Abstract
Hypertension (HTN) affects about 1 billion people worldwide and the lack of a single identifiable cause complicates its treatment. Blood pressure (BP) levels are influenced by environmental factors, but there is a strong genetic component. Linkage analysis has identified several genes involved in Mendelian forms of HTN and the associated pathophysiological mechanisms have been unravelled, leading to targeted therapies. The majority of these syndromes are due to gain-of-function or loss-of-functions mutations, resulting in an alteration of mineralocorticoid, glucocorticoid, or sympathetic pathways. The diagnosis of monogenic forms of HTN has limited practical implications on the population and a systematic genetic screening is not justifiable. Genome-wide linkage and association studies (GWAS) have identified single nucleotide polymorphisms (SNPs), which influence BP. Forty-three variants have been described with each SNP affecting systolic and diastolic BP by 1.0 and 0.5 mmHg, respectively. Taken together Mendelian inheritance and all GWAS-identified HTN-associated variants explain 2–3% of BP variance. Epigenetic modifications, such as DNA methylation, histone modification and non-coding RNAs, have become increasingly recognized as important players in BP regulation and may justify a further part of missing heritability. In this review, we will discuss how genetics and genomics may assist clinicians in managing patients with HTN.
Keywords: monogenic hypertension, genomics, genome-wide association studies, epigenetics, pharmacogenomics
1. Introduction
Hypertension (HTN) is the most frequent modifiable risk factor for cardiovascular disease that affects about 1 billion people worldwide [1,2]. Notwithstanding the major advances in understanding the pathophysiology of HTN and the introduction of new treatment options, HTN remains a major public health problem with a high socio-economic and health burden. Essential HTN is a multifactorial disease and the lack of a single identifiable cause complicates the treatment of this condition.
Blood pressure (BP) levels are affected by modifiable factors such as salt, alcohol consumption, obesity, physical activity and chronic stress. However, twin and family studies have demonstrated that 30–50% of the individual risk comes from genetic factors [3,4] and a family history of HTN increases the risk of developing HTN by four times [5,6]. Over the last few decades a wealth of studies was designed to understand the genetic basis of HTN and the associated molecular alterations. Three different approaches have been used.
The identification of genes involved in Mendelian forms of HTN by linkage analysis and next generation sequencing, with description of rare variants with a major impact on BP levels. This approach may identify the pathophysiological mechanism of a specific disorder and lead to a targeted therapy [7,8].
An approach based on families, twins and adopted children, or population studies using genome-wide linkage and association studies (GWAS). The latter can identify single nucleotide polymorphisms (SNPs), which influence the risk of developing high BP [8]. GWAS has also been applied to pharmacogenomic studies to identify genetic variants that regulate pharmacokinetics and pharmacodynamics of anti-hypertensive drugs [9].
More recently the study of epigenetic modifications has aroused growing interest; DNA methylation, histone modification and non-coding RNAs have become recognized as important players in several pathophysiological processes, including BP regulation [10].
Genetics is the study of the basis of heredity: how phenotypic features are transmitted from one generation to the next and how a specific gene causes a particular disease, whereas genomics is the study of the entire genome. In this review we discuss how genetics and genomics may assist clinicians in managing patients with HTN. For this purpose, Pubmed database, Medline, Cochrane Library and Biomed Central were searched for articles related to the key words “hypertension” (combined with “genetics”, “genomics”, “pharmacogenomics” and “epigenetics”) and “monogenic hypertension”. We selected the most relevant manuscripts pertinent to our review design, which comprised monogenic forms of hypertension, large-scale genome-wide associations studies, epigenetic modifications in hypertensive patients and pharmacogenomics.
2. Monogenic Forms of Hypertension
Over the last century, an increasing number of syndromes associated with high BP that are caused by single rare gene mutations have been identified [11].
The discovery of genes responsible for monogenic forms of HTN identified the kidney and adrenal glands as important players in the regulation of BP levels [12]. The majority of these syndromes are due to gain-of-function or loss-of-function mutations, which result in an alteration of mineralocorticoid, glucocorticoid, or sympathetic pathways (see Table 1).
Table 1.
Hypertension (HTN)-related monogenic syndromes.
| Monogenic Syndrome | Inheritance | Gene | Locus | Phenotype | Therapeutic Indications |
|---|---|---|---|---|---|
| Pheochromocytomas/Paragangliomas | Autosomal dominant |
SDHA
SDHB SDHC SDHD SDHAF2 TMEM127 MAX |
5p15.3 1p36.13 1q23.3 11q23.1 11q12.2 2q11.2 14q23.3 |
Paragangliomas or pheochromocytomas. | Surgery/α adrenergic blockers |
| von Hippel–Lindau syndrome | Autosomal dominant | VHL | 3p25.3 | Retinal, cerebellar and spinal hemangioblastoma, renal cell carcinoma, pheochromocytomas, pancreatic tumours. | Surgery/α adrenergic blockers (for pheochromocytoma) |
| Multiple endocrine neoplasia, type 2A | Autosomal dominant | RET | 10q11.2 | Medullary thyroid carcinoma, parathyroid adenomas, pheochromocytoma. | Surgery/α adrenergic blockers (for pheochromocytoma) |
| Neurofibromatosis type 1 | Autosomal dominant | NF1 | 17q11.2 | Skin pigmentation, skin neurofibromas and brain tumours. Pheochromocytoma. | Surgery/α adrenergic blockers (for pheochromocytoma) |
| GRA–familial hyperaldosteronism type 1 | Autosomal dominant | CYP11B1 CYP11B2 | 8q24.3 | Familial form of PA | Glucocorticoids |
| Familial hyperaldosteronism type 2 | Autosomal dominant | N.A. | 7p22.3-7p22.1 | Familial form of PA | Mineralocorticoid receptor antagonist/unilateral adrenalectomy (for APA) |
| Familial hyperaldosteronism type 3 | Autosomal dominant | KCNJ5 | 8q24.3 | Severe form of PA with bilateral adrenal hyperplasia | Bilateral adrenalectomy in drug-resistant patients |
| Familial hyperaldosteronism type 4 | Autosomal dominant | CACNA1H | 16p13.3 | Familial form of PA | Mineralocorticoid receptor antagonist |
| PASNA syndrome | N.A. | CACNA1D | 3p21.3 | PA and complex neurological disorders (seizures and functional neurological abnormalities, resembling cerebral palsy). | N.A. |
| Sporadic APA | N.A. |
KCNJ5
ATP1A1 ATP2B3 CACNA1D |
11q24.3 1p31.1 Xq28 3p21.3 |
Sporadic forms of PA. | Adrenalectomy |
| Pseudohypoaldosteronism, type 2 (Gordon’s syndrome) | Autosomal dominant (*dominant/recessive) |
WNK1
WNK4 CUL3 KLHL3 * |
12p12.3 17q21.2 2q36.2 5q31.2 |
HyperK+ hyperCl− metabolic acidosis. Low PRA and low-normal AC. | Thiazide diuretics |
| Apparent mineralocorticoid excess (AME) Syndrome | Autosomal recessive | HSD11B2 | 16q22.1 | Hypokalemia. Low PRA and AC. Increased cortisol/cortisone ratio. | Mineralocorticoid receptor antagonist |
| Liddle’s syndrome | Autosomal dominant | SCNN1B, SCNN1G | 16p12.2 | ENaC constitutive activation. Hypokalemia. Low PRA and AC. | ENaC blockers (amiloride, triamterene) |
| 11β-hydroxylase deficiency | Autosomal recessive | CYP11B1 | 8q24.3 | Virilisation, short stature. Low PRA and AC. HypoK+ alkalosis. |
Glucocorticoids to inhibit ACTH-driven adrenal hyperpasia |
| 17α-hydroxylase deficiency | Autosomal recessive | CYP17A1 | 10q24.3 | HypoK+ alkalosis. Absent sexual maturation. Androgen deficiency. | Glucocorticoids to inhibit ACTH-driven adrenal hyperpasia |
| Hypertension with brachydactyly Type E | Autosomal dominant | PDE3A | 12p12.3 12p12.1 |
Brachydactyly, short phalanges and metacarpals. | N.A. |
| Hypertension exacerbated by pregnancy | Autosomal dominant | NR3C2 | 4q31.23 | Early onset hypertension exacerbated during pregnancy. | N.A. |
GRA, glucocorticoid remediable aldosteronism. PA, primary aldosteronism. N.A., not available. PASNA, primary aldosteronism, seizures and neurologic abnormalities. APA, aldosterone producing adenoma. HyperK+, hyperkalemic. HyperCl−, hyperchloraemic. PRA, plasma renin activity. AC, aldosterone concentration. HypoK+, hypokalemic. ENaC, epithelial sodium channel. Modified from Padmanabhan et al. [7].
