Abstract
Protein ubiquitinylation plays a key role in many important cellular processes. Ubiquitinylation requires the E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and, frequently, a substrate-specific E3 ubiquitin-protein ligase. In one class of E3 ubiquitin ligases, the catalytic domain contains a zinc-binding RING finger motif. ARD1 (ADP-ribosylation factor domain protein 1), with a RING finger domain in the N-terminal region, two predicted B-Boxes, and a coiled-coil protein interaction motif immediately preceding an ADP-ribosylation factor domain at the C terminus, belongs to the TRIM (Tripartite motif) or RBCC (RING, B-Box, coiled-coil) family. The region containing the B-Boxes and the coiled-coil motif acts as a GTPase-activating protein for the ADP-ribosylation factor domain of ARD1. We report here that full-length ARD1 or the RING finger domain (residues 1–110) produced polyubiquitinylated proteins in vitro in the presence of mammalian E1, an E2 enzyme (UbcH6 or UbcH5a, -5b, or -5c), ATP, and ubiquitin. Deletion of the RING region or point mutations within the RING sequence abolished ARD1 E3 ligase activity. All data are consistent with a potential function for ARD1 as an E3 ubiquitin ligase in cells.
Keywords: RBCC, TRIM protein, ARF, ARF-GAP
ARD1 (ADP-ribosylation factor domain protein 1) appears to be a unique member of the ADP-ribosylation factor (ARF) family, which otherwise comprises three classes of typical (≈20 kDa) mammalian ARFs that are grouped by similarities of amino acid sequence and gene structure, as well as phylogenetic relationships (1, 2). ARFs, identified and named for their ability to accelerate cholera toxin-catalyzed ADP-ribosylation of Gαs (3), play major roles in regulating intracellular vesicular trafficking through interaction with coat proteins, as well as cytoskeletal and membrane remodeling through activation of phosphatidylinositol 4-phosphate 5-kinase and phospholipase D (4, 5). ARD1, like all ARFs, alternates between active GTP-bound and inactive GDP-bound states (6). Cytohesin-1 is the only guanine nucleotide-exchange protein (GEP) known to activate ARD1 (7). Unlike the ARFs that require GTPase-activating proteins (GAPs) for inactivation, the ARD1 molecule contains an N-terminal GAP domain that enhances the otherwise undetectable GTPase activity of its C-terminal ARF domain (8). In addition, a 15-aa sequence immediately preceding the ARF domain, which corresponds to the 17-aa N-terminal α-helix of a typical ARF, acts as a GDP-dissociation inhibitor (GDI) that maintains the protein in its GDP-bound state (9).
The 64-kDa ARD1 protein is a member of the tripartite motif (TRIM) family (10), also termed RBCC (11), for RING (12), B-Box zinc finger (13), coiled-coil (14) (Fig. 1 Upper). In ARD1, the RBCC motifs that characterize essentially all TRIM family proteins are near the N terminus, whereas at the C terminus, unique to ARD1 among all of the TRIM family members, is an ARF domain. The RING finger domain, initially described by Freemont and coworkers (12), is a cysteine-rich motif that forms a cross-brace structure that can chelate two divalent zinc ions. Additional work showed that proteins containing a RING finger motif can act as E3 ligases in ubiquitinylation pathways (15, 16). Modification of proteins by ubiquitinylation occurs in a series of three reactions in which a molecule of ubiquitin is transferred from the activating enzyme (E1), to a conjugating enzyme (E2), and, with the involvement of an E3 ligase, to a substrate or to the E3 itself. Two known families of E3 ubiquitin ligases (which are named for their molecular structures) are RING (for Really Interesting New Gene) and HECT (Homologue of E6-AP C Terminus) (17). Mechanisms of ubiquitinylation catalyzed by RING finger and HECT domains appear to differ. The HECT domain can form an intermediate thioester with ubiquitin before it is transferred to an acceptor protein (18), whereas the RING domain is thought to serve as a molecular scaffold that brings E2 and its substrate protein into proximity.
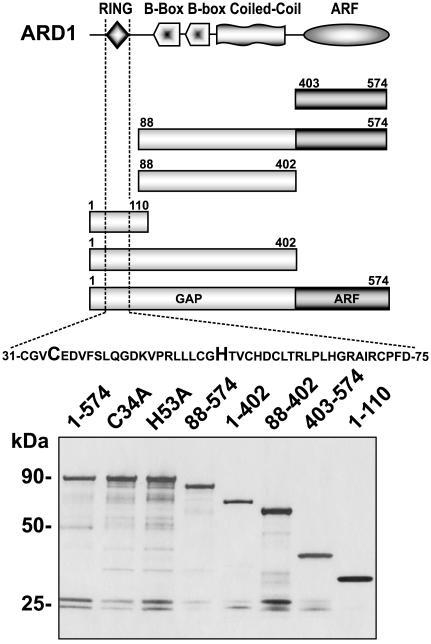
Fig. 1.
ARD1 and its mutant forms: structure and SDS/PAGE of recombinant GST-proteins. (Upper) Predicted domain structures of ARD1, a TRIM/RBCC family member, and related GST-tagged recombinant proteins (6, 9). (Lower) Samples (0.5 μg) of purified recombinant GST proteins were subjected to SDS/PAGE in 4–20% Tris-glycine gels and detected by silver staining. The doublet at 26 kDa probably represents GST generated via proteolysis of bacterially synthesized GST-fusion proteins and was present in all GST-ARD1 preparations.
