Abstract
Distinguishing between follicular thyroid cancer (FTC) and follicular thyroid adenoma (FTA) constitutes a long-standing diagnostic problem resulting in equivocal histopathological diagnoses. There is therefore a need for additional molecular markers. To identify molecular differences between FTC and FTA, we analyzed the gene expression microarray data of 52 follicular neoplasms. We also performed a meta-analysis involving 14 studies employing high throughput methods (365 follicular neoplasms analyzed). Based on these two analyses, we selected 18 genes differentially expressed between FTA and FTC. We validated them by quantitative real-time polymerase chain reaction (qRT-PCR) in an independent set of 71 follicular neoplasms from formaldehyde-fixed paraffin embedded (FFPE) tissue material. We confirmed differential expression for 7 genes (CPQ, PLVAP, TFF3, ACVRL1, ZFYVE21, FAM189A2, and CLEC3B). Finally, we created a classifier that distinguished between FTC and FTA with an accuracy of 78%, sensitivity of 76%, and specificity of 80%, based on the expression of 4 genes (CPQ, PLVAP, TFF3, ACVRL1). In our study, we have demonstrated that meta-analysis is a valuable method for selecting possible molecular markers. Based on our results, we conclude that there might exist a plausible limit of gene classifier accuracy of approximately 80%, when follicular tumors are discriminated based on formalin-fixed postoperative material.
Keywords: follicular thyroid adenoma, follicular thyroid cancer, gene expression, microarray, meta-analysis
1. Introduction
Follicular neoplasms are the most controversial area in the thyroid pathology. According to World Health Organization (WHO) follicular adenoma is a benign, encapsulated tumor of the thyroid showing follicular cell differentiation [1]. This tumor demonstrates no evidence of capsular or vascular invasion. Follicular carcinoma is a malignant tumor showing evidence of follicular cell differentiation. The distinction between follicular adenoma and carcinoma is based on the presence of capsular and/or vascular invasion. Capsular invasion is defined by tumor penetration through the entire thickness of the capsule [1]. The invading tumor nests should present a connection with main tumor mass. The interpretation of capsular invasion may be sometimes problematic. According to the literature data and our experience there is a group of patients with only partial capsular invasion but presenting metastases of follicular carcinoma [2]. Yamashina analyzed entire circumference of tumor capsules of follicular neoplasms and observed that tumors with only capsular invasion in initial sections also presented vascular invasion on additional slices adjacent to tumor capsule [3]. Therefore it would be advisable to evaluate gene expression of follicular adenomas and follicular carcinomas.
Between 2000 and 2014, numerous studies have investigated the gene expression (mRNA) profile that would differentiate follicular thyroid adenoma (FTA) from follicular thyroid cancer (FTC) to improve the diagnostic process and to find features of follicular thyroid tumours important for malignant potential (Table S1) [4,5,6,7,8,9,10,11,12,13,14,15,16,17]. However, reproducibility of results obtained between mentioned publications was rather low. This could be a consequence of slight molecular differences between FTC and FTA [18,19] or the insufficient sample size used in these studies. Genetic alterations, such as RAS gene family somatic mutations or PAX8/PPARG translocations, although very promising in initial studies, were not found to be specific for follicular carcinoma, as these genetic alterations occurred in both FTCs and FTAs with similar frequencies [20,21,22]. These doubts stimulated us to carry on a meta-analysis.
In our study, we also raised the problem of oncocytic tumors. WHO involves oncocytic thyroid carcinoma (OTC) to FTC and respectively oncocytic adenoma to FTA. Oncocytic tumors (Hurthle cell tumors) are believed to have a different gene expression profile [23,24]. Ganly et al. demonstrated on the basis of mutational, transcriptional, and copy number profiles that Hurthle cell carcinoma was a unique thyroid cancer distinct from papillary thyroid cancer (PTC) and FTC [24].
In the present study we decided to base on FTC definition, proposed by the WHO. Nevertheless, we tried to check whether an inclusion of oncocytic follicular carcinoma does not influence on molecular markers selection. OTC is composed predominantly of oncocytic cells. These tumors are associated with a higher frequency of extrathyroidal extension, local recurrence, nodal metastases in more than 30% of cases and occasionally distant lung and bone metastases [1]. Compared with conventional follicular carcinomas, oncocytic follicular carcinomas are more aggressive [1]. Therefore, it may be reasonable to involve oncocytic feature in our analysis.
Most recent thyroid studies have focused on identifying molecular markers supporting pre-operative FNAB examination to exclude malignancy [25,26]. In 2010, Chudova et al. published a study focused on determining the general preoperative distinction between benign and malignant thyroid nodules, which appeared promising and resulted in the establishment of the Afirma classifier [25]. Our approach, used in the present study, is different.
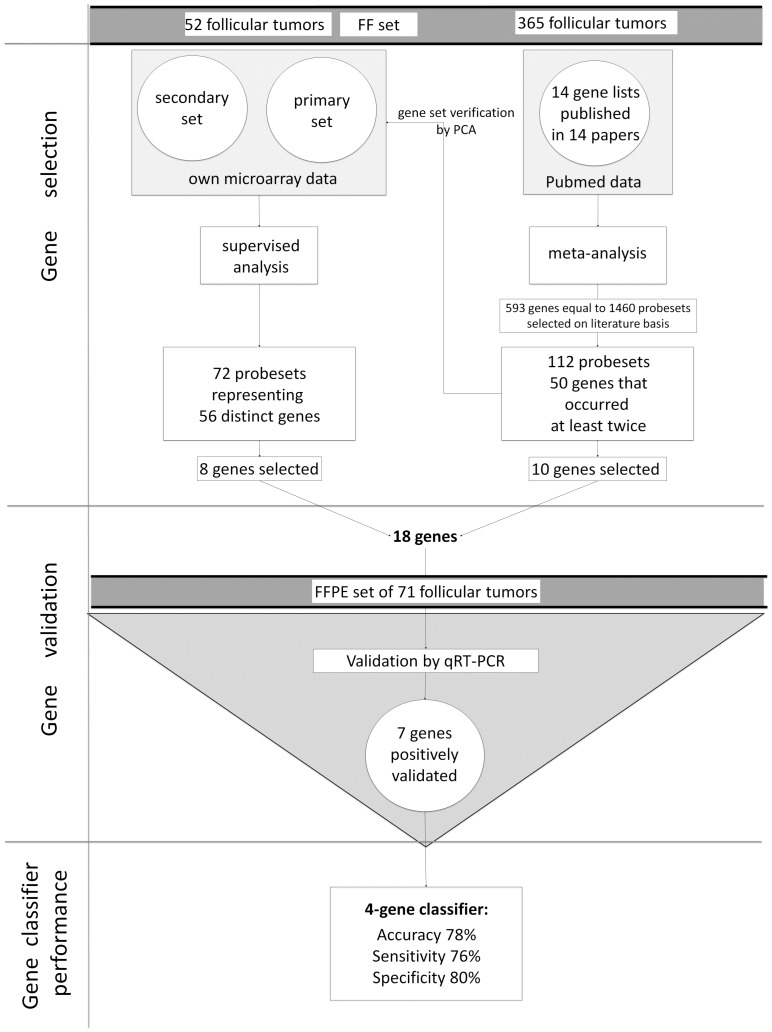
In our study, we utilised two different approaches to select new gene-expression markers for differentiating between FTC and FTA tumours. We performed a two-step analysis: first a statistical testing of a large gene expression microarray dataset of FTC and FTA previously generated in our laboratory [18,27], and next, a meta-analysis of all available datasets, to select the most robustly represented markers [4,5,6,7,8,9,10,11,12,13,14,15,16,17] (Figure 1). Such approach allowed us to select independent genes coming from own dataset and from a meta-analysis. Meta-analysis by combining the results of various studies enabled us to draw common conclusions. The results of both analyses were further validated by quantitative real-time polymerase chain reaction (qRT-PCR) using an independent dataset of follicular tumours.
Figure 1.
Presentation of a study scheme.
2. Results
2.1. Supervised Analysis of Gene Expression Microarrays
Fresh-frozen (FF) material from 52 tumors (27 FTC, 25 FTA) was used for our gene expression microarray experiment and divided into primary and secondary sets. The primary one was considered as highly reliable dataset and contained all samples that were independently and concordantly diagnosed by two thyroid pathology experts. The secondary set contained samples that were diagnosed by only one expert, equivocal samples diagnosed by two experts and a one sample that was discordantly diagnosed according to malignancy.
