Abstract
Milk proteins are a complex and diverse source of biological activities. Beyond their function intact, milk proteins also act as carriers of encrypted functional sequences that when released as peptides exert biological functions, including antimicrobial and immunomodulatory, which could contribute to the infant’s competitive success. Research has now revealed that the release of these functional peptides begins within the mammary gland itself. A complex array of proteases produced in mother’s milk have been shown to be active in the milk, releasing these peptides. Moreover, our recent research demonstrates that these milk proteases continue to digest milk proteins within the infant’s stomach, possibly even to a larger extent than the infant’s own proteases. As the neonate has relatively low digestive capacity, the activity of milk proteases in the infant may provide important assistance to digesting milk proteins. The coordinated release of these encrypted sequences is accomplished by selective proteolytic action provided by an array of native milk proteases and infant-produced enzymes. The task for scientists is now to discover the selective advantages of this protein-protease based peptide release system.
Introduction
As scientists have interrogated mammalian lactation and its remarkable product milk as a complete, comprehensive and sustaining diet for mammalian infants it has become increasingly clear that the simple term ‘milk’ does not do justice to this complex process. Using the term ‘milk’ implies that there is such a thing as a single, ideal sample of this biofluid with a sole definable composition of nutrients. In fact, milk is highly dynamic with many aspects changing constantly throughout the duration of lactation. Scientists must now develop research models that address this dynamic nature of milk. It is also as important to annotate the consequences of the changes in milk as well as the simple compositional diversity. Ultimately, lactation’s dynamic dimension must be understood in terms of its relevance to the nourishment of infants and the protection of the maternal investment. In addition to understanding the temporal dimension, milk is also biologically active containing enzymes, antibodies, cells and microorganisms. Unfortunately, this dimension too is poorly understood and the ‘proof’ of this is the way milk is processed today. The vast majority of these biologically active components, structures and organisms are irreversibly inactivated by the storage and stabilization processes that are applied to human milk if not consumed immediately and of course, are unavailable in current generation infant formula.
The biological processes that constitute the full dimension of mammalian lactation evolved under the relentless pressure of Darwinian evolution through the genetic success of the mother – infant pair. This evolutionary perspective of milk is considering the unique set of selective pressures that have shaped lactation over more than 200 million years [1]. Every milk component costs the mother. Without benefit to the infant, all components, especially those that are essential nutrients, are under negative selective pressure due to the cost to the mother’s survival. Thus, while most research has focused on the nutritional value of milk to infants as a complete diet, lactation properties that supported acute protection of the maternal – infant dyad and the successful long term development of the infant are also selectable traits. Research on the proteins of milk are beginning to reveal a more complex nutrition and protection system than previously considered, tailored to individual infants targeted to specific stages of development. Furthermore, proteins are emerging as a much more complex and diverse source of biological activities. Proteins as biopolymers possess distinct properties intact, act also as carriers of encrypted functional sequences that when released as peptides add distinct properties. The coordinated release of these encrypted sequences is accomplished by selective proteolytic action provided by an array of native milk proteases and infant-produced enzymes. Now the task for scientists is to discover the selective advantages of this protein-protease based peptide release system. The evidence to date suggests that these peptides have antimicrobial, immunomodulatory and other functions that protect the infant from pathogens, guide development and contribute in diverse ways to the infant’s competitive success.
A New Model for Protein Nutrition
The current paradigm for protein nourishment in humans is that intact proteins are denatured by stomach acid and attacked by endogenous proteases in the stomach beginning a digestive process that continues with hydrolysis by neutral proteases in the small intestine ultimately leading to the release and complete absorption of amino acids by the intestinal epithelia. Young infants however are developmentally naïve, produce relatively little gastric acid, and express relatively low protease activity. Nonetheless all evidence suggests that infants digest and absorb milk proteins effectively. How is this possible? The explanation for this apparent inconsistency comes from recent research showing that milk itself contains an array of proteases that are activated within the infant and contribute considerable catalytic activity and specificity to the entire process of protein digestion within the infant. This research is also revealing that in addition to aiding the infant in protein digestion per se, the specificity of digestion is so precise that there is in practice a further role for milk proteins: that they serve as carriers of encrypted peptide fragments that, upon release by selective proteases, exert functions that contribute to infant protection, health and development.
