Abstract
Genera of Phytopathogenic Fungi (GOPHY) is introduced as a new series of publications in order to provide a stable platform for the taxonomy of phytopathogenic fungi. This first paper focuses on 21 genera of phytopathogenic fungi: Bipolaris, Boeremia, Calonectria, Ceratocystis, Cladosporium, Colletotrichum, Coniella, Curvularia, Monilinia, Neofabraea, Neofusicoccum, Pilidium, Pleiochaeta, Plenodomus, Protostegia, Pseudopyricularia, Puccinia, Saccharata, Thyrostroma, Venturia and Wilsonomyces. For each genus, a morphological description and information about its pathology, distribution, hosts and disease symptoms are provided. In addition, this information is linked to primary and secondary DNA barcodes of the presently accepted species, and relevant literature. Moreover, several novelties are introduced, i.e. new genera, species and combinations, and neo-, lecto- and epitypes designated to provide a stable taxonomy. This first paper includes one new genus, 26 new species, ten new combinations, and four typifications of older names.
Key words: DNA barcodes, Fungal systematics, Phytopathogenic fungi, Plant pathology, Taxonomy, Typifications
Taxonomic novelties: New genus: Verkleyomyces Y. Marín & Crous
New species: Bipolaris saccharicola Y. Marín & Crous; Bi. variabilis Y. Marín, Y.P. Tan & Crous; Boeremia trachelospermi Q. Chen & L. Cai; Calonectria ecuadorensis L. Lombard & Crous; Ca. longiramosa L. Lombard & Crous; Ca. nemoralis L. Lombard & Crous; Ca. octoramosa L. Lombard & Crous; Ca. parvispora L. Lombard & Crous; Ca. tucuruiensis L. Lombard & Crous; Cladosporium chasmanthicola Bensch, U. Braun & Crous; Cl. kenpeggii Bensch, U. Braun & Crous; Cl. welwitschiicola Bensch, U. Braun & Crous; Colletotrichum sydowii Damm; Curvularia pisi Y. Marín & Crous; Cu. soli Y. Marín & Crous; Neofusicoccum italicum Dissanayake & K.D. Hyde; Nm. pistaciicola Crous; Nm. pruni Crous; Pilidium septatum Giraldo & Crous; Pleiochaeta carotae Hern.-Rest., van der Linde & Crous; Plenodomus deqinensis Q. Chen & L. Cai; Protostegia eucleicola Crous; Saccharata leucospermi Crous; S. protearum Crous; Thyrostroma franseriae Crous; Venturia phaeosepta Y. Zhang ter & J.Q. Zhang
New combinations: Coniella hibisci (B. Sutton) Crous, Monilinia mumeicola (Y. Harada et al.) Sandoval-Denis & Crous, M. yunnanensis (M.J. Hu & C.X. Luo) Sandoval-Denis & Crous, Pseudopyricularia bothriochloae (Crous & Cheew.) Y. Marín & Crous, Puccinia dianellae (Dietel) McTaggart & R.G. Shivas, Pu. geitonoplesii (McAlpine) McTaggart & R.G. Shivas, Pu. merrilliana (Syd. & P. Syd.) McTaggart & R.G. Shivas, Pu. rhagodiae (Cooke & Massee) McTaggart & R.G. Shivas, Venturia martianoffiana (Thüm.) Y. Zhang ter & J.Q. Zhang, Verkleyomyces illicii (X. Sun et al.) Y. Marín & Crous
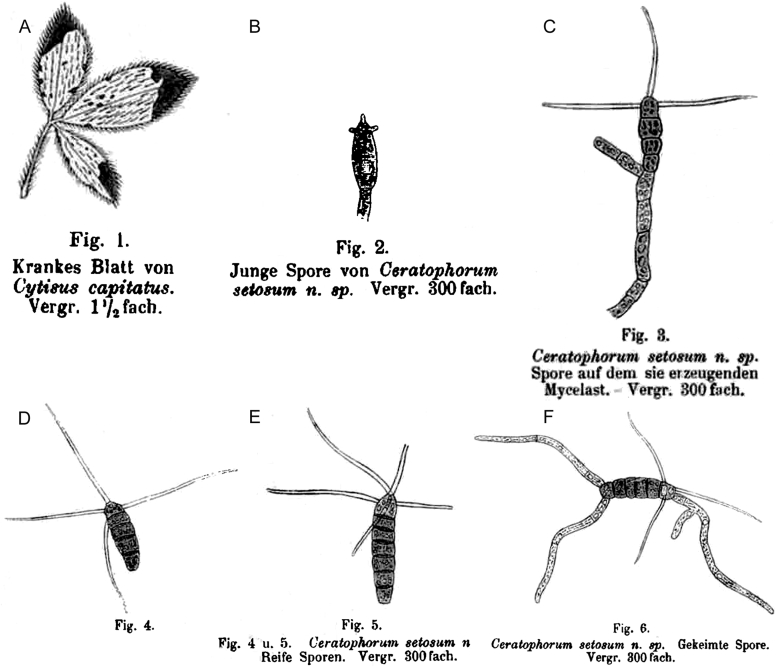
Typification: Epitypification: Ceratophorum setosum Kirchn., Coniella musaiaensis var. hibisci B. Sutton, Helminthosporium carpophilum Lév.
Lectotypification: Ceratophorum setosum Kirchn
Introduction
Since the advent of molecular DNA techniques, many species of phytopathogenic fungi have been shown to represent species complexes or to be included in genera that are poly- or paraphyletic (Crous et al. 2015b). Resolving these generic and species concepts is thus of the utmost importance for plant health and global trade in food and fibre (Crous et al., 2015b, Crous et al., 2016a). The present project focused on genera of fungi that have members causing plant diseases (phytopathogenic), links to a larger initiative called the “The Genera of Fungi project” based on Clements & Shear (1931) (www.GeneraOfFungi.org, Crous et al., 2014a, Crous et al., 2015a, Giraldo et al., 2017), which aims to revise the generic names of all currently accepted fungi (Kirk et al. 2013).
Of the approximately 18 000 fungal genera that have thus far been described, only around 8 000 are in current use. However, the majority of these were described before the DNA era. To validate the application of these names, their type species need to be recollected and designated as epi- or neotypes with a MycoBank Typification (MBT) number to ensure traceability of the nomenclatural act (Robert et al. 2013). Furthermore, to move to a single nomenclature for fungi (Wingfield et al., 2012, Crous et al., 2015b), their sexual–asexual links also need to be confirmed.
The present initiative forms part of the activities of the International Subcommission for the Taxonomy of Phytopathogenic Fungi [Pedro Crous and Amy Rossman (co-chairs), of the International Committee for the Taxonomy of Fungi (www.fungaltaxonomy.org/)].
The aims of this project are to:
-
1.
Establish a new website, www.plantpathogen.org, to host a database that will link metadata to other databases such as MycoBank, Index Fungorum, FacesofFungi, U.S. National Fungus Collections Databases, etc., and associated DNA barcodes (ITS, LSU and other loci as needed) to GenBank (Schoch et al. 2014).
-
2.
Source type specimens and cultures of the type species of genera from fungaria and Biological Resource Centres (BRCs), and generate the required metadata as explained below.
-
3.
Recollect fresh material of the type species if not already available, and as far as possible derive DNA barcodes and cultures from this material.
-
4.
Designate type species, and type specimens of those species, for those genera where this has not been indicated in the original publications.
-
5.
Fix the application of the type species of generic names by means of lecto-, neo-, or epitypification as appropriate, and at the same time deposit cultures in at least two Microbial Biological Resource Centres (M-BRCs) from which they would be widely available to the international research community.
-
6.
Publish modern generic descriptions, and provide DNA barcodes for all accepted species, with reference to appropriate literature.
Authors with new submissions should ensure that all new species and typification events are registered in MycoBank (MB and MBT numbers), respectively. It is recommended that the following issues are addressed in each genus:
-
1.
Modern generic description, and phylogenetic placement of the type species of the genus.
-
2.
Higher order phylogeny.
-
3.
New nomenclature merging asexual and sexual generic names (see Rossman et al., 2013, Johnston et al., 2014).
-
4.
Description of novel taxa, with a reference collection (e.g. fungarium), and MycoBank and GenBank sequence accession numbers.
-
5.
Name changes that result from the new phylogenetic placement.
-
6.
Notes discussing the relevance and implications of the phylogeny, and importance of the genus.
Authored generic contributions will be combined into scientific papers to be published online in Studies in Mycology, and also placed in a database displayed on www.plantpathogen.org. Preference will be given to genera that include novel DNA data and/or novel species or typifications. Authors that wish to contribute to future issues of this project are encouraged to first contact Pedro Crous (p.crous@westerdijkinstitute.nl) before final submission, to ensure there is no potential overlap with activities arising from other research groups. The genera chosen in the first paper were randomly selected, based on the fact that their phylogenetic position was resolved, DNA data were available for those species known from culture, and novel species or typifications were available for inclusion.
Material and methods
Isolates and morphological analysis
Descriptions of the new taxa and typifications are based on cultures obtained from the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands (CBS), the working collection of P.W. Crous (CPC), housed at the Westerdijk Institute, Herbarium Mycologicum Academiae Sinicae (HMAS), BIOTEC Culture Collection (BCC), the Queensland Plant Pathology Herbarium (BRIP), the Chinese General Microbiological Culture Collection Center (CGMCC), the Mae Fah Luang University Culture Collection (MFLUCC), and the Victorian Plant Pathogen Herbarium (VPRI). For fresh collections, we followed the procedures previously described in Crous et al. (1991). Colonies were transferred to different media, i.e. carnation leaf agar (CLA), cornmeal agar (CMA), 2 % malt extract agar (MEA), 2 % potato-dextrose agar (PDA), synthetic nutrient-poor agar (SNA), oatmeal agar (OA), water agar (WA) (Crous et al. 2009c), autoclaved pine needles on 2 % tap water agar (PNA) (Smith et al. 1996), and incubated at different conditions depending on the taxon to induce sporulation (requirements of media and conditions of incubations specified in each genus). Reference strains and specimens are maintained at the BCC, CBS, CGMCC, HMAS and MFLUCC.
Vegetative and reproductive structures were mounted in clear lactic acid, Shear's mounting fluid and lactophenol cotton blue, either directly from specimens or from colonies sporulating on CLA, MEA, OA, PDA, PNA, or SNA. Sections of conidiomata were made by hand for examination purposes. For cultural characterisation, isolates were grown and incubated on different culture media and temperatures as stipulated for each genus. Colour notations were rated according to the colour charts of Rayner (1970). For some taxa, NaOH pot test was carried out on MEA cultures to detect the production of metabolite E (Boerema et al. 2004). Taxonomic novelties were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004b).
DNA isolation, amplification and analyses
Fungal DNA was extracted and purified directly from the colonies or host material according to the Wizard® Genomic DNA purification kit protocol (Promega, Madison, USA). Primers and protocols for the amplification and sequencing of gene loci can be found in the bibliography related to the phylogeny presented for each genus. Phylogenetic analyses consisted of Maximum-Likelihood (ML), Bayesian Inference (BI), and Maximum Parsimony (MP). The ML was carried out using methods described by Hernández-Restrepo et al. (2016), and the MP using those described by Crous et al. (2006b). The BI was inferred as described by Hernández-Restrepo et al. (2016), or on the CIPRES portal (www.phylo.org) using MrBayes on XSEDE v. 3.2.6. Sequence data generated in this study were deposited in GenBank and ENA databases, and the alignments and trees in TreeBASE (http://www.treebase.org).
Results
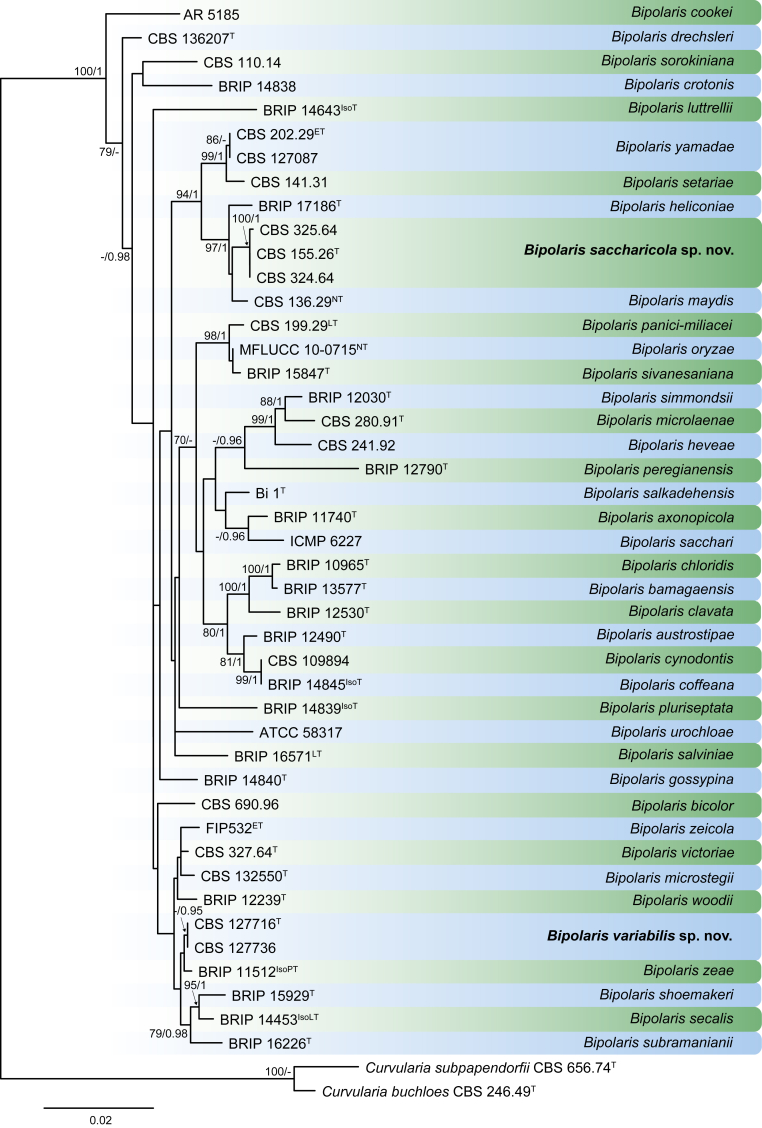
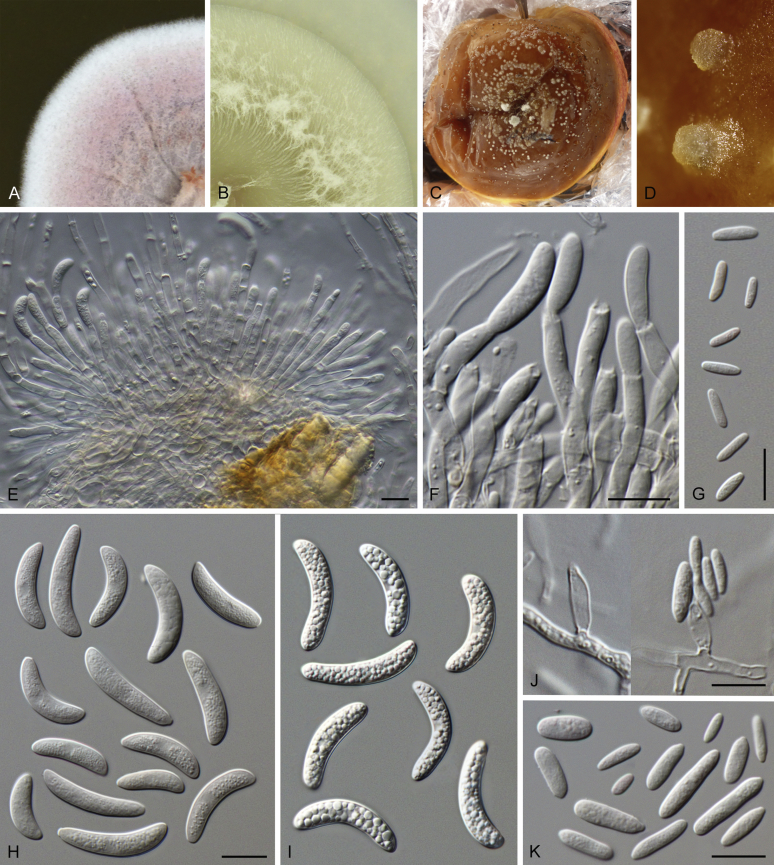
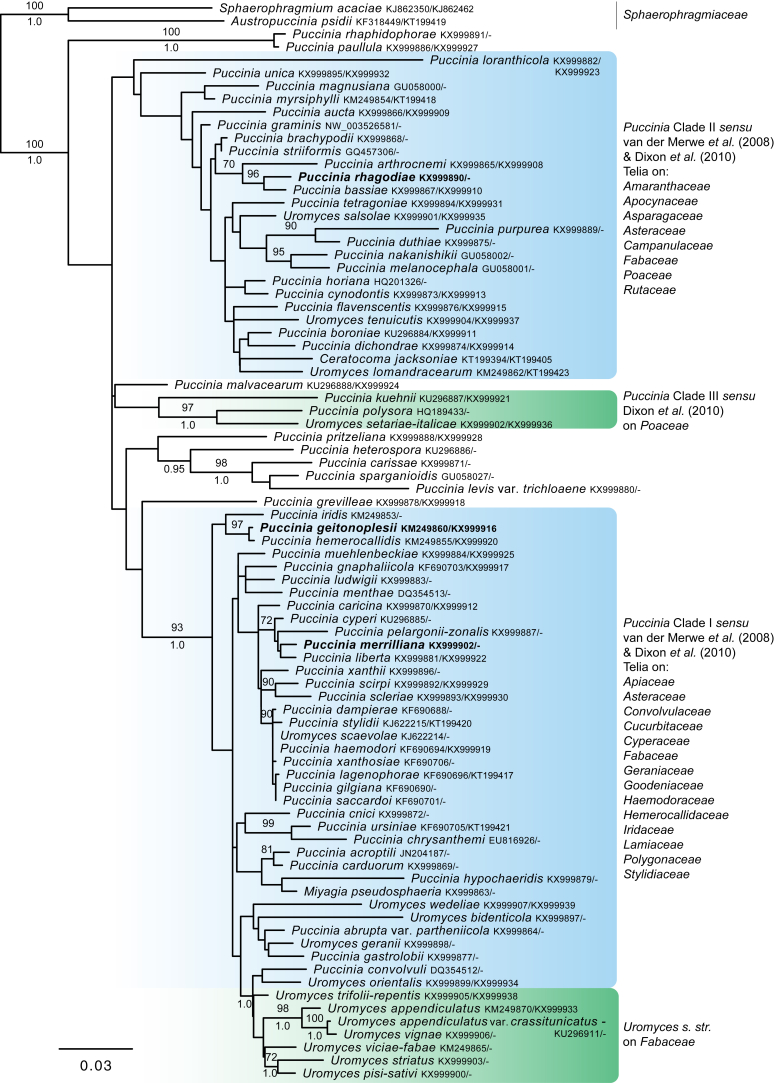
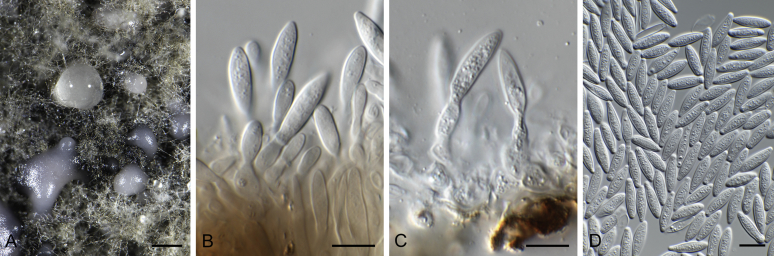
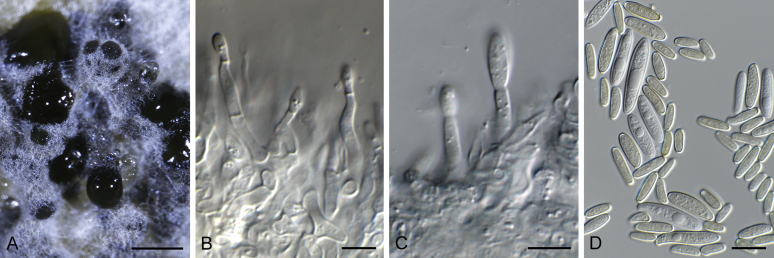
Bipolaris Shoemaker, Canad. J. Bot. 37: 882. 1959. Fig. 1.
Fig. 1.