Among the syndromes that affect the mineralocorticoid pathway, familial hyperaldosteronism is the most frequent. Glucocorticoid-Remediable Aldosteronism (GRA), or familial hyperaldosteronism type 1 (FH-1) results from an unequal crossing over between CYP11B1 (encoding for 11 β-hydroxylase) and CYP11B2 (encoding for aldosterone synthase), leading to ACTH-dependent aldosterone secretion, HTN, hypokalemia, low renin and high aldosterone levels. Low-dose glucocorticoids suppress aldosterone production and normalize BP and potassium levels [13].
For familial hyperaldosteronism type 2 (FH-2) the causative gene has not yet been identified. FH-2 is indistinguishable from sporadic primary aldosteronism (PA) except for the presence of more members affected by PA within the same family. The diagnosis of this condition is made after exclusion of other familial forms of PA [14].
Familial hyperaldosteronism type 3 (FH-3) is caused by mutations in KCNJ5, which encodes the inwardly rectifying potassium channel Kir3.4. Described mutations are located near or within the selectivity filter and are responsible for loss of ion selectivity, resulting in Na+ entry, membrane cell depolarization, opening of voltage-dependent calcium channels, increase in calcium intracellular concentration and aldosterone production [15]. Patients display severe hypokalemia, difficult-to-treat HTN and bilateral adrenal hyperplasia. Bilateral adrenalectomy is necessary in most cases.
Recently, a fourth type of familial hyperaldosteronism (FH-4) has been described [16]. FH-4 is caused by a germline gain-of-function mutation of CACNA1H, encoding for a T-type calcium channel. CACNA1H constitutive activation leads to calcium entry and increased aldosterone production [17].
Of note, somatic mutations in genes encoding ion pumps and channels (KCNJ5, ATP1A1, ATP2B3, CACNA1D) in adrenal cells have been identified in more than 50% of patients with sporadic APA [18,19]. Germline mutations in CACNA1D have been described in two patients with unexplained PA and complex neurological disorders (seizures and functional neurological abnormalities, resembling cerebral palsy) [20]. This syndrome was called PASNA (Primary Aldosteronism, Seizures and Neurologic Abnormalities). Patients with PASNA are not able to transmit the mutation to their offspring because of the severe neurological impairment.
Another Mendelian form of low-renin HTN is the Apparent Mineralocorticoid Excess (AME) syndrome. Cortisol has a strong agonist activity on mineralocorticoid receptor (MR) and is present in a 100× higher concentration in bloodstream. HSD11B2 (type 2, 11β-hydroxysteroid dehydrogenase), converting cortisol in cortisone, prevents its binding to MR. The loss-of-function mutation of HSD11B2 leads to cortisol-dependent activation of the MR resulting in sodium retention, hypokalemia, metabolic alkalosis, suppressed renin and aldosterone levels and increased cortisol/cortisone ratio [21,22]. An acquired deficiency of this enzyme is determined by excessive liquorice intake (glycyrrihizic acid from liquorice inhibit HSD11B2).
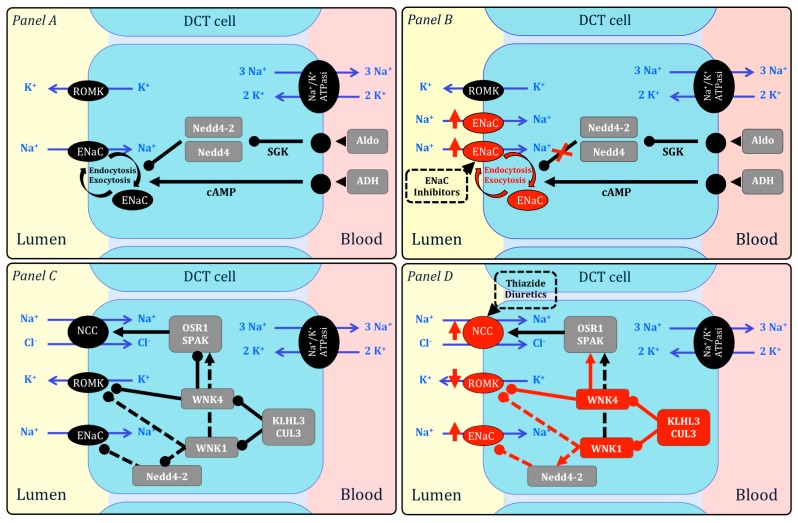
Gordon’s syndrome, also known as type 2 pseudo-hypoaldosteronism or familial hyperkaliemic hypertension is characterized by HTN, hyperkalemia and hyperchloraemic metabolic acidosis [23] (see Figure 1). The diagnosis is mainly clinical with subsequent identification of the causal mutation, that however, is not obtained in all cases, indicating that as yet unidentified genes are associated with this condition [24]. To date, mutations of 4 genes have been described: mutations in WNK1 and WNK4 kinases [25], and more recently KLHL3 and CUL3 mutations [26]. The net effect of gain-of-function mutations in WNK1 and loss-of-function mutations in WNK4, KLHL3 and CUL3 is the excessive activation of sodium-chloride co-transporter (NCC) and epithelial sodium channel (ENaC) and the inhibition of the potassium channel ROMK, with increased reabsorption of sodium and reduced excretion of potassium [27]. The identification of molecular mechanisms underlying the pathology allows a targeted therapy with thiazide diuretics, which inhibit NCC, revert hyperkalaemia and normalize BP. Recently, GILZ (glucocorticoid induced leucine zipper protein) has been demonstrated to modulate renal potassium homeostasis; GILZ-knockout mice had hyperkalemia due to hyperstimulation of NCC, representing a reliable model of Gordon syndrome (even if mice had normal BP values) [28].
Figure 1.
Gordon and Liddle syndromes. Panel A—Epithelial Na+ Channel (ENaC) is expressed in the distal convoluted tubule (DCT) at the apical membrane, where it allows Na+ in the lumen to enter the cell. At the baso-lateral membrane, Na+ is pumped outwards by Na+-K+ ATPase. ENaC membrane expression is regulated through membrane trafficking. Aldosterone (Aldo), vasopressin (ADH), Nedd4 and Nedd4-2 regulate membrane endocytosis and exocytosis, through cAMP-dependent pathways and aldosterone/serum and glucocorticoid-regulated kinase (SGK). Panel B—ENaC gain-of-function mutations determine resistance to Nedd-mediated ubiquitination with ENaC over-expression. ENaC inhibitors (such as amiloride) block ENaC and normalize BP. Panel C—Na+-Cl− co-transporter (NCC), potassium channel (ROMK) and ENaC are responsible for Na+ and K+ homeostasis in DCT. Different intracellular factors regulate the activity of these transporters: WNK1 and WNK4 (kinases which inhibit NCC and ROMK), KLHL3 and CUL3 (ubiquitin ligases which mediate WNK kinases degradation). Panel D—The net effect of gain-of-functions mutations in WNK1 and loss-of-function mutations in WNK4, KLHL3 and CUL3 is the excessive activation of NCC and ENaC and the inhibition of ROMK, with increased reabsorption of sodium and reduced excretion of potassium. Thiazide diuretics block NCC and normalize BP. Arrows indicate up-regulation whereas lines ending in closed circles indicate down-regulation. Fine lines indicate established pathways, whereas dotted lines indicate pathways observed in in vitro models. The effects of mutations are indicated in red. Modified from Pathare 2013 [27] and Snyder 2002 [29].
Liddle’s syndrome leads to hypokalemia, suppressed renin and aldosterone levels and a severe increase of BP, often resistant to anti-hypertensive agents, during childhood (see Figure 1). The disease is determined by gain-of-function mutations in the β- or γ- subunit of ENaC, with a consequent aldosterone-independent sodium retention. The mutations result in the inability to remove ENaC from the cell surface [29,30,31]. The treatment with ENaC inhibitors, such as amiloride, is highly effective in lowering BP and normalizing potassium levels.
