SUMMARY
Fungi and oomycetes are filamentous microorganisms that include a diversity of highly developed pathogens of plants. These are sophisticated modulators of plant processes that secrete an arsenal of effector proteins to target multiple host cell compartments and enable parasitic infection. Genome sequencing revealed complex catalogues of effectors of filamentous pathogens, with some species harboring hundreds of effector genes. Although a large fraction of these effector genes encode secreted proteins with weak or no sequence similarity to known proteins, structural studies have revealed unexpected similarities amid the diversity. This article reviews progress in our understanding of effector structure and function in light of these new insights. We conclude that there is emerging evidence for multiple pathways of evolution of effectors of filamentous plant pathogens but that some families have probably expanded from a common ancestor by duplication and diversification. Conserved folds, such as the oomycete WY and the fungal MAX domains, are not predictive of the precise function of the effectors but serve as a chassis to support protein structural integrity while providing enough plasticity for the effectors to bind different host proteins and evolve unrelated activities inside host cells. Further effector evolution and diversification arise via short linear motifs, domain integration and duplications, and oligomerization.
KEYWORDS: plant pathology
INTRODUCTION
Filamentous pathogens (fungi and oomycetes) are the causative agents of some of the world's most notorious plant diseases. Left unchecked, they can devastate crop harvests, destroy managed and wild forests, affect the supply of ornamental plants, and disturb natural ecosystems (1–3). Perhaps the most famous plant disease outbreak was caused by the oomycete Phytophthora infestans, which spread to Europe and triggered the 19th-century Irish potato famine (4). This pathogen remains relevant in agriculture today, infecting potato and tomato crops throughout the world (5). Diseases caused by fungal pathogens, such as rice and wheat blast and wheat stem and stripe rust, are of immediate concern for global food security (1, 6, 7). Major factors in the ability of these filamentous microbes to cause disease on their hosts are effectors, pathogen-encoded proteins that are secreted to either the apoplast or specialized biotrophic interfaces (both are spaces outside plant cells) or are translocated inside host cells (8–11).
Effectors act to modulate host cell physiology to promote susceptibility to pathogens. In turn, plants have evolved cell surface and intracellular receptors to detect the presence of pathogen signatures and mount an immune response to restrict the progression of disease. Cell surface receptors typically recognize microbe-associated molecular patterns (MAMPs), derived from abundant structural components of microbes' cell walls, or secreted proteins that function as virulence effectors. Intracellular receptors respond to the presence of translocated effectors and/or their activity on host cell targets. These intracellular receptors are nucleotide-binding domain- and leucine-rich repeat-containing (NLR) proteins that mediate innate immunity to pathogens in both plants and animals (recently reviewed in reference 12).
One of the defining features of effector proteins, be they of bacterial or filamentous pathogen origin, is the lack of clear sequence similarity to proteins of known function. This is thought to be the consequence of evolutionary pressure that drives the rapid diversification of effector activities in host cells to optimize function and/or avoid recognition by the innate immune system. The frequent difficulty in recognizing common motifs that indicate the function or activity of effectors may be due to few of them having enzymatic activity or the absence of known domains for direct interaction with host factors. In addition, many effectors are small proteins of <15 kDa, and thus, their rapid diversification would result in a loss of sequence similarity. With a few notable exceptions (the RXLR motif of effectors in some oomycetes being the most prominent), this sequence diversity has meant that it is challenging to confidently produce catalogues of effectors from filamentous plant pathogen genomes despite many of these now being available. In some cases, bioinformatic approaches have been useful in predicting and classifying candidate effectors from filamentous plant pathogens (13–23) (Table 1). However, it can be challenging to pick the most relevant proteins to select for further investigation from these lists. These bioinformatic approaches use some of the commonalities identified among effectors from different organisms, such as genomic context, the presence of a secretion signal, the absence of predicted transmembrane domains, expression patterns, and the lack of similarity to known protein domains. Recent advances in the computational prediction of effectors have employed machine-learning approaches, which are proving useful for prioritizing effectors for further study (24). There are also examples of effectors of filamentous plant pathogens that share common sequence motifs with known enzymes, enzyme inhibitors, sugar-binding proteins, and toxins, with some being shown to possess such activities.
TABLE 1.
Effectors of filamentous plant pathogens that have sequence similarities with enzymes or enzyme inhibitors
| Effector class | Hyphal pathogen | Example(s) | Reference(s) |
|---|---|---|---|
| Chorismate mutases | Ustilago maydis | Cmu1 | 45 |
| Lipase effector | Fusarium graminearum | FGL1 | 112 |
| Enzyme inhibitors | |||
| Protease inhibitors | Cladosporium fulvum | Avr2 | 41 |
| Cystatin-like protease inhibitor domains | Phytophthora infestans | EPIC1, EPIC2B | 42 |
| Chitinase inhibitor | Cladosporium fulvum | Avr4 | 56 |
| Proteases and peptidases | |||
| Proteases | Zymoseptoria tritici (Mycosphaerella graminicola) | 33 | |
| Colletotrichum sp. | 34 | ||
| Secreted peptidases | Zymoseptoria tritici (Mycosphaerella graminicola) | Astacin (peptidase family M12A), serine carboxypeptidase S28 | 113 |
| Serine protease | Fusarium oxysporum f. sp. lycopersici | Sep1 | 35 |
| Alkaline serine protease alp1 | Sclerotinia sclerotiorum | Peptidase inhibitor I9 | 23 |
| Metalloproteases | |||
| Zinc metalloprotease | Magnaporthe oryzae | AVR-Pita (AVR2-YAMO) | 36, 114 |
| Deuterolysin metalloprotease | Sclerotinia sclerotiorum | Deuterolysin metalloprotease (M35) family (PF02102) homolog of M. oryzae AVR-Pita | 23 |
| Metalloprotease | Fusarium oxysporum f. sp. lycopersici | Mep1 | 35 |
| Nudix hydrolases | Phytophthora sojae | Avr3b | 46 |
| Colletotrichum truncatum | CtNUDIX | 115 | |
| Melampsora lini | AvrM14 | 48 | |
| Crinklers | |||
| Kinase activity | Phytophthora infestans | CRN8 | 50 |
It is well established that protein structure is more conserved than amino acid sequence, and in many cases, this is due to the evolutionary relationship between structure and function (25). The fact that structural conservation can be a powerful method for the functional annotation of proteins is a fundamental concept that has driven the development of structure determination as a tool to understand the effector biology of both mammalian and plant pathogens (26, 27). In particular, this has been important where the lack of sequence similarity to known functional proteins has prevented the prediction of the molecular mechanism.
In this review, we focus on recent advances that highlight commonalities shared by effectors of filamentous plant pathogens, focusing on functional similarities with known proteins, on effectors that cluster into large structurally common but sequence-divergent families comprising novel folds, or on those that share structural similarity with proteins of known function. It is timely to review progress in this area in light of new insights. We conclude that there is emerging evidence for multiple pathways of evolution of effectors of filamentous plant pathogens, including that some families appear to have evolved from a common ancestor by duplication and diversification in the pathogen.
EFFECTORS OF FILAMENTOUS PLANT PATHOGENS THAT ENCODE ENZYMES AND PROTEASE INHIBITORS
Structural studies of a number of bacterial plant-pathogenic type III secreted effectors (T3SEs) have revealed similarity with proteins of known function, which suggested both how these proteins act and experiments to test mechanisms (28–31). Remarkably, many of these proteins appear to be enzymes with the potential to catalyze a wide variety of different reactions, such as E3 ligation, ADP ribosylation, and proteolysis. In several cases, specific enzymatic activities have been demonstrated for these proteins (32). In contrast, a number of effectors of filamentous plant pathogens have been predicted to have enzymatic activity, but only a few have had such activities confirmed experimentally. To date, there are no structures of enzymes of effectors of filamentous plant pathogens, so these predictions typically rely primarily on sequence comparisons.
Proteases and Protease Inhibitors
Analyses of fungal genomes, including those of Zymoseptoria tritici (33), Colletotrichum sp. (34), and Sclerotinia sclerotiorum (23), identified families of secreted proteases whose expression pattern supports a putative role as effectors, to promote the colonization and growth of the pathogen. Fusarium oxysporum f. sp. lycopersicum secretes a serine protease, Sep1, and a metalloprotease, Mep1, that act synergistically to cleave host chitinases, preventing their activity in degrading fungal cell walls (35). A double mutant of Sep1 and Mep1 showed reduced disease on tomato, highlighting the importance of these proteins for full virulence.
The rice blast fungus Magnaporthe oryzae produces AVR-Pita, an effector with features typical of zinc metalloproteases, including conserved residues known to mediate zinc coordination and catalysis in homologues from other organisms (9, 36). However, to date, actual protease activity for AVR-Pita has not been demonstrated.
A remarkable case is the glucanase inhibitor proteins (GIPs), which are proteins secreted by Phytophthora spp. to inhibit the degradation of pathogen β-1,3/1,6-glucans and the release of defense-eliciting oligosaccharides by host β-1,3-endoglucanases (37, 38). GIPs share significant sequence similarity with trypsin serine proteases but are predicted to be proteolytically nonfunctional because they carry mutated catalytic residues.
