Abstract
The membrane-bound transcription factors, SREBPs (sterol regulatory element-binding proteins), play a central role in regulating lipid metabolism. The transcriptional activation of SREBPs requires the key protein SCAP (SREBP-cleavage activating protein) to translocate their precursors from the endoplasmic reticulum to the Golgi for subsequent proteolytic activation, a process tightly regulated by a cholesterol-mediated negative feedback loop. Our previous work showed that the SCAP/SREBP-1 pathway is significantly upregulated in human glioblastoma (GBM), the most deadly brain cancer, and that glucose-mediated N-glycosylation of SCAP is a prerequisite step for SCAP/SREBP trafficking. More recently, we demonstrated that microRNA-29 (miR-29) mediates a previously unrecognized negative feedback loop in SCAP/SREBP-1 signaling to control lipid metabolism. We found that SREBP-1, functioning as a transcription factor, promotes the expression of the miR-29 family members, miR-29a, -29b and -29c. In turn, the miR-29 isoforms reversely repress the expression of SCAP and SREBP-1. Moreover, treatment with miR-29 mimics effectively suppressed GBM tumor growth by inhibiting SCAP/SREBP-1 and de novo lipid synthesis. These findings, recently published in Cell Reports, strongly suggest that delivery of miR-29 in vivo may be a promising approach to treat cancer and metabolic diseases by suppressing SCAP/SREBP-1-regulated lipid metabolism.
Keywords: Lipid metabolism, SCAP, SREBP-1, miR-29, glioblastoma, cancer
Lipids are essential components of the cells, particularly phospholipids and cholesterol, constituting the basic structure of cell membrane system [1, 2]. Furthermore, lipids also serve as important signaling molecules, regulating various cellular functions[3]. Dysregulation of lipid metabolism contributes to the pathogenesis of various metabolic syndromes, i.e., atherosclerosis, steatosis, obesity and diabetes [4]. Therefore, interfering with the dysregulated lipid metabolism in metabolic diseases has been a long-term focus of basic research and pharmacological development [4, 5]. Nevertheless, the still incomplete understanding of the molecular mechanisms underlying the alteration of lipid metabolism significantly hinders progress.
The family of basic helix-loop-helix transcription factors, SREBPs (sterol regulatory element-binding proteins), plays a central role in lipid metabolism by controlling the de novo synthesis of fatty acids, phospholipids, cholesterol, and also cholesterol uptake, which were initially discovered by Nobel Laureates Brown & Goldstein around 20 years ago [3–7]. SREBPs are comprised of three members, SREBP-1a, -1c and -2. SREBP-1a and -1c that are encoded by the same gene using different transcriptional start sites, resulting in a distinct exon 1 and around a 20 amino acid longer N-terminus in SREBP-1a than in -1c [8, 9]. SREBP-2 is encoded from a different gene and mainly controls cholesterol synthesis and uptake while SREBP-1c regulates fatty acid synthesis. In contrast, SREBP-1a is able to execute all three functions, i.e., fatty acid synthesis, cholesterol synthesis and cholesterol uptake [6, 10–14].
Brown & Goldstein put forth an elegant model of the regulation of SREBP activation through a cholesterol-mediated negative feedback loop (Fig. 1) [4, 6]. After translation, SREBPs immediately bind to the key protein, SCAP (SREBP-cleavage activating protein), to form a complex. SCAP further binds to Insig (insulin-induced gene protein), an endoplasmic reticulum (ER)-anchored protein, resulting in the formation of the Insig/SCAP/SREBP complex, which is retained in the ER by high levels of cholesterol [15–17]. When cholesterol levels decrease, SCAP dissociates from Insig, resulting in the degradation of Insig. SCAP then interacts with COPII proteins that translocate the SCAP/SREBP complex from the ER to the Golgi, where SREBPs are sequentially cleaved by site-1 and site-2 proteinases to release their transcriptionally active N-terminal fragments that enter into the nucleus to promote the transcription of lipogenic genes including Insig-1 [4, 6, 10, 11, 17, 18]. Consequently, the levels of cholesterol and Insig are restored to bind again to the SCAP/SREBP complex, which is then retained in the ER, resulting in the reduction of lipid synthesis and uptake (Fig. 1) [19–22].
Figure 1. miR-29 mediates a novel negative feedback loop in SCAP/SREBP-1 signaling and regulates lipid metabolism.