To select potential molecular markers useful in the distinction between FTA and FTC, we considered genes that were differentially expressed in the primary and secondary microarray datasets. We compared the lists of genes obtained in the analysis of the primary and the secondary sets and selected only those that were significant in both sets. Our secondary microarray set contained borderline and ambiguous cases, and we established genes as valuable and characteristic when they were also differentially expressed in this set.
There were 72 differentially expressed probe sets (representing 56 distinct genes) and 6 non-annotated probe sets. Eight genes were selected (ACVRL1, CLEC3B, DIP2B, GABARAPL2, ZFYVE21, LIMK2, ZMYND11, and MAFB) for validation by qRT-PCR (Table 1). Those genes were characterised by low false discovery rate (FDR) value, high fold-change, and from our point of view, they could be biologically interesting. Another selection criterion was that these genes were not previously validated as markers differentiating FTCs from FTAs.
Table 1.
Differentially expressed genes selected based on analysis of our own microarray dataset.
| No. | Gene Symbol | Gene Name | Affy ID | Primary Dataset | Primary Dataset—Evaluation of Non-Oncocytic Samples Only | |||
|---|---|---|---|---|---|---|---|---|
| FDR Corrected p-Value | Mean Expression in FTC | Mean Expression in FTA | Fold-Change | FDR Corrected p-Value | ||||
| 1 | ACVRL1 | activin A receptor type II-like 1 | 226950_at | 0.07 | 5.52 | 7.02 | 0.35 | 0.12 |
| 2 | CLEC3B | C-type lectin domain family 3, member B | 205200_at | 0.08 | 7.54 | 9.52 | 0.25 | 0.13 |
| 3 | GABARAPL2 | GABA(A) receptor-associated protein-like 2 | 209046_s_at | 0.08 | 11.05 | 11.84 | 0.58 | 0.15 |
| 4 | ZFYVE21 | zinc finger, FYVE domain containing 21 | 219929_s_at | 0.07 | 7.39 | 8.67 | 0.41 | 0.04 |
| 5 | LIMK2 | LIM domain kinase 2 | 217475_s_at | 0.07 | 4.32 | 5.84 | 0.35 | 0.12 |
| 6 | ZMYND11 | zinc finger, MYND domain containing 11 | 1554159_a_at | 0.10 | 6.60 | 8.05 | 0.37 | 0.15 |
| 7 | DIP2B | DIP2 disco-interacting protein 2 homolog B (Drosophila) | 224872_at | 0.11 | 8.23 | 7.40 | 1.78 | 0.16 |
| 8 | MAFB | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) | 222670_s_at | 0.08 | 8.23 | 9.78 | 0.34 | 0.13 |
The genes were selected for validation from the genes differentially expressed both in primary and secondary microarray set. Values represented in the table are from analysis of the primary microarray data set.
As it has been shown that the oncocytic FTC is a unique thyroid cancer distinct from non-oncocytic FTC [24] we decided to perform an additional analysis. We excluded oncocytic samples from microarray dataset (just for the sake of this particular analysis) and evaluated the significance of eight selected genes in the dataset comprising of non-oncocytic samples only to investigate the differences between FTC and FTA (7 FTC and 11 FTA). All these genes showed significant differential expression between FTC and FTA in this dataset (Table 1).
2.2. Meta-Analysis
We included 14 papers in which the difference in gene expression between FTC and FTA was assessed by a high throughput method (expression microarrays, serial analysis of gene expression (SAGE), high-throughput differential screening by serial analysis of gene expression (HDSS), adapter-tagged competitive polymerase chain reaction (ATAC-PCR)) (Table S1). The papers were published during the years 2000–2014 and in total 365 samples (201 FTA and 164 FTC) were analyzed.
All reported genes differentiating FTC and FTA were extracted from these publications. We identified 600 genes reported in at least one publication, while 57 genes were reported in more than one publication. Fifty out of those 57 genes were reported with concordant direction of change (Table 2). Seven genes (CA4, EGR2, FAM189A2, KCNAB1, CPQ, SLC26A4, TFF3) were reported in 3 publications. Two of these genes (CA4, and KCNAB1) were already evaluated by qRT-PCR as described in our previous study [27].
Table 2.
The results of a meta-analysis of 14 papers, in which differences in gene expression profile between follicular thyroid cancers (FTC) and follicular thyroid adenomas (FTA) were assessed by a high throughput method. Ten genes (highlighted in bold) were selected for our qRT-PCR validation.
| No. | Entrez Gene ID | Symbol | Name | Number of Papers | References | Gene Regulation |
|---|---|---|---|---|---|---|
| 1 | 762 | CA4 | carbonic anhydrase IV | 3 | [5,9,16] | down |
| 2 | 1959 | EGR2 | early growth response 2 | 3 | [5,14,16] | down |
| 3 | 9413 | FAM189A2 | family with sequence similarity 189, member A2 | 3 | [5,9,12] | down |
| 4 | 7881 | KCNAB1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | 3 | [6,9,16] Confirmed by us [27] |
down |
| 5 | 10404 | CPQ | carboxypeptidase Q | 3 | [9,11,14] | down |
| 6 | 5172 | SLC26A4 | solute carrier family 26 (anion exchanger), member 4 | 3 | [6,14,16] | down |
| 7 | 7033 | TFF3 | trefoil factor 3 (intestinal) | 3 | [5,6,10] | down |
| 8 | 185 | AGTR1 | angiotensin II receptor, type 1 | 2 | [13,16] | down |
| 9 | 822 | CAPG | capping protein (actin filament), gelsolin-like | 2 | [14,17] | down |
| 10 | 1306 | COL15A1 | collagen, type XV, alpha 1 | 2 | [5,13] | down |
| 11 | 1363 | CPE | carboxypeptidase E | 2 | [9,17] | down |
| 12 | 3491 | CYR61 | cysteine-rich, angiogenic inducer, 61 | 2 | [8,16] | down |
| 13 | 1733 | DIO1 | deiodinase, iodothyronine, type I | 2 | [6,12] | down |
| 14 | 11072 | DUSP14 | dual specificity phosphatase 14 | 2 | [5,16] | down |
| 15 | 129080 | EMID1 | EMI domain containing 1 | 2 | [5,7] | down |
| 16 | 953 | ENTPD1 | ectonucleoside triphosphate diphosphohydrolase 1 | 2 | [9,14] | down |
| 17 | 8857 | FCGBP | Fc fragment of IgG binding protein | 2 | [5,17] | down |
| 18 | 2354 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 2 | [16,17] | down |
| 19 | 2697 | GJA1 | gap junction protein, alpha 1, 43 kDa | 2 | [5,11] | down |
| 20 | 55830 | GLT8D1 | glycosyltransferase 8 domain containing 1 | 2 | [5,11] | down |
| 21 | 221395 | GPR116 | G protein-coupled receptor 116 | 2 | [5,9] | down |
| 22 | 3043 | HBB | hemoglobin, beta | 2 | [12,15] | down |
| 23 | 3309 | HSPA5 | heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | 2 | [9,17] | down |
| 24 | 3400 | ID4 | inhibitor of DNA binding 4, dominant negative helix-loop-helix protein | 2 | [5,8] | down |
| 25 | 3590 | IL11RA | interleukin 11 receptor, alpha | 2 | [5,11] | down |
| 26 | 9452 | ITM2A | integral membrane protein 2A | 2 | [9,16] | down |
| 27 | 3708 | ITPR1 | inositol 1,4,5-trisphosphate receptor, type 1 | 2 | [5,11] | down |
| 28 | 3725 | JUN | jun proto-oncogene | 2 | [5,16] | down |
| 29 | 3912 | LAMB1 | laminin, beta 1 | 2 | [5,11] | down |
| 30 | 744 | MPPED2 | metallophosphoesterase domain containing 2 | 2 | [16,17] | down |
| 31 | 22795 | NID2 | nidogen 2 (osteonidogen) | 2 | [5,7] | down |
| 32 | 3164 | NR4A1 | nuclear receptor subfamily 4, group A, member 1 | 2 | [12,16] | down |
| 33 | 22925 | PLA2R1 | phospholipase A2 receptor 1, 180 kDa | 2 | [12,16] | down |
| 34 | 83483 | PLVAP | plasmalemma vesicle associated protein | 2 | [9,13] | down |
| 35 | 5583 | PRKCH | protein kinase C, eta | 2 | [9,14] | down |
| 36 | 23180 | RFTN1 | raftlin, lipid raft linker 1 | 2 | [5,9] | down |
| 37 | 8490 | RGS5 | regulator of G-protein signaling 5 | 2 | [9,13] | down |
| 38 | 6414 | SEPP1 | selenoprotein P, plasma, 1 | 2 | [5,14] | down |
| 39 | 7038 | TG | Thyroglobulin | 2 | [10,17] | down |
| 40 | 4982 | TNFRSF11B | tumor necrosis factor receptor superfamily, member 11b | 2 | [5,11] | down |
| 41 | 7173 | TPO | thyroid peroxidase | 2 | [10,17] | down |
| 42 | 440 | ASNS | asparagine synthetase (glutamine-hydrolyzing) | 2 | [5,9] | up |
| 43 | 771 | CA12 | carbonic anhydrase XII | 2 | [5,12] | up |
| 44 | 1164 | CKS2 | CDC28 protein kinase regulatory subunit 2 | 2 | [16,17] | up |
| 45 | 1649 | DDIT3 | DNA-damage-inducible transcript 3 | 2 | [5,7] | up |
| 46 | 2358 | FPR2 | formyl peptide receptor 2 | 2 | [5,11] | up |
| 47 | 9518 | GDF15 | growth differentiation factor 15 | 2 | [9,17] | up |
| 48 | 2896 | GRN | Granulin | 2 | [4,8] | up |
| 49 | 3486 | IGFBP3 | insulin-like growth factor binding protein 3 | 2 | [5,10] | up |
| 50 | 23089 | PEG10 | paternally expressed 10 | 2 | [5,11] | up |
Table 2 shows the Entrez ID, gene symbol, gene name, number of papers in which a particular gene occurs, references to the papers, regulation direction (up–up-regulated in FTC; down–down-regulated in FTC).
