Abstract
Most bacterial cells are surrounded by a peptidoglycan (PG) cell wall that is essential for their integrity. Major synthases of this exoskeleton are called penicillin-binding-proteins (PBPs) 1,2. Surprisingly little is known about how cells control these enzymes given their importance as drug targets. In the model gram-negative bacterium Escherichia coli, outer membrane lipoproteins are critical activators of the class A PBPs (aPBPs) 3,4, bifunctional synthases capable of polymerizing and crosslinking PG to build the exoskeletal matrix 1. Regulators of PBP activity in gram-positive bacteria have yet to be discovered but are likely to be distinct due to the absence of an outer membrane. To uncover gram-positive PBP regulatory factors, we used transposon-sequencing (Tn-Seq) 5 to screen for mutations affecting the growth of Streptococcus pneumoniae cells when the aPBP synthase PBP1a was inactivated. Our analysis revealed a set of genes that were essential for growth in wild-type cells yet dispensable when pbp1a was deleted. The proteins encoded by these genes included the conserved cell wall elongation factors MreC and MreD 2,6,7 as well as a membrane protein of unknown function (SPD_0768) that we have named CozE (coordinator of zonal elongation). Our results indicate that CozE is a novel member of the MreCD complex of S. pneumoniae that directs the activity of PBP1a to the midcell plane where it promotes zonal cell elongation and normal morphology. CozE homologues are widely distributed among bacteria, suggesting they represent a new family of morphogenic proteins controlling cell wall biogenesis by the PBPs.
To investigate PBP regulation in gram-positive organisms we used the ellipsoid-shaped bacterium S. pneumoniae as a model system. In addition to its interesting morphology, this bacterium is an important human pathogen and the causative agent of many invasive diseases. Antibiotic resistance in S. pneumoniae is on the rise worldwide 8. New drugs to combat resistance in this and other bacterial pathogens are therefore needed. A better understanding of the regulation and cellular function of proven target enzymes like the PBPs will aid the development of such therapies.
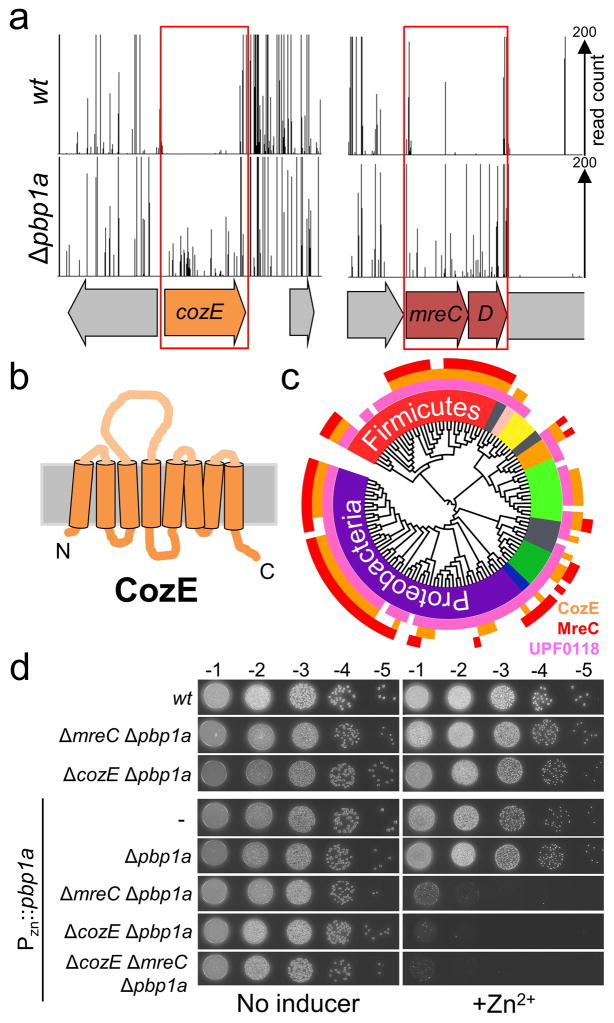
S. pneumoniae encodes three aPBPs (pbp1a, pbp1b and pbp2a), with pbp1a and pbp2a forming an essential pair. Either gene can be deleted individually, but attempts to inactivate both genes have been unsuccessful 9. We reasoned that the lethal phenotype of pbp1a pbp2a double mutants could form the basis of a screen for gram-positive PBP regulators analogous to previous work that identified the Lpo regulators of the E. coli aPBPs 4. The set of mutants synthetically lethal with a deletion of pbp1a is predicted to include factors required for the in vivo function of PBP2a. Similarly, a screen for mutants synthetically lethal with Δpbp2a should identify factors required for PBP1a activity. To identify synthetic interactions, we performed Tn-Seq (IN-Seq, HITS, TraDIS) 5,10,11 using transposon libraries generated in strain D39 lacking its capsule (Δcps) 7 and derivatives inactivated for PBP1a and PBP2a. This approach revealed several factors, which will be investigated in a separate report. Here, we focus on the characterization of an unexpected class of factors with a distinct and intriguing phenotype related to PBP1a, an aPBP that is associated with high-level antibiotic resistance 12 and is indispensable for host colonization 13. The genes encoding these proteins were found to be virtually devoid of insertions in the wild-type transposon library, indicating they are likely essential for growth (Fig. 1A, Supplementary Fig. 1). Strikingly, however, the same genes appeared to be readily inactivated in the Δpbp1a library but not the Δpbp2a library (Fig. 1A, Supplementary Fig. 1), suggesting that pbp1a disruption suppresses their essentiality. Two of the genes encode MreC and MreD, conserved members of the PG biogenesis machinery that promotes cell elongation in rod- and ellipsoid-shaped bacteria 2,6,7. The third gene, spd_0768, encodes CozE, a conserved polytopic membrane protein of unknown function that belongs to the widely-distributed UPF0118 protein family 14 (Fig. 1A–C and Supplementary Fig. 2–3). Like MreC, CozE homologs are absent from the Mollicutes, which lack a cell wall, suggesting a role for CozE in PG biosynthesis (Supplementary Fig. 3).
