Abstract
Microscopy could be an important tool for characterizing stem cell products if quantitative measurements could be collected over multiple spatial and temporal scales. With the cells changing states over time and being several orders of magnitude smaller than cell products, modern microscopes are already capable of imaging large spatial areas, repeat imaging over time, and acquiring images over several spectra. However, characterizing stem cell products from such large image collections is challenging because of data size, required computations, and lack of interactive quantitative measurements needed to determine release criteria. We present a measurement web system consisting of available algorithms, extensions to a client-server framework using Deep Zoom, and the configuration know-how to provide the information needed for inspecting the quality of a cell product. The cell and other data sets are accessible via the prototype web-based system at http://isg.nist.gov/deepzoomweb.
1 Introduction
While bio-manufacturing of stem cell therapies has a great promise for health care, clinical use of stem cell products requires quality measurements that capture dynamic behavior of stem cell populations. Without high confidence quality measurements in bio-manufacturing, products cannot reach the market (e.g., 515 mesenchymal stem cell (MSC) clinical trials are completed or ongoing, yet no MSC-based products have reached the market [1]). Microscopy imaging is one of the tools that can be used for characterizing colony growth rates and spatial heterogeneity of various bio markers which are indicators of quality. High confidence measurements have to be derived from thousands of Mega-pixel microscopy images that spatially cover the entire cell population, and are repeatedly acquired over several days.
One of the challenges is the size of acquired images needed for quality measurements. One field of view (FOV) represents approximately 0.0626 % of the spatial area for a circular region with 3.494 cm diameter imaged at 10X magnification. By spatially stitching Mega-pixel FOV images, we obtain one Giga-pixel 2D image per time point. A stack of Giga-pixel image frames over five days forms a terabyte (TB)-sized 3D volume with the spatial [x, y] and time [t] dimensions. Although the microscopy imaging technology is available, there is no off-the-shelf solution that would allow scientists to interactively inspect, subset, measure, and analyze TB-sized 3D volumes. The reasons lie in the computationally intensive pre-processing steps (e.g., image calibration, stitching, segmentation, feature extraction, and modeling), a limited random access memory (RAM) size of a desktop computer preventing software solutions from loading images to RAM, and a limited bandwidth that is restricting the interactivity of TB-sized image exploration.
In practice, images of stem cell products are captured at low resolution and may be accompanied by small numbers of high-resolution, limited FOV samples. This approach is less than ideal, and has led to the following scientific problems in stem cells [2], very low reproducibility of published work [3], and inadequate conclusions on biological variability. Recent advances in automated capture combined with computational image stitching and visualization have enabled high-resolution imaging while retaining large field of views for cell scientists. The stitched 2D images led to Virtual Nanoscopy [4] with pan and zoom capabilities. This approach is a step forward, but requires computational capabilities to enable viewing and measurements over Terabyte-sized image collections per imaging modality.
As microscopists transition from small to large coverage in a typical scientific discovery process, they encounter three main roadblocks. The first roadblock is in automating microscopy acquisition over extended periods of time and collecting a large number of fields of view using several spectral bands and instrument modalities. It also includes the challenge of automating all calibration and image quantification steps. The second roadblock lies in transitioning from single desktop computations to commercially available cloud computer environments to accommodate for data size and computational requirements. The third roadblock is in sharing image data and performing interactive measurements on images of unprecedented specimen coverage.
As a consequence of these technical roadblocks, “To Measure or Not To Measure Terabyte-Sized Images?” is a dilemma for many scientists that operate imaging instruments capable of acquiring very large quantities of images. Large quantities of images improve statistical significance of scientific findings, address the need for measurement completeness, and enable identifying rare but significant events. However, the current open source solutions do not provide direct quantitative measurement capability, lack the accuracy and uncertainty evaluations of the image processing steps used, and require unprecedented computational resources to enable interactive quantitative measurements.
