Abstract
This study presents pioneering data concerning the human pregnancy-associated glycoprotein-Like family, identified in the genome, of the term placental transcriptome and proteome. RNA-seq allowed the identification of 1364 bp hPAG-L/pep cDNA with at least 56.5% homology with other aspartic proteinases (APs). In silico analyses revealed 388 amino acids (aa) of full-length hPAG-L polypeptide precursor, with 15 aa-signal peptide, 47 aa-blocking peptide and 326 aa-mature protein, and two Asp residues (D), specific for a catalytic cleft of the APs (VVFDTGSSNLWV91-102 and AIVDTGTSLLTG274-285). Capillary sequencing identified 9330 bp of the hPAG-L gene (Gen Bank Acc. No. KX533473), composed of nine exons and eight introns. Heterologous Western blotting revealed the presence of one dominant 60 kDa isoform of the hPAG-L amongst cellular placental proteins. Detection with anti-pPAG-P and anti-Rec pPAG2 polyclonals allowed identification of the hPAG-L proteins located within regions of chorionic villi, especially within the syncytiotrophoblast of term singleton placentas. Our novel data extend the present knowledge about the human genome, as well as placental transcriptome and proteome during term pregnancy. Presumably, this may contribute to establishing a new diagnostic tool for examination of some disturbances during human pregnancy, as well as growing interest from both scientific and clinical perspectives.
Keywords: cDNA, gDNA, human PAG-L, placenta, trophectoderm
1. Introduction
Pregnancy-associated glycoproteins (PAGs) belong to a superfamily of aspartic proteinases (AP), which also include mammalian pepsins (A, C and F), cathepsins (D and E), renin and numerous other enzymes such as parasite plasmepsins and retroviral enzymes [1,2]. All AP members possess a two-bilobe structure with a cleft capable of short peptide binding and are classified into two subfamilies: catalytically active or potentially inactive due to several amino acid (aa) substitutions within two domains creating the binding cleft [3,4]. Among APs, pepsins fulfil digestive functions outside the cells, whereas cathepsin D and E are typical intracellular enzymes generally localized in the lysosomal compartment that provides the acidic environment necessary to accomplish their catalytic functions [5,6]. On the other hand, PAG-Like (PAG-L) family products revealed properties as various chorionic signaling ligands interacting with gonadal and extra-gonadal gonadotropin receptors of early pregnant pigs [7], as well as cyclic pigs and cows [8].
In humans, various AP members are involved in the development of a variety of diseases, e.g., hypertension, gastric ulcers, acquired immunodeficiency syndrome, malaria, lysosomal muscular dystrophy and neoplastic diseases, etc. [2,9]. APs are also involved in defense against infections, tumor cells, cancer and in the development of atopic dermatitis [10,11,12].
During gestation, APs may also play important roles during implantation and in the establishment of early pregnancy, since chorionic expression of cathepsins B and L differ in normal and abnormal deciduas [13], whereas an imbalance of the cathepsin–cystatin system causes miscarriages [14]. Decreased activity of cathepsin E might also be responsible for induction of miscarriages by decreased decidual expression, especially in macrophages of patients with recurrent pregnancy loss [15].
Decreased PAG family expression also occurs during gestation disorders. The PAG family originates from a progene duplication or its fragments and positive selection of these genes [16]. To date, the entire exon-intron structures of only four PAG genes have been identified within some genomes, bovine—bPAG1 [17], bPAG2 [18], porcine—pPAG2 [19] and beaver—CfPAG-L [20]. The PAGs are characterized by a conserved structure that includes nine exons and eight introns [1,2]. All mammalian PAGs and related PAG-L genes are the most closely homologous to the pepsins [18,21].
Mammalian placenta is a unique organ essential for fetal growth, development and survival in the uterus [22], with complex of biomolecular interactions between the fetus and mother that provide structural and biochemical barriers between both compartments [23]. The human placenta is hemochorial (maternal blood is in direct contact with fetal trophoblast) and discoidal in shape with villous materno-fetal interdigitations [24]. Within each placenta type developed in various eutherians, trophoblast forms the outer layer of a blastocyst, then expands into the trophectoderm—chorionic epithelium, which together with the endometrium, forms the placenta [25]. Within a very precise feto-maternal interface [26,27], specific expression of the PAGs is cell- and pregnancy stage-dependent [1,2].
Many purified native or several recombinant proteins, required for anti-PAG sera production, have led to the establishment of various pregnancy diagnoses, based on PAG-L detection in maternal blood or milk by radioimmunological (RIA) and immunoenzymatic (ELISA) tests [1]. These PAG tests are useful for detecting abnormalities during pregnancy in cattle [28,29] and to predict miscarriages after embryo transfers [30]. Since the varying PAG concentration depends on the number of healthy embryos/fetuses, it is higher in females with twin than single pregnancies and can also differ due to the fetal sex and breed in many domestic and some wild ruminants [1,31].
In view of both the commitment of these chorionic proteins in the course of pregnancy and evolutionary persistence of the PAG genes in various eutherian species [1,2], there is growing interest in examining whether they are also present in humans. The subsistence of this unique PAG/PAG-L family has not yet been studied in humans.
The objective of this study was to identify the existence of the PAG-L family in humans: (1) placental transcriptome; (2) genome; (3) placental proteome, including immuno-detection of protein profiles and cellular localization in the term placenta.
2. Results
2.1. Identification of cDNA Sequence Originating from Term Placental Transcriptome
The hPAG-L sequence was identified by two methods. The performed RNA-seq generated a total of 71,271,470 pairs of raw reads and 58,547,248 trimmed pair reads (82%) obtained after removing TruSeq adaptors and low quality reads. TRINITY software enabled de novo assembly of 102,357 contigs. The reconstructed contigs were analyzed for similarity to the AP superfamily, which allowed identification of a 1364 bp cDNA sequence of the placental hPAG-L transcript.
In addition, cDNA evaluation by capillary sequencing firmly confirmed the nucleotide sequence originating from the RNA-seq. Five pairs of homological PAG-L primers applied for PCRs allowed obtaining the entire cDNA sequence (1364 bp), named hPAG-L, and deposited in GenBank database (Acc. No. KX856064). Among the 109 electrophoresed, gel-out purified and sequenced cDNA amplicons, 80 high quality chromatograms (HQ range: 50–93.9%) were applied to estimate the coding (CDS) and non-coding untranslated regions (UTR). Among the identified 1364 bp of cDNA sequence, 1167 bp were determined as CDS, 20 bp as 5′UTR and 177 bp as 3′UTR (Figure 1).
Figure 1.
Identified cDNA sequence of 1364 bp human Pregnancy-Associated Glycoprotein-like (hPAG-L) encoding full-length 388 amino acids (aa) of polypeptide precursor. 5′- and 3′-untranslated regions (5′UTR and 3′UTR), 15 aa-signal peptide, 47 aa-blocking peptide, 326 aa-mature protein, and active sites creating catalytic clefts are indicated.
A megablast of the hPAG-L cDNA revealed 85–99% homology with mammalian pepsinogens A, (peps) whereas Query Cover (QC) ranged from 68% to 100% in various species (e.g., Homo sapiens, Gorilla gorilla, Pan troglodytes, Pongo abeli, Nomascus leucogenys, Macaca Fascicularis, Colobus angolensis, Rhinopithecus roxellana, Camelus ferus, Ursus maritimus, Mustela putorius, Canis lupus, Leptonychotes weddellii).
Pair-wise alignment (Geneious® 8.1.7) of the hPAG-L cDNA sequence with various AP members indicated the highest identity with: human pep A (99%; NM_001079808.3), zebra PAG (66.2%; AF036952); mouse pep F (65.3%; AF240776.1); equine PAG (ePAG; 65.9%; L38511); human pep C (64.2%; J04443.1); with pPAG2 (64%; L34361.1); beaver PAG-L (CfPAG-L; 63.1%; KU245742.1); and also human cathepsin E (61.4%; NM_001910.3); cathepsin D (59.1%; NM_001909.4); napsin A (56.5%; NM_004851.2) and renin (56.5%; NM_000537.3). Due to the highest identity of hPAG-L with human pepsinogens (PGPGA4, PGA3 and PGA5), the identified placental AP can be also named as hPAG-L/pep.
The hPAG-L/pep cDNA allowed identification of a 388 aa-polypeptide precursor (Geneious® 8.1.7). The entire placental AP precursor revealed the highest aa identity with: human pep A (99.2%); mouse pep F (56.2%); pPAG2 (51.2%); cathepsin E (53%); pep C (50%); cathepsin D (44%); napsin A (40.5%) and renin (36.3%).
2.2. Identification of cDNA Sequence Originating from Term Placental Transcriptome
The hPAG-L/pep polypeptide precursor retains two highly conserved domains (NH2 and COOH), specific to other members of AP superfamily. Geneious® 8.1.7 allowed identification of 15 aa-signal peptide (SP), 47 aa-blocking peptide and 326 aa-mature hPAG-L/pep precursor (Figure 1).
The identified SP aa sequence of the hPAG-L/pep shared the highest similarity with SP of the human pep A and it varied with the other members of the AP family in different species (Table 1).
Table 1.
Signal peptide (SP) sequence homology of the hPAG-L/pep polypeptide precursor to various aspartic proteinases.
| Gene Name a | SP Sequence (aa) b | Identity (%) | Positive aa (%) |
|---|---|---|---|
| hPAG-L/pep | MKWLLLLGLVALSEC | this study | this study |
| hPepsinogen A | ............... | 100 | 100 |
| bPAG2 | ....V.......... | 93.3 | 100 |
| pPAG1 | ....VI......... | 86.7 | 100 |
| mPepsinogen F | ....WV......... | 86.7 | 93.3 |
| fPAG | ....WV......... | 86.7 | 93.3 |
| pPAG2 | ....VI.......D. | 80 | 100 |
| ePAG | ...FGV....T.... | 73.3 | 80 |
| CfPAG-L | ...IVVA.LC.P.L.A | 37.5 | 62.5 |
| hCathepsin E | ..T....L..L.ELGEAQG | 60 | 60 |
| hPepsinogen C | ...MVVV-..C.QLLEA | 40 | 66.7 |
| hNapsin A | QPL....P.LNVEPSGA | 33.3 | 46.7 |
| hRenin | PR.G..--.LLWGS.TFG | 33.3 | 46.7 |
| hCathepsin D | .QPSS..P.ALCLLAAPASA | 26.7 | 33.3 |
a aa—Amino acids, b—bovine, e—equine, f—feline, h—human, m—mouse, p—porcine, Cf—beaver; b Identical aa are dotted. Gaps (–) have been inserted to provide maximal alignments.
