ABSTRACT
Neuroblastoma (NB) is the most common extracranial solid tumor occurring in childhood. Amplification of the MYCN oncogene is associated with poor prognosis. Downregulation on NB cells of ligands recognized by Natural Killer (NK) cell-activating receptors, involved in tumor cell recognition and lysis, may contribute to tumor progression and relapse. Here, we demonstrate that in human NB cell lines MYCN expression inversely correlates with that of ligands recognized by NKG2D and DNAM1 activating receptors in human NB cell lines. In the MYCN-inducible Tet-21/N cell line, downregulation of MYCN resulted in enhanced expression of the activating ligands MICA, ULBPs and PVR, which rendered tumor cells more susceptible to recognition and lysis mediated by NK cells. Conversely, a MYCN non-amplified NB cell line transfected with MYCN showed an opposite behavior compared with control cells. Consistent with these findings, an inverse correlation was detected between the expression of MYCN and that of ligands for NK-cell-activating receptors in 12 NB patient specimens both at mRNA and protein levels.
Taken together, these results provide the first demonstration that MYCN acts as an immunosuppressive oncogene in NB cells that negatively regulates the expression of ligands for NKG2D and DNAM-1 NK-cell-activating receptors. Our study provides a clue to exploit MYCN expression levels as a biomarker to predict the efficacy of NK-cell-based immunotherapy in NB patients.
KEYWORDS: Immunosuppressive oncogene, MYCN oncogene, neuroblastoma, NK-cell-activating receptor ligands, tumor immune escape
Introduction
High-risk neuroblastoma (NB) remains a highly challenging pediatric tumor, since 3-y event-free survival remains lower than 40% despite intensive, multimodal therapies.1 One well-established predictor of poor prognosis in NB is MYCN amplification, found in almost 25% of cases. In healthy conditions, MYCN is scarcely expressed in differentiated adult tissues, while playing a key role in several pathways involved in the maintenance of pluripotency.2
Besides downregulation of MHC class I molecules, rendering tumor cells resistant to cytotoxic T lymphocytes,3 an additional mechanism adopted by NB, allowing tumor to escape innate antitumor immune responses, is downregulation of ligands for Natural Killer (NK) cell-activating receptors. Indeed, this downregulation prevents tumor cell recognition and killing by NK cells4 and, together with releasing of NKG2D ligands, has been proposed as a mechanism contributing to the immune evasion strategy in NB.5 The mechanisms that regulate the expression of such ligands on cellular targets are only partially defined. ULBP1, ULBP2 and ULBP3 genes are regulated by c-MYC and p53 transcription factors.6,7 At variance with the majority of tumors, p53 gene is mutated only in 2% of NB, especially in relapse.8
On the other hand, a complex network of regulators, including MDM2,9,10 inhibits p53 function. Remarkably, both p53 and MDM2 are direct transcriptional targets of MYCN and are co-expressed at high levels in MYCN-amplified NB cells.11,12 Of note, NB patients with MYCN-amplified tumors initially respond to therapy, but rapidly develop drug resistance and frequently suffer relapses13 with tumor cells characterized by abnormalities in p53/MDM2 pathway.8 Indeed, p53 is functionally suppressed by MDM2 in MYCN-amplified NB cells. By contrast, MYCN expression inversely correlates with that of c-MYC.14
In this study, we show an inverse correlation between the expression of MYCN and that of ligands for NK-cell NKG2D- and DNAM1-activating receptors both in a panel of human NB cell lines and in primary tumor samples. Downregulation of MYCN in NB cell lines resulted in upregulation of the surface expression of ligands for such activating receptors, thereby rendering tumor cells susceptible to recognition and lysis by NK cells. Taken together, these findings delineate a novel mechanism of NB-driven immune evasion operated by the MYCN oncogene. These data suggest that MYCN expression level in NB cells represents a potential biomarker capable of predicting the susceptibility to therapeutic efficacy of NK-cell-mediated immunotherapy in high-risk NB patients.
Results and discussion
The levels of MYCN expression are inversely correlated with those of ligands for NK-cell-activating receptors in NB cell lines
NK-cell cytotoxicity is triggered by the interaction of NK-cell-activating receptors with their specific ligands. In particular, NKG2D receptor recognizes MICA, MICB, ULBP1, ULBP2/5/6 and ULBP3 ligands,15 while DNAM1 receptor interacts with PVR and Nectin-2 ligands.16 In addition, certain activating killer-immunoglobulin-like receptors (KIRs) interact with MHC class I molecules17 and TRAIL with TRAIL-R2.18
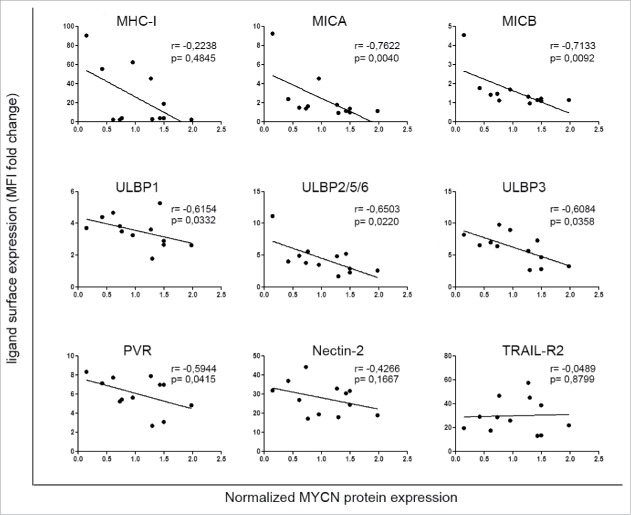
We asked whether the expression levels of the MYCN oncogene correlated with a mechanism of immune escape involving downregulation of NK-cell-activating receptor ligands on NB cells. To investigate this possible correlation, we assessed the protein levels of MYCN by western blotting (Fig. S1A) and the surface expression of activating ligands by flow cytometry (Table S1) in a panel of 12 NB cell lines, including 6 MYCN non-amplified (non-MNA) and 6 MYCN amplified (MNA). Regression analysis of normalized densitometric values of western blotting and mean fluorescence intensity values of flow cytometry analysis revealed a significant inverse correlation between the expression of MYCN and that of the activating ligands MICA, MICB, ULBP1, ULBP2/5/6, ULBP3 and PVR. A trend toward an inverse correlation was also found between expression of MYCN and that of MHC class I and of Nectin-2. On the other hand, MYCN expression did not correlate with that of TRAIL-R2 (Fig. 1).