Our technical work has been motivated by the challenges and roadblocks of quantitative measurements over large coverage, live cell microscopy imaging. The contributions of our technical work lie (a) in designing a system with new software components for off-line and interactive quantitative measurements over TB-sized images and (b) in a set of tradeoffs that are important to consider when configuring this type of a system in application environments with varying hardware, network, and software. The software components consist of off-line image processing algorithms running on a computer cluster and a powerful desktop, and plugins and extensions to the OpenSeadragon project [5] to support interactive web-based sub-setting and computation on 3D images in addition to the currently supported viewing of high-resolution zoomable 2D images. The tradeoffs include configurations for co-localization of data and computational resources in desktop, computer cluster, and client-and-server environments.
2 Transition of image processing algorithms from desktop to computer cluster environments
In order to respond to the technology need for characterizing stem cell product quality, we collected images from three live stem cell preparations. Most of the advanced microscopes provide the capability of automating image acquisition. This requires working with company specific scripting languages or exploring open source frameworks such as the Micro-Manager Open Source Microscopy Software [6]. In our case, the images were acquired by using Zeiss 200M microscope controlled by the Zeiss Axiovision software. Each FOV was acquired by two channels (phase contrast and green fluorescent protein) over 5 days every 15 to 45 min. After pre-processing the individual FOV images, each frame of a time sequence consists of around 23 000 × 21 000 pixels with 16 bits per pixel (bpp).
One 2D frame represents approximately 19.08 % of the area for the round well with 3.494 cm diameter. One 2D frame requires close to 1 GB of RAM to load it into memory. The three cell preparations are sampled for 161, 157, and 136 time points respectively to yield 77.8 Giga-pixel, 75.8 Giga-pixel, and 65.7 Giga-pixel [x, y, t] volumes. The total number of image files is 359 568 which corresponds to approximately 0.9 TB.
The image pre-processing can leverage a number of existing libraries (e.g., ImageJ/Fiji, OpenCV, Matlab, Java advanced imaging). In our case, the list of image processing algorithms includes flat field and background correction, stitching, segmentation, tracking, image feature extraction, pyramid building, and re-projection. The sheer size of the three 4D volumes makes any off-line image processing very time-consuming on a desktop computer. Unfortunately, the majority of image processing libraries are not written for cluster or cloud computing and have to be re-designed. Licensed software has to be purchased for all cluster nodes used and then managed over distributed computational resources. To transition from a desktop to a computer cluster, we created and re-designed algorithms to leverage distributed computational resources.
The re-designed algorithms included flat field correction, segmentation based on convolution kernels, image feature extraction, and pyramid building. The re-design is based on the widely used framework called Hadoop [7]. The middleware assists in distributing images across many cluster nodes, provides a programmatic interface to parallelize algorithms into two steps (Map and Reduce), and offers capabilities for managing hardware failures and monitoring executions.
Each algorithm was assessed for efficiency on configurations of computer clusters [8]. The execution benchmarks with Hadoop were compared against baseline measurements, such as the Terasort/Teragen (number/text sorting computations), executions on a multiprocessor desktop and on a computer cluster using Java Remote Method Invocation (RMI) with multiple configurations. By calculating efficiency metrics for execution times over an increasing number of nodes, we could rank the suitability of each computation for cluster computing. Based on the ranking, the majority of the tested algorithms using Hadoop outperformed their corresponding benchmarks run on Java RMI clusters or compared against Terasort/Teragen. Specific considerations had to be given to RAM requirements per node (one time frame had to be loaded without sub-dividing), and to data transmission packaging and I/O tasks (hundreds of thousands of images as inputs for flat field correction and several millions of images as outputs from pyramid computation). Such efficiency benchmarks can aid scientists in transitioning image processing computations from desktops to Hadoop computer clusters.
3 Interactive viewing, sub-setting and measurements in a browser
In a collaborative research environment, a large 3D volume has to be shared among scientific collaborators and it is too large for the RAM of a single desktop or laptop. It is desirable that the image data reside on a server and are served to a set of clients via a local area network (LAN) or the Internet. Our effort to build such client-server configurations aims at (1) maximizing efficient retrieval, transmission and viewing of large images, (2) pre-computing information to achieve interactivity, and (3) optimizing configurations to support various types of scientific applications.
