Abstract
The Information Age transformed our lives but it has had surprisingly little impact on the way chemical information (e.g., from our biological world) is acquired, analyzed and communicated. Sensor systems are poised to change this situation by providing rapid access to chemical information. This access will be enabled by technological advances from various fields: biology enables the synthesis, design and discovery of molecular recognition elements as well as the generation of cell-based signal processors; physics and chemistry are providing nano-components that facilitate the transmission and transduction of signals rich with chemical information; microfabrication is yielding sensors capable of receiving these signals through various modalities; and signal processing analysis enhances the extraction of chemical information. The authors contend that integral to the development of functional sensor systems will be materials that (i) enable the integrative and hierarchical assembly of various sensing components (for chemical recognition and signal transduction) and (ii) facilitate meaningful communication across modalities. It is suggested that stimuli-responsive self-assembling biopolymers can perform such integrative functions, and redox provides modality-spanning communication capabilities. Recent progress toward the development of electrochemical sensors to manage schizophrenia is used to illustrate the opportunities and challenges for enlisting sensors for chemical information processing.
1. The Importance of Chemical Information
Microelectronics transformed the way we interface with our world: the world’s knowledge is at our fingertips, long distances no longer prevent conversations with loved-ones, and our vehicles are flying/driving themselves. The information age however has had a comparatively limited impact on the way we acquire and understand chemical information that underpins our natural world. And this limitation is important when we recognize that biology routinely uses chemicals (and not electromagnetic modalities) to perform its various functions. This is illustrated by considering the circulation system which provides a conduit for the trafficking of the body’s function-performing chemicals. For instance, blood is the medium for distributing the body’s chemical energy resources (glucose and O2) as well as the medium for sorting wastes for disposal through the lungs, kidneys and liver. In addition, blood vessels are also the transmission lines that carry instruction-containing chemicals (hormones) that coordinate activities to maintain homeostasis. Further, blood is the medium for the immune system to dispatch its emergency response molecules (antibodies) and cells (e.g., monocytes and T-cells). Thus blood is a rich source of chemical information, and as a result blood tests are routinely used to assess a person’s health status.
More broadly, information of chemical compositions, concentrations and activities is essential to many fields of study whether it is to: assess environmental sites, control food quality, detect threats to public health, or manage human diseases. Because of the importance of chemical information, analytical chemistry has become an integral part of many scientific disciplines and commercial enterprises, and a wide variety of analytical chemistry methods have been developed. For a variety of reasons, sensor-based approaches have been developed to displace conventional chemical analysis with two approaches being most common. Figure 1a illustrates a semipermeable membrane that separates away unwanted chemical information to detect a desired signal: the classic example of this approach is the pH electrode. Figure 1b shows a sensor coated with a molecular recognition element that assays for a specific “target”: this approach is common to biosensors. While the sensor approaches in Figure 1 represent the vanguard, we contend that sensors of the future will be much more diverse in their approaches to access and process chemical information.
Figure 1.
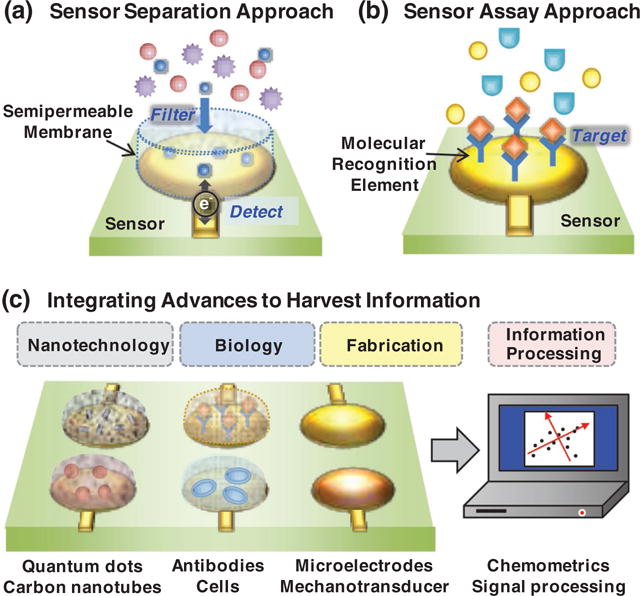
Acquiring chemical information. Historically, sensor approaches were similar to conventional chemical analysis where a membrane/coating was used to (a) separate away unwanted components from a signal, or (b) recognize/assay a specific chemical component. (c) Technological advances from diverse fields are providing entirely new opportunities for acquiring chemical information.
2. Sensors to Acquire Chemical Information
The motivation for developing sensor-based analytical methods can result from a combination of driving forces. Probably the most obvious driving force for the emergence of sensor systems is a market need (i.e., a demand). End-users like sensor systems because they can allow chemical analysis to be performed simply, rapidly and sometimes with low cost and in portable devices. In these cases, a non-chemist can make a relatively complex measurement easily with limited instrumentation needs. Examples include the use of dissolved oxygen probes for biotechnology process-monitoring, or the in-home use of glucometers to enable diabetics to measure their blood glucose.
Alternative drivers of sensor systems are advances in the enabling technologies that make possible what was previously impossible. Importantly, these enabling technological advances for sensor systems have come from many disparate fields as illustrated in Figure 1c.
2.1. Enabling Technological Advances
2.1.1. Biology
The advances in modern biology allow: the production of biology’s molecular recognition elements (e.g., enzymes, antibodies and nucleic acids); the design/discovery of entirely new recognition elements (e.g., aptamers and binding peptides); and the application of synthetic biology to access biology’s information processing capabilities.[1–4]
2.1.2. Nanotechnology
Remarkable developments in physics and chemistry provide novel opportunities to collect and convert chemical information into signals that can be detected through convenient modalities: optical (e.g., from quantum dots), electrical (e.g., from carbon nanotubes) or magnetic (e.g., from iron oxide nanoparticles).[5–7]
2.1.3. Fabrication
Microfabrication allows electronics to be integrated into increasingly complex systems that include smart fabrics, artificial skin, capsular endoscopes and lab-on-a-chip devices.[8,9]
2.1.4. Information Processing
In addition to “hardware” advances, there have also been advances in signal processing through complex algorithms to maximize information extraction (e.g., broad band impedance spectroscopy).[10–13]
2.2. Critical Need
We anticipate that advances from many individual fields will contribute to ever expanding capabilities for acquiring chemical information. We contend that a critical need is the development of robust and generic methods that enable the effective integration of these disparate technological advances into sensor systems. The purpose of this progress report is to illustrate how stimuli-responsive hydrogel-forming biopolymers enable such technology integration. As an illustrative example, we consider recent efforts to develop sensor systems to access relevant chemical information needed to understand and manage mental health disorders. To provide a framework, we first consider common strategies (paradigms) for acquiring chemical information using sensors.
3. Paradigms for Accessing Chemical Information
3.1. Detecting Specific Chemical Information: The One-Sensor One-Analyte Paradigm
A vast diversity of sensor systems have been investigated to access chemical information. For simplicity, we group these approaches into the three broad paradigms illustrated in Figure 2. Figure 2a illustrates the classic paradigm used for biosensing. In this paradigm, a molecular recognition element (traditionally an antibody or enzyme) is used to selectively bind/react with the analyte of interest and some mechanism is used to transduce this molecular recognition event into a convenient, device-compatible modality (typically electrical or optical). In the ideal, this biosensor can detect the analyte of interest with high selectivity and appropriate sensitivity, and thus this single sensor can detect all the necessary chemical information (i.e. the analyte’s concentration) that is required for the application.
Figure 2.
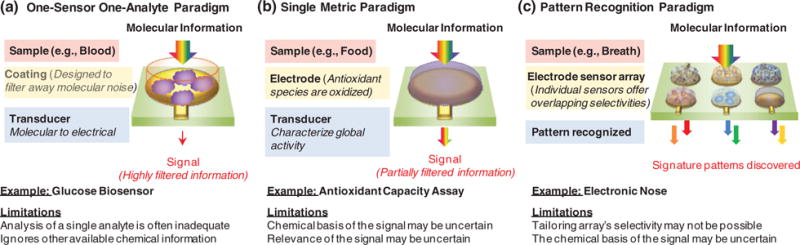
Paradigms for accessing chemical information by electrochemical sensors.
The historical example of this one-sensor one-analyte paradigm is the glucose biosensor where a glucose-specific redox enzyme (e.g., glucose oxidase) recognizes and reacts with glucose (Reaction (1)) and this reaction can be readily detected. For instance, the 1st generation glucose sensors detected the formation of the H2O2 product electrochemically as illustrated by Reaction (2). The technical advances that enabled sensor-based glucose detection coincided with an emerging demand for simple, rapid and reliable methods to measure glucose for managing diabetes. The remarkable technical and market successes of the glucose biosensor have served as a landmark for personalized medicine and also established a one-sensor one-analyte paradigm for sensing.
| (1) |
| (2) |
Despite the success of the glucose biosensor, it has been difficult to extend this one-sensor one-analyte paradigm for the generic management of other diseases. We contend that diabetes may be a unique example in which the analytical challenge is well-posed. Specifically, management of diabetes requires information of a single analyte and this glucose concentration measurement is sufficient to provide the necessary information to determine the patient’s status and indicate the required therapeutic intervention (e.g., insulin injection). While there have been extensive efforts to define single “biomarkers” that characterize various diseases, these efforts have not been entirely successful. Thus, despite the success of the glucose sensor, the one-sensor one-analyte paradigm may not be an entirely generic paradigm because the analytical questions for other diseases may not be as well-posed as for the case of diabetes management.
When information of one analyte is insufficient, the one-sensor one-analyte paradigm can be extended to multiplexed analysis such that multiple sensors are enlisted to simultaneously detect multiple analytes. Probably the most successful example of multiplexed analysis is the DNA microarray that allows an extensive sampling of genetic information. Because the sampling is so extensive, analytical questions don’t need to be well-posed, rather the DNA microarray can be used to discover correlations (e.g., links between gene mutations and disease). The fabrication of multiplexed sensor arrays for acquiring genomic information is facilitated by the comparative simplicity of nucleic acids: nucleic acids tend to be relatively stable; generic coupling chemistries can be used to assemble nucleic acid probes at individual sensor addresses; and molecular recognition through nucleic acid hybridization follows well-known rules. Multiplexed sensor arrays to collect proteomic or metabolomic information are more challenging.
In summary, the one-sensor one-analyte paradigm appears to have become the primary paradigm that guides the development of sensors to access chemical information. Presumably, the prevalence of this paradigm is because of the belief that chemical composition and the concentration of individual chemical components are the key measures of chemical information. While the one-sensor one-analyte paradigm is important, it is not the only paradigm available and we contend that these alternative paradigms have much to contribute to accessing and analyzing chemical information.
3.2. Global vs Specific Chemical Information
Molecular level specificity is a hallmark of biological chemical information processing – enzymes, antibodies and receptors all recognize specific chemical entities. Thus, the idea that biology would use a non-specific modality to perform critical functions seems counter-intuitive. However, the neural and neuromuscular systems use ionic currents to convey information and to initiate actions, and the identity of the specific ions seems less important than the global electrical activity. Even purposeful interventions (e.g. defibrillation) can use non-specific electrical inputs. While ionic electrical activities are integral to neural and neuromuscular systems, these activities may not be as relevant to other systems (e.g., immune system). Thus, a key question is: does biology use other global signaling modalities that could be accessed by sensors to acquire relevant chemical information from biology?
Emerging research suggests that redox may provide an additional global modality for biological signaling. Biology uses several small molecule redox couples to shuttle electrons in various biological processes: NAD(P)H carries reducing equivalents for energy harvesting (e.g., respiration) and biosynthesis; H2O2 is generated during an immune response to pathogen invasion (e.g., oxidative burst) and glutathione (GSH) serves as a protective antioxidant. Importantly, these redox couples are not in equilibrium with each other[14,15] and thus can perform independent functions. More recently, it is recognized that redox reactions, often involving reactive oxygen species (ROS), are integral to biological communication. Unlike typical biological signal transduction pathways that employ receptors and ligands to confer molecular selectivity, the targets of ROS signaling are believed to be atomic – the sulfur atom of cysteine and methionine residues.[16,17] Because these sulfur switches can control protein function (e.g., by the interconversion of free thiol residues to disulfide crosslinks) emerging research suggests redox is an independent and somewhat global cell-signaling modality.[18] The link between ROS and redox signaling has further suggested a need to re-consider oxidative stress in terms of redox dysfunction and not simply in terms of reactive species (i.e., free radicals)[19,20] and possibly such redox dysfunctions may provide clues (potentially measurable clues) of the environmental exposures that contribute to diseases[21,22] (e.g., mental health diseases).[23]
With the increasing understanding that redox plays an integral role in biological communication, we anticipate that electrochemistry will emerge as an increasingly important tool that is well suited to study questions in redox biology.[24–29] Electrochemical sensing systems are well known for their sensitivity, spatial-temporal precision, low cost and potential for portability. As will be discussed electrochemistry also provides data in an electronic format that can be readily analyzed through complex chemometric or signal processing methodologies. Thus, electrochemistry may provide access to global systems-level chemical information of importance to redox biology.
3.3. Detect Global Chemical Activity: The Single Metric Paradigm
The idea that redox may be a global biological modality suggests that a sensor which yields a single integrated measurement of redox activity may provide valuable biological information (Figure 2b). The classic example of this approach comes from food science where standardized antioxidant capacity assays have been developed to provide a single numerical value that characterizes a sample’s ability to donate electrons[30,31] (analogous to a single pH measurement that characterize a sample’s acidity). Typically, these antioxidant capacity assays are colorimetric although electrochemical methods have been explored.[32,33] Recently, medical researchers have begun adapting these methods in the hope that such a global measurement from a blood sample could provide information on oxidative stress that is believed to contribute to a range of human diseases.[34–36]
3.4. Detect Global Chemical Information: Pattern Recognition Paradigm
An alternative analytical paradigm is illustrated by the electronic nose or electronic tongue in Figure 2c which uses an array of electrochemical sensors to access chemical information more broadly (less specifically). In particular, each sensor reacts with multiple chemical species in a sample but the reactivities of the individual sensors are all somewhat different. If the sensor array offers sufficient sensitivity and the sensor selectivities overlap appropriately, then signature patterns can be recognized. Typically, this approach relies less on the design of the sensors and more on analyzing the responses through pattern recognition methodologies.[37,38]
In principle, this pattern recognition paradigm offers two important capabilities. First, sensor arrays can be applied to ill-posed questions and thus can serve as a tool to discover previously unknown patterns. For instance, sensor arrays are being investigated to detect signature patterns from a “breath print” that can be correlated to pathologies that range from lung cancer to Alzheimer’s disease.[39–44] The discovery of such patterns could potentially assist medical researchers by providing new information that could drive transformational advances in understanding and managing diseases. Second, sensor arrays can more broadly sample chemical information and thus may provide robust global signatures (e.g., biomarkers) for complex analysis. In such cases, it may be possible to develop portable devices to provide rapid information at the pharmacy, physician’s office or bedside.
