104. doi: 10.3389/fmicb.2015.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trivedi P, Anderson IC, Singh BK. 2013. Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction. Trends Microbiol 21:641–651. doi: 10.1016/j.tim.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Uroz S, Buée M, Deveau A, Mieszkin S, Martin F. 2016. Ecology of the forest microbiome: highlights of temperate and boreal ecosystems. Soil Biol Biochem 103:471–488. doi: 10.1016/j.soilbio.2016.09.006. [DOI] [Google Scholar]
- 19.Baldrian P. 2017. Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol Rev 41:109–130. doi: 10.1093/femsre/fuw040. [DOI] [PubMed] [Google Scholar]
- 20.Graham EB, Knelman JE, Schindlbacher A, Siciliano S, Breulmann M, Yannarell A, Beman JM, Abell G, Philippot L, Prosser J, Foulquier A, Yuste JC, Glanville HC, Jones DL, Angel R, Salminen J, Newton RJ, Burgmann H, Ingram LJ, Hamer U, Siljanen HM, Peltoniemi K, Potthast K, Baneras L, Hartmann M, Banerjee S, Yu RQ, Nogaro G, Richter A, Koranda M, Castle SC, Goberna M, Song B, Chatterjee A, Nunes OC, Lopes AR, Cao Y, Kaisermann A, Hallin S, Strickland MS, Garcia-Pausas J, Barba J, Kang H, Isobe K, Papaspyrou S, Pastorelli R, Lagomarsino A, Lindstrom ES, Basiliko N, Nemergut DR. 2016. Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front Microbiol 7:214. doi: 10.3389/fmicb.2016.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voříšková J, Brabcová V, Cajthaml T, Baldrian P. 2014. Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol 201:269–278. doi: 10.1111/nph.12481. [DOI] [PubMed] [Google Scholar]
- 22.Sterkenburg E, Bahr A, Brandstrom Durling M, Clemmensen KE, Lindahl BD. 2015. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol 207:1145–1158. doi: 10.1111/nph.13426. [DOI] [PubMed] [Google Scholar]
- 23.Eichlerová I, Homolka L, Žifčáková L, Lisá L, Dobiášová P, Baldrian P. 2015. Enzymatic systems involved in decomposition reflects the ecology and taxonomy of saprotrophic fungi. Fungal Ecol 13:10–22. doi: 10.1016/j.funeco.2014.08.002. [DOI] [Google Scholar]
- 24.van den Brink J, de Vries RP. 2011. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol 91:1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Wal A, Geydan TD, Kuyper TW, de Boer W. 2013. A thready affair: linking fungal diversity and community dynamics to terrestrial decomposition processes. FEMS Microbiol Rev 37:477–494. doi: 10.1111/1574-6976.12001. [DOI] [PubMed] [Google Scholar]
- 26.van der Heijden MG, Martin FM, Selosse MA, Sanders IR. 2015. Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol 205:1406–1423. doi: 10.1111/nph.13288. [DOI] [PubMed] [Google Scholar]
- 27.Peršoh D. 2015. Plant-associated fungal communities in the light of meta'omics. Fungal Divers 75:1–25. doi: 10.1007/s13225-015-0334-9. [DOI] [Google Scholar]
- 28.Averill C, Turner BL, Finzi AC. 2014. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature 505:543–545. doi: 10.1038/nature12901. [DOI] [PubMed] [Google Scholar]
- 29.Clemmensen KE, Finlay RD, Dahlberg A, Stenlid J, Wardle DA, Lindahl BD. 2015. Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol 205:1525–1536. doi: 10.1111/nph.13208. [DOI] [PubMed] [Google Scholar]
- 30.Berlemont R, Martiny AC. 2015. Genomic potential for polysaccharides deconstruction in bacteria. Appl Environ Microbiol 81:1513–1519. doi: 10.1128/AEM.03718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Štursová M, Žifčáková L, Leigh MB, Burgess R, Baldrian P. 2012. Cellulose utilization in forest litter and soil: identification of bacterial and fungal decomposers. FEMS Microbiol Ecol 80:735–746. doi: 10.1111/j.1574-6941.2012.01343.x. [DOI] [PubMed] [Google Scholar]
- 32.Eichorst SA, Kuske CR. 2012. Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl Environ Microbiol 78:2316–2327. doi: 10.1128/AEM.07313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Mondéjar R, Zühlke D, Becher D, Riedel K, Baldrian P. 2016. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep 6:25279. doi: 10.1038/srep25279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown ME, Chang MC. 2014. Exploring bacterial lignin degradation. Curr Opin Chem Biol 19:1–7. doi: 10.1016/j.cbpa.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Tian JH, Pourcher AM, Bouchez T, Gelhaye E, Peu P. 2014. Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Appl Microbiol Biotechnol 98:9527–9544. doi: 10.1007/s00253-014-6142-4. [DOI] [PubMed] [Google Scholar]
- 36.Reed SC, Cleveland CC, Townsend AR. 2011. Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annu Rev Ecol Evol Syst 42:489–512. doi: 10.1146/annurev-ecolsys-102710-145034. [DOI] [Google Scholar]
- 37.Uroz S, Oger P, Lepleux C, Collignon C, Frey-Klett P, Turpault MP. 2011. Bacterial weathering and its contribution to nutrient cycling in temperate forest ecosystems. Res Microbiol 162:820–831. doi: 10.1016/j.resmic.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Walker JK, Cohen H, Higgins LM, Kennedy PG. 2014. Testing the link between community structure and function for ectomycorrhizal fungi involved in a global tripartite symbiosis. New Phytol 202:287–296. doi: 10.1111/nph.12638. [DOI] [PubMed] [Google Scholar]
- 39.Podila GK, Sreedasyam A, Muratet MA. 2009. Populus rhizosphere and the ectomycorrhizal interactome. Crit Rev Plant Sci 28:359–367. doi: 10.1080/07352680903241220. [DOI] [Google Scholar]
- 40.Bonito G, Reynolds H, Robeson MS II, Nelson J, Hodkinson BP, Tuskan G, Schadt CW, Vilgalys R. 2014. Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants. Mol Ecol 23:3356–3370. doi: 10.1111/mec.12821. [DOI] [PubMed] [Google Scholar]
- 41.Uroz S, Calvaruso C, Turpault MP, Pierrat JC, Mustin C, Frey-Klett P. 2007. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl Environ Microbiol 73:3019–3027. doi: 10.1128/AEM.00121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izumi H, Anderson IC, Alexander IJ, Killham K, Moore ER. 2006. Diversity and expression of nitrogenase genes (nifH) from ectomycorrhizas of Corsican pine (Pinus nigra). Environ Microbiol 8:2224–2230. doi: 10.1111/j.1462-2920.2006.01104.x. [DOI] [PubMed] [Google Scholar]
- 43.Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urbanová M, Šnajdr J, Baldrian P. 2015. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol Biochem 84:53–64. doi: 10.1016/j.soilbio.2015.02.011. [DOI] [Google Scholar]
- 46.Prescott CE, Grayston SJ. 2013. Tree species influence on microbial communities in litter and soil: current knowledge and research needs. For Ecol Manage 309:19–27. doi: 10.1016/j.foreco.2013.02.034. [DOI] [Google Scholar]
- 47.Chemidlin Prevost-Boure N, Maron P-A, Ranjard L, Nowak V, Dufrene E, Damesin C, Soudani K, Lata J-C. 2011. Seasonal dynamics of the bacterial community in forest soils under different quantities of leaf litter. Appl Soil Ecol 47:14–23. doi: 10.1016/j.apsoil.2010.11.006. [DOI] [Google Scholar]
- 48.Baldrian P, Merhautová V, Cajthaml T, Petránková M, Šnajdr J. 2010. Small-scale distribution of extracellular enzymes, fungal, and bacterial biomass in Quercus petraea forest topsoil. Biol Fertil Soils 46:717–726. doi: 10.1007/s00374-010-0478-4. [DOI] [Google Scholar]
- 49.Baldrian P. 2014. Distribution of extracellular enzymes in soils: spatial heterogeneity and determining factors at various scales. Soil Sci Soc Am J 78:11. doi: 10.2136/sssaj2013.04.0155dgs. [DOI] [Google Scholar]
- 50.Kuzyakov Y, Blagodatskaya E. 2015. Microbial hotspots and hot moments in soil: concept & review. Soil Biol Biochem 83:184–199. doi: 10.1016/j.soilbio.2015.01.025. [DOI] [Google Scholar]
- 51.Austin AT, Vivanco L, Gonzalez-Arzac A, Perez LI. 2014. There's no place like home? An exploration of the mechanisms behind plant litter-decomposer affinity in terrestrial ecosystems. New Phytol 204:307–314. doi: 10.1111/nph.12959. [DOI] [PubMed] [Google Scholar]
- 52.Hatton PJ, Castanha C, Torn MS, Bird JA. 2015. Litter type control on soil C and N stabilization dynamics in a temperate forest. Glob Chang Biol 21:1358–1367. doi: 10.1111/gcb.12786. [DOI] [PubMed] [Google Scholar]