The building blocks for designing and deploying a client-server web-based system can leverage several technologies for hosting images on a server (e.g., SQLite, MySQL, Apache Tomcat), for rendering content on a client side (e.g., OpenSeadragon, D3, XTK), and for communicating between clients and a server (e.g., RESTful web services, HTML5, and JavaScript. One can customize and integrate the appropriate but diverse technologies, and optimize system configuration settings with respect to specific application domain requirements. In our work, we used open-source components, and our design of a client-server system does not assume any specialized hardware to achieve interactivity.
These types of client-server systems have been built for astronomy images using Aladin Sky Atlas, brain images using Collaborative Annotation Toolkit for Massive Amounts of Image Data (CATMAID) [9], or satellite images using the USGS Global Visualization Viewer (GloVis) and NASA Global Imagery Browse Services (GIBS) with Worldview. At this time, it is also possible to use client-server systems for data sizes that reach Tera- and Peta-pixels, for instance, in the case of selective-plane illumination and electron microscopy [10], [11]. However, the interactivity goal dictates the use of LAN and specialized hardware and software, such as the GPU-based volume ray-casting framework [11].
Given the large number of image files, multiple experts cannot rapidly inspect stem cell product quality from the raw image files. After preprocessing raw image tiles into a large 2D image frame, the image dimensions are larger than a typical computer screen. Thus, the inspection has to be enabled at multiple zoom levels and with panning like in Google Maps (denoted as Deep Zoom). It must also support 3D navigation. To address the visual inspection need, we extended the Deep Zoom based visualization by using the OpenSeadragon project [5]. Our extensions are both measurement plugins to OpenSeadragon (i.e., image measurements in physical units) and measurement widgets on top of OpenSeadragon (i.e., sub-setting, intensity and distance measurements, hyperlinking with spatial statistics and temporal lineage). The final web-based Deep Zoom system allows visual inspections and measurements of (a) measured green fluorescent protein (GFP) and phase contrast (PC) intensities, (b) their side views (i.e., orthogonal projections of 3D volumes), (c) background-corrected GFP intensities, and (d) the segmentation masks.
3.1 Extensions to Deep Zoom
The Deep Zoom technology uses a multi-resolution representation of each 2D image (i.e., one time frame) partitioned into tiles of 256 × 256 pixels. The pyramid representation is used for on-demand access of 3D subsets during (1) viewing (image tile access and retrieval), (2) downloading of regions of interest (image tile selection, re-construction of the requested image area, zip compression, and retrieval), and (3) computing (image tile access, retrieval, and pixel manipulation). The downloading and computing functions are different from other existing client-server solutions using the pyramid representation. The region of interest, colony identification, resolution and time frames of downloaded images are specified by a user. All interactive computations are performed on the 8bpp image tiles retrieved by a browser.
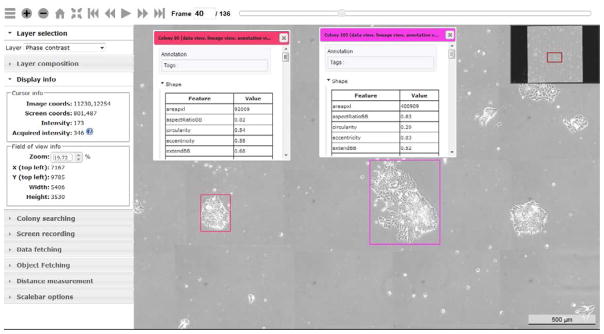
The pyramid representation of these GFP, PC, and mask image layers consists of approximately 11 million files located in a half million folders. These files and folders include 8bpp, 16bpp and 32bpp representations of the data due to floating point operations during the background correction and calibration pre-processing steps. Due to the current 8bpp support of images in browsers, the bit depth is 8bpp for downloading in an interactive mode (i.e., download what you view in a browser), and 16bpp or 32bpp for downloading in a fetching mode (i.e., specify parameters of raw or processed image subsets). These sub-setting options allow further measurements and scientific research with the downloaded images. The neighboring contextual image information must be viewed in three orthogonal planes: [x, y], [x, t] or [y, t]. Any other plane that is not parallel to the three orthogonal planes is considered oblique. The measurements obtained from oblique planes are not useful due to a lack of meaningful units for interpreting such measurements. Figure 1 shows the Deep Zoom interface for visual inspection of cells in [x, y] plane.
