Abstract
We have previously presented the histone methyltransferase enhancer of zeste homolog 2 (EZH2) of the polycomb repressive complex 2 (PRC2) as a potential therapeutic target in Multiple Myeloma (MM). In a recent article in Oncotarget by Alzrigat. et al. 2017, we have reported on the novel finding that EZH2 inhibition using the highly selective inhibitor of EZH2 enzymatic activity, UNC1999, reactivated the expression of microRNA genes previously reported to be underexpressed in MM. Among these, we have identified miR-125a-3p and miR-320c as potential tumor suppressor microRNAs as they were predicted to target MM-associated oncogenes; IRF-4, XBP-1 and BLIMP-1. We also found EZH2 inhibition to reactivate the expression of miR-494, a previously reported regulator of the c-MYC oncogene. In addition, we could report that EZH2 inhibition downregulated the expression of a few well described oncogenic microRNAs in MM. The data from our recent article are here highlighted as it shed a new light onto the oncogenic function of the PRC2 in MM. These data further strengthen the notion that the PRC2 complex may be of potential therapeutic interest.
Keywords: Multiple Myeloma, PRC2, EZH2, H3K27me3, microRNA
Multiple myeloma (MM) is a malignancy of plasmablasts/plasma cells (PCs) characterized by the accumulation of monoclonal antibody producing PCs in the bone marrow (BM). Clinically, MM is a heterogeneous disease and MM patients’ show multiple clinical symptoms including lytic bone lesions, anemia, hypercalcemia, renal failure and immunodeficiency [1–3]. More importantly, MM is a biologically complex disorder characterized by a large clonal heterogeneity as reflected by a wide range of genetic alterations and manifested in a patient-to-patient variation in overall survival and response to treatment [4–7]. This MM associated heterogeneity has certainly limited the clinical benefits of current as well as personalized treatment strategies. Therefore, MM remains a fatal disease making development of new targeted therapeutic approaches imperative.
Several reports from genetic sequencing and gene expression studies in MM have documented a cross-talk between genetic lesions and aberrant epigenetic profiles i.e. DNA methylation [8, 9], histone modifications [10, 11] and non-coding RNA [12–14] in the pathogenesis and prognosis of MM. An emerging notion is now that deregulation of epigenetic modifiers is an important factor contributing to the development of MM [15–18]. For example, the chromosomal translocation t(4;14) results in the overexpression of the multiple myeloma set domain (MMSET) histone methyltransferase leading to an increase in histone 3 lysine 36 di-methylation (H3K36me2) levels and a concomitant decrease in histone 3 lysine 27 tri-methylation (H3K27me3) levels [10, 11]. The enhancer of zeste homolog 2 (EZH2) is an epigenetic modifier that has been shown by us and others to be commonly overexpressed in MM [16, 19, 20]. EZH2 is the enzymatic subunit of the polycomb repressive complex 2 (PRC2), an important regulator of both normal development as well as disease [21–23]. Through EZH2, the PRC2 complex establishes the H3K27me3 mark, a transcriptional repressive histone mark involved in the regulation of transcriptional programs during normal development as well as cellular transformation [21–23]. EZH2 was found to be overexpressed in malignant PCs as compared to normal BM PCs, and to enhance MM cell growth [19]. Recently, we have shown that a common set of PRC2/H3K27me3 targeted genes are underexpressed in MM patients [20, 24]. Stressing the clinical relevance of gene silencing by PRC2, we found that the repression of PRC2 target genes (H3K27me3 targets) in MM correlates with gene silencing in advanced stages of MM and in patients presenting with poor survival [24]. The development of epigenetic inhibitors that specifically dampen the EZH2 enzymatic activity has recently made the evaluation of the therapeutic potential of EZH2 in MM possible. We and others have demonstrated the anti-MM effects mediated by EZH2 inhibition by using highly selective inhibitors of the EZH2 enzymatic activity [24–26]. All these studies reported on the anti-MM effects of EZH2 inhibitors via reactivation of a set of PRC2 target genes with anti-tumor functions such as genes involved in apoptosis, cell differentiation, cell adhesion and migration.