Multiple alignments of the various APs enabled the prediction of 47 aa-blocking peptide (16–62 aa) of the hPAG-L/pep precursor that shared the highest homology with human pep A, whereas identity is equal/similar with peps C and F as well as other PAGs (Table 2).
Table 2.
Blocking peptide aa sequence homology of the human PAG-L/pep polypeptide precursor to various aspartic proteinases.
| Gene Name a | Blocking Peptide Sequence (aa) b | Identity (%) | Positive aa (%) |
|---|---|---|---|
| hPAG-L/pep | IMYKVPLIRKKSLRRTLSERGLLKDFLKKHNLNPARKYFPQWEAPTL | this study | this study |
| hPepsinogen A | ............................................... | 100 | 100 |
| hPepsinogen C | AVV....KKF..I.E.MK.K...GE..RT.KYD..W..R.GDL | 46.5 | 65.1 |
| fPAG | -LVTI..T.V..M.EN.R.KDR.....EN.PY.L.Y.FVD | 43.6 | 59 |
| pPAG2 | -LVMI..TKV..V.ES.R.K....N...E.PY.MIQNL | 43.2 | 67.6 |
| CfPAG-L | AISRI..RKA..V.Q..K.K...EE...T.KYD..Q..LANNFGDF | 41.3 | 65.2 |
| pPAG1 | -LVII..TKV..I.EN.R.KD..LN...E.PY.MIQ.F | 40.5 | 64.9 |
| ePAG | -LVTI..VKI....EN.R.KDM..EY.E.YPFRL | 36.4 | 66.7 |
| mPepsinogen F | -LV.I..MKI..M.EN.R.SQV...Y.E.YPRSR.HVLLE.RRN. | 36.4 | 59.1 |
| bPAG2 | .VIL-..KKM.T..E..R.KN..NN..EEQAYRLSKNDS | 33.3 | 56.4 |
a aa—Amino acids, b—bovine, e—equine, f—feline, h—human, m—mouse, o—ovine, p—porcine, Cf—beaver; b Identical aa are dotted. Gaps (–) have been inserted to provide maximal alignments.
A putative cleavage position was predicted between PTL60–62 of the blocking pro-piece and VDE63–65 of the mature hPAG-L/pep precursor (Figure 1). Two Asp residues (D), specific for the catalytic cleft of AP were predicted at positions 94 aa in the NH2-terminus (VVFDTGSSNLWV91–102) and 277 aa in the COOH-terminus (AIVDTGTSLLTG274–285 Figure 1) of the hPAG-L precursor. The sequences of the NH2- and COOH-terminal domain of the hPAG-L/pep are identical to human pep A and very homologous to many other APs (Table 3). Surprisingly, no potential N-glycosylation site was predicted in the hPAG-L/pep precursor. In addition, in silico analyses permitted the identification of the molecular mass of the hPAG-L/pep polypeptide precursor (41.993 kDa) and its electrostatic property (pI 3.93).
Table 3.
Comparison of the aa sequence of NH2- and COOH-terminal domains in human PAG-L/pep polypeptide precursor to various aspartic proteinases.
| Gene Name a | NH2-Domain b | Identity (%) | COOH-Domain b | Identity (%) |
|---|---|---|---|---|
| hPAG-L/pep. | VVFDTGSSNLWV | this study | AIVDTGTSLLTG | this study |
| hPepsinogen A | ............. | 100 | ............ | 100 |
| hCathepsin D | ............ | 100 | .........MV. | 83.3 |
| hPepsinogen C | .L.......... | 91.7 | ...........V | 91.7 |
| hCathepsin E | .I.......... | 91.7 | .........I.. | 91.7 |
| CfPAG-L | .L.......... | 91.7 | G..........V | 83.3 |
| TrNothepsin | ........D... | 91.7 | .........IA. | 83.3 |
| hNapsin A | .A.......... | 91.7 | ..L......I.. | 83.3 |
| pPAG2, 4, 6, 10 | ........D... | 91.7 | ........MLH. | 75 |
| oPAG2 | ........D... | 91.7 | .L.......IH. | 75 |
| bPAG2 | .......A.... | 91.7 | .LL.....MIY. | 58.3 |
| hRenin A | .........V.. | 91.7 | .L....A.YIS. | 58.3 |
| mPepsinogen F | ..L.....V... | 83.3 | G.M......... | 83.3 |
| fPAG | .I......D... | 83.3 | ..I.......I. | 83.3 |
| ePAG | .I.....AD... | 75 | ..........L. | 91.7 |
| zPAG | .I.....AD... | 75 | ..........L. | 91.7 |
| pPAG1, 3, 5 | .I...A..D... | 75 | ..L.S.SAF.L. | 50 |
a aa—Amino acids, b—bovine, e—equine, f—feline, h—human, m—mouse, o—ovine, p—porcine, z—zebra, Tr—pufferfish, Cf—beaver; b Identical aa are dotted and aspartic acid (D) located within domain creating the substrate binding cleft is underlined.
2.3. Identification of the hPAG-L/pep Sequence within the Human Genome
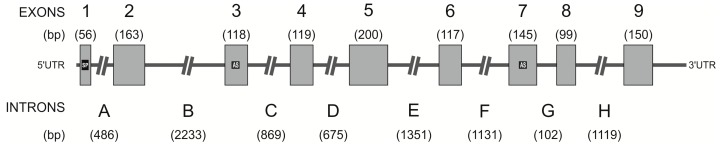
Homological primers (19 pairs) amplified six human gDNA templates, producing multiple hPAG-L/pep amplicons. Approximately 300 hPAG-L/pep gDNA sequenced amplicons were obtained. Only 197 chromatograms revealed high quality (HQ range: 40–98.2%) that were analyzed with Geneious® 8.1.7. A thorough analysis led to the identification of a 9330-bp genomic sequence of the entire hPAG-L/pep and deposition in the GenBank database (Acc. No. KX533473). The novel hPAG-L/pep gDNA sequence is composed of nine exons (56–200 bp) and eight introns (A–H; 102–2233 bp; Figure 2).
Figure 2.
Structural organization of the hPAG-L/pep gene. Exons (1–9) are boxed with their sizes shown in parenthesis above each box. The introns (A–H) are represented as lines with their sizes shown below each. Exons 3 and 7, which contain the catalytic aspartic acids at the active site, are shaded. The untranslated regions are represented by lines labeled 5′UTR and 3′UTR. Abbreviations: SP—signal peptide; AS—active site sequences coding domains 1 and 2 of the catalytic cleft.
The lengths of the hPAG-L/pep exons were generally very similar, or even the same, as in other PAGs (bPAG1: Acc. No. AH003454.1, bPAG2: Acc. No. NM_176614.1, pPAG2: Acc. Nos.: U39198–9; U41421–4; U39762–3; KF471015.1; KF492695.1; KF500427.1; KF527576.1; KF537535.1 and CfPAG-L: Acc. No. KX377932). However, the intron lengths varied, except for E and G introns, which were comparable (Table 4).
Table 4.
Exonic and intronic length of the hPAG-L/pep compared to bPAG1, bPAG2, pPAG2 and CfPAG-L genes.
| Sequence Length (bp) | |||||
|---|---|---|---|---|---|
| Gene Segment | hPAG-L/pep | bPAG1 | bPAG2 | pPAG2 | CfPAG-L |
| Exon 1 | 56 | 53 | 53 | 53 | 59 |
| Intron A | 486 | 1100 | 1300 | 1093 | 1937 |
| Exon 2 | 163 | 151 | 151 | 166 | 160 |
| Intron B | 2233 | 1000 | 1000 | 1324 | 385 |
| Exon 3 | 118 | 118 | 118 | 118 | 118 |
| Intron C | 869 | 100 | 100 | 90 | 917 |
| Exon 4 | 119 | 119 | 119 | 119 | 119 |
| Intron D | 675 | 1200 | 1200 | 1124 | 451 |
| Exon 5 | 200 | 194 | 194 | 200 | 200 |
| Intron E | 1351 | 900 | 1100 | 927 | 1138 |
| Exon 6 | 117 | 117 | 117 | 117 | 123 |
| Intron F | 1131 | 1900 | 1700 | 1455 | 288 |
| Exon 7 | 145 | 142 | 142 | 136 | 148 |
| Intron G | 102 | 100 | 100 | 85 | 681 |
| Exon 8 | 99 | 99 | 99 | 99 | 99 |
| Intron H | 1119 | 1700 | 1700 | 292 | 603 |
| Exon 9 | 150 | 150 | 150 | 156 | 147 |
| Total length | 9133 | 9143 | 9343 | 8031 | 7573 |
Two coded D residues within both domains creating a catalytic cleft (a feature of the AP members) were localized within exons 3 and 7 of the hPAG-L/pep (Figure 2). All exon-intron junctions with 5′ donor and 3′ acceptor sites were identified (Table 5). The sequences between each of the intron-exon junctions were determined and conformed to the standard gt-ag rule for 5′ donor and 3′ acceptor sites. The de novo identified hPAG-L gene is composed of: 24.7% A; 27.4% C; 25.6% G; 22.3% T and 53.0% G+C (Geneious® 8.1.7.).
Table 5.
Characteristics of exon-intron junctions within the hPAG-L/pep gene.
| Donor Splice Sites | Acceptor Splice Sites | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Exon | 5′→3′ | Phase | Intron | 5′→3′ | Intron | 5′→3′ | Phase | Exon | 5′→3′ |
| 1 | TCATGTACAA | 0 | A | GTGAGTCCGG | A | CAAACCACAG | 2 | 2 | GGTCCCCCTC |
| 2 | CTACCTGGAT | 0 | B | GTGAGTGTGC | B | GCCTGGACAG | 0 | 3 | ATGGAGTACT |
| 3 | CTTGCCTGCA | 1 | C | GTAAGTGCCC | C | GTCCTTGCAG | 1 | 4 | CCAACCACAA |
| 4 | CACTGTCCAG | 0 | D | GTGGGCACCT | D | CCCCACCCAG | 0 | 5 | GTTGGAGGCA |
| 5 | ACCTCAGCGC | 2 | E | GTAAGTTGAG | E | CTTTCCACAG | 2 | 6 | CGATGACCAG |
| 6 | CCGTGGACAG | 2 | F | GTGAGACTGC | F | TTGCCCTCAG | 2 | 7 | CATCACCATG |
| 7 | AGATGGCGAC | 0 | G | GTGAGTCCAG | G | CTCTTTCCAG | 0 | 8 | ATGGTGGTCA |
| 8 | CATCCTGCAG | 0 | H | GTGAGGAGGC | H | TTTTCTCCAG | 0 | 9 | AGCGAGGGGA |
Megablast alignments of the hPAG-L/pep gDNA revealed the highest homology with human sequences with various identity (ID) and QC: pepsin-L (e.g., pep 3, 4 and pepsin A; ID 98–100%, 3–15% QC), chromosome 11 BAC and FOSMID clones (99%), pepsin/pep A–L sequences in different species: e.g., Pan troglodytes (98% ID, 20% QC), Macaca fuscata (92% ID, 26% QC) and Papio anubis (92% ID, 18% QC). Furthermore, pairwise alignments (Geneious® 8.1.7) of the hPAG-L/pep indicated higher homology of each exon (52.1–78.6%) and lower intronic homology (25.4–58.5%) with other PAGs identified previously in different species (Table 6).