Figure 1.
Extended Deep Zoom controls for image interactions in a regular browser view. The panels in the left upper corner and the slider at the top provide information about image intensities, zoom level, frame selection, and information layer. Additional panels on the left enable colony searching, screen recording (movie player), sub-setting, distance measurements, and scale bar inclusion.
3.2 Hyperlinking image colony characteristics
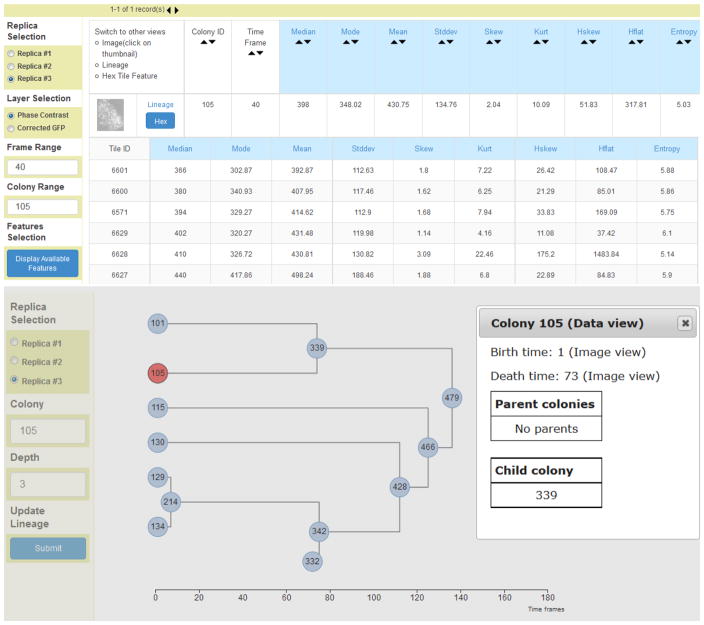
Enabling web browser-based visual inspections is still insufficient to determine quality of stem cell products in bio-manufacturing because of the number of colonies. In our experiments, the spatial coverage increased the number of monitored stem cell colonies to about 300 colonies per initial time point in comparison to typical 3 to 4 colonies being spot-checked by biologists. As the colonies merge over time, we counted more than 1000 unique colonies in each preparation. Thus, automation is needed to extract and compare intensity, shape, and texture characteristics of stem cell colonies in order to determine the product quality. We segmented and extracted automatically 75 image characteristics (features) per colony. Furthermore, we partitioned each colony into hexagonal regions and extracted the same number of features per hexagon to study spatially local properties. The feature extraction yielded approximately 2.6 GB of data per cell preparation. All spatial, temporal, and channel-specific colony information was hyperlinked so that web user interfaces could be re-configured depending on the quality aspect of each stem cell colony under scrutiny. Figure 2 illustrates colony features and temporal lineage information that are hyperlinked with the image information shown in Figure 1.
Figure 2.
A tabular view of colony features (top) and temporal lineage view of colony information (bottom). The image (see Figure 1), tabular and lineage views are hyperlinked. Interactive sub-setting by colony ID and frame ID results in a CSV file with the requested tabular feature information.