Although control of protein turnover via ubiquitinylation leading to proteasome-catalyzed degradation is its best understood function, the regulation of diverse cellular processes by ubiquitinylation, e.g., endocytosis and gene expression, is becoming increasingly recognized (19). We report here that human ARD1, via its RING finger domain, can function with specific human E2 enzymes in vitro to catalyze the attachment of ubiquitin to protein acceptors.
Materials and Methods
Materials. Bovine ubiquitin was purchased from Sigma; reduced glutathione-Sepharose 4B, and ECL Western blotting detection reagent were purchased from Amersham Biosciences; antiubiquitin mouse monoclonal antibody P4D1, anti-GST(B-14)-horseradish peroxidase-conjugated mouse monoclonal antibody, and recombinant GST were purchased from Santa Cruz Biotechnology; purified rabbit E1 and recombinant human ubiquitin-conjugating enzymes for in vitro ubiquitinylation assays were purchased from Biomol; recombinant human ubiquitin and mutant ubiquitin that lacks lysines were purchased from Boston Biochem (Cambridge, MA); the QuikChange site-directed mutagenesis kit and BL21(DE3) gold competent cells were purchased from Stratagene; and primers for sequencing, as well as Hypur-purified primers for site-directed mutagenesis, were purchased from MWG Biotech (High Point, NC).
Constructs and Primers. Bacterial expression vectors (Fig. 1) for synthesis of GST fusion proteins with the N terminus of human ARD1wt (amino acids 1–574, accession no. L04510, GI:292069), ARD1 N-terminal domain (amino acids 1–402), ARD1 ARF domain (amino acids 403–574), ARD1 N-terminal deletion mutant (amino acids 88–574), ARD1 central fragment (amino acids 88–402), and expression vector for 6× His-ARD1wt were constructed as described in refs. 6, 7, and 9. Point mutations that replaced either Cys-34 or His-53 with Ala in the RING domain of GST-ARD1, and GST-ARD1 (1–110), were prepared by using the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions, using specific primers (Table 1). All point mutant plasmids were sequenced completely to verify integrity, using the following sequencing primers: R1(406–387), F1(2–21), F2(358–377), F3(533–552), F4(947–966), F5(1286–1305), and F6(1516–1535), as reported in Table 1.
Table 1. DNA primers for mutagenesis and sequencing.
| ARD1(C34A) | F | 5′-GTGCTAGAGTGTGGAGTTGCTGAAGATGTC-3′ |
| R | 5′-GACATCTTCAGCAACTCCACACTCTAGCAC-3′ | |
| ARD1(H53A) | F | 5′-TTGCTTTGTGGCGCTACCGTCTGTCATGACTGTCTC-3′ |
| R | 5′-GAGACAGTCATGACAGACGGTAGCGCCACAAAGCAA-3′ | |
| ARD1(1-110) | F | 5′-ATTGGTCAGTATTGAGCTGCAGAAGAATCC-3′ |
| R | 5′-GGATTCTTCTGCAGCTCAATACTGACCAAT-3′ | |
| Sequencing primers | R1 | 5′-AGGCAAGGTGAGCTTCATCT-3′ |
| F1 | 5′-TGGCTACCCTGGTTGTAAAC-3′ | |
| F2 | 5′-TCTGGAGAGAGCATCATTCG-3′ | |
| F3 | 5′-CCAGGGTCACAAGCATTCAG-3′ | |
| F4 | 5′-TTGGCTCAGGCAGCAACAAG-3′ | |
| F5 | 5′-TCATGCAGCCCATTCCAACA-3′ | |
| F6 | 5′-GCTCTGCTCCTGATTTTTGC-3′ |
Mutated bases are underlined and in bold.
Fusion Protein Synthesis and Purification. Single colonies of Escherichia coli XL1 blue or BL21(DE3) gold (Stratagene) containing plasmids with inserts encoding GST-tagged ARD1 or related proteins were added to 5 ml of Luria–Bertani broth containing ampicillin at 100 μg/ml. After incubation overnight at 37°C with shaking, the culture was added to 500 ml of the same medium and incubated at 37°C with shaking until A600 = 0.6. Isopropyl β-d-thiogalactoside was added (0.2 mM final concentration), and after incubation at 37°C for 4 h, cells were sedimented by centrifugation (5,000 × g for 10 min) and stored at –20°C.