4. Managing Mental Health Disorders: The Schizophrenia Example
Sensors are poised to make transformational contributions to medicine by providing rapid, reliable and real-time information that characterize diseases both to accelerate our understanding of underlying pathologies and to enhance our abilities to personalize treatment. Our focus here is on sensors that acquire chemical information. We use the example of schizophrenia to illustrate how clinical needs are converging with technological advances to drive the development of sensor systems for managing schizophrenia. We also use this example to illustrate the technical challenges which will further drive a fusion of the sensing paradigms to maximize the extraction of chemical information. We should note that one system will not likely meet all needs, but rather different systems may be required to meet the divergent needs and constraints of specific circumstances. For instance: an in-patient clinic may have access to centralized laboratory facilities and a dedicated staff, and thus the speed-of-analysis may be less important; first-responders may need simple systems that can transmit initial diagnostic information to clinicians at a receiving hospital; and care-givers in a home (e.g., a parent) may need dedicated, inexpensive and portable systems that reliably access a subset of chemical information to adjust dosages.
4.1. Schizophrenia
Schizophrenia is a devastating chronic mental health disorder that requires lifelong treatment.[45] The onset of symptoms typically occurs when a person is in their late teens and early 20s. While the causes of schizophrenia are unclear, there appears to be a combination of genetic and environmental factors that contribute to the disease. Diagnosis of psychiatric disorders is based primarily on interviews and behavioral (and not biochemical) evaluations.
Management of schizophrenia is challenging given the poorly-understood nature of the disease and the absence of readily detectable measures that provide objective real-time information to assess a patient’s status, guide treatment decisions, or evaluate their response to therapeutic interventions. Potentially, advances in sensor technology can contribute to providing such real-time objective measures[46–49] of functional biomarkers.[50] For instance, a common symptom of schizophrenia is disorganized or abnormal motor behavior[51,52] which could be detected or even continuously monitored by wearable sensor systems.[53] Additionally, sensors that analyze a patient’s voice may be able to detect the monotone speech patterns that are sometimes associated with schizophrenia.[54] Our long term goal is to employ sensor technology to access chemical information both to assist in managing mental health disorders and to assist in clarifying the chemical basis of these diseases.
An important medication for managing schizophrenia is the antipsychotic clozapine. Clozapine was developed in the 1970s and is one of the most clinically effective antipsychotics available[55] and is the gold standard antipsychotic for treatment of refractory schizophrenia, violent behavior, or those at high risk of suicide.[56] However, clozapine has some significant side effect risks that make it one of the most challenging antipsychotics to prescribe.[56] Because of its problems, clozapine was once pulled from the market but its therapeutic benefits led to its subsequent re-introduction.[55] There is a growing belief that people with schizophrenia and the mental health system would benefit if this medication was more broadly and effectively utilized, yet clinicians face many challenges in prescribing clozapine.[57–61] A major challenge illustrated in Figure 3a is inter-individual variation: it may be impossible to control serum clozapine levels within its narrow therapeutic range by simply controlling the dose.[62–64] Portable devices that measure clozapine could provide the feedback necessary to enable caregivers to adjust dose just as diabetics adjust their insulin dose in response to measured blood glucose.[63,65–69] Thus, the one-sensor one-analyte paradigm that has been so successful for managing diabetes would seem ideally suited for managing medication treatment for people with schizophrenia.
Figure 3.
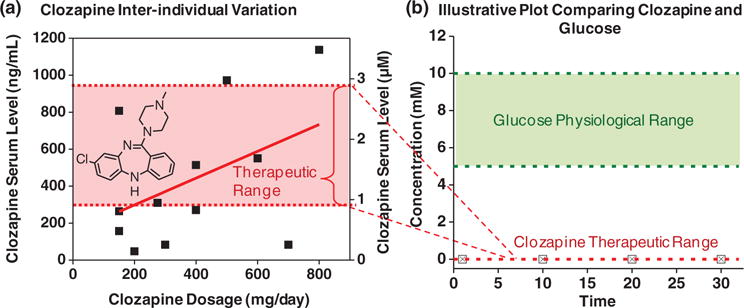
Clozapine is an important antipsychotic medication that is under-utilized in part because of the difficulty of maintaining serum levels within the therapeutic range. (a) Inter-individual variations make it difficult to control serum levels solely by controlling dosage. (b) Illustrative plot comparing clozapine’s therapeutic range with physiological range for serum glucose.
However, the analogy between clozapine monitoring and glucose monitoring breaks down for two broad reasons. First is the challenge of analysis. Clozapine’s therapeutically relevant concentration is small: three-orders-of magnitude less than that for glucose as illustrated in Figure 3b. Thus, any clozapine sensor will need high sensitivities. In addition, clozapine’s low signal exists in a very noisy background. The signal-to-noise problem is less significant for glucose analysis, both because glucose’s signal is high and because enzymes are available to selectively recognize the glucose signal. Analogous enzymes for clozapine detection are not available: while cytochrome P450 enzymes that react with clozapine are known,[70] they have broad substrate ranges and will not likely provide the necessary selectivity. Potentially, this technical challenge is surmountable and an appropriate molecular recognition element (e.g., an antibody, peptide or aptamer) could be developed for monitoring serum clozapine levels through a one-sensor one-analyte approach.
A second challenge to controlling serum clozapine levels (vs glucose) is the uncertainties in its metabolism. Specifically, precisely controlling serum levels within such a narrow therapeutic window may require both information of the current serum level and how rapidly this medication is being metabolized. Knowledge of metabolism is important because one metabolite (N-desmethylclozapine) is believed to have biological activity[55] and because clozapine’s metabolism can vary dramatically.[71] For instance, other commonly-prescribed medications (e.g., the antidepressant fluvoxamine[72,73] and the antibiotic ciprofloxacin[74]) can inhibit clozapine’s metabolism. Also, the patient’s health can affect clozapine’s metabolism (e.g., inflammation suppresses metabolism[71]). Further a patient’s lifestyle can alter metabolism as cigarette smoking induces metabolism[75–77] and coffee drinking may inhibit metabolism.[62,78] Potentially, serum contains information on these factors and accessing this information could better guide decisions of clozapine dosing, however accessing this information from a serum sample will require more than a single sensor measurement. Thus, sensor arrays and a pattern recognition paradigm may be most appropriate for acquiring and analyzing the ill-defined chemical information needed to adjust dosages.
Over the longer term, sensor systems that broadly access global chemical information may serve as a tool not just to personalize treatment but also to provide new information to assist in understanding schizophrenia. Growing evidence indicates that inflammation,[79–81] oxidative stress[82–86] and redox dysregulation[83,87,88] play an integral role in this devastating disease, and oxidative stress has been suggested as a potential biomarker in the pathophysiology and clinical course of schizophrenia.[84] However, the exact molecular mechanisms of oxidative stress have yet to be determined,[86] while the “nonspecific nature of oxidative stress” and the “shortage of techniques for monitoring oxidative stress” deter research.[82] In particular, what should be measured[89–92] and where (the brain, peripheral tissue or blood)?[93] Possibly measurements of individual chemical components (one sensor one-analyte paradigm) could be useful as reports indicate that schizophrenia patients often have lowered serum levels of specific antioxidants (e.g., GSH[94,95] and uric acid[96]). There have also been efforts to develop a single global measure (single metric paradigm) of oxidative stress such as total antioxidant status (TAS),[35,96] total oxidative stress (TOS) or oxidative stress index (OSI).[34] And recent efforts suggest an array-based approach (pattern recognition paradigm) to access the broad range of information of antioxidants and therapeutics.[97]
While the detailed needs and opportunities for schizophrenia may be unique, we believe the broader point is general. Our ability to understand, detect, and manage diseases is limited by our ability to access and interpret the chemical information that underpins the disease’s etiology, symptoms and progression. Thus sensors could fulfill a critical need by opening access to the needed chemical information.
5. Fusing Sensor Paradigms: Opinions, Vision and Needs
One underlying contention of this progress report is that the rate of scientific and technological progress in many cases is limited by the speed with which we can acquire and understand chemical information. The remarkable advances that drove the Information Age have had surprisingly little impact on our ability to access chemical information. This point is illustrated by considering that the initial developments of the glucose sensor (a landmark in chemical information processing) and the personal computer (a landmark in information processing) occurred at about the same time (1960s) but the subsequent trajectories diverged markedly. More powerful systems for information processing became smaller, cheaper and generic (e.g., cell phones), while more powerful systems for chemical information processing became larger, more expensive and specialized (e.g., microarrays). We believe sensors provide the opportunity to apply some of the capabilities of information technology for enhanced access to chemical information.
A second underlying contention is that the one-sensor one-analyte paradigm greatly constrains the potential of sensors to acquire chemical information. This paradigm relegates sensors to a supporting role: this paradigm requires that previous studies have already defined the analyte(s) of importance and the sensor’s role is “simply” to achieve the necessary sensitivity and selectivity. We believe that sensors could be used as a tool for discovery and as a means to cull through the vast amount of chemical information to assist in determining what information is important and how it should be measured. For comparison, imaging (magnetic resonance imaging, MRI)[98] and electrophysiological measurements[99,100] are reasonably well accepted approaches for discovering the structural and electrical activity changes associated with schizophrenia. Potentially, sensors could be used to discover the chemical changes associated with schizophrenia.
A third underlying contention is that sensor systems that provide access to chemical information will serve diverse functions. Sometimes the typical metrics from analytical chemistry (sensitivity and selectivity) will be appropriate, but sometimes the typical metrics from information theory (content and uncertainty) will be appropriate. Sometimes the chemical information needed will be well-defined (medical knowledge may have identified key biomarkers), but sometimes a less-defined sampling of the chemical information will identify signature patterns that will assist in discovering new knowledge. Sometimes the sensor’s capabilities for sensitivity, speed and portability will be important, but sometimes the ability of sensors to acquire data in an electronic format for subsequent processing of the information will be important. We believe that enlisting sensors for chemical information processing will require considerable intellectual creativity to match technological capabilities to application-specific needs.
We envision that various applications would benefit from sensor systems that employed a small set of “semi-smart” sensors. Figure 4 illustrates our vision for fusing the strengths of these electrochemical sensing paradigms: we envision using an electrode array in which the individual electrodes are coated with self-assembling hydrogel films. These individually functionalized, semi-selective films allow a partial filtering of a sample’s (bio)chemical information. For this sensor array to broadly access chemical information, we suggest several requirements must be satisfied:
individual sensors must have appropriate sensitivities to access information at the relevant concentrations;
individual sensors must be semi-selective to differentially filter the sample’s information and the selectivities should be overlapping to provide redundancy and robustness;
some of the individual sensors should interact with the sample to reveal information known to be relevant (e.g., information of important chemical components or biological mechanisms);
some of the sensors should interact with samples in ways that are not entirely defined to permit the discovery of new information.
Figure 4.
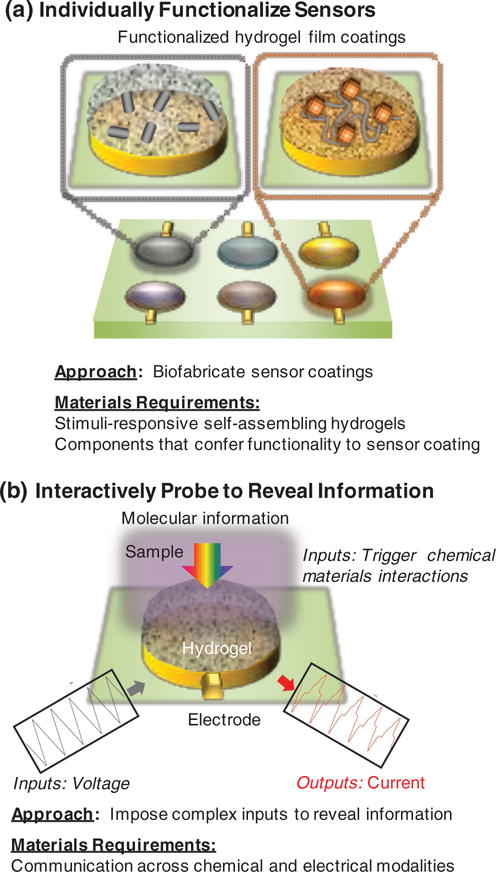
Our vision. (a) Biofabrication to assemble coatings for an array of “semi-smart” sensors. (b) Interactive probing to trigger chemical-material interactions that reveal a sample’s molecular information.
Further, the number of sensors in the array must balance the need for simplicity (favoring a small number of sensors) and the desire for diversity to broadly access the sample’s information content (favoring a large number of sensors).
As suggested in Figure 4, the success of this vision hinges on the ability to create the sensor coatings that can offer the functionalities to both interact with the chemical information in a sample and to communicate this information to the underlying sensor. These sensor coatings must be easy to assemble and enable meaningful signal transmission/transduction. Materials will be integral to achieving these dual goals (i) to hierarchically assemble the various sensing components into the sensor coatings and (ii) to establish meaningful communication that connects chemical and sensor modalities. The remainder of this progress report will focus on these materials requirements.
6. Self-assembling Biopolymer Hydrogels as Integrative Materials
6.1. Biofabrication to Assemble and Functionalize Sensor Coatings
Figure 4a suggests the need for simple, rapid and generic methods to assemble functional sensor coatings. Many approaches to create such functional coating are based on advances in chemistry (e.g., electropolymerization of conducting polymers or click chemistry) or microfabrication (e.g., photolithography or 3D printing). In addition to enlisting advances from physics and chemistry, it is also desirable to enlist the remarkable advances in biology for fabricating functional films.[101,102] Specifically, biology is expert at: fabricating materials at the nano-scale (e.g., proteins); self-assembling nano-components into complex hierarchical structures (e.g., tissue); and accessing, analyzing and responding to chemical information (e.g., through intracellular signal transduction pathways). Thus, biology has much to offer for the construction of functional sensor coatings and our aim is to enlist these biological fabrication capabilities. Specifically, we focus on stimuli-responsive self-assembling biopolymeric hydrogels as base matrix materials for sensor coating because they offer diverse and important capabilities for the fabrication of functional films. Figure 5 provides an overview of these capabilities).
Figure 5.
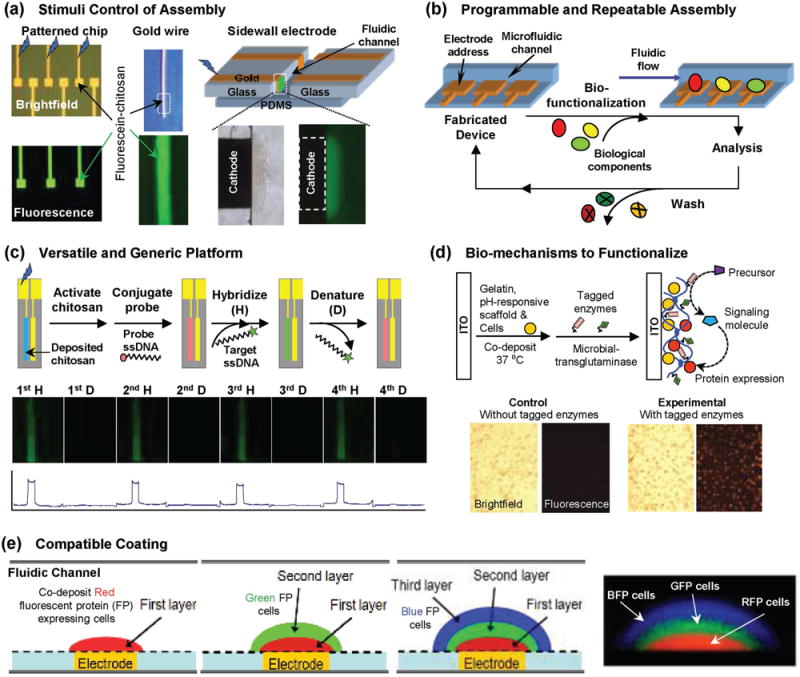
Broad capabilities enabled by enlisting biological materials and mechanisms to biofabricate sensor coatings. Reproduced with permission.[127] Copyright 2015, Elsevier. (a) Stimuli-responsive self-assembly allows simple, rapid and reagentless assembly of the sensor coating. (b) Reversible coating mechanisms allow generic and re-usable electronic systems to be fabricated while application-specific coatings can be applied on-demand. (c) Electrodeposited coatings can be functionalized using diverse methods (e.g., chemical conjugation to create a nucleic acid hybridization assay). Reproduced with permission.[121] Copyright 2004, American Chemical Society. (d) Enzymes can be enlisted to confer functionality. Reproduced with permission.[124] (e) Compatibility of assembly mechanisms and coating systems allows integration of complex cell-based functionalities. Reproduced with permission.[125] Adapted with permission.[180] Copyright 2013, The Royal Society of Chemistry.