Figure 1. The essential genes cozE and mreCD can be deleted in cells lacking PBP1a.
a, Mariner transposon libraries were generated in wt (D39 Δcps) and Δpbp1A mutant strains. Transposon insertions within each library were identified by deep sequencing and mapped onto the D39 genome. Two regions of the genome are shown with ORFs indicated by coloured arrows. The two boxes highlight regions significantly enriched for transposon insertions in the Δpbp1A library compared to the wt (p<0.002 in each case). Full Tn-Seq and statistical data are shown in Supplementary Fig. 1.
b, Schematic of the predicted membrane topology of CozE. Adapted from topological predictions using the Phobius web server. Phobius raw data output is shown in Supplementary Fig. 2a.
c, CozE and CozE-like proteins are widely conserved in the bacterial kingdom and co-occur with MreC. The tree shows 129 diverse bacterial species. The presence of UPF0118 family members are indicated in pink on each leaf. Organisms with a S. pneumoniae CozE/MreC homologue are indicated in orange/red (e-value cut off = 1x10−4). A version of the tree with all phyla and leaves labeled is shown in Supplementary Fig. 2b. A larger tree with 1,576 species showing CozE and MreC homologues are not present in the Mollicutes Class is shown in Supplementary Fig. 3.
d, cozE, mreC and mreD are essential in D39 pbp1a+ backgrounds. The indicated S. pneumoniae strains were grown to exponential phase and normalized to an OD600 of 0.2. Resulting cultures were serially diluted and 5 μl of each dilution spotted onto TSAII 5%SB plates in the presence or absence of 600 μM ZnCl2. Plates were incubated at 37°C in 5% CO2 cabinet and imaged. Spot dilutions showing ΔmreD and ΔmreCD essentiality in pbp1a+ backgrounds are shown in Supplementary Fig. 4.
The essentiality of mreC and mreD and its suppression by PBP1a inactivation were expected from prior work of Winkler and colleagues 7. We confirmed these results using a Δpbp1a strain with an ectopic copy of pbp1a under control of a zinc-regulated promoter 15 (PZn-pbp1a). In this strain background, deletion mutants of mreC, mreD, cozE, or both mreC and cozE were viable in the absence of zinc (Fig. 1D, Supplementary Fig. 4). However, the viability of these strains was severely compromised on solid medium supplemented with zinc (Fig. 1D, Supplementary Fig. 4). As an additional confirmation, we deleted mreC or cozE in strain R6, which harbors a hypomorphic pbp1a allele 16 and found that both mutants were viable displaying only mild morphological defects (Supplementary Fig. 5A) 7. Furthermore, expression of the pbp1a gene from strain D39 was lethal in mreC or cozE R6 deletion mutants (Supplementary Fig. 5B). In liquid culture, pbp1a induction was tolerated in wild-type or Δpbp1a cells of strain D39, but caused cell lysis in Δpbp1a ΔmreC and Δpbp1a ΔcozE double mutants as well as the Δpbp1a ΔmreC ΔcozE triple mutant (Fig. 2A–B and Supplementary Fig. 6B). Upon pbp1a induction, these mutants first displayed a cell chaining phenotype followed by significant rounding and swelling of cells in the chains before most cells in the culture lysed (Fig. 2B and Supplementary Fig. 6D). Similar phenotypes were observed upon CozE or MreCD depletion in an otherwise wild-type background (Supplementary Fig. 7) 7. The PBP1a-induced lysis phenotype appeared to be more pronounced in cells lacking both CozE and MreC (Fig. 2A). However, the drop in viability for single mreC or cozE mutants was similar to that of the double mutant (Supplementary Fig. 6C), suggesting these factors function in the same pathway. Deletion of lytA or cbpD encoding the major S. pneumoniae autolysins did not dramatically alter the lytic effect of PBP1a production in the absence of CozE or MreC, indicating that the growth and lysis phenotypes did not result from misactivation of these autolysins 17,18 (Supplementary Fig. 8).
Figure 2. Phenotype of cozE inactivation.