Recombinant proteins were purified essentially as described by Frangioni and Neel (20). Briefly, cells were dispersed in 20 ml of STE buffer (10 mM Tris·HCl/150 mM NaCl, pH 8/1 mM EDTA) containing lysozyme at 1 mg/ml. After incubation for 1 h on ice and the addition of 1% Triton X-100, cells were sonified and incubated (1 h at room temperature, shaking) with DNase I (Roche Applied Science) at 44 units/ml. Inclusion bodies were collected by centrifugation (15,000 × g for 15 min) and dispersed in 20 ml of STE. After the addition of 1 ml of 20% Sarkosyl and intermittent mixing (vortex mixer, 5 s every 30 s) for 5–10 min, 1 ml of 20% Triton X-100 was added, insoluble material was discarded after centrifugation (15,000 × g for 10 min at 4°C), and the clear supernatant was incubated (2 h at 4°C) with 0.25 ml of reduced glutathione-Sepharose. The mixture was transferred to a column, and beads were washed three times with 10 ml of STE buffer. Bound proteins were eluted with three 0.5-ml portions of 10 mM reduced glutathione in 50 mM Tris·HCl (pH 8) and concentrated by using Microcon centrifugal filter devices (10,000 or 100,000 molecular weight cut-off; Millipore). The protein concentration was determined by the Bradford method (21). Purity assessed by silver staining after SDS/PAGE was >90%. After addition of propylene glycol (35% final concentration), protein (0.1–1 mg/ml) was stored in small portions at –20°C. For ubiquitinylation assays, at least two different preparations of GST- or His-ARD1 protein were used.
Anti-ARD1 Antibodies. Affinity-purified polyclonal anti-ARD1 antibodies were obtained from rabbits immunized with an octadecapeptide (CVDSSHRDRISEAHSELAK), corresponding to amino acids 479–496 of ARD1, with a cysteine added at the N terminus to facilitate conjugation to keyhole limpet hemocyanin.
Ubiquitinylation Assay. Standard assays (total volume of 30 μl) contained 0.1 μg (0.8 pmol) of purified rabbit ubiquitin-activating enzyme (116 kDa, E1), 0.5 μg (19 pmol) of recombinant human UbcH6 (26 kDa, E2), 5 μg (0.6 nmol) of purified bovine or recombinant human ubiquitin (8.5 kDa), 4 mM ATP, and the indicated amount of ARD1 or related protein, in ubiquitinylation buffer (20 mM Tris·HCl, pH 7.4/2 mM MgCl2/0.1 mM DTT). After incubation (usually 60 min at 30°C), reactions were terminated by adding 10 μl of 4× Laemmli sample buffer (22). Proteins were separated by SDS/PAGE in 4–20% Tris-glycine precast gels (Invitrogen), transferred to Hybond-C extra nitrocellulose membranes (Amersham Biosciences), stained with Ponceau S, and, after destaining, reacted with anti-ubiquitin antibodies unless otherwise indicated. Signal was generated by ECL detection reagent and detected either with film or an LAS-3000 imaging system (FUJIFILM Medical Systems). A mutant human recombinant ubiquitin in which arginine replaced all seven lysines was used in some assays. Because reactivity of the mutant ubiquitin in colorimetric or dye-binding protein assays (e.g., Bradford, silver staining) differed from that of native ubiquitin (data not shown), the concentration specified by the supplier was used for those experiments. Reactivity of mutant ubiquitin with the anti-ubiquitin monoclonal antibody appeared to be altered also, but that was not systematically investigated (see Fig. 6).
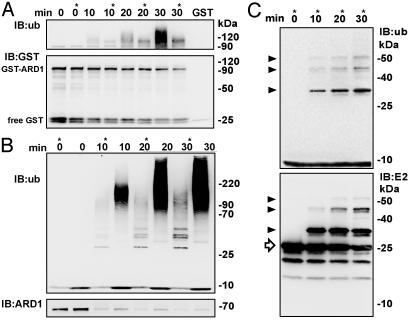
Fig. 6.
Ubiquitinylation of ARD1, free GST, and UbcH6 (E2) in vitro. Standard assay mixture (150 μl) containing 75 nM GST-ARD1 (2.2 pmol/30 μl) (A) or 300 nM His-tagged ARD1 (9 pmol/30 μl) (B and C) was incubated at 30°C with 26 nM E1 (0.8 pmol/30 μl), 0.53 μM UbcH6 (E2, 16 pmol/30 μl), and 3.9 μM recombinant human ubiquitin (117 pmol/30 μl) or mutant human ubiquitin in which arginine replaced all lysines (asterisks) (see methods regarding mutant ubiquitin concentration). At the indicated times, 30-μl samples were removed and added to 10 μl of 4× Laemmli sample buffer. (A) Blot was reacted with anti-GST antibodies before stripping and reaction with anti-ubiquitin antibodies. Free GST was present in all GST-ARD1 preparations, because it was bound along with GST-ARD1 to glutathione-Sepharose during purification. (B and C) Reactions contained His-tagged ARD1 and either wild-type or lysine-free ubiquitin (B, asterisk), or only lysine-free ubiquitin (C). Blots were reacted with anti-ubiquitin, anti-ARD1, or anti-UbcH6 antibodies as indicated. Arrow and arrowheads indicate, respectively, the unmodified and modified forms of the UbcH6 (E2). Data were replicated twice.