Interestingly, filamentous plant pathogens also secrete protease inhibitors, which act on host pathogenesis-related proteases to prevent their activities. Examples include EPI1 and EPI10 of P. infestans, which carry multiple domains with similarity to the Kazal family of serine protease inhibitors (39, 40). In addition, the Avr2 effector of the fungal pathogen Cladosporium fulvum (41) and the P. infestans effectors EPIC1 and EPIC2 (42) are unrelated in sequence but have convergently evolved to target the same host proteases (43, 44). The oomycete EPIC family of protease inhibitor effectors has similarity to the widespread cystatin domain (42), whereas C. fulvum Avr2 is a small cysteine-rich protein without any notable sequence similarity to other proteins (41).
Fungal Cmu1, an Enzyme Interfering with Metabolic Flux
The maize smut fungus Ustilago maydis translocates a chorismate mutase, Cmu1, into plant cells. Cmu1 appears to benefit the pathogen by redirecting the metabolic flux of chorismate away from the biosynthesis of salicylic acid, suppressing the accumulation of this defense-related hormone during infection. Intriguingly, there is evidence to suggest that Cmu1 can move out of infected cells into neighboring cells, where the enzyme's activity can “prime” the host tissue for infection (45).
Translocated Oomycete Effectors Include Enzymes
Oomycete plant pathogens encode putative enzymes in their effector repertoires. Phytophthora species have ∼300 to 550 RXLR-type effectors that rarely have sequence similarity to know enzyme folds. However, P. infestans and Phytophthora sojae contain a sequence signature suggestive of Nudix hydrolase (phosphorylase) activity. The P. sojae effector Avr3b has been shown to possess ADP-ribose/NADH pyrophosphorylase activity when expressed and epitope purified from plant tissue (46). Furthermore, the virulence activity of Avr3b was dependent on the conserved Nudix motif. Interestingly, the activity of Avr3b as a Nudix hydrolase is dependent on its modification by plant cyclophilins; when produced in Escherichia coli, the protein is not active (47). Recently, a putative Nudix hydrolase effector (AvrM14) was identified in the flax rust fungus Melampsora lini (48), but catalytic activity for this protein has yet to be shown.
In addition to RXLR effectors, Phytophthora species also contain hundreds of “Crinkler” effectors (CRNs) (13, 16, 49). CRNs are modular proteins, some of which induce cell death upon expression in plant cells (13, 16). One C-terminal CRN domain has significant sequence similarity to protein Ser/Thr kinases of the RD (arginine-aspartate) class. Indeed, P. infestans CRN8 was shown to be an active kinase present in an autophosphorylated state in plant cells (50). In planta expression of CRN8 enhanced the growth of P. infestans, and this required the intact RD motif, suggesting that the enzymatic activity of this kinase is relevant for virulence.
EFFECTORS OF FILAMENTOUS PLANT PATHOGENS CAN SHARE FOLDS WITH FUNCTIONALLY SIMILAR PROTEINS
Chitin-Binding LysM Effectors
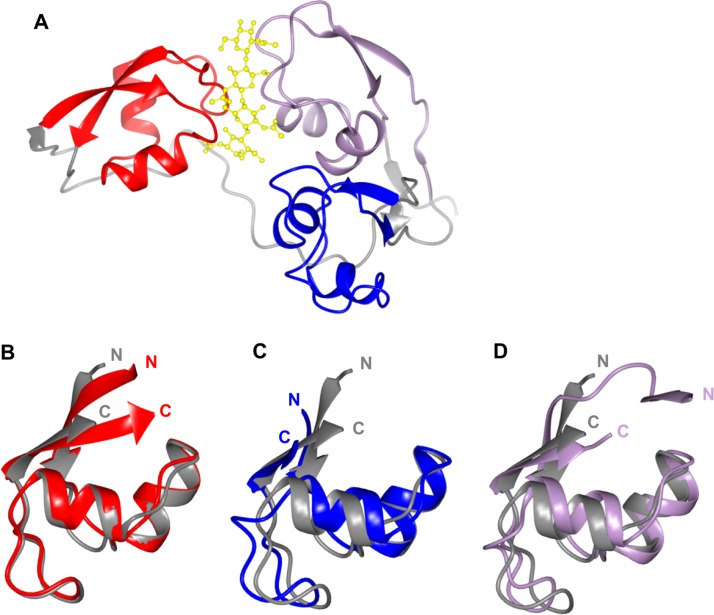
Chitin is a major component of fungal cell walls, and the detection of this homopolymer in the apoplast is used by plants as a strategy for initiating immune responses (51). Plants detect chitin-derived oligosaccharides via cell surface receptors that contain extracellular lysine motif (LysM) domains. Plant LysM domains comprise ∼50 amino acids and adopt a βααβ structural fold (52, 53) (Fig. 1). To protect themselves from detection by the plant immune system, fungi use LysM effectors to sequester chitin oligomers in the apoplast, outcompeting binding by host receptor domains. The crystal structure of Cladosporium fulvum Ecp6 confirmed that this protein contained 3 modular LysM domains (54) (Fig. 1 and Table 2). In a strategy to deliver high-affinity ligand interactions, two of the Ecp6 LysM domains (LysM1 and LysM3) dimerize to “sandwich” a chitin oligomer in a groove via multiple hydrogen bonds and hydrophobic interactions (Fig. 1A). To date, this ligand-induced LysM dimerization to increase binding affinity is unique to Ecp6 and highlights the propensity of pathogen effectors to adapt protein folds to acquire new activities (51). Interestingly, the ligand-binding capability of the LysM2 domain of Ecp6 was also shown to interfere with chitin-triggered immunity in planta, but the underlying mechanistic basis remains unclear (55).
FIG 1.
The crystal structure of the LysM effector Ecp6 shows how modularity can be used by effectors to generate new functions (the three LysM domains are shown in red, blue, and lilac, respectively). (A) Two Ecp6 LysM domains combine to bind to a chitin oligomer (shown in yellow). (B to D) Superposition of the Ecp6 LysM domains on the plant (rice) LysM receptor protein MoCVNH3 (in gray) (LysM domains are colored as described above). The amino (N) and carboxyl (C) termini of the proteins are labeled.
TABLE 2.
Effectors of filamentous plant pathogens that have had their structures determined
| Protein | Origin | Targeted processc | Immune receptor(s) | Fold | Comparison to known structure |
PDB accession no. | Reference | |
|---|---|---|---|---|---|---|---|---|
| RMSD (Å) (no. of residues in overlay)a | Sequence identity (%) | |||||||
| Avr3a11 | P. capsici | Unknown | WY | ND | ND | 3ZR8 | 74 | |
| Avr3a4 | P. capsici | Unknown | WY | 1.26 (42) | 79.0 | 2LC2 | 77 | |
| PexRD2 | P. infestans | MAPKKKε-mediated immune signaling | WY | 1.41 (40) | 27.8 | 3ZRG | 74 | |
| PexRD54 | P. infestans | Autophagy | WY | 1.73 (41) | 20.0 | 5L7S | 78 | |
| ATR1 | H. arabidopsidis | Unknown | RPP1 | WY | 2.37 (36) | 23.7 | 3RMR | 76 |
| AvrL567-D | M. lini | Unknown | L6 | ToxA-like | 2.74 (82) | 22.2 | 2QVT | 105 |
| AvrL567-A | M. lini | Unknown | L5 and L6 | ToxA-like | 2.58 (81) | 19.7 | 2OPC | 105 |
| avrM | M. lini | Unknown | WY-like | ND | 26.1 | 4BJM | 106 | |
| AvrM-A | M. lini | Unknown | M | WY-like | ND | 23.9 | 4BJN | 106 |
| AVR-PikD (in complex) | M. oryzae | Unknown | Pik1/Pik2 | MAX | ND | ND | 5A6W | 82 |
| Avr1-CO39 | M. oryzae | Unknown | RGA5/RGA4 | MAX | 1.36 (55) | 17.2 | 2MYV | 80 |
| AVR-Pia | M. oryzae | Unknown | RGA5/RGA4 | MAX | 2.24 (52) | 16.4 | 2MYW | 80 |
| AVR-Pizt | M. oryzae | E3 ligase-mediated immunity | Piz-t | MAX | 2.33 (58) | 15.6 | 2LW6 | 84 |
| Avr4 | P. fuligena | Chitin-mediated immunity/fungally derived chitin perception | Cf-4 | CBM14-like | 1.98 (52) | 22.2 | 4Z4A | 61 |
| Ecp6 | C. fulvum | Chitin-mediated immunity/fungally derived chitin perception | LysM1 | 0.8 (45) | 35.9 | 4B8V | 54 | |
| Ecp6 | C. fulvum | LysM2 | 1.17 (43) | 37.1 | 4B8V | 54 | ||
| Ecp6 | C. fulvum | LysM3 | 1.51 (45) | 20.8 | 4B8V | 54 | ||
| AvrLm4-7 | L. maculans | Production of plant hormones and hydrogen peroxide/plant hormone-mediated immunity | Rlm4 and Rlm7 | Unique | ND | ND | 4FPR | 110 |
| ToxA | P. tritici-repentis | Photosynthesis | Tsn1b | ToxA-like | ND | ND | 1ZLE | 103 |
| ToxB | P. tritici-repentis | Photosynthesis | MAX | 2.25 (58) | 25.4 | 2MM0 | 81 | |
| toxb | P. tritici-repentis | Inactive allele | MAX | 2.33 (57) | 19.7 | 2MM2 | 81 | |
| NLP | P. aphanidermatum | Plasma membrane integrity | Actinoporin-like | 2.34 (68) | 21.9 | 3GNZ | 64 | |
| NLP | M. perniciosa | Plasma membrane integrity | Actinoporin-like | 2.24 (68) | 19.3 | 3ST1 | 70 | |
Template proteins used for comparison are Avr3a11 (WY and WY-like), AVR-PikD (MAX), tachycitin (CBM14-like), MoCVNH3 (LysM), ToxA (ToxA-like), and sticholysin II (actinoporin-like). RMSD, root mean square deviation; ND, not determined (either to avoid comparison with self or because the comparison is not meaningful).