Δpbp1a ΔcozE and Δpbp1a ΔmreC strains have no morphological phenotype Supplementary Fig. 6a. However, expression of pbp1a in ΔcozE or ΔmreC backgrounds results in growth arrest and lysis. a, Δpbp1a strains containing a zinc inducible pbp1A allele were grown in THY to mid-exponential phase. Cultures were diluted in fresh THY to an OD600 of 0.025 in the presence or absence of 600 μM ZnCl2 and incubated at 37°C in a 5% CO2 cabinet. OD600 were recorded approximately every 30 min for 6 h. A wt growth curve is shown for reference. Additional deletion strain controls and Pzn::pbp1a overexpression controls can be found in Supplementary Fig. 6b. Viable counts of cultures at 6 h and 8 h post-induction can be found in Supplementary Fig. 6c.
b, Induction of pbp1a in ΔcozE or ΔmreC backgrounds lead to morphological defects and lysis. Representative images of ΔcozE, ΔmreC and ΔcozE ΔmreC strains upon induction of Pzn::pbp1a. Strains were grown as in panel (a). At indicated time points cells were removed and placed on a THY 2% agarose pad and immediately imaged. Control images showing wt morphology of uninduced strains are shown in Supplementary Fig. 6a. n = 3, scale bar = 3 μm.
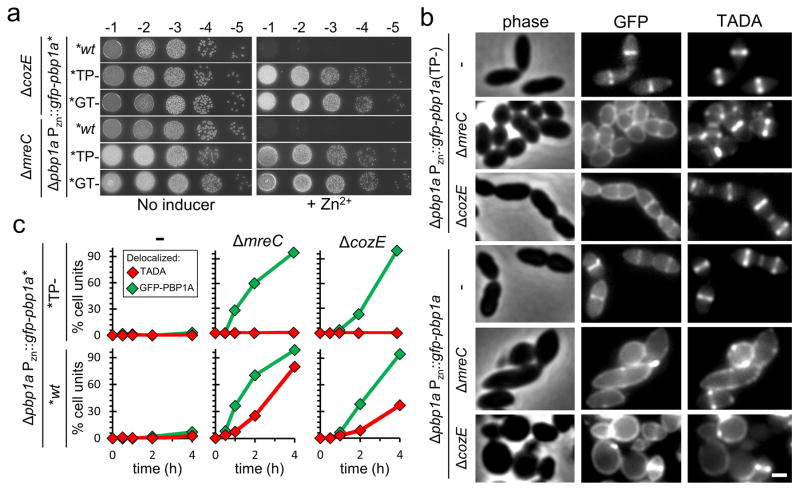
The genetic results suggest a model in which CozE works with the MreCD complex to control PG synthesis by PBP1a and that in their absence, deranged PBP1a activity causes cell lysis. To test this possibility, we monitored PG biogenesis activity using the fluorescent D-amino acid TADA (tetramethylrhodamine 3-amino-D-alanine) 19,20 and the localization of a functional GFP-PBP1a fusion (Supplementary Fig. 9) in cells inactivated for CozE or MreC. As with the untagged version, production of GFP-PBP1a in cells lacking MreC or CozE resulted in a severe growth defect (Fig. 3A and Supplementary Fig. 10A). This phenotype was accompanied by a change in GFP-PBP1a localization and TADA labelling from their normally tightly restricted zone at midcell to a widely distributed pattern throughout the cell periphery. The ΔmreC cells displayed a more severe labelling defect versus ΔcozE cells as expected from the above morphological analysis (Fig. 3B–C and Supplementary Fig. 11). Similar alterations in TADA labelling were observed following the production of untagged PBP1a in the mutant strains (Supplementary Fig. 12). Importantly, variants of GFP-PBP1a inactivated for either PG polymerase/glycosyltransferase activity [GFP-PBP1a(GT−)] or PG crosslinking/transpeptidase activity [GFP-PBP1a(TP−)] similarly lost their midcell localization in cells lacking CozE or MreC, but this delocalization was not associated with a change in TADA labelling nor did it cause a significant growth defect (Fig. 3B–C and Supplementary Fig. 10A). Cells lacking CozE or MreC did not affect midcell localization of GFP-PBP2a, suggesting a specific role in PBP1a recruitment (Supplementary Fig. 13).
Figure 3. Loss of CozE and MreC function results in delocalized PBP1a-dependent PG synthesis.
a, PBP1a enzymatic activity is required to induce lethality in CozE- or MreC-defective cells. The indicated S. pneumoniae strains were grown to mid-exponential phase and normalised to an OD600 of 0.2. Resulting cultures were serially diluted and 5 μl of each dilution spotted onto TSAII 5%SB plates in the presence or absence of 600 μM ZnCl2. Plates were incubated at 37°C in a 5% CO2 cabinet and imaged. Spot dilutions show Pzn::pbp1a induced lethality in ΔcozE or ΔmreC strains can be supressed in cells expressing PBP1a glycosyltransferase (GT−) or transpeptidase defective (TP−) variants.