Congenital adrenal hyperplasia (CAH) syndromes are a group of diseases caused by mutations in enzymes involved in glucocorticoid synthesis. In particular mutations in CYP11B1 and CYP17A1 (encoding for 11β-hydroxylase and 17α-hydroxylase, respectively) are associated with early onset HTN and hypokalemia. In these patients, ACTH is increased due to the absence of cortisol negative feedback resulting in adrenal hyperplasia and accumulation of cortisol precursors with mineralocorticoid activity (such as 11-deoxycorticosterone, DOC), which leads to HTN and hypokalemic alkalosis. Virilisation in affected females due to excessive androgen synthesis is also present in patients with CYP11B1 deficiency [32] and pseudo-hermaphroditism in affected males with CYP17A1 deficiency due to a deficit of androgen synthesis [33]. A CYP11B1 deficiency phenotype can also result from recombination between CYP11B2 and CYP11B1 genes thereby producing a hybrid CYP11B2/B1 gene [34].
Hypertension with brachydactyly Type E is associated with a missense mutation of the PDE3A gene, encoding for phosphodiesterase 3A. This syndrome is characterized by severe salt-independent but age-dependent HTN and short phalanges and metacarpals [35,36]. The mutation increases protein kinase A-mediated PDE3A phosphorylation in vitro, which results in enhanced cell proliferation and contraction in mesenchymal stem cell-derived vascular smooth muscle cells. The mechanism underlying HTN development is not fully elucidated, but authors suggest that these alterations may produce vascular hypertrophy and increase of peripheral vascular resistances [37].
A form of HTN that is exacerbated by pregnancy is caused by a Ser810Leu mutation in the MR. The mutation results in constitutive activation of the receptor and is proposed to impair MR specificity such that some steroid hormone antagonists (such as progesterone and cortisone) act as agonists to the mutated receptor [38,39]. Affected women display severe HTN during pregnancy, with elevated progesterone levels; delivery improves HTN but there is no specific treatment for men and non-pregnant women. Spironolactone increases BP by activating the MR [38,39].
Among syndromes that affect sympathetic pathways about 30% of pheochromocytoma and paragangliomas are associated to germline mutations. The described mutations affect the equilibrium between oncogenes and onco-suppressor genes. In particular, mutations of VHL, SDHA, SDHB, SDHC, SDHD and SDHAF2 alter the pathway related to hypoxia cellular responses mediated by hypoxia inducible factors, whereas mutations of RET and NF1 are associated to a constitutive activation of RAS/RAF/MEK and PI3KT/AKT/mTOR signalling [40]. Patients harbouring VHL, RET or NF1 mutations show syndromic diseases, such as von Hippel-Lindau syndrome (retinal, cerebellar and spinal hemangioblastoma, renal cell carcinoma, pancreatic tumours and pheochromocytoma), type IIA multiple endocrine neoplasia (medullary thyroid carcinoma, parathyroid adenomas and pheochromocytoma) and type 1 neurofibromatosis (skin pigmentation, neurofibromas, brain tumours, and pheochromocytoma) [41]. Because of the impact on the diagnosis, prognosis and the possibility of early detection of the disease in relatives, the systematic targeted genetic screening should be considered in clinical practice for patients with a diagnosis of pheochromocytoma or paragangliomas.
These are some examples in which the description of a rare genetic mutation has enabled the understanding of disease pathogenesis and in some cases the identification of a molecular pharmacologically-targetable pathway. However the diagnosis of these syndromes has limited practical implications on the population, even in cases of a systematic genetic screening of all young hypertensives, which is not justifiable unless in the presence of features that suggest syndromic diseases.
3. Large-Scale Genome-Wide Associations Studies
The study of monogenic forms of HTN improved the knowledge of physiological mechanisms regulating BP. Nevertheless, Mendelian disorders are rare and do not explain the genetic component in patients with essential HTN. Indeed, most human genome variability is determined by SNPs. SNPs are structural variations of a single nucleotide, which occur in a specific position of the genome [42].
The large-scale identification of these polymorphisms has been made possible by linkage and association mapping [43]. Genetic linkage is the tendency of close alleles on a chromosome to be inherited together. Linkage analysis is based on this trend and identifies genetic regions of interest by mapping the genome of families with rare syndromes. This method has been highly successful for the identification of monogenic disorders, but it has also been applied in patients with essential HTN. The BRIGHT study supported the linkage of a locus on chromosome 5q13 with essential HTN susceptibility [44], whereas two other studies demonstrated a linkage with multiple loci on chromosome 1q [45,46].
Association mapping is a technique of mapping expression quantitative trait loci (eQTL), which link phenotypes and genotypes of a population, taking advantage of linkage disequilibrium [47]. Association mapping studies are better suited for complex genetic traits such as HTN but require a higher number of patients compared to linkage analysis and the extensive knowledge of all SNPs within the genome of the organism of interest [48].
Recent technological advances have opened the door to genome-wide association studies (GWAS). GWAS methodology is based on the determination of the statistical association between each SNP and the pressure phenotype through a linear regression analysis [12]. Pressure phenotype is influenced by environmental factors, including anti-hypertensive medications. In GWAS, BP levels are adjusted for age, sex and BMI; subsequently by convention, +15/+10 mmHg are added respectively to systolic blood pressure (SBP) and to diastolic blood pressure (DBP) in the presence of pharmacological therapy with at least one anti-hypertensive drug [49].
The two (rarely three or more) alleles for each SNP have different prevalence in the population; the least frequent allele is defined as the minor allele. The minor allele frequency (MAF) is inversely proportional to the total number of SNPs in the population [50].
Therefore, the statistical power of GWAS is proportional to sample size, MAF and each SNP-effect on SBP/DBP and inversely proportional to the number of independent tests performed (which is equal to the number of SNPs analysed). Therefore, SNPs with an allele frequency less than 5% have a too low predictive power, whereas more frequent variants may represent attractive targets for multifactorial diseases because of the expected stronger impact [51].
The first attempt to identify variants associated with HTN by GWAS was performed in 2007 on 2000 patients with HTN, but failed to identify any association probably because of the small sample size (GWAS by the Wellcome Trust Case Control Consortium) [52]. Later studies with larger sample sizes were subsequently performed: in 2011 the International Consortium for Blood Pressure GWAS (ICBP) completed the analysis of over 200,000 subjects [49], identifying 29 SNPs associated with HTN. Using candidate gene focused arrays, the described variants increased to 43 [53] but the effect size for each SNP was demonstrated to be about 1 mmHg for SBP and 0.5 mmHg for DBP (see Table 2) [12]. A risk score combining the effects of each variation was built with all the identified SNPs: the resulting score proved to be predictive of HTN if applied to different populations, but explained only a small part of the phenotypic variance among patients. Furthermore, it was also associated with the risk of stroke, coronary artery disease and left ventricular hypertrophy, but not with renal failure, indicating that the relationship between HTN and kidney function is more complex than expected [49].
Table 2.
Genetic variants associated with hypertension.