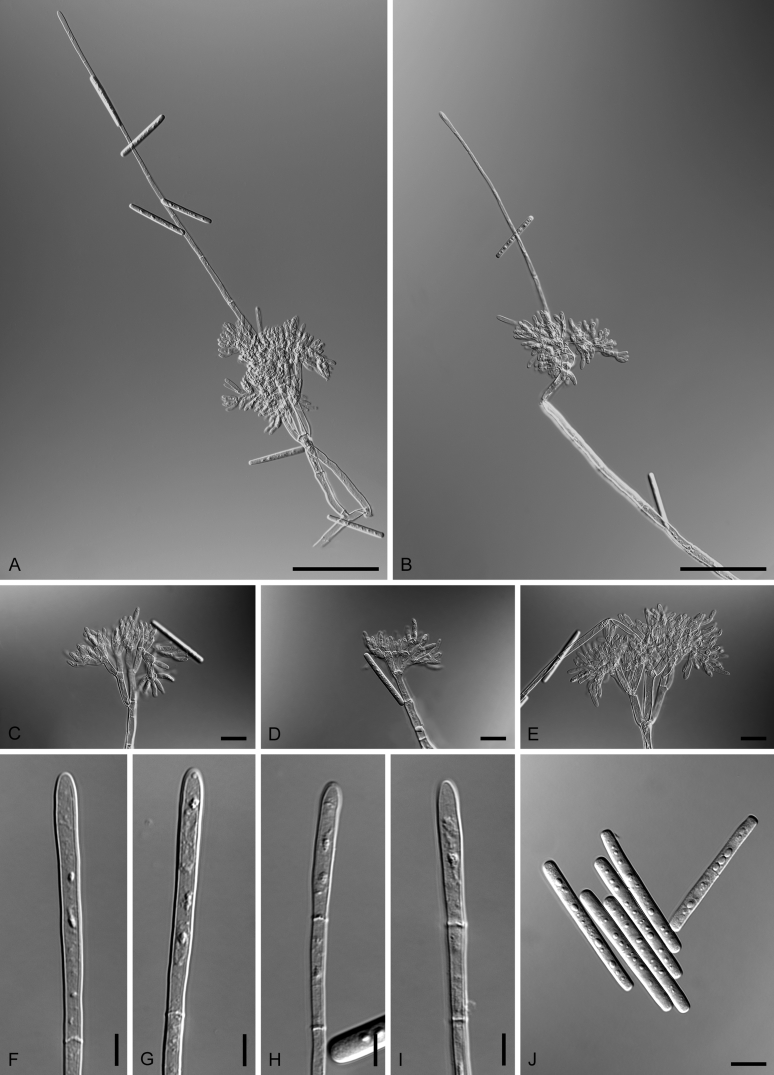
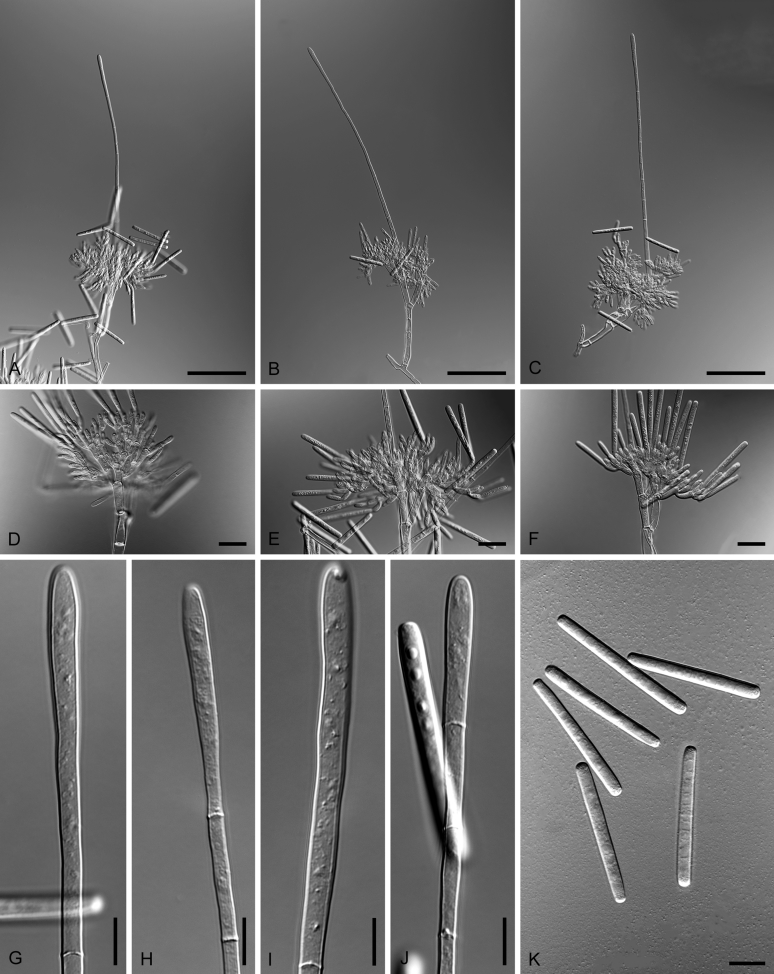
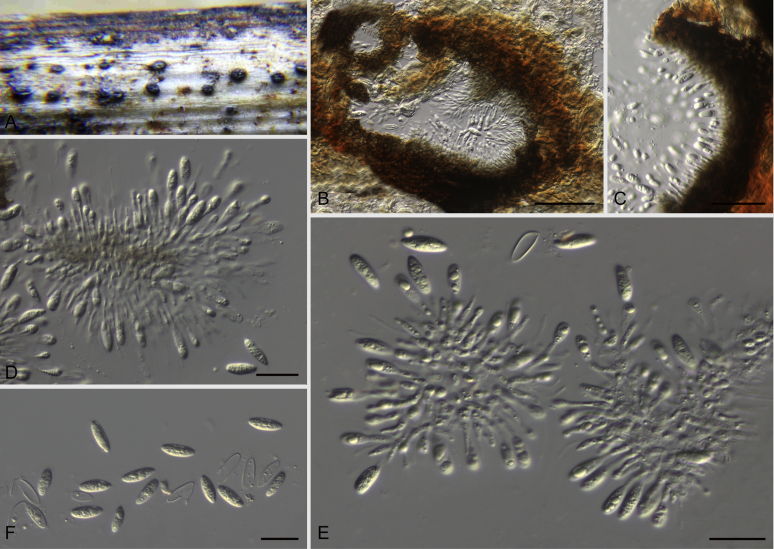
Bipolaris spp. A–F. Disease symptoms. A. Symptoms caused by Bipolaris eragrostiellae (ex-type IMI 155931). B. Symptoms caused by Bipolaris gossypina (IMI 123377). C. Symptoms caused by Bipolaris halepensis (ex-type BPI 1103129). D. Symptoms caused by Bipolaris microstegii. E. Symptoms caused by Bipolaris musae-sapientium (ex-type K (M) 181466). F. Symptoms caused by Bipolaris oryzae (ex-neotype MFLUCC 10-0715). G–L. Sexual morphs. G. Ascoma of Bipolaris luttrellii (IMI 345516). H–K. Asci. H.Bipolaris chloridis (ex-type IMI 213865). I.Bipolaris luttrellii (IMI 345516). J.Bipolaris maydis (CBS 241.92). K.Bipolaris microlaenae (IMI 338218). L. Ascospores of Bipolaris maydis (CBS 241.92). M–Z. Asexual morphs. M–R. Conidiophores and conidia. M.Bipolaris setariae (BPI 880305B). N.Bipolaris zeae (ex-type IMI 202085). O.Bipolaris bicolor (CBS 690.96). P.Bipolaris heveae (CBS 241.92). Q.Bipolaris sorokiniana (ex-type CBS 110.14). R.Bipolaris zeicola (ex-type BPI 626668). S–Z. Conidia. S.Bipolaris cookei (ex-type BPI 428852). T.Bipolaris costina (ex-type IMI 256417). U.Bipolaris crotonis (ex-type IMI 223682). V.Bipolaris gossypina (IMI 123377). W.Bipolaris obclavata (ex-type IMI 331725). X.Bipolaris oryzae (ex-neotype MFLUCC 10-0715). Y.Bipolaris salviniae (DAR 35056). Z.Bipolaris sorokiniana (ex-type CBS 110.14). Scale bars: A, N = 100 μm; B, E, F = 500 μm; C = 1 cm; G, H = 20 μm; I–L, O–Z = 10 μm; M = 50 μm. All pictures except for D taken from Manamgoda et al. (2014).
Synonym: Cochliobolus Drechsler, Phytopathology 24: 973. 1934.
Classification: Dothideomycetes, Pleosporomycetidae, Pleosporales, Pleosporaceae.
Type species: Bipolaris maydis (Y. Nisik. & C. Miyake) Shoemaker. Neotype and ex-neotype culture: ATCC 48332, CBS 137271.
DNA barcodes (genus): LSU, ITS.
DNA barcodes (species): ITS, gapdh, tef1. Table 1. Fig. 2.
Table 1.
DNA barcodes of accepted Bipolaris spp.
AR, FIP: Isolates housed in Systematic Mycology and Microbiology Laboratory, United States Department of Agriculture, Agricultural Research Service, Beltsville, Maryland, USA; Bi: Isolates housed in the Department of Plant Protection, Faculty of Agricultural Sciences and Engineering, University College of Agriculture and Natural Resources, University of Tehran, Karaj, Iran (TUPP); ATCC: American Type Culture Collection, Virginia, USA; BRIP: Queensland Plant Pathology Herbarium, Brisbane, Australia; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; ICMP: International Collection of Micro-organisms from Plants, Landcare Research, Private Bag 92170, Auckland, New Zealand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Ria, Thailand. T, ET, IsoT, IsoLT, IsoPT, LT and NT indicate ex-type, ex-epitype, ex-isotype, ex-isolectotype, ex-isoparatype, ex-lectotype and ex-neotype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; gapdh: partial glyceraldehyde-3-phosphate dehydrogenase gene; tef1: partial translation elongation factor 1-alpha gene.
Fig. 2.
RAxML phylogram obtained from the combined ITS (478 bp), gapdh (472 bp) and tef1 (892 bp) sequences of all the accepted species of Bipolaris. The tree was rooted to Curvularia buchloës CBS 246.49 and Curvularia subpapendorfii CBS 656.74. The novel species described in this study are shown in bold. RAxML bootstrap support (BS) values above 70 % and Bayesian posterior probability scores ≥ 0.95 are shown at the nodes. GenBank accession numbers are indicated in Table 1. T, ET, IsoT, IsoLT, IsoPT, LT and NT indicate ex-type, ex-epitype, ex-isotype, ex-isolectotype, ex-isoparatype, ex-lectotype and ex-neotype strains, respectively. TreeBASE: S20877.
Ascomata pseudothecial, mostly globose to ellipsoidal, sometimes flask-shaped or flattened on hard substrata, brown or black, immersed, erumpent, partially embedded or superficial, free, smooth or covered with vegetative hyphae; ostiole central, papillate or with a sub-conical, conical, paraboloid or cylindrical neck; ascomatal wall comprising pseudoparenchymatous cells of equal thickness or slightly thickened at apex of the ascoma. Hamathecium comprising septate, filiform, branched pseudoparaphyses. Asci bitunicate, clavate, cylindrical-clavate or broadly fusoid, straight or slightly curved, thin-walled, fissitunicate, often becoming more or less distended prior to dehiscence, short pedicellate, rounded at apex. Ascospores multiseriate, filiform or flagelliform, hyaline or sometimes pale yellow or pale brown at maturity, septate, helically coiled within ascus, ascospore coiling moderate to strongly, often with a mucilaginous sheath. Conidiophores single, sometimes arranged in small groups, straight to flexuous or geniculate, pale to dark brown, branched, thick-walled, septate. Conidiogenous nodes smooth to slightly verruculose. Conidia canoe-shaped, fusoid or obclavate, mostly curved, hyaline, pale or dark brown, reddish brown or pale to deep olivaceous, thick-walled, smooth-walled, 3–14-distoseptate, germinating by production of one or two germination tubes by polar cells; hila often slightly protruding or truncate, sometimes inconspicuous; septum ontogeny first septum median to sub-median, second septum delimits basal cell and third delimits distal cell (adapted from Manamgoda et al. 2014).
Culture characteristics: Colonies on PDA white or pale grey when young, brown or dark grey when mature, fluffy, cottony, raised or convex with papillate surface, margin lobate, undulate, entire or sometimes rhizoid.
Optimal media and cultivation conditions: Sterilised Zea mays leaves placed on 1.5 % WA or slide cultures of PDA under near-ultraviolet light (12 h light, 12 h dark) at 25 °C to induce sporulation of the asexual morph, while for the sexual morph Sach's agar with sterilised rice or wheat straw at 25 °C is used.
Distribution: Worldwide.
Hosts: Mainly pathogens of grasses, but some also on non-grass hosts, causing devastating diseases on cereal crops in the Poaceae, including rice, maize, wheat and sorghum and on various other host plants. Moreover, this genus can occur on at least 60 other genera in Anacardiaceae, Araceae, Euphorbiaceae, Fabaceae, Malvaceae, Rutaceae and Zingiberaceae as either saprobes or pathogens.
Disease symptoms: Leaf spots, leaf blight, melting out, root rot, and foot rot, among others.
Notes: Species delimitation based on morphology alone is limited since many species have overlapping characters. Moreover, the morphology of the sexual morph is of limited value due to difficulties to induce this morph in culture, or find it in nature. The genus is morphologically similar to Curvularia, and distinguishing these genera can be problematic. Both genera contain species with straight or curved conidia, but in Bipolaris the curvature is continuous throughout the length of the conidium, while the conidia of Curvularia have intermediate cells inordinately enlarged which contributes to their curvature. Moreover, conidia in Bipolaris are usually longer than in Curvularia. Another morphological difference is the presence of stromata in some species of Curvularia, a feature not observed in species of Bipolaris. In order to properly delineate both genera, phylogenetic studies using ITS, gapdh and tef1 sequences were recently performed (Manamgoda et al., 2014, Manamgoda et al., 2015).
References: Ellis, 1971, Sivanesan, 1987 (morphology and pathogenicity); Manamgoda et al., 2011, Tan et al., 2016 (morphology, phylogeny and pathogenicity); Manamgoda et al. 2014 (morphology, phylogeny, pathogenicity and key of all Bipolaris spp.).
Bipolaris saccharicola Y. Marín & Crous, sp. nov. MycoBank MB820809. Fig. 3.
Fig. 3.
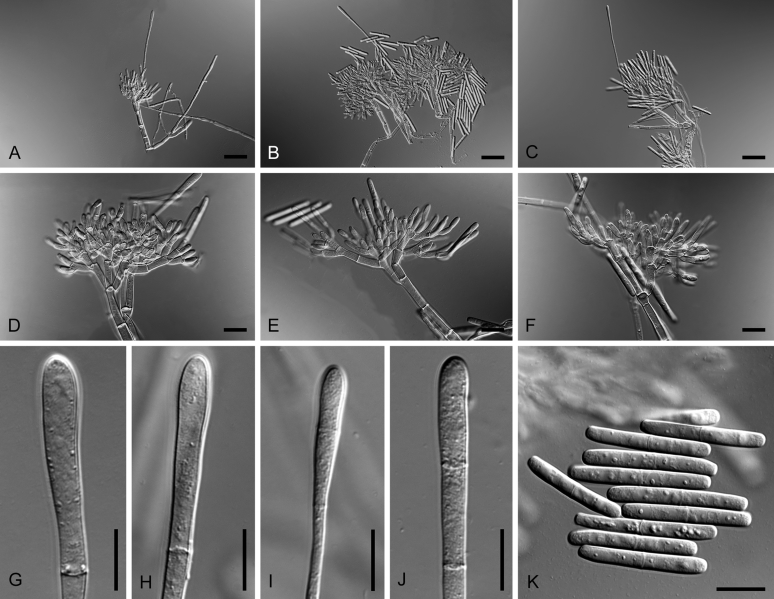
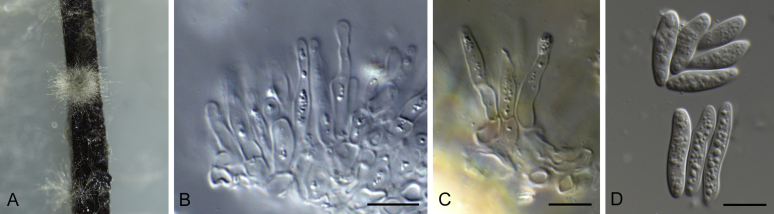
Bipolaris saccharicola (ex-type CBS 155.26). A–C. Conidiophores and conidia. D–H. Conidia. Scale bars: A–C = 20 μm; H applies to D–H = 10 μm.
Etymology: Name refers to the host genus it was isolated from, Saccharum.
Hyphae hyaline to pale brown, branched, septate, thin-walled, 2.5–5.5 μm. Conidiophores arising in smalls groups, septate, straight or flexuous, smooth-walled, sometimes branched, cell walls thicker than those of vegetative hyphae, mononematous, semi- to macronematous, pale brown to brown, paler towards apex, rarely swollen at base, up to 900 μm tall. Conidiogenous cells smooth-walled, terminal or intercalary, subhyaline to pale brown or brown, subcylindrical to swollen, 10–27(–47) × 4–8 μm. Conidia verruculose, curved, rarely straight, fusiform, subhyaline to pale brown or brown, (2–)4–9(–11)-distoseptate, (30–)45–120 × 10.5–20(–21.5) μm; hila inconspicuous, brown, slightly protuberant, flat, darkened, slightly thickened, 2–4 μm. Chlamydospores and sexual morph not observed.
Culture characteristics: Colonies on PDA reaching 41–53 mm diam after 1 wk, moderate aerial mycelium giving a cottony appearance, margin lobate; surface olivaceous grey to olivaceous black; reverse olivaceous black.
Material examined: Unknown country, unknown substratum, 1926, H. Atherton (holotype CBS H-23114, culture ex-type CBS 155.26 = MUCL 9693); Unknown country, from Saccharum officinarum, unknown date, R.R. Nelson, CBS 324.64; CBS 325.64 = DSM 62597 = MUCL 18220 = MUCL 9694 = NRRL 5241.
Notes: This species is closely related to Bi. maydis. However, Bi. saccharicola can easily be distinguished by the absence of a sexual morph, longer conidiophores and verruculose, more prominently curved conidia. Both species can be found on the same host, Saccharum officinarum. Other species of Bipolaris isolated from this host include Bi. cynodontis, Bi. sacchari, Bi. setariae, Bi. stenospila and Bi. yamadae (Manamgoda et al. 2014). Bipolaris saccharicola is morphologically similar to Bi. sacchari, but Bi. saccharicola can be distinguished by its much longer and non-geniculate conidiophores and wider and more septate conidia.
Bipolaris variabilis Y. Marín, Y.P. Tan & Crous, sp. nov. MycoBank MB820810. Fig. 4.
Fig. 4.
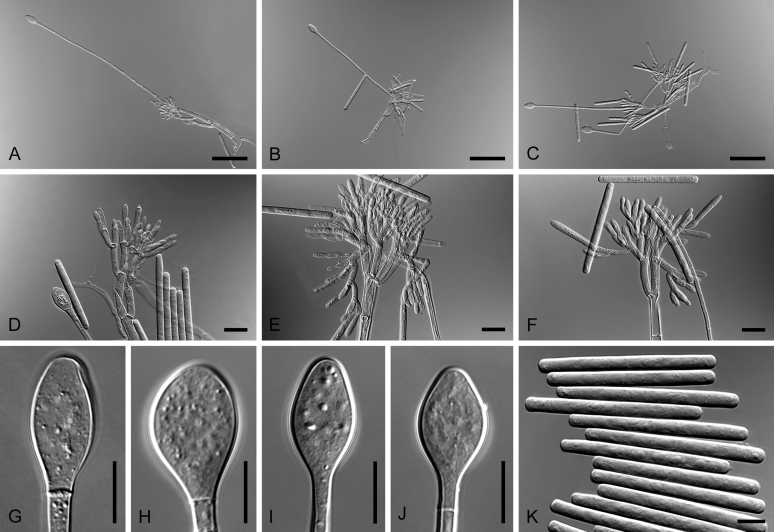
Bipolaris variabilis (ex-type CBS 127716). A–C. Conidiophores and conidia. D–M. Conidia. Scale bars: A = 20 μm; B, C = 15 μm; H applies to D–H, M applies to I–M = 5 μm.
Etymology: Name refers to the highly variable conidial morphology.
Leaf spots brown to reddish, elongated, often confluent and following veins, some with central part brown, 2.5 × 1–2 mm. Hyphae subhyaline to pale brown, branched, septate, thin-walled, 3–6 μm. Conidiophores arising in groups, septate, straight or flexuous, sometimes geniculate at upper part, smooth to verruculose, branched, cells walls thicker than those of vegetative hyphae, mononematous, semi- to macronematous, pale brown to brown, paler towards apex, slightly swollen at base, up to 1 600 μm tall. Conidiogenous cells smooth-walled, terminal or intercalary, proliferating sympodially, subhyaline or pale brown to brown, subcylindrical to swollen, (6.5–)8–26(–35) × 5.5–11 μm. Conidia verruculose, straight or slightly curved, globose, subglobose, ellipsoidal to obclavate, pale brown to brown, apical and basal cells paler than middle cells being subhyaline to pale brown, (1–)3–7(–9)-distoseptate, 13.5–77 × 10–19.5 μm; hila inconspicuous, slightly protuberant, flat, darkened, thickened, 3–6 μm diam. Chlamydospores and sexual morph not observed.
Culture characteristics: Colonies on PDA reaching 90 mm diam within 1 wk, with sparse to moderate aerial mycelium giving a cottony appearance, margin lobate; surface olivaceous grey to iron-grey; reverse olivaceous black.
Material examined: Argentina, from leaf spots on Pennisetum clandestinum, 28 Jul. 1986, col. M.N. Sisterna, isol. J.L. Alcorn (holotype CBS H-23115, culture ex-type CBS 127716 = BRIP 15349). Brazil, from Pennisetum clandestinum, Apr. 1987, J.J. Muchovej, CBS 127736 = BRIP 15702 = ATCC 62423.
Notes: Bipolaris variabilis can easily be distinguished based on its highly variable conidial size, shape and septation. Hitherto, this species has only been found on Pennisetum clandestinum in South America. Other species of Bipolaris can be found on Pennisetum spp., i.e. Bi. bicolor, Bi. colocasiae, Bi. cynodontis, Bi. maydis, Bi. mediocris, Bi. sacchari, Bi. setariae, Bi. sorokiniana, Bi. stenospila, Bi. urochloae and Bi. zeae; however, only Bi. mediocris is restricted to that host (Manamgoda et al. 2014). Bipolaris mediocris and Bi. variabilis are morphologically similar, but Bi. variabilis produces smaller, verruculose conidia. Moreover, Bi. mediocris is characterised by much shorter conidiophores (up to 150 μm tall), and has only been reported in Africa (Farr & Rossman 2017). Bipolaris variabilis is closely related to Bi. zeae, but the latter is characterised by shorter conidiophores (up to 370 μm tall), and less septate conidia that are less variable in shape than those of Bi. variabilis.
Bipolaris yamadae (Y. Nisik.) Shoemaker, Canad. J. Bot. 37: 884. 1959. Fig. 5.
Fig. 5.
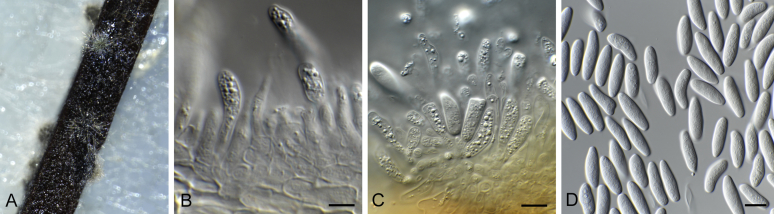
Bipolaris yamadae (CBS 127087). A–C. Conidiophores and conidia. D–G. Conidia. Scale bars: A–C = 20 μm; D–G = 10 μm.
Basionym: Helminthosporium yamadae Y. Nisik., Rept. Ohara. Inst. Agr. Research 4: 120. 1928.
Synonyms: Drechslera yamadai (Y. Nisik.) Subram. & B.L. Jain, Curr. Sci. 35: 355. 1966.
Helminthosporium euphorbiae Hansf., Proc. Linn. Soc. London 155: 49. 1943.
Bipolaris euphorbiae (Hansf.) J.J. Muchovej & A.O. Carvalho, Mycotaxon 35: 160. 1989.
Drechslera euphorbiae (Hansf.) M.B. Ellis, Dematiaceous Hyphomycetes (Kew): 440. 1971.
Notes: Bipolaris euphorbiae was originally described in Helminthosporium (Hansford 1943), then transferred to Drechslera (Ellis 1971), and finally placed in Bipolaris based on the bipolar germination and hilum structure (Muchovej & Carvalho 1989). In their revision of Bipolaris, Manamgoda et al. (2014) accepted this species in the genus despite the lack of molecular data. In the present study, the neotype strain of Bi. euphorbiae CBS 127087 (=BRIP 16567; see Fig. 5), which was designated by Muchovej & Carvalho (1989), clustered with the ex-epitype strain of Bi. yamadae. Both species are morphologically similar differing only in the size of the structures that are usually overlapping. Based on these data, we propose to reduce Bi. euphorbiae to synonymy under Bi. yamadae. Moreover, we emended the description of Bi. yamadae to include the morphology of its new synonym, as well as the new host and distribution records.
Leaf spots on Panicum sp. ovoid, oblong, pale brown at margin and pale brown at centre, with an irregular concentric zone. Hyphae hyaline, branched, septate, anastomosing, thin-walled, 1.5–4.5 μm. Conidiophores arising singly or in small groups, septate, rarely branched, straight or flexuous, sometimes geniculate at upper part, smooth walled, mononematous, semi- to macronematous, olive-brown to pale brown, sometimes paler towards apex, swollen at base, 40–650 × 3–10.5 μm. Conidiogenous cells smooth-walled, sometimes slightly verruculose, terminal or intercalary, subhyaline to pale brown or dark brown, subcylindrical to slightly swollen, 7–30(–40) × 5.5–9.5 μm. Conidia smooth-walled, straight or curved, ellipsoidal, cylindrical, fusiform or obclavate, sometimes obovoid, with rounded ends, subhyaline to pale brown or olive-brown, (3–)5–7(–11)-distoseptate, 27–100(–120) × 11.5–20 μm; hila 2.5–4.5 μm, non or slightly protuberant, flat, darkened; germination at both ends.