Results
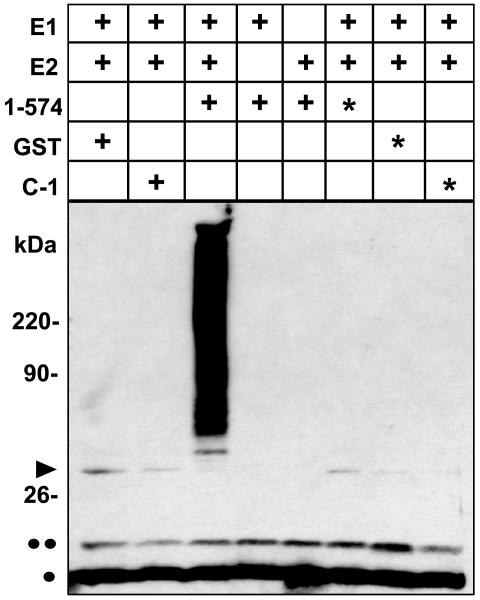
ARD1-Catalyzed Formation of Multiubiquitinylated Products. As seen in Fig. 2, incubation of GST-ARD1 (1–574) with rabbit E1, human recombinant UbcH6 (E2), bovine ubiquitin, and ATP resulted in conspicuous accumulation of polyubiquitinylated products after 60 min, indicative of ARD1-associated E3 ligase activity. Replacement of GST-ARD1 with GST, GST-cytohesin-1 (C-1, an ARD1-activating protein), or heat-inactivated GST-ARD1 yielded no detectable ubiquitinylated products. Similarly, reactions containing GST-ARD1 and either E1 or E2 alone yielded no ubiquitinylated products (Fig. 2), demonstrating an absolute dependence of the reaction on all three enzymes (E1, E2, and E3-ARD1). Proteins migrating at ≈36 kDa (arrowhead) reacted also with antibodies against UbcH6, consistent with ubiquitinylated forms of E2 (cf. Fig. 6).
Fig. 2.
GST-ARD1-catalyzed formation of ubiquitinylated proteins in vitro required E1 and E2. Samples (2.2 pmol) of native or heat-inactivated (asterisk) GST-ARD1 (1–574), GST-cytohesin-1 (C-1), or GST were incubated with 0.8 pmol of E1 and/or 16 pmol of E2 (UbcH6) in standard ubiquitinylation assays (total volume of 30 μl) for 60 min at 30°C before separation of proteins by SDS/PAGE. Blots were reacted with anti-ubiquitin monoclonal antibody. Dots indicate positions of mono- or diubiquitin; the arrowhead indicates ubiquitinylated E2. Data were replicated at least three times.
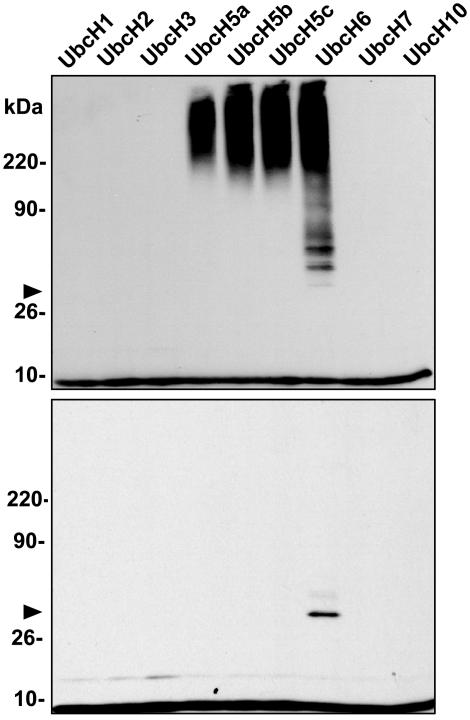
ARD1 E3 Ligase Activity Is Specific for a Subset of E2s in Vitro. In ubiquitinylation assays with nine different His-tagged human E2s (UbcH1, UbcH2, UbcH3/CDC34, UbcH5a, UbcH5b, UbcH5c, UbcH6, UbcH7, and UbcH10), GST-ARD1 catalyzed ubiquitinylation only in the presence of UbcH5a, UbcH5b, UbcH5c, and UbcH6 (Fig. 3 Upper). No products were detected with the other E2s or when GST replaced GST-ARD1 (Fig. 3 Lower). Arrowheads indicate the position of ubiquitinylated UbcH6 (E2), which is not well seen when ARD1 is present.
Fig. 3.
Specificity of E2 requirement for in vitro ubiquitinylation with recombinant GST-ARD1. Standard ubiquitinylation assays contained 0.8 pmol of E1, 2.2 pmol of GST-ARD1 (Upper) or GST (Lower), and 0.4 μg of the indicated human E2 ubiquitin-conjugating enzyme. Ubiquitinylated proteins were detected with anti-ubiquitin antibodies; arrowhead indicates ubiquitinylated UbcH6 (E2). The experiment was repeated twice with similar results. Activities of UbcH1 and UbcH3/CDC34 were demonstrated by the production of E2-ubiquitin conjugates that were stable under reducing conditions (data not shown).
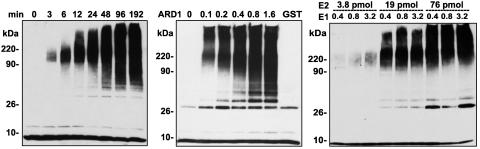
GST-ARD1 E3 Ubiquitin Ligase Activity Depends on Time and Enzyme Concentration. In assays containing UbcH6 as the E2 component, 90- to 220-kDa ubiquitinylated proteins were detected within 3 min of incubation (Fig. 4 Left). With time, progressively larger (>220 kDa) ubiquitinylated products accumulated; smaller products (<90 kDa) began to accumulate as well. By 48 min, the largest products were accumulated at the top of the gel. In assays incubated for 60 min, the sizes of ubiquitinylated products that accumulated clearly differed depending on the concentration of GST-ARD1 (Fig. 4 Center). At the lowest concentration of ARD1, the size of ubiquitinylated products was almost exclusively large (>220 kDa). Smaller products also accumulated with increasing ARD1 concentrations. When the concentrations of E1 or E2 were varied in the presence of 2.2 pmol of GST-ARD1, similar ranges in product sizes were observed (Fig. 4 Right). In these assay conditions, an increasing UbcH6 (E2) concentration appeared to have larger effects on ubiquitinylation than did increases of similar magnitude in E1.