- 53.Becher D, Bernhardt J, Fuchs S, Riedel K. 2013. Metaproteomics to unravel major microbial players in leaf litter and soil environments: challenges and perspectives. Proteomics 13:2895–2909. doi: 10.1002/pmic.201300095. [DOI] [PubMed] [Google Scholar]
- 54.Kim M, Kim WS, Tripathi BM, Adams J. 2014. Distinct bacterial communities dominate tropical and temperate zone leaf litter. Microb Ecol 67:837–848. doi: 10.1007/s00248-014-0380-y. [DOI] [PubMed] [Google Scholar]
- 55.Žifčáková L, Větrovský T, Howe A, Baldrian P. 2016. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ Microbiol 18:288–301. doi: 10.1111/1462-2920.13026. [DOI] [PubMed] [Google Scholar]
- 56.López-Mondéjar R, Voříšková J, Větrovský T, Baldrian P. 2015. The bacterial community inhabiting temperate deciduous forests is vertically stratified and undergoes seasonal dynamics. Soil Biol Biochem 87:43–50. doi: 10.1016/j.soilbio.2015.04.008. [DOI] [Google Scholar]
- 57.Purahong W, Schloter M, Pecyna MJ, Kapturska D, Daumlich V, Mital S, Buscot F, Hofrichter M, Gutknecht JL, Kruger D. 2014. Uncoupling of microbial community structure and function in decomposing litter across beech forest ecosystems in Central Europe. Sci Rep 4:7014. doi: 10.1038/srep07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urbanová M, Šnajdr J, Brabcová V, Merhautová V, Dobiášová P, Cajthaml T, Vaněk D, Frouz J, Šantrůčková H, Baldrian P. 2014. Litter decomposition along a primary post-mining chronosequence. Biol Fertil Soils 50:827–837. doi: 10.1007/s00374-014-0905-z. [DOI] [Google Scholar]
- 59.Augusto L, Ranger J, Binkley D, Rothe A. 2002. Impact of several common tree species of European temperate forests on soil fertility. Ann For Sci 59:233–253. doi: 10.1051/forest:2002020. [DOI] [Google Scholar]
- 60.Gessner MO, Swan CM, Dang CK, McKie BG, Bardgett RD, Wall DH, Hattenschwiler S. 2010. Diversity meets decomposition. Trends Ecol Evol 25:372–380. doi: 10.1016/j.tree.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 61.Thoms C, Gleixner G. 2013. Seasonal differences in tree species' influence on soil microbial communities. Soil Biol Biochem 66:239–248. doi: 10.1016/j.soilbio.2013.05.018. [DOI] [Google Scholar]
- 62.Brooks DD, Chan R, Starks ER, Grayston SJ, Jones MD. 2011. Ectomycorrhizal hyphae structure components of the soil bacterial community for decreased phosphatase production. FEMS Microbiol Ecol 76:245–255. doi: 10.1111/j.1574-6941.2011.01060.x. [DOI] [PubMed] [Google Scholar]
- 63.Nordén U. 1994. Influence of tree species on acidification and mineral pools on deciduous forest soils of south Sweden. Water Air Soil Pollut 76:363–381. doi: 10.1007/BF00482713. [DOI] [Google Scholar]
- 64.Langenbruch C, Helfrich M, Flessa H. 2011. Effects of beech (Fagus sylvatica), ash (Fraxinus excelsior) and lime (Tilia spec.) on soil chemical properties in a mixed deciduous forest. Plant Soil 352:389–403. doi: 10.1007/s11104-011-1004-7. [DOI] [Google Scholar]
- 65.Cesarz S, Fender A-C, Beyer F, Valtanen K, Pfeiffer B, Gansert D, Hertel D, Polle A, Daniel R, Leuschner C, Scheu S. 2013. Roots from beech (Fagus sylvatica L.) and ash (Fraxinus excelsior L.) differentially affect soil microorganisms and carbon dynamics. Soil Biol Biochem 61:23–32. doi: 10.1016/j.soilbio.2013.02.003. [DOI] [Google Scholar]
- 66.Landesman WJ, Nelson DM, Fitzpatrick MC. 2014. Soil properties and tree species drive β-diversity of soil bacterial communities. Soil Biol Biochem 76:201–209. doi: 10.1016/j.soilbio.2014.05.025. [DOI] [Google Scholar]
- 67.Scheibe A, Steffens C, Seven J, Jacob A, Hertel D, Leuschner C, Gleixner G. 2015. Effects of tree identity dominate over tree diversity on the soil microbial community structure. Soil Biol Biochem 81:219–227. doi: 10.1016/j.soilbio.2014.11.020. [DOI] [Google Scholar]
- 68.Thoms C, Gattinger A, Jacob M, Thomas FM, Gleixner G. 2010. Direct and indirect effects of tree diversity drive soil microbial diversity in temperate deciduous forest. Soil Biol Biochem 42:1558–1565. doi: 10.1016/j.soilbio.2010.05.030. [DOI] [Google Scholar]
- 69.Rasche F, Knapp D, Kaiser C, Koranda M, Kitzler B, Zechmeister-Boltenstern S, Richter A, Sessitsch A. 2011. Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME J 5:389–402. doi: 10.1038/ismej.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baldrian P, Šnajdr J, Merhautová V, Dobiášová P, Cajthaml T, Valášková V. 2013. Responses of the extracellular enzyme activities in hardwood forest to soil temperature and seasonality and the potential effects of climate change. Soil Biol Biochem 56:60–68. doi: 10.1016/j.soilbio.2012.01.020. [DOI] [Google Scholar]
- 71.Šnajdr J, Cajthaml T, Valaskova V, Merhautova V, Petrankova M, Spetz P, Leppanen K, Baldrian P. 2011. Transformation of Quercus petraea litter: successive changes in litter chemistry are reflected in differential enzyme activity and changes in the microbial community composition. FEMS Microbiol Ecol 75:291–303. doi: 10.1111/j.1574-6941.2010.00999.x. [DOI] [PubMed] [Google Scholar]
- 72.Tláskal V, Voříšková J, Baldrian P. 2016. Bacterial succession on decomposing leaf litter exhibits a specific occurrence pattern of cellulolytic taxa and potential decomposers of fungal mycelia. FEMS Microbiol Ecol 92:fiw177. doi: 10.1093/femsec/fiw177. [DOI] [PubMed] [Google Scholar]
- 73.Fierer N, Bradford MA, Jackson R. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 74.Voříšková J, Baldrian P. 2013. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J 7:477–486. doi: 10.1038/ismej.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brabcová V, Nováková M, Davidová A, Baldrian P. 2016. Dead fungal mycelium in forest soil represents a decomposition hotspot and a habitat for a specific microbial community. New Phytol 210:1369–1381. doi: 10.1111/nph.13849. [DOI] [PubMed] [Google Scholar]
- 76.Freschet GT, Cornwell WK, Wardle DA, Elumeeva TG, Liu W, Jackson BG, Onipchenko VG, Soudzilovskaia NA, Tao J, Cornelissen JHC, Austin A. 2013. Linking litter decomposition of above- and below-ground organs to plant-soil feedbacks worldwide. J Ecol 101:943–952. doi: 10.1111/1365-2745.12092. [DOI] [Google Scholar]
- 77.Ekblad A, Wallander H, Godbold DL, Cruz C, Johnson D, Baldrian P, Björk RG, Epron D, Kieliszewska-Rokicka B, Kjøller R, Kraigher H, Matzner E, Neumann J, Plassard C. 2013. The production and turnover of extramatrical mycelium of ectomycorrhizal fungi in forest soils: role in carbon cycling. Plant Soil 366:1–27. doi: 10.1007/s11104-013-1630-3. [DOI] [Google Scholar]
- 78.de Boer W, Folman LB, Summerbell RC, Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 79.Kielak AM, Scheublin TR, Mendes LW, van Veen JA, Kuramae EE. 2016. Bacterial community succession in pine-wood decomposition. Front Microbiol 7:231. doi: 10.3389/fmicb.2016.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rinta-Kanto JM, Sinkko H, Rajala T, Al-Soud WA, Sørensen SJ, Tamminen MV, Timonen S. 2016. Natural decay process affects the abundance and community structure of Bacteria and Archaea in Picea abies logs. FEMS Microbiol Ecol 92:fiw087. doi: 10.1093/femsec/fiw087. [DOI] [PubMed] [Google Scholar]
- 81.Johnston SR, Boddy L, Weightman AJ. 2016. Bacteria in decomposing wood and their interactions with wood-decay fungi. FEMS Microbiol Ecol 92:fiw179. doi: 10.1093/femsec/fiw179. [DOI] [PubMed] [Google Scholar]
- 82.Hoppe B, Krüger K, Kahl T, Arnstadt T, Buscot F, Bauhus J, Wubet T. 2015. A pyrosequencing insight into sprawling bacterial diversity and community dynamics in decaying deadwood logs of Fagus sylvatica and Picea abies. Sci Rep 5:9456. doi: 10.1038/srep09456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Folman LB, Gunnewiek P, Boddy L, de Boer W. 2008. Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil. FEMS Microbiol Ecol 63:181–191. doi: 10.1111/j.1574-6941.2007.00425.x. [DOI] [PubMed] [Google Scholar]
- 84.Valášková V, de Boer W, Gunnewiek PJ, Pospíšek M, Baldrian P. 2009. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J 3:1218–1221. doi: 10.1038/ismej.2009.64. [DOI] [PubMed] [Google Scholar]