6.1.1. Electrodeposition for Bottom-up Self-assembly
In early 2000, several groups discovered that the stimuli-responsive self-assembling aminopolysaccharide chitosan could be electrodeposited in response to devise-imposed electrical signals.[103–106] Subsequent work discovered mechanisms to electrodeposit alginate,[107–109] and then mechanisms to electrodeposit other self-assembling biopolymers. Details of the electrodeposition mechanism are provided below and a broader discussion of electrodeposition can be found elsewhere.[110] We should note that biopolymer electrodeposition was preceded by earlier studies on redox hydrogels.[111–115] Electrodeposition is important because it allows the programmable electroaddressing of sensor coatings: film coatings can be deposited at electrode surfaces on-demand with spatial selectivity.[116] Further, electrodeposition uses electrical stimuli to trigger self-assembly and thus allows the coating of complex surfaces without the need for line-of-sight (required for photolithographic methods) or direct contact (required for printing methods). Figure 5a shows the electrodeposition allows the conformal coating of diverse surface with hydrogel films (patterned electrodes, wires or meshes). Electrodeposition enlists the smart properties of the biological polymers to trigger self-assembly of the sensor coating and in most cases electrodeposition is simple, rapid and reagentless.
6.1.2. Reversible Coating for Post-fabrication (Bio)functionalization and Device Re-use
Many biopolymer electrodeposition mechanisms rely on reversible stimuli-responsive self-assembly mechanisms and thus these electrodeposited films can be readily removed. Even for biopolymeric hydrogel films that are electrodeposited through covalent crosslinks, the films can be removed using treatments (thermal or acid/base treatments) that do not damage the underlying sensors (e.g., conventional electrode cleaning methods can remove a biopolymeric sensor coating). The ability to reversibly assemble and disassemble the sensor coatings allows the fabrication of the electronic device (with the embedded sensor systems) to be separated from the sensor coating steps that confer (bio)function as illustrated in Figure 5b.
The capability for post-fabrication (bio)functionalization has two important features.[117] First, it allows reusability of the electronic device (vs single use). By eliminating the severe cost constraints of single-use systems, it is possible to employ complex, generic and reusable electronic systems that can fully access the power of electronics for data collection, analysis, and wireless transmission. Second, post-fabrication biofunctionalization accommodates the inherent differences in lifetimes between electronic systems (operational lives of years) and the biological components that are typically used to confer function to the sensor coatings (operational lives of hours-days). We envision the electronic systems could be fabricated in the most sophisticated fabrication laboratories (fab-labs) and only near the place/time of use these devices could be functionalized by assembling the sensor coatings. For instance, functionalization could be performed each day in a centralized hospital laboratory, or on-demand before deploying personnel to perform testing in remote or resource-poor locations.
6.1.3. Facile Functionalization
Electrodeposition of stimuli-responsive hydrogels involves a sol-gel transition such that an imposed electrical signal induces a spatially localized gelation at the electrode surface (see below for details of mechanisms). There are many examples in which components have been suspended in the deposition solution and then co-deposited in the hydrogel film.[118] Examples include hard nanoparticles such as carbon nanotubes and quantum dots, and softer structures such as vesicles and cells. (Co-deposition studies to create composite films such as implantable materials[110,119,120] are beyond the scope of this progress report.) Co-deposition could also include other polymers to confer other network functions (e.g., dual-responsiveness). In addition to co-deposition, coatings could be functionalized after deposition using standard chemical conjugation (e.g., carbodiimide and click chemistries) or microfabrication (e.g., photolithographic) methods. For instance, Figure 5c shows a chip with a DNA probe covalently tethered to an electrodeposited chitosan film. This probe can hybridize (H) to its fluorescently-labeled complementary target and after hybridization the duplex could be denatured (D) by treatment with 4 M urea at 65 °C for 30 min. This sequence of hybridization and denaturation could be repeated multiple times.[121]
6.1.4. Enzymatic Functionalization/Dis-assembly
Biopolymers can be acted upon by enzymes which provide unique opportunities to enlist these biological catalysts. There are many examples of biopolymer-degrading enzymes which typically de-polymerize the biopolymers through hydrolytic mechanisms (e.g., proteases, nucleases and cellulases). Such enzymes could be used to selectively dis-assemble a sensor coating or to selectively release components from a coating.[122]
It would be desirable if enzymes could also be used to graft functionality to sensor coating, but fewer enzymes are known that can build macromolecular structure (i.e. to increase molecular weight).[123] Obviously, cells routinely synthesize biopolymers and perform such biopolymer grafting reactions, but these intracellular biopolymer synthesis mechanisms typically employ complex cofactors (e.g., ATP and NADH) that are less convenient for in situ fabrication. However, biology does use a handful of cofactor-independent enzymes to build macromolecular structures outside the cell – to coagulate blood or harden cuticles. These enzymes could be enlisted to functionalize sensor coatings. Figure 5d shows the use of a microbial transglutaminase (mTG) enzyme to simultaneously crosslink an electrodeposited gelatin film and to graft proteins to this film: the entrapped cells confer cellular function while the grafted proteins confer molecular functions to this gelatin coating.[124]
6.1.5. Compatibility
The electrodeposition of self-assembling biopolymers occurs from aqueous solution and various methods to functionalize these films (e.g., by co-deposition or enzymatic reactions) also are performed in water. Aqueous-based processing is compatible with the labile biological components (e.g., antibodies and cells) that would typically be used to confer bio-function to sensor coatings. Further, the hydrogel nature of these coatings is also important to retain “viability” of the embedded biocomponents. For instance, Figure 5e shows an assembled “biofilm” with a stratified population of bacteria that was assembled by sequential co-deposition of Ca2+-alginate hydrogel layers.[125] This example illustrates the broad potential of hydrogel electrodeposition: it allows the assembly of cells within biocompatible matrices for biosensing applications and for in vitro study (e.g., for animal-on-a-chip applications). In the longer term, sensors with biofunctionalized hydrogel coating may be able to report chemical information in in vivo applications with implanted or ingested systems (e.g., integrated into a capsular endoscope).
6.2. Biopolymeric Materials and Mechanisms for Electrodeposition
As illustrated in Figure 6, several stimuli-responsive biopolymers have been electrodeposited. To date a handful of electro-deposition mechanisms have been identified.
Figure 6.
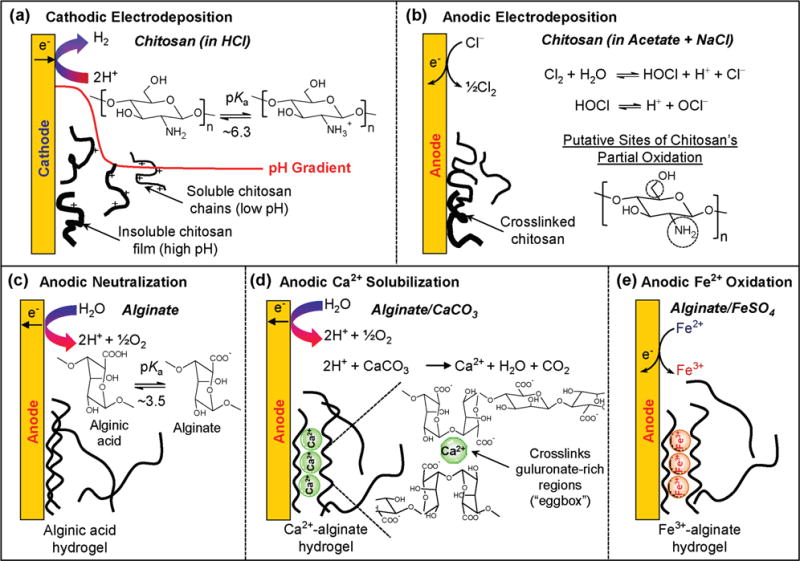
Summary of various known biopolymer electrodeposition mechanisms. Chitosan deposition by (a) cathodic neutralization[196] and (b) anodic oxidation.[129] Alginate’s deposition by (c) anodic neutralization,[109] (d) anodic Ca2+-liberation from CaCO3,[108] and (e) anodic Fe2+ oxidation.[138]
6.2.1. Chitosan
To our knowledge the first biopolymer to be electrodeposited was the aminopolysaccharide chitosan. Chitosan is a weak base that is water-soluble at low pH and undergoes a sol-gel transition as the pH is increased above its pKa (≈6.3). As illustrated in Figure 6a, chitosan’s cathodic electrodeposition occurs because electrode reactions consume protons to generate a localized region of high pH[126] that triggers chitosan’s deprotonation and gel formation. This deposition mechanism is reversible such that the electrodeposited film can be re-dissolved in weak acid. Cathodic chitosan electrodeposition is the best-studied biopolymer deposition system and it has been used to entrap various nanocomponents, and graft a range of function-conferring biomacromolecules (e.g., nucleic acids and proteins). Progress with this system has been extensively reviewed elsewhere.[127,128]
In addition to the cathodic neutralization mechanism, a separate anodic electrodeposition mechanism was discovered.[129] As illustrated in Figure 6b, anodic deposition requires salt (NaCl), and the mechanism has been hypothesized to involve several chemical reactions: anodic oxidation of Cl− to generates Cl2, hydrolysis of Cl2 to yield the HOCl oxidant, partial oxidation of chitosan to generate aldehyde moieties, and Schiff base formation between the generated aldehydes and primary amines of chitosan. There are two important points of anodically-deposited chitosan. First, the Schiff bases serve as covalent crosslinks and thus this anodic deposition mechanism is not readily reversible; the anodically deposited chitosan films do not readily dissolve upon immersion in weak acid. However, Schiff base linkages are slowly reversible in that these linkages can be hydrolyzed back to aldehydes and amines, and thus the anodically deposited films have been observed to re-dissolve under acidic conditions over long times. Second, aldehydes can form Schiff bases with various primary amines including the primary amines of proteins.[130] Thus, anodic deposition can be used as a single step method to both electrodeposit a crosslinked chitosan film and to simultaneously conjugate proteins to this film.[129]
6.2.2. Alginate
To our knowledge, the second biopolymer to be electrodeposited was the weakly acidic polysaccharide alginate.[109] Three mechanisms have been reported for alginate’s electrodeposition. First, is an anodic neutralization mechanism of Figure 6c that involves the electrochemical generation of a low pH, and the localized protonation of alginate that neutralizes this poly-saccharide and induces its gelation as alginic acid (pKa ≈3.5). This mechanism has been reported for the co-deposition of a biosensing enzyme,[131] however the majority of interest in the anodic electrodeposition of alginic acid appears to be to create composite coatings for non-sensor applications.[132,133]
A second mechanism is the anodic deposition of Ca2+-alginate which is illustrated in Figure 6d.[108] This mechanism employs anodic reactions to solubilize CaCO3 and generate the soluble Ca2+ ions that trigger the Ca2+-responsive alginate to undergo gelation through reversible electrostatic crosslinking. Importantly, CaCO3 is a buffer and thus this mechanism does not require significant pH excursions and can be used to co-deposit prokaryotic and eukaryotic cells.[134–137] For instance, the spatially segregated bacterial populations of Figure 5e were generated by the sequential co-deposition of 3 separate Escherichia coli strains.[125] As suggested, the Ca2+-alginate electrodeposition mechanism is useful for conferring cellular function to sensor coating (note: Ca2+-alginate hydrogels are routinely used as a matrix for entrapping microbes and a scaffold for tissue engineering). Cell-based sensing systems are poised to have a significant impact with the development synthetic biology systems capable of detecting chemical signals and converting this recognition through intracellular signal transduction mechanisms to generate outputs that are amplified and/or transduced into device-compatible modalities (e.g., optical and electrical).[1,2,4]
The third alginate electrodeposition mechanism is illustrated in Figure 6e, which shows alginate electrostatically crosslinked by Fe3+ ions. The Fe3+ ions are generated electrochemically by Fe2+ oxidation. This electrochemical reaction is reversible and electrochemical reduction of Fe3+ (back to Fe2+) can trigger dissolution of this alginate gel.[138] Thus, this electrochemical deposition-dissolution mechanism can be used to assemble components into an alginate hydrogel (i.e., to load and store “cargo” in the gel) and to subsequently release these components in response to user-imposed electrical signals.[139] Interestingly, the reducing stimulus that triggers Fe3+ reduction, dissolution of the Fe3+-alginate matrix, and cargo release does not need to be supplied externally but can be supplied by biological reducing reactions (e.g., enzyme-catalyzed NADH oxidation can supply the reducing equivalents). This redox-responsive dissolution mechanism can thus serve to create logic gates to process chemical and biochemical information.[107,140–142]
6.2.3. Gelatin
Recently, an anodic mechanism was reported for the electro-deposition of the protein gelatin.[122] This mechanism requires NaCl and appears to be similar to the mechanism for the anodic deposition of chitosan in Figure 6b. In particular, gelatin’s electrodeposition appears to yield a covalently crosslinked network and this gel-forming process can simultaneously conjugate other proteins to the matrix consistent with a partial oxidation to generate aldehydes and a subsequent Schiff base forming reaction. Interestingly, these electrodeposited gelatin films also retain some thermal responsiveness that allow the creation of a thermally-responsive switch of enzymatic activity.[122] Further, electrodeposited gelatin films can be enzymatically degraded to release grafted components.
6.2.4. Proteins
The anodic deposition of gelatin described above uses salt to generate diffusible oxidants that partially oxidize the protein and induce the formation of covalent crosslinks. We anticipate that it may be possible to extend this anodic oxidation mechanism for the electrodeposition of other proteins, however this mechanism is not reversible. The reversible electrodeposition mechanisms that do not chemically alter the biopolymer’s structure all involve biopolymers with polyelectrolyte properties. Chitosan and alginate are weak polyelectrolytes that can be electrodeposited through the pH-responsive neutralization mechanisms of Figures 6a and 6c. Alginate is ionic-responsive (e.g., to Ca2+ and Fe3+) and can be electrodeposited to form the electrostatically crosslinked networks of Figure 6d and 6e. It would be desirable if reversible mechanisms were available to deposit proteins.