Among the genes selected based on the meta-analysis, ten genes were chosen for qRT-PCR validation. We chose five down-regulated genes that occurred in three papers (EGR2, FAM189A2, SLC26A4, TFF3, CPQ), four up-regulated genes that occurred in two papers (CKS2, GDF15, ASNS, DDIT3), and one down-regulated gene that occurred in two papers and simultaneously showed significant differences in expression in our primary microarray dataset (PLVAP).
2.3. Principal Component Analysis
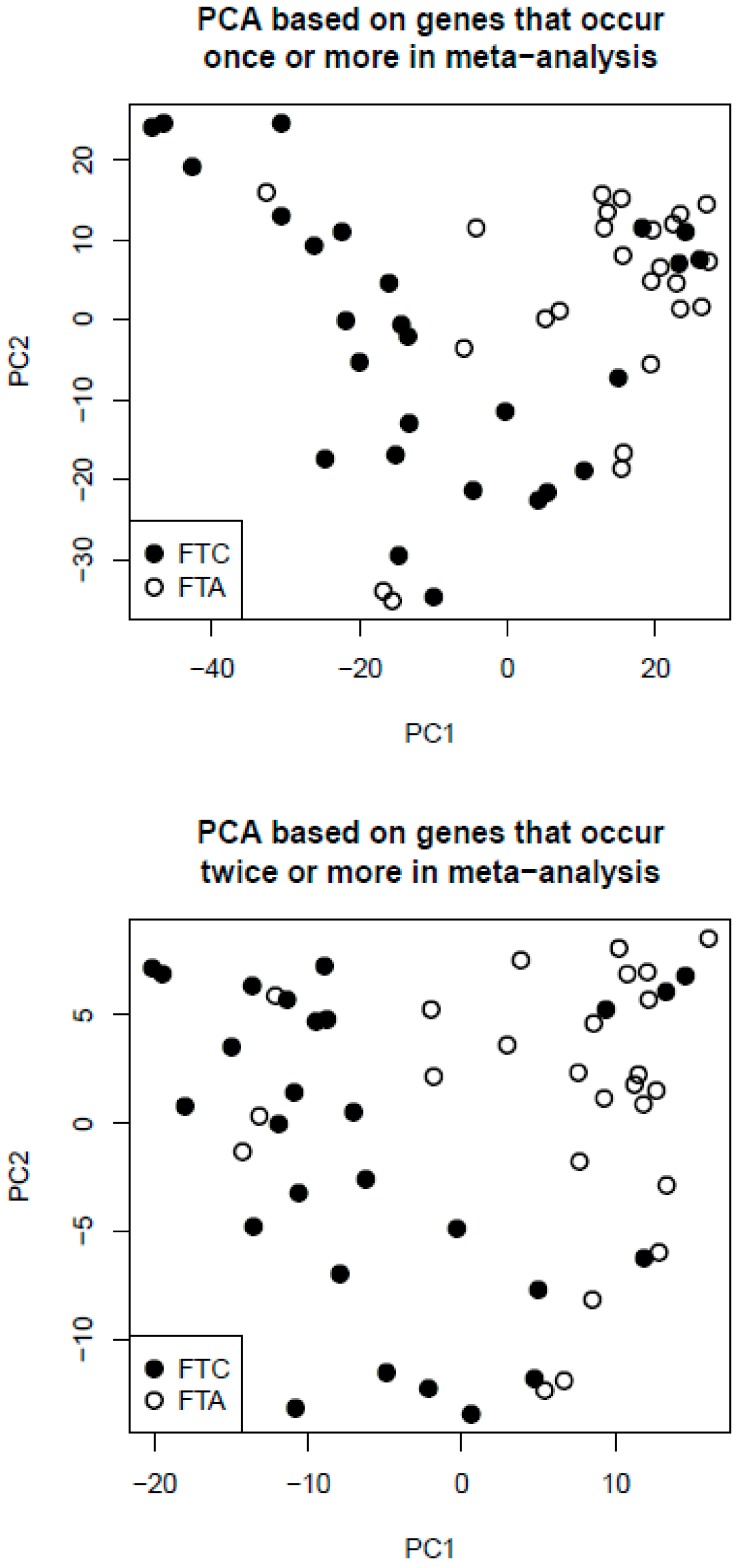
We selected 593 genes that occurred at least once in the meta-analysis (excluding seven genes with discordant direction of change). We identified HG-U133 PLUS 2 Affymetrix microarray probe sets for these genes. There were 1460 such probe sets (for some genes there was more than one probe set). Next, we performed PCA of our own microarray samples (combined primary and secondary dataset) based on these 1460 probe sets (Figure 2, upper plot). Similarly, we selected 50 genes that occurred at least twice in investigated papers (excluding the genes with discordant direction of change). We identified HG-U133 PLUS 2 Affymetrix microarray probe sets for these genes. There were 112 such probe sets. We performed PCA based on these 112 probe sets (Figure 2, lower plot). Although gene selection was independent of the microarray dataset, we achieved good discrimination of benign and malignant tumors in both analyses. However, the discrimination was not perfect, because a few FTA samples clustered with the FTC group, and a few FTC samples clustered with the FTA group.
Figure 2.
Principal component analysis (PCA) results. PCA plots of samples from our own microarray dataset, based on genes selected in the meta-analysis that occurred in at least one paper (upper plot) or at least two papers (lower plot).
2.4. qRT-PCR Validation
qRT-PCR was used to validate 18 genes selected based on the analysis of our own microarray dataset and the meta-analysis (Table 3). GABARAPL2, DDIT3, and SLC26A4 amplification was not possible in the FFPE samples (probably due to low endogenous expression), and therefore, it was excluded from validation.
Table 3.