b, Aberrant GFP-PBP1a and TADA labelling in ΔcozE and ΔmreC strains.Strains were grown to mid-exponential phase, back diluted to an OD600 of 0.025 in THY + 600 μM ZnCl2 and incubated at 37°C in a 5% CO2 cabinet for 3 h 45 min. Where necessary cultures were further back diluted at 2 h to avoid high cell densities and autolysis. Cells were labelled with TADA for 15 min prior to imaging on 2% agarose pads. n = 2, scale bar = 1 μm. Complementation analysis and growth curves demonstrating GFP-PBP1a functionality can be found in Supplementary Fig. 9. Growth curves for all strains demonstrating GFP-PBP1a lethality in ΔcozE and ΔmreC strain backgrounds is similar to untagged PBP1a can be found in Supplementary Fig. 10a. Immunoblot analysis of GFP-PBP1a fusion protein stability and expression levels in all strain backgrounds can be found in Supplementary Fig. 10b. The GFP-PBP1a (TG−) fusion shows a similar phenotype to the GFP-PBP1a (TP−) fusion shown here, see Supplementary Fig. 10c.
c, Quantification of GFP-PBP1a localization and TADA incorporation patterns in ΔcozE and ΔmreC strains. Identical Pzn::gfp-pbp1A constructs expressing either wt or catalytically inactive TP− variants were treated as described in b. Cells were imaged and scored for localization/incorporation patterns at time 0, 30 min, 1 h, 2 h and 4 h post induction. For this analysis each S. pneumoniae cell in the joined diplococcus was scored as a single cell unit. Cell units were sorted into two categories: wt mid-cell localized signal and delocalized signal (examples of which are as shown in b). Graphs comparing the percentage of cells with delocalized signal for each variant are shown side by side in two columns for ΔcozE and ΔmreC compared to the Δpbp1a control strain with >700 cell units scored per time point, n = 2. Demograms of GFP-PBP1a and TADA signal profiles at the 4 h time point are shown in Supplementary Fig. 11. Similar results to those shown in b and c, using untagged PBP1a can be found in Supplementary Fig. 12.
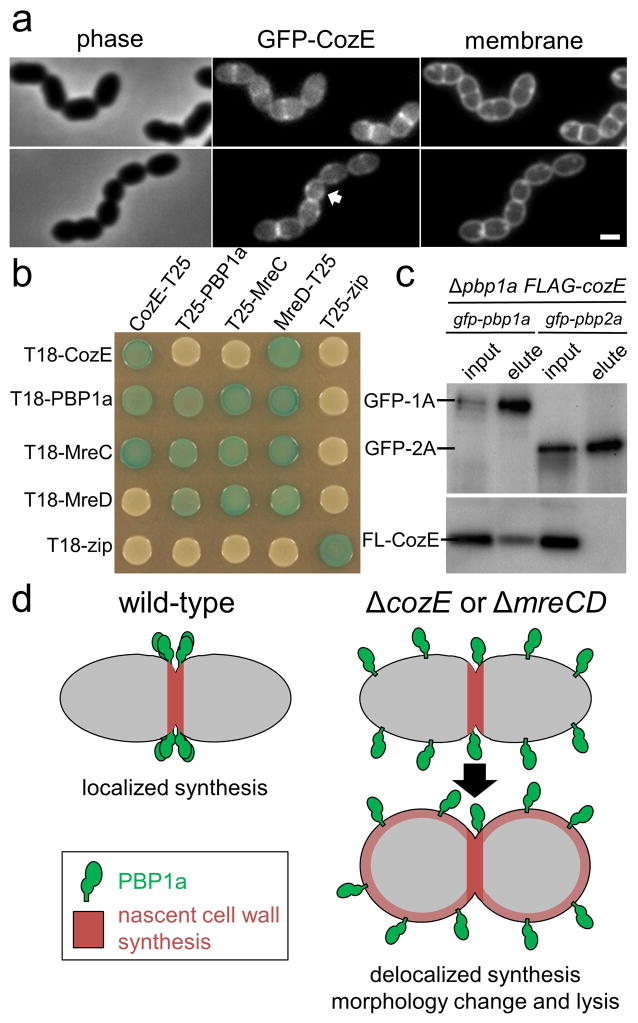
A functional GFP-CozE fusion (Supplementary Fig. 7A) displayed a septal localization pattern that was dependent upon MreC (Fig. 4A and Supplementary Fig. 14). Reciprocally, the midcell localization of GFP-MreC required CozE (Supplementary Fig. 14). Moreover, bacterial two-hybrid analysis 21 in E. coli indicates that CozE forms a complex with MreCD and PBP1a (Fig. 4B and Supplementary Fig. 15). Finally, a functional FLAG-CozE fusion was coimmunoprecipitated with GFP-PBP1a but not GFP-PBP2a (Fig. 4C and Supplementary Fig. 16). Altogether, these data indicate that CozE is a member of the MreCD morphogenic complex in S. pneumoniae and that this complex coordinates cell elongation in part by interacting with and restricting PBP1a to midcell (Fig. 4D).