Figure 1.
Expression of MYCN inversely correlates with that of ligands for NK-cell-activating receptors in NB cell lines. Scatter plots showing the correlation between MYCN and the surface expression of activating ligands for NK-cell receptors in 12 NB cell lines. The means of four independent immunoblotting analyses of MYCN expression in NB cell lines are plotted against the means of 15 independent flow cytometric analyses of surface expression of the indicated activating ligands (see Table S1 for data); Spearman correlation r and p value are shown for each plot.
MYCN expression in NB cells is positively correlated with that of p5312 and inversely with that of c-MYC,14,19 in agreement with the finding that both transcription factors are involved in the induction of ULBP ligands.6,7 The function of p53 protein is inhibited by the MDM2,10 which is overexpressed in many human cancers,20 including NB.8 Of note, p53 and MDM2 genes are both direct transcriptional targets of MYCN in NB.11,12
To assess whether p53 and c-MYC could be involved in the expression of activating ligands in NB, the status of MYCN, c-MYC, p53 and MDM2 was evaluated in NB cell lines in terms of amplification, gain, deletion and protein expression by high-resolution array CGH analysis and western blotting, respectively (Table 1, Fig. S1). p53 gene was lost in four NB cell lines tested [SK-N-AS, ACN, SK-N-BE(2)c and LA-N-1, due to a mutation that has been previously reported],21,22 three of them [SK-N-AS, ACN and SK-N-BE(2)c] also lost MDM2 gene. c-MYC gene gain was detected in two NB cells lines (SK-N-AS and ACN), whereas MYCN gene amplification or gain were found in five [SK-N-BE(2)c, LA-N-1, LA-N-5, SMS-KCNR, IMR-32] and two (GICAN, SK-N-SH) NB cell lines, respectively (Table 1). All these genetic aberrations were confirmed at the protein level, except for SK-N-BE(2)c, which expressed high levels of mutant p53, due to a previously reported missense mutation.22 Consistently with previously published studies,11,12,19 MYCN expression was inversely correlated with c-MYC expression. In contrast, MYCN expression directly correlated with both p53 and MDM2 expression, as evaluated in p53 wt NB cell lines (SH-EP, GICAN, SH-SY-5Y, SK-N-SH, LA-N-5, SMS-KCNR, IMR-32 and Tet-21/N) (Fig. S1B). This indicates that p53 is more functional in non-MNA NB cells, even if expressed at low levels, than in MNA NB cells in which it is inhibited by MDM2.
Table 1.
Status and chromosome coordinates of MYCN, c-MYC, p53 and MDM2 genes in NB cell lines.
| NB cell lines | MYCN | Chr. coordinates of MYCN gain/amp | c-MYC | Chr. coordinates of c-MYC gain | p53 | Chr. coordinates of p53 loss | MDM2 | Chr. coordinates of MDM2 loss |
|---|---|---|---|---|---|---|---|---|
| SK-N-AS | single copy | — | gain | Chr8: 46924418–129841304 | loss | Chr17: 51885–22242373 | loss | Chr12: 37944373–112034509 |
| Cytoband: 8q11.1-q24.21 | Cytoband: 17p13.3-p11.1 | Cytoband: 12q11-q24.12 | ||||||
| Size: 82.9 Mb | Size: 22.1 Mb | Size: 74 Mb | ||||||
| GICAN | gain | Chr2: 16066442–18776030 | single copy | — | single copy | — | single copy | |
| Cytoband: 2p24.3-p24.2 | ||||||||
| Size: 2.7 Mb | ||||||||
| ACN | single copy | — | gain | Chr8: 59043056–146280020 | loss | Chr17: 84287–22080868 | loss | Chr12: 38766104–133767986 |
| Cytoband: 8q12.1-q24.3 | Cytoband: 17p13.3-p11.2 | Cytoband: 12q12-q24.33 | ||||||
| Size: 87.2 Mb | Size: 21.9 Mb | Size: 95 Mb | ||||||
| SH-SY5Y | single copy | — | single copy | — | single copy | — | single copy | — |
| SH-EP | single copy | — | single copy | — | single copy | — | single copy | — |
| SK-N-SH | gain | Chr2: 17019–48571447 | single copy | — | single copy | — | single copy | — |
| Cytoband: 2p25.3-p16.3 | ||||||||
| Size: 48.5 Mb | ||||||||
| SK-N-BE(2)c | amp | Chr2: 16082217–16469668 | single copy | — | loss | Chr17: 29169–22242373 | loss | Chr12: 37873948–131685128 |
| Cytoband: 2p24.3 | Cytoband: 17p13.3-p11.1 | Cytoband: 12q11-q24.33 | ||||||
| Size: 387 Kb | Size: 22.2 Mb | Size: 93.8 Mb | ||||||
| LA-N-1 | amp | Chr2: 16066442–16487029 | single copy | — | loss | Chr17: 183662–22205821 | single copy | — |
| Cytoband: 2p24.3 | Cytoband: 17p13.3-p11.1 | |||||||
| Size: 420 Kb | Size: 22 Mb | |||||||
| LA-N-5 | amp | Chr2: 15496660–17046138 | single copy | — | single copy | — | single copy | — |
| Cytoband: 2p24.3-p24.2 | ||||||||
| Size: 1.4 Mb | ||||||||
| SMS-KCNR | amp | Chr2: 16036272–16428878 | single copy | — | single copy | — | single copy | — |
| Cytoband: 2p24.3 | ||||||||
| Size: 392 Kb | ||||||||
| IMR32 | amp | Chr2: 14773079–16086291 | single copy | — | single copy | — | single copy | — |
| Cytoband: 2p24.3 | ||||||||
| Size: 1.3 Mb | ||||||||
| Tet-21/N | single copy | — | single copy | — | single copy | — | single copy | — |
Chr: chromosome: amp: amplification.