Comparison of gene expression between FTC (29 samples) and FTA (40 samples) in qRT-PCR dataset (t-test and two-way ANOVA calculated p-values corrected for multiple tests by FDR method). FDR corrected p-values below 0.05 are highlighted in bold.
| No. | Gene | Gene Selection | t-Test—FDR Corrected p-Value | Fold Change (FTC/FTA) | Two-Way ANOVA—FDR Corrected p-Value |
|---|---|---|---|---|---|
| 1 | ACVRL1 | Microarrays | 0.0017 | 0.58 | 0.0036 |
| 2 | ZFYVE21 | Microarrays | 0.0024 | 0.69 | 0.0036 |
| 3 | CLEC3B | Microarrays | 0.027 | 0.75 | 0.045 |
| 4 | ZMYND11 | Microarrays | 0.068 | 0.81 | 0.17 |
| 5 | LIMK2 | Microarrays | 0.093 | 0.79 | 0.17 |
| 6 | DIP2B | Microarrays | 0.23 | 0.86 | 0.04 |
| 7 | MAFB | Microarrays | 0.44 | 0.89 | 0.56 |
| 8 | GABARAPL2 | Microarrays | Amplification not possible in FFPE samples | ||
| 9 | CPQ | Meta-analysis | 0.000001 | 0.49 | 0.0004 |
| 10 | PLVAP | Meta-analysis | 0.00001 | 0.51 | 0.0001 |
| 11 | TFF3 | Meta-analysis | 0.0004 | 0.48 | 0.0036 |
| 12 | FAM189A2 | Meta-analysis | 0.0094 | 0.68 | 0.016 |
| 13 | GDF15 | Meta-analysis | 0.058 | 1.49 | 0.99 |
| 14 | CKS2 | Meta-analysis | 0.69 | 1.07 | 0.94 |
| 15 | ASNS | Meta-analysis | 0.90 | 1.02 | 0.17 |
| 16 | EGR2 | Meta-analysis | 0.90 | 0.97 | 0.89 |
| 17 | DDIT3 | Meta-analysis | Amplification not possible in FFPE samples | ||
| 18 | SLC26A4 | Meta-analysis | Amplification not possible in FFPE samples | ||
Two-way analysis of variance (ANOVA) was used in order to adjust for oncocytic feature and the results are shown in column “two-way ANOVA—FDR corrected p-value”.
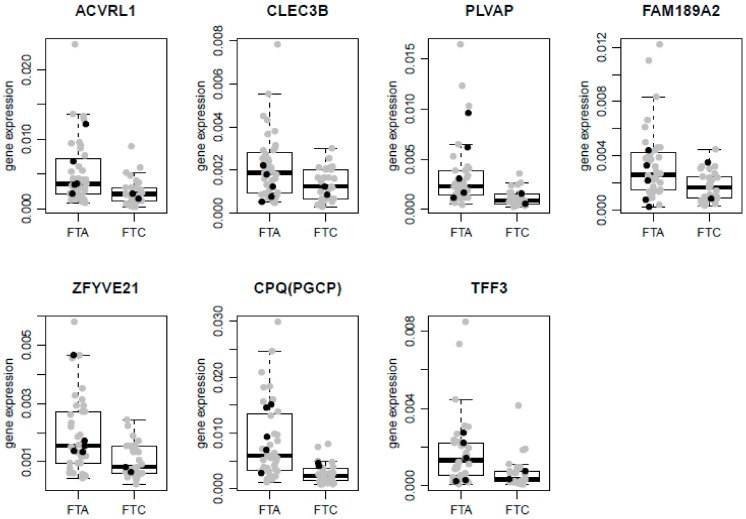
Log-transformed expression levels of the remaining 15 genes were analysed using the Student’s t-test (Table 3). Two FTC samples were extreme outliers (the expression was higher than third quartile (Q3) + 6 × interquartile range (IQR)) in two distinct genes. These samples were excluded from further analysis. Differential expression of CPQ, PLVAP, TFF3, ACVRL1, ZFYVE21, FAM189A2, and CLEC3B was confirmed by qRT-PCR contrary to the expression of ZMYND11, LIMK2, DIP2B, MAFB, CKS2, ASNS, EGR2, and GDF15. All confirmed genes were downregulated in FTC and the direction of change agreed between qRT-PCR data and microarray/meta-analysis data. Boxplots of qRT-PCR results for significantly differentially expressed genes are shown on Figure 3. Based on our results, the following genes that most significantly differentiated between FTC/FTA were selected by a meta-analysis: CPQ (PGCP), PLVAP, and TFF3.
Figure 3.
The normalized relative expression levels of positively validated genes in the FFPE dataset of 69 samples. Boxplots superimposed with scatterplots are shown. The line inside each box corresponds to median. Upper and lower edges of boxes correspond to first (Q1) and third (Q3) quartiles, respectively. The whiskers extend to smallest and largest observations within 1.5 times interquartile range (IQR) from the box. Black dots represent RAS mutation carrying samples, and grey dots represent samples without RAS mutation.
A multivariate ANOVA with two factors: malignancy and oncocytic feature was also performed, in order to evaluate the differential expression between FTC and FTA after adjusting for the effect of oncocytic feature. All seven genes significant in the Student’s t-test were also significant in this ANOVA analysis (Table 3). Adding the additional variables such as age, gender, and RAS mutation status did not substantially modify the ANOVA results.
2.5. Classifier Performance
To evaluate the usefulness of selected genes as diagnostic support, we performed sample classification based on the FFPE dataset. Log-transformation of the gene expression values and a leave-one-out cross-validation of the classifier was performed. In each iteration, the samples were divided into two independent sets: all but one sample were used for significance threshold tuning, gene selection, and classifier training, and the remaining sample was used for testing. Diagonal linear discrimination analysis (DLDA) algorithm was used for the classifier training. After performing all iterations, the classifier’s performance was calculated. The accuracy, sensitivity, and specificity were 78% (95% confidence interval (CI): 67–87%), 76% (95% CI: 56–90%), and 80% (95% CI: 64–91%), respectively. The classifier involved 4 genes with p-value below 0.0005 in the Student’s t-test, namely CPQ, PLVAP, TFF3, and ACVRL1. When accuracy was calculated for non-oncocytic (45 tumors) and oncocytic (24 tumors) tumors separately it was 84% (95% CI: 71–94%) and 67% (95% CI: 45–84%), respectively.
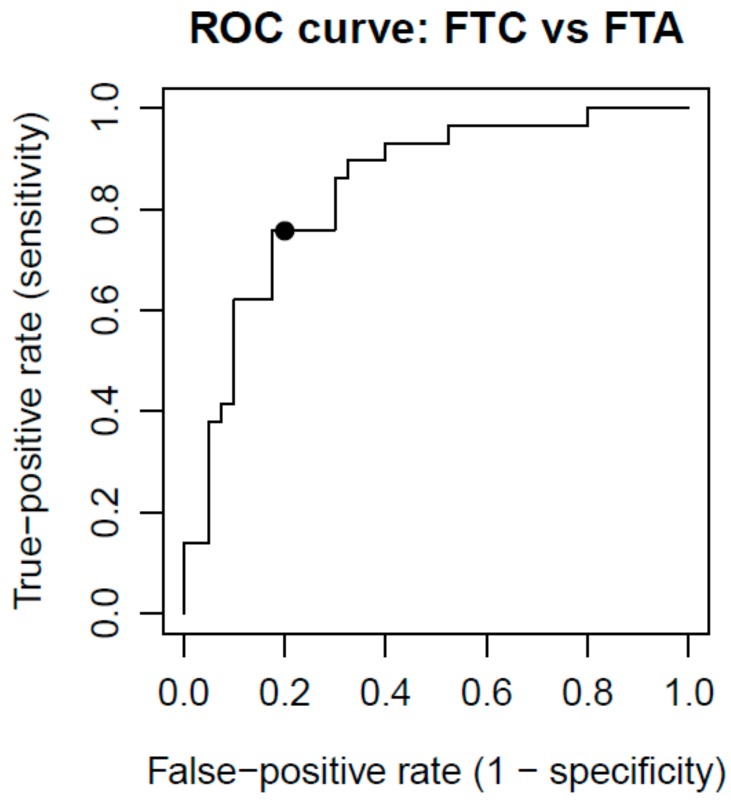
A receiver operating characteristic (ROC) curve was also created in order to assess the diagnostic efficacy of the classifier (Figure 4). The area under the ROC curve (AUC) equals 0.84.
Figure 4.
Receiver operating characteristc (ROC) curve analysis for the predictive power of 4-gene classifier, estimated in qRT-PCR dataset. Using a cutoff probability of 50% (marked with black dot), we obtained sensitivity of 76% and specificity of 80%. The calculated area under the ROC curve was 0.84.
2.6. RAS Mutation Status
The presence of the RAS gene mutation was investigated in freshly frozen FTC (27) and FTA (25) samples. We identified 3 FTC samples with NRAS codon 61 mutation and 1 with KRAS codon 61 mutation (in total 14.8%). In the FTA samples, we identified only 1 mutated sample with NRAS codon 61 mutation (4%) (Table S2). The frequencies of RAS gene mutations in malignant and benign samples did not differ significantly.