3.3 Toward stem cell product release criteria
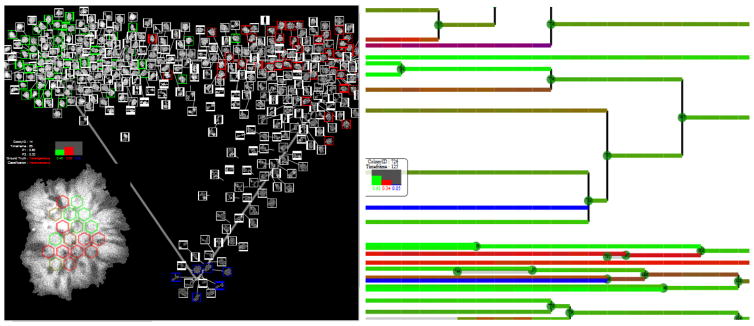
To derive quality criteria from colony features, we classified colonies into heterogeneous, homogeneous, or dark categories. The quality is hypothesized to relate to the purity of each colony with respect to these three categories, the overall distribution of colonies in each category, and any temporal changes of category type over a life time of each colony. Supervised classification was initiated by collecting 143 examples of colony categories provided by biologists using the web tools (i.e., the training data). Next, we trained a logistic regression classification model to assign categories automatically to all colonies in the three data sets. The accuracy of this classification model was 98 %, 87.5 %, and 100 % for the three data sets respectively. Figure 3 illustrates interactive visualizations of the overall distribution (left) and temporal changes of category type (right). Visualizations serve as tools for assessing the stem cell quality visually. The visualization coordinate system (see the projection of categories on a triangle (i.e., 2-simplex)) is also used for extracting quantitative measurements supporting visual quality assessments.
Figure 3.
Left – Interactive visualization of colony images with their hexagonal partitions and classification categories (homogeneous-green, heterogeneous-red and dark-blue). Right – Zoom-able temporal lineage visualization with assigned classification category and probabilities of each category per colony. The three probabilities are color-coded the same way as the lineage lines.
4 Tradeoffs in Off-line and Web-based Interactive Computations
While designing a quantitative microscopy solution for characterizing stem cell products, one has to consider configuration tradeoffs during off-line computations and web-based system configuration. We already mentioned a couple of tradeoffs between open and licensed software when it comes to running algorithms on a computer cluster, and between image depths (bits per pixel) suitable for viewing in a browser and for scientific analysis. Here we describe the tradeoffs between co-localization of data and computational resources in the context of desktop, computer cluster, and client-server environments. To achieve interactivity in a client-server environment, we present the tradeoffs between computing image thumbnails on the fly and pre-computing them at the cost of higher disk storage, as well as interactive and non-interactive options for sub-setting and image filtering.
4.1 Co-localization of data and computational resources during off-line computations
When running all off-line image processing computations, every algorithmic execution will have to be optimized with respect to the parameters of available hardware and network. Thus, there is a high value in knowing the tradeoffs between moving the data to a computer cluster or to a powerful desktop, and the corresponding labor and hardware costs to achieve the computational speed benefit.
We have experimented with existing open-source and newly created Java-based algorithms running on a Hadoop computer cluster (850 Intel/AMD 64-bit nodes) and with custom Matlab-based algorithms running on a personal computer (six physical cores, Intel(R) Xeon(R) CPU E5-2620 0 @ 2.00GHz, 64.0 GB of RAM, and a NVIDIA® Quadro 4000 graphics card). The Hadoop Java-based algorithms were computing image pyramids, flat field correction, segmentation, re-projections, and extracting image features. The MATLAB-based algorithms calculated flat field and background correction, stitching, segmentation, and colony tracking.
We placed the raw images for the three biological preparations (R1, R2, and R3) on (1) a Network Attached Storage (NAS) accessible via local area network (LAN) and (2) a NIST cluster connected via LAN. Our measurements of transfer rates illustrate the following relative performance: (1) to NAS: 60MB/s, (2) from NAS: 40MB/s, (3) to NIST cluster 5.3MB/s, and (4) from NIST cluster: 2.7MB/s. The ratios of transfer speeds (NAS over NIST cluster) are approximately 11.3 and 14.8 and can be used in assessing the cost-benefit of executing computations by attaching NAS with data or transferring data to a computational resource.
For the cluster computations that generate large collections of output file such as multi-resolution pyramid building, there is a cost of transferring the data from the cluster to a web server. The cost depends on whether the data are transferred compressed or uncompressed. In our case, we packaged the pyramid files into a tar file to expedite the file transfer. There were 502 971 input files (192 085 in R1, 166 580 in R2, and 144 306 in R3) processed into 11 million output files located in a half million folders for pyramid representations of all layers. We have measured the time needed to move pyramid files for seven pyramid layers per biological preparation from NIST cluster to NAS. The time measurements included transfer and tar file extraction (R1: 19.61 h, R2: 14.90 h, and R3: 14.82 h).