Figure 4. CozE is a member of the MreCD cell elongation complex.
a, CozE is recruited to mid-cell. S. pneumoniae cells expressing a GFP-CozE under the control of a fucose-inducible promoter were grown to mid-exponential phase in THY + 0.4% fucose. Cells were spotted onto 2% THY agarose pads and imaged. GFP-CozE signal is always enriched at mid-cell positions. In rare cases GFP-CozE can be detected at mid-cell before the onset of membrane invagination (arrow). Two representative images are shown, n = 3, scale bar = 1 μm.
b, Bacterial two-hybrid interactions of CozE with known members of the PG biosynthetic complex. E. coli strain BTH101 (Δcya) cells expressing two protein fusions to either T25(top) or T18(side). T25 and T18 are domains of an adenylate cyclase enzyme. If the target proteins interact the resulting complex complements Δcya resulting in lacZ expression. Thus, if the target proteins interact a blue colony is observed on a LB plate containing X-gal. Strains shown here were grown to stationary phase in LB at 30°C. 5 μl of these cultures were spotted on LB agar containing Xgal 40 μg/ml, incubated 30°C and imaged. The ‘zip’ fusions are to a leucine zipper domain derived from the yeast protein GCN4. This serves as a positive control, but also a negative control against all other target proteins. Representative image of 3 biological replicates is shown. The position of the ‘T25’ and ‘T18’ on the data labels reflect the terminus used for the protein fusion. Additional controls are provided in Supplementary Fig. 15.
c, A functional FLAG-CozE tag co-immunoprecipitates with GFP-PBP1a but not GFP-PBP2a. Membrane preparations of indicated strains were incubated with anti-GFP sepharose resin, washed and eluted in sample buffer. Immunoblot shows matched volumes of solubilized membranes (input) and the elution fraction (elute) probing with anti-GFP and anti-FLAG antibodies. n = 4. Evidence of FLAG-functionality, antibody specificity and a full immunoblot analysis of the co-immunoprecipitation are provided in Supplementary Fig. 16. An untagged PBP1a control showing FLAG-CozE does not precipitate with the resin is included in Supplementary Fig. 16c.
d, CozE and the MreCD complex retains PBP1a at mid-cell in S. pneumoniae. Model for the role of CozE and MreCD in maintaining PBP1a at mid-cell. In wt (cozE+) cells, the cell wall biosynthetic enzyme PBP1a (green) is held at the mid cell position, with all nascent cell wall synthesis occurring exclusively at this location (red shading). In the absence of CozE or MreCD, PBP1a is released from its mid-cell position and catalyzes delocalized PG synthesis causing lethal morphological defects.
In rod-shaped bacteria, MreC and MreD are part of the Rod system that elongates the cylindrical portion of the cell wall 22. The system is organized by dynamic filaments of MreB that facilitate the incorporation of PG at dispersed locations throughout the cylinder 23. In contrast, S. pneumoniae and other ovococci elongate in a restricted zone by incorporating PG at the periphery of the cytokinetic ring 24. These bacteria lack MreB, but retain the other components of the Rod system, including MreC and MreD. It was recently shown that the SEDS-family protein RodA is the core PG polymerase within the Rod system of Bacillus subtilis and E. coli 25,26. Additionally, it was found that although aPBP polymerases principally work outside of the MreB-directed machinery, the two systems display some interdependence through an as yet ill-defined coordination mechanism 26. The results presented in this report suggest the possibility that CozE and related proteins might serve as part of this coordination mechanism by connecting PBP1a with RodA and other components of the elongation machinery via its interactions with PBP1a and the MreCD complex. In this case, CozE may be essential in S. pneumoniae because the spatial localization of aPBPs and their potential coordination with SEDS-family PG polymerases is especially critical for proper PG biogenesis in organisms where zonal cell wall expansion is the principal mode of growth. Such a localized mode of cell elongation is not unique to the ovococci. In addition to the dispersed mode of growth promoted by the Rod system, rod-shaped bacteria like E. coli and Caulobacter cresentus have also been found to undergo zonal elongation for a portion of the cell cycle preceding division 27–29. The broad conservation of CozE suggests that it could more generally recruit and coordinate PG synthetic functions during zonal growth in a range of bacteria. Further characterization of CozE in S. pneumoniae and other organisms will provide deeper mechanistic insight into its function and reveal novel strategies for disrupting PG biogenesis for antibiotic development.
METHODS
Strains, plasmids and routine growth conditions
Unless otherwise indicated, all S. pneumoniae strains in this study were derived from D39 Δcps 16. Cells were grown in Todd Hewitt broth containing 0.5% Yeast Extract (THY) at 37°C in an atmosphere containing 5% CO2. Strains were grown on pre-poured Tryptic Soy Agar 5% sheep blood plates (TSAII 5%SB, Becton Dickinson; BD) with a 5 ml overlay of 1% Nutrient Broth (NB) agar containing additives. When finer control of media components was required, TSA plates containing 5% defibrinated sheep blood were used. A table of all strains, plasmids, and oligonucleotides and a description of strain and plasmid constructions are provided as supplementary material (Supplementary Methods).