Table 6.
Homology of the hPAG-L/pep exons and introns with bPAG1, pPAG2 and CfPAG-L genes.
| Pairwise Identity (%) | |||
|---|---|---|---|
| hPAG-L/pep | bPAG1 | pPAG2 | CfPAG-L |
| Exon 1 | 78.6 | 75.5 | 63.3 |
| Intron A | 49.8 | 51.1 | 50.3 |
| Exon 2 | 52.1 | 58.8 | 59.6 |
| Intron B | 51.7 | 50.7 | 52.7 |
| Exon 3 | 69.5 | 74.6 | 71.2 |
| Intron C | 55.6 | 54.5 | 25.4 |
| Exon 4 | 59.7 | 58.4 | 64.7 |
| Intron D | 52.5 | 52.2 | 52.7 |
| Exon 5 | 58.5 | 65.3 | 65.5 |
| Intron E | 51.3 | 53.3 | 50.9 |
| Exon 6 | 61.9 | 69.7 | 56.9 |
| Intron F | 51.2 | 51.0 | 50.3 |
| Exon 7 | 62.0 | 63.3 | 64.7 |
| Intron G | 58.5 | 54.2 | 54.7 |
| Exon 8 | 67.7 | 66.7 | 70.3 |
| Intron H | 52.9 | 52.3 | 51.8 |
| Exon 9 | 58.7 | 61.4 | 61.5 |
2.4. Identification of Placental hPAG-L/pep Protein
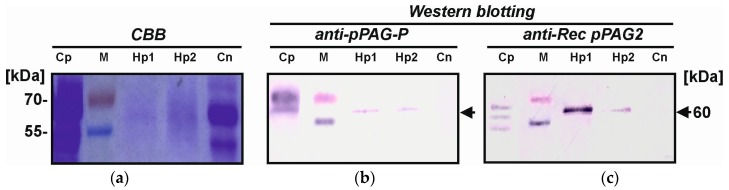
Isolated cellular placental proteins (2.61–5.15 µg/µL) allowed identification of hPAG-L/pep profiles specific for the term gestation. Heterologous Western blotting with either anti-pPAG-P or anti-Rec pPAG2 polyclonals revealed only one dominant 60 kDa hPAG-L protein, similar in molecular mass to a positive control of multiple porcine placental proteins originated from in vitro culture of 77 dpc-chorionic explants (Figure 3). A lack of any signals in the negative control (secretory endometrial proteins; 10 day of cycle) confirmed the correctness of the immunoblotting.
Figure 3.
Identification of cellular human hPAG-L term placental proteins (10 μg/sample) separated by (a) SDS-PAGE and stained by Coomassie Brilliant Blue (CBB); Heterologous Western blottings with (b) anti-porcine PAG-P polyclonals (1:300); (c) Western analysis with anti-Rec pPAG2 (1:50). Abbreviations: Cp—positive control (porcine secretory chorionic proteins; 77 dpc); M—molecular marker; Hp1 and Hp2—human placental proteins; Cn—negative control (secretory endometrial proteins; 10 day of cycle). Arrow indicates a dominant hPAG-L isoform.
2.5. Identification of Cellular hPAG-L/pep Localization
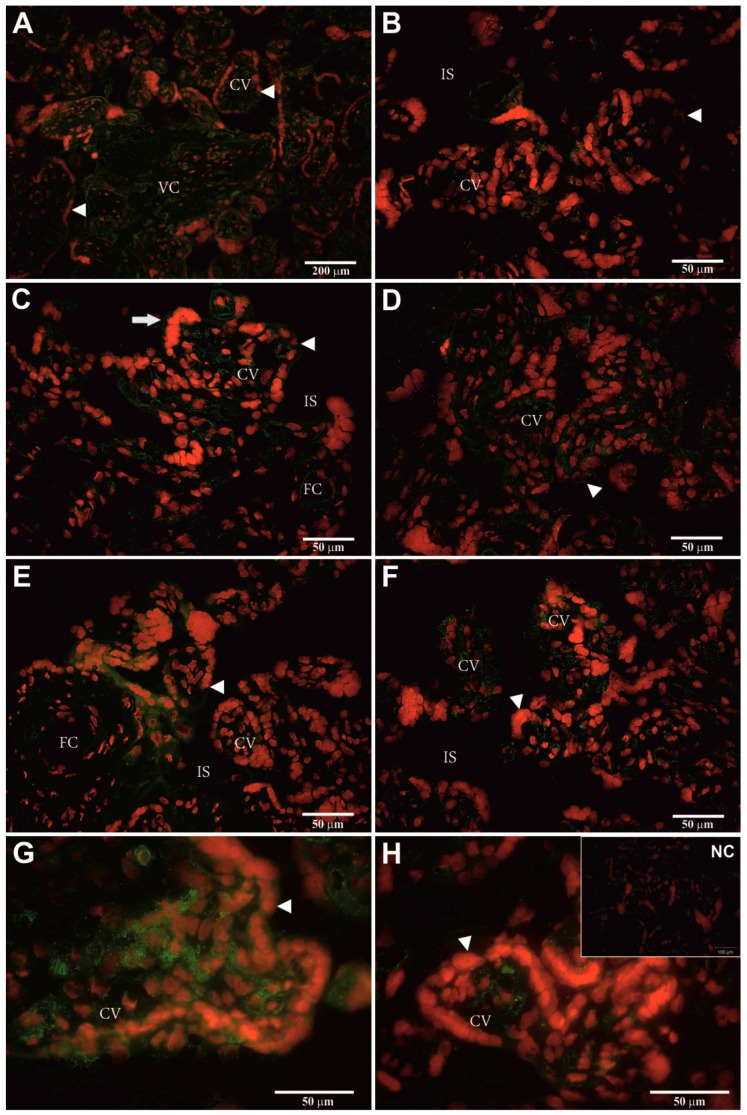
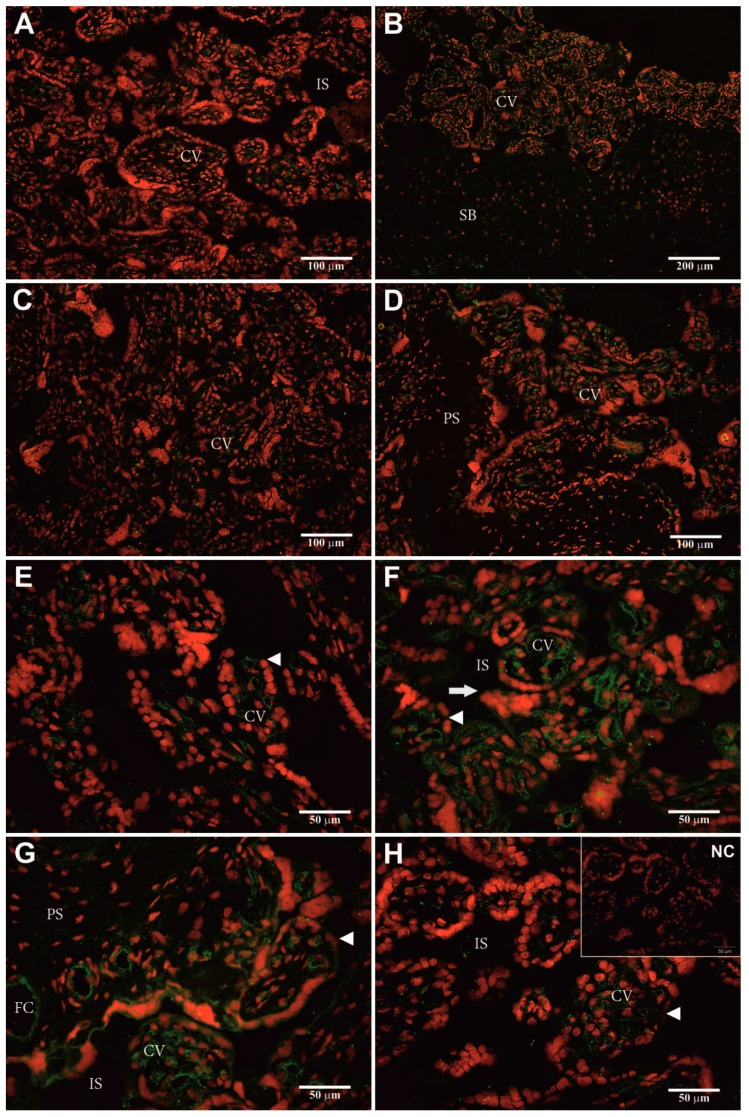
Heterologous dF-IHC with anti-pPAG-P (Figure 4A–H) and anti-Rec pPAG2 polyclonals (Figure 5A–H) allowed localization of the hPAG-L/pep proteins within term placental cells. Generally, stronger immuno-positive signals of the hPAG-L/pep (green) were identified with anti-Rec pPAG2 than anti-pPAG-P polyclonals. The strongest immune-positive hPAG-L/pep signals were observed within the analyzed regions of chorionic villi (CV) and villous core (VC), especially within the syncytiotrophoblast nuclei (red) covering the surface of the terminal villous tree (arrowheads in Figure 4A–H and Figure 5E–H). The hPAG-L/pep signals were also detectable (Figure 4E and Figure 5G) within VC surrounding fetal capillaries (FC) close to the intervillous space (IS). No hPAG-L/pep signals were immuno-detected within placental septa (PS), which were infiltrated by decidual cells (Figure 5D) or within the maternally-originated stratum basale (SB; Figure 5B). A negative control did not generate any signal (Figure 4H and Figure 5H).
Figure 4.