Tsn1 is a susceptibility factor.
MAPKKKε, mitogen-activated protein kinase kinase kinase ε.
Multidomain LysM effectors are also found in other fungal plant pathogens, including the wheat pathogen Zymoseptoria tritici and the rice blast pathogen Magnaporthe oryzae, suggesting that they represent a widespread mechanism for the suppression of detection by the plant immune system. However, unlike Ecp6, Z. tritici LysM effectors protect fungal hyphae against hydrolysis by host chitinases, although the mechanism by which they achieve this is not understood (55).
CBM14-Like Avr4 Effectors
In a second strategy to evade chitin-mediated recognition by the plant immune system, fungi can secrete effector proteins that bind to chitin in their cell wall and prevent the action of host chitinases in generating chito-oligosaccharide fragments. The Cladosporium fulvum effector Avr4 was predicted to adopt a carbohydrate-binding module family 14 (CBM14)-like structure, based on its disulfide bond pattern, and in vitro, Avr4 protects chitin from hydrolysis by plant chitinases (56, 57). CBM14 proteins are defined as having chitin-binding activity, with one being characterized as having antimicrobial properties (58). The structure of the CBM14 member tachycitin, from the horseshoe crab Tachypleus tridentatus, revealed a distorted β-sandwich fold flanked by short loops and turns, stabilized by disulfide bonds (59). Tachycitin was described as sharing some structural similarity to a domain found in the plant chitin-binding protein hevein (60).
Avr4 homologues are found in a number of plant-pathogenic fungal species. Recently, the crystal structure of Avr4 from the tomato pathogen Pseudocercospora fuligena confirmed that the Avr4 family of effectors adopts the CBM14-like fold (Fig. 2), and this enabled the investigation of structure-function relationships in chitin binding by these proteins (61). As predicted for tachycitin, the chitin-binding site of Avr4 is located between two β-strands and the connecting β-hairpin and is mediated by aromatic amino acids and adjacent polar residues (Fig. 2).
FIG 2.
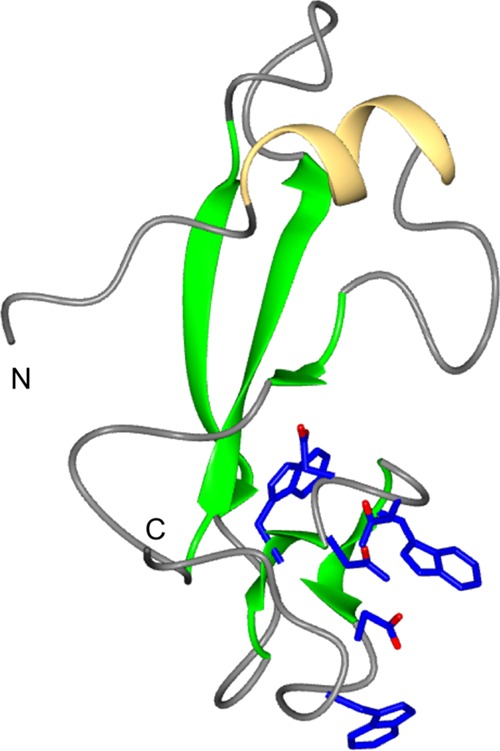
CBM14 family structure of P. fuligena Avr4. The structures comprise an alpha helix (yellow) and five beta strands (green). The residues predicted to be involved in the interaction with chitin are shown in blue.
The evolutionary dynamics of CBM14 family proteins are complex (62). While chitin binding is a critical feature of this fold for fungal defense against the plant immune system, it is clear that other functions can be attributed to the wider family given that CBM14 proteins occur in nonpathogenic species and were previously shown to have antimicrobial properties.
NLPs
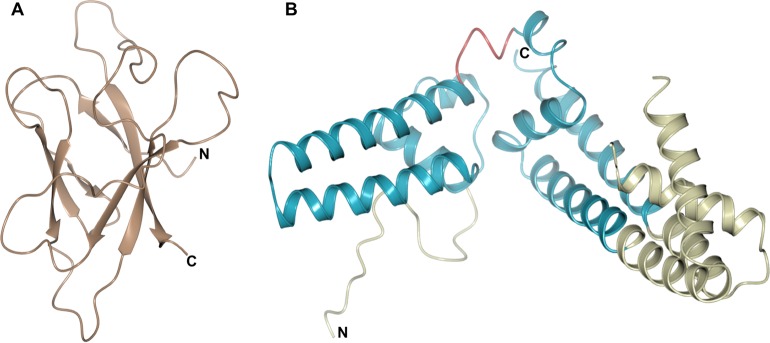
NLPs (necrosis- and ethylene-inducing peptide 1-like proteins) are a large family of secreted proteins found in plant-associated fungi, oomycetes, and bacteria. NLPs were initially characterized by their ability to induce necrotic cell death in dicotyledonous plants (63), which is thought to be dependent on toxin-induced host cell damage (64). However, it is now well established that not all NLPs share this activity (65, 66). Despite this, both cytotoxic and noncytotoxic NLPs can trigger cell surface-dependent immune responses in plant cells, and this activity has been localized to a 24-amino-acid peptide (67, 68) recognized by a receptor complex comprising RLP23/SOBIR-1/BAK1 (69). Clues to the mechanism of the cytolytic activity of NLPs came from the crystal structures of NLPs from Pythium aphanidermatum and Moniliophthora perniciosa (Fig. 3), which showed that this family of proteins shares a fold with the actinoporin pore-forming toxin stichoysin (64, 70). However, there is no experimental evidence for pore-forming activity by NLPs, and their toxicity may be the result of the NLP-induced release of membrane damage factors that are then sensed by the plant (68). Interestingly, the 24-amino-acid peptide, which acts as a MAMP for the activation of plant immunity, is largely buried within the core of the intact structure, with only a small number of residues being displayed on the surface (67). This suggests that the protein is probably unfolded and/or digested for recognition by the receptor.
FIG 3.
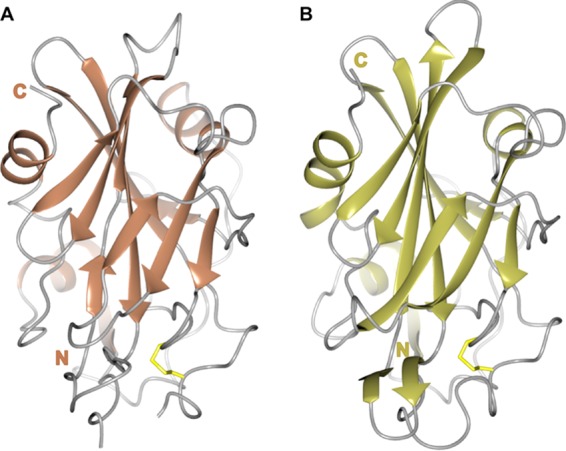
Crystal structures of the NLP family members NLPPya (A) and MpNEP2 (B), showing the central β-sandwich surrounded by 3 helices. The conserved structural elements are shown in a cartoon representation, with residues contributing to disulfide bridges shown as sticks (in yellow) and loops shown in gray.
THE THREE-DIMENSIONAL STRUCTURES OF EFFECTORS OF FILAMENTOUS PLANT PATHOGENS SHOW CONSERVED FOLDS WITHIN FAMILIES
Oomycete Effectors and the WY Fold
The RXLR class of host-translocated oomycete effector proteins is defined by the presence of a conserved N-terminal RXLR motif and a diverse C-terminal domain that exerts effector activity inside the host cell (16, 71, 72). Analysis of the sequences of the RXLR repertoires of Phytophthora sojae and Phytophthora ramorum identified conserved motifs, which were named “W” (Trp), “Y” (Tyr), and “L” (Leu), after the single-letter amino acid code for a highly conserved residue in each sequence (73). Protein structural analysis subsequently revealed that the amino acids at the conserved W and Y positions were buried in the hydrophobic core of a three-α-helical bundle and stacked against one another in an energetically favorable interaction (74) (Fig. 4). Intriguingly, except for the Hyaloperonospora arabidopsidis effector ATR13 (75), all of the structures of oomycete RXLR effectors that have been determined to date adopt the “WY domain” fold. Nonetheless, these proteins display significant primary sequence differences. They also show diverse structural adaptations, including N- and C-terminal extensions, loop regions, and domain duplication, that give rise to very different overall structures (74, 76–78) (Fig. 4). Hidden Markov model (HMM) sequence searches, based on the knowledge of the WY domain structure, predicted that nearly half of the RXLR effector complement of Phytophthora species would adopt this fold (74).
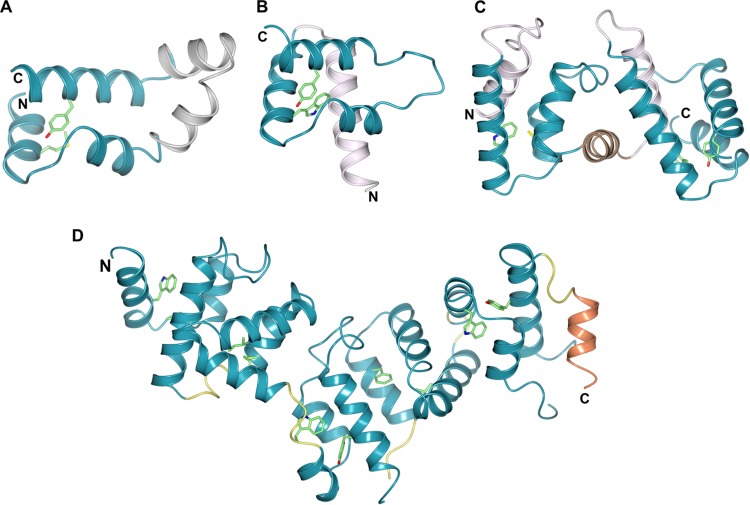
FIG 4.