A long and successful history of research has defined the intestinal enzymes capable of digesting proteins within the human gut. Studies have also been conducted on this repertoire of enzymes and their actions on milk proteins and have identified a large number of peptides released from milk proteins [2, 3]. Parallel studies have examined the actions of milk peptides, including, antimicrobial, anti-inflammatory, mucosal healing, blood pressure-lowering, antithrombotic and calcium delivery [4]. A new set of tools are emerging from chemistry, genomics, computational biology and clinical medicine that are being directed to understanding digestion within the gastrointestinal tract of actual infants, to identifying the peptides that are actually present within breast fed infants, the enzymes responsible for releasing them and the biological properties that they can potentially provide to preterm infants. This principle has been demonstrated using samples from full term and premature infants with indwelling gastric feeding tubes combined with state-of-the-art analytics capable of determining peptide sequences with mass spectrometry and database searching and predict their functions with bioinformatic tools [5, 6].
The identification of hundreds of novel peptides was made possible by revolutionary breakthroughs in analytical chemistry of peptides [7]. Mapping these peptides to biological actions will be a major challenge for biologists as they seek to annotate their functional roles, including antimicrobial and immunomodulatory activities [3, 7]. These peptides have already been proposed to have a range of roles in protecting, nourishing and guiding the development of the infant and also protecting the mother from mammary infection.
This exciting research field that is continuing to reveal structures and functions of milk proteins provide insights into far more than how to nourish infants. The targets of action, mechanisms of health protection and candidate ingredients are envisioned to assist with the support of premature infants whose enzyme repertoires are naive, the elderly whose digestive capacities are diminished and clinical patients whose intestinal functions are pharmacologically impaired. There is a new optimism the lessons from milk’s proteins and peptides will contribute to the health and development of people of all ages.
Research Discovery
The proteolysis of milk proteins was originally documented and perceived as a defect in individual milk samples. For example, the high protease activity in milk from cows with mastitis was associated with casein degradation and lower cheese yields [8]. This endogenous ‘defect’ concept continued to dominate the scientific perspective of milk proteolysis in part because of the lack of tools to interrogate the degree of specificity of the process. The emergence of highly sensitive and accurate analytical tools capable of analyzing peptides is making it now possible to examine milk in much greater detail and quantitative accuracy [9]. These tools have been applied to examining human milk intact and as it is digested within the infant stomach [3, 5]. The first question that was asked was simple: are peptides released from human milk proteins within the infant intestine? The results (Figure 1) graphically document first, that ‘native’ milk, i.e. human milk samples prior to consumption by the infant contain a diverse array of peptides and second, that peptides diversity and abundance rises rapidly within the infant stomach.
Figure 1.
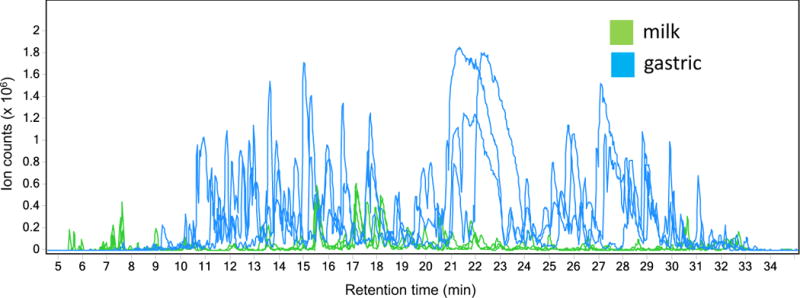
Ion abundances in a) intact human milk and b) gastric aspirates from human infants one hour after consuming the same human milk. Samples were run on an Agilent (Santa Clara, CA) nano-LC-chip-Q-TOF MS/MS (Chip-Q-TOF) with an Agilent chip C18 column. Total ion abundance extracted chromatograms are shown for representative samples.
Enzymatic Proteolysis of milk
The specificity and reproducibility of the digestion of milk proteins provides convincing evidence of a complex, yet controlled active proteolytic system. The origin of that proteolysis began to emerge from studies that showed that hydrolysis begins of specific milk proteins within the mammary gland itself [3]. Once reaching the stomach of the infant proteolysis and release of peptides continues to show considerable protein selectivity and linkage specificity. It is now possible to convert peptide release patterns to sequence specificity of proteolytic cleavage [7, 10]. When this computational strategy is applied to the peptides formed within the stomach of human infants ingesting human milk proteins it is possible to tentatively identify the enzymes that are responsible [10]. Surprisingly, these are not enzymes from the infant, but rather enzymes from the milk itself. Transcriptional profiling of mammary epithelial cells documented that the enzymes predicted by computational analyses of peptides were actively expressed as mRNA in the mammary gland [11]. Thus, milk also contains a mixture of proteases, zymogens (protease precursors), protease activators and protease inhibitors, including components of various proteolytic systems, including plasmin, cathepsin, elastase, kalikrein and amino and carboxypeptidase systems [12]. The basic mechanisms by which enzymes reach milk within the mammary gland are shown in figure 3.