Based on these observations, we hypothesize that the inverse correlation between the expression of MYCN and that of c-MYC or the p53 functional status could explain differences in expression of ULBPs ligands in NB cell lines. Whether MICA, MICB and PVR, whose expression was inversely correlated with that of MYCN, share the same mechanism of regulation of the ULBPs ligands will be matter of future investigation. Moreover, since higher levels of soluble MICA have been reported to be found in sera of NB patients compared with healthy donors,5 the putative role of MYCN in regulating cell surface expression of activating ligands by non-transcriptional mechanisms cannot be excluded and needs further investigation.
Altogether, these data demonstrate that MYCN acts as a negative regulator of the expression of NK-cell-activating receptor ligands. Our findings support the hypothesis that the functional suppression of MYCN may represent a potential strategy to render NB cells more susceptible to NK-cell-mediated recognition and killing.
The modulation of MYCN expression affects that of NK-cell-activating receptor ligands and the susceptibility of NB cells to NK-cell-mediated lysis
To investigate whether modulation of MYCN expression could affect that of activating ligands, we used the conditionally MYCN-expressing Tet-21/N cell line that loses MYCN expression following treatment with doxycycline. Doxycycline treatment caused a drastic downregulation of MYCN expression, early induction of c-MYC expression (at 8 h) and delayed reduction of p53 and MDM2 expression (at 24 h) (Fig. 2A). Following 16-h treatment with doxycycline, the expression of MICA, ULBP2/5/6, ULBP3 and PVR was significantly higher in Doxy-treated Tet-21/N than in untreated Tet-21/N cells. Upregulation of NK-cell-activating receptor ligands was detected up to 24 h after doxycycline treatment, with a peak at 16 h (Fig. 2B), followed by a decrease at 3 d (Fig. S2A, left panel). By contrast, after 10 d of doxycycline treatment downregulation of NK-cell-activating receptor ligands was detected (Fig. S2A, right panel). As reported by other authors,23 doxycycline at the concentration of 1 μg/mL, 100-fold higher than the dose used in this study, enhanced the surface expression of MICA and MICB in different tumor cell lines. To exclude that doxycycline could affect the expression of activating ligands in a MYCN-modulation independent manner, SH-EP and LA-N-5 cell lines, which lack the Tet response element, were treated with 10 ng/mL of doxycycline. No change was detected in both cell lines after 16 and 24 h of treatment (Fig. S3). Indeed, in Tet-21/N cell line a state of cell senescence was detected 16 h after doxycycline treatment, as evaluated by cell count at different time points showing stable cell number due to a block in cell proliferation (data not shown) and by senescence associated-β-galactosidase (SA-β-Gal) staining (Fig. S2B, upper panel). Three days after doxycycline treatment, the occurrence of cell differentiation, revealed by changes of cell morphology, became evident (Fig. S2B, lower panel). This finding is consistent with results reported by other authors24 on the expression of activating ligands in senescent cells, but not in mature and differentiated cells.
Figure 2.
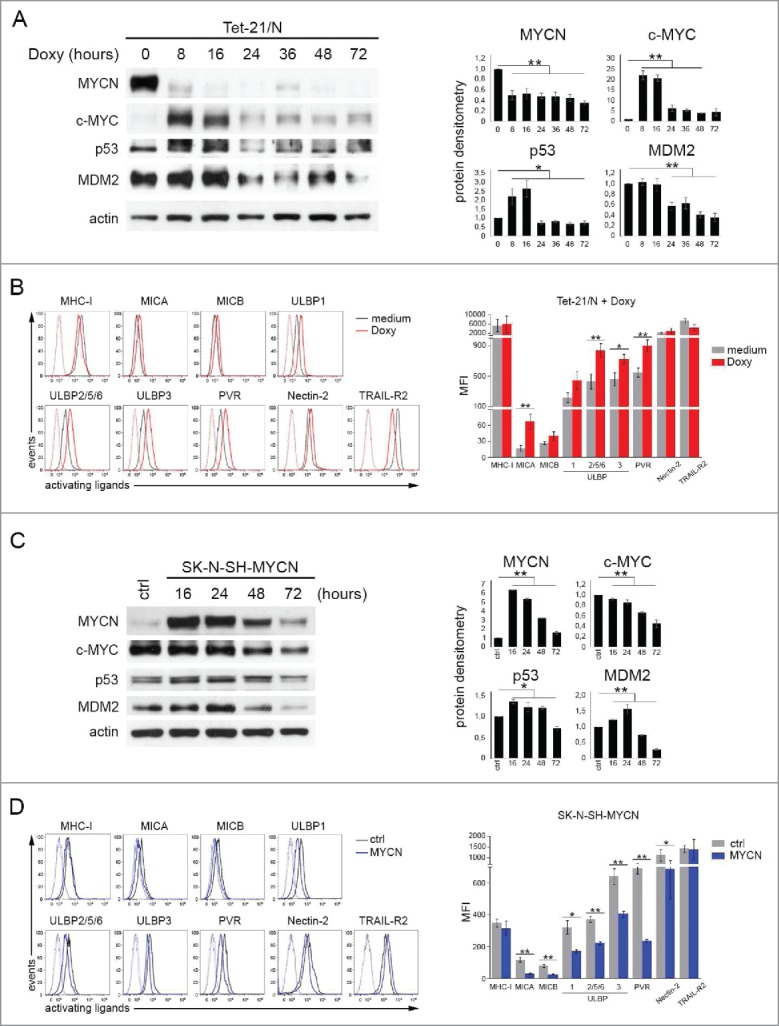
Modulation of MYCN affects the expression of ligands for NK-cell-activating receptors. (A) Representative example of immunoblot analysis of MYCN, c-MYC, p53 and MDM2 in Tet-21/N either left untreated (0) or treated with doxycycline for the indicated time (left panel). An anti-Actin Ab was used for normalization. Densitometry analysis of Actin-normalized proteins values of three independent experiments are shown (right panel). Mean ± SD; *p < 0.05, **p < 0.01. (B) Representative flow cytometric analysis of surface expression of activating ligands for NK-cell receptors in Tet-21/N either left untreated (medium, gray line) or treated with doxycycline for 16 h (Doxy, red line); dotted lines, isotype-matched negative controls (left panel). Summary of five independent flow cytometric analyses (right panel). p values, compared with untreated and Doxy-treated Tet-21/N cells (two-tailed paired Student's t-test); *p < 0.05, **p < 0.01. (C) Representative example of immunoblot analysis as in A of SK-N-SH either left transfected with empty vector (ctrl) or with piRVneoSV-MYCN (MYCN) for the indicated time (left panel). Densitometry analysis of Actin-normalized proteins values of three independent experiments are shown (right panel). Mean ± SD; *p < 0.05, **p < 0.01. (D) Representative flow cytometric analysis of surface expression of activating ligands for NK-cell receptors in SK-N-SH transfected for 48 h with control vector (ctrl, gray line) or with MYCN cDNA vector (MYCN, blue line); dotted lines, isotype-matched negative controls (left panel). Summary of five independent flow cytometric analyses (right panel). p values, compared with ctrl and MYCN cDNA vector transfected cells (two-tailed paired Student's t-test); *p < 0.05, **p < 0.01.