Heterologous immuno-localization of the hPAG-L/pep proteins within sections of a human term placenta identified with polyvalent anti-porcine PAG (anti-pPAG-P) polyclonals (A–H), visualized by goat anti-rabbit IgG-conjugated with Alexa 488 fluorophore (green) among all nuclei stained by propidium iodide (red). Human placenta section—used as negative control (NC; insert in H) with omitted polyvalent anti-pPAG-P polyclonals. The size bars are 50 µm (B–H), 100 µm (NC) and 200 µm (A). Abbreviations: arrowheads—syncytiotrophoblast; arrows—clusters of the syncytiotrophoblast nuclei; CV—chorionic villi; VC—villous core; IS—intervillous space; FC—fetal capillary.
Figure 5.
Heterologous immuno-localization of the hPAG-L/pep proteins within sections of human term placenta identified with recombinant anti-porcine PAG (anti-Rec pPAG2) polyclonals (A–H), visualized by goat anti-rabbit IgG-conjugated with Alexa 488 fluorophore (green) among all nuclei stained by propidium iodide (red). Human placenta section—used as a negative control (NC; insert in H) with omitted polyvalent anti-pPAG-P polyclonals. The size bars are 50 µm (E–H and NC), 100 µm (A,C,D) and 200 µm (B). Abbreviations: arrowheads—syncytiotrophoblast; arrows—clusters of the syncytiotrophoblast nuclei; IS—intervillous space; CV—chorionic villi; SB—stratum basale; PS—placental septa; FC—fetal capillary; NC—negative control.
3. Discussion
This study presents pioneering data concerning identification of the human placental PAG-L/pep cDNA (Acc. No. KX856064) and protein (60 kDa). In addition, the exonic-intronic structure of entire hPAG-L/pep gene has been identified (Acc. No. KX533473). Direct comparison of our data is impossible because similar data are not available. Therefore, our data can only be compared to animal species in which the PAGs have already been identified.
3.1. Identification of hPAG-L Transcript
The novel 1364 bp hPAG-L/pep cDNA, identified with term placental mRNA, allowed identification of nucleotide homology (at least 56.5%) with other AP members. Previously, nucleotide sequences of the PAG cDNAs have only been identified in cattle, sheep, pig, goat, horse, zebra, white-tailed deer, water buffalo, American bison, wapiti, giraffe and the Eurasian beaver [1]. Such a limited number of cloned cDNAs resulted from difficulties during the wild eutherian placenta harvesting required for high-quality RNA and cDNA library. The numbers of the PAG-L cDNAs vary between species and are multiple in cattle, sheep, goats and pigs, while a single PAG-L cDNA has been identified in the horse, zebra, mouse, cat and beaver [1,2,20]. A possible explanation of this fact might be various placenta types, different requirements for the development of the fetus and special environmental needs in different species.
The identified cDNA allowed an encoded 388 aa hPAG-L/pep polypeptide precursor (Figure 1; Table 1, Table 2 and Table 3) to be characterized, which contains 15 aa-signal peptide, 47 aa-blocking peptide and 326 aa-mature polypeptide, making it similar to other PAG precursors. Among the identified PAGs, the length of the 15 aa-signal peptides, as well as the 33–38 aa-blocking pro-pieces are very conservative in various species [21,32,33,34]. Different PAG precursors [1] vary in their entire length (375–389 aa), molecular mass (30–90 kDa) and electrostatic properties (4.0–9.08 pI). Our in silico analyses of the hPAG-L/pep precursor (41.977 kDa; Ip = 3.93 pH) also contributed to the enlargement of the diversity and confirmed membership in the PAG family. The identified hPAG-L/pep precursor was also similar to peps that are composed of 15–16 aa signal peptides, 42–46 aa activation segments and 321–332 aa of mature proteins [5,35].
We are aware that the identified placental hPAG-L/pep transcript is identical to another human AP (pep A); thus, our hPAG-L should be classified as an catalytic active form. Presently, we can expect that the hPAG-L/pep and pep A are similarly activated by degradation of placental or gastric polypeptide precursors due to an identical blocking peptide sequence (Table 2). Such expectation may confirm equal/similar aa homology (56.4–67.6%) of hPAG-L/pep with peps C and F as well as other catalytically active PAGs (fPAG, pPAG2, CfPAG, ePAG and bPAG2).
High N-glycodiversity is very common in the PAGs [1], but in pepsinogens it occurs occasionally and no more than two N-glycosylation sites are generally present [36,37]. Surprisingly, within the aa sequence of the hPAG-L/pep precursor, no potential sites of N-glycosylation were predicted. Thus, it confirms that the hPAG-L/pep precursor is different from pep A.
Due to the conserved sequences of two aspartic acids (D) located within two domains (NH2- and COOH-terminal), creating the substrate binding cleft, the hPAG-L/pep precursor was classified as a catalytically active AP member, similar to human pep A (Figure 1; Table 3). The PAG-Ls identified in the mouse [38], horse, zebra, cat [21,39] and beaver are also classified as active APs [20]. Within the diversified PAG family in species with multiple PAG members, either potentially active as well as potentially inactive forms exist [1,2,32,40,41]. Multiple aa substitutions within both domains contribute to a loss of catalytic activity of many PAGs [1,3,33,42].
Presumably, some similarities of the PAG-L family in the human and some Rodentia species (beaver or mouse) resembled discoid placenta type and potentially comparable requirements of developing fetuses in those taxa.
Most PAG/PAG-L cDNAs share relatively a higher nucleotide homology with each other than to pepsinogens [2]. Interestingly, the hPAG-L/pep shares higher homology with pepsinogens than other PAGs, similar to CfPAG-L and ePAG [20,21]. Pepsins were initially considered to be restricted to the stomach of many vertebrates [5]; however, in lower vertebrates, progastricsin (also known as pep C) was also found in the esophageal mucosa of the frog [43] as well as larval pepsinogen cDNA in whole bodies of the pufferfish [44]. Phylogenetically, peps F and PAGs belong to the same cluster [2,5]. The high (99%) homology of the identified hPAG-L/pep cDNA to the human pep A indicates that both genes are very similar but are two related AP genes with completely different expression.
3.2. Identification of hPAG-L Exonic-Intronic Structure
Because data concerning PAGs in human genome are not available, the presently obtained results can only be compared to studies performed in some animal species. The entire identified hPAG-L/pep gene sequence (9330 bp; Acc. No. KX533473) comprises a structure of nine exons and eight introns (Figure 2). The location of the two D residues, within exons 3 and 7 of hPAG-L/pep, is similar to other PAGs, which is specific for the catalytic cleft of all APs. To date, the entire structure of the PAG genes has only been identified in three species (cattle, pig and beaver). The length of the hPAG-L/pep exons (1–9) is greatly similar to exon lengths of bPAG1, bPAG2, pPAG2 and CfPAG (Table 4) or even the same (especially exons: 3, 4, 8), whereas other exons vary. The gDNA alignments of the hPAG-L/pep exons with bPAG1, pPAG2 and CfPAG-L revealed high homology in the range of 52.1–78.6% (Table 6). However, the length of the hPAG-L/pep introns (A–H) varied from previously discovered PAGs, except in the length of intron G for hPAG/pep and bPAG1 or bPAG2, as well as their total lengths (Table 4). Furthermore, the pairwise sequence alignment of intronic regions in the aforementioned PAGs revealed generally lower homology (25.4–58.5%; Table 6).
Previously, Southern hybridization of gDNA (with selected restrictases) revealed a diversified number of the PAG-L genes in some eutherian species, e.g., the elk, yak, wildebeest, impala, several other antelopes [33], the pig, goat, horse, cow, sheep, deer and wild boar and bisons [1]. Southern blot of amplicons also revealed the PAG-L family in the alpaca, the dromedary and the Bactrian [45]. Sequence identification and comparison of cDNA and gDNA enabled defining the exonic-intronic boundaries for only four PAGs. So far, multiple bovine PAG cDNAs [33,42] have allowed the identification of the bPAG1 gene (8095 bp) as the first representative with an identified exon-intron structure, with an intron length ranging from 87 bp to 1.8 kbp [17]. Identification of the pPAG1 and pPAG2 cDNAs [32] has also led to identification of the pPAG2 gene structure [19]. The pPAG2 belongs to the pPAG2-L subfamily together with other members: pPAG4, 6, 8, 10 and they constitute catalytically active APs. However, potentially inactive members of the pPAG1-L subfamily, pPAG3 and 5, have also been identified [40]. The pPAG2 structure [19] encompasses nine exons (99–200 bp) and eight introns (A–H; 85–1.8 kbp). The entire length of the pPAG2 with a promoter region is equal to 8755 bp [19,46]. Recently, CfPAG-L (Acc. No. KX377932) was discovered in the Eurasian beaver (7657 bp) as an AP member containing nine exons and eight introns [20]. The lengths of the hPAG-L/pep (56–200 bp), as well as CfPAG-L (59–200 bp) exons, are similar to the other known bPAG1, bPAG2 and pPAG2, although the length of the introns differ from previously identified PAG.
Since the results obtained in this study are consistent with the exon-intron structures (length and homology alignment) of four previously described PAGs and other APs, the identified hPAG-L/pep was assuredly classified as a new AP member. However, the high homology of the hPAG-L/pep to the pep A family in various species is also a novel finding. The multigenic AP family is widely distributed in various taxa and emerged from duplication or fusion of the paralogous progene [5,6]. In mammals, the major AP members are well-known pepsinogen genes classified as A, B, C and fetal forms known as pepsinogen F [35,47]. Complete gene structures have been determined, e.g., for human pep A [48], C [49] and prochymosin [50]. The structure is conserved among APs, including PAGs, pepsinogens, cathepsins D, E, and renin, suggesting evolution from a common ancestral gene [51].
3.3. Identification of hPAG-L Proteins
Western blotting (Figure 3), with anti-pPAG-P and anti-Rec pPAG2 polyclonals, identified a uniform cellular protein profile of native hPAG-L/pep isoform (60 kDa) in term singleton placentas. Similar data are unfeasible in humans. In animals, multiple heterogeneous secretory PAG isoforms, 43–70 and 45–85 kDa released by placental explants, have been found in the pig and cattle, respectively [34,52]. In the pig, gestation-stage dependent diversity of glycosylated forms of the pPAG proteins occurs, which contain an average of 9.66% of N-linked oligosaccharides [53]. In the bPAG family, oligosaccharide heterogeneity is caused by diversified tetra-antennary glycans [54]. In addition, three purified PAG isoforms (72, 74 and 76 kDa) secreted by the placenta of the American bison have been sequenced [55]. However, in the European bison, among two major groups (43–45 and 67–69 kDa) of immuno-detected secretory PAG isoforms [52], eleven novel diversified pregnancy-stage dependent (45–129 day post coitum–dpc) EbPAGs (50–71 kDa) have been sequenced [56]. Various PAG isoforms also exist in species of the Cervidae order: 33–55 kDa in the white-tailed deer [41], 39–62 kDa in the fallow deer [57], and dominant 55 kDa fraction-specific isoforms for different pregnancy stages (50–200 dpc) in the European moose [58]. It seems that such diversity of multiple PAGs originated from gene duplication during the evolution of different species.