Culture characteristics: Colonies on PDA reaching 30–65 mm diam after 1 wk, cottony, with irregular margins; surface pale olivaceous grey to olivaceous grey; reverse olivaceous black.
Distribution: Brazil, Cuba, China, India, Japan, Sudan, Tanzania, USA (IA, ID, ND, WI).
Hosts: Panicum capillare, Pa. implicatum, Pa. maximum, Pa. miliaceum, Euphorbia sp., Oryza sp., Saccharum sp., Setaria plicata (Farr & Rossman 2017).
Authors: Y. Marin-Felix, P.W. Crous & Y.P. Tan
Boeremia Aveskamp et al., Stud. Mycol. 65: 36. 2010. Fig. 6.
Fig. 6.
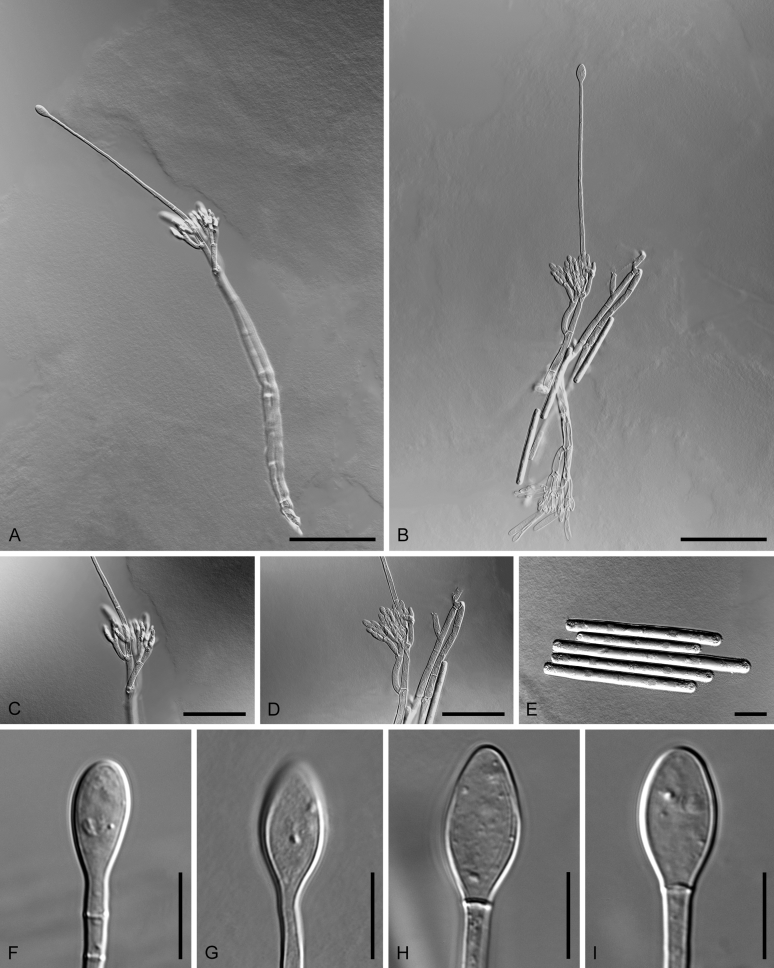
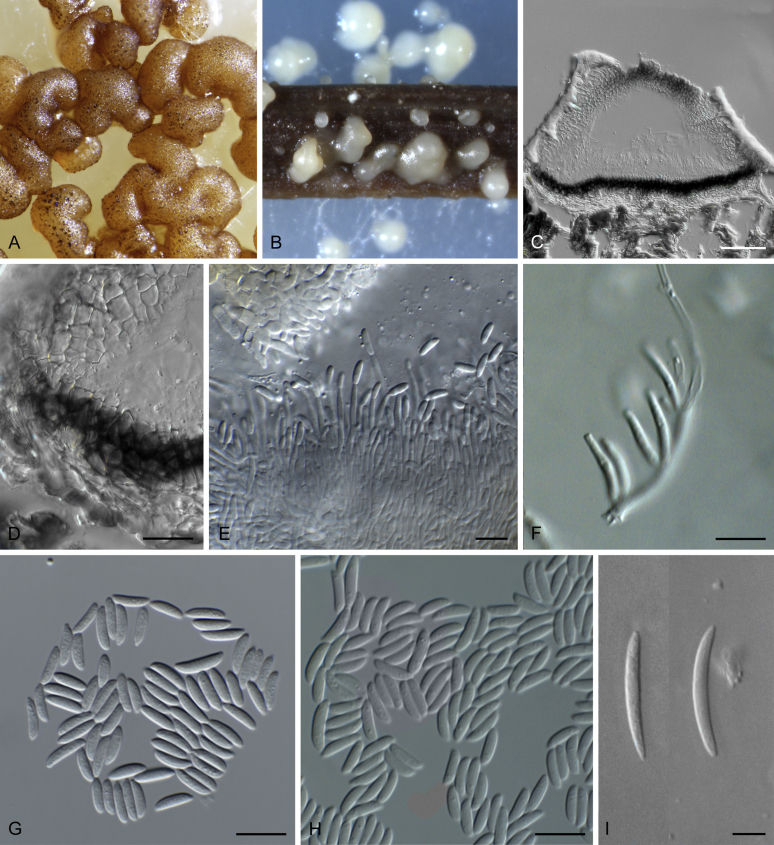
Boeremia spp. A. Symptoms of Boeremia lilacis (LC 8116) on Ocimum sp. B. Symptoms of Boeremia exigua var. rhapontica (ex-type CBS 113651) on Rhaponticum repens. C. Symptoms of Boeremia lilacis (LC 5178) on Lonicera sp. D. Ostiole configuration of Boeremia exigua var. exigua (CBS 431.74). E. Section of young pycnidium of Boeremia exigua var. pseudolilacis (ex-type CBS 101207). F. Conidia of Boeremia exigua var. pseudolilacis (ex-type CBS 101207). G. Conidia of Boeremia exigua var. heteromorpha (ex-neotype CBS 443.94). Scale bars: D–E = 20 μm; F = 5 μm; G = 10 μm. Picture B taken from Berner et al. (2015); D–F from Aveskamp et al. (2010); G from Chen et al. (2015a).
Classification: Dothideomycetes, Pleosporomycetidae, Pleosporales, Didymellaceae.
Type species: Boeremia exigua (Desm.) Aveskamp et al. Representative strain: CBS 431.74.
DNA barcodes (genus): LSU, ITS.
DNA barcodes (species): act, cal, rpb2, tef1, tub2. Table 2. Fig. 7.
Table 2.
DNA barcodes of accepted Boeremia spp.
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CGMCC: Chinese General Microbiological Culture Collection Center, Beijing, China. T and NT indicate ex-type and ex-neotype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; act: partial actin gene; cal: partial calmodulin gene; rpb2: partial RNA polymerase II second largest subunit gene; tef1: partial translation elongation factor 1-alpha gene; tub2: partial β-tubulin gene.
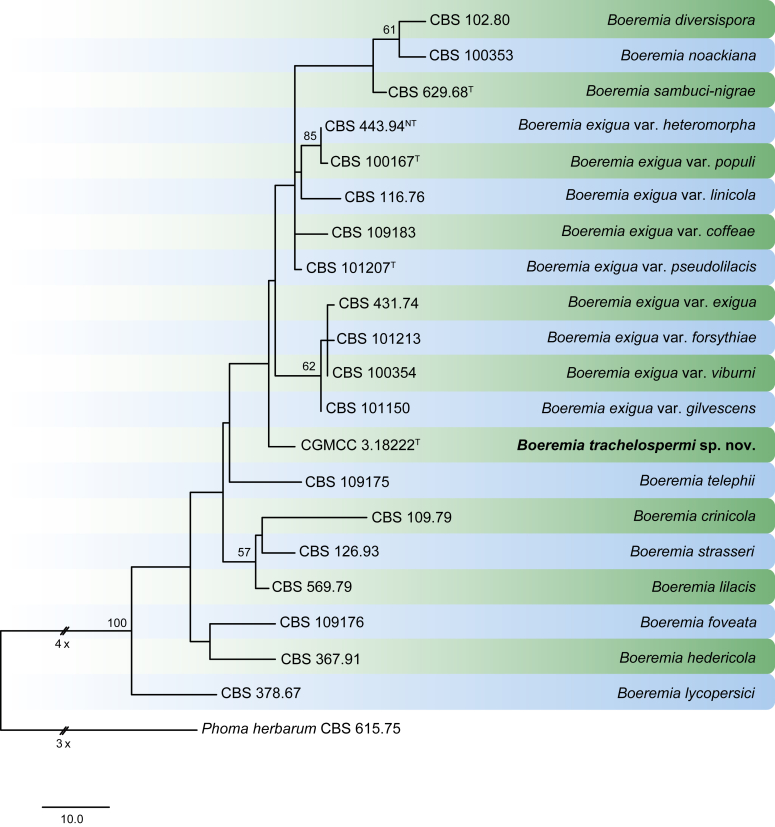
Fig. 7.
Phylogenetic tree generated from a maximum parsimony analysis based on the combined LSU, ITS, tub2 and rpb2 sequences. Values above the branches represent parsimony bootstrap support values (> 50 %). The novel species are shown in bold. The tree is rooted with Phoma herbarum CBS 615.75. GenBank accession numbers are indicated in Table 2. T and NT indicate ex-type and ex-neotype strains, respectively. TreeBASE: S21048.
Ascomata pseudothecial, subglobose. Asci cylindrical or subclavate, 8-spored, biseriate. Ascospores ellipsoidal, 1-septate. Conidiomata pycnidial, variable in shape and size, mostly globose to subglobose, superficial or immersed into agar, solitary or confluent; ostiole non-papillate or papillate, lined internally with hyaline cells when mature; conidiomatal wall pseudoparenchymatous, multi-layered, outer wall brown pigmented. Conidiophores reduced to conidiogenous cells. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform. Conidia variable in shape, hyaline, smooth- and thin-walled, mainly aseptate, but 1(–2)-septate larger conidia may be found (adapted from Aveskamp et al. 2010).
Culture characteristics: Colonies on OA white to dull green, grey olivaceous to olivaceous or smoke-grey, velvety, floccose to woolly, margin often regular, sometimes lobate and irregular scalloped.
Optimal media and cultivation conditions: OA or PNA at 25 °C under near-ultraviolet light (12 h light, 12 h dark) to promote sporulation.
Distribution: Worldwide.
Hosts: Seed-borne pathogens of Phaseolus vulgaris (Fabaceae) and noxious pathogens of Coffea arabica (Rubiaceae). Species on more than 200 host genera including Amaryllidaceae, Apocynaceae, Araliaceae, Asteraceae, Caprifoliaceae, Chenopodiaceae, Crassulaceae, Fabaceae, Lamiaceae, Linaceae, Oleaceae, Salicaceae, Solanaceae, Ulmaceae, Umbelliferae.
Disease symptoms: Leaf spots, stem lesions, black node, bulb rot, root rot, shoot dieback.
Notes: The genus Boeremia was established by Aveskamp et al. (2010) to accommodate phoma-like species that are morphologically similar and closely related to Ph. exigua. Taxa in this genus are characterised by having ostioles with a hyaline inner layer of cells and producing aseptate and septate conidia (Aveskamp et al. 2010). To date only Bo. lycopersici has been reported to have a sexual morph. Recently, Chen et al. (2015a) and Berner et al. (2015) further examined the phylogenetic relationships of taxa in Boeremia in two combined multilocus analyses, the first one based on LSU, ITS, tub2 and rpb2 sequences, and the second on ITS, act, cal, tef1 and tub2 sequences.
References: Boerema et al. 2004 (morphology and pathogenicity); Aveskamp et al., 2010, Chen et al., 2015a (morphology and phylogeny); Berner et al. 2015 (morphology, pathogenicity and phylogeny).
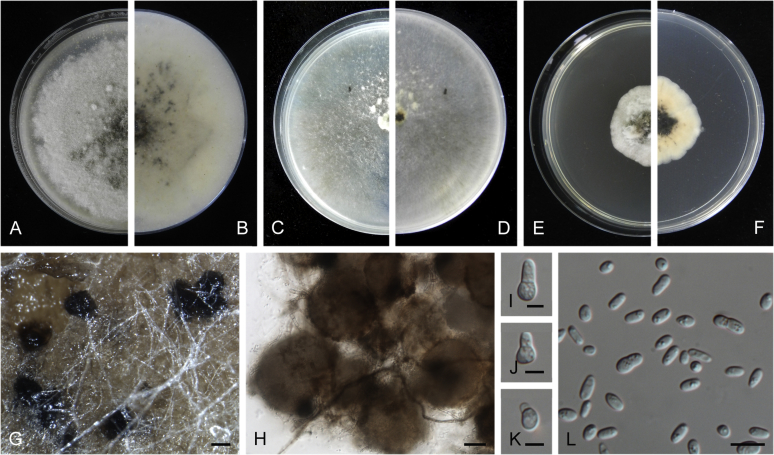
Boeremia trachelospermi Q. Chen & L. Cai, sp. nov. MycoBank MB818819. Fig. 8.
Fig. 8.
Boeremia trachelospermi (ex-type CGMCC 3.18222). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E, F. Colony on PDA (front and reverse). G. Pycnidia forming on OA. H. Pycnidia. I–K. Conidiogenous cells. L. Conidia. Scale bars: G = 200 μm; H = 50 μm; I–K = 5 μm; L = 10 μm.
Etymology: Named for the host genus from which the holotype was collected, Trachelospermum.
Conidiomata pycnidial, solitary or aggregated, globose to subglobose, glabrous or with few hyphal outgrowths, superficial, with a short neck, 75–255 × 60–225 μm; ostiole single, papillate or non-papillate; conidiomatal wall pseudoparenchymatous 2–4-layered, 16.5–37 μm thick, composed of isodiametric cells. Conidiophores reduced to conidiogenous cells. Conidiogenous cells phialidic, hyaline, smooth, ampulliform to doliiform, 4.5–12.5 × 4.5–6 μm. Conidia variable in shape, mostly ovoid, ellipsoidal to cylindrical, smooth- and thin-walled, hyaline, mainly aseptate, occasionally 1-septate large conidia, 4.5–9.5 × 2.5–4.5 μm, with 1–8 guttules. Conidial matrix cream-coloured.
Culture characteristics: Colonies on OA, reaching 47–55 mm diam after 1 wk, margin regular, floccose, white, dark grey near centre; reverse white to buff, dark grey near centre. Colonies on MEA 40–60 mm diam after 1 wk, margin regular, woolly, pale olivaceous grey; reverse concolourous. Colonies on PDA, reaching 20–25 mm diam after 1 wk, margin regular, floccose, compact, white to olivaceous; reverse white to buff, olivaceous near centre. NaOH test negative.
Material examined: USA, on seedlings of Trachelospermum jasminoides, 2014, W.J. Duan (holotype HMAS 246706, culture ex-type CGMCC 3.18222 = LC 8105).
Notes: Boeremia trachelospermi represents the first report of a Boeremia species on Trachelospermum (Apocynaceae). Phylogenetically, it forms a distinct lineage separate from Bo. diversispora, the Bo. exigua varieties, Bo. noackiana and Bo. sambuci-nigrae (Fig. 7), and morphologically it often produces longer conidiogenous cells and conidia than the other taxa.
Authors: Q. Chen & L. Cai
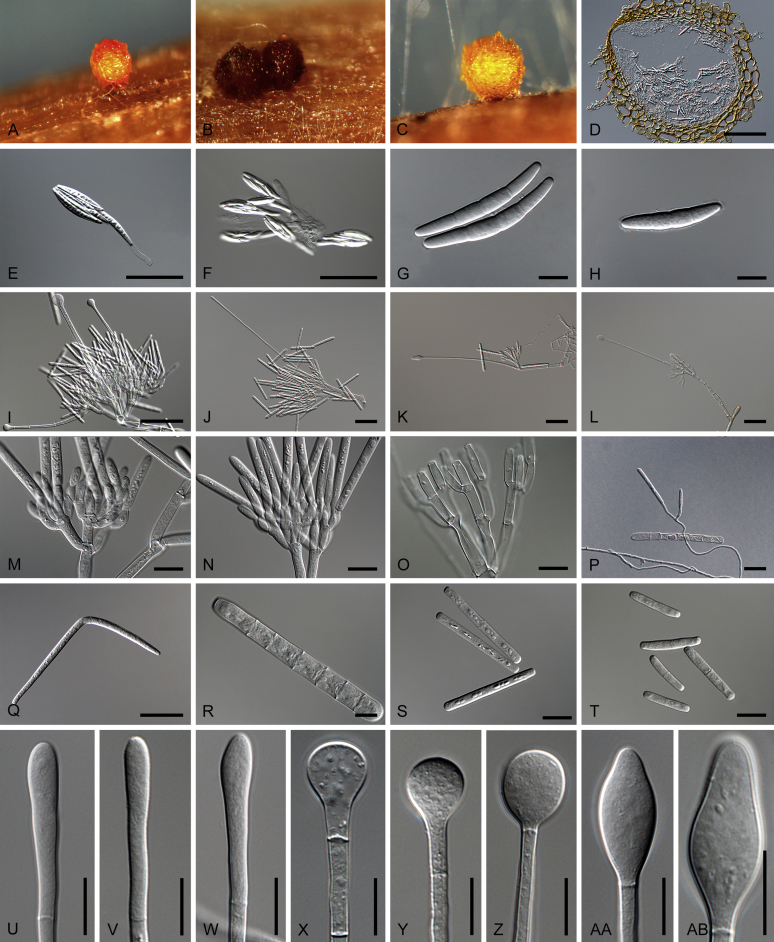
Calonectria De Not., Comm. Soc. crittog. Ital. 2(fasc. 3): 477. 1867. Fig. 9, Fig. 10.
Fig. 9.
Calonectria spp. A−H. Sexual morphs. A−D. Perithecia. A.Calonectria asiatica (ex-type CBS 114073). B.Calonectria braziliensis (ex-type CBS 230.51 × CBS 114257). b Calonectria fujianensis (ex-type CBS 127201). D. Section through perithecium of Calonectria asiatica (ex-type CBS 114073). E−F. Asci. E.Calonectria crousiana (ex-type CBS 127198). F.Calonectria asiatica (ex-type CBS 114073). G−H. Ascospores. G.Calonectria fujianensis (ex-type CBS 127201). H.Calonectria acicola (ex-type CBS 114813). I−AB. Asexual morphs. I−L. Macroconidiophores. I.Calonectria malesiana (ex-type CBS 112752). J.Calonectria macroconidialis (ex-type CBS 114880). K.Calonectria spathulata (ex-type CBS 555.92). L.Calonectria ovata (CBS 111307). M−O. Conidiogenous apparatus. M.Calonectria brachiatica (ex-type CBS 123700). N.Calonectria ecuadoriae (ex-type CBS 111406). O.Calonectria hurae (CBS 114551). P. Microconidiophore of Calonectria reteaudii (ex-type CBS 112144). Q. Megaconidia of Calonectria hurae (CBS 114551). R, S. Macroconidia. R.Calonectria angustata (ex-type CBS 109065). S.Calonectria chinensis (ex-type CBS 114827). T. Microconidia of Calonectria pteridis (ex-type CBS 111793). U−AB. Terminal vesicles of stipe extensions. U.Calonectria brassicae (ex-type CBS 111869). V.Calonectria rumohrae (CBS 109062). W.Calonectria cylindrospora (CBS 119670). X.Calonectria hongkongensis (ex-type CBS 114828). Y.Calonectria chinensis (ex-type CBS 114827). Z.Calonectria humicola (ex-type CBS 125251). AA.Calonectria mexicana (ex-type CBS 110918). AB.Calonectria spathulata (ex-type CBS 555.92). Scale bars: A−C = 500 μm; D−F = 100 μm; G, H, M−P, R−AB = 10 μm; I−L, Q = 20 μm.
Fig. 10.
Disease symptoms associated with Calonectria spp. A−B. Root and collar rot of Pinus spp. C. Cutting rot of Eucalyptus sp. D. Calonectria leaf blight of Eucalyptus sp. E. Calonectria leaf blight of Metrosideros thomasii. F. Calonectria leaf blight of Myrtus communis. G. Seedling blight of Drosera sp. H. Buxus blight. I. Root rot of Persea americana.J. Potato tuber rot. K−L. Calonectria black rot of Arachis hypogaea.
Synonyms: Cylindrocladium Morgan, Bot. Gaz. 17: 191. 1892.
Candelospora Rea & Hawley, Proc. R. Ir. Acad., Sect. B, Biol. Sci. 13: 11. 1912.
Classification: Sordariomycetes, Hypocreomycetidae, Hypocreales, Nectriaceae.
Type species: Calonectria pyrochroa (Desm.) Sacc. Holotype: Italy, leaves of Magnolia grandiflora, Daldini (as Ca. daldiniana); Lectotype: France, litter of Platanus, Autumn. Desm., Pl. Crypt. France Ed. 2 (2) # 372 (fide Rossman 1979); no culture or DNA data available.
DNA barcodes (genus): LSU, ITS.
DNA barcodes (species): cmdA, his3, tef1, tub2, and rpb2. Table 3. Fig. 11.
Table 3.
DNA barcodes of accepted Calonectria spp.
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CPC: Culture collection of Pedro Crous, housed at Westerdijk Fungal Biodiversity Institute. T indicates ex-type strains.
tub2: partial β-tubulin gene; cmdA: partial calmodulin gene; tef1: partial translation elongation factor 1-alpha gene; his3: partial histone H3 gene; rpb2: RNA polymerase II second largest subunit; ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: 28S large subunit RNA gene.
Fig. 11.
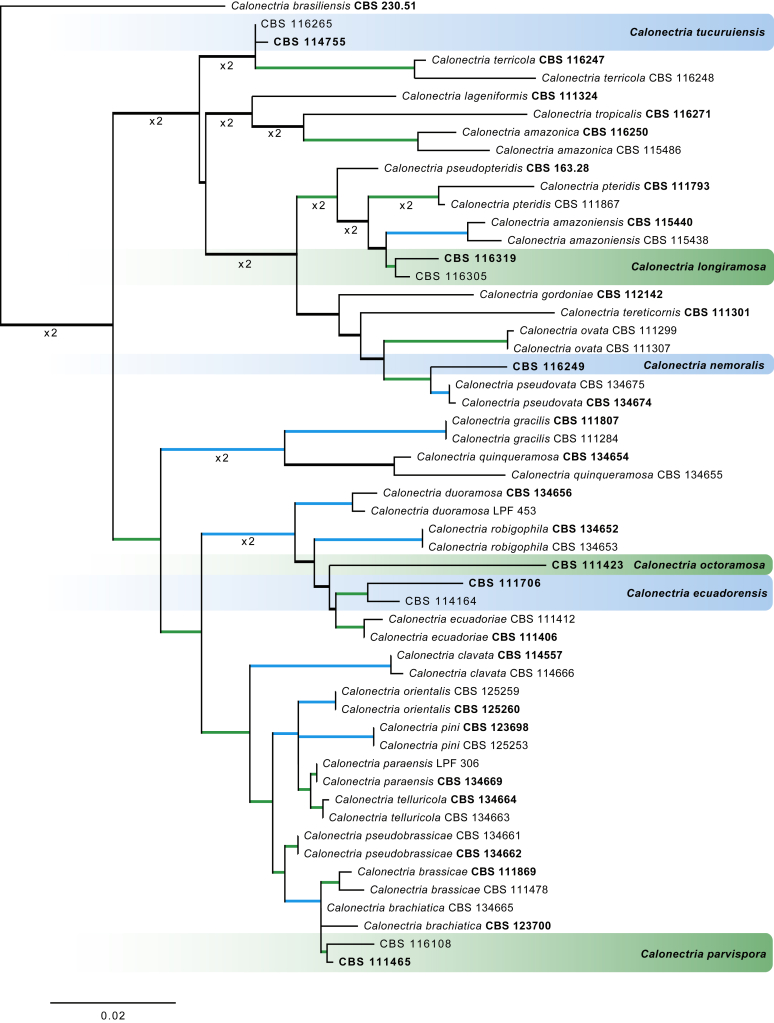
The Maximum Likelihood (ML) consensus tree inferred from the combined cmdA, tef1 and tub2 sequence alignments. Thickened lines indicate branches present in the ML, Maximum parsimony (MP) and Bayesian consensus trees. Branches with ML-bootstrap (BS) & MP-BS = 100 % and posterior probabilities (PP) = 1.00 are in blue. Branches with ML-BS & MP-BS ≥ 75 % and PP ≥ 0.95 are in green. The scale bar indicates 0.02 expected changes per site. The tree is rooted to Calonectria braziliensis (CBS 230.51). Ex-type strains are indicated in bold. GenBank accession numbers are indicated in Lombard et al., 2010a, Lombard et al., 2015, Lombard et al., 2016 and Alfenas et al. (2015). TreeBASE: S20877.