Fig. 4.
Ubiquitinylation catalyzed by E1, E2, and GST-ARD1 (E3) as a function of time and enzyme concentrations. (Left) Standard assay mixture (270 μl) containing 75 nM ARD1 (2.2 pmol/30 μl) was incubated at 30°C with 26 nM E1 (0.8 pmol/30 μl) and 0.53 μM UbcH6 (E2, 16 pmol/30 μl). At the indicated times, 30-μl samples were removed and added to 10 μl of 4× Laemmli sample buffer. (Center) Standard assays (30 μl) containing 0, 0.1, 0.2, 0.4, 0.8, or 1.6 pmol of GST-ARD1 or 2.2 pmol of GST were incubated for 60 min. (Right) Assays (30 μl) containing 2.2 pmol of GST-ARD1 with the indicated amounts (pmol) of E1 and E2 were incubated for 60 min. Data in Left and Center were replicated at least twice.
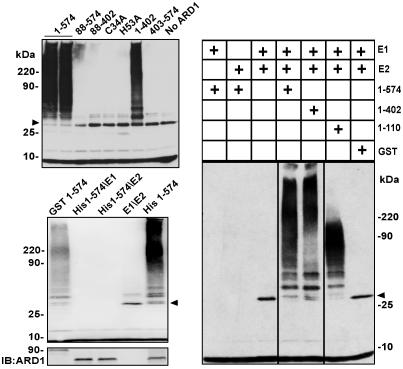
ARD1 RING Finger Domain Is Responsible for the E3 Activity. To determine whether the RING finger domain of ARD1 is responsible for its E3 ubiquitin ligase activity, the activity of GST-ARD1 was compared with that of several recombinant ARD1-related molecules (Fig. 1 Upper), each containing a specific deletion, or a point mutation in the RING finger domain (Fig. 5). No ubiquitinylation activity was detected with GST-ARD1 mutants that lacked the RING domain (88–574, 88–402, and 403–574) or with those in which one of the RING finger metal-binding amino acids, either C34A or H53A, had been replaced by alanine (Fig. 5). Conversely, deletions from GST-ARD1 at the C terminus (1–402 and 1–110, which removed the ARF or the ARF and the GAP domains, respectively) had little effect on ubiquitinylation activity. The sizes of products of GST-ARD1 (1–110) clearly differed, however, from those seen when either full-length GST-ARD1 (1–574) or GST-ARD1 (1–402) supplied the E3 activity. No differences in the patterns of ubiquitinylated products were observed when His-tagged ARD1 was substituted for GST-ARD1 in the assays (Fig. 5 Bottom Left).
Fig. 5.
Requirement of intact ARD1 RING finger domain for E3 ubiquitin ligase activity. (Top Left) Ubiquitinylation activity of GST-ARD1 or the indicated mutant proteins (5 pmol each). (Right) Standard assays (30 μl) contained, as indicated, 0.8 pmol of E1 and 16 pmol of UbcH6 (E2), with 2.2 pmol of the indicated recombinant GST-ARD1 or GST protein. (Middle Left) Standard assays contained, as indicated, His-tagged ARD1 protein (9 pmol) or GST-ARD1 (2.2 pmol) with E1 and/or E2; E1/E2 lane contained no ARD1. In Bottom Left, the same blot reacted with anti-ARD1 antibodies is showing unmodified GST- and His-ARD1. Arrowheads indicate ubiquitinylated E2. Data were replicated at least twice.
ARD1 Ubiquitinylates Itself, Free GST, and UbcH6 in vitro. In the in vitro ubiquitinylation assays containing only ATP and free ubiquitin plus three other purified proteins (E1, E2/UbcH6, and E3/ARD1), products should be only ubiquitinylated E1, E2, E3, and/or heterogeneous “free,” unanchored polyubiquitin chains (23–25), and/or contaminants in the protein preparations. Immunoblotting with antibodies specific for GST, ARD1, E1, or UbcH6 was used to try to detect ubiquitinylated forms of these proteins. As seen in Fig. 6A Lower, the amount of unmodified GST-ARD1 (as well as its fragments or free GST) decreased during ubiquitinylation assays by ≈50% (quantified by densitometry; data not shown) after 10 min, and continued to decrease thereafter. No ubiquitinylated (i.e., slower migrating) GST-ARD1 was detected with anti-GST antibodies; as shown in Fig. 6A Upper (lanes without an asterisk) the same blot, stripped and incubated with anti-ubiquitin antibody, showed the time-dependent accumulation of ubiquitinylated proteins, of ≈100 kDa (≈10 kDa larger than unmodified GST-ARD1) and larger. As also shown in Fig. 6 (asterisks), ubiquitinylation assays were performed in parallel, using a recombinant ubiquitin in which all seven lysines had been replaced by arginine. With this mutant ubiquitin, only monoubiquitinylated or multi-monoubiquitinylated products can be formed, because no ubiquitin lysine is present to serve as an acceptor for chain elongation; thus, the population of modified products might be less complex and/or heterogeneous than that seen in the previous experiments. As in reactions with wild-type ubiquitin, in those with the mutant ubiquitin (Fig. 6, asterisks) amounts of unmodified GST-ARD1 decreased during the 30-min incubation (Fig. 6A Lower) with simultaneous accumulation of ubiquitinylated product(s) ≈10 kDa larger than GST-ARD1 (Fig. 6A Upper). As expected, ubiquitinylated proteins appeared less heterogeneous, and no accumulation of the largest products was seen, although the antibody apparently reacted less well with the mutant than the native ubiquitin.