The structures of oomycete WY domain effectors reveal how modularity and domain repeats give rise to different overall structures. For each panel, the region of the protein comprising the WY domain fold is shown in blue, and the residues at the W and Y positions are shown as sticks (green carbon atoms). Shown are PexRD2 (monomer) (A), Avr3a11 (Avr3a4 is essentially identical and not shown) (B), ATR1 (the region toward the N terminus that does not form a WY domain is not shown) (C), and PexRD54 (D), with amino (N) and carboxyl (C) termini labeled. Avr3a11/4 and ATR1 carry an additional N-terminal helix (pink). The tandem WY domains of ATR1 and PexRD54 are separated by a helix (brown) in ATR1 and loops (yellow) in PexRD54. PexRD54 carries a short helix (coral) at the C-terminal end prior to the ATG8-interacting motif (AIM) (not shown, as it was disordered in the crystals). All structure figures were prepared with ccp4 mg (111).
The structure of the P. infestans effector PexRD2 is comprised of five α-helices, three of which contribute to the WY domain three-α-helical bundle (Fig. 4A). The additional helices (present between two helices of the core WY domain) are instrumental in forming an extensive homodimeric interface in the PexRD2 structure, consistent with the observation that PexRD2 self-associates in planta. The structures of Phytophthora capsici AVR3a4 and AVR3a11 comprise monomeric four-helical bundles (Fig. 4B), with an N-terminal helical extension to the WY domain fold (74). It is possible that the N-terminal helix is important for maintaining the stability of monomeric, single-WY-domain proteins, although this has not been explicitly tested.
The HMM-based sequence searches mentioned above revealed that these effectors could also comprise tandemly repeated WY domains encoded by a single gene. The first crystal structure of a tandem WY domain effector was that of ATR1 from Hyaloperonospora arabidopsidis (76) (Fig. 4C). In ATR1, two WY domains (each with an N-terminal helical extension) are connected through an additional helix, which acts as a linker. Recently, the crystal structure of PexRD54 revealed how five WY domains can pack together in a stable structure with diverse domain-domain interactions (78) (Fig. 4D). Within each of these tandem WY domain structures, the individual domains can be overlaid with high confidence despite limited sequence identity (76, 78). Interestingly, PexRD54 employs a short linear motif known as the ATG8-interacting motif (AIM) to engage a host protein and to exert its virulence activity (79). The AIM is presented at the C terminus of PexRD54 and is linked to the last WY domain via a short helix. The structure of PexRD54 suggests that one function of tandem WY domains is to serve as a scaffold to present functional motifs for interaction with host proteins.
The WY domain fold serves as a chassis for the evolution of novel functions in oomycete effectors while maintaining their structural integrity. The fold presents a flexible platform that supports effector evolution and diversification via the acquisition of short linear motifs, domain duplications, and dimerization. Thus, the WY domain structure is not predictive of the precise function of the effectors but appears to provide enough plasticity for the effectors to bind different host proteins and evolve unrelated activities inside host cells.
MAX Effectors of Magnaporthe
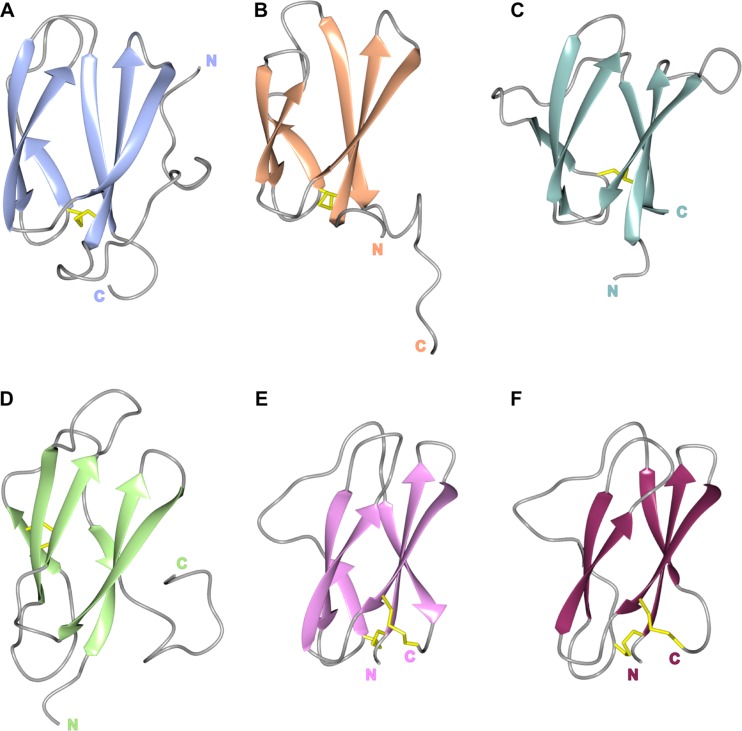
Recently, a new family of effectors of filamentous plant pathogens has been described, which also shares a conserved common structure but displays a diverse protein sequence. The Magnaporthe Avrs and ToxB-like (MAX) family was defined following structural work on effectors from the fungal pathogen M. oryzae, the causal agent of rice blast disease (80). Despite typically sharing less than 25% sequence identity, each member of this family that has had a structure determined (80–84) shares a characteristic six-stranded β-sandwich fold (Fig. 5). This fold is stabilized by at least one disulfide bond, generally with Cys residues present in β1 and in, or immediately before, β5. In most cases, one of the β-sheets is formed by strands β1, β2, and β6, and the second is formed by strands β3, β4, and β5. The length and orientation of the different structural elements are variable, in particular for strand β5 and for the various connecting loops, giving rise to proteins with distinct shapes and surface properties (80). In addition, the M. oryzae effector AVR-PikD contains an N-terminal extension to the six-stranded β-sandwich structure (Fig. 5A), and this region contains polymorphic residues that contribute to the evasion of recognition by the plant innate immune system (82, 85). Interestingly, the M. oryzae effectors AVR-Pik, AVR-Pia, and AVR1-CO39 all bind to heavy metal-associated (HMA) domains that have been integrated in intracellular plant immune receptors (NLRs) throughout evolution. This suggests that the conserved MAX effector family fold is well suited to interact with such domains and may suggest a putative virulence target in host cells for these effectors.
FIG 5.
The structures of MAX effectors reveal the shared β-sandwich fold. The conserved β-strands are shown in a cartoon representation for each protein, with residues contributing to disulfide bridges shown as sticks (in yellow) and loops in gray. Shown are AVR-PikD (A), AVR1-CO39 (B), AVR-Pia (C), AVR-Pizt (D), ToxB (E), and toxb (F), with amino (N) and carboxyl (C) termini labeled.
Intriguingly, the MAX effector family includes ToxB, a proteinaceous toxin from the fungus Pyrenophora tritici-repentis (86). This toxin shares the common three-dimensional structure of MAX effectors (Fig. 5E and F), but its mode of action is unclear, and no interacting partner has been identified. However, the N-terminal region of ToxB has been shown to be essential for activity, while both the central and C-terminal parts are required for full activity (87), suggesting that the conserved structure is important for function. A naturally occurring nontoxic version of ToxB (toxb) shares 78% sequence identity with the active protein. These proteins share essentially the same structure, although toxb may overall be less stable than ToxB (81).
PSI-BLAST followed by HMM-based profile searches revealed that the majority of MAX effectors are found in Magnaporthe species (80). However, a small number of hits were detected in other fungal species such as Colletotrichum (80). Thus, the discovery of the MAX effectors enables a more robust prediction of candidate effectors in these fungal pathogens.
RALPH Effectors of Powdery Mildew
Nearly 500 candidate effectors of the barley powdery mildew fungus Blumeria graminis f. sp. hordei were predicted from the genome sequence using bioinformatic tools by searching for genes with characteristics of effectors, particularly those encoding small secreted proteins. Many of these candidate effectors have been shown to be expressed during infection (88–90).
To further characterize B. graminis candidate effectors, their sequences were subjected to structural annotation using protein fold recognition methods. A subset of these candidate effectors are predicted to have structural similarities with ribonucleases and were named RALPHs (RNase-like proteins expressed in haustoria) (91). Although confirmation that RALPHs adopt RNase-like folds awaits the determination of an experimentally derived structure, it is intriguing that many B. graminis effectors may share a structural scaffold with each other, a feature common in other families of effectors of filamentous plant pathogens. In another parallel with the MAX effectors, RALPHs have been predicted to contain a disulfide bond, with Cys residues being largely conserved toward both the N terminus (contained within a “YxC” motif) and the C terminus of the proteins.