Ascomata perithecial, solitary or in groups, globose to subglobose to ovoid, yellow to orange to red or red-brown to brown, turning dark red to red-brown in KOH, rough-walled; ascomatal apex consisting of flattened, thick-walled hyphal elements with rounded tips forming a palisade, discontinuous with warty wall, gradually becoming thinner towards ostiolar canal, and merging with outer periphyses; ascomatal base consisting of dark brown-red, angular cells, merging with an erumpent stroma, cells of outer wall layer continuing into pseudoparenchymatous cells of erumpent stroma. Asci 8-spored, clavate, tapering to a long thin stalk. Ascospores aggregated in upper third of ascus, hyaline, smooth, fusoid with rounded ends, straight to sinuous, unconstricted, or constricted at septa. Megaconidiophores, if present, borne on agar surface or immersed in agar; stipe extensions mostly absent; conidiophores unbranched, terminating in 1–3 phialides, or sometimes with a single subterminal phialide; phialides straight to curved, cylindrical, seemingly producing a single conidium, periclinal thickening and an inconspicuous, divergent collarette rarely visible. Megaconidia hyaline, smooth, frequently remaining attached to phialide, multi-septate, widest in middle, bent or curved, with a truncated base and rounded apical cell. Macroconidiophores consist of a stipe, a penicillate arrangement of fertile branches, a stipe extension, and a terminal vesicle; stipe septate, hyaline or slightly pigmented at base, smooth or finely verruculose; stipe extensions septate, straight to flexuous, mostly thin-walled, terminating in a thin-walled vesicle of characteristic shape. Conidiogenous apparatus with 0–1-septate primary branches, up to eight additional branches, mostly aseptate, each terminal branch producing 1–6 phialides; phialides cylindrical to allantoid, straight to curved, or doliiform to reniform, hyaline, aseptate, apex with minute periclinal thickening and inconspicuous divergent collarette. Macroconidia cylindrical, rounded at both ends, straight or curved, widest at base, middle, or first basal septum, 1- to multi-septate, lacking visible abscission scars, held in parallel cylindrical clusters by colourless slime. Microconidiophores consist of a stipe and a penicillate or subverticillate arrangement of fertile branches; primary branches 0–1-septate, subcylindrical; secondary branches 0–1-septate, terminating in 1–4 phialides; phialides cylindrical, straight to slightly curved, apex with minute periclinal thickening and marginal frill. Microconidia cylindrical, straight to curved, rounded at apex, flattened at base, 1(–3)-septate, held in asymmetrical clusters by colourless slime.
Culture characteristics: Colonies on MEA white to pale brick when young, becoming pale brick to dark sepia when mature, fluffy, cottony, effuse to convex with papillate surface, margin entire, undulate, lobate, or fimbriate, sometimes with abundant chlamydospores forming microsclerotia within medium.
Optimal media and cultivation conditions: CLA to induce sporulation of the asexual morph at 25 °C, while for the sexual morph sterile toothpicks placed on SNA is used at 20 °C.
Distribution: Worldwide.
Hosts: Soil-borne pathogens of forestry, agricultural and horticultural crops representing approximately 100 plant families and 340 plant host species (Crous, 2002, Lombard et al., 2010c).
Disease symptoms: Leaf spots, leaf and shoot blights, cutting rot, stem cankers, damping-off and root rot.
Notes: The genus Calonectria presently includes 151 species of which only Ca. hederae and Ca. pyrochroa are not supported by ex-type cultures and supplementary DNA barcodes. Species delimitation based on morphology alone is complicated by the large number of cryptic taxa recognised in this genus (Lombard et al. 2016). The perithecia of several Calonectria spp. are morphologically similar. The cylindrocladium-like asexual morph, the life phase most commonly found in nature, is extensively used for taxon identification, although it is complicated by the morphological overlap of some cryptic species. For accurate species delimitation, phylogenetic inference of the cmdA, tef1 and tub2 (or combinations of these) is required.
References: Crous 2002 (morphology, pathogenicity and monograph); Lombard et al., 2010a, Lombard et al., 2010b, Lombard et al., 2010c, Lombard et al., 2010d, Lombard et al., 2010a, Lombard et al., 2010b, Lombard et al., 2010c, Lombard et al., 2010d, Lombard et al., 2010a, Lombard et al., 2010b, Lombard et al., 2010c, Lombard et al., 2010d (morphology, phylogeny and key of Calonectria spp.); Alfenas et al. 2015 (morphology and phylogeny).
Calonectria ecuadorensis L. Lombard & Crous, sp. nov. MycoBank MB820849. Fig. 12.
Fig. 12.
Calonectria ecuadorensis (ex-type CBS 111706). A, B. Macroconidiophores. C−E. Conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides. F−I. Clavate vesicles. J. Macroconidia. Scale bars: A, B = 50 μm; C−J = 10 μm.
Etymology: Name refers to Ecuador, the country where this fungus was collected.
Macroconidiophores consist of a stipe bearing a penicillate arrangement of fertile branches, and a stipe extension terminating in a vesicle; stipe septate, hyaline, smooth, 55–70 × 6–10 μm; stipe extension septate, straight to flexuous, 130–280 μm long, 3–6 μm wide at apical septum, terminating in a clavate vesicle, 4–6 μm diam. Conidiogenous apparatus 45–90 μm wide, and 20–90 μm long; primary branches aseptate, 13–31 × 4–6 μm; secondary branches aseptate, 13–23 × 4–5 μm; tertiary branches aseptate, 9–15 × 3–4 μm, each terminal branch producing 2–6 phialides; phialides doliiform to reniform, hyaline, aseptate, 6–11 × 2–4 μm, apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (34–)35–39(–44) × (3–)3.5–4.5(–5) μm (av. 37 × 4 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Mega- and microconidia not observed.
Culture characteristics: Colonies moderately fast growing (35–55 mm diam) on MEA after 1 wk at room temperature; surface rosy-buff to cinnamon with sparse white woolly aerial mycelium and abundant sporulation on aerial mycelium and colony surface; reverse rosy-buff to cinnamon to sepia with abundant chlamydospores throughout medium, forming microsclerotia.
Material examined: Ecuador, from soil, 20 Jun. 1997, M.J. Wingfield (holotype CBS H-23134, culture ex-type CBS 111706 = CPC 1636); ibid., culture CBS 114164 = CPC 1634.
Notes: Calonectria ecuadorensis can be distinguished from Ca. ecuadoriae (Crous et al. 2006a) by its fewer branches in the conidiogenous apparatus. Also, the conidia of Ca. ecuadorensis [(34–)35–39(–44) × (3–)3.5–4.5(–5) μm (av. 37 × 4 μm)] are smaller than those of Ca. ecuadoriae [(45–)48–55(–65) × (4–)4.5(–5) μm (av. 51 × 4.5 μm); Crous et al. 2006a].
Calonectria longiramosa L. Lombard & Crous, sp. nov. MycoBank MB820843. Fig. 13.
Fig. 13.
Calonectria longiramosa (ex-type CBS 116319). A−C. Macroconidiophores. D−F. Conidiogenous apparatus with conidiophore branches and elongate doliiform to allantoid phialides. G−J. Clavate vesicles. K. Macroconidia. Scale bars: A−C = 50 μm; D−K = 10 μm.
Etymology: Name refers to the characteristic long fertile branches of the conidiogenous apparatus in this fungus.
Macroconidiophores consist of a stipe bearing a penicillate arrangement of fertile branches, and a stipe extension terminating in a vesicle; stipe septate, hyaline, smooth, 100–245 × 6–9 μm; stipe extension septate, straight to flexuous, 155–310 μm long, 4–6 μm wide at apical septum, terminating in a clavate vesicle, 5–8 μm diam. Conidiogenous apparatus 50–85 μm wide, and 60–140 μm long; primary branches aseptate to 1-septate, 22–42 × 4–6 μm; secondary branches aseptate, 15–35 × 3–6 μm; tertiary branches aseptate, 12–30 × 3–6 μm; quaternary branches aseptate, 11–19 × 3–6 μm each terminal branch producing 2–4 phialides; phialides elongate doliiform to allantoid, hyaline, aseptate, 8–16 × 2–4 μm, apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight to slightly curved, (57–)66–76(–84) × (3–)4.5–5.5(–6) μm (av. 71 × 5 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Mega- and microconidia not observed.
Culture characteristics: Colonies moderately fast growing (35–70 mm diam) on MEA after 1 wk at room temperature; surface amber with moderate white, woolly aerial mycelium and moderate sporulation on the aerial mycelium and colony surface; reverse amber with abundant chlamydospores throughout the medium, forming microsclerotia.
Material examined: Brazil, Amazon, from Eucalyptus sp., 1993, P.W. Crous & A.C. Alfenas (holotype CBS H-22759, culture ex-type CBS 116319 = CPC 3761); ibid., cultures CBS 116305 = CPC 3765.
Notes: Calonectria longiramosa is a new species in the Ca. pteridis complex. This species is characterised by the long fertile branches of the conidiogenous apparatus distinguishing it from the other species in this complex (Alfenas et al. 2015).
Calonectria nemoralis L. Lombard & Crous, sp. nov. MycoBank MB820850. Fig. 14.
Fig. 14.
Calonectria nemoralis (ex-type CBS 116319). A, B. Macroconidiophores. C−D. Conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides. E. Macroconidia. F−I. Fusiform to ovoid vesicles. Scale bars: A, B = 50 μm; C−I = 10 μm.
Etymology: Name refers to the environment, a Eucalyptus plantation, from where this fungus was collected.
Macroconidiophores consist of a stipe bearing a penicillate arrangement of fertile branches, and a stipe extension terminating in a vesicle; stipe septate, hyaline, smooth, 40–165 × 6–8 μm; stipe extension septate, straight to flexuous, 140–210 μm long, 3–5 μm wide at the apical septum, terminating in a fusiform to ovoid vesicle, 7–9 μm diam. Conidiogenous apparatus 20–45 μm wide, and 40–55 μm long; primary branches aseptate, 18–24 × 3–6 μm; secondary branches aseptate, 11–19 × 3–5 μm, each terminal branch producing 2–4 phialides; phialides elongate doliiform to reniform, hyaline, aseptate, 6–14 × 2–4 μm, apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (44–)47–59(–71) × (3–)3.5–4.5(–6) μm (av. 53 × 4 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Mega- and microconidia not observed.
Culture characteristics: Colonies moderately fast growing (35–55 mm diam) on MEA after 1 wk at room temperature; surface sienna with sparse buff to white, woolly aerial mycelium with moderate sporulation on the aerial mycelium and colony surface; reverse sienna with abundant chlamydospores throughout the medium, forming microsclerotia.
Material examined: Brazil, from soil in Eucalyptus plantation, 1996, P.W. Crous (holotype CBS H-23135, culture ex-type CBS 116249 = CPC 3533).
Notes: Calonectria nemoralis is closely related to Ca. pseudovata. The macroconidia of Ca. nemoralis [(44–)47–59(–71) × (3–)3.5–4.5(–6) μm (av. 53 × 4 μm)] are smaller than those of Ca. pseudovata [(55–)67–70(–80) × (4–)5 (–7) μm (av. 69 × 5 μm); Alfenas et al. 2015]. Furthermore, no microconidiophores and microconidia were observed in Ca. nemoralis, although they are readily produced by Ca. pseudovata (Alfenas et al. 2015).
Calonectria octoramosa L. Lombard & Crous, sp. nov. MycoBank MB820851. Fig. 15.
Fig. 15.
Calonectria octoramosa (ex-type CBS 111423). A−C. Macroconidiophores. D−F. Conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides. G−J. Clavate vesicles. K. Macroconidia. Scale bars: A−C = 20 μm; D−K = 10 μm.
Etymology: Name refers to the eight levels of branching of the conidiogenous apparatus.
Macroconidiophores consist of a stipe bearing a penicillate arrangement of fertile branches, and a stipe extension terminating in a vesicle; stipe septate, hyaline, smooth, 34–170 × 6–10 μm; stipe extension septate, straight to flexuous, 118–262 μm long, 3–8 μm wide at the apical septum, terminating in a clavate vesicle, 4–8 μm diam. Conidiogenous apparatus 58–128 μm wide, and 50–90 μm long; primary branches aseptate, 14–31 × 5–8 μm; secondary branches aseptate, 10–23 × 4–6 μm; tertiary branches aseptate, 7–19 × 3–5 μm; quaternary branches and additional branches (–8) aseptate, 8–14 × 3–5 μm, each terminal branch producing 2–6 phialides; phialides doliiform to reniform, hyaline, aseptate, 6–12 × 3–5 μm, apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (32–)34–38(–39) × 4–5 μm (av. 36 × 4 μm), 1(–3)-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Mega- and microconidia not observed.
Culture characteristics: Colonies fast growing (60–75 mm diam) on MEA after 1 wk at room temperature; surface cinnamon to brick with abundant white woolly aerial mycelium and abundant sporulation on the aerial mycelium and colony surface; reverse brick to sepia with abundant chlamydospores throughout the medium, forming microsclerotia.
Material examined: Ecuador, from soil, 20 Jun. 1997, M.J. Wingfield (holotype CBS H-23136, culture ex-type CBS 111423 = CPC 1650).
Notes: Calonectria octoramosa is a new species in the Ca. brassicae complex. It can be distinguished from other species in this complex by having eight levels of branching in its conidiogenous apparatus.
Calonectria parvispora L. Lombard & Crous, sp. nov. MycoBank MB820844. Fig. 16.
Fig. 16.
Calonectria parvispora (ex-type CBS 111465). A−C. Macroconidiophores. D−F. Conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides. G−J. Clavate vesicles. K. Macroconidia. Scale bars: A−C = 20 μm; D−K = 10 μm.
Etymology: Name refers to the small macroconidia of this fungus.
Macroconidiophores consist of a stipe bearing a penicillate arrangement of fertile branches, and a stipe extension terminating in a vesicle; stipe septate, hyaline, smooth, 36–152 × 7–9 μm; stipe extension septate, straight to flexuous, 137–277 μm long, 3–6 μm wide at the apical septum, terminating in a clavate vesicle, 4–8 μm diam. Conidiogenous apparatus 56–92 μm wide, and 50–70 μm long; primary branches aseptate, 16–34 × 4–7 μm; secondary branches aseptate, 11–20 × 4–6 μm; tertiary branches aseptate, 7–15 × 3–5 μm; quaternary branches and additional branches (–6) aseptate, 8–16 × 3–5 μm, each terminal branch producing 2–6 phialides; phialides doliiform to reniform, hyaline, aseptate, 7–12 × 3–5 μm, apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (24–)26–32(–36) × (3–)3.5–4.5(–5) μm (av. 29 × 4 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Mega- and microconidia not observed.
Culture characteristics. Colonies fast growing (50–75 mm diam) on MEA after 1 wk at room temperature; surface umber to sepia with abundant buff to white, woolly aerial mycelium and abundant sporulation on the aerial mycelium and colony surface; reverse amber to sepia with abundant chlamydospores throughout the medium, forming microsclerotia.
Material examined: Brazil, from soil, Jun. 1998, A.C. Alfenas (holotype CBS H-22765, culture ex-type CBS 111465 = CPC 1902). Colombia, La Paz, Rodal Seuiller, from soil, Jan. 1994, P.W. Crous, CBS 116108 = CPC 726.
Notes: Calonectria parvispora is a new species in the Ca. brassicae complex (Lombard et al., 2009, Alfenas et al., 2015). The macroconidia of Ca. parvispora [(24–)26–32(–36) × (3–)3.5–4.5(–5) μm (av. 29 × 4 μm)] are smaller than those of Ca. clavata [(44–)50–70(–80) × (4–)5–6 μm (av. 65 × 5 μm); Crous 2002], Ca. brachiatica [(37–)40–48(–50) × 4–6 μm (av. 44 × 5 μm); Lombard et al. 2009], Ca. brassicae [(38–)40–55(–65) × (3.5–)4–5(–6) μm (av. 53 × 4.5 μm); Crous 2002], Ca. ecuadoriae [(45–)48–55(–65) × (4–)4.5(–5) μm (av. 51 × 4.5 μm); Crous et al. 2006a], Ca. gracilipes [(35–)40–48(–60) × 4–5(–6) μm (av. 45 × 4.5 μm); Crous 2002] and Ca. gracilis [(40–)53–58(–65) × (3.5–)4–5 μm (av. 56 × 4.5 μm); Crous 2002].
Calonectria tucuruiensis L. Lombard & Crous, sp. nov. MycoBank MB820845. Fig. 17.
Fig. 17.
Calonectria tucuruiensis (ex-type CBS 114755). A−C. Macroconidiophores. D−F. Conidiogenous apparatus with conidiophore branches and elongate doliiform to reniform phialides. G−J. Fusiform to ovoid to ellipsoid vesicles. K. Macroconidia. Scale bars: A−C = 50 μm; D−K = 10 μm.
Etymology: Name refers to Tucuruí, the region in Brazil from which this fungus was collected.
Macroconidiophores consist of a stipe bearing a penicillate arrangement of fertile branches, and a stipe extension terminating in a vesicle; stipe septate, hyaline, smooth, 35–105 × 6–9 μm; stipe extension septate, straight to flexuous, 165–290 μm long, 4–6 μm wide at the apical septum, terminating in a fusiform to ovoid to ellipsoid vesicle, 9–12 μm diam. Conidiogenous apparatus 40–95 μm wide, and 40–90 μm long; primary branches aseptate, 19–32 × 4–7 μm; secondary branches aseptate, 10–28 × 3–5 μm; tertiary branches aseptate, 11–16 × 3–6 μm; quaternary branches aseptate, 8–14 × 3–4 μm each terminal branch producing 2–4 phialides; phialides elongate doliiform to reniform, hyaline, aseptate, 8–17 × 3–5 μm, apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (51–)57–69(–71) × (4–)4.5–5.5(–6) μm (av. 63 × 5 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Mega- and microconidia not observed.
Culture characteristics. Colonies fast growing (55–75 mm diam) on MEA after 1 wk at room temperature; surface cinnamon to amber with sparse, buff to white, woolly aerial mycelium and abundant sporulation on the aerial mycelium and colony surface; reverse sienna to amber with abundant chlamydospores throughout the medium, forming microsclerotia.
Material examined: Brazil, Tucuruí, from leaves of Eucalyptus tereticornis, 8 Aug. 1996, P.W. Crous (holotype CBS H-22777, culture ex-type CBS 114755 = CPC 1403); ibid., CBS 116265 = CPC 3552.
Notes: Calonectria tucuruiensis is closely related to Ca. terricola (Fig. 11). The macroconidia of Ca. tucuruiensis [(51–)57–69(–71) × (4–)4.5–5.5(–6) μm (av. 63 × 5 μm)] are larger than those of Ca. terricola [(40–)43–49(–53) × (3–)4–5(–6) μm (av. 46 × 4.5 μm); Lombard et al. (2016)].
Authors: L. Lombard & P.W. Crous
Ceratocystis Ellis & Halst., New Jersey Agric. Coll. Exp. Sta. Bull. 76: 14. 1890. Fig. 18.
Fig. 18.
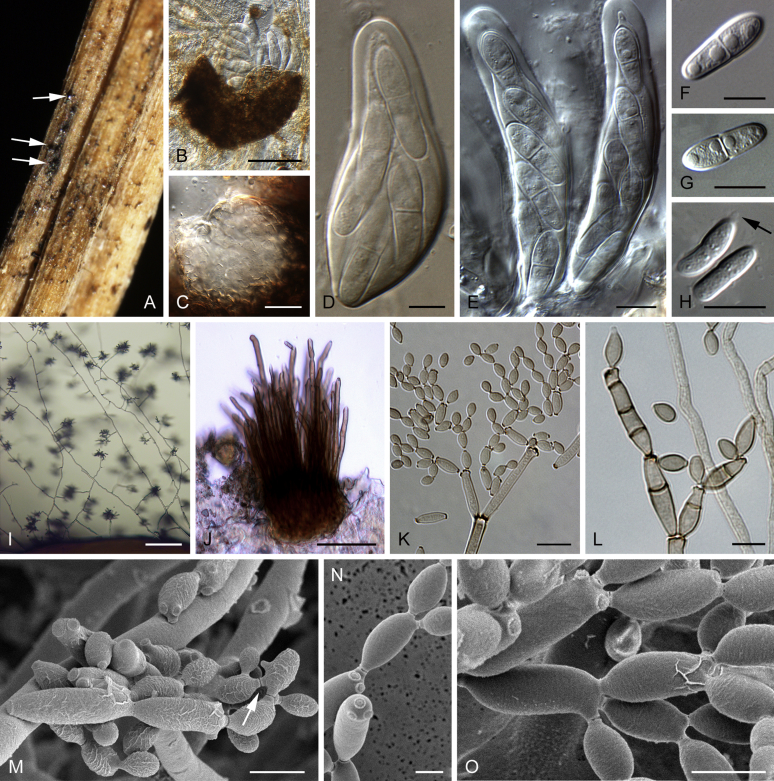
A. Sweet potatoes (Ipomea batatas) infected with Ceratocystis fimbriata. B−O. Microscopic features of Ceratocystis fimbriata (CBS 114723 = CMW 14799) on 2 % MEA. B. Ascomata with yellowish droplets of ascospores at tips of necks, with asexual state (white background). C. Young ascoma. D. Mature ascoma. E. Ostiolar hyphae. F, G. Ascospores. H, I. Aleurioconidia. J. Conidiogenous cells producing aleurioconidia (black arrow) and cylindrical-shape conidia (white arrow). K–O. Conidia of various shapes in chains. Scale bars: B = 500 μm; C, D = 100 μm; E = 50 μm; F, G, K–O = 10 μm; H = 50 μm; I, J = 25 μm.
Synonym: Rostrella Zimm., Meded. Lands Plantentuin 37: 24, 41. 1900.
Classification: Sordariomycetes, Hypocreomycetidae, Microascales, Ceratocystidaceae.
Type species: Ceratocystis fimbriata Ellis & Halst. Neotype: BPI 595863.
DNA barcodes (genus): 60S, LSU, mcm7.
DNA barcodes (species): ITS, bt1, tef1, rpb2, ms204. Table 4.
Table 4.
DNA barcodes of accepted Ceratocystis spp.