Transformation
Cells in mid-exponential phase were grown in THY and back diluted to an OD600 of 0.03. Competence was induced by treating cells with 500 pg ml−1 Competence Stimulating Peptide (CSP-1), 0.2% BSA and 1 mM CaCl2. Typically 1 ml of culture containing 100 ng of gDNA or plasmid DNA was used. Transformants were selected on TSAII overlay plates containing: 5 μg ml−1 chloramphenicol, 0.2 μg ml−1 erythromycin, 250 μg ml−1 kanamycin, 200 μg ml−1 spectinomycin or 0.2 μg ml−1 tetracycline as appropriate.
Transposon insertion sequencing
Transposon insertion sequencing (Tn-Seq) was performed as described previously by van Opijnen, Cammilli and co-workers 5 with minor modifications. A total of four independently generated libraries were used in this study: two in a wt strain and two in the Δpbp1A strain backgrounds. Briefly, a Magellan6 transposon library DNA was transformed into competent S. pneumoniae cells. Approximately 440,000 (wt) and 70,000 (Δpbp1A) transformants were pooled for each strain and genomic DNA isolated. Samples were digested with MmeI, followed by adapter ligation. Transposon–chromosome junctions were amplified in 18 PCR cycles. PCR products were gel-purified and sequenced on the Illumina HiSeq 2500 platform using TruSeq Small RNA reagents (Tufts University Core Facility Genomics). Reads were de-multiplexed and trimmed using the CLC workbench software (Qiagen, Version 6.0.1). Sequences representing transposon-insertion sites were mapped onto the D39 genome using the short read aligner tool Bowtie 30 with a Python script (2.7.6, PSF). Bowtie aligned each sequencing run to a position on the genome, this identified the ‘TA’ insertion site used by the original Mariner transposon in the libraries. The number of times each TA site was identified was scored and used to generate a read count profile across all TA sites in the genome. The read counts are sensitive to the initial number of reads from the sequencing run, therefore the data was normalised using a customised R-Script, normalising the data to the library with the lowest number of reads. After normalisation, a Mann-Whitney U test was used to identify genomic regions with significant differences in transposon insertion profiles using Python scripts (2.7.6, PSF) with both NumPy (© 2005–2013, NumPy Developers) and SciPy (© 2014 SciPy developers) packages. The Mann-Whitney U test treats the TA insertion profile of each ORF independently, comparing them between datasets to identify genes with altered profiles. Insertion data was visualized graphically using the Artemis genome browser (version 10.2) 31. The wt vs Δpbp1a Tn-Seq data is provided in Supplementary Table 1.
Phylogenetic Analysis
CozE and MreC homologues were identified using NCBI BLASTp. S. pneumoniae CozE and MreC protein sequences were used as queries against a database of bacterial genomes with an e-value cut off of 1x10−4. BLAST analysis was carried out using the Harvard Medical School research computing cluster Orchestra (https://rc.hms.harvard.edu/#orchestra).
CozE is a member of the UPF0118 family and contains no other identifiable domains. The UPF0118 family of proteins was identified by the Pfam database (version 29.0) 14. Pfam uses multiple protein alignments to first make a seed alignment, this is used to construct a profile hidden Markov model utilizing the HMMER software. This profile is used to search sequence databases and cluster related proteins into families 15. This approach was taken as a means of non-biased identification of CozE homologues.
In all cases phylogenetic trees displaying the presence or absence of homologues identified by theses analyses were constructed using the Interactive Tree Of Life (v3) web-based tool 32.
Fluorescent Microscopy
S. pneumoniae cells were concentrated by centrifugation at 10,000 g for 1 min and immobilized on 2% agarose pads. Fluorescence microscopy was performed on a Nikon Eclipse Ti-E inverted microscope through a Nikon Plan Apo 100X oil objective (NA 1.4). For fluorescent imaging, a SPECTRA X light engine (Lumencor) was used for excitation in combination with the following filter sets for each fluorophore; GFP: Ex:475/28 Em:500–545 Dichroic: 495, for TADA: Ex:438/24 Em:600–660 Dichroic: 595 and for TMA-DPH: Ex:390/18, Em:435–485, Dichroic: 400. Images were acquired with a CoolSnapHQ2 CCD camera (Photometrics) without gain using Nikon Elements Software (version 4.30). A neutral density 8 (ND8) filter was used to reduce the intensity of excitation light by 87.5%. Typical acquisition times were: 4–7 s GFP-PBP1A/GFP-CozE, 100–150 ms TADA labelled cells and 0.7–1 s TMA-DPH stained samples.
Cell wall labelling using fluorescent D-amino acids (TADA)
The Tetramethylrhodamine(TAMRA) 3-amino-D-alanine (TADA) used in this study was synthesized by Tocris. TADA was used to label nascent S. pneumoniae cell wall material similar to methods described previously 19,20. S. pneumoniae mid-exponential cultures (500 μl) were stained with 50 μM TADA for 15 min at 37°C in a 5% CO2 atmosphere. Cells were centrifuged at 10,000 g for 1 min, washed with 500 μl phosphate-buffered saline (PBS) to remove unincorporated TADA, concentrated, immobilized on 2% PBS agarose pads and imaged immediately.