It is well known that proteins respond to electric fields and pH gradients and a neutralization mechanism allows their isoelectric focusing. However, a protein’s neutralization will be more complex than the neutralization mechanisms used to electrodeposit chitosan and alginate. Specifically, proteins are polyampholytes and are only neutral at their isoelectric point (no net charge). Thus a protein only has a neutral charge at an intermediate pH, as pH is increased or decreased away from the isoelectric point the protein becomes more negatively or more positively charged. In contrast, alginate has only anionic groups and becomes neutral as the pH is lowered below its pKa, and chitosan has only cationic groups and becomes neutral as the pH is raised above its pKa. Thus an effort to electrode-posit a protein may not lead to the formation of a film at an electrode surface, but rather the formation of a band “focused” some distance from the electrode. This appears to be the case for collagen.[143,144] Specifically, several reports show that electrochemical methods can generate collagen films and, these films form some distance from the electrode. Importantly, the electric field is believed to align the collagen[145] and induce it’s self-assembly[146] to yield collagen matrices that are being investigated for applications in regenerative medicine.[147–149]
More recently electrodeposition has been extended to silk fibroin protein-based gels (termed e-gels)[150–154] and this method provides a means to prepare silk-based materials for emerging medical applications.[155] These studies report a reversible electrogelation of silk films at an anode surface[156,157] with the low localized pH adjacent to the anode being important for the deposition mechanism.[157,158] Additional studies indicate that an electric field can align silk supramolecular assemblies.[159] Interestingly, films were also prepared using two proteins, regenerated silk and hydrophobically-modified tropoelastin. These proteins were first enzymatically conjugated using horseradish peroxidase (HRP) to create the dityrosine crosslinks between proteins and then the protein conjugates were electrodeposited.[160]
6.2.5. Co-deposition of Other Biopolymers
There are various thermally-responsive hydrogel-forming biopolymers that would be useful to electrodeposit (e.g., gelatin and agarose). However, it is difficult to provide the low temperature trigger in a spatially controlled manner to induce these hydrogels to form at a sensor surface. Co-deposition provides a means induce spatially-controlled deposition of these thermally-responsive biopolymers. For instance, gelatin has been co-deposited by blending a warm gelatin solution with low molecular weight, pH-responsive Fmoc-peptide hydrogela-tors that can be electrodeposited.[161–163] After deposition, the co-deposited films are cooled allowing gelatin to undergo its physical crosslinking to form a stable network. In fact, after deposition and cooling, the pH can be adjusted to disassemble the Fmoc-peptides which can diffuse out of the film leaving behind this physically-crosslinked gelatin hydrogel. Co-deposition thus provides a mechanism to electrodeposit gelatin into spatially-organized films. Subsequent enzymatic reactions can crosslink this network to confer thermal stability and also to graft proteins to confer function (e.g., the film in Figure 5d was prepared by gelatin’s co-deposition). Gelatin[164] and silk[165] proteins have also been co-deposited using chitosan.
Co-deposition has also been used to generate films with the non-ionic and thermally-responsive polysaccharide agarose. Agarose’s co-deposition has been performed using both the Fmoc-peptide hydrogelators described above[166] and the Ca2+-alginate deposition mechanism. In the latter case, anodic deposition was performed from a warm suspension of agarose, sodium alginate and CaCO3, and after co-deposition, the films were cooled to allow the agarose network to form. Agarose is a useful sensor coating because charged macromolecules can readily pass through this neutral matrix by simple diffusion or by an electric-field driven migration (note: agarose is commonly used in biotechnology laboratories as a matrix for gel electrophoresis). For instance, in one study, cells were co-deposited and cultivated in an agarose-alginate network and then exposed to conditions that affected the cell’s gene expression. The phenotypic changes in cell-surface protein expression were then immuno-analyzed using an electric field to drive the probing antibodies through the agarose-alginate network.[167]
6.3. Assembling a Sensor Coating for Clozapine Detection
To illustrate the potential of biopolymer electrodeposition to functionalize electrochemical sensors, we consider recent work to detect the antipsychotic clozapine from serum samples. As mentioned clozapine is an antipsychotic with both considerable benefit and significant side effects, which has led several groups to conclude the need for therapeutic drug monitoring (TDM).[63,65–69] Electrochemical analysis is commonly considered for clozapine’s TDM[168–170] but there are significant technical challenges for detecting the low therapeutic concentrations in blood (the therapeutic range is 1–3 μM) and overcoming the complex matrix effects.[171] These matrix effects include interference from other chemical species that are present at considerably higher concentrations (e.g., ascorbic and uric acids) and the fact the clozapine is strongly bound to blood proteins. Figure 7a illustrates the difficulty for the electrochemical detection of clozapine. In this control, a gold electrode was used with a sensitive electrochemical method (differential pulse voltammetry, DPV). DPV imposes a sequence of input potentials (insert illustrates these input voltages) and current outputs are detected. When a simple buffer was used, minimal currents were detected in the voltage range where clozapine would be detected. The inclusion of typical serum levels of uric acid (≈400 μM) and ascorbic acid (≈100 μM) resulted in considerable overlap in the voltage region for clozapine detection. This overlap makes it difficult to quantify, or even detect the small peaks associated with clozapine.
Figure 7.
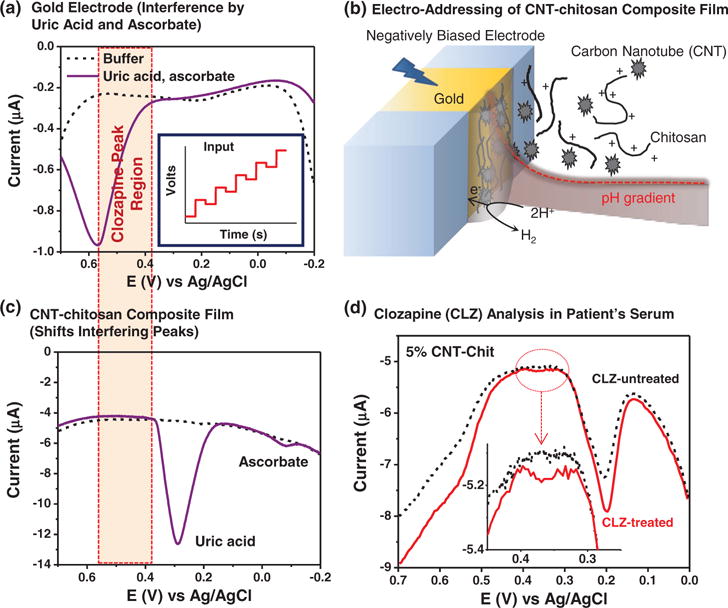
Clozapine detection by gold electrode coated with a chitosan film containing carbon nanotubes (CNTs).[172] (a) Control studies with gold electrode show common blood components uric acid (400 μM) and ascorbate (200 μM) interfere with clozapine’s detection. (b) Cathodic co-deposition of CNT-chitosan film. (c) CNT’s electrocatalytic properties shift interfering peaks for uric acid. (d) Detection of clozapine from serum of a schizophrenia patient. Reproduced with permission.[172]
Figure 7b illustrates the functionalization of the gold electrode by co-deposition of multi-walled carbon nanotubes (CNT) with the chitosan film. The CNTs confer electrocatalytic properties to the deposited electrode coating which has two beneficial effects. First, Figure 7c shows that the CNTs shift the electrochemical oxidation peaks closer to their equilibrium positions (i.e., it diminishes the need for an overpotential) which fortuitously shifts the competing peaks (especially the uric acid peak) outside the window where clozapine is detected (this effect improves selectivity). Second, CNT’s electrocatalytic properties amplify clozapine’s oxidation signal which improves sensitivity. As a result of these two features, Figure 7d shows that the CNT-chitosan coating allowed detection of clozapine from the serum of a schizophrenia patient being treated with this medication (no signal appears for the patient not undergoing clozapine treatment).[172]
6.4. Opportunities and Challenges
We believe the above discussion demonstrates the broad opportunities of self-assembling biopolymers for creating functional films and these opportunities extend beyond sensor applications. The mechanisms outlined in Figure 6 give the impression that the major fundamental questions associated with the triggered self-assembly of these hydrogel matrices have been answered. This impression is incorrect and surprisingly little is known of the detailed mechanistic interactions responsible for self-assembly of these biopolymer systems. To illustrate the profound limitations to our current knowledge we consider the case of chitosan, the best-studied electrodeposited biopolymer.
Cathodic electrodeposition induces chitosan to undergo a reversible sol-gel transition to generate a hydrogel. Traditionally, the structure of hydrogels is considered to be a homogenous polymeric network that traps water. Several years ago, a procedure was reported that systematically interrupted chitosan’s gelation and multi-layer hydrogel structures were generated.[173,174] Different groups performed variations of this interrupted gelation process and also obtained multilayer structures.[175–177] Chitosan’s cathodic electrodeposition can also be interrupted by supplying a sequence of electrical on-off inputs (vs a single on-off input) and the SEM images in Figure 8a show multilayers are also generated.[178] This interrupted electrodeposition process controls the hydrogel structure qualitatively: the thickness of the layers is controlled by the “on” signal while the “off” signal generates the interface between layers. The electrical inputs also control electrodeposition quantitatively: the film thickness is controlled by the total charge transfer (i.e., the number of protons consumed) and is semi-quantitatively described by a moving front mathematical model.[178] Incredibly, there are no detailed theories to explain why and how interrupted gelation processes cause the formation of these complex multilayer structures.
Figure 8.
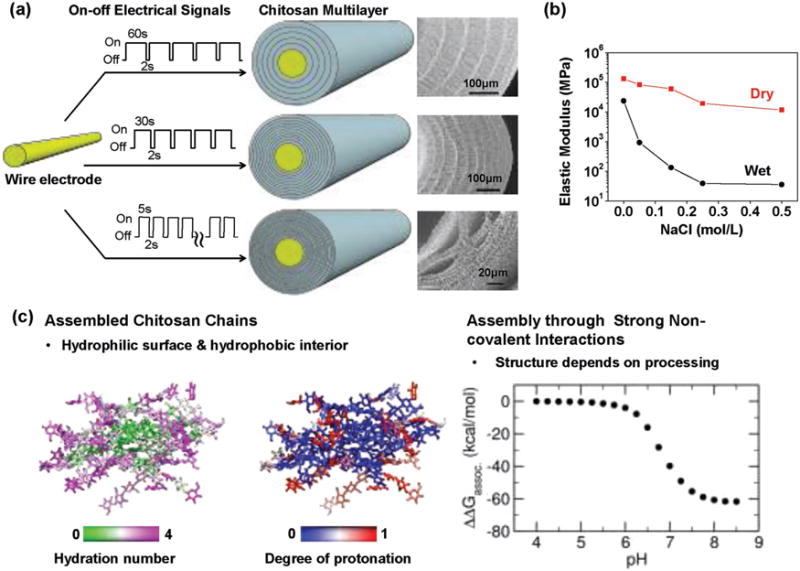
Challenges and opportunities to understand biopolymer self-assembly. (a) A sequence of on-off inputs to a wire electrode can interrupt chitosan’s cathodic electrodeposition and yield a complex multilayer film.[178] (b) The presence of salt in the deposition solution significantly alters the deposited film’s properties.[180] (c) Molecular modeling suggests chitosan self-assembles into crystalline domains with considerable hydrophobic character and that this pH-dependent self-assembly is highly favorable indicating that meta-stable states may be effectively “locked” in place.[181] Adapted from Ref. [178,180] with permission from The Royal Society of Chemistry. Reproduced with permission.[181] Copyright 2015, American Chemical Society.
A second example that illustrates the limitations to our knowledge of chitosan’s electrodeposition is shown in Figure 8b. In this study, the cathodic deposition process was performed from chitosan solutions containing different levels of added salt (i.e., NaCl). Salt addition should screen electrostatic repulsions of the protonated chitosan chains. Because chitosan chains are somewhat rigid, the salt is expected to have little impact on chain conformation[179] yet this screening of repulsions should facilitate the chain-chain self-assembly that results in film formation. Experimentally, when deposition was performed under increasing salt concentrations, the resulting films differed between strong transparent films (no salt) to a weak opaque network of aggregates (high salt).[180] The quartz crystal microbalance (QCM-D) measurements in Figure 8b show orders-of-magnitude differences in the mechanical properties (i.e., the elastic modulus) of the wet films depending on the salt concentration of the initial deposition solution.[180] This result demonstrates how processing conditions (salt concentration) dramatically affects structure (transparent film vs network of aggregates) and properties, yet the fundamental framework to understand processing, structure and properties is lacking.
To better understand the mechanistic details of biopolymer (e.g., polysaccharide) self-assembly, we have begun to enlist molecular modeling.[181] Specifically, we used all-atom molecular dynamics (MD)[182] and the all-atom version[183–185] of continuous constant pH molecular dynamics (CpHMD)[186,187] with the pH replica exchange (pH-REX).[188] To simulate chitosan’ self-assembly, we modeled the reverse process – the dis-assembly (dissociation) of 16 10-mer chains of chitosan that had initially been assembled into a “cystallite” structure. There are two important features from the simulation results in Figure 8c. First, these simulations indicate that the partially assembled chitosan structure has a hydrophobic interior (with little hydration and neutral deprotonated amines), while the water-exposed exterior is hydrophilic (with considerable hydration and cationic protonated amines). This feature is reminiscent of the tertiary structure of globular proteins with their hydrophobic core and hydrophilic solvent-exposed shell. Second, the free energy diagram in Figure 8c shows that the energetics of self-assembly (ΔΔGassoc) vary considerably with pH and at a high pH, chitosan’s self-assembly is highly favorable (ΔΔGassoc ≪ 0). The rugged free energy landscape observed in these polysaccharide self-assembly simulations are reminiscent of those observed for RNA folding.[189,190]
The experimental and simulation results in Figure 8 indicate that chitosan self-assembles through strong non-covalent interactions. Self-assembly through strong non-covalent interactions yields structures that tend to be meta-stable and do not “anneal” to a more thermodynamically-favored state.[191–195] This situation is analogous to a traditional materials science view that a material’s structure and properties are controlled by processing conditions (not by thermodynamics). Thus, these results demonstrate the broad design space available to control the deposited film’s structure and properties, but these results also raise concerns of repeatability: small, seemingly insignificant changes in deposition conditions can yield large variations in the resulting structure and properties.
7. Redox-capacitor for Interactive Redox Probing of Chemical Information
7.1. Electrochemical Acquisition of Redox Information
Figure 4b illustrates that the second part of the broad vision is to have some sensors that can interactively probe for chemical information, and especially for redox information. As mentioned, we believe redox measurements may provide global systems-level chemical information relevant to biology because (i) redox is emerging as an important biological communication modality and (ii) redox measurements may provide insights on the oxidative stress that contributes to various human diseases. Because biological redox-communication does not follow the traditional lock-and-key model for molecular specificity, we envision the possibility that global redox information can be accessed using a single sensor analogous to the single metric paradigm of Figure 2b. However, we extend this paradigm to actively probe the local environment interactively. Figure 9a illustrates that we envision an interactive electrochemical approach that is approximately analogous to sonar’s ability to use sound waves to probe a local environment for the presence of objects in water. At first glance, this may not seem like a particularly useful analogy: the physical interactions of the sound waves with objects are not obviously related to the underlying chemical interactions that are measured by electrochemical analyses. Yet we believe there are three important features in Figure 9b that can be used to extend electrochemical analysis to enhance its ability to probe for the ill-defined chemical information.
Figure 9.