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; MAFF: Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Ibaraki, Japan. T and PT indicate ex-type and ex-paratype, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; bt1: partial β-tubulin gene; tef1: partial translation elongation factor 1-alpha gene, ms204: partial guanine nucleotide-binding protein subunit beta-like protein gene; rpb2: partial RNA polymerase II second largest subunit gene. ∗Multiple ITS types reported.
Ascomata perithecial, scattered or gregarious, immersed, partially embedded or superficial on the substrate; bases subglobose to globose or obpyriform, brown to black, covered with undifferentiated hyphae; ostiolar necks central, long, tapering towards apex; ascomatal apex straight or undulate, unbranched or branched, brown to black and becoming paler; ostiolar hyphae divergent or convergent, non-septate, straight, tapering towards apex, hyaline to light brown. Asci evanescent. Ascospores hyaline, 1-celled, ellipsoidal with gelatinous sheath which gives hat-shaped impression, accumulating in white, creamy to yellow masses at tips of necks. Conidiophores branched, straight or flexuous, hyaline to pale brown. Conidiogenous cells endophialidic, flask-shaped (lageniform) producing various shapes of cylindrical conidia or tubular-form producing barrel-shaped (doliiform) conidia, either lageniform alone or both forms present. Conidia hyaline, 1-celled, doliiform to cylindrical. Aleurioconidia (in some literature as chlamydospores) absent or present, pale brown to dark brown, pyriform, ellipsoidal to globose, singly or in chains.
Culture characteristics: Colonies showing circular growth with undulate margins, mycelium submerged to aerial, colour ranging from moderate yellowish brown to greyish or brownish olive when mature, releasing sweet fruity aroma. No growth on cycloheximide.
Optimal media and cultivation conditions: 2 % MEA incubated at 25 °C. Addition of thiamin stimulates the development of sexual morph.
Distribution: Worldwide.
Hosts: Herbaceous root crops, Ipomea batatas (sweet potato), wounds or larval tunnels of woody angiosperms, Acacia, Annona, Carya, Citrus, Coffea, Colocacia, Colophospermum, Combretum, Corymbia, Cunninghamia, Dalbergia, Eucalyptus, Coffea, Mangifera, Platanus, Populus, Prosopis, Punica, Quercus, Rapanea, Saccharum, Schizolobium, Schotia, Styrax, Syzygium, Terminalia, Theobroma. Some known to be vectored by flies (Diptera), non-specific ambrosia beetles (Scolytinae), or nitidulid beetles (Nitidulidae), but without specific insect associates.
Disease symptoms: Root rot, cankers and vascular stain.
Notes: Ceratocystis sensu lato included a heterogeneous group of fungi classified under this generic name due to similar morphology resulting from convergent evolution, despite their diverse ecological and biological features (Upadhyay 1981). The group has recently been divided into seven more narrowly defined homogeneous genera, supported by multigene phylogenies, morphological similarities and ecological commonality (Wingfield et al., 2013a, De Beer et al., 2014). The family Ceratocystidaceae includes nine genera, namely Ambrosiella, Ceratocystis, Chalaropsis, Davidsoniella, Endoconidiophora, Huntiella, Thielaviopsis, Meredithiella and Phialophoropsis (De Beer et al., 2014, Mayers et al., 2015). Ceratocystis sensu stricto is now restricted to those species producing ascomata with smooth bases, ascospores with hat-shaped sheaths, and thielaviopsis-like asexual morphs, which differentiate them from other genera (De Beer et al. 2014). Within Ceratocystis, morphological differences between species are insignificant and phylogenies based on multiple gene regions are used to distinguish them from each other (Fourie et al. 2015). The ITS region has been widely used for delimiting species of Ceratocystis (Schoch et al. 2012). However, discovery of multiple ITS types within single species in the genus (Al Adawi et al., 2013, Naidoo et al., 2013, Harrington et al., 2014) raised an awareness that the ITS region alone should not be applied to delimit species in Ceratocystis, and that additional gene regions should also be considered. Loci such as bt1 and tef1 do not provide good species resolution on their own, but provide strong support in combination with ITS (Fourie et al. 2015). The loci rpb2 and ms204 give stronger resolution than tef1 and bt1, but also need to be used in combination with ITS (Fourie et al. 2015).
References: Hunt, 1956, Upadhyay, 1981 (morphology); Nag Raj and Kendrick, 1975, Paulin-Mahady et al., 2002 (asexual morphs and species); Kile, 1993, Van Wyk et al., 2013 (pathogenicity); De Beer et al. 2013a (higher classification); De Beer et al. 2013b (nomenclator); Wilken et al., 2013, Van der Nest et al., 2014a, Van der Nest et al., 2014b, Wingfield et al., 2015, Wingfield et al., 2016a, Wingfield et al., 2016b (genomes); Wingfield et al., 2013a, De Beer et al., 2014 (generic definitions and phylogenetic relationships); Wingfield et al. 2013b (international spread).
Authors: I. Barnes, S. Marincowitz, Z.W. de Beer, & M.J. Wingfield
Cladosporium Link, Mag. Gesell. naturf. Freunde, Berlin 7: 37. 1816 (1815). Fig. 19.
Fig. 19.
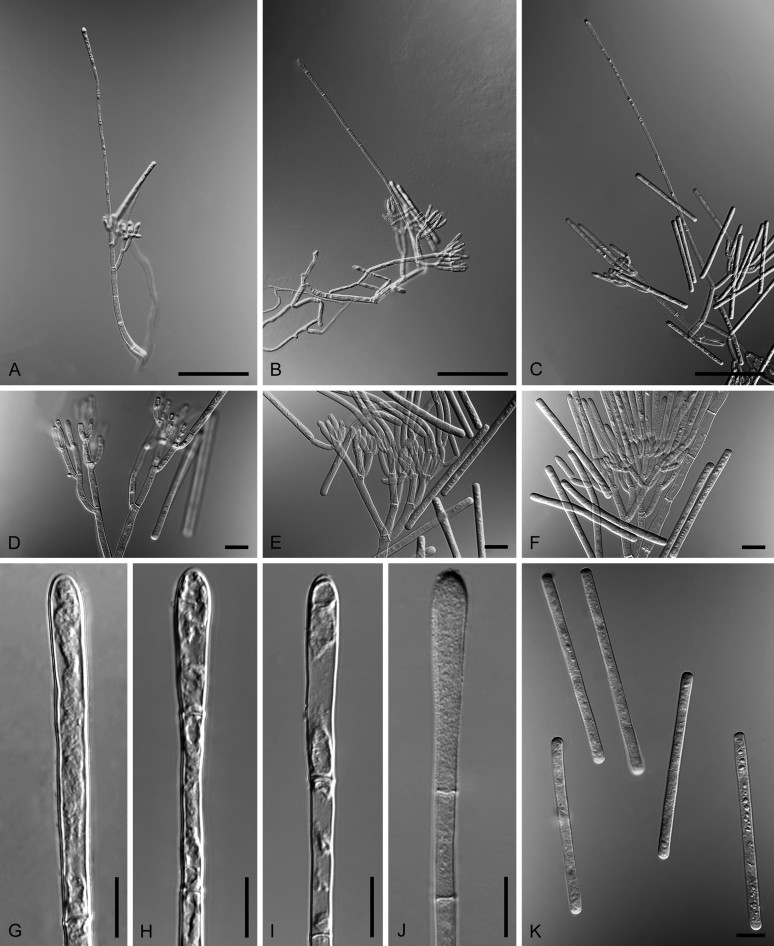
Cladosporium spp. A–H. Sexual morphs. A. Ascomata on host tissue (arrows) of Cladosporium silenes (holotype CBS H-19874). B. Ascoma and asci of Cladosporium herbarum (CPC 11600). C. Ostiole of Cladosporium macrocarpum (CBS 299.67). D, E. Asci of Cladosporium herbarum (CPC 11600). F, G. Ascospores of Cladosporium herbarum (CPC 11600). H. Ascospores (arrow denotes mucoid appendage) of Cladosporium silenes (holotype CBS H-19874). I–O. Asexual morphs. I. Conidiophores of Cladosporium halotolerans (ex-type CBS 119416). J. Fascicle of conidiophores of Cladosporium soldanellae (BPI 427476). K. Macronematous conidiophores and conidial chains of Cladosporium cladosporioides (ex-neotype CBS 112388). L. Conidial chains, septa of secondary ramoconidia distinctly darkened of Cladosporium paracladosporioides (ex-type CBS 171.54). M. CryoSEM of different types of conidia on aerial structures of Cladosporium exile (ex-type CBS 125987). Note a remarkable pattern of blastoconidium formation (backwards) (arrow). N. Secondary ramoconidia, conidia and scars of Cladosporium perangustum (ex-type CBS 125996). O. Whorls of secondary ramoconidia and conidia of Cladosporium scabrellum (ex-type CBS 126358). Scale bars: B, C, M, O = 5 μm; D–H, K, L = 10 μm; I = 100 μm; J = 50 μm; N = 2 μm. Pictures taken from Bensch et al. (2012).
For synonyms see Bensch et al. (2012).
Classification: Dothideomycetes, Dothideomycetidae, Capnodiales, Cladosporiaceae.
Type species: Cladosporium herbarum (Pers. : Fr.) Link. Lectotype: L 910.225-733. Epitype and ex-epitype culture: CBS H-19853, CPC 12100 = CBS 121621.
DNA barcodes (genus): LSU.
DNA barcodes (species): act and tef1; in a few cases tub2. Table 5. Fig. 20.
Table 5.
DNA barcodes of accepted Cladosporium spp.
ATCC: American Type Culture Collection, Virginia, USA; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CPC: Culture collection of Pedro Crous, housed at Westerdijk Fungal Biodiversity Institute. T, ET, NT and RS indicate ex-type, ex-epitype, ex-neotype and reference strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; act: partial actin gene; tef1: partial translation elongation factor 1-alpha gene.
Fig. 20.
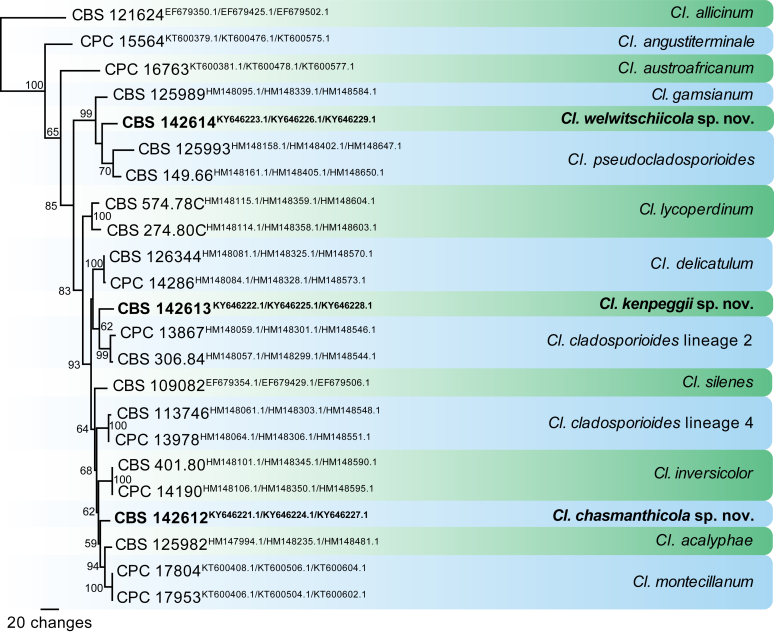
The first of two equally most parsimonious trees obtained from a heuristic search of the combined ITS/tef1/actA alignment. The tree was rooted to Cladosporium allicinum CBS 121624 and the novel species described in this study are shown in bold. Bootstrap support values from 1000 replicates are shown at the nodes and the scale bar represents the number of changes. GenBank accession numbers are indicated in superscript (ITS/tef1/actA). TreeBASE: S20877.
Ascomata pseudothecial, black to red-brown, globose, inconspicuous and immersed beneath stomata to superficial, situated on a reduced stroma, with 1(–3) short, periphysate ostiolar necks; periphysoids frequently growing down into cavity; ascomatal wall consisting of 3–6 layers of textura angularis. Pseudoparaphyses frequently present in mature ascomata, hyaline, septate, subcylindrical. Asci fasciculate, short-stalked or not, subsessile, bitunicate, obovoid to broad ellipsoid or subcylindrical, straight to slightly curved, 8-spored. Ascospores bi- to multiseriate, hyaline, obovoid to ellipsoid-fusiform, with irregular luminar inclusions, mostly thick-walled, straight to slightly curved, frequently becoming brown and verruculose in asci, at times covered in mucoid sheath. Dematiaceous hyphomycetes. In vivo: Mycelium internal or external, superficial; hyphae branched, septate, subhyaline to usually pigmented, smooth, sometimes slightly rough-walled to verruculose. Stromata absent to sometimes well-developed. Conidiophores mononematous, usually macronematous, solitary, fasciculate, in small to large fascicles, loosely to densely caespitose, usually erect, occasionally subdecumbent, decumbent or repent, straight to flexuous, unbranched or branched, continuous to septate, subhyaline to usually distinctly pigmented, smooth to verruculose, proliferation holoblastic, occasionally enteroblastic (after a period when growth has stopped and then resumed), usually sympodial, rarely monopodial (sometimes leaving coarse annellations from repeated enteroblastic proliferation). Conidiogenous cells integrated, terminal or intercalary, monoblastic or usually polyblastic, mostly sympodially proliferating, more or less cylindrical, geniculate-sinuous or nodulose, sometimes with unilateral swellings; conidiogenous loci usually conspicuous, protuberant, composed of a central convex dome surrounded by a more or less raised periclinal rim (coronate), thickened, refractive or barely to distinctly darkened; conidial formation holoblastic. Conidia solitary or catenate, in unbranched or branched acropetal chains, amero- to phragmosporous, shape and septation variable, usually subglobose, ovoid, obovoid, ellipsoid, fusiform, limoniform to cylindrical, aseptate or with several transverse eusepta, rarely with a single longitudinal septum, subhyaline to usually pigmented, smooth, verruculose, verrucose, echinulate, cristate; hila protuberant, coronate, with a central convex dome and raised periclinal rim, thickened, refractive to darkened; microcyclic conidiogenesis often occurring. In vitro: Stromata usually lacking. Conidiophores usually solitary, arising terminally or laterally from plagiotropous or ascending hyphae, often longer than in vivo. Micronematous conidiophores, lacking in vivo, are often formed in culture. Conidial chains often longer than in vivo (species with solitary conidia are often capable of forming conidial chains in culture).
Culture characteristics: Colonies on SNA often grey olivaceous or olivaceous grey, reverse leaden-grey or black, flat, velvety with fluffy or cottony patches, margin irregular or undulate, aerial mycelium loose diffuse or more abundantly formed, often with abundant submerged mycelium.
Optimal media and cultivation conditions: For morphological examinations SNA incubated under continuous near-ultraviolet light at 25 °C proved to be best suited to promote sporulation. The sexual morph can be induced by inoculating plates of 2 % WA onto which autoclaved stem pieces of Urtica dioica (European stinging nettle) are placed. Inoculated plates have to be incubated on the laboratory bench for 1 wk, after that period they have to be further incubated at 10 °C in the dark for 1–2 mo to stimulate sexual morph development.
Distribution: Worldwide.
Hosts: Asparagaceae, Asteraceae, Fabaceae, Myrtaceae, Orchidaceae, Poaceae and many other hosts, including fungi and insects.
Disease symptoms: Leaf spots, leaf blight, discolourations, necrosis, or shot-hole symptoms, on stems and fruits, but also saprobic, endophytic or isolated from numerous substrates and environments, e.g. indoor environments, salterns and human and animal infections.
Notes: The monophyletic genus Cladosporium is well characterised by the coronate structure of its conidiogenous loci and conidial hila, consisting of a central convex dome surrounded by a raised periclinal rim (David, 1997, Braun et al., 2003). At the moment it comprises about 200 species. Cladosporium was previously extremely heterogeneous and encompassed 772 names assigned to this genus (Dugan et al. 2004). Heuchert et al. (2005) examined Cladosporium spp. dwelling on other fungi, and Schubert (2005) provided a comprehensive treatment of foliicolous species. Crous et al. (2007a) encompassed a series of papers dealing with a reassessment and new circumscription of Cladosporium s. str. and treatments of several cladosporioid genera. Bensch et al. (2012) published a taxonomic monograph of the genus Cladosporium which can be consulted for further information on the history and many other aspects of this genus.
Species delimitation in Cladosporium based on morphology alone is limited since many species have overlapping characters. Some key differential features have been identified and detailed in a series of monographic papers (Schubert et al., 2007, Zalar et al., 2007, Bensch et al., 2010, Bensch et al., 2012). The most relevant differential morphological traits are the shape, width and complexity of conidiophores, the presence of ramoconidia, and the formation and ornamentation of conidia. However, given the overlapping of these features, and the need for standardisation using special culture media and scanning electron microscopy procedures, the use of a molecular approach should be mandatory for correct identification of the species in this complex fungal group (Sandoval-Denis et al. 2016).
Three different species complexes are recognised within the genus, mainly based on morphology, and used for practical purposes. The Cl. cladosporioides species complex is characterised by mainly narrowly cylindrical or cylindrical-oblong, non-nodulose, mostly non-geniculate conidiophores and conidia with a quite variable surface ornamentation ranging from smooth to irregularly verrucose-rugose or rough-walled (reticulate or embossed stripes under SEM); the Cl. herbarum species complex includes species mainly having nodulose conidiophores, with conidiogenesis confined to swellings, and verruculose, verrucose or echinulate conidia; and the Cl. sphaerospermum complex is most remarkable due to forming numerous globose or subglobose terminal and intercalary conidia with variable surface ornamentation and often poorly differentiated conidiophores in most of the species (Bensch et al., 2012, Bensch et al., 2015). Morphologically similar genera have been treated in Crous et al. (2007b).
Members of Cladosporiaceae: Cladosporium, Graphiopsis, Neocladosporium, Rachicladosporium, Toxicocladosporium, Verrucocladosporium.
References: Braun et al. 2003 (sexual morph); Crous et al., 2007a, Crous et al., 2007b (cladosporium-like genera); Schubert et al. 2007 (morphology, phylogeny Cl. herbarum complex); Zalar et al. 2007 (morphology, phylogeny Cl. sphaerospermum complex); Bensch et al. 2010 (morphology, phylogeny Cl. cladosporioides complex); Bensch et al. 2012 (morphology, phylogeny and key of all Cladosporium species); Bensch et al. 2015 (morphology, additions to the three species complexes); Sandoval-Denis et al. 2016 (morphology, phylogeny of clinical samples).
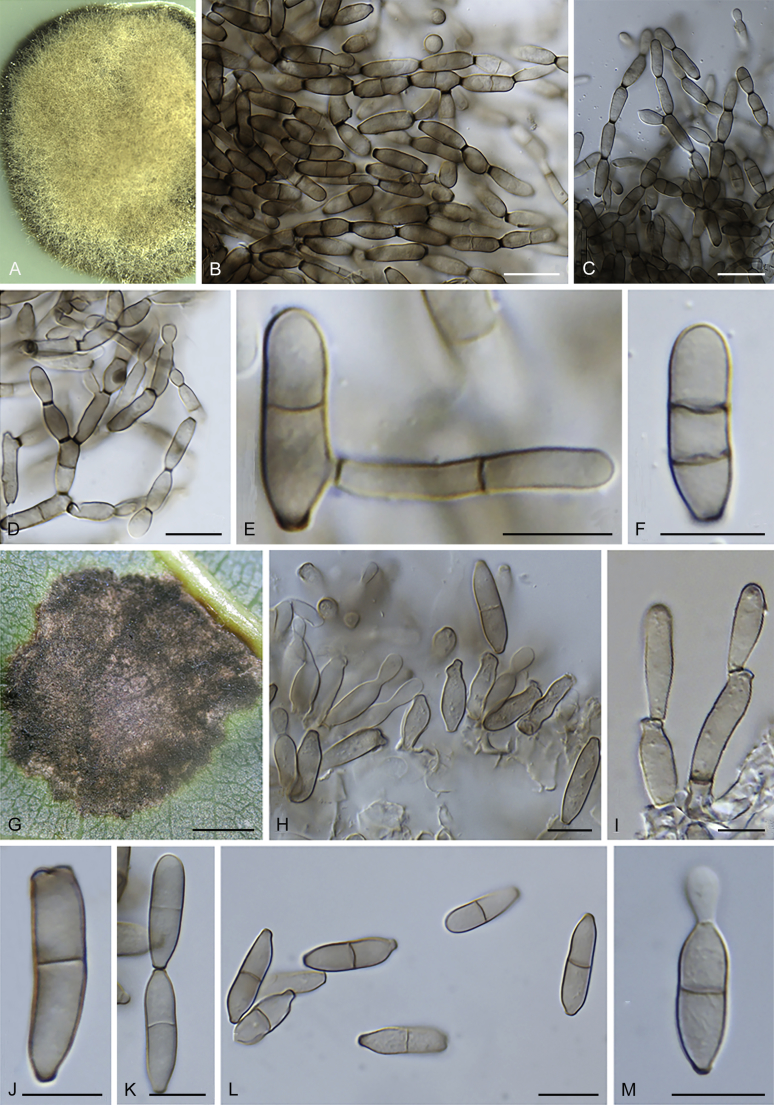
Cladosporium chasmanthicola Bensch, U. Braun & Crous, sp. nov. MycoBank MB819978. Fig. 21.
Fig. 21.
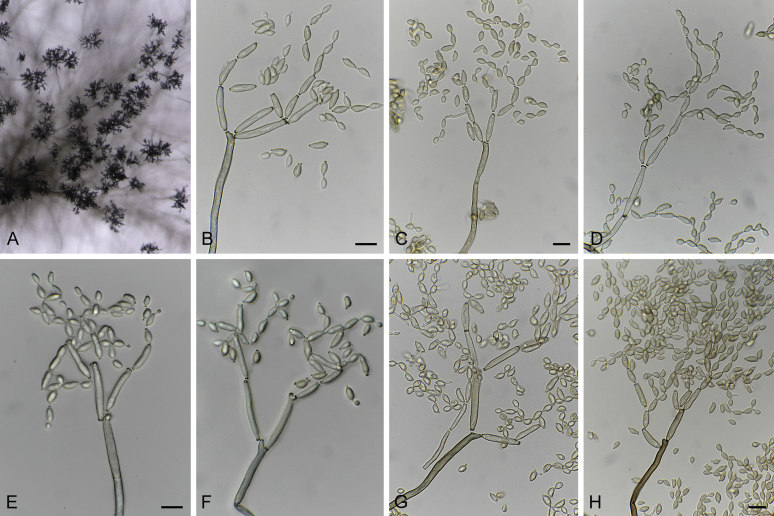
Cladosporium chasmanthicola (ex-type CBS 142612). A–H. Conidiophores and conidial chains. Scale bars = 10 μm; C applies to C–G.
Etymology: Epithet composed of the name of the host genus, Chasmanthe, and -cola, dweller.