Figure 3.
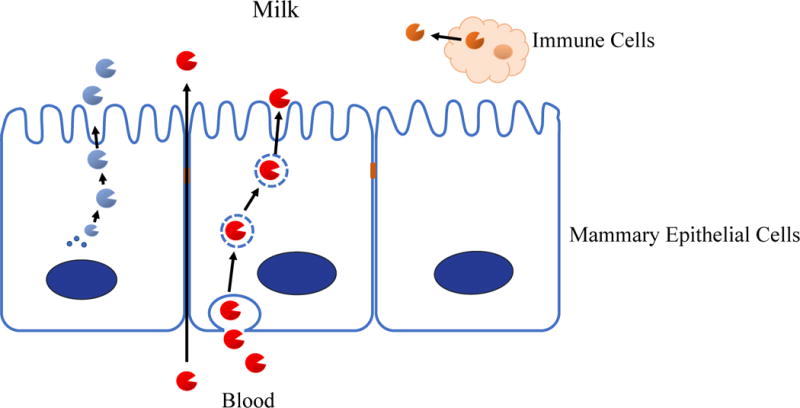
Schematic of production and transfer of enzymes into milk within the lactating mammary gland. Enzymes can reach milk by direct protein synthesis by the epithelial cell, by translocation from blood and from secretion from immune cells within the mammary gland and milk itself.
These studies are confirming that enzymes within the milk contribute to the timing, selectivity, intermediate products and overall success of protein digestion in the infant stomach and intestine. The advantages to the infant and maternal dyad that drove these aspects of milk through evolutionary pressure remain largely unknown. The value of secreting this complex array of proteolytic system components with milk is a tantalizing example of a dimension of food that is not appreciated, but provides insights into mechanisms by which diet supports the infant’s digestive capacity and guides the release of specific, and potentially functional peptides.
Implications to Infant health
A primary role of milk that is well accepted is providing the neonate with high nutritional value proteins. Indeed research over the past century has documented that milk contains hundreds of proteins, many with identified actions, from antibacterial to immunomodulatory. These components are present in milk due to mammary epithelial cell expression, active or passive transport from blood or secretion by host immune cells. Now scientific research must incorporate this dimension of selective hydrolysis of milk proteins and the release of specific peptides into our understanding of milk and its value to infant health.
The control of the catalytic activity of the active components of milk including the proteases appears to be complex and multifactorial including the balance of inhibitors, activators, zymogens, proteases, pH, micro-location and specific protein structure of substrates. These milk proteases act on certain proteins, like the caseins but not on others, notably lactoferrin and immunoglobulin. This selectivity means that in vivo some of the milk proteins remain intact to exert their well known bioactive functions within the infant and others deliver distinct peptides to sites along the complex biogeography of the infant intestine.
The next phase of research is to understand all of the roles of the milk proteases in vivo. In experiments carried out on infants, they are quantitatively important, and by computation are responsible for the majority of protein degradation within the infant stomach [5, 6]. Studies to date provoke the hypothesis that milk proteases are an evolutionarily driven, beneficial class of milk components that contributes to infant digestive capacity and releases functional peptides that affect infant health. While we have previously assumed that the slow development of the human gastric proteolytic system is a liability to infants, even this assumption may be naïve. As infants have lower digestive capacity than adults [13], the abundance and activity of the proteases of milk exert more control over the digestion of milk itself. The transfer of milk digestion to milk and maternal control implies that the apparent digestive insufficiency of infants has selective advantage as well.
CONCLUSIONS
A series of enabling scientific technologies have highlighted the paradoxical properties of milk as a dynamic and biologically active fluid. The breakthroughs that are now emerging on milk emphasize that future research must view the mother – infant dyad as a system of nourishment. Milk needs to be understood for its dynamic nature, its diversity and its biological activities. Specific peptides are released at specific times into specific sites along the mammary gland and infant intestinal tract. This is possible by virtue of the protein structures in milk, the enzymes that are present and the ability of the infant to activate those enzymes at specific places and times. Understanding this magnificent system of nourishment is likely to provide insights for nourishing humans of all ages and all health conditions.