3.4. Identification of Cellular hPAG-L Localization
Double-labeling heterologous immuno-detection revealed the strongest positive hPAG-L signals within the chorionic villi, localized especially within the syncytiotrophoblast cells (Figure 4 and Figure 5). Similar data are unachievable in humans. Localization of the hPAG-L expression may be directly compared only with results for the beaver (a discoid-placenta type species) in which CfPAG-L signals are found either in regular or giant trophectodermal cells [59].
In other animals, cellular expression of the PAGs was previously mostly localized in embryo-originated chorionic cells in some species of the Artiodactyla order, with cotyledonary (bovine, bison, white-tailed deer, moose) or the diffuse (porcine, alpaca, camels) placenta types, as described below. In the pig, pPAG expression is restricted to diversified chorionic cell layers throughout (16–61 dpc) placenta development [60]. In ruminants, multi-nucleated, enlarged and multi-granulated trophectodermal cells expressing PAGs have been observed in the white-tail deer [1], while in placentomes of the European bison, the EbPAGs were localized in apical regions of cotyledonary villi folds [61]. Similarly, in camelids during alpaca pregnancy (150–347 dpc), Lama pacos–LpPAGs are present in the trophectoderm cell layer and within very rare giant cells [62]. In the term placenta of both camels, CdPAG (Camelus dromedarius—dromader) and CfPAG (Camelus ferrus—Bactrian) are present in the cytoplasm of the outer folded layer of the mononuclear trophectodermal cells, mostly at the apex of the placental folds [63]. In the moose (Alces alces), AaPAG-L signals are related to placentome growth (50–200 dpc) and are localized in the trophectodermal cells, especially within secretory granules [58]. Despite the morphological and developmental divergences of various placenta types, the localization of the hPAG-L/pep resembled chorionic expression previously determined in other mammals.
This study describes pioneering identification of novel aspartic proteinase named hPAG-L/pep in the genome (Acc. No. KX533473; 9330 bp), placental transcriptome (Acc. No. KX856064; 1364 nt mRNA) and proteome (60 kDa) of the human. The expression of the hPAG-L/pep in chorionic cells can influence the regulation of placental development. The identified placental glycoprotein presumably can be used as a novel biomarker for prenatal pregnancy diagnosis of embryo/fetus well-being by a noninvasive test based on concentration measurement in peripheral maternal blood, similar to β-hCG test in the human as well as various PAG tests in the ruminants. In addition, the identified cDNA (ORF) and 9-exonic and 8-intronic gDNA sequences provide a major pattern of SNPs/InDels required for a novel marker preparation profitable for genotyping and detection of various genomic disorders in embryo/fetus and mother, similar to our report on SNPs/InDels for the pig [46,64] and the European moose [58]
4. Materials and Methods
4.1. Ethics Statement and Collection of Samples
All clinical samples (placentas and blood) were collected at the Clinical Ward for Gynecology, Obstetrics and Oncological Gynecology at the Regional Specialist Hospital in Olsztyn, following informed written consent from the parturient women. The study protocol was approved by the Bioethics Committee of the Warmia-Mazury Medical Chamber (OIL.164/15/Bioet; 2 April 2015) in Olsztyn, Poland. Only healthy mothers, after uncomplicated single pregnancy and without diagnosed medical conditions were selected for this study. Term placentas (n = 2) were collected from the women who underwent scheduled Caesarean section. Whole blood samples from both men (n = 3) and women (n = 3) were also collected from jugular veins. Placental tissues were immediately preserved in liquid nitrogen and blood samples were placed on ice and transported directly to the laboratory. Samples of blood were centrifuged (3500× g) for 30 min at 4 °C, plasma was discarded and the buffy coat of the white cells, as well as placental tissues were stored at −70 °C until further analyses.
4.2. Total RNA Extraction
Total RNA was isolated from term placental fragments, using a QiagenRNeasy Kit in conjunction with the QiagenRNase-Free DNase Set (Qiagen, Hilden, Germany), according to the manufacturer’s recommendations. RNA quality was evaluated via microfluidic electrophoresis (2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA, USA). Only a high RNA integrity number (RIN > 8.0) of each sample was accepted for high throughput mRNA sequencing (RNA-seq).
4.3. High Throughput mRNA and Bioinformatics
Complementary DNA (cDNA) libraries were constructed using the protocol of TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) involving the following steps: RNA purification and fragmentation, synthesis of the first and the second strand of cDNA, 3′ adenylation and adaptor ligation. After amplification and quantification (KAPA Library Quantification Kit, Illumina), the cDNA libraries were indexed, diluted and pooled in equimolar ratios.
The paired-end sequencing was performed on the HiSeq2500 (Illumina). The quality of raw reads (2 × 100 bp reads) was controlled by FASTQC software v.0.11.2 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). FLEXBAR software v.2.5 (https://github.com/seqan/flexbar) was used for trimming the Illumina adaptors and poly(A) stretches. All reads shorter than 32 bp and reads with a PHRED quality score lower than 10 were then disqualified from the dataset. The trimmed reads were used for de novo assembly with TRINITY software v.r20140717 (https://github.com/trinityrnaseq/trinityrnaseq/releases). To select potential human PAG-L (hPAG-L) sequences, reconstructed contigs were searched against a database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequencing data from this study have been submitted (http://www.ncbi.nlm.nih.gov/sra) to the NCBI Sequence Read Archive (SRA) under Accession no. BioProject ID: PRJNA326064.
4.4. Capillary Sequencing
Capillary sequencing was performed to confirm the coding sequence of the placental AP, identified by RNA-Seq. Briefly, total RNA (from the same samples that were used for RNA-Seq) was transcribed to cDNA in two-step RT-PCR using an Enhanced Avian HS RT-PCR Kit (Sigma-Aldrich, USA). The first strand cDNA was synthesized with dNTPs, and random hexamers were used as primers. PCR amplification of target cDNA templates to obtain hPAG-L amplicons was performed with specific primers (Table 7) designed by applying Geneious® 8.1.7 software (Biomatters Ltd., Auckland, New Zealand) and Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html), basing on PAG-L sequence identified using RNA-Seq.
Table 7.
Specific primers applied for the amplification of human PAG-L cDNA.
| Primers Name | Sequence (5′–3′) | Position (bp) a | Amplicon Length (bp) a | |
|---|---|---|---|---|
| 1 | MMstart | AGTTGGGACCCGGGAAGA | 1–18 | 1363 |
| MMutrR | TCCACAAAACCTGTTTCAGTG | 1343–1364 | ||
| 2 | MM2s | TCATCAGAAAGAAGTCCTTGAG | 85–106 | 496 |
| MM5as | TAGGCCAGSCCCAKGATGCCATC | 558–580 | ||
| 3 | MM3s | GCTCCTCCAACCTGTGGGT | 307–325 | 560 |
| MM7as | CAGAGAGGTGCCKGTGTCMACAA | 844–866 | ||
| 4 | MM5s | GATGGCATCMTGGGSCTGGCCTA | 558–580 | 564 |
| MM9as | GAAGACATCWCCMAGGATCCAA | 1100–1121 | ||
| 5 | MM7s | TTGTKGACACMGGCACCTCTCTG | 844–866 | 520 |
| MMutrR | TCCACAAAACCTGTTTCAGTG | 1343–1364 | ||
a Position and amplicon length was estimated according to the human PAG-L cDNA sequence identified using RNA-seq.
The obtained amplicons of examined hPAG-L, parallel to porcine PAG10 (pPAG10) cDNA—used as a positive control and negative control (without templates)—were separated in 1.5% agarose gels, along with a marker (100–3000 bp; Thermo Fisher Scientific, Waltham, MA, USA), UV-visualized using Midori Green Nucleic Acid Staining Solution (NIPPON Genetics Europe GmbH, Dueren, Germany) and archived (G:Box, SynGen, Sacramento, CA, USA). Gel-out purified hPAG-L amplicons were used as templates for capillary sequencing (3130 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA) in both sense and antisense directions. Amplicon labeling was performed with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), under the following conditions: initial denaturation (at 96 °C for 1 min) and 30 cycles of amplification (96 °C/10 s, 50 °C/5 s, 60 °C/4 min). Each labeling mix (20 μL) was composed of 12 μL (5–10 ng) of amplicon template, 1.2 μL Ready Reaction Mix, 4 μL BigDye Terminator v1.1/3.1 Sequencing buffer (5×), 2 μL of each primer and 0.8 μL H2O. The labeled hPAG-L amplicons were purified with the BigDye X Terminator Purification Kit (Applied Biosystems) and separated in capillaries filled with POP-7™ polymer (Applied Biosystems). The obtained hPAG-L sequences were analyzed by Geneious® 8.1.7. In addition, in silico analyses of the cDNAs were performed applying the following online tools: http://www.cbs.dtu.dk/services/SignalP/; http://prosite.expasy.org; http://www.cbs.dtu.dk/services/NetNGlyc/.
4.5. Genomic Identification of the hAP/PAG-L Sequence
Genomic DNA (gDNA) templates (n = 6) were isolated from the leukocytes with the use of a commercially available kit (Sherlock AX, A&A Biotechnology, Gdynia, Poland). Only high quality gDNA templates (700 ng) were used for PCR amplifications of the hPAG-L gene fragments. In order to identify either initial or partial nucleotide sequence of the hAP/hPAG-L, the gDNA amplicons were produced with 19 pairs of specific homologous primers (Table 8), designed on the basis of the hPAG-L cDNA sequence originating from the aforementioned RNA-seq. JumpStart™ Taq ReadyMix™ (Sigma-Aldrich, St. Louis, MO, USA) was used for efficient PCR amplification, under the following conditions: initial activation (95 °C/2 min), followed by 40 following cycles: 95 °C/1 min for the denaturation of gDNA templates, 60 °C for primer annealing (1 min) and 72 °C/4.5 min for amplicon synthesis. The obtained hPAG-L gDNA amplicons were electrophoresed, gel-out purified, subjected to capillary sequencing and analyzed as described above (see Capillary sequencing). Identified cDNA and gDNA sequences of hPAGL-L have been deposited in GenBank (Accession nos: KX856064 and KX533473, respectively
Table 8.