Leaf spots solitary, distributed over leaf surface, amphigenous, ellipsoid, 1–2 mm diam, pale brown with dark red-brown margin, some spots also associated with uredinia of Uromyces kentaniensis. On SNA: Mycelium loosely branched, filiform, narrowly cylindrical-oblong or irregular in outline due to swellings and constrictions, 0.5–4 μm wide, septate, subhyaline or pale olivaceous or olivaceous brown, almost smooth, verruculose, distinctly verrucose or irregularly rough-walled. Conidiophores solitary, formed terminally or laterally from hyphae, straight or somewhat flexuous, macro- and micronematous; macronematous conidiophores cylindrical, sometimes geniculate, often irregular in outline due to lateral outgrowths, swellings and constrictions (not connected with conidiogenesis), mostly unbranched, 20–100(–140) × 3.5–5(–6) μm, up to 6 μm wide at the base, 1–6-septate, septa sometimes in short succession, not constricted at septa, pale olivaceous or pale to medium olivaceous brown, smooth, walls slightly thickened; micronematous conidiophores shorter, narrower and paler than macronematous ones, 15–30(–80) × 2–3 μm, 0–2-septate, subhyaline or pale olivaceous. Conidiogenous cells integrated, terminal and intercalary, 8–24 μm long, short cylindrical or often irregular in outline due to lateral prolongations and shoulders and numerous conidiogenous loci often crowded at or towards the apex, up to eight loci in terminal cells, 1–3 loci in intercalary cells, loci conspicuous, subdenticulate, 1–2 μm diam. Ramoconidia commonly formed, subcylindrical or irregular due to numerous loci at the distal end, 15–33 × 3–4.5 μm, 0–1(–3)-septate, base broadly truncate, 2.5(–3.5) μm wide. Conidia numerously formed, especially small terminal and intercalary conidia, in branched chains, branching in all directions with 1–3 conidia in the terminal unbranched part of the chain; terminal conidia very small, ovoid or obovoid, very pale, subhyaline or pale olivaceous brown, 2.5–4.5 × 2–2.5(–3) μm (av. ± SD: 3.4 ± 0.6 × 2.2 ± 0.3), apex rounded; intercalary conidia ovoid, limoniform, ellipsoid or irregular due to lateral outgrowths, 4–10.5 × (2–)3–3.5(–4) μm (av. ± SD: 7.2 ± 2.0 × 3.1 ± 0.5), aseptate, with 1–4 distal hila; secondary ramoconidia ellipsoid, subcylindrical or irregular in outline due to numerous hila crowded at or towards the distal end, sometimes located on lateral shoulders or lateral prolongations, those formed on micronematous conidiophores shorter and narrower, (5–)8–23 × (2.5–)3–4.5 μm (av. ± SD: 13.3 ± 5.4 × 3.5 ± 0.6), 0–1(–3)-septate, very pale olivaceous or pale olivaceous brown, smooth, walls unthickened, with (2–)3–6(–7) distal scars; hila conspicuous, 0.5–2 μm diam, darkened-refractive and somewhat thickened; conidia sometimes germinating.
Culture characteristics: Colonies on PDA reaching 28–35 mm diam after 2 wk, olivaceous grey, grey olivaceous with several smoke-grey patches of dense, felty aerial mycelium, reverse leaden-grey to olivaceous grey, powdery, margin white, broad, glabrous, colony centre somewhat folded and wrinkled, growth flat. Colonies on MEA attaining 29–35 mm diam, whitish, smoke-grey to pale olivaceous grey, reverse greyish sepia or olivaceous grey, velvety; margin glabrous, to somewhat feathery, radially furrowed, colony centre elevated, wrinkled and folded; aerial mycelium abundant, covering large parts of the colony surface, dense, fluffy. Colonies on OA reaching 20–28 mm diam, olivaceous grey with patches of smoke-grey, grey olivaceous or glaucous-grey towards margins, reverse leaden-grey to iron-grey, fluffy-felty; margin glabrous, undulate, colony centre somewhat elevated; aerial mycelium loose, diffuse to dense and fluffy in a few spots. On all media without prominent exudates, sporulation profuse.
Material examined: South Africa, Western Cape Province, Cape Town, Brackenfell, Bracken Nature Reserve, isol. from leaf spots on Chasmanthe aethiopica, 25 Sep. 2012, A.R. Wood (holotype CBS H-23117, culture ex-type CBS 142612 = CPC 21300).
Note: Cladosporium chasmanthicola is closely related to Cl. acalyphae, but the latter species has much longer and narrower conidiophores (150–430 × 3–4 μm) and smooth to loosely verruculose, irregularly verruculose-rugose or rough-walled conidia (Bensch et al. 2010).
Cladosporium kenpeggii Bensch, U. Braun & Crous, sp. nov. MycoBank MB819979. Fig. 22.
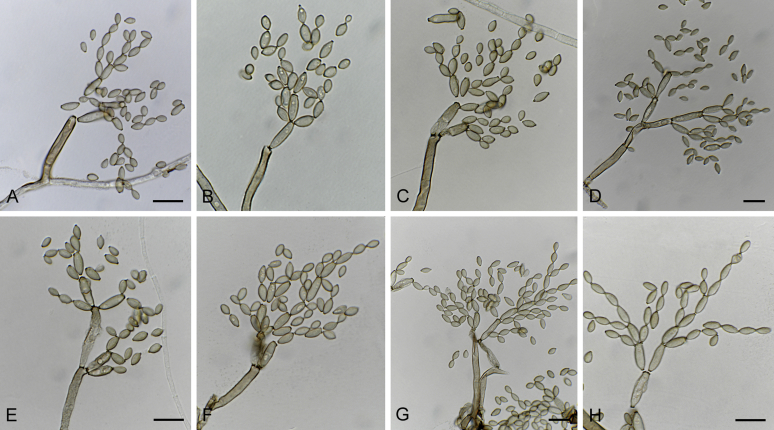
Fig. 22.
Cladosporium kenpeggii (ex-type CBS 142613). A. Part of the colony on SNA. B–H. Conidiophores and conidial chains. Note the microcyclic conidiogenesis in C, forming a secondary conidiophore at a still attached conidium with giving rise to secondary conidia and the germinating conidia in C and G. Scale bars = 10 μm; C applies to C, D; E applies to E–G.
Etymology: Named after Dr Ken Pegg (Agri-Science and Biosecurity Queensland, Australia), the collector of the species, who celebrates his 80th birthday this year.
On SNA: Superficial mycelium sparingly formed, unbranched, occasionally branched, 2.5–3.5 μm wide, septate, without swellings and constrictions, pale olivaceous brown, almost smooth to verruculose. Conidiophores macronematous, solitary, arising mostly terminally, rarely laterally from hyphae, narrowly cylindrical-oblong, usually unbranched, non-nodulose, sometimes slightly geniculate towards the apex, 15–100(–150) × 2.5–4 μm, 0–2(–5)-septate, pale to medium olivaceous brown, smooth or minutely verruculose, walls unthickened or slightly thickened. Conidiogenous cells integrated, mainly terminally, narrowly cylindrical-oblong, 16–60 μm long, with (1–)2–3(–4) distal conidiogenous loci, crowded at or towards the apex, sometimes slightly geniculate due to sympodial proliferation, conidiogenous loci conspicuous, 1–2 μm diam, thickened and darkened-refractive, sometimes cells germinating. Ramoconidia frequently formed, (17–)25–55 × 3–4(–5) μm, 0–1(–2)-septate, base broadly truncate, 2–4 μm wide, unthickened, somewhat darkened-refractive. Conidia numerous, formed in branched chains, branching in all directions, up to eight conidia in the terminal unbranched part of the chain; small terminal conidia obovoid or ellipsoid, 4.5–6 × (2–)2.5–3(–3.5) μm (av. ± SD: 5.0 ± 0.5 × 2.7 ± 0.5), apex rounded; intercalary conidia ovoid or ellipsoid, 5.5–15 × (2–)2.5–3.5 μm (av. ± SD: 8.9 ± 3.2 × 3.0 ± 0.4), aseptate, with 1–2 distal hila, attenuated towards apex and base; secondary ramoconidia subcylindrical or cylindrical, 14.5–35 × 3–4(–5) μm (av. ± SD: 22.4 ± 5.8 × 3.8 ± 0.6), 0–1(–2)-septate, with 2–3 distal hila, pale olivaceous or pale olivaceous brown, smooth, walls slightly thickened; hila conspicuous, subdenticulate, 1–2 μm diam, somewhat thickened and darkened-refractive; microcyclic conidiogenesis occurring, conidia often germinating, often with more than one germination tube, tubes sometimes even branched, filiform or irregular in outline.
Culture characteristics: Colonies on PDA attaining 35–47 mm diam after 2 wk, olivaceous to olivaceous grey, dull green towards margins, reverse iron-grey, greyish blue towards margins, powdery to fluffy, margin feathery, growth flat, aerial mycelium loose, diffuse, dense, fluffy and high in a few spots, pale olivaceous grey, sporulation profuse, without prominent exudates. Colonies in MEA reaching 39–48 mm diam, grey olivaceous, reverse iron-grey, velvety, margin white, broad, feathery, colony centre elevated, wrinkled and folded, radially furrowed, aerial mycelium loose, diffuse to denser and fluffy, several small but prominent exudates formed, sporulation profuse. Colonies on OA grey olivaceous when sporulating profusely, whitish or smoke-grey due to aerial mycelium, reverse lead-grey or iron-grey, some parts with a cinnamon margin (both on top and reverse), powdery to fluffy-felty, aerial mycelium forming high strains, growth flat, without exudates.
Material examined: Australia, New South Wales, Upper Dungay, 28°15′ S 153°21′ E, isol. from leaves of Passiflora edulis, 20 Oct. 1999, K.G. Pegg & J. Dawes, FP 24737 (holotype BRIP 26701a, isotype CBS H-23118, culture ex-type CBS 142613 = CPC 19248 = BRIP 26701a).
Notes: The smooth conidia formed in long branched chains and the frequently formed ramoconidia remind one of Cl. cladosporioides and Cl. iranicum. However, compared with Cl. cladosporioides, Cl. kenpeggii possesses much shorter macronematous conidiophores, micronematous conidiophores are not formed and the conidia are very often germinating and forming secondary conidiophores. In Cl. iranicum the conidia also germinate quite often, but the conidiophores are longer, ramoconidia are shorter and somewhat wider with a narrower base and intercalary conidia are shorter and narrower (Bensch et al. 2012).
Cladosporium maracuja, described from Passiflora in Brazil in 1935, is morphologically quite similar in having smooth, catenate, 0–1-septate conidia and short conidiophores but since it is only known from the type specimen it is kept separate. The conidia of this species are shorter and wider and the conidiophores wider in vivo.
Cladosporium welwitschiicola Bensch, U. Braun & Crous, sp. nov. MycoBank MB819980. Fig. 23.
Fig. 23.
Cladosporium welwitschiicola (ex-type CBS 142614). A–G. Conidiophores and conidial chains. H. Conidial chain. Scale bars = 10 μm; A applies to A–C; E applies to E, F.
Etymology: Epithet composed of the name of the host genus, Welwitschia, and -cola, dweller.
On SNA: Superficial mycelium abundantly formed, filiform to cylindrical-oblong, unbranched or loosely branched, (0.5–)1–4 μm wide, sometimes slightly swollen or constricted, septate, subhyaline, pale olivaceous or pale olivaceous brown, surface ornamentation variable, smooth or almost so, asperulate, verruculose or sometimes even verrucose, walls unthickened, sometimes forming ropes of several hyphae. Conidiophores macronematous, solitary, erect, straight or slightly flexuous, terminally or laterally formed from hyphae, narrowly cylindrical-oblong, non-nodulose, occasionally once geniculate towards the apex due to sympodial proliferation, 25–90 × (2.5–)3–4.5(–5.5) μm, 0–3(–4)-septate, not constricted at septa, pale to medium olivaceous brown, smooth, sometimes verruculose or irregularly rough-walled towards the base, walls thickened. Conidiogenous cells integrated, usually terminal, cylindrical, 12.5–42 μm long, with 2–4 conidiogenous loci crowded at the apex, conspicuous, subdenticulate, 1–2 μm diam, somewhat thickened and darkened-refractive. Ramoconidia not observed. Conidia catenate, in branched chains, branching in all directions, (1–)2–5(–6) conidia in the terminal unbranched part of the chain; small terminal conidia obovoid, ellipsoid, 4–5 × 2.5–3.5 μm (av. ± SD: 4.6 ± 0.6 × 3.0 ± 0.4), rugulose, broadly rounded at the apex; intercalary conidia ellipsoid, limoniform or fusiform, sometimes irregular in outline due to surface ornamentation, slightly to distinctly attenuated towards apex and base, 5–11 × (2.5–)3–3.5(–4) μm (av. ± SD: 7.4 ± 1.9 × 3.2 ± 0.4), 0–1-septate, with 1–3 distal hila, rugulose to distinctly rugose; secondary ramoconidia ellipsoid or subcylindrical, often 3–4 formed at the apex of conidiophores, 8.5–21 × 3–4(–4.5) μm (av. ± SD: 14.6 ± 3.6 × 3.5 ± 0.4), 0–2(–3)-septate, mostly 1-septate, septum median or somewhat in the lower half, pale to medium olivaceous brown or dingy brown, smooth or almost so to rugulose, walls somewhat thickened; hila conspicuous, 0.5–2 μm diam; microcyclic conidiogenesis not occurring.
Culture characteristics: Colonies on PDA reaching up to 78 mm diam after 2 wk, olivaceous grey, fawn at margins, reverse mouse-grey, vinaceous-buff at margins, fluffy; margins feathery, growth low convex. Colonies on MEA reaching up to 80 mm diam, smoke-grey, pale olivaceous grey to olivaceous grey, reverse iron-grey, fluffy; margin feathery. Colonies on OA reaching up to 72 mm diam, smoke-grey and pale olivaceous grey, reverse iron-grey, fluffy. On all three media aerial mycelium abundantly formed covering large parts of the colony, loose to dense, high, fluffy; without prominent exudates, sporulation profuse.
Material examined: Namibia, isol. from dead leaf of Welwitschia mirabilis, 1 Oct. 2010, M.J. Wingfield (holotype CBS H-23119, culture ex-type CBS 142614 = CPC 18648).
Notes: With its rugulose or distinctly rugose conidia and relatively short conidiophores, Cl. welwitschiicola reminds one of Cl. exasperatum and Cl. verrucocladosporioides, but the latter two species differ in forming ramoconidia and in having longer and slightly wider small, intercalary and secondary ramoconidia. Phylogenetically, it is closest to Cl. gamsianum and Cl. pseudocladosporioides, but these species are easily distinguishable in having smooth and narrower conidia (Bensch et al. 2012).
Authors: K. Bensch, U. Braun, J.Z. Groenewald & P.W. Crous
Colletotrichum Corda, in Sturm, Deutschl. Fl., 3 Abt. (Pilze Deutschl.) 3: 41, tab. 21. 1831. Fig. 24, Fig. 25.
Fig. 24.
Colletotrichum spp. A–AA. Asexual morphs. A–C. Conidiomata. A.Colletotrichum acutatum (ex-type CBS 112996). B.Colletotrichum destructivum (ex-type CBS 136228). C.Colletotrichum cymbidiicola (ex-type IMI 347923). D. Seta of Colletotrichum torulosum (ex-type CBS 128544). E. Tip of a seta of Colletotrichum gloeosporioides (ex-type CBS 112999). F. Basis of a seta of Colletotrichum gloeosporioides (ex-type CBS 112999). G–K. Conidiogenous cells. G. Colletotrichum brasiliense (ex-type CBS 128501). H.Colletotrichum scovillei (ex-type CBS 126529). I.Colletotrichum tofieldiae (CBS 495.85). J.Colletotrichum petchii (ex-type CBS 378.94). K.Colletotrichum gloeosporioides (ex-type CBS 112999). L–R. Appressoria. L.Colletotrichum americae-borealis (CBS 136855). M.Colletotrichum graminicola (ex-epitype CBS 130836). N.Colletotrichum gloeosporioides (ex-type CBS 112999). O.Colletotrichum laticiphilum (ex-type CBS 112989). P.Colletotrichum phormii (ex-type CBS 118194). Q.Colletotrichum liriopes (ex-type CBS 119444). R.Colletotrichum truncatum (ex-type CBS 151.35). S–AA. Conidia of the ex-type strains of the name-giving species of nine Colletotrichum species complexes. S.Colletotrichum dematium (ex-type CBS 125.25). T.Colletotrichum acutatum (ex-type CBS 112996). U.Colletotrichum truncatum (ex-type CBS 151.35). V.Colletotrichum gloeosporioides (ex-type CBS 112999). W.Colletotrichum graminicola (ex-epitype CBS 130836). X.Colletotrichum boninense (ex-type CBS 123755). Y.Colletotrichum destructivum (ex-type CBS 136228). Z.Colletotrichum orbiculare (ex-type CBS 570.97). AA.Colletotrichum gigasporum (ex-type CBS 133266). A–C, E–H, K. from Anthriscus stem. D, I, J, L–AA. from SNA. Scale bars: A = 200 μm; B applies to B, C = 100 μm; G applies to D–AA = 10 μm. A–AA Pictures taken by U. Damm; A, H, O–P, T from Damm et al. (2012b); B, L, Y from Damm et al. (2014); C, D, G, J, X from Damm et al. (2012a); I, Q–S, U from Damm et al. (2009); Z from Damm et al. (2013).
Fig. 25.
A–F. Disease symptoms caused by Colletotrichum spp. A. Anthracnose on fruit of Cucurbita maxima cv. Red Hokkaido caused by Colletotrichum coccodes. B. Leaf spot of red clover caused by Colletotrichum utrechtense. C. Anthracnose on bean hypocotyl caused by Colletotrichum lindemuthianum. D. Leaf spot of Paphiopedilum sp. caused by Colletotrichum arxii. E. Anthracnose on strawberry fruit caused by Colletotrichum nymphaeae. F. Leaf spot of Mahonia aquifolium caused by Colletotrichum godetiae. G–R. Sexual morphs of Colletotrichum spp. G, H. Ascomata. G. Colletotrichum petchii (ex-type CBS 378.94). H.Colletotrichum karstii (CBS 127597). I. Peridium in cross section of Colletotrichum karstii (CBS 127597). J. Outer surface of peridium of Colletotrichum constrictum (ex-type CBS 128504). K–N. Ascospores. K.Colletotrichum salicis (ex-type CBS 607.94). L. Colletotrichum constrictum (ex-type CBS 128504). M.Colletotrichum cymbidiicola (ex-type IMI 347923). N.Colletotrichum parsonsiae (ex-type CBS 128525). O–Q. Asci. O.Colletotrichum cymbidiicola (ex-type IMI 347923). P.Colletotrichum salicis (ex-type CBS 607.94). Q.Colletotrichum constrictum (ex-type CBS 128504). R. Paraphyses of Colletotrichum salicis (ex-type CBS 607.94). G, K, M, O–R. from Anthriscus stem. H–J, L, N. from SNA. Scale bars: G = 100 μm; H = 50 μm; I applies to I–R = 10 μm. A–R Pictures taken by U. Damm; E from Cannon et al. (2012); G–J, L–O, Q from Damm et al. (2012a); K, P, R from Damm et al. (2012b).
Synonyms: Glomerella Spauld. & H. Schrenk, Science, N.Y. 17: 751. 1903.
For additional synonyms see Sutton (1980).
Classification: Sordariomycetes, Hypocreomycetidae, Glomerellales, Glomerellaceae.
Type species: Colletotrichum lineola Corda. Holotype: PRM 155463. Epitype and ex-epitype culture: CBS H-20362, CBS 125337.
DNA barcodes (genus): ITS.
DNA barcodes (species): act, ApMat, apn2, cal, chs-1, gapdh, gs, his3, sod2, tub2. Table 6, Table 7. Fig. 26.
Table 6.
DNA barcodes of accepted Colletotrichum spp. except for species in the Col. graminicola and caudatum complexes.
ATCC: American Type Culture Collection, Virginia, USA; BCC: BIOTEC Culture Collection, National Center for Genetic Engineering and Biotechnology (BIOTEC), Khlong Luang, Pathumthani, Thailand; BRIP: Queensland Plant Pathology Herbarium, Brisbane, Australia; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CGMCC: Chinese General Microbiological Culture Collection Center, Beijing, China; COAD: Coleção Octávio Almeida Drummond, Universidade Ferderal de Viçosa, Viçosa, Brazil; CPC: Culture collection of Pedro Crous, housed at Westerdijk Fungal Biodiversity Institute; GZAAS: Guizhou Academy of Agricultural Sciences, Guizhou Province, China. ICMP: International Collection of Micro-organisms from Plants, Landcare Research, Private Bag 92170, Auckland, New Zealand; IMI: International Mycological Institute, Kew, UK; MAFF: Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Ibaraki, Japan; MFLU: Herbarium of Mae Fah Luang University, Chiang Rai, Thailand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Ria, Thailand. HT and T indicate holotype specimens and ex-type strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; gapdh: partial glyceraldehyde-3-phosphate dehydrogenase gene; chs-1: partial chitin synthase-1 gene; his: partial histone H3 gene; act: partial actin gene; tub2: partial beta-tubulin gene; cal: partial calmodulin gene; gs: partial glutamine synthetase gene; sod2: partial manganese superoxide dismutase gene; ApMat: partial Apn2-Mat1-2 intergenic spacer and partial mating type (Mat1-2) gene.
Table 7.
DNA barcodes of accepted Colletotrichum spp. in the Col. graminicola and caudatum complexes.
BPI: US National Fungus Collections, Beltsville, Maryland, USA; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CGMCC: Chinese General Microbiological Culture Collection Center, Beijing, China; IMI: International Mycological Institute, Kew, UK; MAFF: Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Ibaraki, Japan. LT, ET and T indicate lectotype specimen and ex-epitype and ex- type strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; gapdh: partial glyceraldehyde-3-phosphate dehydrogenase gene; chs-1: partial chitin synthase-1 gene; his: partial histone H3 gene; act: partial actin gene; tub2: partial beta-tubulin gene; apn2: partial DNA lyase gene; ApMat: partial Apn2-Mat1-2 intergenic spacer and partial mating type (Mat1-2) gene; sod2: partial manganese superoxide dismutase gene.
Fig. 26.
One of the 100 equally most parsimonious trees obtained from a heuristic search of the combined ITS, gapdh, chs-1, act and tub2 sequence data of the currently accepted species of Colletotrichum. Parsimony and likelihood bootstrap support values ≥ 50 % are indicated at the nodes and branches with Bayesian posterior probabilities above 0.80 given in bold. The tree is rooted with Monilochaetes infuscans CBS 869.96. GenBank accession numbers are listed in Table 6, Table 7. The ex-type strains are in bold. TreeBASE: S21045.
Ascomata solitary or gregarious, globose to subglobose, dark brown to black, ostiole periphysate; ascomatal wall composed of pale to medium brown flattened cells of textura angularis. Hamathecium composed of hyaline, septate paraphyses, branched at the bases, rounded at the tips. Asci 8-spored, unitunicate, cylindrical to subfusoid, short pedicellate, with an inamyloid, refractive ring at the apex. Ascospores uni- to biseriate, aseptate, hyaline, smooth-walled, cylindrical, oval, fusiform or rhomboid, straight or curved, one end ± acute and one end rounded or both ends rounded. Conidiomata acervular, conidiophores and setae formed on cushions of pale to medium brown, roundish to angular cells, but very variable in culture, ranging from sporodochia-like aggregations of conidiophores directly on hyphae to closed conidiomata that open by rupture. Setae may or may not be present, straight, pale to dark brown, sometimes hyaline towards the tip, smooth-walled, verruculose to verrucose, 1–8-septate, base cylindrical, conical or slightly inflated, tip ± rounded to ± acute. Conidiophores hyaline to pale brown, simple or septate, branched or unbranched, smooth-walled, sometimes verruculose. Conidiogenous cells enteroblastic, hyaline to pale brown, smooth-walled, discrete, cylindrical, ellipsoidal, doliiform or ampulliform, collarette usually distinct, periclinal thickening visible to conspicuous, sometimes extending to form new conidiogenous loci (percurrent) or surrounded by a gelatinous coating. Conidia hyaline, smooth-walled, aseptate, cylindrical, clavate, fusiform, sometimes ellipsoidal to ovoid, straight or curved, apex rounded to acute, sometimes with a filiform appendage, base rounded to truncate, sometimes with a prominent hilum. Appressoria single or in small groups, pale to dark brown, with a globose, elliptical, clavate, navicular or irregular outline and an entire, undulate or lobate edge.