- 85.Nacke H, Engelhaupt M, Brady S, Fischer C, Tautzt J, Daniel R. 2012. Identification and characterization of novel cellulolytic and hemicellulolytic genes and enzymes derived from German grassland soil metagenomes. Biotechnol Lett 34:663–675. doi: 10.1007/s10529-011-0830-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Šnajdr J, Valášková V, Merhautová V, Herinková J, Cajthaml T, Baldrian P. 2008. Spatial variability of enzyme activities and microbial biomass in the upper layers of Quercus petraea forest soil. Soil Biol Biochem 40:2068–2075. doi: 10.1016/j.soilbio.2008.01.015. [DOI] [Google Scholar]
- 87.Uroz S, Ioannidis P, Lengelle J, Cebron A, Morin E, Buéé M, Martin F. 2013. Functional assays and metagenomic analyses reveals differences between the microbial communities inhabiting the soil horizons of a Norway spruce plantation. PLoS One 8:e55929. doi: 10.1371/journal.pone.0055929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eilers KG, Lauber CL, Knight R, Fierer N. 2010. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol Biochem 42:896–903. doi: 10.1016/j.soilbio.2010.02.003. [DOI] [Google Scholar]
- 89.Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Daniel R. 2011. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6:e17000. doi: 10.1371/journal.pone.0017000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuffner M, Hai B, Rattei T, Melodelima C, Schloter M, Zechmeister-Boltenstern S, Jandl R, Schindlbacher A, Sessitsch A. 2012. Effects of season and experimental warming on the bacterial community in a temperate mountain forest soil assessed by 16S rRNA gene pyrosequencing. FEMS Microbiol Ecol 82:551–562. doi: 10.1111/j.1574-6941.2012.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lipson DA. 2007. Relationships between temperature responses and bacterial community structure along seasonal and altitudinal gradients. FEMS Microbiol Ecol 59:418–427. doi: 10.1111/j.1574-6941.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 92.Kurth F, Zeitler K, Feldhahn L, Neu TR, Weber T, Krištůfek V, Wubet T, Herrmann S, Buscot F, Tarkka M. 2013. Detection and quantification of a mycorrhization helper bacterium and a mycorrhizal fungus in plant-soil microcosms at different levels of complexity. BMC Microbiol 13:205. doi: 10.1186/1471-2180-13-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ferrenberg S, O'Neill SP, Knelman JE, Todd B, Duggan S, Bradley D, Robinson T, Schmidt SK, Townsend AR, Williams MW, Cleveland CC, Melbourne BA, Jiang L, Nemergut DR. 2013. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J 7:1102–1111. doi: 10.1038/ismej.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jeanbille M, Buee M, Bach C, Cebron A, Frey-Klett P, Turpault MP, Uroz S. 2016. Soil parameters drive the structure, diversity and metabolic potentials of the bacterial communities across temperate beech forest soil sequences. Microb Ecol 71:482–493. doi: 10.1007/s00248-015-0669-5. [DOI] [PubMed] [Google Scholar]
- 95.Högberg MN, Högberg P, Myrold DD. 2007. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia 150:590–601. [DOI] [PubMed] [Google Scholar]
- 96.Shen C, Xiong J, Zhang H, Feng Y, Lin X, Li X, Liang W, Chu H. 2013. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211. doi: 10.1016/j.soilbio.2012.07.013. [DOI] [Google Scholar]
- 97.Norman JS, Barrett JE. 2016. Substrate availability drives spatial patterns in richness of ammonia-oxidizing bacteria and archaea in temperate forest soils. Soil Biol Biochem 94:169–172. doi: 10.1016/j.soilbio.2015.11.015. [DOI] [Google Scholar]
- 98.Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL. 2008. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci U S A 105(Suppl 1):S11505–S11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lauber CL, Strickland MS, Bradford MA, Fierer N. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- 101.Naether A, Foesel BU, Naegele V, Wust PK, Weinert J, Bonkowski M, Alt F, Oelmann Y, Polle A, Lohaus G, Gockel S, Hemp A, Kalko EK, Linsenmair KE, Pfeiffer S, Renner S, Schoning I, Weisser WW, Wells K, Fischer M, Overmann J, Friedrich MW. 2012. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl Environ Microbiol 78:7398–7406. doi: 10.1128/AEM.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garcia-Fraile P, Benada O, Cajthaml T, Baldrian P, Lladó S. 2015. Terracidiphilus gabretensis gen. nov., sp. nov., an abundant and active forest soil acidobacterium important in organic matter transformation. Appl Environ Microbiol 82:560–569. doi: 10.1128/AEM.03353-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Větrovský T, Baldrian P. 2013. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e0057923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lladó S, Žifčáková L, Větrovský T, Eichlerová I, Baldrian P. 2016. Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biol Fertil Soils 52:251–260. doi: 10.1007/s00374-015-1072-6. [DOI] [Google Scholar]
- 105.Kielak AM, Barreto CC, Kowalchuk GA, van Veen JA, Kuramae EE. 2016. The ecology of acidobacteria: moving beyond genes and genomes. Front Microbiol 7:744. doi: 10.3389/fmicb.2016.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uroz S, Buéé M, Murat C, Frey-Klett P, Martin F. 2010. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2:281–288. doi: 10.1111/j.1758-2229.2009.00117.x. [DOI] [PubMed] [Google Scholar]
- 107.Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem 43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun H, Terhonen E, Koskinen K, Paulin L, Kasanen R, Asiegbu FO. 2014. Bacterial diversity and community structure along different peat soils in boreal forest. Appl Soil Ecol 74:37–45. doi: 10.1016/j.apsoil.2013.09.010. [DOI] [Google Scholar]
- 109.Eilers KG, Debenport S, Anderson S, Fierer N. 2012. Digging deeper to find unique microbial communities: the strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol Biochem 50:58–65. doi: 10.1016/j.soilbio.2012.03.011. [DOI] [Google Scholar]
- 110.Collignon C, Uroz S, Turpault MP, Frey-Klett P. 2011. Seasons differently impact the structure of mineral weathering bacterial communities in beech and spruce stands. Soil Biol Biochem 43:2012–2022. doi: 10.1016/j.soilbio.2011.05.008. [DOI] [Google Scholar]
- 111.Leveau JH, Uroz S, de Boer W. 2010. The bacterial genus Collimonas: mycophagy, weathering and other adaptive solutions to life in oligotrophic soil environments. Environ Microbiol 12:281–292. doi: 10.1111/j.1462-2920.2009.02010.x. [DOI] [PubMed] [Google Scholar]
- 112.Uroz S, Oger P, Tisserand E, Cebron A, Turpault MP, Buee M, De Boer W, Leveau JH, Frey-Klett P. 2016. Specific impacts of beech and Norway spruce on the structure and diversity of the rhizosphere and soil microbial communities. Sci Rep 6:27756. doi: 10.1038/srep27756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.O'Brien SL, Gibbons SM, Owens SM, Hampton-Marcell J, Johnston ER, Jastrow JD, Gilbert JA, Meyer F, Antonopoulos DA. 2016. Spatial scale drives patterns in soil bacterial diversity. Environ Microbiol 18:2039–2051. doi: 10.1111/1462-2920.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martiny JB, Eisen JA, Penn K, Allison SD, Horner-Devine MC. 2011. Drivers of b-diversity depend on spatial scale. Proc Natl Acad Sci U S A 108:7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Spielvogel S, Prietzel J, Kögel-Knabner I. 2016. Stand scale variability of topsoil organic matter composition in a high-elevation Norway spruce forest ecosystem. Geoderma 267:112–122. doi: 10.1016/j.geoderma.2015.12.001. [DOI] [Google Scholar]
- 116.Izumi H, Finlay RD. 2011. Ectomycorrhizal roots select distinctive bacterial and ascomycete communities in Swedish subarctic forests. Environ Microbiol 13:819–830. doi: 10.1111/j.1462-2920.2010.02393.x. [DOI] [PubMed] [Google Scholar]
- 117.Brockett BFT, Prescott CE, Grayston SJ. 2012. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol Biochem 44:9–20. doi: 10.1016/j.soilbio.2011.09.003. [DOI] [Google Scholar]
- 118.Lennon JT, Aanderud ZT, Lehmkuhl BK, Schoolmaster DR. 2012. Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 93:1867–1879. doi: 10.1890/11-1745.1. [DOI] [PubMed] [Google Scholar]
- 119.Pollierer MM, Langel R, Korner C, Maraun M, Scheu S. 2007. The underestimated importance of belowground carbon input for forest soil animal food webs. Ecol Lett 10:729–736. doi: 10.1111/j.1461-0248.2007.01064.x. [DOI] [PubMed] [Google Scholar]