Recently, data have emerged showing that RALPH effectors function as both virulence and avirulence determinants in B. graminis-barley and -wheat interactions. Using host-induced gene silencing, five RALPHs were shown to be involved in the formation of haustoria (92, 93). AVRA1 and AVRA13 were shown to be required for disease resistance in barley mediated by the powdery mildew resistance loci Mla1 and Mla13, respectively (94), and AvrPm2 was recently cloned as the cognate effector of the wheat Pm2 gene (95). Furthermore, the B. graminis f. sp. tritici effector SvrPm3a1/f1 (formerly Bcg1vir) has been shown to suppress avirulence triggered by the interaction of effector AvrPm3a2/f2 (svrPm3a1/f1, formerly Bcg1avr) with its receptor Pm3a/f (96, 97). As with other host-translocated effectors, the ability of RALPHs to activate plant immune responses may help explain the strong diversifying selection seen in these proteins.
STRUCTURES OF OTHER NOTABLE EFFECTORS OF FILAMENTOUS PLANT PATHOGENS
Flax Rust Effectors Show Divergent Structures
Melampsora lini causes rust disease on crop plants such as flax and linseed. Genomic analyses of M. lini predicted that this fungus has a large repertoire of putative effector proteins (22). Unlike oomycete RXLR and CRN effectors, but similar to effectors from other fungal species, no widely conserved sequence-based motifs have been identified for flax rust effectors thus far. To date, six M. lini effector proteins have been validated experimentally, based on their avirulence activity (AvrL567, AvrM, AvrP4, AvrP123, AvrL2, and AvrM14) (48, 98–101). These effectors trigger specific immune responses mediated by NLRs in the host cell. AvrL567, AvrM, and their cognate NLRs exhibit polymorphisms giving rise to allelic variants of the effector and receptor with specific recognition profiles (98, 102). For example, AvrL567-A is recognized by the NLRs L5 and L6, whereas AvrL567-D is recognized by L6 but not L5.
Crystal structures of the AvrL567 alleles AvrL567-D and AvrL567-A revealed that the two proteins share the same architecture, adopting a β-sandwich fold comprising seven antiparallel β-strands (Fig. 6A). Interestingly, the structures share some homology with ToxA (103), a host-selective toxin of Pyrenophora tritici-repentis which induces cell death in sensitive wheat cultivars. ToxA was described as having a distant relationship with mammalian fibronectin proteins, and an Arg-Glu-Asp (RGD) motif was found in a loop region of the protein that may mediate interactions with plant cell integrin-like receptors (103). This motif was subsequently shown to be required for protein internalization (104), although the precise mechanism remains unclear. AvrL567 lacks the RGD motif, implying that it is internalized by a different mechanism. Both AvrL567-D and -A display two positively charged patches on the protein surface and have been shown to bind nucleic acid in vitro (105). However, the biological relevance of nucleic acid binding remains unknown. Structure-led mutagenesis revealed that multiple contacts mediate the interaction between AvrL567 alleles and their cognate receptors (105).
FIG 6.
Divergent structures obtained for flax rust effectors. (A) Cartoon representation of AvrL567-A (the D allele is essentially identical and not shown), showing the β-sandwich fold. (B) Cartoon diagram of avrM, where the helical repeats, which have some resemblance to the oomycete WY domain fold, are shown in blue and separated by a loop (red). The amino (N) and carboxyl (C) termini of the proteins are labeled.
Crystal structures of C-terminal domains of two allelic variants of AvrM (AvrM-A and avrM) revealed an L-shaped α-helical fold comprising two helical repeats (106) (Fig. 6B). The structural repeat, another example of modularity in effectors of filamentous plant pathogens, was not evident from sequence analysis and was revealed only after the structure was determined.
AvrLm4-7, a Lone Effector Structure with a Novel Fold
AvrLm4-7 is a Cys-rich protein that is recognized by oilseed rape cultivars harboring Rlm4 and Rlm7 resistances (107). The loss of AvrLm4-7 in the pathogen strongly impacts pathogen fitness (108, 109). The crystal structure of AvrLm4-7 does not share significant homology with other structures in the Protein Data Bank, and as such, it has proven challenging to infer putative protein function (110). The crystal structure identified the positions of the four disulfide bonds in the protein, which, as for other effectors, are probably involved in stabilizing the structure. In addition, a strongly positive patch was identified on the protein surface, which may represent a functionally relevant surface of the protein, although it has not been possible to show that this region binds a negatively charged ligand. A single amino acid polymorphism that perturbs the recognition of the effector by Rlm4 is located on a loop of the protein, exposed to the surface. It is therefore unlikely that this polymorphism affects the overall structure of the protein, but it may be important for a specific recognition site.
CONCLUSION
The high complexity of the secretomes of filamentous plant pathogens points to a multitude of independent evolutionary pathways to generate effector proteins that target a diversity of host molecules and processes. However, despite this extraordinary sequence diversity, it is now evident that some conserved protein folds, such as the WY and MAX domains, define widespread families of effector proteins that occur across different plant pathogen taxa. There are both practical and theoretical implications of this finding. Structure-guided sequence similarity searches enable more precise and sensitive annotation of effector catalogues, notably of fungal effectors, which have proven more difficult to annotate than their oomycete counterparts. This should enable prioritization of effectors for further study, thus accelerating their functional characterization. In addition, the conserved structures provide a framework to unravel how the rapid evolution of effector proteins has resulted in new host targeting activities and tease out the physical and physiological constraints that these proteins face. In this regard, the next phase of research should go beyond the analyses of individual structures of effectors of individual filamentous pathogens and consider the structures of effectors in complex with host proteins (78, 82). In the future, we need to further improve our understanding of the biophysical properties of effector-host protein complexes to gain comprehensive knowledge of effector structures and functions.
ACKNOWLEDGMENTS
M.J.B. is supported by the BBSRC (UK) (relevant grants J004553 and M02198), the ERC (proposals 294608 [acronym NGRB] and SEP-210218966 [acronym ImmunityByPairDesign]), and the John Innes Foundation. S.K. is funded by the Biotechnology and Biological Sciences Research Council, the European Research Council (NGRB), and the Gatsby Charitable Foundation.
We thank Amey Redkar for discussions.
REFERENCES
- 1.Pennisi E. 2010. Armed and dangerous. Science 327:804–805. doi: 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- 2.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer V, Andersen MR, Brakhage AA, Braus GH, Caddick MX, Cairns TC, de Vries RP, Haarmann T, Hansen K, Hertz-Fowler C, Krappmann S, Mortensen UH, Peñalva MA, Ram AFJ, Head RM. 2016. Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: a white paper. Fungal Biol Biotechnol 3:6. doi: 10.1186/s40694-016-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erwin DC, Ribeiro OK. 1996. Phytophthora diseases worldwide. APS Press, St Paul, MN. [Google Scholar]
- 5.Fry WE, Goodwin SB. 1997. Re-emergence of potato and tomato late blight in the United States. Plant Dis 81:1349–1357. doi: 10.1094/PDIS.1997.81.12.1349. [DOI] [PubMed] [Google Scholar]
- 6.Hovmoller MS, Walter S, Justesen AF. 2010. Escalating threat of wheat rusts. Science 329:369. doi: 10.1126/science.1194925. [DOI] [PubMed] [Google Scholar]
- 7.Islam MT, Croll D, Gladieux P, Soanes DM, Persoons A, Bhattacharjee P, Hossain MS, Gupta DR, Rahman MM, Mahboob MG, Cook N, Salam MU, Surovy MZ, Sancho VB, Maciel JL, NhaniJunior A, Castroagudin VL, Reges JT, Ceresini PC, Ravel S, Kellner R, Fournier E, Tharreau D, Lebrun MH, McDonald BA, Stitt T, Swan D, Talbot NJ, Saunders DG, Win J, Kamoun S. 2016. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol 14:84. doi: 10.1186/s12915-016-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodds PN, Rafiqi M, Gan PHP, Hardham AR, Jones DA, Ellis JG. 2009. Effectors of biotrophic fungi and oomycetes: pathogenicity factors and triggers of host resistance. New Phytol 183:993–999. doi: 10.1111/j.1469-8137.2009.02922.x. [DOI] [PubMed] [Google Scholar]
- 9.Giraldo MC, Valent B. 2013. Filamentous plant pathogen effectors in action. Nat Rev Microbiol 11:800–814. doi: 10.1038/nrmicro3119. [DOI] [PubMed] [Google Scholar]
- 10.Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R. 2015. Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66:513–545. doi: 10.1146/annurev-arplant-043014-114623. [DOI] [PubMed] [Google Scholar]