TMA-DPH membrane staining
Where TMA-DPH was used, 1 ml of culture was centrifuged at 10,000 g for 1 min. Cells were re-suspended in 10 μl of PBS and TMA-DPH added to a final concentration of 50 μM. Cells were imaged on 2% PBS agarose pads.
Microscopy image analysis
Acquired images were processed using the Metamorph image analysis software (version 7.7.0.0). Images were cropped and scale bars added using appropriate tools. Images were corrected for background fluorescence using the subtraction feature.
Bacterial two-hybrid analysis
Competent BTH101 (Δcya) E. coli cells were transformed with two 100 ng plasmid aliquots containing ‘T25’ and ‘T18’ protein fusions in one step. Transformants were selected on LB agar plates containing: 50 μg ml−1 ampicillin (Amp50), 25 μg ml−1 kanamycin (Kan25) and 40 μg ml−1 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Xgal40). Plates were incubated at 30° and checked for signal heterogeneity. Single colonies were picked into 150 μl LB Amp50 Kan25 containing 500 μg ml−1 Isopropyl β-D-1-thiogalactopyranoside (IPTG500) in 96 deep-well plates and incubated at 30°. 5 μl of resulting cultures were spotted onto LB Amp50 Kan25 IPTG500 Xgal40 plates. Plates were incubated at 30°C in an environment protected from light and imaged. Plates were picked in triplicate with selected images representative of three biological replicates.
Immunoblot analysis
S. pneumoniae cultures were normalized to an OD600 of 0.3 and 3 ml harvested. Cell extracts were prepared by resuspension of cell pellets in 100 μl lysis buffer (20 mM Tris pH 7.5, 10 mM EDTA, 1 mg ml−1 lysozyme, 10 μg ml−1 DNase I, 100 μg ml−1 RNase A, with protease inhibitors: 1 mM PMSF, 1 μg ml−1 leupeptin, 1 μg ml−1 pepstatin) and incubation at 37°C for 10 min, followed by addition of 10 μl 10% Sarcosyl for 5 min. 100 μl SDS sample buffer (0.25 M Tris pH 6.8, 4% SDS, 20% glycerol, 10 mM EDTA) containing 10% 2-mercaptoethanol was added to each prep and samples were heated for 15 min at 50°C prior to loading 10 μl per lane. Proteins were separated by SDS-PAGE on 12.5% polyacrylamide gels, electroblotted onto a PVDF membrane and blocked in 5% non-fat milk in PBS-0.5% Tween-20. The blocked membranes were probed with rabbit anti-FtsE (1:20,000) 33 and affinity-purified rabbit anti-GFP (1:10,000) diluted into 3% BSA in 1x PBS-0.05% Tween-20. Primary antibodies were detected using horseradish peroxidase-conjugated goat anti-rabbit IgG (1:20,000, BioRad) and the Western Lighning Plus ECL reagent as described by the manufacturer (PerkinElmer). Membrane chemiluminescence was imaged on a FluorChem R system (ProteinSimple).
Co-immunoprecipitation Assay
S. pnemoniae strains were grown in 60 ml THY in the presence of 400 μM ZnCl2 at 37°C in 5% CO2. 50 ml of these cultures was matched to a starting OD600 of 0.5. Cells were harvested by centrifugation at 5,000 g for 5 min and cell pellets re-suspended in 25 ml SMM (1 M Sucrose, 40 mM Maleic acid, 40 mM MgCl2, pH 6.5). Cells were washed a second time and finally re-suspended in 2 ml SMM. Cell protoplasts were generated by enzymatic digestion of the cell wall with 8 mg ml−1 lysozyme, cells do not lyse due to the osmotic potential of the SMM. Protoplasts were pelleted at 5,000 g for 5 min and re-suspended in 5 ml cold Hypotonic Buffer ‘Buffer-H’ (20 mM HEPES [Na+], 100 mM NaCl, 1mM dithiothreitol [DTT], 1 mM MgCl2, 1 mM CaCl2, 1 mM phenylmethane sulfonyl fluoride [PMSF], 1 μM leupeptin, 1 μM pepstatinA). The protoplasts lysed in H-buffer in due to the change in buffer osmolarity. Lysates were further treated with: DNase 6 μg ml−1, RNaseA 12 μg ml−1 and 8 mg ml−1 lysozyme to form a crude extract. These extracts were pelleted by ultracentrifugation at 100,000 g for 1 h at 4°C. Crude membrane pellets were dispersed in 420 μl Glycerol Buffer ‘Buffer-G’ (20 % glycerol, 20 mM HEPES [Na+], 100 mM NaCl, 1mM dithiothreitol [DTT], 1 mM MgCl2, 1 mM CaCl2, 1 mM phenylmethane sulfonyl fluoride [PMSF], 1 μM leupeptin, 1 μM pepstatinA) and stored at −80°C. 50 μl aliquots of crude membranes were solubilized with 450 μl Buffer-G-DIG (Buffer-G, 5% digitonin) for 1 h at 4°C with gentle agitation followed by ultracentrifugation at 100,000 g for 1 h at 4°C. 400 μl of digitonin-solubilized membrane proteins were incubated with anti-GFP sepharose resin for 4 h at 4°C with slow agitation. The unbound material was removed (Flow Through) and the resin washed four times with 400 μl buffer Buffer-G-DIG (decreasing to a final concentration of 0.05% digitonin). Proteins were eluted from the resin with 100 μl SDS-PAGE sample buffer heated to 50°C for 15 min. Samples were denatured for 15 min at 50°C prior to loading. Proteins were separated by SDS-PAGE on 15% polyacrylamide gels, electroblotted onto a PVDF membrane and blocked in 5% non-fat milk in PBS-0.5% Tween-20. The blocked membranes were probed with mouse monocolonal anti-GFP (1:5000, Sigma) and mouse monocolonal anti-FLAG M2 (1:1000, Sigma) diluted into 3% BSA in 1x PBS-0.05% Tween-20. Primary antibodies were detected using horseradish peroxidase-conjugated goat anti-mouse IgG (1:10000, Biorad) and the Western Lightning Plus ECL reagent as described by the manufacturer (PerkinElmer). Membrane chemiluminescence was imaged on a FluorChem R system (ProteinSimple).