Specific primers applied for identification of the human PAG-L genomic sequence.
| Primers Name | Sequence (5′–3′) | Position (bp) a | Amplicon Length (bp) a | |
|---|---|---|---|---|
| 1 | MMstart | AGTTGGGACCCGGGAAGA | 1–18 | 725 |
| MM2as | ATCCAGGTAGTTCTCCAGGG | 706–725 | ||
| 2 | MM2s | TCATCAGAAAGAAGTCCTTGAG | 571–592 | 1789 |
| MMintronBr | ATTCTCCTGCCTCAACCTCCCAA | 2337–2359 | ||
| 3 | MMintronB | CTCCGCATAGCCTGATCCCTT | 1180–1200 | 1180 |
| MMintronBr | ATTCTCCTGCCTCAACCTCCCAA | 2337–2359 | ||
| 4 | MMintronB3 | CCTCCTGCAGATATTGTATGTCC | 1429–1451 | 1616 |
| MM3as | ACCCACAGGTTGGAGGAGCC | 3025–3044 | ||
| 5 | MMintronB2 | TGTGAGGAATGAAGGAAAAGATGG | 2840–2863 | 1533 |
| MMintronDr | GGTGCTGCATGTCGGGAGAA | 4353–4372 | ||
| 6 | MMintronC | GCTGTAGAATAGCCCACCAGG | 3381–3401 | 992 |
| MMintronDr | GGTGCTGCATGTCGGGAGAA | 4353–4372 | ||
| 7 | MMintronC | GCTGTAGAATAGCCCACCAGG | 3381–3401 | 1663 |
| MMintronEr | AAGACCCTCTCCATCGCACCCA | 5022–5043 | ||
| 8 | MMintronD(N) | AGTCCTGCATGAGATGAACCA | 4636–4656 | 1284 |
| MMintronEr3 | CTTAAGGACTTGAGGGTGGAGGTC | 5896–5919 | ||
| 9 | MMintronE3 | GCACAACTCAAATGTCATCAGCCA | 5178–5201 | 742 |
| MMintronEr3 | CTTAAGGACTTGAGGGTGGAGGTC | 5896–5919 | ||
| 10 | MMintronE3 | GCACAACTCAAATGTCATCAGCCA | 5178–5201 | 1325 |
| MMintronFr2 | CTGGGGGGATTCTGGAAAGCTGA | 6480–6502 | ||
| 14 | MM6sens | AGTGGCAGCGTGGTGATCTTTG | 6301–6322 | 1311 |
| MM7as | CAGAGAGGTGCCKGTGTCMACAA | 7589–7611 | ||
| 15 | MMintronF2 | TCAGCTTTCCAGAATCCCCCCAG | 6480–6502 | 1132 |
| MM7as | CAGAGAGGTGCCKGTGTCMACAA | 7589–7611 | ||
| 16 | MMintronF | TGGATGGGTGGGGAAGAAATGT | 7438–7459 | 1650 |
| MM9as | GAAGACATCWCCMAGGATCCAA | 9066–9087 | ||
| 17 | MM8s | GACATCGTCTTCACCATCAAT | 7822–7842 | 1266 |
| MM9as | GAAGACATCWCCMAGGATCCAA | 9066–9087 | ||
| 18 | MMintronF | TGGATGGGTGGGGAAGAAATGT | 7438–7459 | 405 |
| MM8as | ATTGATGGTGAAGACGATGTC | 7822–7842 | ||
| 19 | MM992 | GAGCTGCATCAGTGGCTTCC | 9012–9031 | 318 |
| MMutrR | TCCACAAAACCTGTTTCAGTG | 9309–9329 | ||
a Position and amplicon length was estimated according to the human PAG-L gDNA identified using capillary sequencing.
4.6. Identification of the Exon-Intron Organization of the hPAG-L
To estimate the length of the introns and exons in the hPAG-L, the identified sequences were analyzed by NetGene2 v. 2.4 software (www.cbs.dtu.dk/services/NetGene2/) to predict a structure, based on multiple alignments (Geneious® 8.1.7, www.geneious.com and BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi) of the entire hPAG-L genomic sequence with the identified cDNA (see above).
4.7. Cellular Placental Protein Extraction
Cellular proteins were isolated as previously described for other species [58,65]. Briefly, the frozen human placental tissues (n = 2) were homogenized on ice and lysed by alkaline buffer (Total Protein Extraction Kit, Genoplast, Rokocin, Poland). The obtained protein homogenates of each placental sample were concentrated (3000 rpm/4 °C) by ultra-filtration in Centriprep-10 cartridges (>MWCO 10 kDa; Amicon, Billerica, MA, USA) until a 0.5 mL of final volume was received. Total protein concentration was determined by the standard Bradford procedure. Concentrated placental proteins (10 µg/line) were separated by denaturing polyacrylamide electrophoresis (SDS-PAGE, 12.5% gels), parallel to porcine placental proteins (positive control for Western blotting), endometrial proteins of cyclic pigs (negative control) and a molecular marker (10–250 kDa; Fermentas, Waltham, MA, USA). Electrophoresed proteins were stained with Coomassie Brilliant Blue (CBB) to identify total human placental protein profiles.
4.8. Western Blotting
Duplicates of PAGE-separated human cellular proteins, porcine secretory chorionic (positive control; 77 dpc) and porcine secretory endometrial proteins (negative control; 10 day of cycle) were transferred onto 0.45 µm nitrocellulose membranes (Optitran BA-S58, Whatman, GE Healtcare Life Science, Issaquah, MA, USA) and then analyzed by Western blotting to identify human placental PAG-L fractions by the heterologous—ht (cross-species) immuno-detection described previously [66,67]. Briefly, blotting was performed with primary rabbit polyvalent anti-pPAG polyclonals, raised against various porcine antigens (anti-pPAG-P; 1:300) and against recombinant pPAG2 antigen (anti-Rec pPAG2; 1:50). Immuno-complexes were identified by secondary mouse anti-rabbit IgG monoclonals–conjugated with alkaline phosphatase (1:100,000). The immuno-complexes were visualized with the use of nitro blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) as standard substrates for alkaline phosphatase activity detection. Gels and blots were photographed and archived (GBox, SynGen, Sacramento, CA, USA). Methodological details, as full description and validation methods of applied polyclonals, are provided in Supplementary material.
4.9. Heterologous Double Fluorescent Immunohistochemistry (htdF-IHC)
Placental explants were cryo-sectioned (−20 °C; 6 μm), fixed, dehydrated and then subjected to htdF-IHC, as previously described [61,62,63]. Briefly, the htdF-IHC was performed with the aforementioned anti-pPAG-P (1:300) and anti-Rec pPAG2 (1:50) polyclonals. Parallel negative controls were performed without the primary antisera. Double immunostainings were visualized with secondary goat anti-rabbit polyclonals (1:1000)–conjugated with Alexa 488 fluorophore (A488; green) and all nuclei of placental cells were stained with propidium iodide (PI; red).
5. Conclusions
Our discerning and comprehensive studies provide novel data identifying the placental hPAG-L/pep transcript, gene structure and chorionic protein in humans. Our pioneering data extend the present knowledge of the human genome, placental transcriptome and proteome, which may contribute to establishing a new diagnostic tool for examination of various disturbances during human pregnancy, with growing interest from both scientific and clinical perspectives.
Acknowledgments
We are very grateful to Jan P. Jastrzebski and Lukasz Paukszto, the Department of Plant Physiology, Genetics and Biotechnology, Faculty of Biology and Biotechnology, University of Warmia and Mazury in Olsztyn, for guidance during the bioinformatics analysis. This study was supported by the Department of Human Physiology, Faculty of Medical Science (25.610.001-300) and by the Department of Animal Physiology, Faculty of Biology and Biotechnology (12.610.005-300), UWM Olsztyn.
Abbreviations
| PAG | Pregnancy-associated glycoprotein family |
| PAG-L | PAG-like |
| hPAG-L | Human PAG-L |
| Pep | Pepsinogen |
| RNA-seq | RNA sequencing |
| aa | Amino acids |
| D | Asparagine (Asp) |
| cDNA | Complementary DNA |
| gDNA | Genomic DNA |
| bPAG | Bovine PAG |
| pPAG | Porcine PAG |
| CfPAG-L | Beaver PAG-L |
| EbPAG | European bison PAG |
| LpPAG | Lama pacos PAG |
| CdPAG | Camelus dromedarius PAG |
| CfPAG | Camelus ferrus PAG |
| AaPAG-L | Alces alces PAG |
| RIA | Radioimmunological test |
| ELISA | Immunoenzymatic test |
| RT-PCR | Reverse transcriptase PCR |
| dNTP | Deoxynucleotide |
| MWCO | Molecular weight cut-off |
| SDS-PAGE | Denaturing polyacrylamide electrophoresis |
| CBB | Coomassie Brilliant Blue |
| NBT | Nitro blue tetrazolium |
| BCIP | 5-Bromo-4-chloro-3-indolyl-phosphate |
| ht | Heterologous (cross-species) |
| anti-pPAG-P | Polyclonals raised against various porcine antigens |
| anti-Rec pPAG2 | Polyclonals raised against recombinant pPAG2 antigen |
| htdF-IHC | Heterologous double fluorescent immunohistochemistry |
| A488 | Alexa 488 fluorophore |
| PI | Propidium iodide |
| HQ | High quality |
| CDS | Coding sequence |
| UTR | Untranslated region |
| QC | Query Cover |
| SP | Signal peptide |
| ID | Sequence identity |
| CV | Chorionic villi |
| VC | Villous core |
| FC | Fetal capillaries |
| IS | Intervillous space |
| PS | Placental septa |
| SB | Stratum basale |
| NC | Negative control |
| dpc | Day post coitum |
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/6/1227/s1.