- 120.Phillips RP, Finzi AC, Bernhardt ES. 2011. Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long-term CO2 fumigation. Ecol Lett 14:187–194. doi: 10.1111/j.1461-0248.2010.01570.x. [DOI] [PubMed] [Google Scholar]
- 121.Beidler KV, Taylor BN, Strand AE, Cooper ER, Schonholz M, Pritchard SG. 2015. Changes in root architecture under elevated concentrations of CO2 and nitrogen reflect alternate soil exploration strategies. New Phytol 205:1153–1163. doi: 10.1111/nph.13123. [DOI] [PubMed] [Google Scholar]
- 122.Fender A-C, Leuschner C, Schützenmeister K, Gansert D, Jungkunst HF. 2013. Rhizosphere effects of tree species—large reduction of N2O emission by saplings of ash, but not of beech, in temperate forest soil. Eur J Soil Biol 54:7–15. doi: 10.1016/j.ejsobi.2012.10.010. [DOI] [Google Scholar]
- 123.Markkola AM. 1996. Resource allocation in ectomycorrhizal symbiosis in Scots pine affected by environmental changes. University of Oulu, Oulu, Finland. [Google Scholar]
- 124.Kluber LA, Smith JE, Myrold DD. 2011. Distinctive fungal and bacterial communities are associated with mats formed by ectomycorrhizal fungi. Soil Biol Biochem 43:1042–1050. doi: 10.1016/j.soilbio.2011.01.022. [DOI] [Google Scholar]
- 125.Churchland C, Grayston SJ. 2014. Specificity of plant-microbe interactions in the tree mycorrhizosphere biome and consequences for soil C cycling. Front Microbiol 5:261. doi: 10.3389/fmicb.2014.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aspray TJ, Eirian Jones E, Whipps JM, Bending GD. 2006. Importance of mycorrhization helper bacteria cell density and metabolite localization for the Pinus sylvestris-Lactarius rufus symbiosis. FEMS Microbiol Ecol 56:25–33. doi: 10.1111/j.1574-6941.2005.00051.x. [DOI] [PubMed] [Google Scholar]
- 127.Vik U, Logares R, Blaalid R, Halvorsen R, Carlsen T, Bakke I, Kolsto AB, Okstad OA, Kauserud H. 2013. Different bacterial communities in ectomycorrhizae and surrounding soil. Sci Rep 3:3471. doi: 10.1038/srep03471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Collignon C, Calvaruso C, Turpault M-P. 2011. Temporal dynamics of exchangeable K, Ca and Mg in acidic bulk soil and rhizosphere under Norway spruce (Picea abies Karst) and beech (Fagus sylvatica L) stands. Plant Soil 349:355–366. doi: 10.1007/s11104-011-0881-0. [DOI] [Google Scholar]
- 129.Drake JE, Darby BA, Giasson MA, Kramer MA, Phillips RP, Finzi AC. 2013. Stoichiometry constrains microbial response to root exudation—insights from a model and a field experiment in a temperate forest. Biogeosciences 10:821–838. doi: 10.5194/bg-10-821-2013. [DOI] [Google Scholar]
- 130.Finzi AC, Abramoff RZ, Spiller KS, Brzostek ER, Darby BA, Kramer MA, Phillips RP. 2015. Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Glob Chang Biol 21:2082–2094. doi: 10.1111/gcb.12816. [DOI] [PubMed] [Google Scholar]
- 131.Brzostek ER, Greco A, Drake JE, Finzi AC. 2012. Root carbon inputs to the rhizosphere stimulate extracellular enzyme activity and increase nitrogen availability in temperate forest soils. Biogeochemistry 115:65–76. doi: 10.1007/s10533-012-9818-9. [DOI] [Google Scholar]
- 132.Loeppmann S, Blagodatskaya E, Pausch J, Kuzyakov Y. 2016. Substrate quality affects kinetics and catalytic efficiency of exo-enzymes in rhizosphere and detritusphere. Soil Biol Biochem 92:111–118. doi: 10.1016/j.soilbio.2015.09.020. [DOI] [Google Scholar]
- 133.Tefs C, Gleixner G. 2012. Importance of root derived carbon for soil organic matter storage in a temperate old-growth beech forest—evidence from C, N and 14C content. For Ecol Manage 263:131–137. doi: 10.1016/j.foreco.2011.09.010. [DOI] [Google Scholar]
- 134.Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. 2013. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 8:e55731. doi: 10.1371/journal.pone.0055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kaiser C, Koranda M, Kitzler B, Fuchslueger L, Schnecker J, Schweiger P, Rasche F, Zechmeister-Boltenstern S, Sessitsch A, Richter A. 2010. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol 187:843–858. doi: 10.1111/j.1469-8137.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shi S, Nuccio E, Herman DJ, Rijkers R, Estera K, Li J, da Rocha UN, He Z, Pett-Ridge J, Brodie EL, Zhou J, Firestone M. 2015. Successional trajectories of rhizosphere bacterial communities over consecutive seasons. mBio 6:e00746. doi: 10.1128/mBio.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mao Y, Li X, Smyth EM, Yannarell AC, Mackie RI. 2014. Enrichment of specific bacterial and eukaryotic microbes in the rhizosphere of switchgrass (Panicum virgatum L.) through root exudates. Environ Microbiol Rep 6:293–306. doi: 10.1111/1758-2229.12152. [DOI] [PubMed] [Google Scholar]
- 138.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. 2013. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM. 2014. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8:1577–1587. doi: 10.1038/ismej.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Donn S, Kirkegaard JA, Perera G, Richardson AE, Watt M. 2015. Evolution of bacterial communities in the wheat crop rhizosphere. Environ Microbiol 17:610–621. doi: 10.1111/1462-2920.12452. [DOI] [PubMed] [Google Scholar]
- 141.Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 142.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ai C, Liang G, Sun J, Wang X, He P, Zhou W, He X. 2015. Reduced dependence of rhizosphere microbiome on plant-derived carbon in 32-year long-term inorganic and organic fertilized soils. Soil Biol Biochem 80:70–78. doi: 10.1016/j.soilbio.2014.09.028. [DOI] [Google Scholar]
- 144.Gschwendtner S, Engel M, Lueders T, Buegger F, Schloter M. 2016. Nitrogen fertilization affects bacteria utilizing plant-derived carbon in the rhizosphere of beech seedlings. Plant Soil 407:203–215. doi: 10.1007/s11104-016-2888-z. [DOI] [Google Scholar]
- 145.Lladó S, Baldrian P. 2017. Community-level physiological profiling analyses show potential to identify the copiotrophic bacteria present in soil environments. PLoS One 12:e0171638. doi: 10.1371/journal.pone.0171638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Izumi H, Cairney JW, Killham K, Moore E, Alexander IJ, Anderson IC. 2008. Bacteria associated with ectomycorrhizas of slash pine (Pinus elliottii) in south-eastern Queensland, Australia. FEMS Microbiol Lett 282:196–204. doi: 10.1111/j.1574-6968.2008.01122.x. [DOI] [PubMed] [Google Scholar]
- 147.Tanaka M, Nara K. 2009. Phylogenetic diversity of non-nodulating Rhizobium associated with pine ectomycorrhizae. FEMS Microbiol Ecol 69:329–343. doi: 10.1111/j.1574-6941.2009.00720.x. [DOI] [PubMed] [Google Scholar]
- 148.Yu HX, Wang CY, Tang M. 2013. Fungal and bacterial communities in the rhizosphere of Pinus tabulaeformis related to the restoration of plantations and natural secondary forests in the Loess Plateau, northwest China. Sci World J 2013:606480. doi: 10.1155/2013/606480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Marupakula S, Mahmood S, Finlay RD. 2016. Analysis of single root tip microbiomes suggests that distinctive bacterial communities are selected by Pinus sylvestris roots colonized by different ectomycorrhizal fungi. Environ Microbiol 18:1470–1483. doi: 10.1111/1462-2920.13102. [DOI] [PubMed] [Google Scholar]
- 150.Labbe JL, Weston DJ, Dunkirk N, Pelletier DA, Tuskan GA. 2014. Newly identified helper bacteria stimulate ectomycorrhizal formation in Populus. Front Plant Sci 5:579. doi: 10.3389/fpls.2014.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Frey-Klett P, Garbaye J, Tarkka M. 2007. The mycorrhiza helper bacteria revisited. New Phytol 176:22–36. doi: 10.1111/j.1469-8137.2007.02191.x. [DOI] [PubMed] [Google Scholar]
- 152.Fransson P, Andersson A, Norström S, Bylund D, Bent E. 2016. Ectomycorrhizal exudates and pre-exposure to elevated CO2 affects soil bacterial growth and community structure. Fungal Ecol 20:211–224. doi: 10.1016/j.funeco.2016.01.003. [DOI] [Google Scholar]