Results were similar when His-tagged ARD1 replaced GST-ARD1 in ubiquitinylation assays (Fig. 6B). As seen in Fig. 6B Lower, detection of His-ARD1 with anti-ARD1 antibodies clearly showed that it decreased in amount with time. The reaction of the blot with anti-ubiquitin antibodies revealed an increasing accumulation of ubiquitinylated products. With lysine-free ubiquitin (Fig. 6B Upper, asterisks), the pattern was, again, less complex, and none of the largest ubiquitinylated products was visible. Finally, as shown in Fig. 6C Lower when ubiquitinylated products of in vitro reaction with His-ARD1 and the mutant lysine-free ubiquitin were reacted with antibodies specific for UbcH6 (E2), the amount of unmodified UbcH6 (arrow) was seen to decrease with time, concomitant with the appearance over time of immunoreactive bands ≈10, 20, and 30 kDa larger than unmodified UbcH6 (arrowheads). When duplicate samples from the same assay were analyzed with anti-ubiquitin antibodies, increasing accumulation of mono-, di-, and triubiquitinylated UbcH6 was clearly detected (Fig. 6C Upper, arrowheads). Similar experiments analyzed for possible ubiquitinylation of E1 showed no detectable changes in either amount or position of the E1 band (data not shown). Taken together, these data are consistent with in vitro ubiquitinylation of ARD1 itself, small GST-containing fragments derived from GST-ARD1, and UbcH6.
Discussion
The E3 ubiquitin ligase activity of ARD1, which contains a RING finger motif, was readily demonstrated in vitro in assays containing purified mammalian E1 and E2. This finding is consistent with previous reports of ubiquitinylation catalyzed by other RING finger proteins for which the intracellular substrate was not identified (15, 23, 26, 27). Two groups of E3 ligases are defined by differences in the structures of the E3 catalytic domains, those with HECT domains and those in which RING finger motifs are responsible for the catalytic activity (28, 29). The involvement of an enzyme thioester intermediate in the HECT E3 ubiquitin ligase catalytic mechanism has been established (18, 30). To date, however, there is no evidence of ubiquitin thioester intermediate formation by any RING finger E3 ligase. RING-type E3 ligases appear to facilitate the transfer of ubiquitin from E2 directly to an acceptor protein or to E3 itself, although the molecular mechanism remains to be established (31–33).
ARD1 is a multifunctional protein, with E3 ligase, GAP, and ARF activities residing in separate protein domains. The activity of each of these domains appeared to be independent of the intact protein structure. The ARD1 recombinant protein comprising amino acids 1–110 exhibited E3 ligase activity, although the ubiquitinylated products were much more limited in size than those of ARD1(1–402) or of full-length ARD1, which contain also the GAP (amino acids 101–333) or the GAP plus ARF (amino acids 403–574) domain activities, respectively. It was not surprising, therefore, that GDP and GTPγS did not affect the ARD1 E3 ligase activity (data not shown), although structural elements outside the RING domain and guanine nucleotide or other ligands may well influence ubiquitinylation of the physiological substrate(s).
RING finger E3 ligases have been reported to catalyze autoubiquitinylation (15) and substrate ubiquitinylation (34), as well as the synthesis of unanchored polyubiquitin chains in vitro (23, 24). Although previously unrecognized substrate(s) for many proteins containing RING finger domain(s) continue to be identified, for the majority, including ARD1, the physiological substrates remain unknown. The decrease in GST-ARD1 with time in our in vitro assays is consistent with GST-ARD1-catalyzed ubiquitinylation of its in-frame fused GST moiety, which would be reminiscent of the finding of Matsuda et al. (35) that the RING finger-containing protein Rma1 ubiquitinylated an in-frame fused maltose-binding protein, or of a GST protein in trans. We failed, however, to detect either ubiquitinylated GST or GST-ARD1 when using anti-GST antibodies. The potential ubiquitinylation of GST by ARD1 might reflect the reported homodimerization of GST (36), which could also be involved in the ubiquitinylation of GST-ARD1, although the His-tagged ARD1 did seem to be similarly ubiquitinylated. The dimerization of ARD1 itself has been described (10) and is consistent with the possibility that ARD1 is autoubiquitinylated even in the absence of the GST moiety. UbcH6, the E2 used in most of our ARD1 ubiquitinylation assays, was apparently modified with the addition of up to three ubiquitin moieties. Both GST-ARD1 and His-tagged ARD1 enhanced UbcH6 ubiquitinylation, yielding similar patterns of ubiquitinylated products. Analysis of the in vitro ubiquitinylated proteins should enable us to identify mono- or polyubiquitinylation and to characterize the sites in substrate protein(s) to which ubiquitin is attached.