Culture characteristics: Colonies on PDA flat, with an entire to irregular margin, grey to dark in centre, aerial mycelium, if present, sparse to cottony, white, buff or pale olivaceous green in colour. Reverse first white, with age turning grey to black, olivaceous green or smoke-grey, concentric rings can be observed. Conidia in mass orange, salmon, pink, white or pale grey. Colonies on SNA flat, with an entire, erose, dentate or undulate margin, aerial mycelium, if present, hyaline, white, honey colour, iron-grey, greenish black or dark olivaceous. Reverse hyaline, honey, pale olivaceous grey to iron-grey. Colonies on OA flat, with an entire to umbonate margin, aerial mycelium, if present, white, buff, rosy-buff, very pale glaucous, hyaline or honey coloured. Reverse buff, rosy-buff, flesh, pale luteous, honey coloured, smoke-grey or olivaceous grey. Conidia in mass salmon, saffron, orange, white or rosy-buff.
Optimal media and cultivation conditions: For morphological examinations of the asexual morphs SNA amended with double autoclaved stems of Anthriscus sylvestris (wild chervil) and autoclaved filter paper placed onto the agar surface and incubated under near-ultraviolet light with a 12 h photoperiod at 20 °C for 10 d proved to be best suited to promote sporulation of most of the species, while for other species, culturing on OA or PDA incubated under the same conditions is more suitable. Plates sometimes need to be incubated for 1–2 mo to allow development of the sexual morph.
Distribution: Worldwide.
Hosts: Occurs on a wide range of plant families.
Disease symptoms: Anthracnose disease symptoms include defined, often sunken necrotic spots on leaves, stems, flowers or fruits. Additionally, crown and stem rots, ripe rot, seedling blights and brown blotch are caused by species of this genus.
Notes: Due to the overlapping morphological characters, species delimitation based on morphology alone is hardly possible in Colletotrichum. Multilocus sequence analyses combined with a polyphasic approach, including the analysis of geographical, ecological and morphological data, is generally suggested for species differentiation within the genus Colletotrichum (Cai et al. 2009). This approach resulted in the differentiation of almost 200 species, most of them belonging to species complexes. Due to simultaneous studies in the genus by different researchers, the sets of loci used for differentiating species vary among the different species complexes. ITS, gapdh, chs-1, act, his3 and tub2 (with some also gs or cal) gene regions have been used for studying species within the Col. acutatum, boninense, dematium, destructivum, gigasporum, orbiculare, spaethianum and truncatum species complexes (Cannon et al., 2012, Damm et al., 2012a, Damm et al., 2012b, Damm et al., 2013, Damm et al., 2014, Liu et al., 2014, Jayawardena et al., 2016b), while gs, cal and sod2 were additionally applied for the species differentiation within the Col. gloeosporioides species complex (Weir et al. 2012) (Table 6). In contrast, Crouch et al. (2009b) and Crouch (2014) applied ITS, sod2, apn2 and Mat1/apn2 (= ApMat), to study the Col. graminicola and Col. caudatum species complexes (Table 7). Silva et al. (2012) and Sharma et al. (2015) emphasised the use of ApMat in Colletotrichum species delimitation because of its high resolution within the Col. gloeosporioides species complex compared to previously used loci. Liu et al., 2015b, Liu et al., 2016 applied different sets of loci and different phylogenetic methods on a large set of closely related Colletotrichum strains/species belonging to this complex and revealed that ApMat should be combined with other loci to achieve satisfactory species delimitation in the Col. gloeosporioides complex.
Because different sets of loci are used in different species complexes and the resolution of species differs depending on both locus and species, there is no agreement among the mycologists on the locus or loci to use for species identification/barcoding. For example, most species in the Col. acutatum complex can be separated by tub2 sequences (Damm et al. 2012b), while species in the Col. gloeosporioides complex can be identified with a combination of ApMat and gs sequences (Liu et al. 2015b). Research to select better genetic markers and the best secondary barcoding gene(s) is still ongoing.
References: Cannon et al. 2012 (species complexes); Crouch et al., 2009b, Crouch, 2014 (phylogeny); Damm et al., 2009, Damm et al., 2012a, Damm et al., 2012b, Damm et al., 2013, Damm et al., 2014, Weir et al., 2012, Liu et al., 2014 (morphology, phylogeny).
Colletotrichum sydowii Damm, sp. nov. MycoBank MB820688. Fig. 27.
Fig. 27.
Colletotrichum sydowii (holotype CBS 135819). A–B. Conidiomata. C. Tip of seta. D. Base of seta. E–F. Conidiophores. G. Tip of seta. H. Base of seta. I–K. Conidiophores. L–P. Appressoria. Q–R. Conidia. A, C–F, Q. from Anthriscus stem. B, G–P, R. from SNA. A–B. DM; C–R. DIC. Scale bars: A applies to A, B = 100 μm; E applies to C–R = 10 μm.
Etymology: The species epithet is derived from Hans Sydow (1879–1946), a German mycologist who described several Colletotrichum species including one on Sambucus, host from which this fungus was isolated.
Sexual morph not observed. Asexual morph on SNA: Vegetative hyphae 1.5–9.5 μm diam, hyaline to pale brown, smooth-walled, septate, branched. Chlamydospores not observed. Conidiomata absent, conidiophores and setae formed directly on hyphae. Setae medium brown, smooth-walled, upper part verrucose, 60–115 μm long, 2–4-septate, base cylindrical, 4–6 μm diam, tip ± acute to ± rounded. Conidiophores hyaline to pale brown, smooth-walled to verrucose, septate, branched, to 50 μm long. Conidiogenous cells hyaline to pale brown, smooth-walled to verrucose, cylindrical to clavate, 13–28 × 4–5 μm, with a gelatinous coating, opening 1–2 μm diam, collarette ≤ 0.5 μm long, periclinal thickening visible. Conidia hyaline, smooth-walled, aseptate, straight, slightly clavate to cylindrical, with one end round and the other truncate, (17–)17.5–19.5(–21) × 5–5.5 μm, mean ± SD = 18.3 ± 0.9 × 5.2 ± 0.2 μm, L/W ratio = 3.5. Appressoria single, medium brown, smooth-walled, subglobose, elliptical or irregular in outline, with a strongly lobate margin, (7.5–)9–14(–17.5) × (5.5–)7–10.5(–12) μm, mean ± SD = 11.4 ± 2.4 × 8.6 ± 1.8 μm, L/W ratio = 1.3, appressoria of strain CBS 132889 shorter, measuring (7.5–)8.5–12.5(–14) × (6.5–)7.5–11(–13) μm, mean ± SD =10.6 ± 1.9 × 9.1 ± 1.8 μm, L/W ratio = 1.2. Asexual morph on Anthriscus stem: Conidiomata, conidiophores and setae formed on pale brown, angular cells, 3.5–8 μm diam. Setae medium brown, verruculose to verrucose, 30–80 μm long, (1–)2–3-septate, base conical to ± inflated, 4.5–7.5 μm diam, tip ± acute to ± rounded. Conidiophores pale brown, smooth-walled, septate, branched, to 20 μm long. Conidiogenous cells pale brown, smooth-walled, cylindrical to doliiform, 6.5–18 × 5–6.5 μm, opening 1.5–2 μm diam, collarette 0.5–1 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, straight, cylindrical, with one end round and the other truncate, (14–)15.5–18.5(–20.5) × 5–6 μm, mean ± SD = 17.0 ± 1.6 × 5.5 ± 0.3 μm, L/W ratio = 3.1, conidia of strain CBS 132889 larger, measuring (15.5–)17–20(–20.5) × (4.5–)5–5.5(–6) μm, mean ± SD = 18.6 ± 1.4 × 5.4 ± 0.3 μm, L/W ratio = 3.5.
Culture characteristics: Colonies on SNA flat with entire margin, hyaline to honey, filter paper and Anthriscus stem partly pale to dark grey, agar medium partly covered with short felty whitish aerial mycelium, reverse similar; growth 27.5–29.5 mm diam in 1 wk (≥ 40 mm diam in 10 d). Colonies on OA flat with entire margin; olivaceous buff to greenish olivaceous, partly covered with short felty whitish aerial mycelium and grey conidiomata, saffron to salmon conidial masses in the centre, reverse primrose, rosy-buff to grey olivaceous, growth 26–28 mm diam in 1 wk (≥ 40 mm in 10 d). Conidia in mass saffron to salmon.
Material examined: Taiwan, from leaves of Sambucus sp., 18 Dec. 2011, P.W. Crous (holotype CBS H-21509, culture ex-type CBS 135819 = CPC 20071); ibid., CBS 132889 = CPC 20070.
Notes: Colletotrichum sydowii is to date only known from Sambucus leaves in Taiwan. The conidia of this species resemble those of several species, e.g. Col. clidemiae, Col. australe and Col. parsonsiae belonging to the Col. gloeosporioides, acutatum and boninense species complexes (Damm et al., 2012a, Damm et al., 2012b, Weir et al., 2012). Based on DNA sequences, Col. sydowii does not belong to any known Colletotrichum species complex; the closest matches in blastn searches of the ex-holotype strain in GenBank with sequences of the different loci resulted in sequences of strains from different species complexes. The ITS sequence is 99 % (1–2 nucleotide difference) identical to those of “Col. gloeosporioides” strain EECC-453 from Ensete ventricosum (GenBank KP942898, from an unpublished study in Ethiopia by Y. Mulugeta et al.) and “Fungal sp.” strain TCPR 106 from a photosynthetic root of Tinospora cordifolia in India (GenBank JX951175, R.N. Kharwar et al., unpubl. data), as well as 93–94 % identical to the ITS sequences of several species of the Col. gigasporum and gloeosporioides complexes and Col. coccodes. The tub2 sequence is 83 % (> 130 nucleotides difference) identical to those of Col. vietnamense strain CBS 125477 (GenBank KF687876), Col. gigasporum strain CBS 109355 (GenBank KF687870), both belonging to the Col. gigasporum complex (Liu et al. 2014), and Col. dracaenophilum isolate DMM 170 (GenBank KJ653227, Macedo & Barreto 2016). The his3 sequence is 90–91 % identical with species from different complexes, including Col. constrictum strain CBS 128503 (GenBank JQ005498, Col. boninense complex, Damm et al. 2012a) and Col. vietnamense strain CBS 125477 (GenBank KF687854, Col. gigasporum complex, Liu et al. 2014) as well as Col. yunnanense strain CBS 132135 (GenBank JX546755, Liu et al. 2014). The chs-1 sequence is 89–91 % identical with e.g. Col. dacrycarpi strain CBS 130241 (GenBank JQ005410, Col. boninense complex, Damm et al. 2012a) and Col. grevilleae strain CBS 132879 (GenBank KC296987, Col. gloeosporioides complex, Liu et al. 2013b). Closest match with the act sequence is Col. magnisporum strain CBS 398.84 with 82 % identity (GenBank KF687803, Col. gigasporum complex, Liu et al. 2014). There is no species with more than 52 % query cover to the gapdh of Col. sydowii.
There is one Colletotrichum species that was previously described from Sambucus, Col. sambuci Syd. 1942, that caused fruit anthracnose of Sa. nigra in Germany. Sydow (1942) regarded Gloeosporium fructigenum f. sambuci Müll.-Thurg. 1922, described from Sa. nigra in Switzerland, as a synonym of Col. sambuci. Conidia of Col. sambuci are cylindrical, elongate ellipsoidal to clavate with one end rounded tapering to the other slightly acute end, measuring 13–20 × 4.5–6 μm. They have similar dimensions as those of Col. sydowii, however it is unlikely that the fungus collected from Sambucus leaves in Taiwan is identical with the fruit anthracnose pathogen of black elderberry in Europe, as the morphological characters apply to many Colletotrichum species and all molecular data suggest a species in the Col. acutatum species complex. Based on ITS sequences, Benduhn et al. (2011) and Michel et al. (2013) identified Col. acutatum (s. lat.) as causal agent of the fruit anthracnose of Sa. nigra in Germany and Switzerland, respectively. As part of the multilocus alignment of the Col. acutatum complex, Col. godetiae was identified from fruits of Sa. nigra in the Netherlands (Damm et al. 2012b). The ITS sequences of “Col. cf. gloeosporioides” strain BBA 67435 (GenBank AJ301931) from Sa. nigra in Germany and of strain BBA 71332 (GenBank AJ301972) also from Sambucus (Nirenberg et al. 2002) are identical with that of strain CBS 862.70; these isolates are probably also Col. godetiae. Conidia of the Col. godetiae strain from the Netherlands (CBS 862.70) measure (8–)14–19(–24) × (4–)4.5–5(–5.5) μm on SNA. The shape of this species can be either fusiform or clavate with only one acute end, depending on the strain (Damm et al. 2012b) and there were no setae observed. Strain BBA 67435 also had conidia pointed only at one end (Nirenberg et al. 2002), which agrees with the shape of Col. sambuci. It is possible that Col. sambuci is an older name of Col. godetiae, however, we cannot confirm this here as we could not locate the type material.
Another species was described from Sambucus in Canada, Vermicularia sambucina (Ellis & Dearness 1897), which however has curved conidia with different dimensions (24 × 3–3.5 μm, Saccardo & Sydow 1899). In contrast, Col. fructicola, a species with considerably shorter conidia belonging to the Col. gloeosporioides complex, was isolated from leaves with anthracnose leaf spot symptoms on Sa. ebulus in Iran (Arzanlou et al. 2015).
Authors: U. Damm, R.S. Jayawardena, L. Cai
Coniella Höhn. Ber. Deutsch. Bot. Ges. 36: 316. 1918. Fig. 28.
Fig. 28.
Coniella spp. A–D. Disease symptoms. A, B.Coniella eucalyptorum on Eucalyptus sp. C.Coniella tibouchinae on Tibouchina granulosa. D.Coniella granati on Punica granatum (pictures taken by M. Mirabolfathy). E–I. Sexual morph of Coniella eucalyptigena (ex-type CBS 139893). E. Ascomata forming on OA. F. Ostiolar área. G, H. Asci. I. Ascospores. J–R. Asexual morphs. J. Conidiomata forming on OA of Coniella diplodiella (ex-epitype CBS 111858). K. Transverse section through a conidioma of Coniella eucalyptorum (ex-type CBS 112640). L, M. Conidiogenous cells giving rise to conidia. L.Coniella diplodiopsis (ex-type CBS 590.84). M.Coniella obovata (CBS 111025). N–R. Conidia. N.Coniella africana (ex-type CBS 114133). O.Coniella diplodiella (ex-epitype CBS 111858). P.Coniella fusiformis (ex-type CBS 141596). Q.Coniella limoniformis (ex-type CBS 111021). R.Coniella obovata (CBS 111025). Scale bars: E = 250 μm, others = 10 μm. Pictures taken from Alvarez et al. (2016).
Synonyms: Schizoparme Shear, Mycologia 15: 120. 1923.
Baeumleria Petr. & Syd., Repert. Spec. Nov. Regni Veg. Beih. 42: 268. 1927.
Pilidiella Petr. & Syd., Repert. Spec. Nov. Regni Veg. Beih. 42: 462. 1927.
Anthasthoopa Subram. & K. Ramakr., Proc. Indian Acad. Sci., Sect. B 43: 173. 1956.
Cyclodomella Mathur et al., Sydowia 13: 144. 1959.
Embolidium Bat., Brotéria, N.S. 33(3–4): 194. 1964 non Sacc. 1978.
Classification: Sordariomycetes, Sordariomycetidae, Diaporthales, Schizoparmaceae.
Type species: Coniella fragariae (Oudem.) B. Sutton (syn. Coniella pulchella Höhn.). Neotype and ex-neotype culture: CBS H-10697, CBS 172.49 = CPC 3930.
DNA barcodes (genus): LSU, rpb2.
DNA barcodes (species): ITS, rpb2, tef1. Table 8.
Table 8.
DNA barcodes of accepted Coniella spp.
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; NFCCI: National Fungal Culture Collection of India, Agharkar Research Institute, Pune, India; VPRI: Victorian Plant Pathogen Herbarium, Bundoora, Australia. T, ET and NT indicate ex-type, ex-epitype and ex-neotype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; rpb2: partial RNA polymerase II second largest subunit gene; tef1: partial translation elongation factor 1-alpha gene.
Ascomata brown to black, collapsed collabent, erumpent, becoming superficial, globose, papillate, with central periphysate ostiole. Paraphyses lacking. Asci clavate to subcylindrical, with distinct apical ring, free at maturity. Ascospores ellipsoid, aseptate, hyaline, at times becoming pale brown at maturity, smooth, with or without mucoid caps. Conidiomata pycnidial, immersed to semi-immersed, unilocular, glabrous, ostiolate; ostiole central, circular or oval, often situated in a conical or rostrate neck; conidiomatal wall brown to dark brown or black, composed of thin, pale brown textura angularis on exterior, and hyaline, thin-walled, textura prismatica in the inner layers except at base, which has a convex, pulvinate tissue of hyaline textura angularis giving rise to conidiophores or conidiogenous cells. Conidiophores mostly reduced to conidiogenous cells, occasionally septate and branched at base, invested in mucus. Conidiogenous cells discrete, cylindrical, subcylindrical, obclavate or lageniform, hyaline, smooth-walled, proliferating percurrently, with visible periclinal thickening. Conidia ellipsoid, fusiform, globose, napiform or naviculate with a truncate base and an obtuse to apiculate apex, unicellular, thin- or thick-walled, smooth, hyaline, pale yellowish, pale yellowish brown, or olivaceous brown to brown, sometimes with a longitudinal germ-slit, with or without a mucoid appendage extending from apex to base on one side; basal hila with or without a short tubular basal appendage. Spermatophores formed in same conidioma, hyaline, smooth, 1-septate with several apical conidiogenous cells, or reduced to conidiogenous cells. Spermatogenous cells hyaline, smooth, lageniform to subcylindrical, with visible periclinal thickening. Spermatia hyaline, smooth, rod-shaped with rounded ends (adapted from Crous et al. 2014a).
Culture characteristics: On PDA produces white aerial mycelium with or without black conidiomata. On OA frequently produces white aerial mycelium with black conidiomata, but sometimes with luteous to orange zones.
Optimal media and cultivation conditions: On 2 % MEA, PDA and OA, at 25 °C under continuous near-ultraviolet light to promote sporulation.
Distribution: Worldwide.
Hosts: Wide variety of hosts, e.g. Eucalyptus (Myrtaceae), Fragaria (Rosaceae), Hibiscus (Malvaceae), Psidium (Myrtaceae), Punica (Lythraceae), Terminalia (Combretaceae) and Vitis (Vitaceae).
Disease symptoms: Foliar, fruit, stem and root lesions, white rot and crown rot.
Notes: In the most recent revision of the members of Schizoparmaceae, Pilidiella and its sexual morph Schizoparme were synonymised under Coniella because the type species of the three genera clustered in a single well-supported clade in a phylogenetic analysis based on four different loci (LSU, ITS, rpb2 and tef1) (Alvarez et al. 2016). Coniella and Pilidiella were initially distinguished by von Arx (1981) based on their conidial pigmentation, being hyaline to pale brown in Pilidiella and dark brown in Coniella. However, Alvarez et al. (2016) demonstrated that conidial colour evolved multiple times throughout the clade representing Coniella, and therefore rejected it as a character for generic delimitation in Schizoparmaceae. Sutton (1980) and Nag Raj (1993) also considered Coniella and Pilidiella synonymous since both genera presented identical conidiomata, conidiogenesis and orientation of conidiophores. However, Castlebury et al. (2002) demonstrated a separation of both genera in a phylogenetic study based on LSU sequences. This was further supported by van Niekerk et al. (2004b) based on their LSU, ITS and tef1 sequence data. Based on these molecular studies, together with the difference in conidial pigmentation reported by von Arx, 1981, Wijayawardene et al., 2016 regarded Coniella and Pilidiella as two separate genera in a recent study of dematiaceous coelomycetes. By adding more loci and expanding the number of isolates studied, Alvarez et al. (2016) resolved the conflict that lasted a few decades regarding the classification of these genera.
References: Van Niekerk et al., 2004b, Crous et al., 2014a, Alvarez et al., 2016 (morphology and phylogeny).
Coniella duckerae H.Y. Yip, Trans. Brit. Mycol. Soc. 89: 587. 1987. Fig. 29.
Fig. 29.
Coniella duckerae (ex-type CBS 142045). A. Conidiomata forming on OA. B, C. Conidiogenous cells giving rise to conidia. D. Conidia. Scale bars: A = 350 μm, others = 10 μm.
Description and illustration: Yip (1987).
Material examined: Australia, Victoria, Wilson's Promontory, Five Mile Road, on rhizosphere of Lepidospermum concavum, unknown collector and date (holotype DAR 55703, isotype VPRI 13689, culture ex-type VPRI 13689 = CBS 142045).
Notes: Coniella duckerae was excluded from the study of Alvarez et al. (2016), as no ex-type culture was available. However, the original culture was recently revived, and DNA barcodes could thus be generated for inclusion in this study.
Coniella hibisci (B. Sutton) Crous, comb. nov. MycoBank MB820811. Fig. 30.
Fig. 30.
Coniella hibisci (ex-epitype CBS 109757). A. Conidiomata forming on OA. B, C. Conidiogenous cells giving rise to conidia. D. Conidia. Scale bars: A = 350 μm, others = 10 μm.
Basionym: Coniella musaiaensis var. hibisci B. Sutton, The Coelomycetes (Kew): 420. 1980.
Plant pathogenic. Conidiomata separate, immersed or superficial, globose to depressed, initially appearing hyaline, becoming olivaceous to black with age, with plate-like structures, up to 350 μm diam; ostiole central, 40–80 μm diam; conidiomatal wall consisting of 2–4 layers of medium brown textura angularis. Conidiophores densely aggregated, slightly thicker, subulate, simple, frequently branched above, reduced to conidiogenous cells, or with 1–2 supporting cells, 25–35 × 3–5 μm. Conidiogenous cells simple, hyaline, smooth, tapering, 8–15 × 2.5–3 μm, 1.5–2 μm wide at apex, surrounded by a gelatinous coating, with visible periclinal thickening. Conidia hyaline to pale yellowish brown with age, fusoid to ellipsoidal, inequilateral, apex acutely rounded, widest at middle tapering to slightly truncate base, smooth-walled, mono- to multiguttulate, germ slits absent, (10–)11–13(–15) × (3–)3.5–4(–5) μm (L/W = 3.4), with a mucoid appendage alongside conidium.
Culture characteristics: Colonies on MEA surface dirty white, with prolific black conidial masses spreading from centre. On OA and PDA surface dirty white with profuse black conidiomata and sparse aerial mycelium.
Material examined: Africa, from Hibiscus sp., unknown date, A.R. Rossman (epitype designated here BPI 748426, MBT376042, culture ex-epitype CBS 109757 = ARS 3534). Nigeria, on leaves of Hibiscus esculentus, 25 Jul. 1967, Arny (holotype IMI 129200).
Notes: The morphology of the present African ex-epitype strain from Hibiscus sp. (CBS 109757 = ARS 3534) compares well with that of the holotype of Coniella musaiaensis var. hibisci, which was described from Hibiscus esculentus collected in Nigeria. A new combination is therefore proposed, elevating it to species rank. Presently there are still no cultures available of Con. musaiaensis, and further collections from Bauhinia reticulata (Sierra Leone) need to be made to resolve its phylogeny. Coniella hibisci is also morphologically similar to Con. javanica (on Hibiscus sabdariffa, Indonesia), although they are phylogenetically divergent (Alvarez et al. 2016).