| SNPs | Nearest Gene(s) | Position | Encoded Protein Function |
|---|---|---|---|
| rs880315 | CASZ | 1p36.22 | Zinc finger transcription factor that acts as tumour suppressor. |
| rs4846049 | MTHFR(3′)-NPPB | 1p36.22 | MTHFR catalyse the conversion of 5,10-MTH in 5-MTH and it is involved in homocysteine metabolism. NPPB encodes for the B natriuretic peptide. |
| rs17367504 | MTHFR(5′)-NPPB | ||
| rs17030613 | ST7L-CAPZA1 | 1p13.2 | ST7L is a tumour suppressor factor. CAPZA1 regulates growth of actin filaments. |
| rs2932538 | MOV10 | 1p13.2 | A component of the RISC complex RNA helicase. |
| rs2004776 | AGT | 1q42.2 | Pre-angiotensinogen. |
| rs16849225 | FIGN-GRB14 | 2q24.3 | FIGN regulates microtubules synthesis. GRB4 is a growth factor receptor-binding protein, which interacts with insulin receptors and insulin-like growth factors. |
| rs13082711 | SLC4A7 | 3p24.1 | Sodium bicarbonate co-transporter in neuronal cells, involved in visual and auditory transmission. |
| rs3774372 | ULK4 | 3p22.1 | Serine/Threonine kinase involved in neurite branching and elongation and neuronal migration. |
| rs319690 | MAP4 | 3p21.31 | Promotion of microtubule assembly. |
| rs419076 | MECOM | 3q26.2 | Transcriptional regulator and oncoprotein involved in apoptosis, hematopoiesis, cell differentiation and proliferation. |
| rs1458038 | FGF5 | 4q21.21 | Fibroblast growth factor 5, involved in embryonic development, cell growth, morphogenesis, tissue repair, tumour growth and invasion. |
| rs13107325 | SLC39A8 | 4q24 | Mitochondrial cellular import of zinc, involved in inflammation. |
| rs6825911 | ENPEP | 4q25 | Glutamyl aminopeptidase; associated with renal neoplasm. |
| rs13139571 | GUCY1A3-1B3 | 4q32.1 | Guanylate cyclase 1 soluble subunit α, involved in nitric oxide pathway transduction. |
| rs1173771 | NPR3-C5orf23 | 5p13.3 | Natriuretic peptide receptor 3, responsible for clearing natriuretic peptides through endocytosis of the receptor. |
| rs11953630 | EBF1 | 5q33.3 | Early B-cell factor 1, associated with central obesity, B-lymphocytes differentiation and Hodgkin lymphoma. |
| rs1799945 | HFE | 6p22.2 | Hemochromatosis protein; regulation of iron absorption. |
| rs805303 | BAT2-BAT5 | 6p21.33 | Genes cluster localized near genes for TNF α and β, involved in inflammatory process and associated with insulin dependent diabetes and rheumatoid arthritis. |
| rs17477177 | PIK3CG | 7q22.3 | A catalytic subunit of PI3K, involved in the immune response. |
| rs3918226 | NOS3 | 7q36.1 | Endothelial nitric oxide synthase. |
| rs2898290 | BLK-GATA4 | 8p23.1 | BLK is a tyrosine kinase involved in cell proliferation and differentiation. GATA4 is a zinc finger transcription factor involved in embryogenesis and myocardial differentiation and function. |
| rs1799998 | CYP11B2 | 8q24.3 | Aldosterone synthase. |
| rs4373814 | CACNB2(5′) | 10p12.33 | Member of a voltage-gated calcium channel superfamily, associated with Brugada and Lambert-Eaton myasthenic syndrome. |
| rs1813353 | CACNB2(3′) | ||
| rs4590817 | C10orf107 | 10q21.2 | Chromosome 10 open reading frame 107. Unknown function. |
| rs932764 | PLCE1 | 10q23.33 | Phospholipase involved in Ras pathway, associated with early onset nephrotic syndrome. |
| rs11191548 | CYP17A1-NT5C2 | 10q24.32 | CYP17A1 is the 17α hydroxylase, involved in the steroidogenic pathway; mutated in congenital adrenal hyperplasia. NT5C2 is a hydrolase involved in purine nucleotides metabolism. |
| rs1801253 | ADRB1 | 10q25.3 | Adrenoreceptor β1, which mediate physiological effects of epinephrine and norepinephrine. |
| rs7129220 | ADM | 11p15.4 | Pre-hormone cleaved in adrenomedullin and pro-adrenomedullin, which act as vasodilator, hormone secretion regulators and angiogenesis promoters. |
| rs381815 | PLEKHA7 | 11p15.2 | Pleckstrin homology domain containing A7 associated with breast carcinomas and glaucoma. |
| rs633185 | FLJ32810-TMEM133 | 11q22.1 | FLJ32810 is a regulator of vascular tone. TMEM133 is a transmembrane protein; unknown function. |
| rs17249754 | ATP2B1 | 12q21.33 | Calcium ATPase with critical role in intracellular Ca2+ homeostasis |
| rs3184504 | SH2B3 | 12q24.12 | Signalling activities by growth factor and cytokine receptors; associated with celiac disease and insulin-dependent diabetes. |
| rs11066280 | ALDH2 | 12q24.12 | Mitochondrial aldehyde dehydrogenase 2, involved in the oxidative pathway of alcohol metabolism. |
| rs10850411 | TBX5-TBX3 | 12q24.21 | T-box protein family encoding for transcriptional factors regulating heart and limbs developmental processes. |
| rs1378942 | CYP1A1-ULK3 | 15q24.1 | CYP1A1 is a mono-oxygenases involved in drug catabolism and synthesis of cholesterol, steroid and other lipids. ULK3 is a serine/threonine kinase; unknown function. |
| rs2521501 | FURIN-FES | 15q26.1 | FURIN is a protease, involved in the catabolism of PTH, TGFβ1 and other growth factors. FES is a tyrosine kinase, involved in hematopoiesis and cytokine receptor signalling. |
| rs13333226 | UMOD | 16p12.3 | Regulation of renal sodium handling. See text. |
| rs17608766 | GOSR2 | 17q21.32 | Trafficking membrane protein. |
| rs12940887 | ZNF652 | 17q21.32 | Zinc finger protein, associated with breast and prostate cancer. |
| rs1327235 | JAG1 | 20p12.2 | Hematopoiesis regulation through notch 1 signalling. |
| rs6015450 | GNAS-EDN3 | 20q13.32 | GNAS is a G-protein that activates adenylyl cyclase with a wide variety of cellular responses. EDN3 encode for endothelin 3, implicated also in neural crest-derived cell lineages differentiation. |
The table reports the 43 SNPs associated with blood pressure and the nearest gene (when there were two flanking genes, we reported both). Only a minority of the reported SNPs are near a gene known to be related to BP. Modified from Ehret et al. [12].
Finally, only a minority of all the 43 described variants were near a gene known to be associated with BP levels [12]. The remaining SNPs were located in regions of the genome which were not thought to be related to HTN and were thus proposed as future sequencing targets to identify new biological pathways associated with pathophysiological alterations and potentially new therapeutic targets.
Taken together all GWAS-identified HTN-associated variants only explain 2–3% of BP variance [8,49], whereas the expected heritability varied between 30% and 50% [3,4]. However, studying a specific cohort under a specific environment overestimates the expected heritability and it is also likely that many more SNPs remain to be discovered [7].
4. Epigenetic Modifications in Hypertensive Patients
Epigenetics includes all heritable changes in gene expression regulation, which do not involve mutations in the DNA sequence [54]. Epigenetic modifications can be determined by many factors such as environmental influences during foetal life and childhood, exposure to chemical agents, aging, diet, medications and others [55]. Research in the field of epigenetic modifications provides insights on how the regulation of BP levels cannot be explained just by Mendelian inheritance [10,56] and could also justify a further part of the missing heritability, unexplained by GWAS. The processes that may cause epigenetic modifications of DNA include methylation, post-translational histone modification and mechanisms based on small non-coding RNAs [10] (see Table 3).
Table 3.
Epigenetic modifications in hypertension.
| Epigenetic Modification | Findings/Effectors | Reference |
|---|---|---|
| DNA methylation | Amount of 5mC in DNA inversely proportional to BP levels | Smolarek et al. [57] |
| HSD11B2 promoter methylation associated with HTN onset at young age | Friso et al. [58] | |
| RAA system gene hypomethylation associated with HTN in offspring, BP regulation and ACE inhibitor response. | Goyal et al. [59] Pei et al. [61] Rivière et al. [62] |
|
| Hypomethylation of SIC2A2 associated with NKCC1 overexpression, Na+ reabsorption and HTN. | Lee et al. [60] | |
| Histones acetylation/deacetylation | Acetylation/deacetylation of endothelial cells nucleosomes regulate eNOS expression | Fish et al. [63] |
| Nucleosomes modifications regulate ACE transcription | Lee et al. [64] | |
| Nucleosomes modifications regulate NKCC1 transcription and sodium renal reabsorption | Cho et al. [65] | |
| WNK4 down-regulation determines histones acetylation and NCC overexpression. | Mu et al. [66] | |
| Non coding RNA | miR-27a and -27b down-regulate ACE1 mRNA | Goyal et al. [59] |
| miR-155 down-regulate AGTR1 mRNA | Cheng et al. [67] | |
| miR-181a and -663 down-regulate renin mRNA and are under-expressed in HTN-patients | Marquez et al. [68] | |
| miR-425 down-regulate NNPA, leading to ANP under-production and salt overload | Arora et al. [69] |
5mC, 5-methil-cytosine. BP, blood pressure. HSD11B2, hydroxysteroid 11-β dehydrogenase 2. HTN, hypertension. RAA, renin-angiotensin-aldosterone system. ACE, angiotensin converting enzyme. NKCC1, Na-K-Cl co-transporter 1. eNOS, endothelial nitric oxide synthase. NCC, NaCl co-transporter. miR, microRNA. AGTR1, angiotensin II type 1 receptor. ANP, Atrial natriuretic peptide. Modified from Wise et al. [10].