The status of the RAS gene mutations was also analysed in FFPE specimens, however due to limitations related to sample quantity, 14 samples were not fully profiled (only NRAS codon 61 was analysed and mutations were excluded in these samples). Among 31 FFPE FTC specimens, 2 samples with NRAS codon 61 mutation and 1 with HRAS codon 12 mutation were identified (9.7%) (Table S3). More RAS mutations were observed in FFPE FTA samples; however, the difference was not significant. Among 40 FFPE FTA specimens, 3 samples with NRAS codon 61 mutations, 1 sample with KRAS codon 12 mutation, and 1 with HRAS codon 61 mutation were detected (12.5%) (Table S3). However, analysis of the total prevalence of RAS mutations in FTC and FTA, regardless of the method used for tissue preservation (FF vs. FFPE) demonstrated that there was no difference in the occurrence of RAS mutations between FTC and FTA: 12% and 9.3%, respectively.
3. Discussion
The differential diagnostics between FTC and FTA is still challenging, particularly because in a molecular sense these lesions lie on a continuum, with similar molecular profiles. Perhaps the 2nd or 3rd molecular hit converts adenoma to carcinoma [28,29] In our study, we performed a meta-analysis of markers differentiating FTC and FTA to summarise the results obtained over a 15-year period (2000–2014), and described in multiple papers.
We obtained a list of 50 genes that were significantly differentially expressed in concordant direction in two or more such papers. We selected 10 genes from the meta-analysis and positively validated 4 of them: CPQ, PLVAP, TFF3, and FAM189A2. While, of the 8 genes selected from our own gene expression microarray dataset, three genes: ACVRL1, CLEC3B, and ZFYVE21, were positively validated by qRT-PCR (Table 3).
Due to small number of RAS mutation positive samples, we were not able to establish its influence on the expression of genes selected for qRT-PCR validation (Figures S1 and S2).
Finally, we created a gene classifier involving 4 genes (CPQ, PLVAP, TFF3, and ACVRL1) that showed a diagnostic accuracy of 78%, sensitivity of 76%, and specificity of 80% for FTC and FTA differentiation. We are aware that our set of genes requires confirmation by an independent clinical study, similar to the study by Alexander et al. [30], which positively verified the clinical utility of a gene classifier proposed by Chudova et al. [25]. However, there are some important differences between Afirma and our approach. While FNAB-based Afirma classifier, used in a preoperative diagnostics, considered all malignant tumors and differed them from benign ones, our classifier was devoted to discriminate only between FTC and FTA on the basis of postoperative material. We did not consider the results of fine-needle aspiration biopsy (FNAB) at any time during our analyses as well as did not link our results to Bethesda Categories. We hope that our classifier may help in such cases where there is a dilemma in a post-operative diagnostics in FTA/FTC distinction. Thus, our work may not be considered as a kind of confirmation of Afirma results.
Transcription profiling, as a method for selection of gene expression markers for distinguishing follicular neoplasms, has been used for over a decade. However, to date, no powerful molecular markers have been established. Similarly, our previous study did not fully accomplish this goal [27]. Therefore, we decided to strengthen our results by performing a meta-analysis of all available studies related to FTC and FTA differentiation [4,5,6,7,8,9,10,11,12,13,14,15,16,17].
The analysis of genes differentially expressed in FTC and FTA in our own gene expression microarray dataset revealed 56 genes. Genes with higher fold-changes and lower p values (Table S4), as well as those related to other types of cancer or tumour aggressiveness were preferably selected for qRT-PCR validation. One of these genes, ACVRL1 correlated with tumour progression in patients with head and neck cancers [31]; whereas two other genes: ZFYVE21, and CLEC3B were related to cancer invasiveness [32,33]. Four genes, obtained from the meta-analysis were subsequently positively validated CPQ, PLVAP, TFF3, and FAM189A2.
Based on the meta-analysis, it appears that building an accurate classifier to differentiate FTCs from FTAs is impossible, even using a large dataset of follicular tumour samples (365 samples in meta-analysis). Therefore, we propose that an accuracy of approximately 80% constitutes a plausible limit of FTC vs. FTA gene classifier performance when analysis is performed in postoperative formalin-fixed material [27].
Possible reason for not satisfying classifier accuracy is that follicular tumours are too similar at the gene expression level. Another hypothetic possible reason is that FTC and FTA classes may have been incorrectly assigned prior to the microarray experiments. Histopathological diagnosis in case of follicular tumours can be influenced by intraobserver variability [34]. To circumvent this, we involved two experienced pathologists in the diagnostic process. It is possible however, that some minimally invasive FTCs did not yet demonstrate any signs of vascular or capsular invasion, and were classified as FTAs.
We assume that FTCs and FTAs are biologically different as they have different clinical outcomes. We are however aware that to date, histopathology constitutes the best option in differential diagnostics of follicular tumours, but a gene-classifier may provide more information in difficult cases. Therefore, we may try to use classifiers ([27], current classifier) to distinguish FTCs and FTAs without histopathological data (unsupervised approach). The results from an unsupervised approach can then be compared to histopathological evaluation, with focus on cases showing discrepancy between the histopathology and classifier data.
It is possible, that we may not reach better classifier performance because of over-simplification that we applied in our analysis. We assumed that both FTC and FTA tumours are internally homogenous, but quite often they are not and they may encompass different zones of differentiation or different histopathological features [35]. Neither FTA nor FTC are completely similar. Considering diversity of biology we cannot expect to cover the whole biological variance with four genes only.
In the present study we decided not to include PTC, because it demonstrated its own, characteristic gene expression profile [36] and the differences between PTC and FTC were quite intense [37]. We believe that an inclusion of PTC to malignant samples may lead to inadequate conclusions, whereas without PTC the study is much cleaner.
The low number of RAS-positive samples did not allow an evaluation of the impact of the RAS gene mutations on the gene expression profile. However, RAS-positive samples did not cluster differentially compared to samples not carrying mutations based on the unsupervised PCA analysis, which suggests small biological differences (Figures S3 and S4). Interestingly, the prevalence of RAS somatic mutations in our own FF FTC dataset was 12%, while other studies show the prevalence of RAS mutation at 60% [38]. This result might be attributable to the population in the studied region of Europe. Unfortunately, we were not able to analyse of the RAS gene mutations in 14 samples due to limited amount of material.
We are aware that our findings would be more robust if we use a single technique of tissue preservation but to a much larger group and the using of FFPE material for validation had a possible limitation. Performing gene expression on FFPE is very challenging and these results could even improve when using cryopreserved samples instead. However, malignant follicular thyroid neoplasms are rare and we had to base on the available material. We did our best to collect as large group as it was possible. We used qRT-PCR with multiple reference genes, to assure that we can amplify sequences coming from reference genes in our tumor samples. Moreover, the results obtained in our study were validated on the independent set of samples. We believe that our results constitute an essential input into the better understanding of molecular biology of follicular thyroid neoplasms.
4. Materials and Methods
4.1. Material
4.1.1. Clinical Materials for Gene Expression Microarray Analysis Using Our Own Thyroid Samples
Fresh-frozen (FF) material from 52 tumours (27 FTC, 25 FTA) was used for our gene expression microarray experiments. The samples and microarray data have been already used in our previous studies and are reused in the current study [18,27]. Surgical procedures on patients were conducted in Polish and German centres, at the MSC Institute—Oncology Center in Gliwice, University of Leipzig, University of Halle, and Mainz University Hospital. Samples collected in hospitals were subsequently sent to our laboratory in Gliwice for microarray molecular profiling. Because the diagnosis of follicular thyroid tumours may be often equivocal [34], we attempted to obtain the evaluation of each pathology slide by two pathologists. However, we had access to the paraffin slides in only a part of the samples. If the slide was available for us, the sample was evaluated by two highly qualified pathologists. If the slide was not available for us, we based on the primary diagnosis, stated in the origin hospital by a single pathologist.
Next, the clinical material was divided into primary and secondary sets of tumors, depending on the concordance in histopathological diagnosis. The primary set contained all samples that were independently and concordantly diagnosed by two thyroid pathology experts (Dariusz Lange, Gliwice, and Steffen Hauptmann, Halle (Saale)). The secondary set contained samples that were diagnosed by only one expert, equivocal samples diagnosed by two experts and one sample that was discordantly diagnosed according to malignancy. A description of the material and the frequency of oncocytic tumors is shown in Table 4 (detailed description is given in Table S2).