Author Contributions
Marta Majewska performed experimental work and drafted the manuscript. Aleksandra Lipka performed experimental work. Grzegorz Panasiewicz assisted in the study design. Placental samples were provided by Marek Gowkielewicz and Marcin Jozwik. The draft manuscript was read by Marcin Jozwik and Mariusz Krzysztof Majewski. Bozena Szafranska conceived the study design and helped in writing the final version of the manuscript. All authors have seen and approved the final version.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Szafranska B., Panasiewicz G., Majewska M. Biodiversity of multiple pregnancy-associated glycoprotein (PAG) family: Gene cloning and chorionic protein purification in domestic and wild eutherians (Placentalia)—A review. Reprod. Nutr. Dev. 2006;46:481–502. doi: 10.1051/rnd:2006034. [DOI] [PubMed] [Google Scholar]
- 2.Wallace R.M., Pohler K.G., Smith M.F., Green J.A. Placental PAGs: Gene origins, expression patterns, and use as markers of pregnancy. Reproduction. 2015;149:R115–R126. doi: 10.1530/REP-14-0485. [DOI] [PubMed] [Google Scholar]
- 3.Guruprasad K., Blundell T.L., Xie S., Green J., Szafranska B., Nagel R.J., McDowell K., Baker C.B., Roberts R.M. Comparative modelling and analysis of amino acid substitutions suggests that the family of pregnancy-associated glycoproteins includes both active and inactive aspartic proteinases. Protein Eng. 1996;9:849–856. doi: 10.1093/protein/9.10.849. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K. Structure and function studies on enzymes with a catalytic carboxyl group(s): From ribonuclease T1 to carboxyl peptidases. Proc. Jpn. Acad. Ser. B. 2013;89:201–225. doi: 10.2183/pjab.89.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kageyama T. Pepsinogens progastricsins and prochymosins: Structure function evolution and development. Cell. Mol. Life Sci. 2002;59:288–306. doi: 10.1007/s00018-002-8423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carginale V., Trinchella F., Capasso C., Scudiero R., Riggio M., Parisi E. Adaptive evolution and functional divergence of pepsin gene family. Gene. 2004;333:81–90. doi: 10.1016/j.gene.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Panasiewicz G., Majewska M., Romanowska A., Dajnowiec J., Szafranska B. Radiocompetition of secretory pregnancy-associated glycoproteins as chorionic ligands with luteal and uterine gonadotrophin receptors of pregnant pigs. Anim. Reprod. Sci. 2007;99:285–298. doi: 10.1016/j.anireprosci.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Szafranska B., Panasiewicz G., Majewska M., Romanowska A., Dajnowiec J. Pregnancy-associated glycoprotein family (PAG)—As chorionic signaling ligands for gonadotropin receptors of cyclic animals. Anim. Reprod. Sci. 2007;99:269–284. doi: 10.1016/j.anireprosci.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen J.T., Hamada Y., Kimura T., Kiso Y. Design of potent aspartic protease inhibitors to treat various diseases. Arch. Pharm. 2008;341:523–535. doi: 10.1002/ardp.200700267. [DOI] [PubMed] [Google Scholar]
- 10.Tsukuba T., Okamoto K., Okamoto Y., Yanagawa M., Kohmura K., Yasuda Y., Uchi H., Nakahara T., Furue M., Nakayama K., et al. Association of cathepsin E deficiency with development of atopic dermatitis. J. Biochem. 2003;134:893–902. doi: 10.1093/jb/mvg216. [DOI] [PubMed] [Google Scholar]
- 11.Kawakubo T., Okamoto K., Iwata J., Shin M., Okamoto Y., Yasukochi A., Nakayama K., Kadowaki T., Tsukuba T., Yamamoto K. Cathepsin E prevents tumor growth and metastasis by catalyzing the proteolytic release of soluble TRAIL from tumor cell surface. Cancer Res. 2007;67:10869–10878. doi: 10.1158/0008-5472.CAN-07-2048. [DOI] [PubMed] [Google Scholar]
- 12.Shin M., Kadowaki T., Iwata J., Kawakubo T., Yamaguchi N., Takii R., Tsukuba T., Yamamoto K. Association of cathepsin E with tumor growth arrest through angiogenesis inhibition and enhanced immune responses. Biol. Chem. 2007;388:1173–1181. doi: 10.1515/BC.2007.154. [DOI] [PubMed] [Google Scholar]
- 13.Wang A.M., Chen S.L., Xing F.Q. Expression of cathepsins B and L in early gestational decidua and chorionic villi. Di Yi Jun Yi Da Xue Xue Bao. 2005;25:1365–1368. (In Chinese) [PubMed] [Google Scholar]
- 14.Nakanishi T., Ozaki Y., Blomgren K., Tateyama H., Sugiura-Ogasawara M., Suzumori K. Role of cathepsins and cystatins in patients with recurrent miscarriage. Mol. Hum. Reprod. 2005;11:351–355. doi: 10.1093/molehr/gah172. [DOI] [PubMed] [Google Scholar]
- 15.Goto S., Ozaki Y., Suzumori N., Yasukochi A., Kawakubo T., Furuno T., Nakanishi M., Yamamoto K., Sugiura-Ogasawara M. Role of cathepsin E in decidual macrophage of patients with recurrent miscarriage. Mol. Hum. Reprod. 2014;20:454–462. doi: 10.1093/molehr/gau008. [DOI] [PubMed] [Google Scholar]
- 16.Hughes A.L., Green J.A., Piontkivska H., Roberts R.M. Aspartic proteinase phylogeny and the origin of pregnancy-associated glycoproteins. Mol. Biol. Evol. 2003;20:1940–1945. doi: 10.1093/molbev/msg217. [DOI] [PubMed] [Google Scholar]
- 17.Xie S., Green J., Beckers J.F., Roberts R.M. The gene encoding bovine pregnancy-associated glycoprotein-1, an inactive member of the aspartic proteinase family. Gene. 1995;159:193–197. doi: 10.1016/0378-1119(94)00928-L. [DOI] [PubMed] [Google Scholar]
- 18.Telugu B.P., Walker A.M., Green J.A. Characterization of the bovine pregnancy-associated glycoprotein gene family—Analysis of gene sequences, regulatory regions within the promoter and expression of selected genes. BMC Genom. 2009;24:185. doi: 10.1186/1471-2164-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szafranska B., Miura R., Ghosh D., Ezashi T., Xie S., Roberts R.M., Green J.A. Gene for porcine Pregnancy-Associated Glycoprotein 2 (poPAG2): Its structural organization and analysis of its promoter. Mol. Reprod. Dev. 2001;60:137–146. doi: 10.1002/mrd.1070. [DOI] [PubMed] [Google Scholar]
- 20.Lipka A., Majewska M., Panasiewicz G., Bieniek-Kobuszewska M., Szafranska B. Gene structure of the pregnancy associated glycoprotein-like (PAG-L) in the Eurasian beaver (Castor fiber L.) Funct. Integr. Genom. 2017 doi: 10.1007/s10142-017-0557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green J., Xie S., Szafranska B., Gan X., Newman A.G., McDowell K., Roberts R.M. Identification of a new aspartic proteinase expressed by the outer chorionic cell layer of the equine placenta. Biol. Reprod. 1999;60:1069–1077. doi: 10.1095/biolreprod60.5.1069. [DOI] [PubMed] [Google Scholar]
- 22.Carter A.M., Enders A.C., Pijnenborg R. The role of invasive trophoblast in implantation and placentation of primates. Philos. Trans. R. Soc. Lond. B. 2015;370:20140070. doi: 10.1098/rstb.2014.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monk D. Genomic imprinting in the human placenta. Am. J. Obstet. Gynecol. 2015;213:S152–162. doi: 10.1016/j.ajog.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 24.Huppertz B., Ghosh D., Sengupta J. An integrative view on the physiology of human early placental villi. Prog. Biophys. Mol. Biol. 2014;114:33–48. doi: 10.1016/j.pbiomolbio.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Carter A.M., Enders A.C. Placentation in mammals: Definitive placenta, yolk sac, and paraplacenta. Theriogenology. 2016;86:278–287. doi: 10.1016/j.theriogenology.2016.04.041. [DOI] [PubMed] [Google Scholar]
- 26.Roberts R.M., Xie S., Mathialagan N. Maternal recognition of pregnancy. Biol. Reprod. 1996;54:294–302. doi: 10.1095/biolreprod54.2.294. [DOI] [PubMed] [Google Scholar]
- 27.Roberts R.M., Ezashi T., Das P. Trophoblast gene expression: Transcription factors in the specification of early trophoblast. Reprod. Biol. Endocrinol. 2004;2:47. doi: 10.1186/1477-7827-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopes-da-Costa L., Chagas e Silva J., Deloche M.C., Jeanguyot N., Humblot P., Horta A.E. Effects of embryo size at transfer (whole versus demi) and early pregnancy progesterone supplementation on embryo growth and pregnancy-specific protein bovine concentrations in recipient dairy heifers. Theriogenology. 2011;76:522–531. doi: 10.1016/j.theriogenology.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 29.García-Ispierto I., Almería S., Serrano B., de Sousa N., Beckers J., López-Gatius F. Plasma concentrations of pregnancy-associated glycoproteins measured using anti-bovine PAG-2 antibodies on day 120 of gestation predict abortion in dairy cows naturally infected with Neospora caninum. Reprod. Domest. Anim. 2013;48:613–618. doi: 10.1111/rda.12134. [DOI] [PubMed] [Google Scholar]
- 30.Breukelman S.P., Perényi Z., Taverne M.A., Jonker H., van der Weijden G.C., Vos P.L., de Ruigh L., Dieleman S.J., Beckers J.F., Szenci O. Characterisation of pregnancy losses after embryo transfer by measuring plasma progesterone and bovine pregnancy-associated glycoprotein-1concentrations. Vet. J. 2012;194:71–76. doi: 10.1016/j.tvjl.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 31.García-Ispierto I., Rosselló-Visa M.A., Serrano-Pérez B., Mur-Novales R., deSousa N.M., Beckers J.F., López-Gatius F. Plasma concentrations of pregnancy-associated glycoproteins I and II and progesterone on day 28 post-AI as markers of twin pregnancy in dairy cattle. Livest. Sci. 2016;192:44–47. doi: 10.1016/j.livsci.2016.09.003. [DOI] [Google Scholar]
- 32.Szafranska B., Xie S., Green J., Roberts R.M. Porcine pregnancy-associated glycoproteins: New members of the aspartic proteinase gene family expressed in trophectoderm. Biol. Reprod. 1995;53:21–28. doi: 10.1095/biolreprod53.1.21. [DOI] [PubMed] [Google Scholar]