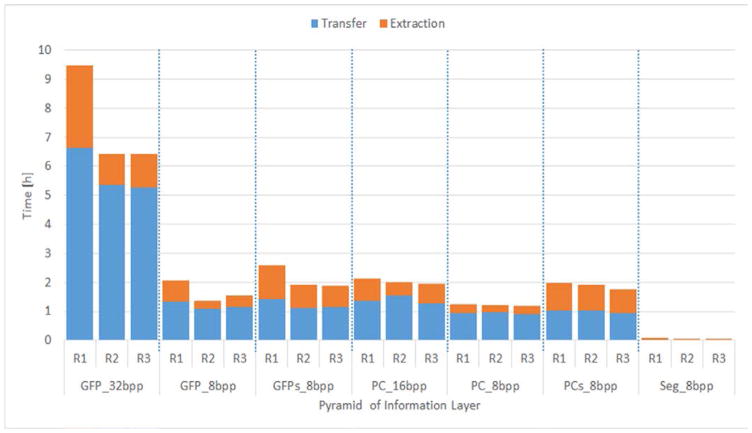
Figure 4 shows the time measurements per pyramid layer. The transfer times correspond to the following three steps. (1) A set of pyramids representing one image layer is compressed into one tar file per pyramid using block-sorting and Huffman compression. (2) A set of tar files is compressed into one big tar file per set of pyramids. (3) One big tar file is transferred. The extraction time consists of the following two steps. (1) A set of tar files is extracted from one big tar file. (2) Files are extracted from each tar file. Based on Figure 4, the ratio of transfer to tar extraction time was on average 2. 5 (R1: 1.86, R2: 3.00, and R3: 2.63). These measurements were also useful to compare the transfer of pyramids with 8bpp, 16bpp and 32bpp image depth.
Figure 4.
Transfer and extraction times to retrieve pyramid sets for three biological replicas (R1, R2, and R3) from a computer cluster to a storage array over LAN. The names of pyramid sets refer to the acquired channel (green fluorescent protein - GFP and phase contrast - PC), re-projected side view (additional s), segmentation (Seg), and the number of bits per pixels (bpp).
4.2 Tradeoffs to achieve Web-based Interactive Computations
While building client-server systems for large images or 3D volumes, the main challenges arise due to limited storage, RAM, processing, and bandwidth resources defined by costs. Optimal design decisions to achieve interactivity are typically based on anticipated system utilization and available resource specifications. Nonetheless, the utilization is unknown (or vaguely defined) in scientific discoveries. In our case, we assumed that the utilization of our client-server systems would be primarily for dissemination of data, extracting basic statistical summaries, simple image filtering, and multiple length scale distance measurements. Thus, our goal was to achieve interactivity of browsing, sub-setting, re-projecting, colony characterization, thumbnail rendering, annotation, sorting, image filtering, and multiple length scale distance measurements.
To achieve an interactive experience with web-based computations and measurements, we opted for higher storage cost on the server side over more powerful computer resources needed to support many on-demand computations (client or server side). We pre-computed off-line as much information as possible (e.g., re-projected 3D volumes, extracted image features, predicted colony labels, and stored them in a database). Next, we evaluated a few tradeoffs between on-demand computation speed and storage costs of pre-computed thumbnail images, and interactive and non-interactive query-based execution times for sub-setting and image filtering.
Thumbnail images
The thumbnail images can be computed on-the-fly or pre-computed and stored in a database for retrieval. In our case, 31 312 cell colonies can be represented as thumbnails of size 50×50, 75×75 and 100×100 by approximately 1.12 GB for two acquired image channels. In return for the extra storage cost and pre-computation time (3.12 h in our case), the thumbnail retrieval from DB is faster 10 to 15 times on average per single request than using the on-the-fly implementation. The retrieval time also has a much smaller standard deviation which leads to better interactivity.