The relationship between functions of the ARF domain at the C terminus of ARD1 and its E3 ubiquitin ligase activity is an obvious and important question. GFP-ARD1 overexpressed in NIH 3T3 cells was seen concentrated in perinuclear structures resembling Golgi and lysosomes (37). Fragments of the ARD1 N terminus up to 300 aa in length expressed as GFP fusion proteins appeared to be cytosolic, whereas GFP-ARD1(101–574), which lacks the first 100 aa, had a distribution similar to that of full-length GFP-ARD1 (38), indicating that the RING motif did not influence localization. The ARD1 molecule contains between the RING and ARF domains, two B-boxes, and a coiled-coiled domain, defining ARD1 as one of the reported mammalian TRIM proteins (10). Numerous TRIMs have been implicated in diverse cellular regulatory processes, including transcription (39), signal transduction (40, 41), vesicular transport (42), exocytosis (43), and protein degradation via ubiquitinylation (44–46). Diamonti et al. (46) established an interesting link among TRIM protein, intracellular trafficking, and ubiquitin in processes that maintain the cell-surface population of neuregulin receptors, which cycle constitutively from synthesis to plasma membrane to proteolysis.
Some of the TRIM proteins lack a C-terminal domain following the RBCC motifs, whereas the different C-terminal sequences of other TRIMs presumably reflect their different roles (47). The known functions of ARF proteins and the ARD1 ARF domain might indicate an association with Golgi structure and lysosomes in processes related to vesicular transport. Relationships among ubiquitin, trans-Golgi network, and endosomal compartments have been reported (48, 49). These studies demonstrated that Golgi-localized, gamma-ear-containing, ARF-binding (GGA) proteins bind ubiquitin through their GAT domains to accomplish protein sorting at the trans-Golgi network and transport toward endosomes.
Our data appear to indicate that ARD1 can catalyze its own autoubiquitinylation, and it is this reaction, plus the ubiquitinylation of UbcH6 (E2), that enabled us to demonstrate its E3 ligase activity. Among many intriguing and important questions that are subjects of continuing investigation is the identity of intracellular substrates, characterization of which should help us to understand the biological role(s) of ARD1 and its multiple catalytic functions.
Acknowledgments
We thank Dr. Vincent C. Manganiello for valuable discussions and manuscript review.
Abbreviations: ARF, ADP-ribosylation factor; ARD1, ARF; domain protein 1.
References
- 1.Moss, J. & Vaughan, M. (1998) J. Biol. Chem. 273, 21431–21434. [DOI] [PubMed] [Google Scholar]
- 2.Moss, J. & Vaughan, M. (1995) J. Biol. Chem. 270, 12327–12330. [DOI] [PubMed] [Google Scholar]
- 3.Kahn, R. A. & Gilman, A. G. (1984) J. Biol. Chem. 259, 6228–6234. [PubMed] [Google Scholar]
- 4.Holthuis, J. C. & Burger, K. N. (2003) Dev. Cell 5, 821–822. [DOI] [PubMed] [Google Scholar]
- 5.Nie, Z., Hirsch, D. S. & Randazzo, P. A. (2003) Curr. Opin. Cell Biol. 15, 396–404. [DOI] [PubMed] [Google Scholar]
- 6.Mishima, K., Tsuchiya, M., Nightingale, M. S., Moss, J. & Vaughan, M. (1993) J. Biol. Chem. 268, 8801–8807. [PubMed] [Google Scholar]
- 7.Vitale, N., Pacheco-Rodriguez, G., Ferrans, V. J., Riemenschneider, W., Moss, J. & Vaughan, M. (2000) J. Biol. Chem. 275, 21331–21339. [DOI] [PubMed] [Google Scholar]
- 8.Vitale, N., Moss, J. & Vaughan, M. (1996) Proc. Natl. Acad. Sci. USA 93, 1941–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitale, N., Moss, J. & Vaughan, M. (1997) J. Biol. Chem. 272, 25077–25082. [DOI] [PubMed] [Google Scholar]
- 10.Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., et al. (2001) EMBO J. 20, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saurin, A. J., Borden, K. L., Boddy, M. N. & Freemont, P. S. (1996) Trends Biochem. Sci. 21, 208–214. [PubMed] [Google Scholar]
- 12.Freemont, P. S., Hanson, I. M. & Trowsdale, J. (1991) Cell 64, 483–484. [DOI] [PubMed] [Google Scholar]
- 13.Reddy, B. A. & Etkin, L. D. (1991) Nucleic Acids Res. 19, 6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupas, A. (1996) Trends Biochem. Sci. 21, 375–382. [PubMed] [Google Scholar]