Figure 2.
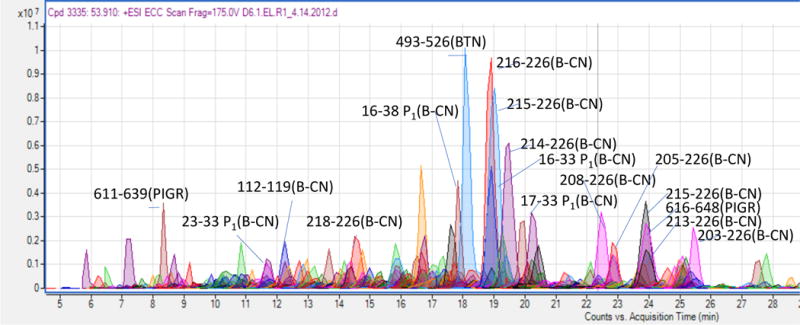
Annotated Ion chromatogram of samples from human stomach aspirates. Peptides were identified by tandem mass spectrometry and mapped to specific proteins according to libraries established through stepwise construction (). Samples were run on an Agilent (Santa Clara, CA) nano-LC-chip-Q-TOF MS/MS (Chip-Q-TOF) with an Agilent chip C18 column.
Acknowledgments
Dr. Dallas’s work is supported by the K99/R00 Pathway to Independence Career Award, Eunice Kennedy Shriver Institute of Child Health & Development of the National Institutes of Health (R00HD079561).
Footnotes
Dr. Dallas reports no conflicts of interest.
References
- 1.Oftedal OT. The mammary gland and its origin during synapsid evolution. J Mammary Gland Biol Neoplasia. 2002;7(3):225–52. doi: 10.1023/a:1022896515287. [DOI] [PubMed] [Google Scholar]
- 2.Lonnerdal B. Human Milk: Bioactive Proteins/Peptides and Functional Properties. Nestle Nutr Inst Workshop Ser. 2016;86:97–107. doi: 10.1159/000442729. [DOI] [PubMed] [Google Scholar]
- 3.Dallas D, et al. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. Journal of Proteome Research. 2013;12(5):2295–2304. doi: 10.1021/pr400212z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clare DA, Swaisgood HE. Bioactive milk peptides: a prospectus. Journal of Dairy Science. 2000;83(6):1187–1195. doi: 10.3168/jds.S0022-0302(00)74983-6. [DOI] [PubMed] [Google Scholar]
- 5.Dallas DC, et al. A peptidomic analysis of human milk digestion in the infant stomach reveals protein-specific degradation patterns. Journal of Nutrition. 2014;144(6):815–820. doi: 10.3945/jn.113.185793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holton TA, et al. Following the digestion of milk proteins from mother to baby. J Proteome Res. 2014;13(12):5777–83. doi: 10.1021/pr5006907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero A, et al. Mechanistic peptidomics: factors that dictate specificity in the formation of endogenous peptides in human milk. Mol Cell Proteomics. 2014;13(12):3343–51. doi: 10.1074/mcp.M113.036194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbano D, Rasmussen R, Lynch J. Influence of milk somatic cell count and milk age on cheese yield. Journal of Dairy Science. 1991;74(2):369–388. [Google Scholar]
- 9.Dallas DC, et al. Current peptidomics: Applications, purification, identification, quantification and functional analysis. Proteomics. 2015;15:1026–1038. doi: 10.1002/pmic.201400310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khaldi N, et al. Predicting the important enzymes in human breast milk digestion. J Agric Food Chem. 2014;62(29):7225–32. doi: 10.1021/jf405601e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickramasinghe S, et al. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genomics. 2012;13(1):45–58. doi: 10.1186/1471-2164-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallas DC, Murray NM, Gan J. Proteolytic Systems in Milk: Perspectives on the Evolutionary Function within the Mammary Gland and the Infant. Journal of mammary gland biology and neoplasia. 2015;20(3–4):1–15. doi: 10.1007/s10911-015-9334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallas DC, et al. Digestion of protein in premature and term infants. Journal of Nutritional Disorders and Therapy. 2012;2(3):112–121. doi: 10.4172/2161-0509.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]