- 11.Kamoun S. 2006. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol 44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 12.Jones JDG, Vance RE, Dangl JL. 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354(6316):aaf6395. doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- 13.Torto TA, Li SA, Styer A, Huitema E, Testa A, Gow NAR, van West P, Kamoun S. 2003. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res 13:1675–1685. doi: 10.1101/gr.910003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyler BM, Tripathy S, Zhang XM, Dehal P, Jiang RHY, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CMB, Dorrance AE, Dou DL, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee MK, McDonald WH, Medina M, Meijer HJG, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JKC, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BWS, Terry A, Torto-Alalibo TA, Win J, Xu ZY, Zhang HB, et al. 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 15.Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HAB, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrugge M, Basse CW, Steinberg G, Ibeas JI, Holloman W, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho ECH, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li WX, et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 16.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AMV, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JIB, Bulone V, Cai GH, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, Fischbach MA, Fugelstad J, Gilroy EM, Gnerre S, Green PJ, Grenville-Briggs LJ, Griffith J, Grunwald NJ, Horn K, Horner NR, Hu CH, Huitema E, Jeong DH, Jones AME, Jones JDG, Jones RW, Karlsson EK, Kunjeti SG, Lamour K, Liu ZY, et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 17.Levesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, Zerillo MM, Beakes GW, Boore JL, Busam D, Dumas B, Ferriera S, Fuerstenberg SI, Gachon CMM, Gaulin E, Govers F, Grenville-Briggs L, Horner N, Hostetler J, Jiang RHY, Johnson J, Krajaejun T, Lin HN, Meijer HJG, Moore B, Morris P, Phuntmart V, Puiu D, Shetty J, Stajich JE, Tripathy S, Wawra S, van West P, Whitty BR, Coutinho PM, Henrissat B, Martin F, Thomas PD, Tyler BM, De Vries RP, Kamoun S, Yandell M, Tisserat N, Buell CR. 2010. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol 11:R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duplessis S, Cuomo CA, Lin YC, Aerts A, Tisserant E, Veneault-Fourrey C, Joly DL, Hacquard S, Amselem J, Cantarel BL, Chiu R, Coutinho PM, Feau N, Field M, Frey P, Gelhaye E, Goldberg J, Grabherr MG, Kodira CD, Kohler A, Kues U, Lindquist EA, Lucas SM, Mago R, Mauceli E, Morin E, Murat C, Pangilinan JL, Park R, Pearson M, Quesneville H, Rouhier N, Sakthikumar S, Salamov AA, Schmutz J, Selles B, Shapiro H, Tanguay P, Tuskan GA, Henrissat B, Van de Peer Y, Rouze P, Ellis JG, Dodds PN, Schein JE, Zhong SB, Hamelin RC, Grigoriev IV, Szabo LJ, Martin F. 2011. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A 108:9166–9171. doi: 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders DGO, Win J, Cano LM, Szabo LJ, Kamoun S, Raffaele S. 2012. Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS One 7:e29847. doi: 10.1371/journal.pone.0029847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stergiopoulos I, Kourmpetis YAI, Slot JC, Bakker FT, de Wit PJGM, Rokas A. 2012. In silico characterization and molecular evolutionary analysis of a novel superfamily of fungal effector proteins. Mol Biol Evol 29:3371–3384. doi: 10.1093/molbev/mss143. [DOI] [PubMed] [Google Scholar]
- 21.Cantu D, Segovia V, MacLean D, Bayles R, Chen XM, Kamoun S, Dubcovsky J, Saunders DGO, Uauy C. 2013. Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics 14:270. doi: 10.1186/1471-2164-14-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemri A, Saunders DGO, Anderson C, Upadhyaya NM, Win J, Lawrence GJ, Jones DA, Kamoun S, Ellis JG, Dodds PN. 2014. The genome sequence and effector complement of the flax rust pathogen Melampsora lini. Front Plant Sci 5:98. doi: 10.3389/fpls.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guyon K, Balague C, Roby D, Raffaele S. 2014. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15:336. doi: 10.1186/1471-2164-15-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sperschneider J, Gardiner DM, Dodds PN, Tini F, Covarelli L, Singh KB, Manners JM, Taylor JM. 2016. EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol 210:743–761. doi: 10.1111/nph.13794. [DOI] [PubMed] [Google Scholar]
- 25.Illergard K, Ardell DH, Elofsson A. 2009. Structure is three to ten times more conserved than sequence—a study of structural response in protein cores. Proteins 77:499–508. doi: 10.1002/prot.22458. [DOI] [PubMed] [Google Scholar]
- 26.Wirthmueller L, Maqbool A, Banfield MJ. 2013. On the front line: structural insights into plant-pathogen interactions. Nat Rev Microbiol 11:761–776. doi: 10.1038/nrmicro3118. [DOI] [PubMed] [Google Scholar]
- 27.Stebbins CE. 2005. Structural microbiology at the pathogen-host interface. Cell Microbiol 7:1227–1236. doi: 10.1111/j.1462-5822.2005.00564.x. [DOI] [PubMed] [Google Scholar]
- 28.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. 2006. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 29.Jeong BR, Lin Y, Joe A, Guo M, Korneli C, Yang H, Wang P, Yu M, Cerny RL, Staiger D, Alfano JR, Xu Y. 2011. Structure function analysis of an ADP-ribosyltransferase type III effector and its RNA-binding target in plant immunity. J Biol Chem 286:43272–43281. doi: 10.1074/jbc.M111.290122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer AU, Desveaux D, Betts L, Chang JH, Nimchuk Z, Grant SR, Dangl JL, Sondek J. 2004. Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure 12:1669–1681. doi: 10.1016/j.str.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Zhu M, Shao F, Innes RW, Dixon JE, Xu Z. 2004. The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site. Proc Natl Acad Sci U S A 101:302–307. doi: 10.1073/pnas.2036536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hann DR, Rathjen JP. 2010. The long and winding road: virulence effector proteins of plant pathogenic bacteria. Cell Mol Life Sci 67:3425–3434. doi: 10.1007/s00018-010-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodwin SB, M'Barek SB, Dhillon B, Wittenberg AHJ, Crane CF, Hane JK, Foster AJ, Van der Lee TAJ, Grimwood J, Aerts A, Antoniw J, Bailey A, Bluhm B, Bowler J, Bristow J, van der Burgt A, Canto-Canche B, Churchill ACL, Conde-Ferraez L, Cools HJ, Coutinho PM, Csukai M, Dehal P, De Wit P, Donzelli B, van de Geest HC, Van Ham RCHJ, Hammond-Kosack KE, Henrissat B, Kilian A, Kobayashi AK, Koopmann E, Kourmpetis Y, Kuzniar A, Lindquist E, Lombard V, Maliepaard C, Martins N, Mehrabi R, Nap JPH, Ponomarenko A, Rudd JJ, Salamov A, Schmutz J, Schouten HJ, Shapiro H, Stergiopoulos I, Torriani SFF, Tu H, de Vries RP, et al. 2011. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet 7:e1002070. doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, Altmuller J, Alvarado-Balderrama L, Bauser CA, Becker C, Birren BW, Chen ZH, Choi J, Crouch JA, Duvick JP, Farman MA, Gan P, Heiman D, Henrissat B, Howard RJ, Kabbage M, Koch C, Kracher B, Kubo Y, Law AD, Lebrun MH, Lee YH, Miyara I, Moore N, Neumann U, Nordstrom K, Panaccione DG, Panstruga R, Place M, Proctor RH, Prusky D, Rech G, Reinhardt R, Rollins JA, Rounsley S, Schardl CL, Schwartz DC, Shenoy N, Shirasu K, Sikhakolli UR, Stuber K, et al. 2012. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jashni MK, Dols IHM, Iida Y, Boeren S, Beenen HG, Mehrabi R, Collemare J, de Wit PJGM. 2015. Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp. lycopersici cleaves chitin-binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Mol Plant Microbe Interact 28:996–1008. doi: 10.1094/MPMI-04-15-0074-R. [DOI] [PubMed] [Google Scholar]
- 36.Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B. 2000. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12:2019–2032. doi: 10.2307/3871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose JK, Ham KS, Darvill AG, Albersheim P. 2002. Molecular cloning and characterization of glucanase inhibitor proteins: coevolution of a counterdefense mechanism by plant pathogens. Plant Cell 14:1329–1345. doi: 10.1105/tpc.002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damasceno CM, Bishop JG, Ripoll DR, Win J, Kamoun S, Rose JK. 2008. Structure of the glucanase inhibitor protein (GIP) family from Phytophthora species suggests coevolution with plant endo-beta-1,3-glucanases. Mol Plant Microbe Interact 21:820–830. doi: 10.1094/MPMI-21-6-0820. [DOI] [PubMed] [Google Scholar]
- 39.Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S. 2004. A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem 279:26370–26377. doi: 10.1074/jbc.M400941200. [DOI] [PubMed] [Google Scholar]
- 40.Tian M, Benedetti B, Kamoun S. 2005. A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol 138:1785–1793. doi: 10.1104/pp.105.061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rooney HCE, van't Klooster JW, van der Hoorn RAL, Joosten MHAJ, Jones JDG, de Wit PJGM. 2005. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- 42.Tian MY, Win J, Song J, van der Hoorn R, van der Knaap E, Kamoun S. 2007. A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol 143:364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ilyas M, Horger AC, Bozkurt TO, van den Burg HA, Kaschani F, Kaiser M, Belhaj K, Smoker M, Joosten MH, Kamoun S, van der Hoorn RA. 2015. Functional divergence of two secreted immune proteases of tomato. Curr Biol 25:2300–2306. doi: 10.1016/j.cub.2015.07.030. [DOI] [PubMed] [Google Scholar]