Table 4.
Fresh-frozen material used for microarray analysis.
| Set | Histotype | Samples | % of Men | Median Age (Years) | Frequency of Oncocytic Tumours | Concordance of Pathologic Diagnosis by 2 Experts |
|---|---|---|---|---|---|---|
| Primary set | FTC | 13 | 38.5% | 66 | 46.2% | 100% |
| FTA | 13 | 0% | 42 | 15.4% | 100% | |
| Secondary set | FTC | 14 | 21.4% | 69 | 7.1% | 28.6% |
| FTA | 12 | 25% | 49.5 | 0% | 75% | |
| Total | - | 52 | 21.2% | 60.5 | 17.3% | 75% |
The study was approved by the local ethics committees (Bioethics Committee of MSC Institute—Oncology Center in Gliwice; approvals: DK/ZMN-493-1-10/09, 20 November 2002 and KB/492-17/11, 9 February 2011), and informed consent was obtained from all patients.
4.1.2. Clinical Materials for Validation Studies
FFPE tissue was used for validation in qRT-PCR experiments. The FFPE tissue consisted of 40 FTA and 31 FTC samples from patients treated in the MSC Institute—Oncology Center in Gliwice. The same set of samples was used in our previous study [27]. Diagnosis of FFPE tumours was based on the independent diagnoses of two pathologists. Material description and frequency of oncocytic tumours is presented in Table 5 (detailed description is given in Table S3 in Supplementary Material). Fresh frozen and FFPE datasets were independent datasets; there was no patient overlap between them.
Table 5.
FFPE material used for qRT-PCR validation.
| Histopathological Diagnosis | Number of Samples | % of Men | Median Age (Years) | Frequency of Oncocytic Tumours |
|---|---|---|---|---|
| FTC | 31 | 32.3% | 59 | 61.3% |
| FTA | 40 | 12.5% | 45 | 15% |
| Total | 71 | 21.1% | 52 | 35.2% |
4.1.3. RAS Mutation Screening
All 123 samples of thyroid follicular tumour used for gene expression microarray (52 samples) and qRT-PCR experiments (71 samples) were screened for RAS mutations using the Sanger sequencing method with the ABI 3130xl Genetic Analyzer. Three RAS genes (H-, K-, N-RAS) sequences in commonly mutated codon sites (12, 13, and 61) were analysed. Different primer sets (different size of amplicon) for FF and FFPE samples were used due to sample degradation in FFPE samples (details Table S5).
4.2. Gene Expression Microarray-Based Analysis of Our Own Follicular Tumours
4.2.1. Gene Expression Microarray Experiment
FF materials from 52 follicular thyroid tumours (27 FTC, 25 FTA) were used for microarray analysis. RNA was isolated using the RNeasy Mini kit (Qiagen, Hilden, Germany). The RNA quality was assessed with capillary electrophoresis (Bioanalyzer 2100) and all the samples had the RNA integrity number (RIN) higher than 7. An Affymetrix (Santa Clara, CA, USA) HG-U133 PLUS 2 array experiment was performed as described previously [27].
4.2.2. Gene Expression Microarray Data Preprocessing
All microarray data analyses were performed in an R/Bioconductor environment. The microarray data preprocessing was performed in the same way as described in our previous study [27]. The quality of the microarray data was analysed using arrayMvout 1.12.0 library [39]. The raw data were preprocessed using the GCRMA method [40]. The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [41], and are accessible through GEO Series accession number GSE82208 (available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82208).
4.2.3. Supervised Analysis of Our Own Gene Expression Microarray Data
The selection of differentially expressed genes was performed independently on the primary and secondary microarray dataset (FF material) (Figure 1), in order to take into account the different levels of diagnosis certainty in the two sets. The following criteria were used for the primary dataset: normalized mean expression of the gene above 4.5, the variance of the gene above the 20th percentile, p-value in Student’s t-test below 0.001, a fold-change above 1.5 in either direction of the change. The following, less strict criterion was used for the secondary dataset: p-value in Student’s t-test < 0.05.
For genes selected for validation study, an additional analysis was performed in order to assess the significance of difference between FTC and FTA in microarray dataset comprised of non-oncocytic samples only. The genes were considered significant if the unadjusted p-value in the Student’s t-test was below 0.005 in primary dataset and below 0.05 in secondary dataset.
In order to adjust p-values for multiple comparisons, false discovery rate (FDR) was estimated by Benjamini and Hochberg procedure [42].
4.3. Meta-Analysis of All Published Papers
The meta-analysis included all 14 papers in which the difference in gene expression between FTC and FTA was assessed by a high throughput method (gene expression microarrays, SAGE, HDSS, ATAC-PCR); which were published during 2000–2014; and found in PubMed, Google Scholar, or by screening the reference lists of selected papers (Table S1). The following criteria were used for the selection of papers: “follicular thyroid carcinoma/cancer/tumour/adenoma AND microarray/gene expression”.
The lists of genes that were reported by the authors as differentially expressed between FTC and FTA, were extracted from each paper. Different types of gene identifiers were used in each study, such as gene symbols, gene names, GenBank accession numbers, cDNA sequences, Affymetrix identifiers, RefSeq accession numbers, and UniGene accession numbers. All gene identifiers were converted to EntrezID, the lists of genes were compared, and common genes were extracted. Finally, ten genes among the most frequently occurring ones were chosen for qRT-PCR validation (Figure 1).
Principal Component Analysis of Microarrays Based on the Meta-Analysis Identified Genes
To visually inspect whether the genes selected in the meta-analysis are able to separate FTC and FTA on an independent dataset, Principal Component Analysis (PCA) was conducted. We performed PCA on our own microarray samples, based on genes that occurred at least once in the meta-analysis (Figure 2 upper plot). We also performed PCA on these samples, based on the genes that occurred at least twice in the meta-analysis (Figure 2 lower plot).
4.4. qRT-PCR Validation
4.4.1. qRT-PCR Experiment
FFPE materials from 71 follicular thyroid tumours (31 FTC, 40 FTA) were used for qRT-PCR analysis. RNA was isolated using the FFPE RNeasy Mini Kit (Qiagen) from 5 slices of paraffin blocks selected by a histopathologist. qRT-PCR was carried out for 18 genes (gene names given in Table S6, primer probe design given in Table S7). This experiment was performed with the 7900HT Fast Real-Time PCR (Life Technologies, Carlsbad, CA, USA) using Universal Probe Library fluorescent probes (Roche, Basel, Switzerland) and the 5′-nuclease assay, starting from 200 ng of total RNA. All experiments were performed twice. Results were normalised using the Pfaffl method [43] and the GeNorm application [44] with a combination of 3 normalisation genes: EIF3A (eukaryotic translation initiation factor 3, subunit A), EIF5 (eukaryotic translation initiation factor 5), and HADHA (hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit). Obtained normalised relative expression levels were further log-transformed (Figure S5).
Differences between FTC and FTA were tested using the Student’s t-test. In addition, two-way analysis of variance (ANOVA) was used in order to adjust for oncocytic feature. False Discovery Rate (FDR) correction was applied and genes with FDR < 0.05 in both analyses were considered as significant.
4.4.2. Classifier Performance
The classifier was created and validated on the FFPE dataset using CMA package [45] in R/Bioconductor environment. The DLDA was used as a classification algorithm. Student’s t-test was used for gene selection with significance level threshold tuned over a grid of significance levels. The performance of the classifier was evaluated by the doubly nested leave-one-out cross validation (LOOCV) approach in order to obtain an unbiased estimate of the accuracy [46]. The outer loop was used for estimating the classifier accuracy, and the inner loop was used for optimising the significance level threshold.
The ROC curve was also created to assess the diagnostic efficacy of the classifier (Figure 4). In the outer leave-one-out loop, for each sample, the probability that the sample belongs to the FTC class was calculated, based on DLDA algorithm. Varying the threshold for the probability, the ROC curve was plotted.
5. Conclusions
In our study, we have demonstrated that meta-analysis is a valuable method for selecting possible molecular markers. We showed that genes CPQ, PLVAP, TFF3, ACVRL1, ZFYVE21, FAM189A2, and CLEC3B are differentially expressed between FTC and FTA. Furthermore, we propose a 4-gene classifier, which discriminates between benign and malignant follicular neoplasms with the accuracy of 78%. Based on our results, we conclude that there might exist a plausible limit of gene classifier accuracy of approximately 80%, when follicular tumors are discriminated based on postoperative formalin-fixed material.