As here highlighted, we have recently reported for the first time that inhibition of EZH2 using the small highly selective inhibitor of EZH2 enzymatic activity, the UNC1999 [27], has an impact on the global expression of microRNA genes in MM. In this study we presented PRC2 as a novel regulator of a set of microRNAs with tumor suppressor or oncogenic function in MM [28]. In the study, we found that EZH2 inhibition by UNC1999 resulted in the upregulation of 118 microRNAs, of which many have been identified as downregulated tumor suppressor microRNAs in MM [28]. We could show that 2 potential tumor suppressor microRNAs, miR-125a-3p and miR-320c, were reactivated upon EZH2 inhibition (Figure 1). We selected these microRNAs based on their predicted binding and function as common regulators of MM important oncogenes i.e. IRF-4, XBP-1 and BLIMP-1 [28]. We also found that UNC1999 upregulated the expression of miR-494 with a previously reported function to negatively regulate the expression of the c-MYC oncogene [29]. Using chromatin immunoprecipitation followed by quantitative real time PCR (ChIP-qPCR), we found that miR-125a and miR-320c were direct targets of PRC2 in MM cell lines and primary MM patient cells and that their reactivation, as predicted, correlated with the downregulation of expression of MM-associated oncogenes IRF-4, XBP-1, BLIMP-1 and c-MYC [28]. The significance of our finding relies on the fact that these oncogenes have been demonstrated to be essential for MM cell growth and survival [30–33] and MM pathogenesis in human and murine models [34–37].
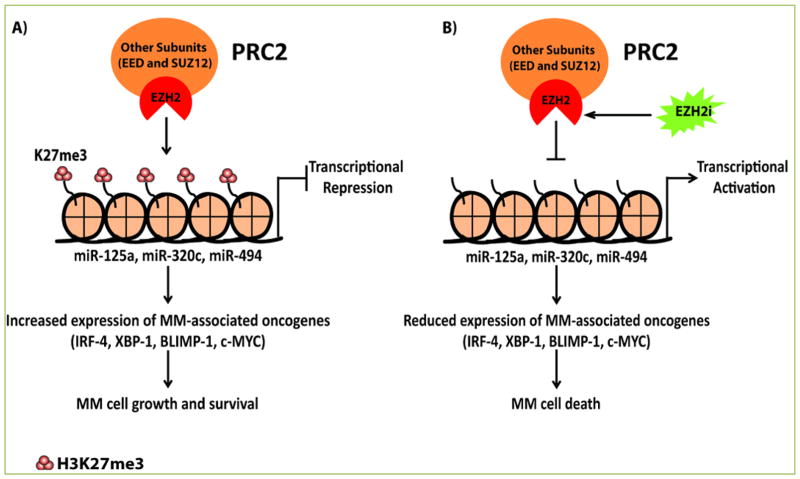
Figure 1. PRC2 represses the expression of tumor suppressor microRNAs in multiple myeloma.
(a) PRC2 via EZH2 enzymatic subunit installs H3K27me3 mark at the genes encoding miR-125a, miR-320c and miR-494 in MM cells leading to their silencing. This leads to sustained expression of MM-associated oncogenes predicted to be targets of the repressed microRNA resulting in enhanced MM cell growth and survival. (b) Pharmacological inhibition of EZH2 methyltransferase activity using highly selective inhibitors such as UNC1999 abolishes the installation of H3K27me3 at the microRNA genes, leading to their transcriptional activation. Expression of miR-125a, miR-320c and miR-494 leads to reduced expression of MM-associated oncogenes thus inducing MM cell death.
We also showed that the inhibition of EZH2 downregulated the expression of microRNAs reported to be overexpressed and to possess oncogenic functions in MM [28]. Among these, the miR-17-92 and miR-106b-25 clusters in MM have been attributed oncogenic functions due to their regulation of MM associated tumor suppressor genes. For example, members of the miR-17-92 cluster have previously been shown to target the tumor suppressors SOCS1 and BIM [38, 39]. Similarly miR-106b-25 in MM has been suggested to modulate the activity of the tumor suppressor P53 [38, 40]. Furthermore, members of the miR-17-92 cluster and Let-7 family were suggested to enhance MM angiogenesis [41], an important step in MM establishment and progression. Interestingly, the expression of miR-17-92 and miR-106b-25 clusters is positively modulated by c-MYC in tumors other than MM [42, 43]. Therefore, we suggest that EZH2 inhibition in MM may indirectly affect the expression of onco-miRNAs via downregulation of MM-associated oncogenes such as c-MYC.
Recent analysis of microRNA expression in MM revealed that deregulation of microRNA expression correlates with molecular subtype, disease progression, patients’ survival and response to treatment [13, 14, 44–47]. Several reports have suggested genetic lesions such as chromosomal translocations and copy number variations [13, 14, 46, 47], but also epigenetic mechanisms e.g. DNA methylation [48, 49] as possible mechanisms leading to aberrant expression of microRNA genes in MM. For example, DNA methylation was shown to epigenetically silence the expression of tumor suppressor microRNAs such as miR-155, miR-198, miR-135a*, miR-200c, miR-663 and miR-483-5p [50]. Our recent findings expand the knowledge concerning the regulation of microRNA expression in MM and suggest also polycomb-mediated gene repression as a mechanism that may deregulate and silence tumor suppressor microRNAs. Overexpression of tumor suppressor microRNAs or their mimics have in some cases been proven to have anti-MM activity inhibiting MM cell growth, migration and colony formation in vitro [48, 50] and in vivo [51, 52]. The possibility of using microRNAs as a novel therapeutic strategy in MM should thus be the subject for further investigation.