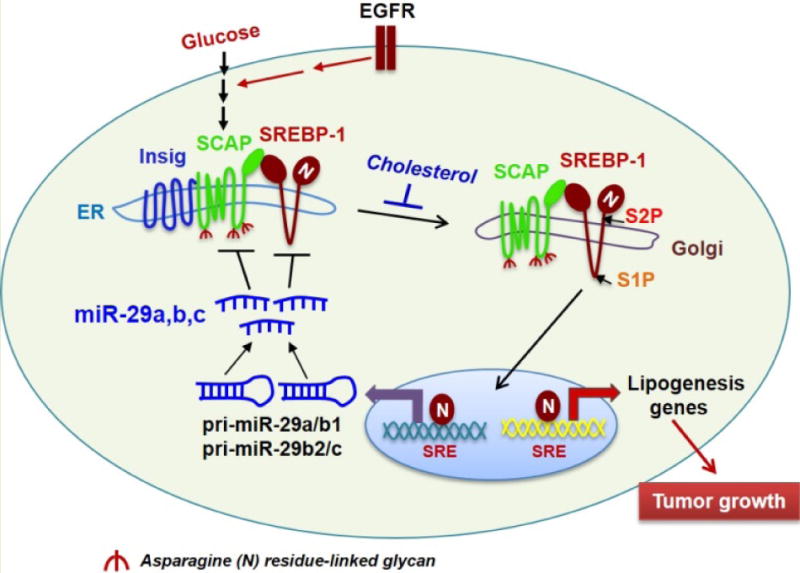
Our previous study showed that glucose-mediated SCAP N-glycosylation enables SCAP/SREBP-1 trafficking from the ER to the Golgi for subsequent proteolytic activation. Furthermore, EGFR signaling enhances glucose uptake, thereby increasing SCAP N-glycosylation and SREBP-1 activation to promote tumor growth [23–25]. High levels of cholesterol increase the association of Insig and SCAP, resulting in the retention of the complex in the ER [6]. Our newly discovered negative feedback loop shows that SREBP-1 transcriptionally activates the expression of pri-miR-29a/b1 and pri-miR-29b2/c, which generate the mature miR-29a, -29b and 29c. In turn, miR-29 reversely inhibits the expression of SCAP and SREBP-1 by binding to their 3′-UTRs, resulting in the downregulation of lipogenesis genes [55]. SRE, sterol regulatory element (SREBP-binding motif present in the promoters of SREBP target genes). S1P, site 1 protease. S2P, site 2 protease.
To explore whether other factors are critical for SCAP/SREBP trafficking, we investigated the role of glucose-mediated N-glycosylation modification of the SCAP protein, and showed that it was a prerequisite step for SCAP/SREBP trafficking and activation upon cholesterol reduction [23–25]. We found that N-glycosylation stabilizes SCAP and reduces its association with Insig-1, allowing SCAP/SREBP movement from the ER to the Golgi (Fig. 1). Our study demonstrated that glucose is an essential activator of SCAP/SREBP trafficking, while cholesterol functions as a key inhibitor of this process [23–25].
Recent evidence shows that lipid metabolism is largely altered in cancer cells [26–33]. Our previous studies were the first to demonstrate that lipid metabolism is reprogrammed in glioblastoma (GBM) [29, 33–36], the most common primary brain tumor and one of the most lethal of all cancers [36–41]. Our data show that GBM tumors bearing amplification of the tyrosine kinase receptor, EGFR, or expressing the constitutively active EGFRvIII, which lacks a portion of the extracellular ligand binding domain due to the deletion of exons 2–7 of the EGFR gene [39, 42, 43], were greatly dependent on SREBP-1-mediated lipogenesis and cholesterol uptake for their rapid growth[34, 36, 44, 45]. We found that EGFR/EGFRvIII activates SREBP-1 via PI3K/Akt signaling to promote lipid synthesis [6, 10, 36], and that EGFR signaling enhances glucose uptake to promote SCAP N-glycosylation and SCAP/SREBP-1 trafficking [23–25].
SREBP proteins were recently reported to be upregulated in various cancers and now emerge as promising molecular targets for cancer treatment [33, 44, 46]. Nevertheless, the pharmacological development to directly target SCAP/SREBP has not been successful so far, and alternative means to block this pathway are needed. Thus, we turned our attention to microRNAs (miRNAs), small non-coding RNAs that greatly affect the expression and translation of a large number of genes [47,48]. miRNAs are involved in many biological processes, i.e., cell growth, development, differentiation, survival, etc. [47–49]. Moreover, miRNAs have been shown to be involved in tumorigenesis where they function as tumor suppressors or oncomiRs [50], and to regulate lipid metabolism [51–53].