Acknowledgments
This study was supported by the Ministry of Science and Higher Education (grant number N N401 072637); Polish National Science Center (decision number DEC-2011/03/N/NZ5/05623); Foundation for Polish Science (MPD Program “Molecular Genomics, Transcriptomics and Bioinformatics in Cancer”); Postgraduate School of Molecular Medicine (fellowships to Bartosz Wojtas and Tomasz Stokowy); National Center for Research and Development project under the program “Prevention practices and treatment of civilisation diseases” STRATEGMED (MILESTONE, project number STRATEGMED2/267398/4/NCBR/2015); and DFG (grant number ES162/4-1 to Markus Eszlinger). The costs to publish in open access were covered by Maria Sklodowska-Curie Institute—Oncology Center, Gliwice Branch.
Abbreviations
| ANOVA | Analysis of variance |
| ATAC-PCR | Adapter-tagged competitive polymerase chain reaction |
| AUC | Area under the ROC curve |
| DLDA | Diagonal linear discrimination analysis |
| FDR | False discovery rate |
| FF | Fresh-frozen |
| FFPE | Formaldehyde-fixed paraffin embedded |
| FNAB | Fine needle aspiration biopsy |
| FTA | Follicular thyroid adenoma |
| FTC | Follicular thyroid cancer |
| HDSS | High-throughput differential screening by serial analysis of gene expression |
| IQR | Interquartile range |
| LOOCV | Leave-one-out cross validation |
| OTC | Oncocytic thyroid carcinoma |
| PCA | Principal component analysis |
| Q1 | First quartile |
| Q3 | Third quartile |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| RIN | RNA integrity number |
| ROC | Receiver operating characteristc |
| SAGE | Serial analysis of gene expression |
| WHO | World Health Organization |
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/6/1184/s1.
Author Contributions
Bartosz Wojtas, Aleksandra Pfeifer, Jolanta Krajewska, Michal Jarzab and Ewa Chmielik contributed to the writing of the manuscript; Michal Jarzab, Markus Eszlinger, Ralf Paschke, and Barbara Jarzab designed and coordinated the study; Agnieszka Czarniecka and Thomas Musholt collected the tissue material; Aleksandra Kukulska and Jolanta Krajewska analyzed patient data; Ewa Stobiecka, Ewa Chmielik, Steffen Hauptmann, and Dariusz Lange performed the histopathological examination of the samples; Bartosz Wojtas, Malgorzata Oczko-Wojciechowska, Dagmara Rusinek, Tomasz Tyszkiewicz and Monika Halczok performed the experiments described in this study; Aleksandra Pfeifer, Tomasz Stokowy and Michal Swierniak performed the bioinformatics analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.DeLellis R., Lloyd R., Heitz P., Eng C. WHO Pathology and Genetics. Tumours of Endocrine Organs. IARC Press; Lyon, France: 2004. [Google Scholar]
- 2.LiVolsi V.A., Baloch Z.W. Follicular-patterned tumors of the thyroid: The battle of benign vs. malignant vs. so-called uncertain. Endocr. Pathol. 2011;22:184–189. doi: 10.1007/s12022-011-9183-6. [DOI] [PubMed] [Google Scholar]
- 3.Yamashina M. Follicular neoplasms of the thyroid. Total circumferential evaluation of the fibrous capsule. Am. J. Surg. Pathol. 1992;16:392–400. doi: 10.1097/00000478-199204000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Takano T., Hasegawa Y., Matsuzuka F., Miyauchi A., Yoshida H., Higashiyama T., Kuma K., Amino N. Gene expression profiles in thyroid carcinomas. Br. J. Cancer. 2000;83:1495–1502. doi: 10.1054/bjoc.2000.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barden C.B., Shister K.W., Zhu B., Guiter G., Greenblatt D.Y., Zeiger M.A., Fahey T.J., III Classification of follicular thyroid tumors by molecular signature: Results of gene profiling. Clin. Cancer Res. 2003;9:1792–1800. [PubMed] [Google Scholar]
- 6.Takano T., Miyauchi A., Yoshida H., Kuma K., Amino N. High-throughput differential screening of mRNAs by serial analysis of gene expression: Decreased expression of trefoil factor 3 mRNA in thyroid follicular carcinomas. Br. J. Cancer. 2004;90:1600–1605. doi: 10.1038/sj.bjc.6601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerutti J.M., Delcelo R., Amadei M.J., Nakabashi C., Maciel R.M., Peterson B., Shoemaker J., Riggins G.J. A preoperative diagnostic test that distinguishes benign from malignant thyroid carcinoma based on gene expression. J. Clin. Investig. 2004;113:1234–1242. doi: 10.1172/JCI19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevillard S., Ugolin N., Vielh P., Ory K., Levalois C., Elliott D., Clayman G.L., El-Naggar A.K. Gene expression profiling of differentiated thyroid neoplasms: Diagnostic and clinical implications. Clin. Cancer Res. 2004;10:6586–6597. doi: 10.1158/1078-0432.CCR-04-0053. [DOI] [PubMed] [Google Scholar]
- 9.Weber F., Shen L., Aldred M.A., Morrison C.D., Frilling A., Saji M., Schuppert F., Broelsch C.E., Ringel M.D., Eng C. Genetic classification of benign and malignant thyroid follicular neoplasia based on a three-gene combination. J. Clin. Endocrinol. Metab. 2005;90:2512–2521. doi: 10.1210/jc.2004-2028. [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi K., Takano T., Miyauchi A., Koizumi K., Ito Y., Takamura Y., Ishitobi M., Miyoshi Y., Taguchi T., Tamaki Y., et al. Differentiation of follicular thyroid adenoma from carcinoma by means of gene expression profiling with adapter-tagged competitive polymerase chain reaction. Oncology. 2005;69:428–435. doi: 10.1159/000089998. [DOI] [PubMed] [Google Scholar]
- 11.Lubitz C.C., Gallagher L.A., Finley D.J., Zhu B., Fahey T.J., III Molecular analysis of minimally invasive follicular carcinomas by gene profiling. Surgery. 2005;138:1042–1048. doi: 10.1016/j.surg.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Fryknas M., Wickenberg-Bolin U., Goransson H., Gustafsson M.G., Foukakis T., Lee J.J., Landegren U., Hoog A., Larsson C., Grimelius L., et al. Molecular markers for discrimination of benign and malignant follicular thyroid tumors. Tumour Biol. 2006;27:211–220. doi: 10.1159/000093056. [DOI] [PubMed] [Google Scholar]
- 13.Stolf B.S., Santos M.M., Simao D.F., Diaz J.P., Cristo E.B., Hirata R., Jr., Curado M.P., Neves E.J., Kowalski L.P., Carvalho A.F. Class distinction between follicular adenomas and follicular carcinomas of the thyroid gland on the basis of their signature expression. Cancer. 2006;106:1891–1900. doi: 10.1002/cncr.21826. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J., Leonard C., Gemsenjager E., Heitz P.U., Moch H., Odermatt B. Differentiation of human follicular thyroid adenomas from carcinomas by gene expression profiling. Oncol. Rep. 2008;19:329–337. doi: 10.3892/or.19.2.329. [DOI] [PubMed] [Google Scholar]
- 15.Hinsch N., Frank M., Doring C., Vorlander C., Hansmann M.L. QPRT: A potential marker for follicular thyroid carcinoma including minimal invasive variant; a gene expression, RNA and immunohistochemical study. BMC Cancer. 2009;9:93. doi: 10.1186/1471-2407-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borup R., Rossing M., Henao R., Yamamoto Y., Krogdahl A., Godballe C., Winther O., Kiss K., Christensen L., Hogdall E., et al. Molecular signatures of thyroid follicular neoplasia. Endocr. Relat. Cancer. 2010;17:691–708. doi: 10.1677/ERC-09-0288. [DOI] [PubMed] [Google Scholar]
- 17.Williams M.D., Zhang L., Elliott D.D., Perrier N.D., Lozano G., Clayman G.L., El-Naggar A.K. Differential gene expression profiling of aggressive and nonaggressive follicular carcinomas. Hum. Pathol. 2011;42:1213–1220. doi: 10.1016/j.humpath.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wojtas B., Pfeifer A., Jarzab M., Czarniecka A., Krajewska J., Swierniak M., Stokowy T., Rusinek D., Kowal M., Zebracka-Gala J., et al. Unsupervised analysis of follicular thyroid tumours transcriptome by oligonucleotide microarray gene expression profiling. Endokrynol. Pol. 2013;64:328–334. doi: 10.5603/EP.2013.0013. [DOI] [PubMed] [Google Scholar]