- 153.Aspray TJ, Frey-Klett P, Jones JE, Whipps JM, Garbaye J, Bending GD. 2006. Mycorrhization helper bacteria: a case of specificity for altering ectomycorrhiza architecture but not ectomycorrhiza formation. Mycorrhiza 16:533–541. doi: 10.1007/s00572-006-0068-3. [DOI] [PubMed] [Google Scholar]
- 154.Lang C, Seven J, Polle A. 2011. Host preferences and differential contributions of deciduous tree species shape mycorrhizal species richness in a mixed Central European forest. Mycorrhiza 21:297–308. doi: 10.1007/s00572-010-0338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Korkama T, Fritze H, Pakkanen A, Pennanen T. 2007. Interactions between extraradical ectomycorrhizal mycelia, microbes associated with the mycelia and growth rate of Norway spruce (Picea abies) clones. New Phytol 173:798–807. doi: 10.1111/j.1469-8137.2006.01957.x. [DOI] [PubMed] [Google Scholar]
- 156.Xia M, Talhelm AF, Pregitzer KS. 2015. Fine roots are the dominant source of recalcitrant plant litter in sugar maple-dominated northern hardwood forests. New Phytol 208:715–726. doi: 10.1111/nph.13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Van Dyk JS, Pletschke BI. 2012. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes—factors affecting enzymes, conversion and synergy. Biotechnol Adv 30:1458–1480. doi: 10.1016/j.biotechadv.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 158.Rytioja J, Hildén K, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR. 2014. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol Mol Biol Rev 78:614–649. doi: 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Berlemont R, Martiny AC. 2013. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol 79:1545–1554. doi: 10.1128/AEM.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Větrovský T, Steffen KT, Baldrian P. 2014. Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria. PLoS One 9:e89108. doi: 10.1371/journal.pone.0089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brumm PJ. 2013. Bacterial genomes: what they teach us about cellulose degradation. Biofuels 4:669–681. doi: 10.4155/bfs.13.44. [DOI] [Google Scholar]
- 162.Lopez-Mondejar R, Zuhlke D, Vetrovsky T, Becher D, Riedel K, Baldrian P. 2016. Decoding the complete arsenal for cellulose and hemicellulose deconstruction in the highly efficient cellulose decomposer Paenibacillus O199. Biotechnol Biofuels 9:104. doi: 10.1186/s13068-016-0518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Llado S, Benada O, Cajthaml T, Baldrian P, Garcia-Fraile P. 2016. Silvibacterium bohemicum gen. nov. sp. nov., an acidobacterium isolated from coniferous soil in the Bohemian Forest National Park. Syst Appl Microbiol 39:14–19. doi: 10.1016/j.syapm.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 164.Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denance N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yokoyama H, Yamashita T, Morioka R, Ohmori H. 2014. Extracellular secretion of noncatalytic plant cell wall-binding proteins by the cellulolytic thermophile Caldicellulosiruptor bescii. J Bacteriol 196:3784–3792. doi: 10.1128/JB.01897-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vodovnik M, Duncan SH, Reid MD, Cantlay L, Turner K, Parkhill J, Lamed R, Yeoman CJ, Miller ME, White BA, Bayer EA, Marinsek-Logar R, Flint HJ. 2013. Expression of cellulosome components and type IV pili within the extracellular proteome of Ruminococcus flavefaciens 007. PLoS One 8:e65333. doi: 10.1371/journal.pone.0065333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wegmann U, Louis P, Goesmann A, Henrissat B, Duncan SH, Flint HJ. 2014. Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota (“Ruminococcus bicirculans”) reveals two chromosomes and a selective capacity to utilize plant glucans. Environ Microbiol 16:2879–2890. doi: 10.1111/1462-2920.12217. [DOI] [PubMed] [Google Scholar]
- 168.Himmel ME, Xu Q, Luo Y, Ding S, Lamed R, Bayer EA. 2010. Microbial enzyme systems for biomass conversion: emerging paradigms. Biofuels 1:323–341. doi: 10.4155/bfs.09.25. [DOI] [Google Scholar]
- 169.Sukharnikov LO, Cantwell BJ, Podar M, Zhulin IB. 2011. Cellulases: ambiguous nonhomologous enzymes in a genomic perspective. Trends Biotechnol 29:473–479. doi: 10.1016/j.tibtech.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang JK, Zhang JJ, Yu HY, Cheng JW, Miao LH. 2014. Community composition and cellulase activity of cellulolytic bacteria from forest soils planted with broad-leaved deciduous and evergreen trees. Appl Microbiol Biotechnol 98:1449–1458. doi: 10.1007/s00253-013-5130-4. [DOI] [PubMed] [Google Scholar]
- 171.Bandounas L, Wierckx NJ, de Winde JH, Ruijssenaars HJ. 2011. Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol 11:94. doi: 10.1186/1472-6750-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kellner H, Luis P, Zimdars B, Kiesel B, Buscot F. 2008. Diversity of bacterial laccase-like multicopper oxidase genes in forest and grassland Cambisol soil samples. Soil Biol Biochem 40:638–648. doi: 10.1016/j.soilbio.2007.09.013. [DOI] [Google Scholar]
- 173.Nacke H, Fischer C, Thurmer A, Meinicke P, Daniel R. 2014. Land use type significantly affects microbial gene transcription in soil. Microb Ecol 67:919–930. doi: 10.1007/s00248-014-0377-6. [DOI] [PubMed] [Google Scholar]
- 174.Baldrian P, Větrovský T, Cajthaml T, Dobiášová P, Petránková M, Šnajdr J, Eichlerová I. 2013. Estimation of fungal biomass in forest litter and soil. Fungal Ecol 6:1–11. doi: 10.1016/j.funeco.2012.10.002. [DOI] [Google Scholar]
- 175.Fernandez CW, Koide RT. 2014. Initial melanin and nitrogen concentrations control the decomposition of ectomycorrhizal fungal litter. Soil Biol Biochem 77:150–157. doi: 10.1016/j.soilbio.2014.06.026. [DOI] [Google Scholar]
- 176.Beier S, Bertilsson S. 2013. Bacterial chitin degradation—mechanisms and ecophysiological strategies. Front Microbiol 4:149. doi: 10.3389/fmicb.2013.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Talamantes D, Biabini N, Dang H, Abdoun K, Berlemont R. 2016. Natural diversity of cellulases, xylanases, and chitinases in bacteria. Biotechnol Biofuels 9:133. doi: 10.1186/s13068-016-0538-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bai Y, Eijsink VG, Kielak AM, van Veen JA, de Boer W. 2016. Genomic comparison of chitinolytic enzyme systems from terrestrial and aquatic bacteria. Environ Microbiol 18:38–49. doi: 10.1111/1462-2920.12545. [DOI] [PubMed] [Google Scholar]
- 179.Tolonen AC, Cerisy T, El-Sayyed H, Boutard M, Salanoubat M, Church GM. 2015. Fungal lysis by a soil bacterium fermenting cellulose. Environ Microbiol 17:2618–2627. doi: 10.1111/1462-2920.12495. [DOI] [PubMed] [Google Scholar]
- 180.Wang X, Sharp CE, Jones GM, Grasby SE, Brady AL, Dunfield PF. 2015. Stable-isotope probing identifies uncultured Planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl Environ Microbiol 81:4607–4615. doi: 10.1128/AEM.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kaiser C, Kilburn MR, Clode PL, Fuchslueger L, Koranda M, Cliff JB, Solaiman ZM, Murphy DV. 2015. Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytol 205:1537–1551. doi: 10.1111/nph.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Högberg MN, Briones MJ, Keel SG, Metcalfe DB, Campbell C, Midwood AJ, Thornton B, Hurry V, Linder S, Nasholm T, Högberg P. 2010. Quantification of effects of season and nitrogen supply on tree below-ground carbon transfer to ectomycorrhizal fungi and other soil organisms in a boreal pine forest. New Phytol 187:485–493. doi: 10.1111/j.1469-8137.2010.03274.x. [DOI] [PubMed] [Google Scholar]
- 183.Kuzyakov Y, Cheng W. 2001. Photosynthesis controls of rhizosphere respiration and organic matter decomposition. Soil Biol Biochem 33:1915–1925. doi: 10.1016/S0038-0717(01)00117-1. [DOI] [Google Scholar]
- 184.Högberg P, Nordgren A, Buchmann N, Taylor AFS, Ekblad A, Högberg MN, Nyberg G, Ottosson-Lofvenius M, Read DJ. 2001. Large-scale forest girdling shows that current photosynthesis drives soil respiration. Nature 411:789–792. doi: 10.1038/35081058. [DOI] [PubMed] [Google Scholar]
- 185.Buée M, de Boer W, Martin F, van Overbeek L, Jurkevitch E. 2009. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212. doi: 10.1007/s11104-009-9991-3. [DOI] [Google Scholar]