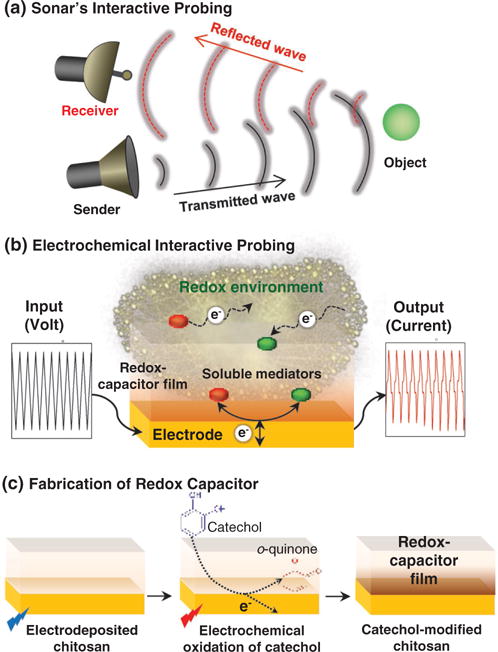
Interactive probing of redox activity. (a) Analogy to sonar. (b) Electrochemical probing using (i) redox mediators to both transduce electrical-redox I/O and transmit redox signals, (ii) complex I/O to tailor interactive probing, and (iii) redox capacitor to modify redox I/O to facilitate interpretation. (c) Fabrication of catechol-chitosan redox-capacitor by (i) cathodic electrodeposition of chitosan, and (ii) anodic oxidative-grafting of catechol moieties.
7.2. Redox Mediators
Standard electrochemical approaches require the redox-active chemical species to diffuse through the medium to access the electrode surface in order to be detected. To extend electrochemical analysis toward a sonar analogy, we add low molecular weight diffusible redox mediators that can actively probe the local environment. Specifically, Figure 9b shows these mediators can: be oxidized/reduced at the electrode surface, diffuse away from the electrode where they can undergo redox interactions with various components in the film-coating or medium, and then return to the electrode for subsequent electron exchange. Electron exchange between the mediators and electrode is detected electrochemically (e.g., as a current) and these signals contain historical information of the mediator’s redox interactions. Basically, the mediators serve two functions: they transduce the electrode’s electrical input/output into redox-signals and they serve as the “particles” that transmit the redox-signal throughout the film-coating and into the surrounding solution.
7.3. Complex Inputs and Outputs to Process Information
The second feature of the sonar analogy in Figure 9b is that an electrochemical probing could employ analogous signal processing methodologies that underpin sonar. Sonar and modern telecommunications encode, transmit and decode information using oscillating signals (e.g., the sound waves for sonar or the radio waves for wireless communication). Electrochemistry already provides inputs/outputs in an electronic format convenient for signal processing, and many electrochemistry techniques employ oscillating inputs and outputs. For instance, impedance spectroscopy employs sinusoidal inputs and outputs and directly enlists the Nyquist and Bode formalisms common to signal processing. Cyclic voltammetry (CV) also uses oscillating inputs and the information embedded in the outputs has analogies to signal processing: the CV inputs and outputs have the same frequency; the amplitude (peak current) contains information of reactions at the solution-electrode interface; and the phase (potential at peak current) contains information of the reacting species and possible mass transfer limitations for these species to diffuse to the electrode interface. Thus, while electrochemistry and signal processing already share some concepts, we suggest that a fuller utilization of signal processing methodologies could extend the reach of electrochemistry for extracting global systems-level chemical information from a complex and noisy matrix. In essence, we envision that interactive probing will allow chemical information to be acquired by hypothesis-free reverse engineering which is an entirely different approach from the standard hypothesis-driven one sensor one-analyte paradigm.
7.4. Redox-Capacitor Film
The third feature of Figure 9b is a redox-capacitor film that serves to manipulate (i.e., process) redox-based signals in ways that facilitate interpretation. This redox-capacitor film is fabricated on an electrode surface in two steps as illustrated in Figure 9c. First, chitosan is cathodically electrodeposited on the gold electrode.[128,181,196] Second, the chitosan-coated electrode is immersed in a solution containing catechol and the underlying gold electrode is biased to anodically oxidize catechol to o-quinones which initiates its covalent grafting to the chitosan film.[197,198] This catechol-modified-chitosan film is non-conducting but redox-active: it cannot directly exchange electrons with the underlying electrode but it can exchange electrons with soluble redox-active species that can diffuse into the film.[199,200] Importantly, this redox-capacitor has been observed to exchange electrons with a broad range of redox active species that include common electrochemical mediators and also common biological reductants (e.g., NADH) and oxidants (e.g., O2).[201,202] As illustrated with results for the analysis of clozapine, the combination of redox-active but non-conducting confers interesting molecular electronic properties to the film.
7.5. Clozapine’s Redox-Cycling with the Catechol-Chitosan Redox-Capacitor
Clozapine is redox active and can be anodically oxidized at the gold surface. As suggested in Figure 10a, anodic oxidation initiates clozapine’s redox-cycling such that the oxidized species diffuses from the underlying electrode into the redox capacitor film where it is re-reduced by accepting electrons from the grafted catechol moieties. Clozapine’s oxidative redox-cycling thus serves to discharge the capacitor by mediating the transfer of electrons from the film to the electrode (i.e., by converting reduced catechol moieties to oxidized o-quinone moieties). Because the redox-capacitor has a finite capacity to store electrons, this oxidative redox-cycling cannot proceed indefinitely. To re-charge the capacitor with electrons, a second mediator (e.g., Ru(NH3)6Cl3; Ru3+: E° = −0.2 V vs Ag/AgCl) is used to accept electrons from the electrode and transfer them to the capacitor through a reductive redox-cycling mechanism. The thermodynamic plot in Figure 10b indicates that the combination of oxidative and reductive redox-cycling in the presence of both electron mediators (clozapine and Ru3+) can occur if (a) the mediators’ redox potentials bracket the redox-potential of the grafted catechol moieties, and (b) the imposed electrode potential is cycled between oxidative potentials (more positive than clozapine’s redox potential) and negative potentials (more negative than Ru3+ redox potential). From a functional standpoint, the redox-cycling interactions with the redox-capacitor substantially alter the measured electrochemical currents.
Figure 10.
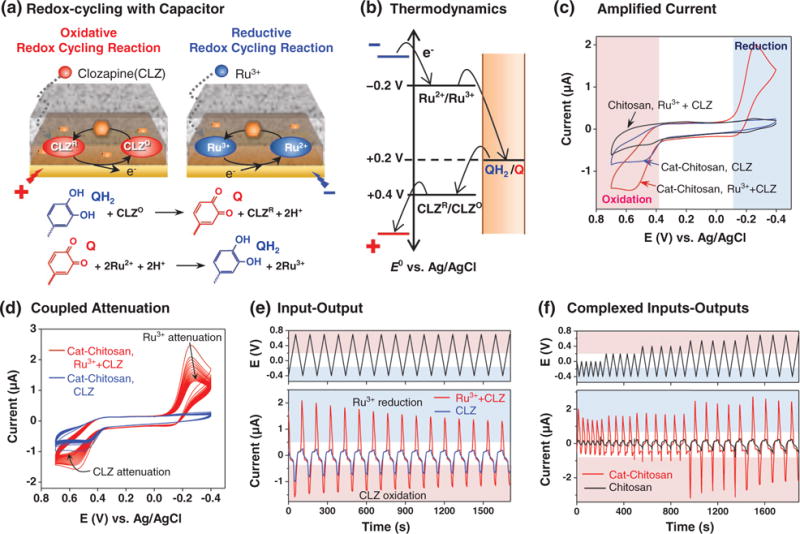
Interactive redox probing of clozapine. (a) Clozapine undergoes oxidative redox-cycling with the catechol-chitosan redox capacitor and this can be balanced by the reductive redox-cycling of an added mediator (Ru3+).[206,207] Redox-cycling interactions (b) are thermodynamically constrained, (c) amplify output currents, and (d) couple output responses. (e) Input-output representation illustrates that irreversibilities in clozapine’s oxidation can be detected in the attenuation of Ru3+ reduction currents. (f) Complex inputs can be imposed to generate complex, but interpretable outputs: an initial reductive input potential range that precludes clozapine oxidation also suppresses Ru3+ reductive redox-cycling, while the expanded input potential range that allows clozapine oxidation also allows Ru3+ reductive redox-cycling.
7.5.1. Amplification
Redox-cycling with the capacitor links clozapine’s oxidation with Ru3+ reduction and leads to an amplification of both electrochemical currents. Such amplification can enhance the sensitivity of electrochemical analysis.[203,204] Amplification of clozapine’s oxidation and Ru3+ reduction currents is illustrated by the cyclic voltammograms (CVs) in Figure 10c. Specifically, when a solution containing both clozapine (25 μM) and Ru3+ (25 μM) was probed using the redox-capacitor film, amplified oxidation and reduction currents were observed (compared to controls in which Ru3+ was absent or the electrode was coated with a control chitosan film).[205–207]
7.5.2. Repeated Cyclic Input/Output
If the oxidative and reductive redox-cycling mechanisms are fully reversible and reasonably balanced, then oscillating voltage inputs can be imposed over long periods to generate time-invariant (i.e., steady) output currents. However, irreversibilities in the underlying chemistries or electrode fouling can result in attenuations in output currents over time. Such irreversibilities occur with clozapine because this molecule can undergo complex irreversible oxidation reactions.[208] The CVs in Figure 10d show attenuation in output currents for an experiment in which the clozapine-Ru3+ solution was probed using the catechol-chitosan redox capacitor for multiple cycles. Figure 10e shows the same results plotted as an input-output representation where time is shown explicitly as the x-axis. This input-output representation is a more common form for analysis by signal processing. Importantly, the observed attenuation provides information of clozapine’s underlying oxidation chemistries and appears as outputs in the Ru3+ reduction region which should facilitate interpretation because (i) Ru3+ electrochemistry is well-understood, and (ii) Ru3+ oxidation/reduction occurs in a less-crowded potential region (compared to the potential region where clozapine is oxidized). While this input-output representation is neither linear nor time-invariant – two key assumptions in the mathematical analysis in signal processing – it does suggest the opportunity to apply the powerful methods of signal processing to enhance the extraction of chemical information.
7.5.3. Complex Input/Outputs
Finally, the redox-capacitor allows novel approaches to be used to access chemical information. Specifically, a sequence of complex input signals can be imposed and the response characteristics can be monitored to acquire information in a way that resembles typical reverse engineering methods.[209] Such a sequence of signals is illustrated in Figure 10f which shows the probing of a clozapine-Ru3+ solution with varying potential amplitudes (i.e., the input voltage range was progressively increased). Initially the solution was probed by cycling the voltage through a limited range of reducing potentials. This initial potential range was sufficient to reduce Ru3+ and charge the film, but insufficiently oxidative to oxidize clozapine and discharge the film. Thus, the initial output current shows small Ru3+ reduction peaks and no clozapine oxidation peaks. Over time, the input voltage range was progressively expanded to allow clozapine’s oxidation which then induces the oxidative redox-cycling that discharges the capacitor. As expected, a clozapine oxidation peaks appears when this expanded voltage range is imposed. In addition, when this enlarged potential range is imposed, clozapine’s oxidative redox-cycling is initiated and the capacitor can be discharged. Upon discharging, the capacitor can then begin accepting electrons via Ru3+ redox-cycling and thus an amplification in Ru3+ reduction is also observed in Figure 10f. The important point is that because of the capacitor’s redox-cycling, an input perturbation in an oxidizing potential region (i.e., by expanding the voltage range to oxidize clozapine) results in an output perturbation in a reducing region (i.e., an amplification of Ru3+ reduction). Mechanistically, the redox capacitor couples the oxidative redox-cycling of clozapine with the reductive redox-cycling of Ru3+. We believe this example illustrates the opportunity for interactively probing for chemical information: by creatively coupling mediators with a complex sequence of voltage inputs it is possible to generate signature outputs that reflect the underlying redox interactions in a sample.
In summary, we believe electrochemistry is uniquely suited to access redox-based chemical information relevant to biology. Here, we use the example of clozapine’s redox-cycling, but various other biologically-relevant molecules (e.g., the bacterial virulence factor pyocyanin,[204] the plant signaling molecule acetosyringone,[202] and the analgesic drug acetaminophen[210]) and cell populations[209] can undergo redox-cycling with the catechol-chitosan redox-capacitor. From a sensor standpoint, we envision this redox-capacitor coating could access information of biologically relevant redox interactions and thus could be a useful element in a sensor array.
7.6. Opportunities and Challenges
Redox mediators are integral to the interactive probing illustrated in Figure 9 as they serve to convert the imposed electrical inputs into redox signals that can be transmitted to undergo interactions with the local environment. Electrochemists routinely use mediators to exchange electrons with electrodes, and mediators are sometimes used to exchange electrons with redox-active components in a local environment to perform functions for bioelectronics (enzyme-based biosensing[211] and microbial fuel cells[212]), bioelectrosynthesis,[213,214] environmental remediation,[214–216] sample characterization,[25,202,217–219] and molecular actuation.[220–222] We extend the use of redox mediators to engage in redox-cycling with the redox capacitor for the purpose of enhancing access to chemical information.[201] We anticipate that a judicious selection of mediators (including biological oxidants and reductants) could enhance the selectivity of information access, while the simultaneous use of multiple mediators may provide broader access to redox information.[25] The ability to enlist mediators to probe with increased selectivity or breadth will require a better understanding of mediator-based redox interactions.
Complex electrical inputs and outputs allow the mediator-based transmissions to be tailored to probe for specific information (e.g., as in Figure 10f). Further, the use of cyclic inputs and outputs to generate time invariant signals also allows access to the formalisms of information processing and control theories for signal analysis. Extending these theories for the interpretation of electrochemically generated chemical information should offer exciting possibilities.[10–13] These theories typically use cyclic inputs of varying frequencies and interpret results using mathematical formalisms that assume linear time invariant responses. Electrochemical responses may not always satisfy these assumptions[223] and it is not clear how much information can be extracted from the frequency-dependence of the response signal (compared the non-sinusoidal morphology of the signal).[25,224] Despite these limitations however, the extension of well-developed communication theories to the acquisition of chemical information from electrochemical signals seems like a fruitful area for future investigation.
The redox capacitor in Figure 9 is fabricated from catechols which are among nature’s most abundant redox-active organic compounds.[225] These catechol-based films have been shown to exchange electrons with a broad range of mediators including biological oxidants and reductants. Further, the film’s redox-potential (E° ≈ +0.2 V vs Ag/AgCl) is within a biologically relevant range thus positioning the films to exchange electrons under physiologically-relevant conditions. Further, these redox-capacitor films are simple to fabricate requiring only two electrochemical steps and no reagents. Potentially, capacitor films could be fabricated using alternative redox-active moieties (with different redox potentials) or even multiple moieties to access different redox information (e.g., information from a different set of molecules).
8. Conclusions and Future Perspectives
Simpler, faster and more reliable access to chemical information could transform our ability study our environment, protect our citizens and manage our health, yet accessing such chemical information has proven to be difficult. There have been remarkable technical advances from diverse fields of science and engineering that can contribute “hardware” (e.g., nanocomponents) and “software” (e.g., signal processing) to access and interpret chemical information. Key to success is the ability to integrate these diverse capabilities into functional systems, and we suggest that biological materials have important integrative capabilities. We summarize results which show biopolymers (polysaccharides and proteins) can recognize device-imposed electrical stimuli to trigger the self-assembly of hydrogel sensor coatings. Through a variety of mechanisms (e.g., co-deposition and enzymatic assembly) these electrodeposited coatings can be equipped with function-conferring components. The speed, simplicity and programmability of biopolymer electrodeposition should allow the generic on-demand creation of a small set of sensors that can provide the overlapping selectivities that allow broad access to chemical information. We further summarize results with a catechol-based redox capacitor that allows interactive probing through a redox modality that is integral to biological signaling yet accessible by electrochemistry. Potentially redox may provide global systems-level access to biological information while enabling the capabilities of signal processing to be applied to understand this information.