- 19.Swierniak M., Pfeifer A., Stokowy T., Rusinek D., Chekan M., Lange D., Krajewska J., Oczko-Wojciechowska M., Czarniecka A., Jarzab M., et al. Somatic mutation profiling of follicular thyroid cancer by next generation sequencing. Mol. Cell. Endocrinol. 2016;433:130–137. doi: 10.1016/j.mce.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Cheung L., Messina M., Gill A., Clarkson A., Learoyd D., Delbridge L., Wentworth J., Philips J., Clifton-Bligh R., Robinson B.G. Detection of the PAX8-PPARγ fusion oncogene in both follicular thyroid carcinomas and adenomas. J. Clin. Endocrinol. Metab. 2003;88:354–357. doi: 10.1210/jc.2002-021020. [DOI] [PubMed] [Google Scholar]
- 21.Sahin M., Allard B.L., Yates M., Powell J.G., Wang X.L., Hay I.D., Zhao Y., Goellner J.R., Sebo T.J., Grebe S.K., et al. PPARγ staining as a surrogate for PAX8/PPARγ fusion oncogene expression in follicular neoplasms: Clinicopathological correlation and histopathological diagnostic value. J. Clin. Endocrinol. Metab. 2005;90:463–468. doi: 10.1210/jc.2004-1203. [DOI] [PubMed] [Google Scholar]
- 22.Kloos R.T., Reynolds J.D., Walsh P.S., Wilde J.I., Tom E.Y., Pagan M., Barbacioru C., Chudova D.I., Wong M., Friedman L., et al. Does addition of BRAF V600E mutation testing modify sensitivity or specificity of the Afirma Gene Expression Classifier in cytologically indeterminate thyroid nodules? J. Clin. Endocrinol. Metab. 2013;98:761–768. doi: 10.1210/jc.2012-3762. [DOI] [PubMed] [Google Scholar]
- 23.Baris O., Savagner F., Nasser V., Loriod B., Granjeaud S., Guyetant S., Franc B., Rodien P., Rohmer V., Bertucci F., et al. Transcriptional profiling reveals coordinated up-regulation of oxidative metabolism genes in thyroid oncocytic tumors. J. Clin. Endocrinol. Metab. 2004;89:994–1005. doi: 10.1210/jc.2003-031238. [DOI] [PubMed] [Google Scholar]
- 24.Ganly I., Ricarte Filho J., Eng S., Ghossein R., Morris L.G., Liang Y., Socci N., Kannan K., Mo Q., Fagin J.A., et al. Genomic dissection of Hurthle cell carcinoma reveals a unique class of thyroid malignancy. J. Clin. Endocrinol. Metab. 2013;98:962–972. doi: 10.1210/jc.2012-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chudova D., Wilde J.I., Wang E.T., Wang H., Rabbee N., Egidio C.M., Reynolds J., Tom E., Pagan M., Rigl C.T., et al. Molecular classification of thyroid nodules using high-dimensionality genomic data. J. Clin. Endocrinol. Metab. 2010;95:5296–5304. doi: 10.1210/jc.2010-1087. [DOI] [PubMed] [Google Scholar]
- 26.Keutgen X.M., Filicori F., Crowley M.J., Wang Y., Scognamiglio T., Hoda R., Buitrago D., Cooper D., Zeiger M.A., Zarnegar R., et al. A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin. Cancer Res. 2012;18:2032–2038. doi: 10.1158/1078-0432.CCR-11-2487. [DOI] [PubMed] [Google Scholar]
- 27.Pfeifer A., Wojtas B., Oczko-Wojciechowska M., Kukulska A., Czarniecka A., Eszlinger M., Musholt T., Stokowy T., Swierniak M., Stobiecka E., et al. Molecular differential diagnosis of follicular thyroid carcinoma and adenoma based on gene expression profiling by using formalin-fixed paraffin-embedded tissues. BMC Med. Genom. 2013;6:38. doi: 10.1186/1755-8794-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans H.L., Vassilopoulou-Sellin R. Follicular and Hurthle cell carcinomas of the thyroid: A comparative study. Am. J. Surg. Pathol. 1998;22:1512–1520. doi: 10.1097/00000478-199812000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Parameswaran R., Brooks S., Sadler G.P. Molecular pathogenesis of follicular cell derived thyroid cancers. Int. J. Surg. 2010;8:186–193. doi: 10.1016/j.ijsu.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Alexander E.K., Kennedy G.C., Baloch Z.W., Cibas E.S., Chudova D., Diggans J., Friedman L., Kloos R.T., LiVolsi V.A., Mandel S.J., et al. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N. Engl. J. Med. 2012;367:705–715. doi: 10.1056/NEJMoa1203208. [DOI] [PubMed] [Google Scholar]
- 31.Chien C.Y., Chuang H.C., Chen C.H., Fang F.M., Chen W.C., Huang C.C., Huang H.Y. The expression of activin receptor-like kinase 1 among patients with head and neck cancer. Otolaryngol. Head Neck Surg. 2013;148:965–973. doi: 10.1177/0194599813479556. [DOI] [PubMed] [Google Scholar]
- 32.Arvanitis D.L., Kamper E.F., Kopeikina L., Stavridou A., Sgantzos M.N., Kallioras V., Athanasiou E., Kanavaros P. Tetranectin expression in gastric adenocarcinomas. Histol. Histopathol. 2002;17:471–475. doi: 10.14670/HH-17.471. [DOI] [PubMed] [Google Scholar]
- 33.Hoshino D., Nagano M., Saitoh A., Koshikawa N., Suzuki T., Seiki M. The phosphoinositide-binding protein ZF21 regulates ECM degradation by invadopodia. PLoS ONE. 2013;8:e50825. doi: 10.1371/journal.pone.0050825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franc B., de la Salmonière P., Lange F., Hoang C., Louvel A., de Roquancourt A., Vilde F., Hejblum G., Chevret S., Chastang C. Interobserver and intraobserver reproducibility in the histopathology of follicular thyroid carcinoma. Hum. Pathol. 2003;34:1092–1100. doi: 10.1016/S0046-8177(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 35.Da S.L., James D., Simpson P.T., Walker D., Vargas A.C., Jayanthan J., Lakhani S.R., McNicol A.M. Tumor heterogeneity in a follicular carcinoma of thyroid: A study by comparative genomic hybridization. Endocr. Pathol. 2011;22:103–107. doi: 10.1007/s12022-011-9154-y. [DOI] [PubMed] [Google Scholar]
- 36.Jarzab B., Wiench M., Fujarewicz K., Simek K., Jarzab M., Oczko-Wojciechowska M., Wloch J., Czarniecka A., Chmielik E., Lange D., et al. Gene expression profile of papillary thyroid cancer: Sources of variability and diagnostic implications. Cancer Res. 2005;65:1587–1597. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- 37.Aldred M.A., Huang Y., Liyanarachchi S., Pellegata N.S., Gimm O., Jhiang S., Davuluri R.V., de la Chapelle A., Eng C. Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. J. Clin. Oncol. 2004;22:3531–3539. doi: 10.1200/JCO.2004.08.127. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigues H.G., de Pontes A.A., Adan L.F. Use of molecular markers in samples obtained from preoperative aspiration of thyroid. Endocr. J. 2012;59:417–424. doi: 10.1507/endocrj.EJ11-0410. [DOI] [PubMed] [Google Scholar]
- 39.Asare A.L., Gao Z., Carey V.J., Wang R., Seyfert-Margolis V. Power enhancement via multivariate outlier testing with gene expression arrays. Bioinformatics. 2009;25:48–53. doi: 10.1093/bioinformatics/btn591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Z.J., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. doi: 10.1198/016214504000000683. [DOI] [Google Scholar]
- 41.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 43.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandesompele J., de Preter K., Pattyn F., Poppe B., van Roy N., de Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slawski M., Daumer M., Boulesteix A.L. CMA: A comprehensive bioconductor package for supervised classification with high dimensional data. BMC Bioinform. 2008;9:439. doi: 10.1186/1471-2105-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varma S., Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinform. 2006;7 doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.