To further confirm that MYCN expression affects that of NK-cell-activating receptor ligands, we transiently transfected a non-MNA NB cell line as SK-N-SH with piRV-neoSV vector bearing MYCN cDNA (SK-N-SH-MYCN) or the empty vector (SK-N-SH-ctrl) as control. The overexpression of MYCN, peaking at 16 h and gradually decreasing until 72 h, induced downregulation of c-MYC (from 16 to 72 h) and upregulation of both p53 (from 16 to 48 h) and MDM2 (from 16 to 24 h), as revealed by western blotting (Fig. 2C). SK-N-SH-MYCN cells showed decreased surface expression of activating ligands as compared with SK-N-SH-ctrl cells, significantly evident 48 h after transfection (Fig. 2D).
To test whether the increased expression of NK-cell-activating receptor ligands induced by MYCN modulation could affect NK-cell-mediated recognition of NB cells, we performed degranulation and cytotoxicity assays using untreated and Doxy-treated Tet-21/N cells, as well as SK-N-SH-ctrl and SK-N-SH-MYCN cell lines as targets. After 16-h treatment with doxycycline, Tet-21/N cells were significantly more susceptible to NK-cell-mediated lysis than untreated cells in both degranulation and cytotoxicity assays (Figs. 3A and B). Conversely, SK-N-SH-MYCN cell lines were less susceptible to NK cell recognition and lysis, compared with control cells, as evaluated in both degranulation and cytotoxicity assays (Figs. 3C and D). Of note, Tet-21/N cells treated with doxycycline for 3 and 10 d were less susceptible to NK-cell-mediated recognition than untreated cells (Fig. S2C).
Figure 3.
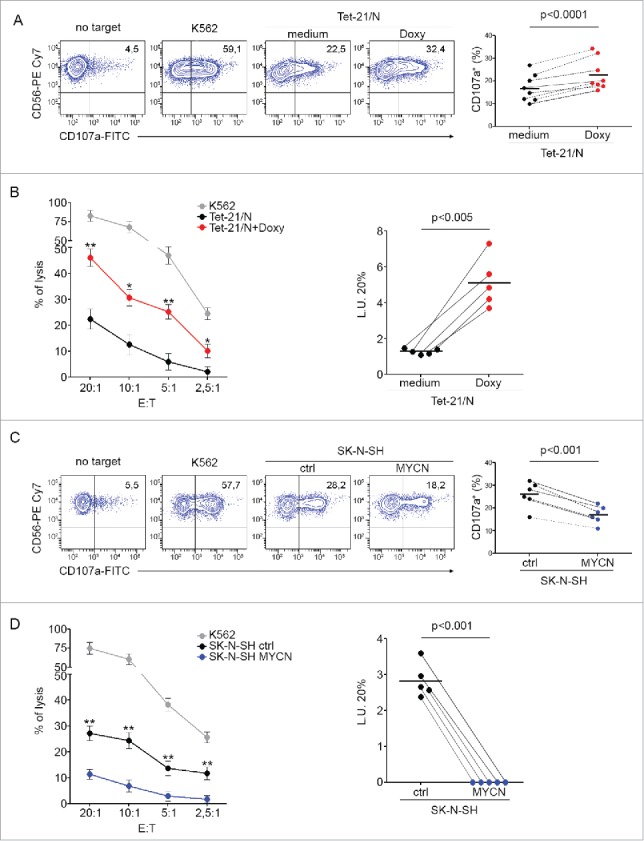
Modulation of MYCN renders NB cells differently susceptible to NK-cell-mediated lysis. (A) Degranulation of human CD3−CD56+ CD45+ NK cells from healthy donors, measured as CD107a cell-surface expression following stimulation with Tet-21/N cells, either left untreated (medium) or treated with doxycycline for 16 h (Doxy). K562 cells were used as positive control. The percentage of CD107a+ NK cells is indicated. A representative experiment out of the eight performed is shown (left panel). Summary of NK-cell degranulation of cells isolated from eight healthy donors is shown (right panel). Dots, percentage of CD107a+ NK cells; horizontal bars, average values. Dashed lines connect percentage of CD107a+ NK cells from each donor cells. p values, compared with untreated and doxycycline-treated Tet-21/N cells (two-tailed paired Student's t-test). (B) Tet-21/N untreated or treated as in (A) were tested as targets for NK cells at the indicated effector:target (E:T) ratios in a standard 51Cr-release assay. One representative experiment out of the five performed is shown (left panel). p values, compared with untreated and doxycycline-treated Tet-21/N cells (two-tailed paired Student's t-test); *p < 0.05, **p < 0.01. Summary of cytotoxic assay of Tet-21/N cells treated as in A and tested as targets of NK cells isolated from five healthy donors in a standard 51Cr-release assay (right panel). Specific lysis was converted to L.U. 20%. Dots, L.U. 20% of the effector/target pairs tested; horizontal bars, average values. Dashed lines connect L.U. 20% from each donor cells obtained with the indicated target. p values, compared with untreated and doxycycline-treated Tet-21/N cells (two-tailed paired Student's t-test). (C) Degranulation assay of human NK cells, as in (A), following stimulation with SK-N-SH cells transfected for 48 h with piRVneoSV vector (ctrl) or with piRVneoSV-MYCN (MYCN). One representative experiment out of the six performed (left panel) and the summary of six experiments (right panel) are shown. (D) Cytotoxic activity of healthy donor NK cells, as in (B), against SK-N-SH cells transfected for 48 h with ctrl or MYCN cDNA vector. One representative experiment out of the five performed (left panel) and the summary of five experiments (right panel) are shown.