- 44.Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, van der Hoorn RA, Kamoun S. 2009. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci U S A 106:1654–1659. doi: 10.1073/pnas.0809201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, Osorio S, Tohge T, Fernie AR, Feussner I, Feussner K, Meinicke P, Stierhof YD, Schwarz H, Macek B, Mann M, Kahmann R. 2011. Metabolic priming by a secreted fungal effector. Nature 478:395–398. doi: 10.1038/nature10454. [DOI] [PubMed] [Google Scholar]
- 46.Dong SM, Yin WX, Kong GH, Yang XY, Qutob D, Chen QH, Kale SD, Sui YY, Zhang ZG, Dou DL, Zheng XB, Gijzen M, Tyler BM, Wang YC. 2011. Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog 7:e1002353. doi: 10.1371/journal.ppat.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong GH, Zhao Y, Jing MF, Huang J, Yang J, Xia YQ, Kong L, Ye WW, Xiong Q, Qiao YL, Dong SM, Ma WB, Wang YC. 2015. The activation of Phytophthora effector Avr3b by plant cyclophilin is required for the Nudix hydrolase activity of Avr3b. PLoS Pathog 11:e1005139. doi: 10.1371/journal.ppat.1005139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson C, Khan MA, Catanzariti AM, Jack CA, Nemri A, Lawrence GJ, Upadhyaya NM, Hardham AR, Ellis JG, Dodds PN, Jones DA. 2016. Genome analysis and avirulence gene cloning using a high-density RADseq linkage map of the flax rust fungus, Melampsora lini. BMC Genomics 17:667. doi: 10.1186/s12864-016-3011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schornack S, van Damme M, Bozkurt TO, Cano LM, Smoker M, Thines M, Gaulin E, Kamoun S, Huitema E. 2010. Ancient class of translocated oomycete effectors targets the host nucleus. Proc Natl Acad Sci U S A 107:17421–17426. doi: 10.1073/pnas.1008491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Damme M, Bozkurt TO, Cakir C, Schornack S, Sklenar J, Jones AME, Kamoun S. 2012. The Irish potato famine pathogen Phytophthora infestans translocates the CRN8 kinase into host plant cells. PLoS Pathog 8:e1002875. doi: 10.1371/journal.ppat.1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Vallet A, Mesters JR, Thomma BPHJ. 2015. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol Rev 39:171–183. doi: 10.1093/femsre/fuu003. [DOI] [PubMed] [Google Scholar]
- 52.Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, Zhou JM, Chai J. 2012. Chitin-induced dimerization activates a plant immune receptor. Science 336:1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 53.Koharudin LM, Debiec KT, Gronenborn AM. 2015. Structural insight into fungal cell wall recognition by a CVNH protein with a single LysM domain. Structure 23:2143–2154. doi: 10.1016/j.str.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Vallet A, Saleem-Batcha R, Kombrink A, Hansen G, Valkenburg DJ, Thomma BPHJ, Mesters JR. 2013. Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife 2:e00790. doi: 10.7554/eLife.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kombrink A, Thomma BP. 2013. LysM effectors: secreted proteins supporting fungal life. PLoS Pathog 9:e1003769. doi: 10.1371/journal.ppat.1003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Burg HA, Westerink N, Francoijs KJ, Roth R, Woestenenk E, Boeren S, de Wit PJGM, Joosten MHAJ, Vervoort J. 2003. Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J Biol Chem 278:27340–27346. doi: 10.1074/jbc.M212196200. [DOI] [PubMed] [Google Scholar]
- 57.van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, de Wit PJ. 2006. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19:1420–1430. doi: 10.1094/MPMI-19-1420. [DOI] [PubMed] [Google Scholar]
- 58.Kawabata S, Nagayama R, Hirata M, Shigenaga T, Agarwala KL, Saito T, Cho J, Nakajima H, Takagi T, Iwanaga S. 1996. Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J Biochem 120:1253–1260. doi: 10.1093/oxfordjournals.jbchem.a021549. [DOI] [PubMed] [Google Scholar]
- 59.Suetake T, Tsuda S, Kawabata S, Miura K, Iwanaga S, Hikichi K, Nitta K, Kawano K. 2000. Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif. J Biol Chem 275:17929–17932. doi: 10.1074/jbc.C000184200. [DOI] [PubMed] [Google Scholar]
- 60.Andersen NH, Cao B, Rodriguezromero A, Arreguin B. 1993. Hevein: NMR assignment and assessment of solution-state folding for the agglutinin-toxin motif. Biochemistry 32:1407–1422. doi: 10.1021/bi00057a004. [DOI] [PubMed] [Google Scholar]
- 61.Kohler AC, Chen LH, Hurlburt N, Salvucci A, Schwessinger B, Fisher AJ, Stergiopoulos I. 2016. Structural analysis of an Avr4 effector ortholog offers insight into chitin binding and recognition by the Cf-4 receptor. Plant Cell 28:1945–1965. doi: 10.1105/tpc.15.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang TC, Stergiopoulos I. 2015. Inter- and intra-domain horizontal gene transfer, gain-loss asymmetry and positive selection mark the evolutionary history of the CBM14 family. FEBS J 282:2014–2028. doi: 10.1111/febs.13256. [DOI] [PubMed] [Google Scholar]
- 63.Pemberton CL, Salmond GPC. 2004. The Nep1-like proteins—a growing family of microbial elicitors of plant necrosis. Mol Plant Pathol 5:353–359. doi: 10.1111/j.1364-3703.2004.00235.x. [DOI] [PubMed] [Google Scholar]
- 64.Ottmann C, Luberacki B, Kufner I, Koch W, Brunner F, Weyand M, Mattinen L, Pirhonen M, Anderluh G, Seitz HU, Nurnberger T, Oecking C. 2009. A common toxin fold mediates microbial attack and plant defense. Proc Natl Acad Sci U S A 106:10359–10364. doi: 10.1073/pnas.0902362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cabral A, Oome S, Sander N, Kufner I, Nurnberger T, Van den Ackerveken G. 2012. Nontoxic Nep1-like proteins of the downy mildew pathogen Hyaloperonospora arabidopsidis: repression of necrosis-inducing activity by a surface-exposed region. Mol Plant Microbe Interact 25:697–708. doi: 10.1094/MPMI-10-11-0269. [DOI] [PubMed] [Google Scholar]
- 66.Dong SM, Kong GH, Qutob D, Yu XL, Tang JL, Kang JX, Dai TT, Wang H, Gijzen M, Wang YC. 2012. The NLP toxin family in Phytophthora sojae includes rapidly evolving groups that lack necrosis-inducing activity. Mol Plant Microbe Interact 25:896–909. doi: 10.1094/MPMI-01-12-0023-R. [DOI] [PubMed] [Google Scholar]
- 67.Oome S, Raaymakers TM, Cabral A, Samwel S, Bohm H, Albert I, Nurnberger T, Van den Ackerveken G. 2014. Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc Natl Acad Sci U S A 111:16955–16960. doi: 10.1073/pnas.1410031111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bohm H, Albert I, Oome S, Raaymakers TM, Van den Ackerveken G, Nurnberger T. 2014. A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog 10:e1004491. doi: 10.1371/journal.ppat.1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Albert I, Bohm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang HQ, Krol E, Grefen C, Gust AA, Chai JJ, Hedrich R, Van den Ackerveken G, Nurnberger T. 2015. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1:15140. doi: 10.1038/nplants.2015.140. [DOI] [PubMed] [Google Scholar]
- 70.Zaparoli G, Barsottini MRD, de Oliveira JF, Dyszy F, Teixeira PJPL, Barau JG, Garcia O, Costa AJ, Ambrosio ALB, Pereira GAG, Dias SMG. 2011. The crystal structure of necrosis- and ethylene-inducing protein 2 from the causal agent of cacao's witches' broom disease reveals key elements for its activity. Biochemistry 50:9901–9910. doi: 10.1021/bi201253b. [DOI] [PubMed] [Google Scholar]
- 71.Schornack S, Huitema E, Cano LM, Bozkurt TO, Oliva R, van Damme M, Schwizer S, Raffaele S, Chaparro-Garcia A, Farrer R, Segretin ME, Bos J, Haas BJ, Zody MC, Nusbaum C, Win J, Thines M, Kamoun S. 2009. Ten things to know about oomycete effectors. Mol Plant Pathol 10:795–803. doi: 10.1111/j.1364-3703.2009.00593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, Whisson SC, Kamoun S, Tyler BM, Birch PRJ, Beynon JL. 2005. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17:1839–1850. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang RHY, Tripathy S, Govers F, Tyler BM. 2008. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci U S A 105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boutemy LS, King SRF, Win J, Hughes RK, Clarke TA, Blumenschein TMA, Kamoun S, Banfield MJ. 2011. Structures of Phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. J Biol Chem 286:35834–35842. doi: 10.1074/jbc.M111.262303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leonelli L, Pelton J, Schoeffler A, Dahlbeck D, Berger J, Wemmer DE, Staskawicz B. 2011. Structural elucidation and functional characterization of the Hyaloperonospora arabidopsidis effector protein ATR13. PLoS Pathog 7:e1002428. doi: 10.1371/journal.ppat.1002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chou S, Krasileva KV, Holton JM, Steinbrenner AD, Alber T, Staskawicz BJ. 2011. Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci U S A 108:13323–13328. doi: 10.1073/pnas.1109791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yaeno T, Li H, Chaparro-Garcia A, Schornack S, Koshiba S, Watanabe S, Kigawa T, Kamoun S, Shirasu K. 2011. Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc Natl Acad Sci U S A 108:14682–14687. doi: 10.1073/pnas.1106002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maqbool A, Hughes RK, Dagdas YF, Tregidgo N, Zess E, Belhaj K, Round A, Bozkurt TO, Kamoun S, Banfield MJ. 2016. Structural basis of host autophagy-related protein 8 (ATG8) binding by the Irish potato famine pathogen effector protein PexRD54. J Biol Chem 291:20270–20282. doi: 10.1074/jbc.M116.744995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dagdas YF, Belhaj K, Maqbool A, Chaparro-Garcia A, Pandey P, Petre B, Tabassum N, Cruz-Mireles N, Hughes RK, Sklenar J, Win J, Menke F, Findlay K, Banfield MJ, Kamoun S, Bozkurt TO. 2016. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5:e10856. doi: 10.7554/eLife.10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Guillen K, Ortiz-Vallejo D, Gracy J, Fournier E, Kroj T, Padilla A. 2015. Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog 11:e1005228. doi: 10.1371/journal.ppat.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nyarko A, Singarapu KK, Figueroa M, Manning VA, Pandelova I, Wolpert TJ, Ciuffetti LM, Barbar E. 2014. Solution NMR structures of Pyrenophora tritici-repentis ToxB and its inactive homolog reveal potential determinants of toxin activity. J Biol Chem 289:25946–25956. doi: 10.1074/jbc.M114.569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maqbool A, Saitoh H, Franceschetti M, Stevenson CE, Uemura A, Kanzaki H, Kamoun S, Terauchi R, Banfield MJ. 2015. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4:e08709. doi: 10.7554/eLife.08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ose T, Oikawa A, Nakamura Y, Maenaka K, Higuchi Y, Satoh Y, Fujiwara S, Demura M, Sone T, Kamiya M. 2015. Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae. J Biomol NMR 63:229–235. doi: 10.1007/s10858-015-9979-7. [DOI] [PubMed] [Google Scholar]