We identified miRNA-29 as a critical mediator of a novel negative feedback loop in the regulation of SCAP/SREBP-1 signaling [54], providing a promising new approach to target GBM. The miRNA-29 family includes 3 members, miR-29a, -29b and -29c, which share the same seed sequence. miR-29b is encoded by pri-miR-29b1 and pri-miR-29b2, which are located on different chromosomes but generate the same mature miR-29b. Interestingly, pri-miR-29a and pri-miR-29b1 are both located on chromosome 7 and share the same promoter. Similarly, pri-miR-29b2 and pri-miR-29c are located on chromosome 1 and are co-transcribed by the same promoter [55,56].
In our study, we found that expression of all 3 mature miR-29s was positively correlated with SREBP-1 gene expression in samples from a large cohort of GBM patients with altered EGFR (amplification or mutation) [55]. Furthermore, activating EGFR/PI3K/Akt signaling via EGF stimulation significantly enhanced the expression of all 3 miR-29s in GBM cell lines. Interestingly, both SREBP-1a and -1c directly bind to the promoter region of miR-29a/b1 and miR-29b2/c, activating their expression and generating mature miRNA-29a, -29b and -29c. We also showed that the 3′-untranslated region (3′-UTR) of SREBP-1 has binding sites for miR-29, and demonstrated that miR-29a, -29b and -29c inhibited the mRNA and protein levels of SREBP-1 by directly binding to these complementary sites. Importantly, our intracranial GBM xenograft studies show that miR-29 treatment significantly suppressed tumor growth via inhibition of SCAP/SREBP-1 and lipid synthesis [54].
miR-29 has been shown to be transcriptionally inhibited by transcription factors such as c-Myc, TGF-β and NF-κB in cancer cells [57–59]. Our study was the first to show that miR-29 expression is controlled by SREBP-1, and that miR-29 is directly involved in the regulation of lipid metabolism. This newly discovered negative feedback loop regulation of SCAP/SREBP-1 by miR-29 further demonstrates that lipid homeostasis is elegantly regulated by multi-layer of mechanisms in addition to cholesterol and glucose regulation [23–25, 54]. Considering the simple synthesis and easy delivery of mature microRNAs, miR-29 treatment may be a feasible and promising approach to treat cancers and other metabolic diseases.
Acknowledgments
This work was supported by the grants NIH NS079701 (DG), American Cancer Society Research Scholar Grant RSG-14-228-01-CSM (DG) and OSUCCC TT-seed grant (DG). The authors wish to thank Dr. Martine Torres for her editorial assistance.
Abbreviations
- miR-29
microRNA-29
- SREBPs
sterol regulatory element-binding proteins
- SCAP
SREBP-cleavage activating protein
- GBM
glioblastoma
- Insig
insulin-induced gene
- ER
endoplasmic reticulum
- 3′-UTR
3′-untranslated region
Footnotes
Conflicting interests
The authors have declared that no conflict of interests exist.
References
- 1.Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 2.Maxfield FR, van Meer G. Cholesterol, the central lipid of mammalian cells. Current opinion in cell biology. 2010;22:422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nohturfft A, Zhang SC. Coordination of lipid metabolism in membrane biogenesis. Annual review of cell and developmental biology. 2009;25:539–566. doi: 10.1146/annurev.cellbio.24.110707.175344. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein JL, Brown MS. A Century of Cholesterol and Coronaries: From Plaques to Genes to Statins. Cell. 2015;161:161–172. doi: 10.1016/j.cell.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends in endocrinology and metabolism: TEM. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Shao W, Espenshade PJ. Expanding Roles for SREBP in Metabolism. Cell metabolism. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. The Journal of clinical investigation. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. The Journal of clinical investigation. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 13.Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, et al. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci U S A. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, et al. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 15.Lee PC, Sever N, Debose-Boyd RA. Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. The Journal of biological chemistry. 2005;280:25242–25249. doi: 10.1074/jbc.M502989200. [DOI] [PubMed] [Google Scholar]