- 186.Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, Stenlid J, Finlay RD, Wardle DA, Lindahl BD. 2013. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 339:1615–1618. doi: 10.1126/science.1231923. [DOI] [PubMed] [Google Scholar]
- 187.Wang Y, Li C, Wang Q, Wang H, Duan B, Zhang G. 2016. Environmental behaviors of phenolic acids dominated their rhizodeposition in boreal poplar plantation forest soils. J Soils Sediments 16:1858–1870. doi: 10.1007/s11368-016-1375-8. [DOI] [Google Scholar]
- 188.Haichar FE, Heulin T, Guyonnet JP, Achouak W. 2016. Stable isotope probing of carbon flow in the plant holobiont. Curr Opin Biotechnol 41:9–13. doi: 10.1016/j.copbio.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 189.Churchland C, Weatherall A, Briones MJI, Grayston SJ. 2012. Stable-isotope labeling and probing of recent photosynthates into respired CO2, soil microbes and soil mesofauna using a xylem and phloem stem-injection technique on Sitka spruce (Picea sitchensis). Rapid Comm Mass Spectrom 26:2493–2501. doi: 10.1002/rcm.6368. [DOI] [PubMed] [Google Scholar]
- 190.Uroz S, Courty PE, Pierrat JC, Peter M, Buéé M, Turpault MP, Garbaye J, Frey-Klett P. 2013. Functional profiling and distribution of the forest soil bacterial communities along the soil mycorrhizosphere continuum. Microb Ecol 66:404–415. doi: 10.1007/s00248-013-0199-y. [DOI] [PubMed] [Google Scholar]
- 191.Izumi H, Anderson IC, Alexander IJ, Killham K, Moore ER. 2006. Endobacteria in some ectomycorrhiza of Scots pine (Pinus sylvestris). FEMS Microbiol Ecol 56:34–43. doi: 10.1111/j.1574-6941.2005.00048.x. [DOI] [PubMed] [Google Scholar]
- 192.Lau E, Nolan EJ IV, Dillard ZW, Dague RD, Semple AL, Wentzell WL. 2015. High throughput sequencing to detect differences in methanotrophic Methylococcaceae and Methylocystaceae in surface peat, forest soil, and sphagnum moss in Cranesville Swamp Preserve, West Virginia, USA. Microorganisms 3:113–136. doi: 10.3390/microorganisms3020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Savi F, Di Bene C, Canfora L, Mondini C, Fares S. 2016. Environmental and biological controls on CH4 exchange over an evergreen Mediterranean forest. Agric For Meteorol 226–227:67–79. [Google Scholar]
- 194.Covey KR, Wood SA, Warren RJ, Lee X, Bradford MA. 2012. Elevated methane concentrations in trees of an upland forest. Geophys Res Lett 39:L15705. doi: 10.1029/2012GL052361. [DOI] [Google Scholar]
- 195.Serrano-Silva N, Sarria-Guzmán Y, Dendooven L, Luna-Guido M. 2014. Methanogenesis and methanotrophy in soil: a review. Pedosphere 24:291–307. doi: 10.1016/S1002-0160(14)60016-3. [DOI] [Google Scholar]
- 196.Shoemaker JK, Keenan TF, Hollinger DY, Richardson AD. 2014. Forest ecosystem changes from annual methane source to sink depending on late summer water balance. Geophys Res Lett 41:673–679. doi: 10.1002/2013GL058691. [DOI] [Google Scholar]
- 197.Esson KC, Lin X, Kumaresan D, Chanton JP, Murrell JC, Kostka JE. 2016. Alpha- and gammaproteobacterial methanotrophs codominate the active methane-oxidizing communities in an acidic boreal peat bog. Appl Environ Microbiol 82:2363–2371. doi: 10.1128/AEM.03640-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nazaries L, Murrell JC, Millard P, Baggs L, Singh BK. 2013. Methane, microbes and models: fundamental understanding of the soil methane cycle for future predictions. Environ Microbiol 15:2395–2417. doi: 10.1111/1462-2920.12149. [DOI] [PubMed] [Google Scholar]
- 199.Okubo T, Tsukui T, Maita H, Okamoto S, Oshima K, Fujisawa T, Saito A, Futamata H, Hattori R, Shimomura Y, Haruta S, Morimoto S, Wang Y, Sakai Y, Hattori M, Aizawa S-I, Nagashima KVP, Masuda S, Hattori T, Yamashita A, Bao Z, Hayatsu M, Kajiya-Kanegae H, Yoshinaga I, Sakamoto K, Toyota K, Nakao M, Kohara M, Anda M, Niwa R, Jung-Hwan P, Sameshima-Saito R, Tokuda S-I, Yamamoto S, Yamamoto S, Yokoyama T, Akutsu T, Nakamura Y, Nakahira-Yanaka Y, Takada Hoshino Y, Hirakawa H, Mitsui H, Terasawa K, Itakura M, Sato S, Ikeda-Ohtsubo W, Sakakura N, Kaminuma E, Minamisawa K. 2012. Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microb Environ 27:306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.VanInsberghe D, Maas KR, Cardenas E, Strachan CR, Hallam SJ, Mohn WW. 2015. Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J 9:2435–2441. doi: 10.1038/ismej.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Berthrong ST, Yeager CM, Gallegos-Graves LV, Steven B, Eichorst SA, Jackson RB, Kuske CR. 2014. Nitrogen fertilization has a stronger effect on soil nitrogen-fixing bacterial communities than elevated atmospheric CO2. Appl Environ Microbiol 80:3103–3112. doi: 10.1128/AEM.04034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hsu SF, Buckley DH. 2009. Evidence for the functional significance of diazotroph community structure in soil. ISME J 3:124–136. doi: 10.1038/ismej.2008.82. [DOI] [PubMed] [Google Scholar]
- 203.Levy-Booth DJ, Prescott CE, Grayston SJ. 2014. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol Biochem 75:11–25. doi: 10.1016/j.soilbio.2014.03.021. [DOI] [Google Scholar]
- 204.Pajares S, Bohannan BJM. 2016. Ecology of nitrogen fixing, nitrifying, and denitrifying microorganisms in tropical forest soils. Front Microbiol 7:1045. doi: 10.3389/fmicb.2016.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jung J, Yeom J, Han J, Kim J, Park W. 2012. Seasonal changes in nitrogen-cycle gene abundances and in bacterial communities in acidic forest soils. J Microbiol 50:365–373. doi: 10.1007/s12275-012-1465-2. [DOI] [PubMed] [Google Scholar]
- 206.Isobe K, Koba K, Otsuka S, Senoo K. 2011. Nitrification and nitrifying microbial communities in forest soils. J For Res 16:351–362. doi: 10.1007/s10310-011-0266-5. [DOI] [Google Scholar]
- 207.Long X, Chen C, Xu Z, Oren R, He J-Z. 2012. Abundance and community structure of ammonia-oxidizing bacteria and archaea in a temperate forest ecosystem under ten-years elevated CO2. Soil Biol Biochem 46:163–171. doi: 10.1016/j.soilbio.2011.12.013. [DOI] [Google Scholar]
- 208.Laverman AM, Speksnijder AG, Braster M, Kowalchuk GA, Verhoef HA, Van Verseveld HW. 2001. Spatiotemporal stability of an ammonia-oxidizing community in a nitrogen-saturated forest soil. Microb Ecol 42:35–45. doi: 10.1007/s002480000038. [DOI] [PubMed] [Google Scholar]
- 209.Szukics U, Hackl E, Zechmeister-Boltenstern S, Sessitsch A. 2012. Rapid and dissimilar response of ammonia oxidizing archaea and bacteria to nitrogen and water amendment in two temperate forest soils. Microbiol Res 167:103–109. doi: 10.1016/j.micres.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hynes HM, Germida JJ. 2012. Relationship between ammonia oxidizing bacteria and bioavailable nitrogen in harvested forest soils of central Alberta. Soil Biol Biochem 46:18–25. doi: 10.1016/j.soilbio.2011.10.018. [DOI] [Google Scholar]
- 211.Malchair S, Carnol M. 2012. AOB community structure and richness under European beech, sessile oak, Norway spruce and Douglas fir at three temperate forest sites. Plant Soil 366:521–535. doi: 10.1007/s11104-012-1450-x. [DOI] [Google Scholar]
- 212.Cong J, Yang Y, Liu X, Lu H, Liu X, Zhou J, Li D, Yin H, Ding J, Zhang Y. 2015. Analyses of soil microbial community compositions and functional genes reveal potential consequences of natural forest succession. Sci Rep 5:10007. doi: 10.1038/srep10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Freedman Z, Eisenlord SD, Zak DR, Xue K, He Z, Zhou J. 2013. Towards a molecular understanding of N cycling in northern hardwood forests under future rates of N deposition. Soil Biol Biochem 66:130–138. doi: 10.1016/j.soilbio.2013.07.010. [DOI] [Google Scholar]
- 214.Shaw LJ, Nicol GW, Smith Z, Fear J, Prosser JI, Baggs EM. 2006. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ Microbiol 8:214–222. doi: 10.1111/j.1462-2920.2005.00882.x. [DOI] [PubMed] [Google Scholar]
- 215.Orellana LH, Rodriguez-R LM, Higgins S, Chee-Sanford JC, Sanford RA, Ritalahti KM, Löffler FE, Konstantinidis KT. 2014. Detecting nitrous oxide reductase (NosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio 5:e01193-14. doi: 10.1128/mBio.01193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Priyanka J, Mukherjee K. 2015. Diversity study of nitrate reducing bacteria from soil samples—a metagenomics approach. J Comp Sci Syst Biol 8:191–198. [Google Scholar]