Supplementary Material
Acknowledgments
The authors would like to thank all members of the Bernhardt and Rudner laboratories for support and helpful comments. Andrew Fenton was a jointly mentored postdoctoral fellow bridging work in both labs. Special thanks to Rachel Yunck, Harvey Kimsey, Malcolm Winkler, Tim van Opijnen, Andy Camilli, Nathalie Campo, Jan-Willem Veening, Thierry Vernet and Don Morrison for strains, reagents, and technical assistance. This work was supported by the National Institutes of Health (R01AI083365 to TGB, CETR U19 AI109764 to TGB and DZR, GM073831 to DZR, and RC2 GM092616 to DZR).
Footnotes
Supplementary Information is available online and includes Supplementary Figures and Supplementary Methods.
AUTHOR CONTRIBUTIONS
AKF performed all experiments, designed part of the experimental program and coauthored the manuscript. LEM carried out essential pilot experiments for the project. DTCL helped adopt the Tn-seq data analysis pipeline and proofread the manuscript. DZR and TGB co-supervised the project and co-authored the manuscript.
The authors declare that they have no competing financial interests.
References
- 1.Lovering AL, Safadi SS, Strynadka NCJ. Structural Perspective of Peptidoglycan Biosynthesis and Assembly. Annu Rev Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 2.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–36. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paradis-bleau C, et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2011;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–72. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinho MG, Kjos M, Veening JW. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol. 2013;11:601–14. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 7.Land AD, Winkler ME. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J Bacteriol. 2011;193:4166–79. doi: 10.1128/JB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakenbeck R. Discovery of β-lactam-resistant variants in diverse pneumococcal populations. Genome Med. 2014;6:72. doi: 10.1186/s13073-014-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paik J, Kern I, Lurz R, Hakenbeck R. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J Bacteriol. 1999;181:3852–3856. doi: 10.1128/jb.181.12.3852-3856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langridge GC, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Job V, Carapito R, Vernet T, Dessen A, Zapun A. Common alterations in PBP1a from resistant Streptococcus pneumoniae decrease its reactivity toward β-lactams: Structural insights. J Biol Chem. 2008;283:4886–4894. doi: 10.1074/jbc.M706181200. [DOI] [PubMed] [Google Scholar]
- 13.Opijnen T, Van Camilli A, Van Opijnen T, Camilli A. A fine scale phenotype –genotype virulence map of a bacterial pathogen A fine scale phenotype – genotype virulence map of a bacterial pathogen. Genome Res. 2012:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2015;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kloosterman TG, Van Der Kooi-Pol MM, Bijlsma JJE, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol Microbiol. 2007;65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 16.Lanie JA, et al. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol. 2007;189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85–109. doi: 10.1128/MMBR.00030-07. table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A. 2005;102:8710–5. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boersma MJ, et al. Minimal Peptidoglycan (PG) Turnover in Wild-Type and PG Hydrolase and Cell Division Mutants of Streptococcus pneumoniae D39 Growing Planktonically and in Host-Relevant Biofilm. J Bacteriol. 2015;197:3472–3485. doi: 10.1128/JB.00541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuru E, et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chemie - Int Ed. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karimova G, Pidoux J, Ullmann A, Ladant D, Adant DAL. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. PNAS. 1998;95:5752–6. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 23.Jones LJ, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–22. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 24.Tsui HCT, et al. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol Microbiol. 2014;94:21–40. doi: 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meeske AJ, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016 doi: 10.1038/nature19331. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho H, et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol. 2016 doi: 10.1038/nmicrobiol.2016.172. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varma A, de Pedro MA, Young KD. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 2007;189:5692–704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–34. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aaron M, et al. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–52. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 33.Meisner J, et al. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol. 2013;89:1069–83. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.