Sub-setting and image filtering
The sub-setting function captures either the images rendered on a client side (JavaScript code) or the images requested to be retrieved on a server side (Java code). The two sub-setting implementations represent the trade-offs in terms of interactivity and capabilities. On one side, image rendering in a browser (client) enables interactivity of sub-setting with the time overhead of rendering. On the other side, the browser capabilities are limited to rendering 8bpp and to saving data in client’s RAM (Note: writing to client’s disk directly is not possible). Furthermore, a client side is typically constrained by limited multi-threaded execution.
Many of the above disadvantages can be avoided on a server side by sacrificing some level of sub-setting interactivity. Based on collected benchmarks for the 3D sub-setting trade-offs using the Chrome 32 browser, we observed the execution time for client-side sub-setting to linearly increase 16.7 times faster than the time for the server side sub-setting (50 s versus 3 s per [x, y] cross section from [x, y, t] volume) in addition to the RAM requirements imposed on the client machine.
Similarly, the image filtering operations performed during image measurements can be executed either on a client side or a server side. The client side execution can operate on retrieved images and provide immediate feedback during parameter optimization and visual verification. On the other hand, the client side is limited in terms of computational resources and operates only on an image sub-area at the original resolution or on a large image at lower resolution. To enable the client side execution, we implemented pixel level manipulations on top of the multi-resolution pyramid representation as a plugin to OpenSeadragon. Thus, a user can optimize parameters of image analyses interactively in a browser and then launch the analyses of the entire image on a more powerful computational resource than the client.
5 Conclusions
We presented a system for monitoring and characterizing stem cell colonies that is leveraging cloud/cluster computing for off-line computations and Deep Zoom extensions for interactive viewing, sub-setting and measurements. Scientists who operate a microscope, process microscope images and deploy computations in a computer cloud and client-server web systems might benefit from the presented components and tradeoffs during system configuration and deployment. The OpenSeadragon [5] plugins and developed measurement widgets are available for downloading from the GitHub repository at https://github.com/NIST-ISG. The majority of off-line algorithms are available from our web page at https://isg.nist.gov/deepzoomweb/activities.” TB-sized 3D volume examples from cell biology and materials science are accessible via the prototype interactive web-based system at http://isg.nist.gov/deepzoomweb.
The technology described in this article can be applied within biology to any time lapse imaging study of live cells, confocal laser scanning study of cell morphology, or coherent anti-Stokes Raman spectroscopy imaging study of fixed cells with any subset of [x, y, z, t, λ] dimensions. While we explored the pipeline of off-line and on-demand image analyses and focused on enabling terabyte-sized image measurements, we identified several pipeline steps that lack standard operating protocols and/or interoperability, and have unknown accuracy and uncertainty. Due to the variety of microscope hardware and software, community-wide meetings and community-consensus efforts can unify interfaces to microscopes (e.g., the Micro-Manager open source microscopy software [6]) and to image data (e.g., the Open Microscopy Environment [12]) which are important for interoperability and research reproducibility. Such efforts are needed to address the missing measurement infrastructure and eliminate today’s dilemma “To Measure or Not To Measure Terabyte-Sized Images?”