These results revealed that MYCN overexpression contributes to protect NB cells from NK-cell-mediated recognition and killing, thus delineating a novel mechanism of tumor escape from the control of the innate immune system, that is mediated by the repression of NK-cell-activating receptor ligands. This immune evasion mechanism adds to another already reported in relation to NB-infiltrating NKT cells, that is mediated by the inhibition of chemokine production.25 These data suggest that a pharmacologic treatment aimed at decreasing MYCN expression in high-risk NB could represent a novel valuable therapeutic strategy to restore the NK-cell-mediated immune response against tumor cells.
The expression of MYCN in primary NB samples is inversely correlated with that of ligands for NK-cell-activating receptors
Next, the expression of MYCN and that of MICA, ULBP1, ULBP2, ULBP3, PVR and Nectin-2 was investigated in 12 different primary NB samples (Table 2) by qPCR. Similarly to NB cell lines, MYCN expression was inversely correlated with that of activating ligands also in NB patient samples (Fig. 4A). A significant inverse correlation between the expression of MYCN and that of MICA and Nectin-2 (Fig. 4B), and a trend toward an inverse correlation between the expression of MYCN and that of the other activating ligands (data not shown), was detected. Thus, high levels of MYCN expression corresponded to low levels of expression of activating ligands and vice-versa, with the exception for some activating ligands in patients 6, 12 and 8. Interestingly, primary sample from patient 12 expressed moderate levels of MYCN and high levels of MICA, ULBP1 and ULBP2. Notably, this patient had 4S NB, a tumor characterized by high rate of spontaneous regression.13
Table 2.
Diagnostic characteristics of NB patients.
| Patient number | Age | Stage | Subtype§ | MYCNβ | 1qDelγ | Diagnostic categoryδ | Differentiation gradeε | Histological classificationθ | Tumor sideλ | Follow-up |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 16y 9m | 1 | GN | un | un | SD | M | fav. | A | alive |
| 2 | 7y | 1 | GN | un | un | SD | M | fav. | EA | alive |
| 3 | 3y 4m | 3 | GN | un | un | SD | M | fav. | EA | alive |
| 4 | 7y 2m | 1 | GNBL | un | un | SR | / | / | M | alive |
| 5 | 1y 6m | 1 | NB | no | no | SP | SD | fav. | A | alive |
| 6 | 3m | 1 | NB | no | no | SP | SD | fav. | A | alive |
| 7 | 8m | 1 | NB | no | no | SP | SD | fav. | EA | alive |
| 8 | 16y 8m | 2 | NB | no | imb. | SP | SD | unf. | EA | alive |
| 9 | 1y 8m | 4 | NB | yes | imb. | SP | SD | fav. | EA | dead |
| 10 | 1y 4m | 4 | NB | gain | no | SP | SD | unf. | A | alive |
| 11 | 3y 11m | 4 | NB | yes | no | SP | SD | unf. | EA | alive |
| 12 | 4m | 4s | NB | no | no | SP | SD | fav. | A | alive |
GN: ganglioneuroma; GNBL: ganglioneuroblastoma; NB: neuroblastoma.
un: undetectable; no: no-amplified; yes: amplified; imb.: imbalance.
SD: stroma dominant; SR: stroma reach; SP: stroma poor.
M: maturing; SD: scarcely differentiated.
fav.: favorable; unf: unfavorable.
A: adrenal; EA: extra-adrenal; M: mediastinal mass.
Figure 4.