In summary, our recent publication [28] demonstrated for the first time PRC2 as a regulator of microRNA expression in MM, thus emphasizing the oncogenic role of EZH2 in MM. Our present findings show that EZH2 inhibition leads to upregulation of a set of tumor suppressor microRNAs targeting important MM-associated oncogenes, and suggest EZH2 inhibitors and the silenced tumor suppressor microRNAs as possible novel therapeutic strategies in MM.
Acknowledgments
We are very grateful to all co-authors of the research highlighted in this article. We specially want to thank Professor Jian Jin from Icahn School of Medicine at Mount Sinai, New York, NY, USA, for providing us with the EZH2 inhibitor UNC1999. The work highlighted in this article was supported by grants from the Swedish Cancer Society, the Hans von Kantzow Foundation, the Swedish Research Council and the NIH grant R01GM103893 from the U.S. National Institutes of Health.
Abbreviations
- BM
bone marrow
- EZH2
enhancer of zeste homolog 2
- H3K27me3
histone 3 lysine 27 tri-methylation
- H3K36me2
histone 3 lysine 36 di-methylation
- miR
microRNA
- MM
multiple myeloma
- MMSET
multiple myeloma set domain
- PCs
plasma cells
- PRC2
polycomb repressive complex 2
Footnotes
Conflicting interests
The authors have declared that no conflicts of interests exist.
Author contributions
MA and HJW wrote the manuscript, revised it and approved it.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Raab MS, Podar K, Breitkreutz I, Richardson PG, Anderson KC. Multiple myeloma. Lancet. 2009;374:324–339. doi: 10.1016/S0140-6736(09)60221-X. [DOI] [PubMed] [Google Scholar]
- 3.Kumar S. Multiple myeloma - current issues and controversies. Cancer Treat Rev. 2010;36(Suppl 2):S3–11. doi: 10.1016/S0305-7372(10)70006-2. [DOI] [PubMed] [Google Scholar]
- 4.Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120:1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28:384–390. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawyer JR, Tian E, Heuck CJ, Johann DJ, Epstein J, Swanson CM, et al. Evidence of an epigenetic origin for high-risk 1q21 copy number aberrations in multiple myeloma. Blood. 2015;125:3756–3759. doi: 10.1182/blood-2015-03-632075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A, et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. 2011;117:553–562. doi: 10.1182/blood-2010-04-279539. [DOI] [PubMed] [Google Scholar]
- 10.Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Zhang Q, Ezponda T, et al. Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation. PLoS Genet. 2014;10:e1004566. doi: 10.1371/journal.pgen.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci U S A. 2010;107:7904–7909. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 14.Chi J, Ballabio E, Chen XH, Kusec R, Taylor S, Hay D, et al. MicroRNA expression in multiple myeloma is associated with genetic subtype, isotype and survival. Biol Direct. 2011;6:23. doi: 10.1186/1745-6150-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 17.De Vos J, Thykjaer T, Tarte K, Ensslen M, Raynaud P, Requirand G, et al. Comparison of gene expression profiling between malignant and normal plasma cells with oligonucleotide arrays. Oncogene. 2002;21:6848–6857. doi: 10.1038/sj.onc.1205868. [DOI] [PubMed] [Google Scholar]
- 18.Pawlyn C, Kaiser MF, Heuck C, Melchor L, Wardell CP, Murison A, et al. The Spectrum and Clinical Impact of Epigenetic Modifier Mutations in Myeloma. Clin Cancer Res. 2016;22:5783–5794. doi: 10.1158/1078-0432.CCR-15-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croonquist PA, Van Ness B. The polycomb group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene. 2005;24:6269–6280. doi: 10.1038/sj.onc.1208771. [DOI] [PubMed] [Google Scholar]
- 20.Kalushkova A, Fryknas M, Lemaire M, Fristedt C, Agarwal P, Eriksson M, et al. Polycomb target genes are silenced in multiple myeloma. PLoS One. 2010;5:e11483. doi: 10.1371/journal.pone.0011483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 23.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal P, Alzrigat M, Parraga AA, Enroth S, Singh U, Ungerstedt J, et al. Genome-wide profiling of histone H3 lysine 27 and lysine 4 trimethylation in multiple myeloma reveals the importance of Polycomb gene targeting and highlights EZH2 as a potential therapeutic target. Oncotarget. 2016;7:6809–6823. doi: 10.18632/oncotarget.6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernando H, Gelato KA, Lesche R, Beckmann G, Koehr S, Otto S, et al. EZH2 Inhibition Blocks Multiple Myeloma Cell Growth through Upregulation of Epithelial Tumor Suppressor Genes. Mol Cancer Ther. 2016;15:287–298. doi: 10.1158/1535-7163.MCT-15-0486. [DOI] [PubMed] [Google Scholar]