DNA methylation is the transfer of a methyl group from S-adenosyl-l-methionine to the 5′-position of a cytosine ring, to form 5-methyl-cytosine (5mC). This occurs in specific genomic regions called CpGs islands, composed of a series of dinucleotides, guanine and cytosine. Of note, CpG islands are often located in promoter regions [10]. DNA methylation down-regulates gene transcription resulting in gene silencing [55]. In patients with HTN the total number of 5mC is decreased with increasing HTN severity [57]. Furthermore, HSD11B2 gene promoter methylation is associated with HTN onset [58], as well as hypomethylation of genes of the renin-angiotensin-aldosterone (RAA) system or NKCC1 (Na-K-Cl co-transporter 1) gene respectively affect the response to ACE (angiotensin-converting enzyme) inhibitors and diuretics [59,60].
Histones package DNA into structural units called nucleosomes. Post-translational DNA modifications include histone acetylation, or less frequently methylation and phosphorylation [70]. Histone acetylation promotes gene transcription, whereas deacetylation produces the opposite effect [71]. Acetylation or deacetylation of eNOS (endothelial nitric oxide synthase), RAA system genes, NKCC1, NCC or WNK4 were demonstrated to affect vascular tone, renal hydro-electrolyte homeostasis and therefore BP [63,64,65,66].
Finally, small non-coding RNAs are also implicated in epigenetic modifications. In particular microRNAs (miRNA/miR) may down-regulate genes by binding the corresponding mRNA, resulting in the degradation of the latter and repression of translation [72]. miRNA with a possible role in BP regulation are listed in Table 3 [59,67,68,73].
Through these mechanisms the environment could influence genotype in each individual and these findings complicate the field of HTN genetics and genomics, adding another factor able to modify the phenotypic expression of patients with HTN [10].
5. Pharmacogenomics
Theoretically genomics can also be applied to predict drug response. The goal of pharmacogenomics is to identify genetic variants associated with a better response to a pharmacological class over another, in the attempt to administer “the right dose of the right drug to the right patient” [74]. In Table 4 we report some significant studies published on how a patient's genotype can affect pharmaco-kinetics or -dynamics of a given drug.
Table 4.
Main genetic associations with response to anti-hypertensive drugs.
| Genetic Association | Effect | Reference |
|---|---|---|
| NEDD4L, PRKCA, GNAS-EDN3, TET2, CSMD1, HSD3B1 | Enhanced response to thiazide diuretics | Dahlberg et al. [85] Duarte et al. [86] Chittani et al. [87] Salvi et al. [88] |
| ADRB1 | Enhanced response to metoprolol | Ganesh et al. [89] |
| ADRB1 | Enhanced response to carvedilol | Mialet-Perez et al. [90] |
| ADRB1 | Greater mortality in patients treated with verapamil than with atenolol | Karlsson et al. [91] |
| CYP2D6 | Decrease in metoprolol clearance | Rau et al. [92] |
| GRK4 | Enhanced response to atenolol | Bhatnagar et al. [93] |
| SIGLEC12, A1BG, F5 | Higher risk of treatment-related adverse CV outcomes in patients treated with CCB | Hannila et al. [94] |
| CAMK1D | Enhanced response to losartan | Frau et al. [75] |
| ADD1 | Enhanced response to rostafuroxin | Staessen et al. [79] |
The table describes the effects of SNPs in genes indicated in the first column. CV, cardiovascular. CCB, calcium channel blockers. Modified from Cooper-DeHoff et al. [9].
An example of this type of study is the SOPHIA study: 372 patients with HTN treated in monotherapy with losartan were included in the analysis. Through a whole-genome approach, the authors identified a significant association between 4 SNPs on CAMK1D gene and BP response. CAMK1D encodes for a calcium-dependent kinase that modulates the transcription of factors NURR1, ARF1, ATF2 and CREB, involved in aldosterone production in the adrenal zona glomerulosa [75].
Pharmacogenomics may also be applied to the identification of new therapeutic targets in patients with HTN. GWAS on Milan hypertensive rats and humans revealed the ADD1 gene (encoding for α-adducin), as an eQTL associated to a significant increase in BP [76,77]. Adducin is a cytoskeletal protein consisting of two subunits: a α-subunit associated to either a β- or a γ-subunit. Mutated α-adducin stimulates the Na+/K+ ATPase in tubular renal cells, increasing Na+ reabsorption. Ouabain, a hormone released by hypothalamus and adrenal glands, is able to modulate Na+/K+ ATPase activity and natriuresis. Rostafuroxin antagonizes ouabain and lowers BP. Plasma ouabain levels are directly correlated with the number of copies of the mutated α-adducin gene [78]. The OASIS-HT [79] and the PEARL (ongoing) randomized controlled trials investigated the BP response to rostafuroxin through a pharmacogenomic approach.
Another example is uromodulin (UMOD) genes. In 2010, its minor allele was demonstrated to be associated with a lower risk of HTN [80] and reduced urinary uromodulin excretion [80]. UMOD encodes for a protein expressed in the kidney, exclusively in the thick ascending limb of the loop of Henle [81]. UMOD knockout models in vivo had lower SBP and were resistant to salt-induced changes of BP [82], whereas UMOD overexpression produced an increase in UMOD renal excretion and BP levels [83]. Furthermore, furosemide significantly increased natriuresis and decreased BP in the latter model compared to wild type controls [83]. These findings suggest that patients with increased urinary UMOD excretion may have an enhanced response to loop diuretics and that the uromodulin pathway may represent a new target for future drug development [81].
Although studies are promising with possible practical applications, these results should be interpreted with caution because their reproducibility was poor, suggesting that the observed associations may not be effective or that they are applicable only within specific sub-populations [84].
6. Conclusions
Genetic and genomics studies have provided major insights into understanding the pathogenesis of HTN. Identification of Mendelian syndromes allows an effective targeted therapy, however, they are relatively rare diseases with limited economic-health implications. On the other hand, essential HTN is very frequent but clinical applications of GWAS are undefined. The challenge lies in moving from the identification of SNPs associated with hypertension to the description of new pharmacologically-targetable causal molecular mechanisms. An example is the discovery of UMOD by GWAS studies and the subsequent description of a new potentially targetable pathway. Considering the complexity of BP regulation, only a minority of all pharmacologically targetable pathways are known. It is possible that further technological innovations will provide tools able to unequivocally define the genetic architecture of HTN and provide new therapeutic options for hypertensive patients. A future perspective could be the definition of scores on HTN-related SNPs to predict which patients will develop HTN and to which therapy they will respond better.
Acknowledgments
No sources of funding were used for this study.
Abbreviations
Hypertension (HTN), Blood Pressure (BP), Genome-Wide Association Studies (GWAS), Single Nucleotide Polymorphisms (SNPs), Glucocorticoid Remediable Aldosteronism (GRA), Familial Hyperaldosteronism (FH), Primary Aldosteronism (PA), Primary Aldosteronism Seizures and Neurologic Abnormalities (PASNA), Apparent Mineralocorticoid Excess (AME), Mineralocorticoid Receptor (MR), 11β-Hydroxysteroid Dehydrogenase type 2 (HSD11B2), Sodium-Chloride co-transporter (NCC), Epithelial Sodium Channel (ENaC), Congenital Adrenal Hyperplasia (CAH), expression Quantitative Trait Loci (eQTL), Systolic Blood Pressure (SBP), Diastolic Blood Pressure (DBP), Minor Allele Frequency (MAF), Renin-Angiotensin-Aldosterone (RAA) system, Na-K-Cl Co-transporter 1 (NKCC1), Angiotensin-Converting Enzyme (ACE), endothelial Nitric Oxide Synthase (eNOS), microRNA (miRNA/miR), Uromodulin (UMOD).