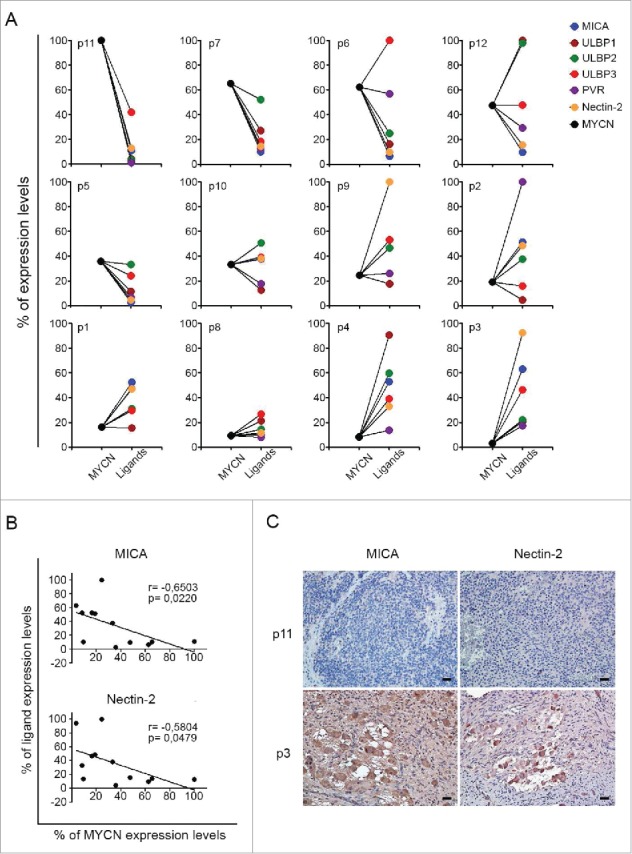
Expression of MYCN and of NK-cell-activating receptor ligands in primary NB cells. (A) qPCR analysis of MYCN and the indicated activating ligands in 12 primary NB samples. GAPDH was used for normalization. Data were normalized as percentages of the highest value obtained for each marker and plotted as percentage of expression levels. Data are representative of four experiments. (B) Scatter plots showing the correlation between MYCN and both MICA and Nectin-2 in 12 NB samples. The means of four independent qPCR analyses, expressed as percentage of expression levels, of MYCN are plotted against those of MICA and Nectin-2; Spearman correlation r and p value are shown for each plot. (C) Expression of MICA and Nectin-2 in primary NB samples by immunohistochemistry assay. MICA and Nectin-2 expressing cells are shown in brown. Nuclei are counterstained with hematoxylin (blue). Original magnification, ×20. Scale bars 30 µm.
Finally, the expression of MICA and Nectin-2 protein was evaluated by immunohistochemical assay in primary NB samples from patient 11 and 3 that displayed markedly different expression of MYCN mRNA and of activating ligands (Fig. 4A). The stroma-poor tumor from patient 11, characterized by MYCN gain and low differentiation grade (Table 2), showed low expression of both MICA and Nectin-2 (Fig. 4C, upper panels). By contrast, the stroma-rich tumor from patient 3, characterized by undetectable MYCN expression, with mature differentiation grade (Table 2), showed high expression levels of both MICA and Nectin-2 in ganglion cells (Fig. 4C, lower panels). Thus, the inverse correlation between the expression of MYCN and MICA and Nectin-2 ligands, detected at the mRNA level, was confirmed at the protein level in primary NB samples.
In conclusion, this study shows that the expression of MYCN is inversely correlated with that of the activating ligands for NKG2D and DNAM1 NK-cell receptors in NB cells. As a consequence, MYCN expression could represent a biomarker allowing to predict the susceptibility of NB cells to NK-cell-mediated immunotherapy. In addition, we suggest that MYCN targeting could represent a novel therapeutic strategy to induce the expression of activating ligands in MNA NB samples. Further investigations exploiting the potential activation of NK cells against NB cells following treatment with drugs able to efficiently downmodulate MYCN levels are warranted.
Materials and methods
Cell lines and reagents
Human NB cell lines were obtained as follow: GICAN and ACN from Interlab Cell Line Collection, Banca Biologica and Cell Factory (www.iclc.it), SK-N-AS, SH-SY5Y, SH-EP, SK-N-SH, SK-N-BE(2)c, IMR-32 from the American Type Culture Collection (ATCC), LA-N-1 from Creative Bioarray, LA-N-5 from the Leibniz-Institut DMSZ, SMS-KCNR from Children's Oncology Group Cell Culture, while the Tet-21/N cell line was kindly provided by Dr M. Schwab (University of Heidelberg, Heidelberg, Germany). All NB cell lines were characterized by (i) HLA class I typing by PCR-SSP sets (Genovision) according to the manufacturer's instructions, and (ii) array CGH (see below). The human erythro-leukemia cell line K562 was purchased from ATCC and used as control target for NK cell functional assays. Cells were grown in RPMI 1640 medium supplemented with 10% FBS (Thermo Fisher Scientific), 2 mM glutamine, 100 mg/mL penicillin and 50 mg/mL streptomycin (Euro Clone S.p.a.). Doxycycline (Sigma Aldrich) was used at 10 ng/mL. Lipofectamine 2000 was used, according to manufacturer's instructions (Invitrogen), to transfect SK-N-SH cells with pIRVneoSV empty vector or pIRVneoSV-MYCN, both kindly provided by G. Giannini (“La Sapienza” University of Rome, Italy).
Patient samples
Tumor samples from 12 NB patients and sections of normal intestinal mucosa and colon carcinoma, diagnosed at the Bambino Gesù Children's Hospital, were used. For each patient, written informed parental consent and approval by the Ethical Committee of the Institution were obtained. Details on clinical information are provided in Supplementary Materials and Methods.
Antibodies, western blotting and flow cytometry
The following antibodies were used: anti-MYCN, anti-p53, anti-Actin (B8.4.B, FL-393 and I-19, respectively, Santa Cruz Biotechnology), anti-MYC (Y69, OriGene), and anti-MDM2 (2A10, Calbiochem-Millipore) for western blotting; anti-CD107a-FITC (H4A3), anti-CD3-Alexa-700 (UCHT1), anti-CD56-PE-Cy7 (B159), anti-CD45 (HI30), FITC-conjugated rat anti-mouse IgG1 (A85–1) and PE-conjugated rat anti-mouse IgM (R6–60.2) purchased from BD Biosciences; anti-ULBP1-PE (170818), anti-ULBP2/5/6-PE (165903), anti-ULBP3-PE (166510), anti-MICA (159227), anti-MICB (236511), anti-TRAIL/R2-APC (17908), anti-CD155/PVR-PE (300907), anti-Nectin-2/CD112-APC (610603) purchased from R&D Systems; W6/32 which recognizes human fully-assembled MHC class I heavy chains and goat F(ab’)2 Fragment anti-mouse IgG FITC (IM1619, Dako) for flow cytometry; anti-MICA and anti-Nectin-2 (62540 and 154895, respectively, Abcam) for immunohistochemistry assay.