- 26.Zeng D, Liu M, Pan J. Blocking EZH2 methylation transferase activity by GSK126 decreases stem cell-like myeloma cells. Oncotarget. 2017;8:3396–3411. doi: 10.18632/oncotarget.13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alzrigat M, Parraga AA, Agarwal P, Zureigat H, Osterborg A, Nahi H, et al. EZH2 inhibition in multiple myeloma downregulates myeloma associated oncogenes and upregulates microRNAs with potential tumor suppressor functions. Oncotarget. 2017;8:10213–10224. doi: 10.18632/oncotarget.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X, Lwin T, Zhang X, Huang A, Wang J, Marquez VE, et al. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia. 2013;27:2341–2350. doi: 10.1038/leu.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holien T, Sundan A. Oncogene addiction to c-MYC in myeloma cells. Oncotarget. 2012;3:739–740. doi: 10.18632/oncotarget.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holien T, Vatsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012;120:2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 33.Bagratuni T, Wu P, Gonzalez de Castro D, Davenport EL, Dickens NJ, Walker BA, et al. XBP1s levels are implicated in the biology and outcome of myeloma mediating different clinical outcomes to thalidomide-based treatments. Blood. 2010;116:250–253. doi: 10.1182/blood-2010-01-263236. [DOI] [PubMed] [Google Scholar]
- 34.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D’Costa K, Emslie D, Metcalf D, Smyth GK, Karnowski A, Kallies A, et al. Blimp1 is limiting for transformation in a mouse plasmacytoma model. Blood. 2009;113:5911–5919. doi: 10.1182/blood-2008-08-172866. [DOI] [PubMed] [Google Scholar]
- 37.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25:1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Li C, Zhang R, Gao X, Qu X, Zhao M, et al. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309:62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Kumar M, Lu Z, Takwi AA, Chen W, Callander NS, Ramos KS, et al. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene. 2011;30:843–853. doi: 10.1038/onc.2010.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S, Yu W, Qu X, Wang R, Xu J, Zhang Q, et al. Argonaute 2 promotes myeloma angiogenesis via microRNA dysregulation. J Hematol Oncol. 2014;7:40. doi: 10.1186/1756-8722-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan W, Li Y, Lim SG, Tan TM. miR-106b-25/miR-17-92 clusters: polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol. 2014;20:5962–5972. doi: 10.3748/wjg.v20.i20.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buendia MA, Bourre L, Cairo S. Myc target miRs and liver cancer: small molecules to get Myc sick. Gastroenterology. 2012;142:214–218. doi: 10.1053/j.gastro.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 44.Bi C, Chng WJ. MicroRNA: important player in the pathobiology of multiple myeloma. Biomed Res Int. 2014;2014:521586. doi: 10.1155/2014/521586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seckinger A, Meissner T, Moreaux J, Benes V, Hillengass J, Castoldi M, et al. miRNAs in multiple myeloma - a survival relevant complex regulator of gene expression. Oncotarget. 2015;6:39165–39183. doi: 10.18632/oncotarget.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corthals SL, Sun SM, Kuiper R, de Knegt Y, Broyl A, van der Holt B, et al. MicroRNA signatures characterize multiple myeloma patients. Leukemia. 2011;25:1784–1789. doi: 10.1038/leu.2011.147. [DOI] [PubMed] [Google Scholar]
- 47.Lionetti M, Musto P, Di Martino MT, Fabris S, Agnelli L, Todoerti K, et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clin Cancer Res. 2013;19:3130–3142. doi: 10.1158/1078-0432.CCR-12-2043. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Wang YE, Zhang Y, Leleu X, Reagan M, Zhang Y, et al. Global Epigenetic Regulation of MicroRNAs in Multiple Myeloma. PLoS One. 2014;9:e110973. doi: 10.1371/journal.pone.0110973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong KY, Huang X, Chim CS. DNA methylation of microRNA genes in multiple myeloma. Carcinogenesis. 2012;33:1629–1638. doi: 10.1093/carcin/bgs212. [DOI] [PubMed] [Google Scholar]
- 50.Bi C, Chung TH, Huang G, Zhou J, Yan J, Ahmann GJ, et al. Genome-wide pharmacologic unmasking identifies tumor suppressive microRNAs in multiple myeloma. Oncotarget. 2015;6:26508–26518. doi: 10.18632/oncotarget.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18:6260–6270. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morelli E, Leone E, Cantafio ME, Di Martino MT, Amodio N, Biamonte L, et al. Selective targeting of IRF4 by synthetic microRNA-125b-5p mimics induces anti-multiple myeloma activity in vitro and in vivo. Leukemia. 2015;29:2173–2183. doi: 10.1038/leu.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]