- 16.Sun LP, Li L, Goldstein JL, Brown MS. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. The Journal of biological chemistry. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 17.Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 19.Espenshade PJ, Li WP, Yabe D. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11694–11699. doi: 10.1073/pnas.182412799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 21.Adams CM, Goldstein JL, Brown MS. Cholesterol-induced conformational change in SCAP enhanced by Insig proteins and mimicked by cationic amphiphiles. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10647–10652. doi: 10.1073/pnas.1534833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, et al. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. The Journal of biological chemistry. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 23.Cheng C, Ru P, Geng F, Liu J, Yoo JY, Wu X, et al. Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer Cell. 2015;28:569–581. doi: 10.1016/j.ccell.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo D. SCAP links glucose to lipid metabolism in cancer cells. Mol Cell Oncol. 2016;3 doi: 10.1080/23723556.2015.1132120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng C, Guo JY, Geng F, Wu X, Cheng X, Li Q, et al. Analysis of SCAP N-glycosylation and Trafficking in Human Cells. J Vis Exp. 2016 doi: 10.3791/54709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 27.Santos CR, Schulze A. Lipid metabolism in cancer. Febs J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 28.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular Fatty Acid Metabolism and Cancer. Cell metabolism. 2013 doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncology. 2013;2:289–299. doi: 10.2217/cns.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo D, Cloughesy TF, Radu CG, Mischel PS. AMPK: A metabolic checkpoint that regulates the growth of EGFR activated glioblastomas. Cell Cycle. 2010;9:211–212. doi: 10.4161/cc.9.2.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo D, Hildebrandt IJ, Prins RM, Soto H, Mazzotta MM, Dang J, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc Natl Acad Sci U S A. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng F, Cheng X, Wu X, Yoo JY, Cheng C, Guo JY, et al. Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP-1-Mediated Lipogenesis. Clin Cancer Res. 2016;22:5337–5348. doi: 10.1158/1078-0432.CCR-15-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo D, Hildebrandt IJ, Prins RM, Soto H, Mazzotta MM, Dang J, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ru P, Williams TM, Chakravarti A, Guo D. Tumor metabolism of malignant gliomas. Cancers (Basel) 2013;5:1469–1484. doi: 10.3390/cancers5041469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo D, Prins RM, Dang J, Kuga D, Iwanami A, Soto H, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal. 2009;2:ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen PY, Kesari S. Malignant gliomas in adults. The New England journal of medicine. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 38.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The lancet oncology. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 39.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 40.Paleologos NA, Merrell RT. Anaplastic glioma. Curr Treat Options Neurol. 2012;14:381–390. doi: 10.1007/s11940-012-0177-6. [DOI] [PubMed] [Google Scholar]
- 41.Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379:1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 42.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 43.Yoshimoto K, Dang J, Zhu S, Nathanson D, Huang T, Dumont R, et al. Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Clin Cancer Res. 2008;14:488–493. doi: 10.1158/1078-0432.CCR-07-1966. [DOI] [PubMed] [Google Scholar]
- 44.Guo D, Bell EH, Mischel P, Chakravarti A. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr Pharm Des. 2014;20:2619–2626. doi: 10.2174/13816128113199990486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo D, Reinitz F, Youssef M, Hong C, Nathanson D, Akhavan D, et al. An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov. 2011;1:442–456. doi: 10.1158/2159-8290.CD-11-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell EH, Guo D. Biomarkers for malignant gliomas. Malignant Gliomas, Radiation Medicine Rounds. 2012;3:389–357. [Google Scholar]
- 47.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 48.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Liu J, Wang X, Feng Z. The regulation of the p53/MDM2 feedback loop by microRNAs. RNA Dis. 2015;2:e502. doi: 10.14800/rd.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeon TI, Esquejo RM, Roqueta-Rivera M, Phelan PE, Moon YA, Govindarajan SS, et al. An SREBP-Responsive microRNA Operon Contributes to a Regulatory Loop for Intracellular Lipid Homeostasis. Cell metabolism. 2013;18:51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeon TI, Osborne TF. miRNA and cholesterol homeostasis. Biochim Biophys Acta. 2016;1861:2041–2046. doi: 10.1016/j.bbalip.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Hernando C, Suarez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011;22:86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ru P, Hu P, Geng F, Mo X, Cheng C, Yoo JY, et al. Feedback Loop Regulation of SCAP/SREBP-1 by miR-29 Modulates EGFR Signaling-Driven Glioblastoma Growth. Cell Rep. 2016;16:1527–1535. doi: 10.1016/j.celrep.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics. 2012;44:237–244. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang H, Zhang G, Wu JH, Jiang CP. Diverse roles of miR-29 in cancer (review) Oncol Rep. 2014;31:1509–1516. doi: 10.3892/or.2014.3036. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R, et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell. 2012;22:506–523. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