Whole-cell extracts were quantified by the bicinchoninic acid assay (Thermo Fisher Scientific), resolved on 8–10% SDS-PAGE and electroblotted. Filters were probed with primary antibodies followed by goat anti-mouse IgG HRP conjugated (Jackson). Flow cytometry was performed on FACSCantoII (BD Bioscences) and analyzed by FlowJo Software.
Senescence-associated β-galactosidase (SA β-Gal) activity
Subconfluent Tet-21/N left untreated or treated with doxycycline, cultured in six-well plates, were fixed using 4% formaldehyde for 10 min at room temperature, washed twice with PBS and then stained for β-gal activity at pH 6.0, according to manufacturer's instructions (Promega). Images were acquired on an Olympus IX51 inverted microscope, and SA β-Gal positive (blue) cells were counted for five fields of view (× 20 magnification) per well.
Array CGH
DNA from NB cell lines was tested by high-resolution array comparative genomic hybridization (CGH). The test involved the use of a 180 K platform with a mean resolution of approximately 40 kb (4 × 180 platform, Agilent Technologies). A copy number variant was defined as a displacement of the normal value of at least three consecutive probes, and the mapping positions refer to the Genome Assembly hg19 (build 37). The quality of the test was assessed on the strength of the QCmetrics values. Polymorphisms (http://projects.trag.ca/variation/) were not included because considered normal variants.
Quantitative mRNA expression
Total RNA was extracted using TRIzol Reagent (Thermo Fisher Scientific). First-strand cDNA was synthesized using the SuperScript II First Strand cDNA synthesis kit (Thermo Fisher Scientific). Quantitative real-time PCR (qPCR) reactions were performed using pre-validated TaqMan gene expression assays from Applied Biosystems, Thermo Fisher Scientific (Hs00792195_m1 for MICA, Hs00360941_m1 for ULBP1, Hs00607609_m1 for ULBP2, Hs00225909_m1 for ULBP3, Hs00197846_m1 for PVR, Hs01071562_m1 for Nectin-2). Relative gene expression was determined using the 2−∆∆Ct method and 2−∆Ct considered as expression level, with GAPDH (Hs02758991_g1) as endogenous control.
NK cell isolation
Human NK cells were isolated from peripheral blood mononuclear cells (PBMCs) of healthy donors with the RosetteSep NK-cell enrichment mixture method (StemCell Technologies) and Ficoll-Paque Plus (Lympholyte Cedarlane) centrifugation. NK cells were routinely checked for the CD3−CD56+ immunophenotype by flow cytometry and those with purity greater than 90% were cultured with 600 IU/mL of recombinant human IL-2 (PeproTech) at 37°C and used up to 5 d after isolation.
Cytotoxicity and degranulation assay
NK cell cytotoxic activity was tested by a standard 4-h 51Cr-release assay. Degranulation assay was performed by co-culturing NK cells with target cells at 1:1 ratio for 3 h, in complete medium in presence of anti-CD107a and in the last 2 h of GolgiStop (BD Bioscence). Then, cells were stained with anti-CD56 and anti-CD45 and expression of CD107a was evaluated by flow cytometry in the CD56+CD45+ subset. Specific lysis was converted to lytic units (L.U.) calculated from the curve of the percentage lysis. One lytic unit is defined as the number of NK cells required to produce 20% lysis of 106 target cells during the 4 h of incubation.
Immunohistochemistry assay
Formaldehyde-fixed paraffin-embedded blocks were cut into 3-μm sections and baked for 60 min at 56° C in a dehydration oven. Antigen retrieval and deparaffinization were performed on a PT-Link (Agilent Technologies) using the EnVision FLEX Target Retrieval Solution kits at high pH (Agilent Technologies) for both MICA and Nectin-2, as per manufacturer's instruction. Slides were then blocked for endogenous peroxidase for 10 min with a peroxidase blocking solution (Agilent Technologies), rinsed in the appropriate wash buffer (Agilent Technologies), and incubated for 30 min with 5% PBS/BSA. Slides were then incubated overnight at 4°C with primary antibodies MICA (1:200) and Nectin-2 (1:300). Twenty minutes incubation with secondary antibody coupled with peroxidase (Agilent Technologies) has been subsequently performed. Bound peroxidase was detected with diaminobenzidine (DAB) solution and EnVision FLEX Substrate buffer containing peroxide (Agilent Technologies). Tissue sections were counterstained with EnVision FLEX Haematoxylin (Agilent Technologies). Sections of normal intestinal mucosa and colon carcinoma were used as positive controls for MICA and Nectin-2, respectively (Fig. S3). Isotype-matched mouse mAbs were used as negative controls.