- 15.Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S. & Weissman, A. M. (1999) Proc. Natl. Acad. Sci. USA 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joazeiro, C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T. & Liu, Y. C. (1999) Science 286, 309–312. [DOI] [PubMed] [Google Scholar]
- 17.Huibregtse, J. M., Scheffner, M., Beaudenon, S. & Howley, P. M. (1995) Proc. Natl. Acad. Sci. USA 92, 5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheffner, M., Nuber, U. & Huibregtse, J. M. (1995) Nature 373, 81–83. [DOI] [PubMed] [Google Scholar]
- 19.Aguilar, R. C. & Wendland, B. (2003) Curr. Opin. Cell Biol. 15, 184–190. [DOI] [PubMed] [Google Scholar]
- 20.Frangioni, J. V. & Neel, B. G. (1993) Anal. Biochem. 210, 179–187. [DOI] [PubMed] [Google Scholar]
- 21.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 23.Boutell, C. & Everett, R. D. (2003) J. Biol. Chem. 278, 36596–36602. [DOI] [PubMed] [Google Scholar]
- 24.Yao, T. & Cohen, R. E. (2000) J. Biol. Chem. 275, 36862–36868. [DOI] [PubMed] [Google Scholar]
- 25.Amerik, A. Y., Swaminathan, S., Krantz, B. A., Wilkinson, K. D. & Hochstrasser, M. (1997) EMBO J. 16, 4826–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang, J. H. (2004) FEBS Lett. 578, 21–25. [DOI] [PubMed] [Google Scholar]
- 27.Jones, J. M. & Gellert, M. (2003) Proc. Natl. Acad. Sci. USA 100, 15446–15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freemont, P. S. (2000) Curr. Biol. 10, R84–R87. [DOI] [PubMed] [Google Scholar]
- 29.Joazeiro, C. A. & Weissman, A. M. (2000) Cell 102, 549–552. [DOI] [PubMed] [Google Scholar]
- 30.Huang, L., Kinnucan, E., Wang, G., Beaudenon, S., Howley, P. M., Huibregtse, J. M. & Pavletich, N. P. (1999) Science 286, 1321–1326. [DOI] [PubMed] [Google Scholar]
- 31.Passmore, L. A. & Barford, D. (2004) Biochem. J. 379, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503–533. [DOI] [PubMed] [Google Scholar]
- 33.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 34.Rajendra, R., Malegaonkar, D., Pungaliya, P., Marshall, H., Rasheed, Z., Brownell, J., Liu, L. F., Lutzker, S., Saleem, A. & Rubin, E. H. (2004) J. Biol. Chem. 279, 36440–36444. [DOI] [PubMed] [Google Scholar]
- 35.Matsuda, N., Suzuki, T., Tanaka, K. & Nakano, A. (2001) J. Cell Sci. 114, 1949–1957. [DOI] [PubMed] [Google Scholar]
- 36.Walker, J., Crowley, P., Moreman, A. D. & Barrett, J. (1993) Mol. Biochem. Parasitol. 61, 255–264. [DOI] [PubMed] [Google Scholar]
- 37.Vitale, N., Horiba, K., Ferrans, V. J., Moss, J. & Vaughan, M. (1998) Proc. Natl. Acad. Sci. USA 95, 8613–8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitale, N., Ferrans, V. J., Moss, J. & Vaughan, M. (2000) Mol. Cell. Biol. 20, 7342–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng, H., Feldman, I. & Rauscher, F. J., III (2002) J. Mol. Biol. 320, 629–644. [DOI] [PubMed] [Google Scholar]
- 40.Toniato, E., Chen, X. P., Losman, J., Flati, V., Donahue, L. & Rothman, P. (2002) J. Biol. Chem. 277, 37315–37322. [DOI] [PubMed] [Google Scholar]
- 41.Dho, S. H. & Kwon, K. S. (2003) J. Biol. Chem. 278, 31902–31908. [DOI] [PubMed] [Google Scholar]
- 42.El Husseini, A. E. & Vincent, S. R. (1999) J. Biol. Chem. 274, 19771–19777. [DOI] [PubMed] [Google Scholar]
- 43.Li, Y., Chin, L. S., Weigel, C. & Li, L. (2001) J. Biol. Chem. 276, 40824–40833. [DOI] [PubMed] [Google Scholar]
- 44.Trockenbacher, A., Suckow, V., Foerster, J., Winter, J., Krauss, S., Ropers, H. H., Schneider, R. & Schweiger, S. (2001) Nat. Genet. 29, 287–294. [DOI] [PubMed] [Google Scholar]
- 45.Urano, T., Saito, T., Tsukui, T., Fujita, M., Hosoi, T., Muramatsu, M., Ouchi, Y. & Inoue, S. (2002) Nature 417, 871–875. [DOI] [PubMed] [Google Scholar]
- 46.Diamonti, A. J., Guy, P. M., Ivanof, C., Wong, K., Sweeney, C. & Carraway, K. L., III (2002) Proc. Natl. Acad. Sci. USA 99, 2866–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venturini, L., You, J., Stadler, M., Galien, R., Lallemand, V., Koken, M. H., Mattei, M. G., Ganser, A., Chambon, P., Losson, R., et al. (1999) Oncogene 18, 1209–1217. [DOI] [PubMed] [Google Scholar]
- 48.Puertollano, R. & Bonifacino, J. S. (2004) Nat. Cell Biol. 6, 244–251. [DOI] [PubMed] [Google Scholar]
- 49.Scott, P. M., Bilodeau, P. S., Zhdankina, O., Winistorfer, S. C., Hauglund, M. J., Allaman, M. M., Kearney, W. R., Robertson, A. D., Boman, A. L. & Piper, R. C. (2004) Nat. Cell Biol. 6, 252–259. [DOI] [PubMed] [Google Scholar]