Acknowledgments
E. K. and Y. L. contributed equally to this work. We gratefully acknowledge financial support from the United States: National Science Foundation (CBET-1435957), Defense Threat Reduction Agency (HDTRA1-13-1-0037), USDA-National Institute of Food and Agriculture (20146702121585), and the National Institutes of Health (R01 MH105571-01).
Biographies

Eunkyoung Kim currently works as a research associate of Institute for Bioscience & Biotechnology Research (IBBR) and Fischell Department of Bioengineering at University of Maryland. She received her B.S. degree in Chemistry from Kyughee University in South Korea in 1997. She obtained her M.S and Ph.D. degrees in Electrochemistry from Korea Advanced Institute of Science and Technology (KAIST) in South Korea in 2004. After Ph.D., she joined into University of Pittsburgh as a post-doctor during 2007-2008. Her research interest is focused on the integration of electrochemistry with biology including understanding redox biology as well as developing electrochemical clinical devices.

Yi Liu is a Research Associate in the Institute for Bioscience & Biotechnology Research (IBBR) and Fischell Department of Bioengineering at University of Maryland. His research activities include electrodeposition of stimuli-responsive biological polymers, enzyme-mediated biofunctionalization, bio-sensing and bioelectrochemistry. Dr. Liu’s current research focuses on building smart biopolymeric hydrogels from which biology and electronics can communicate. Yi Liu holds a bachelor’s degree from Peking University and a Ph.D. degree from Lehigh University.
Contributor Information
Dr. Eunkyoung Kim, Institute for Biosystems and Biotechnology Research, University of Maryland, College Park, MD 20742, USA Fischell Department of Bioengineering, University of Maryland, College Park, MD 20742, USA.
Dr. Yi Liu, Institute for Biosystems and Biotechnology Research, University of Maryland, College Park, MD 20742, USA Fischell Department of Bioengineering, University of Maryland, College Park, MD 20742, USA.
Dr. Hadar Ben-Yoav, Department of Biomedical Engineering, Ben-Gurion University of the Negev, Beer Sheva 8410501, Israel
Thomas E. Winkler, Institute for Systems Research, University of Maryland, College Park, MD 20742, USA Department of Electrical and Computer Engineering, University of Maryland, College Park, MD 20742, USA.
Kun Yan, School of Resource and Environmental Science, Hubei Biomass-Resource Chemistry Environmental, Biotechnology Key Laboratory, Wuhan University, Wuhan 430079, China.
Prof. Xiaowen Shi, School of Resource and Environmental Science, Hubei Biomass-Resource Chemistry Environmental, Biotechnology Key Laboratory, Wuhan University, Wuhan 430079, China
Prof. Jana Shen, Department of Pharmaceutical Sciences, School of Pharmacy, University of Maryland, Baltimore, MD 21201, USA
Prof. Deanna L. Kelly, Maryland Psychiatric Research Center, University of Maryland School of Medicine, Baltimore, MD 21228, USA
Prof. Reza Ghodssi, Institute for Systems Research, University of Maryland, College Park, MD 20742, USA Department of Electrical and Computer Engineering, University of Maryland, College Park, MD 20742, USA.
Prof. William E. Bentley, Institute for Biosystems and Biotechnology Research, University of Maryland, College Park, MD 20742, USA Fischell Department of Bioengineering, University of Maryland, College Park, MD 20742, USA.
Prof. Gregory F. Payne, Institute for Biosystems and Biotechnology Research, University of Maryland, College Park, MD 20742, USA Fischell Department of Bioengineering, University of Maryland, College Park, MD 20742, USA.
References
- 1.Tschirhart T, Zhou XY, Ueda H, Tsao CY, Kim E, Payne GF, Bentley WE. ACS Synth Biol. 2016;5:28. doi: 10.1021/acssynbio.5b00073. [DOI] [PubMed] [Google Scholar]
- 2.Aurand TC, March JC. Biotechnol Bioeng. 2016;113:492. doi: 10.1002/bit.25832. [DOI] [PubMed] [Google Scholar]
- 3.Terrell JL, Wu HC, Tsao CY, Barber NB, Servinsky MD, Payne GF, Bentley WE. Nat Commun. 2015;6:12. doi: 10.1038/ncomms9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng J, Jester BW, Tinberg CE, Mandell DJ, Antunes MS, Chari R, Morey KJ, Rios X, Medford JI, Church GM, Fields S, Baker D. eLife. 2015;4:e10606. doi: 10.7554/eLife.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rana S, Le NDB, Mout R, Saha K, Tonga GY, Bain RES, Miranda OR, Rotello CM, Rotello VM. Nat Nanotechnol. 2015;10:65. doi: 10.1038/nnano.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rana S, Le NDB, Mout R, Duncan B, Elci SG, Saha K, Rotello VM. ACS Central Sci. 2015;1:191. doi: 10.1021/acscentsci.5b00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyle S, Saha S. Adv Healthcare Mater. 2014;3:1703. doi: 10.1002/adhm.201400009. [DOI] [PubMed] [Google Scholar]
- 8.Gao W, Emaminejad S, Nyein HYY, Challa S, Chen K, Peck A, Fahad HM, Ota H, Shiraki H, Kiriya D, Lien DH, Brooks GA, Davis RW, Javey A. Nature. 2016;529:509. doi: 10.1038/nature16521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yildiz UH, Inci F, Wang S, Toy M, Tekin HC, Javaid A, Lau DTY, Demirci U. Biotechnol Adv. 2015;33:178. doi: 10.1016/j.biotechadv.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giner-Sanz JJ, Ortega EM, Perez-Herranz V. Electrochim Acta. 2015;186:598. [Google Scholar]
- 11.Macia L Fernandez, Petrova M, Hauffman T, Muselle T, Doneux T, Hubin A. Electrochim Acta. 2014;140:266. [Google Scholar]
- 12.Sanchez B, Vandersteen G, Bragos R, Schoukens J. Meas Sci Technol. 2012;23:14. [Google Scholar]
- 13.Van Ingelgem Y, Tourwe E, Blajiev O, Pintelon R, Hubin A. Electroanalysis. 2009;21:730. [Google Scholar]
- 14.Jones DP, Go YM. Diabetes Obes Metab. 2010;12:116. doi: 10.1111/j.1463-1326.2010.01266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp M, Go YM, Jones DP. Free Radic Biol Med. 2008;44:921. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nathan C, Cunningham-Bussel A. Nat Rev Immunol. 2013;13:349. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray PD, Huang BW, Tsuji Y. Cellular Signalling. 2012;24:981. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Go YM, Jones DP. J Biol Chem. 2013;288:26512. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Circu ML, Aw TY. Semin Cell Dev Biol. 2012;23:729. doi: 10.1016/j.semcdb.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go YM, Jones DP. Free Radic Biol Med. 2011;50:495. doi: 10.1016/j.freeradbiomed.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Go YM, Chandler JD, Jones DP. Free Radic Biol Med. 2015;84:227. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Go YM, Jones DP. Redox Biol. 2014;2:358. doi: 10.1016/j.redox.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiavone S, Jaquet V, Trabace L, Krause KH. Antioxid Redox Signal. 2013;18:1475. doi: 10.1089/ars.2012.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellin DL, Sakhtah H, Zhang YH, Price-Whelan A, Dietrich LEP, Shepard KL. Nat Commun. 2016;7:10. doi: 10.1038/ncomms10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim E, Panzella L, Micillo R, Bentley WE, Napolitano A, Payne GF. Sci Rep. 2015;5:18447. doi: 10.1038/srep18447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trouillon R, Passarelli MK, Wang J, Kurczy ME, Ewing AG. Anal Chem. 2013;85:522. doi: 10.1021/ac303290s. [DOI] [PubMed] [Google Scholar]
- 27.Santos RM, Rodrigues MS, Laranjinha J, Barbosa RM. Biosens Bioelectron. 2013;44:152. doi: 10.1016/j.bios.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Ganesana M, Erlichman JS, Andreescu S. Free Radic Biol Med. 2012;53:2240. doi: 10.1016/j.freeradbiomed.2012.10.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillard EA, de Abreu FC, Ferreira DCM, Jaouen G, Goulart MOF, Amatore C. Chem Commun. 2008:2612. doi: 10.1039/b718116g. [DOI] [PubMed] [Google Scholar]
- 30.Prior RL, Wu XL, Schaich K. J Agric Food Chem. 2005;53:4290. doi: 10.1021/jf0502698. [DOI] [PubMed] [Google Scholar]
- 31.Huang D, Ou B, Prior RL. J Agric Food Chem. 2005;53:1841. doi: 10.1021/jf030723c. [DOI] [PubMed] [Google Scholar]
- 32.Kilmartin PA, Hsu CF. Food Chem. 2003;82:501. doi: 10.1021/jf030086q. [DOI] [PubMed] [Google Scholar]
- 33.Kilmartin PA, Zou HL, Waterhouse AL. J Agric Food Chem. 2001;49:1957. doi: 10.1021/jf001044u. [DOI] [PubMed] [Google Scholar]
- 34.Guney E, Ceylan MF, Tektas A, Alisik M, Ergin M, Goker Z, Dinc GS, Ozturk O, Korkmaz A, Eker S, Kizilgun M, Erel O. J Affect Disord. 2014;156:62. doi: 10.1016/j.jad.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XY, Chen DC, Xiu MH, Tang W, Zhang FX, Liu LJ, Chen YL, Liu JH, Yao JK, Kosten TA, Kosten TR. Schizophr Res. 2012;139:66. doi: 10.1016/j.schres.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Erel O. Clinical Biochemistry. 2004;37:112. doi: 10.1016/j.clinbiochem.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Ceto X, Voelcker NH, Prieto-Simon B. Biosens Bioelectron. 2016;79:608. doi: 10.1016/j.bios.2015.12.075. [DOI] [PubMed] [Google Scholar]
- 38.Smyth H, Cozzoino D. Chem Rev. 2013;113:1429. doi: 10.1021/cr300076c. [DOI] [PubMed] [Google Scholar]
- 39.Kurada S, Alkhouri N, Fiocchi C, Dweik R, Rieder F. Aliment Pharmacol Ther. 2015;41:329. doi: 10.1111/apt.13050. [DOI] [PubMed] [Google Scholar]
- 40.Adiguzel Y, Kulah H. Biosens Bioelectron. 2015;65:121. doi: 10.1016/j.bios.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 41.Wlodzimirow KA, Abu-Hanna A, Schultz MJ, Maas MAW, Bos LDJ, Sterk PJ, Knobel HH, Soers RJT, Chamuleau RAFM. Biosens Bioelectron. 2014;53:129. doi: 10.1016/j.bios.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 42.van de Kant KDG, van der Sande L, Jobsis Q, van Schayck OCP, Dompeling E. Respir Res. 2012;13:117. doi: 10.1186/1465-9921-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tisch U, Schlesinger I, Ionescu R, Nassar M, Axelrod N, Robertman D, Tessler Y, Azar F, Marmur A, Aharon-Peretz J, Haick H. Nanomedicine. 2012;8:43. doi: 10.2217/nnm.12.105. [DOI] [PubMed] [Google Scholar]
- 44.Santonico M, Pennazza G, Capuano R, Falconi C, Vink TJ, Knobel HH, Van der Schee MP, Sterk PJ, Montuschi P, D’Amico A. Sens Actuator B-Chem. 2012;173:555. [Google Scholar]
- 45.van Os J, Kapur S. Lancet. 2009;374:635. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 46.Naslund JA, Marsch LA, McHugo GJ, Bartels SJ. J Ment Heal. 2015;24:320. doi: 10.3109/09638237.2015.1019054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zao JK, Gan TT, You CK, Chung CE, Wang YT, Mendez SJR, Mullen T, Yu C, Kothe C, Hsiao CT, Chu SL, Shieh CK, Jung TP. Front Hum Neurosci. 2014;8:16. doi: 10.3389/fnhum.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gramann K, Jung TP, Ferris DP, Lin CT, Makeig S. Front Hum Neurosci. 2014;8:3. doi: 10.3389/fnhum.2014.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kane JM, Perlis RH, DiCarlo LA, Au-Yeung K, Duong J, Petrides G. J Clin Psychiatry. 2013;74:E533. doi: 10.4088/JCP.12m08222. [DOI] [PubMed] [Google Scholar]
- 50.Sun JF, Tang YY, Lim KO, Wang JJ, Tong SB, Li H, He B. IEEE Trans Biomed Eng. 2014;61:1756. doi: 10.1109/TBME.2014.2306424. [DOI] [PubMed] [Google Scholar]
- 51.Lallart E, Jouvent R, Herrmann FR, Perez-Diaz F, Lallart X, Beauchet O, Allali G. J Neural Transm. 2014;121:443. doi: 10.1007/s00702-013-1111-0. [DOI] [PubMed] [Google Scholar]
- 52.Stensdotter AK, Loras HW, Flovig JC, Djupsjobacka M. Gait Posture. 2013;38:918. doi: 10.1016/j.gaitpost.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 53.Glenn T, Monteith S. Curr Psychiatry Rep. 2014;16:10. doi: 10.1007/s11920-014-0494-4. [DOI] [PubMed] [Google Scholar]
- 54.Rapcan V, D’Arcy S, Yeap S, Afzal N, Thakore J, Reilly RB. Med Eng Phy. 2010;32:1074. doi: 10.1016/j.medengphy.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 55.Wenthur CJ, Lindsley CW. ACS Chem Neurosci. 2013;4:1018. doi: 10.1021/cn400121z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fakra E, Azorin JM. Expert Opin Pharmacother. 2012;13:1923. doi: 10.1517/14656566.2012.709235. [DOI] [PubMed] [Google Scholar]