- 217.Rosch C, Mergel A, Bothe H. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl Environ Microbiol 68:3818–3829. doi: 10.1128/AEM.68.8.3818-3829.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nelson MB, Martiny AC, Martiny JBH. 2016. Global biogeography of microbial nitrogen-cycling traits in soil. Proc Natl Acad Sci U S A 113:8033–8040. doi: 10.1073/pnas.1601070113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Szukics U, Hackl E, Zechmeister-Boltenstern S, Sessitsch A. 2009. Contrasting response of two forest soils to nitrogen input: rapidly altered NO and N2O emissions and nirK abundance. Biol Fertil Soils 45:855–863. doi: 10.1007/s00374-009-0396-5. [DOI] [Google Scholar]
- 220.Richardson AE, Simpson RJ. 2011. Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol 156:989–996. doi: 10.1104/pp.111.175448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Barton LL, Northup DE. 2011. Microbial ecology. Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 222.Bergkemper F, Scholer A, Engel M, Lang F, Kruger J, Schloter M, Schulz S. 2016. Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ Microbiol 18:1988–2000. doi: 10.1111/1462-2920.13188. [DOI] [PubMed] [Google Scholar]
- 223.Bergkemper F, Kublik S, Lang F, Kruger J, Vestergaard G, Schloter M, Schulz S. 2016. Novel oligonucleotide primers reveal a high diversity of microbes which drive phosphorous turnover in soil. J Microbiol Methods 125:91–97. doi: 10.1016/j.mimet.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 224.Singh BK, Bardgett RD, Smith P, Reay DS. 2010. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- 225.Mouri G, Nakano K, Tsuyama I, Tanaka N. 2016. The effects of future nationwide forest transition to discharge in the 21st century with regard to general circulation model climate change scenarios. Environ Res 149:288–296. doi: 10.1016/j.envres.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 226.Ruosch M, Spahni R, Joos F, Henne PD, van der Knaap WO, Tinner W. 2016. Past and future evolution of Abies alba forests in Europe—comparison of a dynamic vegetation model with palaeo data and observations. Glob Chang Biol 22:727–740. doi: 10.1111/gcb.13075. [DOI] [PubMed] [Google Scholar]
- 227.Ma W, Liang J, Cumming JR, Lee E, Welsh AB, Watson JV, Zhou M. 2016. Fundamental shifts of central hardwood forests under climate change. Ecol Model 332:28–41. doi: 10.1016/j.ecolmodel.2016.03.021. [DOI] [Google Scholar]
- 228.Crabbe RA, Dash J, Rodriguez-Galiano VF, Janous D, Pavelka M, Marek MV. 2016. Extreme warm temperatures alter forest phenology and productivity in Europe. Sci Total Environ 563–564:486–495. doi: 10.1016/j.scitotenv.2016.04.124. [DOI] [PubMed] [Google Scholar]
- 229.Zhang Z, Qiao M, Li D, Yin H, Liu Q. 2016. Do warming-induced changes in quantity and stoichiometry of root exudation promote soil N transformations via stimulation of soil nitrifiers, denitrifiers and ammonifiers? Eur J Soil Biol 74:60–68. doi: 10.1016/j.ejsobi.2016.03.007. [DOI] [Google Scholar]
- 230.Vanhala P, Bergström I, Haaspuro T, Kortelainen P, Holmberg M, Forsius M. 2016. Boreal forests can have a remarkable role in reducing greenhouse gas emissions locally: land use-related and anthropogenic greenhouse gas emissions and sinks at the municipal level. Sci Total Environ 557–558:51–57. doi: 10.1016/j.scitotenv.2016.03.040. [DOI] [PubMed] [Google Scholar]
- 231.Vanhala P, Karhu K, Tuomi M, Björklöf K, Fritze H, Hyvärinen H, Liski J. 2011. Transplantation of organic surface horizons of boreal soils into warmer regions alters microbiology but not the temperature sensitivity of decomposition. Glob Chang Biol 17:538–550. doi: 10.1111/j.1365-2486.2009.02154.x. [DOI] [Google Scholar]
- 232.Bonan GB. 2008. Forests and climate change: forcings, feedbacks, and the climate benefits of forests. Science 320:1444–1449. doi: 10.1126/science.1155121. [DOI] [PubMed] [Google Scholar]
- 233.Dooley SR, Treseder KK. 2011. The effect of fire on microbial biomass: a meta-analysis of field studies. Biogeochemistry 109:49–61. doi: 10.1007/s10533-011-9633-8. [DOI] [Google Scholar]
- 234.Pold G, Melillo JM, DeAngelis KM. 2015. Two decades of warming increases diversity of a potentially lignolytic bacterial community. Front Microbiol 6:480. doi: 10.3389/fmicb.2015.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Norby RJ, Delucia EH, Gielen B, Calfapietra C, Giardina CP, King JS, Ledford J, McCarthy HR, Moore DJ, Ceulemans R, De Angelis P, Finzi AC, Karnosky DF, Kubiske ME, Lukac M, Pregitzer KS, Scarascia-Mugnozza GE, Schlesinger WH, Oren R. 2005. Forest response to elevated CO2 is conserved across a broad range of productivity. Proc Natl Acad Sci U S A 102:18052–18056. doi: 10.1073/pnas.0509478102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Luo Y, Su B, Currie WS, Dukes JS, Finzi A, Hartwig U, Hungate BA, McMurtrie RE, Oren R, Parton WJ, Pataki DE, Shaw MR, Zak DR, Field CB. 2004. Progressive N limitation of ecosystem responses to rising CO2. Bioscience 54:731–739. doi: 10.1641/0006-3568(2004)054[0731:PNLOER]2.0.CO;2. [DOI] [Google Scholar]
- 237.Meier IC, Pritchard SG, Brzostek ER, McCormack ML, Phillips RP. 2015. The rhizosphere and hyphosphere differ in their impacts on carbon and nitrogen cycling in forests exposed to elevated CO2. New Phytol 205:1164–1174. doi: 10.1111/nph.13122. [DOI] [PubMed] [Google Scholar]
- 238.Dunbar J, Eichorst SA, Gallegos-Graves LV, Silva S, Xie G, Hengartner NW, Evans RD, Hungate BA, Jackson RB, Megonigal JP, Schadt CW, Vilgalys R, Zak DR, Kuske CR. 2012. Common bacterial responses in six ecosystems exposed to 10 years of elevated atmospheric carbon dioxide. Environ Microbiol 14:1145–1158. doi: 10.1111/j.1462-2920.2011.02695.x. [DOI] [PubMed] [Google Scholar]
- 239.Sulman BN, Phillips RP, Oishi AC, Shevliakova E, Pacala SW. 2014. Microbe-driven turnover offsets mineral-mediated storage of soil carbon under elevated CO2. Nat Clim Chang 4:1099–1102. doi: 10.1038/nclimate2436. [DOI] [Google Scholar]
- 240.Dunbar J, Gallegos-Graves LV, Steven B, Mueller R, Hesse C, Zak DR, Kuske CR. 2014. Surface soil fungal and bacterial communities in aspen stands are resilient to eleven years of elevated CO2 and O3. Soil Biol Biochem 76:227–234. doi: 10.1016/j.soilbio.2014.05.027. [DOI] [Google Scholar]
- 241.Lesaulnier C, Papamichail D, McCorkle S, Ollivier B, Skiena S, Taghavi S, Zak D, van der Lelie D. 2008. Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ Microbiol 10:926–941. doi: 10.1111/j.1462-2920.2007.01512.x. [DOI] [PubMed] [Google Scholar]
- 242.Garcia-Palacios P, Vandegehuchte ML, Shaw EA, Dam M, Post KH, Ramirez KS, Sylvain ZA, de Tomasel CM, Wall DH. 2015. Are there links between responses of soil microbes and ecosystem functioning to elevated CO2, N deposition and warming? A global perspective. Glob Chang Biol 21:1590–1600. doi: 10.1111/gcb.12788. [DOI] [PubMed] [Google Scholar]
- 243.Schleppi P, Bucher-Wallin I, Hagedorn F, Körner C. 2012. Increased nitrate availability in the soil of a mixed mature temperate forest subjected to elevated CO2 concentration (canopy FACE). Glob Chang Biol 18:757–768. doi: 10.1111/j.1365-2486.2011.02559.x. [DOI] [Google Scholar]
- 244.Reich PB, Sendall KM, Stefanski A, Wei X, Rich RL, Montgomery RA. 2016. Boreal and temperate trees show strong acclimation of respiration to warming. Nature 531:633–636. doi: 10.1038/nature17142. [DOI] [PubMed] [Google Scholar]
- 245.Zhao C, Zhu L, Liang J, Yin H, Yin C, Li D, Zhang N, Liu Q. 2014. Effects of experimental warming and nitrogen fertilization on soil microbial communities and processes of two subalpine coniferous species in Eastern Tibetan Plateau, China. Plant Soil 382:189–201. doi: 10.1007/s11104-014-2153-2. [DOI] [Google Scholar]