Author Contributions
All the authors contributed extensively to the work presented in this manuscript. Jacopo Burrello, Martina Tetti and Fabrizio Buffolo performed the literature search and wrote the manuscript. Tracy A. Williams and Paolo Mulatero conceived the work and critically revised it. Silvia Monticone and Franco Veglio revised the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Piepoli M.F., Hoes A.W., Agewall S., Albus C., Brotons C., Catapano A.L., Cooney M.T., Corrà U., Cosyns B., Deaton C., et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice. Developed with the special contribution of the European Association for Cardiovascular Prevention. Eur. Heart J. 2016;37:2315–2381. doi: 10.1093/eurheartj/ehw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mozaffarian D., Benjamin E.J., Go A.S., Arnett D.K., Blaha M.J., Cushman M., de Ferranti S., Després J.P., Fullerton H.J., Howard V.J., et al. Heart disease and stroke statistics—2015 Update: A report from the American Heart Association. Circulation. 2015;131:e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 3.Hottenga J.J., Boomsma D.I., Kupper N., Posthuma D., Snieder H., Willemsen G., de Geus E.J. Heritability and stability of resting blood pressure. Twin. Res. Hum. Genet. 2005;8:499–508. doi: 10.1375/twin.8.5.499. [DOI] [PubMed] [Google Scholar]
- 4.Miall W.E., Oldham P. The hereditary factor in arterial blood-pressure. Br. Med. J. 1963;1:75–80. doi: 10.1136/bmj.1.5323.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown M.J. The causes of essential hypertension. Br. J. Clin. Pharmacol. 1996;42:21–27. doi: 10.1046/j.1365-2125.1996.03785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams R.R., Hunt S.C., Hopkins P.N., Wu L.L., Hasstedt S.J., Berry T.D., Barlow G.K., Stults B.M., Schumacher M.C., Ludwig E.H., et al. Genetic basis of familial dyslipidemia and hypertension: 15-year results from Utah. Am. J. Hypertens. 1993;6:319–327. doi: 10.1093/ajh/6.11.319S. [DOI] [PubMed] [Google Scholar]
- 7.Padmanabhan S., Caulfield M., Dominiczak A.F. Genetic and molecular aspects of hypertension. Circ. Res. 2015;116:937–959. doi: 10.1161/CIRCRESAHA.116.303647. [DOI] [PubMed] [Google Scholar]
- 8.Munroe P.B., Barnes M.R., Caulfield M.J. Advances in blood pressure genomics. Circ. Res. 2013;112:1365–1379. doi: 10.1161/CIRCRESAHA.112.300387. [DOI] [PubMed] [Google Scholar]
- 9.Cooper-DeHoff R.M., Johnson J.A. Hypertension pharmacogenomics: In search of personalized treatment approaches. Nat. Rev. Nephrol. 2016;12:110–122. doi: 10.1038/nrneph.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wise I.A., Charchar F.J. Epigenetic Modifications in Essential Hypertension. Int. J. Mol. Sci. 2016;17:451. doi: 10.3390/ijms17040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lifton R.P., Gharavi A.G., Geller D.S. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/S0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 12.Ehret G.B., Caulfield M.J. Genes for blood pressure: An opportunity to understand hypertension. Eur. Heart J. 2013;34:951–961. doi: 10.1093/eurheartj/ehs455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lifton R.P., Dluhy R.G., Powers M., Rich G.M., Cook S., Ulick S., Lalouel J.M. A chimaeric 11β-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992;355:262–265. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- 14.Stowasser M. Update in primary aldosteronism. J. Clin. Endocrinol. Metab. 2015;100:1–10. doi: 10.1210/jc.2014-3663. [DOI] [PubMed] [Google Scholar]
- 15.Choi M., Scholl U.I., Yue P., Björklund P., Zhao B., Nelson-Williams C. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331:768–772. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholl U.I., Stölting G., Nelson-Williams C., Vichot A.A., Choi M., Loring E., Prasad M.L., Goh G., Carling T., Juhlin C.C., et al. Recurrent gain of function mutation in calcium channel CACNA1H causes early-onset hypertension with primary aldosteronism. eLife. 2015;4:e06315. doi: 10.7554/eLife.06315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniil G., Fernandes-Rosa F.L., Chemin J., Blesneac I., Beltrand J., Polak M. CACNA1H Mutations Are Associated With Different Forms of Primary Aldosteronism. EBioMedicine. 2016;13:225–236. doi: 10.1016/j.ebiom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandes-Rosa F.L., Williams T.A., Riester A., Steichen O., Beuschlein F., Boulkroun S. Genetic spectrum and clinical correlates of somatic mutations in aldosterone-producing adenoma. Hypertension. 2014;64:354–361. doi: 10.1161/HYPERTENSIONAHA.114.03419. [DOI] [PubMed] [Google Scholar]
- 19.Monticone S., Else T., Mulatero P., Williams T.A., Rainey W.E. Understanding primary aldosteronism: Impact of next generation sequencing and expression profiling. Mol. Cell. Endocrinol. 2015;399:311–320. doi: 10.1016/j.mce.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholl U.I., Goh G., Stolting G., de Oliveira R.C., Choi M., Overton J.D., Fonseca A.L., Korah R., Starker L.F., Kunstman J.W., et al. Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat. Genet. 2013;45:1050–1054. doi: 10.1038/ng.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulick S., Levine L.S., Gunczler P., Zanconato G., Ramirez L.C., Rauh W. A syndrome of apparent mineralocorticoid excess associated with defects in the peripheral metabolism of cortisol. J. Clin. Endocrinol. Metab. 1979;49:757–764. doi: 10.1210/jcem-49-5-757. [DOI] [PubMed] [Google Scholar]
- 22.Funder J.W. Apparent mineralocorticoid excess. J. Steroid. Biochem. Mol. Biol. 2017;65:151–153. doi: 10.1016/j.jsbmb.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Arnold J.E., Healy J.K. Hyperkalemia, hypertension and systemic acidosis without renal failure associated with a tubular defect in potassium excretion. Am. J. Med. 1969;47:461–472. doi: 10.1016/0002-9343(69)90230-7. [DOI] [PubMed] [Google Scholar]
- 24.Stowasser M., Gordon R.D. Primary Aldosteronism: Changing Definitions and New Concepts of Physiology and Pathophysiology Both Inside and Outside the Kidney. Physiol. Rev. 2016;96:1327–1384. doi: 10.1152/physrev.00026.2015. [DOI] [PubMed] [Google Scholar]
- 25.Wilson F.H., Disse-Nicodème S., Choate K.A., Ishikawa K., Nelson-Williams C., Desitter I., Gunel M., Milford D.V., Lipkin G.W., Achard J.M., et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 26.Boyden L.M., Choi M., Choate K.A., Nelson-Williams C.J., Farhi A., Toka H.R., Tikhonova I.R., Bjornson R., Mane S.M., Colussi G., et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482:98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathare G., Hoenderop J.G., Bindels R.J., San-Cristobal P. A molecular update on pseudohypoaldosteronism type II. Am. J. Physiol. Renal. Physiol. 2013;305:1513–1520. doi: 10.1152/ajprenal.00440.2013. [DOI] [PubMed] [Google Scholar]
- 28.Rashmi P., Colussi G., Ng M., Wu X., Kidwai A., Pearce D. Glucocorticoid-induced leucine zipper protein regulates sodium and potassium balance in the distal nephron. Kidney Int. 2017;91:1159–1177. doi: 10.1016/j.kint.2016.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snyder P.M. The epithelial Na+ channel: Cell surface insertion and retrieval in Na+ homeostasis and hypertension. Endocr. Rev. 2002;23:258–275. doi: 10.1210/edrv.23.2.0458. [DOI] [PubMed] [Google Scholar]
- 30.Hansson J.H., Nelson-Williams C., Suzuki H., Schild L., Shimkets R., Lu Y. Hypertension caused by a truncated epithelial sodium channel gamma subunit: Genetic heterogeneity of Liddle syndrome. Nat. Genet. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- 31.Shimkets R.A., Warnock D.G., Bositis C.M., Nelson-Williams C., Hansson J.H., Schambelan M. Liddle’s syndrome: Heritable human hypertension caused by mutations in the β subunit of the epithelial sodium channel. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-X. [DOI] [PubMed] [Google Scholar]