Statistical analysis
Digital images of western blotting were analyzed by Image J (http://rsbweb.nih.gov/ij/index.html) and statistical significance of densitometric values was evaluated by the two-tailed paired Student's t-test. Normalized values were analyzed for correlation by the regression analysis using GraphPad software. p values lower than 0.05 were considered to be statistically significant.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the Italian Ministry of Health (Rome, Italy) Grant GR-2011–02352151 to Loredana Cifaldi and from Fondazione Neuroblastoma to Franco Locatelli.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Eng J Med 2010; 362:2202-11; PMID:20558371; https://doi.org/ 10.1056/NEJMra0804577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, Nau MM, Witte ON, Toran-Allerand D, Gee CE et al. Differential expression of MYC family genes during murine development. Nature 1986; 319:780-3; PMID:2419762; https://doi.org/ 10.1038/319780a0 [DOI] [PubMed] [Google Scholar]
- 3.Raffaghello L, Prigione I, Bocca P, Morandi F, Camoriano M, Gambini C, Wang X, Ferrone S, Pistoia V. Multiple defects of the antigen-processing machinery components in human neuroblastoma: immunotherapeutic implications. Oncogene 2005; 24:4634-44; PMID:15897905; https://doi.org/ 10.1038/sj.onc.1208594 [DOI] [PubMed] [Google Scholar]
- 4.Marcus A, Gowen BG, Thompson TW, Iannello A, Ardolino M, Deng W, Wang L, Shifrin N, Raulet DH. Recognition of tumors by the innate immune system and natural killer cells. Adv Immunol 2014; 122:91-128; PMID:24507156; https://doi.org/ 10.1016/B978-0-12-800267-4.00003-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raffaghello L, Prigione I, Airoldi I, Camoriano M, Levreri I, Gambini C, Pende D, Steinle A, Ferrone S, Pistoia V. Downregulation and/or release of NKG2D ligands as immune evasion strategy of human neuroblastoma. Neoplasia 2004; 6:558-68; PMID:15548365; https://doi.org/ 10.1593/neo.04316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Textor S, Fiegler N, Arnold A, Porgador A, Hofmann TG, Cerwenka A. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res 2011; 71:5998-6009; PMID:21764762; https://doi.org/ 10.1158/0008-5472.CAN-10-3211 [DOI] [PubMed] [Google Scholar]
- 7.Nanbakhsh A, Pochon C, Mallavialle A, Amsellem S, Bourhis JH, Chouaib S. c-Myc regulates expression of NKG2D ligands ULBP1/2/3 in AML and modulates their susceptibility to NK-mediated lysis. Blood 2014; 123:3585-95; PMID:24677544; https://doi.org/ 10.1182/blood-2013-11-536219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr-Wilkinson J, O'Toole K, Wood KM, Challen CC, Baker AG, Board JR, Evans L, Cole M, Cheung NK, Boos J et al. High frequency of p53/MDM2/p14ARF pathway abnormalities in relapsed neuroblastoma. Clin Cancer Res 2010; 16:1108-18; PMID:20145180; https://doi.org/ 10.1158/1078-0432.CCR-09-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992; 69:1237-45; PMID:1535557; https://doi.org/ 10.1016/0092-8674(92)90644-R [DOI] [PubMed] [Google Scholar]
- 10.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumor suppressor p53. Nature 1993; 362:857-60; PMID:8479525; https://doi.org/ 10.1038/362857a0 [DOI] [PubMed] [Google Scholar]
- 11.Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, Shohet JM. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci U S A 2005; 102:731-6; PMID:15644444; https://doi.org/ 10.1073/pnas.0405495102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Iraci N, Gherardi S, Gamble LD, Wood KM, Perini G, Lunec J, Tweddle DA. p53 is a direct transcriptional target of MYCN in neuroblastoma. Cancer Res 2010; 70:1377-88; PMID:20145147; https://doi.org/ 10.1158/0008-5472.CAN-09-2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norris MD, Bordow SB, Marshall GM, Haber PS, Cohn SL, Haber M. Expression of the gene for multidrug-resistance-associated protein and outcome in patients with neuroblastoma. N Eng J Med 1996; 334:231-8; PMID:8532000; https://doi.org/ 10.1056/NEJM199601253340405 [DOI] [PubMed] [Google Scholar]
- 14.Westermann F, Muth D, Benner A, Bauer T, Henrich KO, Oberthuer A, Brors B, Beissbarth T, Vandesompele J, Pattyn F et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol 2008; 9:R150; PMID:18851746; https://doi.org/ 10.1186/gb-2008-9-10-r150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mistry AR, O'Callaghan CA. Regulation of ligands for the activating receptor NKG2D. Immunology 2007; 121:439-47; PMID:17614877; https://doi.org/ 10.1111/j.1365-2567.2007.02652.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, Marcenaro S, Reymond N et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med 2003; 198:557-67; PMID:12913096; https://doi.org/ 10.1084/jem.20030788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, Bottino C, Moretta L. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med 1995; 182:875-84; PMID:7650491; https://doi.org/ 10.1084/jem.182.3.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamai L, Ahmad M, Bennett IM, Azzoni L, Alnemri ES, Perussia B. Natural killer (NK) cell-mediated cytotoxicity: differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J Exp Med 1998; 188:2375-80; PMID:9858524; https://doi.org/ 10.1084/jem.188.12.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breit S, Schwab M. Suppression of MYC by high expression of NMYC in human neuroblastoma cells. J Neurosci Res 1989; 24:21-8; PMID:2810395; https://doi.org/ 10.1002/jnr.490240105 [DOI] [PubMed] [Google Scholar]
- 20.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9:749-58; PMID:19776744; https://doi.org/ 10.1038/nrc2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Tweddle DA. p53, SKP2, and DKK3 as MYCN target genes and their potential therapeutic significance. Front Oncol 2012; 2:173; PMID:23226679; https://doi.org/ 10.3389/fonc.2012.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Maerken T, Rihani A, Dreidax D, De Clercq S, Yigit N, Marine JC, Westermann F, De Paepe A, Vandesompele J, Speleman F. Functional analysis of the p53 pathway in neuroblastoma cells using the small-molecule MDM2 antagonist nutlin-3. Mol Cancer Therapeutics 2011; 10:983-93; PMID:21460101; https://doi.org/ 10.1158/1535-7163.MCT-10-1090 [DOI] [PubMed] [Google Scholar]
- 23.Tang H, Sampath P, Yan X, Thorne SH. Potential for enhanced therapeutic activity of biological cancer therapies with doxycycline combination. Gene Therapy 2013; 20:770-8; PMID:23282955; https://doi.org/ 10.1038/gt.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan CJ, Smyth MJ, Martinet L. Molecular mechanisms of natural killer cell activation in response to cellular stress. Cell Death Differentiation 2014; 21:5-14; PMID:23579243; https://doi.org/ 10.1038/cdd.2013.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song L, Ara T, Wu HW, Woo CW, Reynolds CP, Seeger RC, DeClerck YA, Thiele CJ, Sposto R, Metelitsa LS. Oncogene MYCN regulates localization of NKT cells to the site of disease in neuroblastoma. J Clin Invest 2007; 117:2702-12; PMID:17710228; https://doi.org/ 10.1172/JCI30751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.