- 33.Xie S., Green J., Bixby J.B., Szafranska B., DeMartini J.C., Hecht S., Roberts R.M. The diversity and evolutionary relationships of the pregnancy-associated glycoproteins an aspartic proteinase subfamily consisting of many trophoblast-expressed genes. Proc. Natl. Acad. Sci. USA. 1997;94:12809–12816. doi: 10.1073/pnas.94.24.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garbayo J.M., Green J., Manikkam M., Beckers J.F., Kiesling D.O., Ealy A.D., Roberts R.M. Caprine Pregnancy-Associated Glycoproteins (PAGs): Their cloning expression and evolutionary relationship to other PAG. Mol. Reprod. Dev. 2000;57:311–322. doi: 10.1002/1098-2795(200012)57:4<311::AID-MRD2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 35.Kageyama T., Ichinose M., Tsukada-Kato S., Omata M., Narita Y., Moriyama A., Yonezawa S. Molecular cloning of neonate/infant-specific pepsinogens from rat stomach mucosa and their expressional change during development. Biochem. Biophys. Res. Commun. 2000;267:806–812. doi: 10.1006/bbrc.1999.2047. [DOI] [PubMed] [Google Scholar]
- 36.Kageyama T., Takahashi K. Monkey pepsinogens and pepsins III Carbohydrate moiety of Japanese monkey pepsinogens and the amino acid sequence around the site of its attachment to protein. J. Biochem. 1978;84:771–778. doi: 10.1093/oxfordjournals.jbchem.a132188. [DOI] [PubMed] [Google Scholar]
- 37.Martin P., Trieu-Cuot P., Collin J.C., Ribadeau-Dumas B. Purification and characterization of bovine gastricsin. Eur. J. Biochem. 1982;122:31–39. doi: 10.1111/j.1432-1033.1982.tb05844.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen X., Rosenfeld C.S., Roberts R.M., Green J.A. An aspartic proteinase expressed in the yolk sac and neonatal stomach of the mouse. Biol. Reprod. 2001;65:1092–1101. doi: 10.1095/biolreprod65.4.1092. [DOI] [PubMed] [Google Scholar]
- 39.Green J., Xie S., Gan X., Roberts R.M. An aspartic proteinase expressed in the equine placenta. Adv. Exp. Med. Biol. 1998;436:163–167. doi: 10.1007/978-1-4615-5373-1_22. [DOI] [PubMed] [Google Scholar]
- 40.Panasiewicz G., Majewska M., Szafranska B. Trophoblastic cDNA cloning of porcine pregnancy-associated glycoprotein genes (pPAG) and in silico analysis of coded polypeptide precursors. Reprod. Biol. 2004;4:131–141. [PubMed] [Google Scholar]
- 41.Brandt G.A., Parks T.E., Killian G., Ealy A.D., Green J.A. A cloning and expression analysis of pregnancy-associated glycoproteins expressed in trophoblasts of the white-tail deer placenta. Mol. Reprod. Dev. 2007;74:1355–1362. doi: 10.1002/mrd.20669. [DOI] [PubMed] [Google Scholar]
- 42.Xie S., Low B.G., Nagel R.J., Kramer K.K., Anthony R.V., Zoli A.P., Beckers J.F., Roberts R.M. Identification of the major pregnancy-specific antigens of cattle and sheep as inactive members of the aspartic proteinase family. Proc. Natl. Acad. Sci. USA. 1991;88:10247–10251. doi: 10.1073/pnas.88.22.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yakabe E., Tanji M., Ichinose M., Goto S., Miki K., Kurokawa K., Ito H., Kageyama T., Takahashi K. Purification, characterization, and amino acid sequences of pepsinogens and pepsins from the esophageal mucosa of bullfrog (Rana catesbeiana) J. Biol. Chem. 1991;266:22436–22443. [PubMed] [Google Scholar]
- 44.Kurokawa T., Uji S., Suzuki T. Identification of pepsinogen gene in the genome of stomachless fish, Takifugu rubripes. Comp. Biochem. Physiol. B. 2005;140:133–140. doi: 10.1016/j.cbpc.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 45.Majewska M., Panasiewicz G., Louis K.K., Olivera V.M., Mamani J.M., Abd-Elnaeim M.M., Szafranska B. Pregnancy-associated glycoprotein (PAG) family: Transcripts and gene amplicons in camelids. Reprod. Biol. 2009;9:127–150. doi: 10.1016/S1642-431X(12)60022-9. [DOI] [PubMed] [Google Scholar]
- 46.Bieniek-Kobuszewska M., Panasiewicz G., Lipka A., Majewska M., Szafranska B. Novel SNPs and InDels discovered in two promoter regions of porcine pregnancy-associated glycoprotein 2-like subfamily (pPAG2-Ls) in crossbreed pigs. Funct. Integr. Genom. 2016;16:705–715. doi: 10.1007/s10142-016-0522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kageyama T., Tanabe K., Koiwai O. Structure and development of rabbit pepsinogens stage-specific zymogens nucleotide sequences of cDNAs molecular evolution and gene expression during development. J. Biol. Chem. 1990;265:17031–17038. [PubMed] [Google Scholar]
- 48.Sogawa K., Fujii-Kuriyama Y., Mizukami Y., Ichihara Y., Takahashi K. Primary structure of human pepsinogen gene. J. Biol. Chem. 1983;258:5306–5311. [PubMed] [Google Scholar]
- 49.Hayano T., Sogawa K., Ichihara Y., Fujii-Kuriyama Y., Takahashi K. Primary structure of human pepsinogen C gene. J. Biol. Chem. 1988;263:1382–1385. [PubMed] [Google Scholar]
- 50.Örd T., Kolmer M., Villems R., Saarma M. Structure of the human genomic region homologous to the bovine prochymosin-encoding gene. Gene. 1990;91:241–246. doi: 10.1016/0378-1119(90)90094-8. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K. Gene structures of pepsinogens A and C. Scand. J. Clin. Lab. Investig. 1992;210:97–110. doi: 10.1080/00365519209104658. [DOI] [PubMed] [Google Scholar]
- 52.Majewska M., Panasiewicz G., Dabrowski M., Gizejewski Z., Beckers J.F., Szafranska B. Multiple forms of pregnancy-associated glycoproteins released in vitro by porcine chorion or placentomal and interplacentomal explants of wild and domestic ruminants. Reprod. Biol. 2005;5:185–203. [PubMed] [Google Scholar]
- 53.Szafranska B., Majewska M., Panasiewicz G. N-glycodiversity of the pregnancy-associated glycoprotein family (PAG) produced in vitro by trophoblast and trophectoderm explants during implantation, placentation and advanced pregnancy in the pig. Reprod. Biol. 2004;4:67–89. [PubMed] [Google Scholar]
- 54.Klisch K., Jeanrond E., Pang P.C., Pich A., Schuler G., Dantzer V., Kowalewski M.P., Dell A. A tetraantennary glycan with bisecting N-acetylglucosamine and the Sd(a) antigen is the predominant N-glycan on bovine pregnancy-associated glycoproteins. Glycobiology. 2008;18:42–52. doi: 10.1093/glycob/cwm113. [DOI] [PubMed] [Google Scholar]
- 55.Kiewisz J., Sousa N.M., Beckers J.F., Vervaecke H., Panasiewicz G., Szafranska B. Isolation of pregnancy-associated glycoproteins from placenta of the American bison (Bison bison) at first half of pregnancy. Gen. Comp. Endocrinol. 2008;155:164–175. doi: 10.1016/j.ygcen.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Kiewisz J., Melo de Sousa N., Beckers J.F., Panasiewicz G., Gizejewski Z., Szafranska B. Identification of multiple pregnancy-associated glycoproteins (PAGs) purified from the European bison (Eb; Bison bonasus L.) placentas. Anim. Reprod. Sci. 2009;112:229–250. doi: 10.1016/j.anireprosci.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Beriot M., Tchimbou A.F., Barbato O., Beckers J.F., de Sousa N.M. Identification of pregnancy-associated glycoproteins and alpha-fetoprotein in fallow deer (Dama dama) placenta. Acta Vet. Scand. 2014;56:4–14. doi: 10.1186/1751-0147-56-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lipka A., Panasiewicz G., Majewska M., Bieniek-Kobuszewska M., Saveljev A.P., Pankratov A.P., Szafranska B. Identification of the pregnancy-associated glycoprotein family (PAGs) and some aspects of placenta development in the European moose (Alces alces L.) Theriogenology. 2016;86:2119–2135. doi: 10.1016/j.theriogenology.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 59.Lipka A., Panasiewicz G., Majewska M., Paukszto L., Bieniek-Kobuszewska M., Szafranska B. Identification of placental aspartic proteinase in the Eurasian beaver (Castor fiber L.) Theriogenology. 2017 doi: 10.3390/ijms19041229. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Majewska M., Panasiewicz G., Majewski M., Szafranska B. Localization of chorionic pregnancy-associated glycoprotein family in the pig. Reprod. Biol. 2006;6:205–230. [PubMed] [Google Scholar]
- 61.Majewska M., Panasiewicz G., Szafranska B., Gizejewski Z., Majewski M., Borkowski K. Cellular localization of the pregnancy-associated glycoprotein family (PAGs) in the synepitheliochorial placenta of the European bison. Gen. Comp. Endocrinol. 2008;155:422–431. doi: 10.1016/j.ygcen.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 62.Majewska M., Panasiewicz G., Szafranska B. Pregnancy-associated glycoprotein (PAG) family localized in chorionic cells within the epitheliochorial/diffuse placenta of the alpaca (Lama pacos) Acta Histochem. 2011;113:570–577. doi: 10.1016/j.acthis.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Majewska M., Panasiewicz G., Szafranska B. Expression of pregnancy-associated glycoprotein family in the epitheliochorial placenta of two Camelidae species (C. dromedarius and C. bactrianus) Acta Histochem. 2013;113:570–577. doi: 10.1016/j.acthis.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Panasiewicz G., Bieniek-Kobuszewska M., Lipka A., Majewska M., Jedryczko R., Szafranska B. Novel effects of identified SNPs within the porcine pregnancy-associated glycoprotein gene family (pPAGs) on the major reproductive traits in Hirschmann hybrid-line sows. Res. Vet. Sci. 2017;114:123–130. doi: 10.1016/j.rvsc.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 65.Panasiewicz G., Zamojska A., Bieniek M., Gizejewski Z., Szafranska B. Persistent Müllerian duct syndrome (PMDS) in the Polish free-ranged bull populations of the European bison (Bison bonasus L.) Anim. Reprod. Sci. 2015;152:123–136. doi: 10.1016/j.anireprosci.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 66.Szafranska B., Panasiewicz G. The placental expression of the porcine pregnancy-associated glycoprotein (pPAG) gene family examined in situ and in vitro. Anim. Reprod. Sci. 2002;72:95–113. doi: 10.1016/S0378-4320(02)00066-0. [DOI] [PubMed] [Google Scholar]
- 67.Szafranska B., Panasiewicz G., Majewska M., Beckers J.F. Chorionic expression of heterogeneous products of the PAG (pregnancy-associated glycoprotein) gene family secreted in vitro throughout embryonic and foetal development in the pig. Reprod. Nutr. Dev. 2003;43:497–516. doi: 10.1051/rnd:2004004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.