- 32.Turcu A.F., Auchus R.J. Adrenal steroidogenesis and congenital adrenal hyperplasia. Endocrinol. Metab. Clin. N. Am. 2015;44:275–296. doi: 10.1016/j.ecl.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biglieri E.G., Kater C.E. Mineralocorticoids in congenital adrenal hyperplasia. J. Steroid. Biochem. Mol. Biol. 1991;40:493–499. doi: 10.1016/0960-0760(91)90268-A. [DOI] [PubMed] [Google Scholar]
- 34.Portrat S., Mulatero P., Curnow K.M., Chaussain J.L., Morel Y., Pascoe L. Deletion hybrid genes, due to unequal crossing over between CYP11B1 (11β-hydroxylase) and CYP11B2 (aldosterone synthase) cause steroid 11β -hydroxylase deficiency and congenital adrenal hyperplasia. J. Clin. Endocrinol. Metab. 2001;86:3197–3201. doi: 10.1210/jcem.86.7.7671. [DOI] [PubMed] [Google Scholar]
- 35.Schuster H., Wienker T.E., Bähring S., Bilginturan N., Toka H.R., Neitzel H. Severe autosomal dominant hypertension and brachydactyly in a unique Turkish kindred maps to human chromosome 12. Nat. Genet. 1996;13:98–100. doi: 10.1038/ng0596-98. [DOI] [PubMed] [Google Scholar]
- 36.Maass P.G., Aydin A., Luft F.C., Schächterle C., Weise A., Stricker S. PDE3A mutations cause autosomal dominant hypertension with brachydactyly. Nat. Genet. 2015;47:647–653. doi: 10.1038/ng.3302. [DOI] [PubMed] [Google Scholar]
- 37.Toka O., Tank J., Schächterle C., Aydin A., Maass P.G., Elitok S. Clinical effects of phosphodiesterase 3A mutations in inherited hypertension with brachydactyly. Hypertension. 2015;66:800–808. doi: 10.1161/HYPERTENSIONAHA.115.06000. [DOI] [PubMed] [Google Scholar]
- 38.Geller D.S., Farhi A., Pinkerton N., Fradley M., Moritz M., Spitzer A., Meinke G., Tsai F.T., Sigler P.B., Lifton R.P. Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science. 2000;289:119–123. doi: 10.1126/science.289.5476.119. [DOI] [PubMed] [Google Scholar]
- 39.Rafestin-Oblin M.E., Souque A., Bocchi B., Pinon G., Fagart J., Vandewalle A. The severe form of hypertension caused by the activating S810L mutation in the mineralocorticoid receptor is cortisone related. Endocrinology. 2003;144:528–533. doi: 10.1210/en.2002-220708. [DOI] [PubMed] [Google Scholar]
- 40.Opocher G., Schiavi F. Genetics of pheochromocytomas and paragangliomas. Best Pract. Res. Clin. Endocrinol. Metab. 2010;24:943–956. doi: 10.1016/j.beem.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Welander J., Söderkvist P., Gimm O. Genetics and clinical characteristics of hereditary pheochromocytomas and paragangliomas. Endocr. Relat. Cancer. 2011;18:253–276. doi: 10.1530/ERC-11-0170. [DOI] [PubMed] [Google Scholar]
- 42.Sachidanandam R., Weissman D., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 43.Altshuler D., Daly M.J., Lander E.S. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munroe P.B., Wallace C., Xue M.Z., Marcano A.C., Dobson R.J., Onipinla A.K., Burke B., Gungadoo J., Newhouse S.J., Pembroke J., et al. Increased support for linkage of a novel locus on chromosome 5q13 for essential hypertension in the British Genetics of Hypertension Study. Hypertension. 2006;48:105–111. doi: 10.1161/01.HYP.0000228324.74255.f1. [DOI] [PubMed] [Google Scholar]
- 45.Chang Y.P., Liu X., Kim J.D., Ikeda M.A., Layton M.R., Weder A.B., Cooper R.S., Kardia S.L., Rao D.C., Hunt S.C., et al. Multiple genes for essential-hypertension susceptibility on chromosome 1q. Am. J. Hum. Genet. 2007;80:253–264. doi: 10.1086/510918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehret G.B., O’Connor A.A., Weder A., Cooper R.S., Chakravarti A. Follow-up of a major linkage peak on chromosome 1 reveals suggestive QTLs associated with essential hypertension: GenNet study. Eur. J. Hum. Genet. 2009;17:1650–1657. doi: 10.1038/ejhg.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Croteau-Chonka D.C., Rogers A.J., Raj T., McGeachie M.J., Qiu W., Ziniti J.P., Stubbs B.J., Liang L., Martinez F.D., Strunk R.C., et al. Expression Quantitative Trait Loci Information Improves Predictive Modelling of Disease Relevance of Non-Coding Genetic Variation. PLoS ONE. 2015;10:e0140758. doi: 10.1371/journal.pone.0140758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 49.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reich D., Lander S. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/S0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 52.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson T., Gaunt T.R., Newhouse S.J., Padmanabhan S., Tomaszewski M., Kumari M., Morris R.W., Tzoulaki I., O’Brien E.T., Poulter N.R., et al. Blood pressure loci identified with a gene-centric array. Am. J. Hum. Genet. 2011;89:688–700. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López-Jaramillo P., Camacho P.A., Forero-Naranjo L. The role of environment and epigenetics in hypertension. Expert Rev. Cardiovasc. Ther. 2013;11:1455–1457. doi: 10.1586/14779072.2013.846217. [DOI] [PubMed] [Google Scholar]
- 55.Raftopoulos L., Katsi V., Makris T., Tousoulis D., Stefanadis C., Kallikazaros I. Epigenetics, the missing link in hypertension. Life Sci. 2015;129:22–26. doi: 10.1016/j.lfs.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Loscalzo J., Handy D.E. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease (2013 grover conference series) Pulm. Circ. 2014;4:169–174. doi: 10.1086/675979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smolarek I., Wyszko E., Barciszewska A.M., Nowak S., Gawronska I., Jablecka A., Barciszewska M.Z. Global DNA methylation changes in blood of patients with essential hypertension. Med. Sci. Monit. Basic. Res. 2010;16:149–155. [PubMed] [Google Scholar]
- 58.Friso S., Pizzolo F., Choi S.-W., Guarini P., Castagna A., Ravagnani V., Carletto A., Pattini P., Corrocher R., Olivieri O., et al. Epigenetic control of 11 β-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis. 2008;199:323–327. doi: 10.1016/j.atherosclerosis.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 59.Goyal R., Goyal D., Leitzke A., Gheorghe C.P., Longo L.D. Brain renin-angiotensin system: Fetal epigenetic programming by maternal protein restriction during pregnancy. Reprod. Sci. 2010;17:227–238. doi: 10.1177/1933719109351935. [DOI] [PubMed] [Google Scholar]
- 60.Lee H.A., Baek I., Seok Y.M., Yang E., Cho H.M., Lee D.Y., Hong S.H., Kim I.K. Promoter hypomethylation upregulates Na+-K+-2Cl− cotransporter 1 in spontaneously hypertensive rats. Biochem. Biophys. Res. Commun. 2010;396:252–257. doi: 10.1016/j.bbrc.2010.04.074. [DOI] [PubMed] [Google Scholar]
- 61.Pei F., Wang X., Yue R., Chen C., Huang J., Huang J., Li X., Zeng C. Differential expression and DNA methylation of angiotensin type 1A receptors in vascular tissues during genetic hypertension development. Mol. Cell. Biochem. 2015;402:1–8. doi: 10.1007/s11010-014-2295-9. [DOI] [PubMed] [Google Scholar]
- 62.Rivière G., Lienhard D., Andrieu T., Vieau D., Frey B.M., Frey F.J. Epigenetic regulation of somatic angiotensin-converting enzyme by DNA methylation and histone acetylation. Epigenetics. 2011;6:478–489. doi: 10.4161/epi.6.4.14961. [DOI] [PubMed] [Google Scholar]
- 63.Fish J.E., Matouk C.C., Rachlis A., Lin S., Tai S.C., D’Abreo C., Marsden P.A. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J. Biol. Chem. 2005;280:4824–4838. doi: 10.1074/jbc.M502115200. [DOI] [PubMed] [Google Scholar]
- 64.Lee H.A., Cho H.M., Lee D.Y., Kim K.C., Han H.S., Kim I.K. Tissue-specific upregulation of angiotensin-converting enzyme 1 in spontaneously hypertensive rats through histone code modifications. Hypertension. 2012;59:621–626. doi: 10.1161/HYPERTENSIONAHA.111.182428. [DOI] [PubMed] [Google Scholar]