Acknowledgments
The stem cell colony images were prepared and acquired courtesy of Dr. Kiran Bhadriraju at the NIST cell biology microscopy facility. We also thank Anne Plant, John Elliott, and Michael Halter from the Biosystems and Biomaterials Division at NIST, and Jing Gao and Mylene Simon from the Software and Systems Division at NIST for their contributions
Biographies
Peter Bajcsy is a computer scientist at NIST working on automatic transfer of image content to knowledge. He received his PhD in electrical and computer engineering from the University of Illinois at Urbana-Champaign in 1997. Peter’s scientific interests include image processing, machine learning, and computer and machine vision. He is a senior IEEE member. peter.bajcsy@nist.gov
Antoine Vandecreme is a computer scientist at NIST working on image processing and big data computations. He received his M.S. degree in computer science from ISIMA (Institut Supérieur d’Informatique de Modélisation et de leurs Applications) a French engineering school in 2008. His research domains include distributed computing, web services and web application design. antoine.vandecreme@nist.gov
Julien Amelot is a data scientist and software engineer at NIST. He has received a master’s degree in computer science from Telecom Nancy (known as ESIAL - “École Supérieure d’Informatique et Applications de Lorraine”) in France. His area of expertise is in machine learning, data mining, interactive visualization and distributed computing for Big data. Current projects involve image processing to support research on stem cells. Julien.amelot@nist.gov
Joe Chalfoun is a research scientist at the National Institute of Standards and Technology (NIST). He received his Doctoral degree in mechanical robotics engineering from the University of Versailles, France, in 2005. Joe’s current interest is in medical robotics field, mainly in cell biology: dynamic behavior, microscope automation, segmentation, real-time tracking, and subcellular feature analysis, classification and clustering. joe.chalfoun@nist.gov
Michael Majurski is a computer science trainee at NIST working on image processing and big data computations. He is completing his masters in Information Systems at University of Maryland Baltimore County. His research domains include image processing, microscope automation, and computer vision. michael.majurski@nist.gov
Mary Brady is the Manager of the Information Systems Group in NIST’s Information Technology Laboratory (ITL). She received a M.S. degree in Computer Science from George Washington University, and a B.S. degree in both Computer Science and Mathematics from Mary Washington College. Her group is focused on developing measurements, standards, and underlying technologies that foster innovation throughout the information life cycle from collection and analysis to sharing and preservation. mary.brady@nist.gov
Footnotes
6 Disclaimer
Commercial products are identified in this document in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the products identified are necessarily the best available for the purpose.
The published paper is accessible on-line from http://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=7503500
References
- 1.ClinicalTrials.gov. mesenchymal stem cell. [Accessed: 03-Jul-2015];database. 2015 [Online]. Available: https://www.clinicaltrials.gov/ct2/results?term=mesenchymal+stem+cell&Search=Search.
- 2.Reuters. Japanese scientist wants own ‘breakthrough’ stem cell study retracted. CBS News. 2014 Mar 10; [Google Scholar]
- 3.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 4.Williams EH, Carpentier P, Misteli T. The JCB DataViewer scales up. J Cell Biol. 2012 Aug;198(3):271–2. doi: 10.1083/jcb.201207117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Open Seadragon. [Accessed: 03-Feb-2015];Open Seadragon project. 2015 [Online]. Available: http://openseadragon.github.io/
- 6.Edelstein AD, Tsuchida Ma, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using μManager software. J Biol Methods. 2014 Nov;1(2):10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White T. Hadoop: The Definitive Guide MapReduce for the Cloud. 3. O’Reilly Media; 2012. p. 528. [Google Scholar]
- 8.Bajcsy P, Vandecreme A, Amelot J, Nguyen P, Chalfoun J, Brady M. Terabyte-sized Image Computations on Hadoop Cluster Platforms. IEEE International Conference on Big Data. 2013 [Google Scholar]
- 9.Saalfeld S, Cardona A, Hartenstein V, Tomančák P. The Collaborative Annotation Toolkit for Massive Amounts of Image Data (CATMAID) [Accessed: 05-Mar-2015];Max Planck Institute of Molecular Cell Biology and Genetics. 2013 [Online]. Available: http://catmaid.org/
- 10.Pietzsch T, Saalfeld S, Preibisch S, Tomancak P. Big Data Viewer: visualization and processing for large image data sets. Nat Methods. 2015 May;12(6):481–483. doi: 10.1038/nmeth.3392. [DOI] [PubMed] [Google Scholar]
- 11.Beyer J, Hadwiger M, Al-Awami A, Jeong WK, Kasthuri N, Lichtman JW, Pfister H. Exploring the connectome: petascale volume visualization of microscopy data streams. IEEE Comput Graph Appl. 2015;33(4):50–61. doi: 10.1109/MCG.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiffmann D, Dikovskaya D, Appleton P, Newton I, Creager D, Allan C, Näthke I, Goldberg I. Open Microscopy Environment and FindSpots: integrating image informatics with quantitative multidimensional image analysis. Biotechniques. 2006 Aug;41(2):199–208. doi: 10.2144/000112224. [DOI] [PubMed] [Google Scholar]