- 84.Zhang ZM, Zhang X, Zhou ZR, Hu HY, Liu M, Zhou B, Zhou J. 2013. Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t. J Biomol NMR 55:219–223. doi: 10.1007/s10858-012-9695-5. [DOI] [PubMed] [Google Scholar]
- 85.Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Alaux L, Fournier E, Tharreau D, Terauchi R. 2012. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J 72:894–907. doi: 10.1111/j.1365-313X.2012.05110.x. [DOI] [PubMed] [Google Scholar]
- 86.Strelkov SE, Lamari L, Ballance GM. 1999. Characterization of a host-specific protein toxin (Ptr ToxB) from Pyrenophora tritici-repentis. Mol Plant Microbe Interact 12:728–732. doi: 10.1094/MPMI.1999.12.8.728. [DOI] [Google Scholar]
- 87.Betts MF, Manning VA, Cardwell KB, Pandelova I, Ciuffetti LM. 2011. The importance of the N-terminus for activity of Ptr ToxB, a chlorosis-inducing host-selective toxin produced by Pyrenophora tritici-repentis. Physiol Mol Plant Pathol 75:138–145. doi: 10.1016/j.pmpp.2011.03.002. [DOI] [Google Scholar]
- 88.Hacquard S, Kracher B, Maekawa T, Vernaldi S, Schulze-Lefert P, Ver Loren van Themaat E. 2013. Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc Natl Acad Sci U S A 110:E2219–E2228. doi: 10.1073/pnas.1306807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bindschedler LV, McGuffin LJ, Burgis TA, Spanu PD, Cramer R. 2011. Proteogenomics and in silico structural and functional annotation of the barley powdery mildew Blumeria graminis f. sp. hordei. Methods 54:432–441. doi: 10.1016/j.ymeth.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 90.Pedersen C, Ver Loren van Themaat E, McGuffin LJ, Abbott JC, Burgis TA, Barton G, Bindschedler LV, Lu X, Maekawa T, Wessling R, Cramer R, Thordal-Christensen H, Panstruga R, Spanu PD. 2012. Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13:694. doi: 10.1186/1471-2164-13-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spanu PD. 2015. RNA-protein interactions in plant disease: hackers at the dinner table. New Phytol 207:991–995. doi: 10.1111/nph.13495. [DOI] [PubMed] [Google Scholar]
- 92.Aguilar GB, Pedersen C, Thordal-Christensen H. 2016. Identification of eight effector candidate genes involved in early aggressiveness of the barley powdery mildew fungus. Plant Pathol 65:953–958. doi: 10.1111/ppa.12476. [DOI] [Google Scholar]
- 93.Pliego C, Nowara D, Bonciani G, Gheorghe DM, Xu R, Surana P, Whigham E, Nettleton D, Bogdanove AJ, Wise RP, Schweizer P, Bindschedler LV, Spanu PD. 2013. Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol Plant Microbe Interact 26:633–642. doi: 10.1094/MPMI-01-13-0005-R. [DOI] [PubMed] [Google Scholar]
- 94.Lu X, Kracher B, Saur IM, Bauer S, Ellwood SR, Wise R, Yaeno T, Maekawa T, Schulze-Lefert P. 2016. Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc Natl Acad Sci U S A 113:E6486–E6495. doi: 10.1073/pnas.1612947113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Praz CR, Bourras S, Zeng F, Sanchez-Martin J, Menardo F, Xue M, Yang L, Roffler S, Boni R, Herren G, McNally KE, Ben-David R, Parlange F, Oberhaensli S, Fluckiger S, Schafer LK, Wicker T, Yu D, Keller B. 2017. AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytol 213:1301–1314. doi: 10.1111/nph.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bourras S, McNally KE, Ben-David R, Parlange F, Roffler S, Praz CR, Oberhaensli S, Menardo F, Stirnweis D, Frenkel Z, Schaefer LK, Fluckiger S, Treier G, Herren G, Korol AB, Wicker T, Keller B. 2015. Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27:2991–3012. doi: 10.1105/tpc.15.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parlange F, Roffler S, Menardo F, Ben-David R, Bourras S, McNally KE, Oberhaensli S, Stirnweis D, Buchmann G, Wicker T, Keller B. 2015. Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal Genet Biol 82:181–192. doi: 10.1016/j.fgb.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 98.Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. 2006. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A 103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. 2006. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18:243–256. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Catanzariti AM, Dodds PN, Ve T, Kobe B, Ellis JG, Staskawicz BJ. 2010. The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant Microbe Interact 23:49–57. doi: 10.1094/MPMI-23-1-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ellis JG, Dodds PN, Lawrence GJ. 2007. Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol 45:289–306. doi: 10.1146/annurev.phyto.45.062806.094331. [DOI] [PubMed] [Google Scholar]
- 102.Ellis JG, Lawrence GJ, Luck JE, Dodds PN. 1999. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11:495–506. doi: 10.2307/3870876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sarma GN, Manning VA, Ciuffetti LM, Karplus PA. 2005. Structure of Ptr ToxA: an RGD-containing host-selective toxin from Pyrenophora tritici-repentis. Plant Cell 17:3190–3202. doi: 10.1105/tpc.105.034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Manning VA, Hamilton SM, Karplus PA, Ciuffetti LM. 2008. The Arg-Gly-Asp-containing, solvent-exposed loop of Ptr ToxA is required for internalization. Mol Plant Microbe Interact 21:315–325. doi: 10.1094/MPMI-21-3-0315. [DOI] [PubMed] [Google Scholar]
- 105.Wang CI, Guncar G, Forwood JK, Teh T, Catanzariti AM, Lawrence GJ, Loughlin FE, Mackay JP, Schirra HJ, Anderson PA, Ellis JG, Dodds PN, Kobe B. 2007. Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell 19:2898–2912. doi: 10.1105/tpc.107.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ve T, Williams SJ, Catanzariti AM, Rafiqi M, Rahman M, Ellis JG, Hardham AR, Jones DA, Anderson PA, Dodds PN, Kobe B. 2013. Structures of the flax-rust effector AvrM reveal insights into the molecular basis of plant-cell entry and effector-triggered immunity. Proc Natl Acad Sci U S A 110:17594–17599. doi: 10.1073/pnas.1307614110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parlange F, Daverdin G, Fudal I, Kuhn ML, Balesdent MH, Blaise F, Grezes-Besset B, Rouxel T. 2009. Leptosphaeria maculans avirulence gene AvrLm4-7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4-mediated recognition through a single amino acid change. Mol Microbiol 71:851–863. doi: 10.1111/j.1365-2958.2008.06547.x. [DOI] [PubMed] [Google Scholar]
- 108.Huang Y-J, Li Z-Q, Evans N, Rouxel T, Fitt BDL, Balesdent M-H. 2006. Fitness cost associated with loss of the AvrLm4 avirulence function in Leptosphaeria maculans (phoma stem canker of oilseed rape). Eur J Plant Pathol 114:77–89. doi: 10.1007/s10658-005-2643-4. [DOI] [Google Scholar]
- 109.Huang Y-J, Balesdent M-H, Li Z-Q, Evans N, Rouxel T, Fitt BDL. 2009. Fitness cost of virulence differs between the AvrLm1 and AvrLm4 loci in Leptosphaeria maculans (Phoma stem canker of oilseed rape). Eur J Plant Pathol 126:279. [Google Scholar]
- 110.Blondeau K, Blaise F, Graille M, Kale SD, Linglin J, Ollivier B, Labarde A, Lazar N, Daverdin G, Balesdent MH, Choi DH, Tyler BM, Rouxel T, van Tilbeurgh H, Fudal I. 2015. Crystal structure of the effector AvrLm4-7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins. Plant J 83:610–624. doi: 10.1111/tpj.12913. [DOI] [PubMed] [Google Scholar]
- 111.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Blumke A, Falter C, Herrfurth C, Sode B, Bode R, Schafer W, Feussner I, Voigt CA. 2014. Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol 165:346–358. doi: 10.1104/pp.114.236737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Plissonneau C, Stürchler A, Croll D. 2016. The evolution of orphan regions in genomes of a fungal pathogen of wheat. mBio 7:e01231-16. doi: 10.1128/mBio.01231-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. 2000. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bhadauria V, Banniza S, Vandenberg A, Selvaraj G, Wei YD. 2013. Overexpression of a novel biotrophy-specific Colletotrichum truncatum effector, CtNUDIX, in hemibiotrophic fungal phytopathogens causes incompatibility with their host plants. Eukaryot Cell 12:2–11. doi: 10.1128/EC.00192-12. [DOI] [PMC free article] [PubMed] [Google Scholar]