- 57.Warnez S, Alessi-Severini S. BMC Psychiatry. 2014;14:5. doi: 10.1186/1471-244X-14-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gee S, Vergunst F, Howes O, Taylor D. Acta Psychiatr Scand. 2014;130:16. doi: 10.1111/acps.12193. [DOI] [PubMed] [Google Scholar]
- 59.Freudenreich O, Henderson DC, Sanders KM, Goff DC. Acad Psych. 2013;37:27. doi: 10.1176/appi.ap.11090159. [DOI] [PubMed] [Google Scholar]
- 60.Nielsen J, Roge R, Schjerning O, Sorensen HJ, Taylor D. Eur Neuropsychopharmacol. 2012;22:818. doi: 10.1016/j.euroneuro.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Kelly DL, Kreyenbuhl J, Dixon L, Love RC, Medoff D, Conley RR. Schizophr Bull. 2007;33:1221. doi: 10.1093/schbul/sbl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajkumar AP, Poonkuzhali B, Kuruvilla A, Jacob M, Jacob KS. Int Clin Psychopharmacol. 2013;28:50. doi: 10.1097/YIC.0b013e32835ac9da. [DOI] [PubMed] [Google Scholar]
- 63.Couchman L, Bowskill SVJ, Handley S, Patel MX, Flanagan RJ. Early Interv Psychiatry. 2013;7:122. doi: 10.1111/j.1751-7893.2012.00374.x. [DOI] [PubMed] [Google Scholar]
- 64.Rostami-Hodjegan A, Amin AM, Spencer EP, Lennard MS, Tucker GT, Flanagan RJ. J Clin Psychopharmacol. 2004;24:70. doi: 10.1097/01.jcp.0000106221.36344.4d. [DOI] [PubMed] [Google Scholar]
- 65.Couchman L, Belsey SL, Handley SA, Flanagan RJ. Anal Bioanal Chem. 2013;405:9455. doi: 10.1007/s00216-013-7361-8. [DOI] [PubMed] [Google Scholar]
- 66.Cohen D, Bogers J, van Dijk D, Bakker B, Schulte PFJ, G. Dutch Clozapine Collaboration J Clin Psychiatry. 2012;73:1307. doi: 10.4088/JCP.11r06977. [DOI] [PubMed] [Google Scholar]
- 67.Couchman L, Morgan PE, Spencer EP, Flanagan RJ. Ther Drug Monit. 2010;32:438. doi: 10.1097/FTD.0b013e3181dad1fb. [DOI] [PubMed] [Google Scholar]
- 68.Ulrich S, Baumann B, Wolf R, Lehmann D, Peters B, Bogerts B, Meyer FP. Int J Clin Pharmacol Ther. 2003;41:3. doi: 10.5414/cpp41003. [DOI] [PubMed] [Google Scholar]
- 69.Freeman DJ, Oyewumi LK. Clin Pharmacokinet. 1997;32:93. doi: 10.2165/00003088-199732020-00001. [DOI] [PubMed] [Google Scholar]
- 70.Zhou SF, Wang B, Yang LP, Liu JP. Drug Metab Rev. 2010;42:268. doi: 10.3109/03602530903286476. [DOI] [PubMed] [Google Scholar]
- 71.de Leon J, Diaz FJ. Prog Neuropsychopharmacol Biol Psych. 2003;27:1059. doi: 10.1016/S0278-5846(03)00148-9. [DOI] [PubMed] [Google Scholar]
- 72.Gahr M, Rehbaum K, Connemann BJ. Pharmacopsychiatry. 2014;47:118. doi: 10.1055/s-0034-1374611. [DOI] [PubMed] [Google Scholar]
- 73.Legare N, Gregoire CA, De Benedictis L, Dumais A. Med Hypotheses. 2013;80:689. doi: 10.1016/j.mehy.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 74.Raaska K, Neuvonen PJ. Eur J Clin Pharmacol. 2000;56:585. doi: 10.1007/s002280000192. [DOI] [PubMed] [Google Scholar]
- 75.Tsuda Y, Saruwatari J, Yasui-Furukori N. BMJ Open. 2014;4:9. doi: 10.1136/bmjopen-2013-004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dratcu L, Grandison A, McKay G, Bamidele A, Vasudevan V. Am J Therapeutics. 2007;14:314. doi: 10.1097/01.pap.0000249958.96498.ce. [DOI] [PubMed] [Google Scholar]
- 77.Haslemo T, Eikeseth PH, Tanum L, Molden E, Refsum H. Eur J Clin Pharmacol. 2006;62:1049. doi: 10.1007/s00228-006-0209-9. [DOI] [PubMed] [Google Scholar]
- 78.Thompson L, Pennay A, Zimmermann A, Cox M, Lubman DI. BMC Psychiatry. 2014;14:10. doi: 10.1186/1471-244X-14-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cox D, Chan MK, Bahn S. J Nerv Ment Dis. 2015;203:393. doi: 10.1097/NMD.0000000000000289. [DOI] [PubMed] [Google Scholar]
- 80.Richard MD, Brahm NC. Am J Health-Syst Pharm. 2012;69:757. doi: 10.2146/ajhp110271. [DOI] [PubMed] [Google Scholar]
- 81.Potvin S, Stip E, Sepehry AA, Gendron A, Bah R, Kouassi E. Biol Psychiatry. 2008;63:801. doi: 10.1016/j.biopsych.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 82.Emiliani FE, Sedlak TW, Sawa A. Curr Opin Psychiatr. 2014;27:185. doi: 10.1097/YCO.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu JQ, Kosten TR, Zhang XY. Prog Neuro-Psychopharmacol Biol Psychiatry. 2013;46:200. doi: 10.1016/j.pnpbp.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 84.Flatow J, Buckley P, Miller BJ. Biol Psychiatry. 2013;74:400. doi: 10.1016/j.biopsych.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boskovic M, Grabnar I, Terzic T, Plesnicar B Kores, Vovk T. Psychiatry Res. 2013;210:761. doi: 10.1016/j.psychres.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 86.Bitanihirwe BKY, Woo TUW. Neurosci Biobehav Rev. 2011;35:878. doi: 10.1016/j.neubiorev.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kulak A, Steullet P, Cabungcal JH, Werge T, Ingason A, Cuenod M, Do KQ. Antioxid Redox Signal. 2013;18:1428. doi: 10.1089/ars.2012.4858. [DOI] [PubMed] [Google Scholar]
- 88.Do KQ, Cabungcal JH, Frank A, Steullet P, Cuenod M. Curr Opin Neurobiol. 2009;19:220. doi: 10.1016/j.conb.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 89.Pickard BS. J Psychopharmacol. 2015;29:138. doi: 10.1177/0269881114566631. [DOI] [PubMed] [Google Scholar]
- 90.Kalia M, Costa e Silva J. Metabolism. 2015;64:S11. doi: 10.1016/j.metabol.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 91.Yang J, Chen T, Sun L, Zhao Z, Qi X, Zhou K, Cao Y, Wang X, Qiu Y, Su M, Zhao A, Wang P, Yang P, Wu J, Feng G, He L, Jia W, Wan C. Mol Psychiatry. 2013;18:67. doi: 10.1038/mp.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andreazza AC. Mol Biosyst. 2012;8:2503. doi: 10.1039/c2mb25118c. [DOI] [PubMed] [Google Scholar]
- 93.Boskovic M, Vovk T, Plesnicar B Kores, Grabnar I. Curr Neuropharmacol. 2011;9:301. doi: 10.2174/157015911795596595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsai MC, Liou CW, Lin TK, Lin IM, Huang TL. Psychiatry Res. 2013;209:284. doi: 10.1016/j.psychres.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 95.Fraguas D, Gonzalez-Pinto A, Mico JA, Reig S, Parellada M, Martinez-Cengotitabengoa M, Castro-Fornieles J, Rapado-Castro M, Baeza I, Janssen J, Desco M, Leza JC, Arango C. Schizophr Res. 2012;137:58. doi: 10.1016/j.schres.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 96.Yao JK, Keshavan MS. Antioxid Redox Signal. 2011;15:2011. doi: 10.1089/ars.2010.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chocron SE, Weisberger BM, Ben-Yoav H, Winkler TE, Kim E, Kelly DL, Payne GF, Ghodssi R. PloS One. 2015;10:21. doi: 10.1371/journal.pone.0116310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fornito A, Zalesky A, Pantelis C, Bullmore ET. Neuroimage. 2012;62:2296. doi: 10.1016/j.neuroimage.2011.12.090. [DOI] [PubMed] [Google Scholar]
- 99.Weickert CS, Weickert TW, Pillai A, Buckley PF. Dis Markers. 2013:3. doi: 10.1155/2013/510402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sturman DA, Moghaddam B. Neurosci Biobehav Rev. 2011;35:1704. doi: 10.1016/j.neubiorev.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Payne GF, Kim E, Cheng Y, Wu HC, Ghodssi R, Rubloff GW, Raghavan SR, Culver JN, Bentley WE. Soft Matter. 2013;9:6019. [Google Scholar]
- 102.Liu Y, Kim E, Ghodssi R, Rubloff GW, Culver JN, Bentley WE, Payne GF. Biofabrication. 2010;2:022002. doi: 10.1088/1758-5082/2/2/022002. [DOI] [PubMed] [Google Scholar]
- 103.Pang X, Zhitomirsky I. Mater Chem Phys. 2005;94:245. [Google Scholar]
- 104.Luo XL, Xu JJ, Du Y, Chen HY. Anal Biochem. 2004;334:284. doi: 10.1016/j.ab.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 105.Redepenning J, Venkataraman G, Chen J, Stafford N. J Biomed Mater Res Part A. 2003;66A:411. doi: 10.1002/jbm.a.10571. [DOI] [PubMed] [Google Scholar]
- 106.Wu LQ, Gadre AP, Yi HM, Kastantin MJ, Rubloff GW, Bentley WE, Payne GF, Ghodssi R. Langmuir. 2002;18:8620. [Google Scholar]
- 107.Mailloux S, Halamek J, Katz E. Analyst. 2014;139:982. doi: 10.1039/c3an02162a. [DOI] [PubMed] [Google Scholar]
- 108.Shi XW, Tsao CY, Yang XH, Liu Y, Dykstra P, Rubloff GW, Ghodssi R, Bentley WE, Payne GF. Adv Funct Mater. 2009;19:2074. [Google Scholar]
- 109.Cheong M, Zhitomirsky I. Colloid Surf A-Physicochem Eng Asp. 2008;328:73. [Google Scholar]
- 110.Boccaccini AR, Keim S, Ma R, Li Y, Zhitomirsky I. J R Soc Interface. 2010;7:S581. doi: 10.1098/rsif.2010.0156.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Heller A. Curr Opin Chem Biol. 2006;10:664. doi: 10.1016/j.cbpa.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 112.Gao ZQ, Binyamin G, Kim HH, Barton SC, Zhang YC, Heller A. Angew Chem Int Ed. 2002;41:810. doi: 10.1002/1521-3773(20020301)41:5<810::aid-anie810>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 113.Shkil H, Schulte A, Guschin DA, Schuhmann W. ChemPhysChem. 2011;12:806. doi: 10.1002/cphc.201000889. [DOI] [PubMed] [Google Scholar]
- 114.Guschin DA, Castillo J, Dimcheva N, Schuhmann W. Anal Bioanal Chem. 2010;398:1661. doi: 10.1007/s00216-010-3982-3. [DOI] [PubMed] [Google Scholar]
- 115.Hasan K, Patil SA, Leech D, Hagerhall C, Gorton L. Biochem Soc Trans. 2012;40:1330. doi: 10.1042/BST20120120. [DOI] [PubMed] [Google Scholar]
- 116.Wu LQ, Yi HM, Li S, Rubloff GW, Bentley WE, Ghodssi R, Payne GF. Langmuir. 2003;19:519. [Google Scholar]
- 117.Park JJ, Luo X, Yi H, Valentine TM, Payne GF, Bentley WE, Ghodssi R, Rubloff GW. Lab Chip. 2006;6:1315. doi: 10.1039/b603101c. [DOI] [PubMed] [Google Scholar]
- 118.Fusco S, Chatzipirpiridis G, Sivaraman KM, Ergeneman O, Nelson BJ, Pane S. Adv Healthcare Mater. 2013;2:1037. doi: 10.1002/adhm.201200409. [DOI] [PubMed] [Google Scholar]
- 119.Chen Q, Li W, Goudouria OM, Ding YP, Cabanas-Polo S, Boccaccini AR. Colloid Surf B-Biointerfaces. 2015;130:199. doi: 10.1016/j.colsurfb.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 120.Seuss S, Boccaccini AR. Biomacromolecules. 2013;14:3355. doi: 10.1021/bm401021b. [DOI] [PubMed] [Google Scholar]
- 121.Yi HM, Wu LQ, Ghodssi R, Rubloff GW, Payne GF, Bentley WE. Anal Chem. 2004;76:365. doi: 10.1021/ac034662t. [DOI] [PubMed] [Google Scholar]
- 122.Peng XH, Liu Y, Bentley WE, Payne GF. Biomacromolecules. 2016;17:558. doi: 10.1021/acs.biomac.5b01491. [DOI] [PubMed] [Google Scholar]
- 123.Shoda S, Uyama H, Kadokawa J, Kimura S, Kobayashi S. Chem Rev. 2016;116:2307. doi: 10.1021/acs.chemrev.5b00472. [DOI] [PubMed] [Google Scholar]
- 124.Liu Y, Terrell JL, Tsao CY, Wu HC, Javvaji V, Kim E, Cheng Y, Wang YF, Ulijn RV, Raghavan SR, Rubloff GW, Bentley WE, Payne GF. Adv Funct Mater. 2012;22:3004. [Google Scholar]
- 125.Cheng Y, Tsao CY, Wu HC, Luo X, Terrell JL, Betz J, Payne GF, Bentley WE, Rubloff GW. Adv Funct Mater. 2012;22:519. [Google Scholar]
- 126.Krabbenborg SO, Huskens J. Angew Chem-Int Edit. 2014;53:9152. doi: 10.1002/anie.201310349. [DOI] [PubMed] [Google Scholar]
- 127.Kim E, Xiong Y, Cheng Y, Wu HC, Liu Y, Morrow BH, Ben-Yoav H, Ghodssi R, Rubloff GW, Shen JN, Bentley WE, Shi XW, Payne GF. Polymers. 2015;7:1. [Google Scholar]
- 128.Suginta W, Khunkaewla P, Schulte A. Chem Rev. 2013;113:5458. doi: 10.1021/cr300325r. [DOI] [PubMed] [Google Scholar]
- 129.Gray KM, Liba BD, Wang Y, Cheng Y, Rubloff GW, Bentley WE, Montembault A, Royaud I, David L, Payne GF. Biomacromolecules. 2012;13:1181. doi: 10.1021/bm3001155. [DOI] [PubMed] [Google Scholar]
- 130.Shi XW, Yang XH, Gaskell KJ, Liu Y, Kobatake E, Bentley WE, Payne GF. Adv Mater. 2009;21:984. [Google Scholar]
- 131.Li Y, Zhitomirsky I. Surf Eng. 2011;27:698. [Google Scholar]
- 132.Zhang TS, Luo D, Wojtal P, Zhitomirsky I. Mater Lett. 2015;159:106. [Google Scholar]
- 133.Ma R, Pang X, Zhitomirsky I. Surf Eng. 2011;27:693. [Google Scholar]
- 134.Cheng Y, Luo XL, Payne GF, Rubloff GW. J Mater Chem. 2012;22:7659. [Google Scholar]
- 135.Cheng Y, Luo XL, Betz J, Payne GF, Bentley WE, Rubloff GW. Soft Matter. 2011;7:5677. [Google Scholar]
- 136.Ozawa F, Ino K, Takahashi Y, Shiku H, Matsue T. J Biosci Bioeng. 2013;115:459. doi: 10.1016/j.jbiosc.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 137.Ozawa F, Ino K, Arai T, Ramon-Azcon J, Takahashi Y, Shiku H, Matsue T. Lab Chip. 2013;13:3128. doi: 10.1039/c3lc50455g. [DOI] [PubMed] [Google Scholar]