- 65.Cho H.M., Lee D.Y., Kim H.Y., Lee H.A., Seok Y.M., Kim I.K. Upregulation of the Na+-K+-2Cl− cotransporter 1 via histone modification in the aortas of angiotensin II-induced hypertensive rats. Hypertens. Res. 2012;35:819–824. doi: 10.1038/hr.2012.37. [DOI] [PubMed] [Google Scholar]
- 66.Mu S., Shimosawa T., Ogura S., Wang H., Uetake Y., Kawakami-Mori F., Marumo T., Yatomi Y., Geller D.S., Tanaka H., et al. Epigenetic modulation of the renal β-adrenergic-WNK4 pathway in salt-sensitive hypertension. Nat. Med. 2011;17:573–580. doi: 10.1038/nm.2337. [DOI] [PubMed] [Google Scholar]
- 67.Cheng W., Liu T., Jiang F., Liu C., Zhao X., Gao Y., Wang H., Liu Z. MicroRNA-155 regulates angiotensin II type 1 receptor expression in umbilical vein endothelial cells from severely pre-eclamptic pregnant women. Int. J. Mol. Med. 2011;27:393–399. doi: 10.3892/ijmm.2011.598. [DOI] [PubMed] [Google Scholar]
- 68.Marques F.Z., Campain A.E., Tomaszewski M., Zukowska-Szczechowska E., Yang Y.H.J., Charchar F.J., Morris B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension. 2011;58:1093–1098. doi: 10.1161/HYPERTENSIONAHA.111.180729. [DOI] [PubMed] [Google Scholar]
- 69.Arora P., Wu C., Khan A.M., Bloch D.B., Davis-Dusenbery B.N., Ghorbani A. Atrial natriuretic peptide is negatively regulated by microRNA-425. J. Clin. Investig. 2013;123:3378–3382. doi: 10.1172/JCI67383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 71.Millis R.M. Epigenetics and hypertension. Curr. Hypertens. Rep. 2011;13:21–28. doi: 10.1007/s11906-010-0173-8. [DOI] [PubMed] [Google Scholar]
- 72.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 73.Sethupathy P., Borel C., Gagnebin M., Grant G.R., Deutsch S., Elton T.S., Hatzigeorgiou A.G., Antonarakis S.E. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 31 untranslated region: A mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am. J. Hum. Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voora D., Ginsburg G.S. Clinical application of cardiovascular pharmacogenetics. J. Am. Coll. Cardiol. 2012;60:9–20. doi: 10.1016/j.jacc.2012.01.067. [DOI] [PubMed] [Google Scholar]
- 75.Frau F., Zaninello R., Salvi E., Ortu M.F., Braga D., Velayutham D. Genome-wide association study identifies CAMKID variants involved in blood pressure response to losartan: The SOPHIA study. Pharmacogenomics. 2014;15:1643–1652. doi: 10.2217/pgs.14.119. [DOI] [PubMed] [Google Scholar]
- 76.Zagato L., Modica R., Florio M., Bihoreau M.T., Bianchi G., Tripodi G. Genetic mapping of blood pressure quantitative trait loci in Milan hypertensive rats. Hypertension. 2000;36:734–739. doi: 10.1161/01.HYP.36.5.734. [DOI] [PubMed] [Google Scholar]
- 77.Bianchi G., Ferrari P., Staessen J.A. Adducin polymorphism: Detection and impact on hypertension and related disorders. Hypertension. 2005;45:331–340. doi: 10.1161/01.HYP.0000156497.39375.37. [DOI] [PubMed] [Google Scholar]
- 78.Staessen J.A., Bianchi G. Adducin and hypertension. Pharmacogenomics. 2005;6:665–669. doi: 10.2217/14622416.6.7.665. [DOI] [PubMed] [Google Scholar]
- 79.Staessen J.A., Thijs L., Stolarz-Skrzypek K., Bacchieri A., Barton J., Espositi E.D., de Leeuw P.W., Dłużniewski M., Glorioso N., Januszewicz A., et al. Main results of the ouabain and adducin for Specific Intervention on Sodium in Hypertension Trial (OASIS-HT): A randomized placebo-controlled phase-2 dose-finding study of rostafuroxin. Trials. 2011;12:13. doi: 10.1186/1745-6215-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Padmanabhan S., Melander O., Johnson T., Di Blasio A.M., Lee W.K., Gentilini D., Global BPgen Consortium Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Padmanabhan S., Graham L., Ferreri N.R., Graham D., McBride M., Dominiczak A.F. Uromodulin, an emerging novel pathway for blood pressure regulation and hypertension. Hypertension. 2014;64:918–923. doi: 10.1161/HYPERTENSIONAHA.114.03132. [DOI] [PubMed] [Google Scholar]
- 82.Graham L.A., Padmanabhan S., Fraser N.J., Kumar S., Bates J.M., Raffi H.S., Welsh P., Beattie W., Hao S., Leh S., et al. Validation of uromodulin as a candidate gene for human essential hypertension. Hypertension. 2014;63:551–558. doi: 10.1161/HYPERTENSIONAHA.113.01423. [DOI] [PubMed] [Google Scholar]
- 83.Trudu M., Janas S., Lanzani C., Debaix H., Schaeffer C., Ikehata M., Swiss Kidney Project on Genes in Hypertension (SKIPOGH) Team Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat. Med. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Menni C. Blood pressure pharmacogenomics: Gazing into a misty crystal ball. J. Hypertens. 2015;33:1142–1143. doi: 10.1097/HJH.0000000000000574. [DOI] [PubMed] [Google Scholar]
- 85.Dahlberg J., Nilsson L.O., von Wowern F., Melander O. Polymorphism in NEDD4L is associated with increased salt sensitivity, reduced levels of P-renin and increased levels of Nt-proANP. PLoS ONE. 2007;2:e432. doi: 10.1371/journal.pone.0000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duarte J.D., Turner S.T., Tran B., Chapman A.B., Bailey K.R., Gong Y., Gums J.G., Langaee T.Y., Beitelshees A.L., Cooper-Dehoff R.M., et al. Association of chromosome 12 locus with antihypertensive response to hydrochlorothiazide may involve differential YEATS4 expression. Pharmacogenom. J. 2013;13:257–263. doi: 10.1038/tpj.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chittani M., Zaninello R., Lanzani C., Frau F., Ortu M.F., Salvi E. TET2 and CSMD1 genes affect SBP response to hydrochlorothiazide in never-treated essential hypertensives. J. Hypertens. 2015;33:1301–1309. doi: 10.1097/HJH.0000000000000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Salvi E., Wang Z., Rizzi F., Gong Y., McDonough CW., Padmanabhan S., Hiltunen T.P., Lanzani C., Zaninello R., Chittani M., et al. Genome-Wide and Gene-Based Meta-Analyses Identify Novel Loci Influencing Blood Pressure Response to Hydrochlorothiazide. Hypertension. 2017;69:51–59. doi: 10.1161/HYPERTENSIONAHA.116.08267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ganesh S.K., Tragante V., Guo W., Guo Y., Lanktree M.B., Smith E.N., Johnson T., Castillo B.A., Barnard J., Baumert J., et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum. Mol. Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mialet Perez J., Rathz D.A., Petrashevskaya N.N., Hahn H.S., Wagoner L.E., Schwartz A. β 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat. Med. 2003;9:1300–1305. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- 91.Karlsson J., Lind L., Hallberg P., Michaëlsson K., Kurland L., Kahan T., Malmqvist K., Ohman K.P., Nyström F., Melhus H. β1-adrenergic receptor gene polymorphisms and response to β1-adrenergic receptor blockade in patients with essential hypertension. Clin. Cardiol. 2004;27:347–350. doi: 10.1002/clc.4960270610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rau T., Heide R., Bergmann K., Wuttke H., Werner U., Feifel N., Eschenhagen T. Effect of the CYP2D6 genotype on metoprolol metabolism persists during long-term treatment. Pharmacogenetics. 2002;12:465–472. doi: 10.1097/00008571-200208000-00007. [DOI] [PubMed] [Google Scholar]
- 93.Bhatnagar V., O’Connor D.T., Brophy V.H., Schork N.J., Richard E., Salem R.M., Nievergelt C.M., Bakris G.L., Middleton J.P., AASK Study Investigators et al. G-protein-coupled receptor kinase 4 polymorphisms and blood pressure response to metoprolol among African Americans: Sex-specificity and interactions. Am. J. Hypertens. 2009;22:332–338. doi: 10.1038/ajh.2008.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hannila-Handelberg T., Kontula K., Tikkanen I., Tikkanen T., Fyhrquist F., Helin K., Fodstad H., Piippo K., Miettinen H.E., Virtamo J., et al. Common variants of the β and gamma subunits of the epithelial sodium channel and their relation to plasma renin and aldosterone levels in essential hypertension. BMC Med. Genet. 2005;6:4. doi: 10.1186/1471-2350-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]