- 246.Schindlbacher A, Rodler A, Kuffner M, Kitzler B, Sessitsch A, Zechmeister-Boltenstern S. 2011. Experimental warming effects on the microbial community of a temperate mountain forest soil. Soil Biol Biochem 43:1417–1425. doi: 10.1016/j.soilbio.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rousk K, Rousk J, Jones DL, Zackrisson O, DeLuca TH. 2013. Feather moss nitrogen acquisition across natural fertility gradients in boreal forests. Soil Biol Biochem 61:86–95. doi: 10.1016/j.soilbio.2013.02.011. [DOI] [Google Scholar]
- 248.Peltoniemi K, Laiho R, Juottonen H, Kiikkila O, Makiranta P, Minkkinen K, Pennanen T, Penttilä T, Sarjala T, Tuittila ES, Tuomivirta T, Fritze H. 2015. Microbial ecology in a future climate: effects of temperature and moisture on microbial communities of two boreal fens. FEMS Microbiol Ecol 91:fiv062. doi: 10.1093/femsec/fiv062. [DOI] [PubMed] [Google Scholar]
- 249.Wu J, Xiong J, Hu C, Shi Y, Wang K, Zhang D. 2015. Temperature sensitivity of soil bacterial community along contrasting warming gradient. Appl Soil Ecol 94:40–48. doi: 10.1016/j.apsoil.2015.04.018. [DOI] [Google Scholar]
- 250.Xue K, Xie J, Zhou A, Liu F, Li D, Wu L, Deng Y, He Z, Van Nostrand JD, Luo Y, Zhou J. 2016. Warming alters expressions of microbial functional genes important to ecosystem functioning. Front Microbiol 7:668. doi: 10.3389/fmicb.2016.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Schütt M, Borken W, Spott O, Stange CF, Matzner E. 2014. Temperature sensitivity of C and N mineralization in temperate forest soils at low temperatures. Soil Biol Biochem 69:320–327. doi: 10.1016/j.soilbio.2013.11.014. [DOI] [Google Scholar]
- 252.Li J, Ziegler SE, Lane CS, Billings SA. 2013. Legacies of native climate regime govern responses of boreal soil microbes to litter stoichiometry and temperature. Soil Biol Biochem 66:204–213. doi: 10.1016/j.soilbio.2013.07.018. [DOI] [Google Scholar]
- 253.Galloway JN, Dentener FJ, Capone DG, Boyer EW, Howarth RW, Seitzinger SP, Asner GP, Cleveland CC, Green PA, Holland EA, Karl DM, Michaels AF, Porter JH, Townsend AR, Vorosmarty CJ. 2004. Nitrogen cycles: past, present and future. Biogeochemistry 70:153–226. doi: 10.1007/s10533-004-0370-0. [DOI] [Google Scholar]
- 254.Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA. 2008. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320:889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- 255.Tamm CO. 1991. Nitrogen in terrestrial ecosystems. Springer, Berlin, Germany. [Google Scholar]
- 256.Högberg MN, Blaško R, Bach LH, Hasselquist NJ, Egnell G, Näsholm T, Högberg P. 2014. The return of an experimentally N-saturated boreal forest to an N-limited state: observations on the soil microbial community structure, biotic N retention capacity and gross N mineralisation. Plant Soil 381:45–60. doi: 10.1007/s11104-014-2091-z. [DOI] [Google Scholar]
- 257.Wallenstein MD, McNulty S, Fernandez IJ, Boggs J, Schlesinger WH. 2006. Nitrogen fertilization decreases forest soil fungal and bacterial biomass in three long-term experiments. For Ecol Manage 222:459–468. doi: 10.1016/j.foreco.2005.11.002. [DOI] [Google Scholar]
- 258.Frey SD, Ollinger S, Nadelhoffer K, Bowden R, Brzostek E, Burton A, Caldwell BA, Crow S, Goodale CL, Grandy AS, Finzi A, Kramer MG, Lajtha K, LeMoine J, Martin M, McDowell WH, Minocha R, Sadowsky JJ, Templer PH, Wickings K. 2014. Chronic nitrogen additions suppress decomposition and sequester soil carbon in temperate forests. Biogeochemistry 121:305–316. doi: 10.1007/s10533-014-0004-0. [DOI] [Google Scholar]
- 259.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. 2012. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6:1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zak DR, Holmes WE, Burton AJ, Pregitzer KS, Talhelm AF. 2008. Simulated atmospheric NO3 deposition increases soil organic matter by slowing decomposition. Ecol Appl 18:2016–2027. doi: 10.1890/07-1743.1. [DOI] [PubMed] [Google Scholar]
- 261.Freedman ZB, Zak DR. 2015. Atmospheric N deposition alters connectance, but not functional potential among saprotrophic bacterial communities. Mol Ecol 24:3170–3180. doi: 10.1111/mec.13224. [DOI] [PubMed] [Google Scholar]
- 262.Freedman ZB, Upchurch RA, Zak DR, Cline LC. 2016. Anthropogenic N deposition slows decay by favoring bacterial metabolism: insights from metagenomic analyses. Front Microbiol 7:259. doi: 10.3389/fmicb.2016.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Maaroufi NI, Nordin A, Hasselquist NJ, Bach LH, Palmqvist K, Gundale MJ. 2015. Anthropogenic nitrogen deposition enhances carbon sequestration in boreal soils. Glob Chang Biol 21:3169–3180. doi: 10.1111/gcb.12904. [DOI] [PubMed] [Google Scholar]
- 264.Ramirez KS, Craine JM, Fierer N. 2012. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Chang Biol 18:1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x. [DOI] [Google Scholar]
- 265.Kamble PN, Rousk J, Frey SD, Bååth E. 2013. Bacterial growth and growth-limiting nutrients following chronic nitrogen additions to a hardwood forest soil. Soil Biol Biochem 59:32–37. doi: 10.1016/j.soilbio.2012.12.017. [DOI] [Google Scholar]
- 266.Frey SD, Knorr M, Parrent JL, Simpson RT. 2004. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. For Ecol Manage 196:159–171. doi: 10.1016/j.foreco.2004.03.018. [DOI] [Google Scholar]
- 267.Long X, Chen C, Xu Z, Linder S, He J. 2012. Abundance and community structure of ammonia oxidizing bacteria and archaea in a Sweden boreal forest soil under 19-year fertilization and 12-year warming. J Soils Sediments 12:1124–1133. doi: 10.1007/s11368-012-0532-y. [DOI] [Google Scholar]
- 268.Turlapati SA, Minocha R, Bhiravarasa PS, Tisa LS, Thomas WK, Minocha SC. 2013. Chronic N-amended soils exhibit an altered bacterial community structure in Harvard Forest, MA, USA. FEMS Microbiol Ecol 83:478–493. doi: 10.1111/1574-6941.12009. [DOI] [PubMed] [Google Scholar]
- 269.Aber J, McDowell WH, Nadelhoffer K, Magill A, Berntson G, Kamakea M, McNulty SG, Currie WS, Rustad L, Fernandez IJ. 1998. Nitrogen saturation in temperate forests. Bioscience 48:921–934. doi: 10.2307/1313296. [DOI] [Google Scholar]
- 270.Zak DR, Holmes WE, Tomlinson MJ, Pregitzer KS, Burton AJ. 2006. Microbial cycling of C and N in northern hardwood forests receiving chronic atmospheric NO3− deposition. Ecosystems 9:242–253. doi: 10.1007/s10021-005-0085-7. [DOI] [Google Scholar]
- 271.Zak DR, Pregitzer KS, Kubiske ME, Burton AJ. 2011. Forest productivity under elevated CO2 and O3: positive feedbacks to soil N cycling sustain decade-long net primary productivity enhancement by CO2. Ecol Lett 14:1220–1226. doi: 10.1111/j.1461-0248.2011.01692.x. [DOI] [PubMed] [Google Scholar]
- 272.Li J, Ziegler S, Lane CS, Billings SA. 2012. Warming-enhanced preferential microbial mineralization of humified boreal forest soil organic matter: interpretation of soil profiles along a climate transect using laboratory incubations. J Geophys Res Biogeosci 117:G2008. [Google Scholar]
- 273.Luo X, Fu X, Yang Y, Cai P, Peng S, Chen W, Huang Q. 2016. Microbial communities play important roles in modulating paddy soil fertility. Sci Rep 6:20326. doi: 10.1038/srep20326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Savage KE, Parton WJ, Davidson EA, Trumbore SE, Frey SD. 2013. Long-term changes in forest carbon under temperature and nitrogen amendments in a temperate northern hardwood forest. Glob Chang Biol 19:2389–2400. doi: 10.1111/gcb.12224. [DOI] [PubMed] [Google Scholar]
- 275.Stone MM, Weiss MS, Goodale CL, Adams MB, Fernandez IJ, German DP, Allison SD. 2012. Temperature sensitivity of soil enzyme kinetics under N-fertilization in two temperate forests. Glob Chang Biol 18:1173–1184. doi: 10.1111/j.1365-2486.2011.02545.x. [DOI] [Google Scholar]
- 276.Schindlbacher A, Schnecker J, Takriti M, Borken W, Wanek W. 2015. Microbial physiology and soil CO2 efflux after 9 years of soil warming in a temperate forest—no indications for thermal adaptations. Glob Chang Biol 21:4265–4277. doi: 10.1111/gcb.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Yue K, Peng Y, Peng C, Yang W, Peng X, Wu F. 2016. Stimulation of terrestrial ecosystem carbon storage by nitrogen addition: a meta-analysis. Sci Rep 6:19895. doi: 10.1038/srep19895. [DOI] [PMC free article] [PubMed] [Google Scholar]