- 138.Jin ZY, Guven G, Bocharova V, Halamek J, Tokarev I, Minko S, Melman A, Mandler D, Katz E. ACS Appl Mater Interfaces. 2012;4:466. doi: 10.1021/am201578m. [DOI] [PubMed] [Google Scholar]
- 139.Jin ZY, Harvey AM, Mailloux S, Halamek J, Bocharova V, Twiss MR, Katz E. J Mater Chem. 2012;22:19523. [Google Scholar]
- 140.Katz E, Pingarron JM, Mailloux S, Guz N, Gamella M, Melman G, Melman A. J Phys Chem Lett. 2015;6:1340. doi: 10.1021/acs.jpclett.5b00118. [DOI] [PubMed] [Google Scholar]
- 141.Gamella M, Guz N, Pingarron JM, Aslebagh R, Darie CC, Katz E. Chem Commun. 2015;51:7618. doi: 10.1039/c5cc01498k. [DOI] [PubMed] [Google Scholar]
- 142.Gamella M, Guz N, Mailloux S, Pingarron JM, Katz E. Electroanalysis. 2014;26:2552. [Google Scholar]
- 143.Uquillas JA, Akkus O. Ann Biomed Eng. 2012;40:1641. doi: 10.1007/s10439-012-0528-1. [DOI] [PubMed] [Google Scholar]
- 144.Baker HR, Merschrod EF, Poduska KA. Langmuir. 2008;24:2970. doi: 10.1021/la703743m. [DOI] [PubMed] [Google Scholar]
- 145.Cheng XG, Gurkan UA, Dehen CJ, Tate MP, Hillhouse HW, Simpson GJ, Akkus O. Biomaterials. 2008;29:3278. doi: 10.1016/j.biomaterials.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 146.Kumar MR, Merschrod EF, Poduska KM. Biomacromolecules. 2009;10:1970. doi: 10.1021/bm900379g. [DOI] [PubMed] [Google Scholar]
- 147.Kishore V, Uquillas JA, Dubikovsky A, Alshehabat MA, Snyder PW, Breur GJ, Akkus O. J Biomed Mater Res Part B. 2012;100B:400. doi: 10.1002/jbm.b.31962. [DOI] [PubMed] [Google Scholar]
- 148.Gendron R, Kumar MR, Paradis H, Martin D, Ho N, Gardiner D, Merschrod EF, Poduska KM. Macromol Biosci. 2012;12:360. doi: 10.1002/mabi.201100341. [DOI] [PubMed] [Google Scholar]
- 149.Abu-Rub MT, Billiar KL, van Es MH, Knight A, Rodriguez BJ, Zeugolis DI, McMahon S, Windebank AJ, Pandit A. Soft Matter. 2011;7:2770. [Google Scholar]
- 150.Elia R, Michelson CD, Perera AL, Brunner TF, Harsono M, Leisk GG, Kugel G, Kaplan DL. J Biomed Mater Res Part B. 2015;103:1602. doi: 10.1002/jbm.b.33351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang HY, Zhang YQ. Sci Rep. 2014;4:6182. doi: 10.1038/srep06182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lu Q, Huang YL, Li MZ, Zuo BQ, Lu SZ, Wang JN, Zhu HS, Kaplan DL. Acta Biomater. 2011;7:2394. doi: 10.1016/j.actbio.2011.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maniglio D, Bonani W, Bortoluzzi G, Servoli E, Motta A, Migliaresi C. J Bioact Compat Polym. 2010;25:441. [Google Scholar]
- 154.Leisk GG, Lo TJ, Yucel T, Lu Q, Kaplan DL. Adv Mater. 2010;22:711. doi: 10.1002/adma.200902643. [DOI] [PubMed] [Google Scholar]
- 155.Omenetto FG, Kaplan DL. Science. 2010;329:528. doi: 10.1126/science.1188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jose RR, Elia R, Tien LW, Kaplan DL. ACS Appl Mater Interfaces. 2014;6:6212. doi: 10.1021/am501242w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bressner JE, Marelli B, Qin GK, Klinker LE, Zhang YJ, Kaplan DL, Omenetto FG. J Mat Chem B. 2014;2:4983. doi: 10.1039/c4tb00833b. [DOI] [PubMed] [Google Scholar]
- 158.Kojic N, Panzer MJ, Leisk GG, Raja WK, Kojic M, Kaplan DL. Soft Matter. 2012;8:6897. doi: 10.1039/C2SM25783A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Servoli E, Maniglio D, Motta A, Migliaresi C. Macromol Biosci. 2008;8:827. doi: 10.1002/mabi.200800057. [DOI] [PubMed] [Google Scholar]
- 160.Lin Y, Wang S, Chen Y, Wang Q, Burke KA, Spedden EM, Staii C, Weiss AS, Kaplan DL. Nanomedicine. 2015;10:803. doi: 10.2217/nnm.14.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Raeburn J, Alston B, Kroeger J, McDonald TO, Howse JR, Cameron PJ, Adams DJ. Mater Horizons. 2014;1:241. [Google Scholar]
- 162.Liu Y, Kim E, Ulijn RV, Bentley WE, Payne GF. Adv Funct Mater. 2011;21:1575. [Google Scholar]
- 163.Johnson EK, Adams DJ, Cameron PJ. J Am Chem Soc. 2010;132:5130. doi: 10.1021/ja909579p. [DOI] [PubMed] [Google Scholar]
- 164.Jiang T, Zhang Z, Zhou Y, Liu Y, Wang ZW, Tong H, Shen XY, Wang YN. Biomacromolecules. 2010;11:1254. doi: 10.1021/bm100050d. [DOI] [PubMed] [Google Scholar]
- 165.Zhang Z, Jiang T, Ma KN, Cai XJ, Zhou Y, Wang YN. J Mater Chem. 2011;21:7705. [Google Scholar]
- 166.Liu Y, Cheng Y, Wu HC, Kim E, Ulijn RV, Rubloff GW, Bentley WE, Payne GF. Langmuir. 2011;27:7380. doi: 10.1021/la201541c. [DOI] [PubMed] [Google Scholar]
- 167.Yang XH, Kim E, Liu Y, Shi XW, Rubloff GW, Ghodssi R, Bentley WE, Pancer Z, Payne GF. Adv Funct Mater. 2010;20:1645. [Google Scholar]
- 168.Shahrokhian S, Kamalzadeh Z, Hamzehloei A. Bioelectrochemistry. 2013;90:36. doi: 10.1016/j.bioelechem.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 169.Qu S, Pei S, Zhang S, Song P. Mater Lett. 2013;102:56. [Google Scholar]
- 170.Mashhadizadeh MH, Afshar E. Electrochim Acta. 2013;87:816. [Google Scholar]
- 171.Rouhollahi A, Kouchaki M, Seidi S. RSC Adv. 2016;6:12943. [Google Scholar]
- 172.Kim E, Chocron SE, Ben-Yoav H, Winkler TE, Liu Y, Glassman M, Wolfram C, Kelly DL, Ghodssi R, Payne GF. Adv Funct Mater. 2015;25:2156. [Google Scholar]
- 173.Ladet SG, Tahiri K, Montembault AS, Domard AJ, Corvol MTM. Biomaterials. 2011;32:5354. doi: 10.1016/j.biomaterials.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 174.Ladet S, David L, Domard A. Nature. 2008;452:76. doi: 10.1038/nature06619. [DOI] [PubMed] [Google Scholar]
- 175.Nie JY, Lu WT, Ma JJ, Yang L, Wang ZK, Qin A, Hu QL. Sci Rep. 2015;5:7635. doi: 10.1038/srep07635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Xiong Y, Yan K, Bentley WE, Deng HB, Du YM, Payne GF, Shi XW. ACS Appl Mater Interfaces. 2014;6:2948. doi: 10.1021/am405544r. [DOI] [PubMed] [Google Scholar]
- 177.Dai HJ, Li XF, Long YH, Wu JJ, Liang SM, Zhang XL, Zhao N, Xu J. Soft Matter. 2009;5:1987. [Google Scholar]
- 178.Yan K, Ding FY, Bentley WE, Deng HB, Du YM, Payne GF, Shi XW. Soft Matter. 2014;10:465. doi: 10.1039/c3sm52405a. [DOI] [PubMed] [Google Scholar]
- 179.Schatz C, Viton C, Delair T, Pichot C, Domard A. Biomacromolecules. 2003;4:641. doi: 10.1021/bm025724c. [DOI] [PubMed] [Google Scholar]
- 180.Liu Y, Zhang B, Gray KM, Cheng Y, Kim E, Rubloff GW, Bentley WE, Wang Q, Payne GF. Soft Matter. 2013;9:2703. [Google Scholar]
- 181.Morrow BH, Payne GF, Shen J. J Am Chem Soc. 2015;137:13024. doi: 10.1021/jacs.5b07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theory Comput. 2008;4:435. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 183.Chen W, Shen JK. J Comput Chem. 2014;35:1986. doi: 10.1002/jcc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chen W, Wallace JA, Yue Z, Shen JK. Biophys J. 2013;105:L15. doi: 10.1016/j.bpj.2013.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wallace JA, Shen JK. J Chem Phys. 2012;137:9. doi: 10.1063/1.4766352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Khandogin J, Brooks CL. Biophys J. 2005;89:141. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lee MS, Salsbury FR, Brooks CL. Proteins. 2004;56:738. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 188.Wallace JA, Shen JK. J Chem Theory Comput. 2011;7:2617. doi: 10.1021/ct200146j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sarkar K, Meister K, Sethi A, Gruebele M. Biophys J. 2009;97:1418. doi: 10.1016/j.bpj.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chen SJ, Dill KA. Proc Natl Acad Sci U S A. 2000;97:646. doi: 10.1073/pnas.97.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang JX, Li XD, Li XH. Prog Polym Sci. 2012;37:1130. [Google Scholar]
- 192.Rybtchinski B. ACS Nano. 2011;5:6791. doi: 10.1021/nn2025397. [DOI] [PubMed] [Google Scholar]
- 193.Carvajal D, Bitton R, Mantei JR, Velichko YS, Stupp SI, Shull KR. Soft Matter. 2010;6:1816. [Google Scholar]
- 194.Moore JS, Kraft ML. Science. 2008;320:620. doi: 10.1126/science.1157225. [DOI] [PubMed] [Google Scholar]
- 195.Capito RM, Azevedo HS, Velichko YS, Mata A, Stupp SI. Science. 2008;319:1812. doi: 10.1126/science.1154586. [DOI] [PubMed] [Google Scholar]
- 196.Cheng Y, Luo XL, Betz J, Buckhout-White S, Bekdash O, Payne GF, Bentley WE, Rubloff GW. Soft Matter. 2010;6:3177. [Google Scholar]
- 197.Wu LQ, McDermott MK, Zhu C, Ghodssi R, Payne GF. Adv Funct Mater. 2006;16:1967. [Google Scholar]
- 198.Wu LQ, Ghodssi R, Elabd YA, Payne GF. Adv Funct Mater. 2005;15:189. [Google Scholar]
- 199.Kim E, Leverage WT, Liu Y, White IM, Bentley WE, Payne GF. Analyst. 2014;139:32. doi: 10.1039/c3an01632c. [DOI] [PubMed] [Google Scholar]
- 200.Kim E, Liu Y, Shi XW, Yang X, Bentley WE, Payne GF. Adv Funct Mater. 2010;20:2683. [Google Scholar]
- 201.Kim E, Liu Y, Bentley WE, Payne GF. Adv Funct Mater. 2012;22:1409. [Google Scholar]
- 202.Kim E, Liu Y, Baker CJ, Owens R, Xiao SY, Bentley WE, Payne GF. Biomacromolecules. 2011;12:880. doi: 10.1021/bm101499a. [DOI] [PubMed] [Google Scholar]
- 203.Li J, Wang PG, Zhang N, Yang Y, Zheng JB. Electrochim Acta. 2015;166:253. [Google Scholar]
- 204.Kim E, Gordonov T, Bentley WE, Payne GF. Anal Chem. 2013;85:2102. doi: 10.1021/ac302703y. [DOI] [PubMed] [Google Scholar]
- 205.Ben-Yoav H, Chocron SE, Winkler TE, Kim E, Kelly DL, Payne GF, Ghodssi R. Electrochim Acta. 2015;163:260. [Google Scholar]
- 206.Winkler TE, Ben-Yoav H, Chocron SE, Kim E, Kelly DL, Payne GF, Ghodssi R. Langmuir. 2014;30:14686. doi: 10.1021/la503529k. [DOI] [PubMed] [Google Scholar]
- 207.Ben-Yoav H, Winkler TE, Kim E, Chocron SE, Kelly DL, Payne GF, Ghodssi R. Electrochim Acta. 2014;130:497. [Google Scholar]
- 208.van Leeuwen SM, Blankert B, Kauffmann JM, Karst U. Anal Bioanal Chem. 2005;382:742. doi: 10.1007/s00216-005-3053-3. [DOI] [PubMed] [Google Scholar]
- 209.Kim E, Gordonov T, Liu Y, Bentley WE, Payne GF. ACS Chem Biol. 2013;8:716. doi: 10.1021/cb300605s. [DOI] [PubMed] [Google Scholar]
- 210.Liu Y, Kim E, White IM, Bentley WE, Payne GF. Bioelectrochemistry. 2014;98:94. doi: 10.1016/j.bioelechem.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 211.Wang J. Chem Rev. 2008;108:814. doi: 10.1021/cr068123a. [DOI] [PubMed] [Google Scholar]
- 212.Logan BE. Nat Rev Microbiol. 2009;7:375. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- 213.Rabaey K, Rozendal RA. Nat Rev Microbiol. 2010;8:706. doi: 10.1038/nrmicro2422. [DOI] [PubMed] [Google Scholar]
- 214.von Canstein H, Ogawa J, Shimizu S, Lloyd JR. Appl Environ Microbiol. 2008;74:615. doi: 10.1128/AEM.01387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Thrash JC, Coates JD. Environ Sci Technol. 2008;42:3921. doi: 10.1021/es702668w. [DOI] [PubMed] [Google Scholar]
- 216.Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. Proc Natl Acad Sci U S A. 2008;105:3968. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sander M, Hofstetter TB, Gorski CA. Environ Sci Technol. 2015;49:5862. doi: 10.1021/acs.est.5b00006. [DOI] [PubMed] [Google Scholar]
- 218.Aeschbacher M, Brunner SH, Schwarzenbach RP, Sander M. Environ Sci Technol. 2012;46:3882. doi: 10.1021/es204496d. [DOI] [PubMed] [Google Scholar]
- 219.Aeschbacher M, Vergari D, Schwarzenbach RP, Sander M. Environ Sci Technol. 2011;45:8385. doi: 10.1021/es201981g. [DOI] [PubMed] [Google Scholar]
- 220.Ranallo S, Amodio A, Idili A, Porchetta A, Ricci F. Chem Sci. 2016;7:66. doi: 10.1039/c5sc03694a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mailloux S, Gerasimova YV, Guz N, Kolpashchikov DM, Katz E. Angew Chem Int Ed. 2015;54:6562. doi: 10.1002/anie.201411148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gordonov T, Kim E, Cheng Y, Ben-Yoav H, Ghodssi R, Rubloff G, Yin JJ, Payne GF, Bentley WE. Nat Nanotechnol. 2014;9:605. doi: 10.1038/nnano.2014.151. [DOI] [PubMed] [Google Scholar]
- 223.Van Gheem E, Pintelon R, Vereecken J, Schoukens J, Hubin A, Verboven P, Blajiev O. Electrochim Acta. 2004;49:4753. [Google Scholar]
- 224.Liu Y, Kim E, Lee ME, Zhang B, Elabd YA, Wang Q, White IM, Bentley WE, Payne GF. Adv Funct Mater. 2014:480. [Google Scholar]
- 225.Gray KM, Kim E, Wu LQ, Liu Y, Bentley WE, Payne GF. Soft Matter. 2011;7:9601. [Google Scholar]