Authors: Y. Marin-Felix, J. Edwards, A.Y. Rossman & P.W. Crous
Curvularia Boedijn, Bull. Jard. Bot. Buitenzorg, 3 Sér. 13: 123. 1933. Fig. 31.
Fig. 31.
Curvularia spp. A–F. Conidiophores and conidia. A.Curvularia geniculata. B.Curvularia neergaardii (CBS 277.91). C.Curvularia portulacea (ex-isotype BRIP 14541). D.Curvularia tropicalis (ex-isotype BRIP 14834). E.Curvularia hominis (ex-type CBS 136985). F.Curvularia muehlenbeckiae (ex-type CBS 144.63). G–I. Conidia. G.Curvularia crustacea (ex-epitype BRIP 13524). H.Curvularia nicotiae (ex-isotype BRIP 11983). I.Curvularia pseudolunata (ex-type CBS 136987). J. Germinating conidium of Curvularia neergaardii (CBS 277.91). K, L. Microconidiation. K.Curvularia americana (ex-type CBS 136983). L.Curvularia chlamydospora (ex-type CBS 136984). M. Chlamydospores of Curvularia pseudolunata (ex-type CBS 136987). Scale bars: A = 50 μm; the others = 10 μm. Picture A taken from Samson et al. (2010); C, D, G, H from Tan et al. (2014); E, F, I, K–M from Madrid et al. (2014).
Synonyms: Malustela Bat. & J.A. Lima, Publ. Inst. Micol. Recife 263: 5. 1960.
Curvusporium Corbetta as “Curvosporium”, Riso 12: 28, 30. 1963.
Pseudocochliobolus Tsuda, et al., Mycologia 69: 1117. 1978.
Classification: Dothideomycetes, Pleosporomycetidae, Pleosporales, Pleosporaceae.
Type species: Curvularia lunata (Wakker) Boedijn. Ex-neotype culture: CBS 730.96.
DNA barcodes (genus): LSU, ITS.
DNA barcodes (species): ITS, gapdh, tef1. Table 9. Fig. 32.
Table 9.
DNA barcodes of accepted Curvularia spp.
ATCC: American Type Culture Collection, Virginia, USA; BRIP: Queensland Plant Pathology Herbarium, Brisbane, Australia; Bp-Zj: Isolate housed in Biotechnology Institute, Zhejiang University, Hangzhou, China; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; DAOM: Plant Research Institute, Department of Agriculture (Mycology), Ottawa, Canada; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Ria, Thailand, UTHSC: Fungus Testing Laboratory, Department of Pathology at the University of Texas Health Science Center, San Antonio, Texas, USA. ET, IsoT, IsoLT, PT,SynT and T indicate ex-epitype, ex-isotype, ex-isolectotype, ex-paratype, ex-syntype and ex-type strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; gapdh: partial glyceraldehyde-3-phosphate dehydrogenase gene; tef1: partial translation elongation factor 1-alpha gene.
Fig. 32.
RAxML phylogram obtained from the combined ITS (504 bp), gapdh (461 bp) and tef1 (893 bp), sequences of all the accepted species of Curvularia. The tree was rooted to Bipolaris panici-miliacei CBS 199.29 and Bipolaris peregianensis DAOM 221998. The novel species described in this study are shown in bold. RAxML bootstrap support (BS) values above 70 % and Bayesian posterior probability scores ≥ 0.95 are shown at the nodes. GenBank accession numbers are indicated in Table 9. ET, IsoT, IsoLT, PT,SynT and T indicate ex-epitype, ex-isotype, ex-isolectotype, ex-paratype, ex-syntype and ex-type strains, respectively. TreeBASE: S20877.
Ascomata pseudothecial, mostly globose to ellipsoidal, sometimes flask-shaped or flattened on hard substrata, brown or black, immersed, erumpent, partially embedded or superficial, free or developing on a basal columnar or flat stroma, smooth or covered with vegetative filaments; ostiole central, papillate or with a sub-conical, conical, paraboloid or cylindrical neck; ascomatal wall comprising pseudoparenchymatous cells of equal thickness or slightly thickened at apex of the ascoma. Hamathecium comprising septate, filiform, branched pseudoparaphyses. Asci bitunicate, clavate, cylindrical-clavate or broadly fusoid, straight or slightly curved, thin-walled, fissitunicate, often becoming more or less distended prior to dehiscence, short pedicellate, rounded at the apex. Ascospores multiseriate, filiform or flagelliform, hyaline or sometimes pale yellow or pale brown at maturity, septate, helically coiled within ascus, degree of ascospore coiling moderate to very strongly coiled, often with a mucilaginous sheath. Conidiophores straight to flexuous, often geniculate, multiseptate, usually simple, sometimes branched, smooth to verruculose, macronematous, mononematous, sometimes nodose, cylindrical. Conidiogenous nodes cylindrical, integrated, terminal and intercalary, proliferating sympodially, cicatrised. Conidia solitary, often curved, acropleurogenous, broadly fusoid, elliptical, obovoid or obpyriform, mostly smooth, sometimes verruculose, echinulate or tuberculate, 3 or more distoseptate, with or without an unequally swollen cell which is more pigmented than the other cells, septa sometimes accentuated with a dark band in some or all the cells, germinating mainly from one or both polar cells with the basal germ tube growing semiaxially, hilum in a slightly protruding truncate basal section of the conidial wall and often visible as two dark lenticular spots in optical section arranged close together with a small obscure narrow separating canal between them or distinctly protuberant, first conidial septum median or submedian, second septum often delimiting the basal cell of the mature conidium, third septum then distal. Microconidiation not common, producing conidia 1–2-celled, pale brown, globose to subglobose (adapted from Sivanesan 1987).
Culture characteristics: Colonies on PDA white or pale grey when young, orange to brown or different shades of grey (mainly dark olivaceous grey) when mature, fluffy, cottony, raised or convex with papillate surface, margin lobate, undulate, entire or sometimes rhizoid.
Optimal media and cultivation conditions: Sterilised Zea mays leaves placed on 1.5 % WA or slide cultures of half-strength PDA under near-ultraviolet light (12 h light, 12 h dark) to induce sporulation of the asexual morph, while for the sexual morph Sach's agar with sterilised rice or wheat straw at 25 °C is used.
Distribution: Worldwide.
Hosts: Wide host range, occurring as pathogens or saprobes. Mainly found on members of the Poaceae, being pathogens of grass and staple crops, including rice, maize, wheat and sorghum. This genus also occurs on genera belonging to Actinidiaceae, Aizoaceae, Caricaceae, Convolvulaceae, Fabaceae, Iridaceae, Lamiaceae, Lythraceae, Oleaceae, Polygonaceae and Rubiaceae.
Disease symptoms: Leaf spots, leaf blight, melting out, root rot, foot rot, among others.
Notes: Species delimitation in Curvularia based on morphology only is difficult due to the morphological complexity within this genus, as also observed in Bipolaris. Furthermore, the differentiation of both genera based on morphology alone is sometimes complicated (see Bipolaris notes for morphological differences between Bipolaris and Curvularia). Therefore, molecular data are essential for an accurate identification of species within these genera, ITS, gapdh and tef1 being the loci selected for this purpose (Manamgoda et al., 2014, Manamgoda et al., 2015).
Curvularia is a rich genus in host range and geographic distribution compared to Bipolaris. Apart from phytopathogenic species, this genus comprises species that are pathogens of humans and other animals, causing respiratory tract, cutaneous, cerebral and corneal infections, mainly in immunocompromised patients (Carter & Boudreaux 2004). Some species can be found in association with both humans and plants, such as Cu. hawaiiensis, Cu. lunata and Cu. spicifera (Manamgoda et al. 2015).
References: Sivanesan 1987 (morphology and pathogenicity); Manamgoda et al. 2011 (pathogenicity), Manamgoda et al. 2015 (morphology, pathogenicity and phylogeny).
Curvularia pisi Y. Marín & Crous, sp. nov. MycoBank MB820814. Fig. 33.
Fig. 33.
Curvularia pisi (ex-type CBS 190.48). A, B. Conidiophores and conidia. C–G. Conidia. Scale bars: A–C = 10 μm; D–G = 5 μm.
Etymology: Name refers to the host genus from which it was isolated, Pisum.
Hyphae hyaline to pale brown, branched, septate, thin-walled, 1.5–5 μm. Conidiophores arising in groups, septate, straight or flexuous, geniculate at upper part, verruculose, tapering towards apex, sometimes branched, cells walls thicker than those of vegetative hyphae, mononematous, semi- to macronematous, pale brown to brown, paler towards apex, not swollen at the base, (35–)50–210 × 2.5–5 μm. Conidiogenous cells verruculose, terminal or intercalary, proliferating sympodially, pale brown to brown, subcylindrical to swollen, (2.5–)5–15.5 × 3–7.5 μm. Conidia verruculose, curved, rarely straight, middle cells unequally enlarged, reniform, rarely ellipsoidal, brown, with apical and basal cells paler than middle cells being subhyaline to pale brown, (2–)3-distoseptate, 16–35 × 9–15.5 μm; hila slightly protuberant, flat, darkened, slightly thickened, 1.5–4 μm. Chlamydospores, microconidiation and sexual morph not observed.
Culture characteristics: Colonies on PDA reaching 90 mm diam within 1 wk, with sparse to moderate aerial mycelium giving a slightly cottony appearance, margin lobate; surface apricot to chestnut; reverse umber to chestnut.
Material examined: Canada, Ontario, Renfrew, on Pisum sativum seeds, 15 Feb. 1943, J.W. Groves (holotype CBS H-11405, culture ex-type CBS 190.48).
Notes: Curvularia pisi is closely related to Cu. muehlenbeckiae and Cu. hominis. Morphologically, these species are similar but Cu. pisi produces shorter conidiophores. Moreover, Cu. muehlenbeckiae produces smaller conidia than Cu. pisi, and Cu. hominis is characterised by 3–4-distoseptate conidia while the conidia in the other two species are 3-distoseptate.
Curvularia pisi is known to occur on Pisum sativum, which is also host to two other species of Curvularia, Cu. inaequalis and Cu. spicifera. Curvularia spicifera produces a sexual morph, while no sexual morph has been observed in the other two species. Moreover, Cu. spicifera differs from Cu. pisi in having smooth-walled conidia. Curvularia inaequalis can be distinguished from Cu. pisi by its longer conidia, which are predominantly 4-distoseptate.
Curvularia soli Y. Marín & Crous, sp. nov. MycoBank MB820816. Fig. 34.
Fig. 34.
Curvularia soli (ex-type CBS 222.96). A–C. Conidiophores and conidia. D–G. Conidia. Scale bars: A = 10 μm; others = 5 μm.
Etymology: Named after its ecology, occurring in soil, “soli”.
Hyphae subhyaline to pale brown, branched, septate, thin-walled, 2.5–5.5 μm. Conidiophores arising in groups, septate, straight or flexuous, geniculate at upper part, smooth to verruculose, unbranched, cells walls thicker than those of vegetative hyphae, mononematous, semi- to macronematous, pale brown to brown, slightly paler towards apex, not swollen at the base, (65–)90–270(–390) × 2.5–5(–6) μm. Conidiogenous cells smooth-walled to finely verruculose, terminal or intercalary, proliferating sympodially, pale brown to brown, subcylindrical to swollen, 4–13 × 2.5–5 μm. Conidia verruculose, curved, rarely straight, middle cells unequally enlarged, reniform, rarely ellipsoidal, pale brown to brown, apical and basal cells paler than middle cells being subhyaline to pale brown, 3–4(–5)-distoseptate, (13.5–)18–28 × 7.5–11 μm; hila protuberant, flat, darkened, thickened, 1.3–3.5 μm. Chlamydospores, microconidiation and sexual morph not observed.
Culture characteristics: Colonies on PDA reaching 75–79 mm diam after 1 wk, velvety to slightly powdery; surface and reverse grey olivaceous to olivaceous black.
Material examined: Papua New Guinea, Madang, Jais Aben, isolated from soil along coral reef coast, Nov. 1995, collected by A. Aptroot, isol. by A. van Iperen (holotype CBS H-23116, culture ex-type CBS 222.96).
Notes: Curvularia soli is closely related to Cu. asianensis, Cu. geniculata and Cu. senegalensis. All three species are characterised by conidia that are predominantly 4-distoseptate. Curvularia geniculata is the only species that produces a sexual morph and has the longest conidia among these taxa (26–48 μm). Curvularia asiatica can be distinguished from Cu. soli by its much longer conidiophores [(75–)100–700(–708) μm] and shorter conidia [(11–)15–23(–23.5) μm]. Curvularia senegalensis is characterised by having shorter conidiophores (up to 150 μm) and wider conidia (10–14 μm) than Cu. soli.
Authors: Y. Marin-Felix, P.W. Crous & Y.P. Tan
Monilinia Honey, Mycologia 20: 153. 1928. Fig. 35.
Fig. 35.
Monilinia spp. A–C. Disease symptoms. A, B.Monilinia fructigena on Malus sp. (A, CBS 348.72) and on Sorbus aucuperiae mummified fruit (B, CBS H-14553). C.Monilinia laxa (CBS H-14556) leaf spot on Prunus padus. D. Sporodochia in vivo of Monilinia fructigena (CBS 348.72). E, F. Conidiophores. E.Monilinia fructigena (CBS 348.72). F.Monilinia fructicola (CBS 101512). G, H. Apothecia. G.Monilinia johnsonii (CBS H-005908) on Crategus sp. mummified fruit. H.Monilinia johnsonii (CBS H-005908) stipitate apothecia. I–K. Asci of Monilinia johnsonii (CBS H-14554). K. Tip of an ascus showing a blue reaction with Meltzer's solution. L. Ascospores of Monilinia johnsonii (CBS H-14554). M, N. Macroconidia. M.Monilinia fructicola (CBS 101512). N.Monilinia fructigena (CBS 348.72). O. Microconidia of Monilinia fructicola (CBS 101512). Scale bars: A–C, G, H = 1 mm; D = 100 μm; E, F, I = 20 μm; J–O = 10 μm.
Synonym: Monilia Bonord., Handb. Mykol.: 7. 1851.
Classification: Leotiomycetes, Leotiomycetidae, Helotiales, Sclerotiniaceae.
Type species: Monilinia fructicola (G. Winter) Honey. Holotype: BPI 1109031.
DNA barcode (genus): ITS.
DNA barcode (species): tef1. Table 10. Fig. 36.
Table 10.
DNA barcodes of accepted Monilinia spp.
| Species | Isolates1 | Genbank accession number2 |
References | |
|---|---|---|---|---|
| ITS | tef1 | |||
| Moniliniaamelanchieris | ATCC 58538 | Z73769 | – | Holst-Jensen et al. (1997) |
| M. aucupariae | ARO 885.2 | Z73771 | – | Holst-Jensen et al. (1997) |
| M. azaleae | ATCC 58539 | AB182266 | – | Takahashi et al. (2005) |
| M. baccarum | CBS 388.93 | KX982694 | LT632532 | Present study |
| M. cassiopes | ARO 1459.S | Z73776 | – | Holst-Jensen et al. (1997) |
| M. fructicola | CBS 329.35 | KX982695 | LT632533 | Present study |
| M. fructigena | CBS 348.72 | KX982697 | LT632535 | Present study |
| M. gaylussaciae | ATCC 64508 | Z73782 | – | Holst-Jensen et al. (1997) |
| M. jezoensis | 4222T * | AB182265 | – | Takahashi et al. (2005) |
| M. johnsonii | ATCC 58542 | Z73783 | – | Holst-Jensen et al. (1997) |
| M. kusanoi | NBRC 9725 | 00972502A | Harada et al. (2004) | |
| M. laxa | CBS 132.21 | KX982699 | LT632537 | Present study |
| M. linhartiana | CBS 150.22 | KX982701 | LT632539 | Present study |
| M. megalospora | ARO 619.2 | Z73788 | – | Holst-Jensen et al. (1997) |
| M. mali | 2769* | AB125619 | – | Harada et al. (2004) |
| M. mespili | CBS 139.23 | KX982702 | LT632540 | Present study |
| M. mumeicola | 3231 01-01* | AB125613 | – | Harada et al. (2004) |
| M. oxycocci | ARO 1087.P | Z73789 | – | Holst-Jensen et al. (1997) |
| M. padi | ARO 923.K | Z73791 | – | Holst-Jensen et al. (1997) |
| M. polycodii | ATCC 58546 | Z73792 | – | Holst-Jensen et al. (1997) |
| M. polystroma | CBS102688T | KX982704 | LT632542 | Present study |
| M. seaveri | CBS 170.24 | KX982705 | – | Present study |
| M. ssiori | HHUF 19771T | AB220062 | – | Harada et al. (2005) |
| M. urnula | ARO 476.1 | Z73794 | – | Holst-Jensen et al. (1997) |
| M. vaccinii-corymbosi | CBS 172.24 | KX982706 | LT632543 | Present study |
| M. yunnanensis | KY-1 | HQ908788 | – | Hu et al. (2011) |
ARO: Ascomycete Systematics Research Group, University of Oslo, Norway; ATCC: American Type Culture Collection, Virginia, USA; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; HHUF: Hirosaki University, Japan; KY: Strain code as stated in GenBank, * Hirosaki University Culture Collection, Japan. T indicates ex-type strain. A Accession number corresponding to the NITE Biological Resource Center, Japan.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; tef1: partial translation elongation factor 1-alpha gene.
Fig. 36.
RaxML phylogram obtained from the combined ITS (428 bp) and tef1 (420 bp) sequences of Monilinia spp. currently known by DNA data. Maximum parsimony and RaxML bootstrap support (BS) values above 70 % are shown at the nodes. Numbers between parentheses correspond to GenBank accession numbers for ITS and tef1 sequences, respectively. T indicates ex-type strain. TreeBASE: S20877.
Ascomata apothecial, cup- or funnel-shaped, long stipitate, pale brown, formed solitary or in groups from pseudosclerotia in aborted or mummified fruits and debris partially or completely buried in soil; stipe cylindrical, flexuous, pale brown, often darker near the base; hymenium comprising filiform, septate, unbranched and hyaline paraphyses. Asci unitunicate, inoperculate, with amyloid apical apparatus, cylindrical to clavate, flattened or rounded at the apex, thin-walled, 8-spored. Ascospores ellipsoid, often with tapered ends, 1-celled, hyaline, sometimes covered with a gelatinous sheet. Conidiophores single or aggregated forming sporodochia, straight or flexuous, hyaline to subhyaline, branched, thin-walled, septate. Macroconidia blastic-acropetal, oval, lemon-shaped or broadly ellipsoidal, rarely doliiform, hyaline to subhyaline, thin-and smooth-walled, 1-celled, sometimes presenting distinct axial connections (disjunctors), formed in chains, simple or dichotomously branched; microconidia (spermatia) sometimes present in old cultures, globose to pyriform, hyaline, smooth- and thin-walled, borne on lageniform, often asymmetric phialides. Arthric conidia occasionally formed, ovoid to ellipsoid, smooth- and thin-walled.
Culture characteristics: Colonies on PDA white, yellow-grey, brown-grey or olive-grey, often zonate or forming concentric rings, felty to velvety, flat or concave, margin entire or lobed giving a rosette-like appearance, brown to black stromata can be present in old cultures.
Optimal media and cultivation conditions: PDA and WA, incubated under near-ultraviolet light (12 h light, 12 h dark) at 22–25 °C to determine growth rates, colour and shape of the colony, and induce sporulation of the asexual morph. The sexual morph is not formed under in vitro culture conditions but can be induced by inoculation on natural substrata and incubated several months partially buried in sterilised soil.
Distribution: Worldwide.
Hosts: Mostly found as crop pathogens or causing post-harvest losses on stone fruits, most commonly on members of Rosaceae, predominantly on Cydonia spp., Malus spp., Prunus spp. and Pyrus spp., but have been reported in at least 11 other genera on this family, linked to some kind of host specialisation. Other known hosts include members of Actinidiaceae, Berberidaceae, Betulaceae, Ebernaceae, Ericaceae, Euphorbiaceae, Moraceae, Myricaceae, Myrtaceae, Solanaceae and Vitaceae.
Disease symptoms: Leaf spots, blossom and twig blight, twig and stem canker, fruit rot.
Notes: Generic identification in vivo or in vitro is easy considering the characteristic monilioid hyphae and sexual-morphs. Monilinia is morphologically similar and closely related to the genus Sclerotinia, from which it can be differentiated by the absence of asexual reproduction and formation of true sclerotia in Sclerotinia. However, species identification in Monilinia is rather difficult by means of morphology alone. A combination of cultural features, physiology and host range is often necessary, including macro and micromorphology, growth rates, conidial dimension and characteristics of the germ tube during sporulation. Other employed techniques include AFLP and RFLP (Gril et al., 2010, Vasić et al., 2016), specific PCR amplification for the three major brown rot pathogens M. fructigena, M. fructicola and M. laxa (Cote et al., 2004, Gell et al., 2007) and amplification of specific introns for rapid identification of M. fructicola (Fulton & Brown 1997). A species delimitation based on molecular phylogeny is currently lacking and no ex-type material is known to exist for most taxa. However, several reference ITS and tef1 sequences are available from a set of curated isolates in Q-bank (http://www.q-bank.eu/Fungi/).
A proposal to protect the generic name Monilinia over Monilia has been recently published based on the complex and often conflicting taxonomic history of the latter name (Johnston et al. 2014). Following this proposal, two new combinations are proposed below.
References: Batra, 1988, Batra, 1991, Honey, 1928, Honey, 1936, Van Leeuwen et al., 2002 (morphology and pathogenicity); van Leeuwen 2000 (morphology, pathogenicity and epidemiology); OEPP/EPPO, 2009, Martini and Mari, 2014 (morphology, pathogenicity and biology).
Monilinia mumeicola (Y. Harada et al.) Sandoval-Denis & Crous, comb. nov. MycoBank MB819176.
Basionym: Monilia mumeicola [as ‘mumecola’] Y. Harada et al., J. Gen. Plant Pathol. 70: 305. 2004.
Notes: This species is only known from its asexual morph. It was described as a pathogen on Japanese apricot (Prunus mume) in Japan (Harada et al. 2004), and later reported causing brown rot of Prunus armeniaca (Yin et al. 2014) and Prunus salicina (Yin et al. 2015) in China. Our phylogeny (Fig. 36) included sequences of two authentic isolates of Monilia numeicola and supported its location in the genus Monilinia, being closely related to the common agents of brown rot M. fructicola, M. fructigena and M. laxa.
Monilinia yunnanensis (M.J. Hu & C.X. Luo) Sandoval-Denis & Crous, comb. nov. MycoBank MB819177.
Basionym: Monilia yunnanensis M.J. Hu & C.X. Luo, PloS ONE 6: 11. 2011.
Notes: This taxon was described as a pathogen of peach (Prunus persica) in China and has subsequently been isolated as the most prevalent pathogen of apple and pear in the southern, northern and western regions of that country (Zhu et al. 2016). Its phylogenetic placement in Monilinia was supported in our phylogeny (Fig. 36) based on sequences from two authentic isolates, showing that it forms a clade basal to the main cluster grouping the most economically relevant species of the genus.
Authors: M. Sandoval-Denis & P.W. Crous
Neofabraea H.S. Jacks., Rep. Oregon Exp. Sta. 1911–1912: 187. 1913. Fig. 37.
Fig. 37.
Neofabraea malicorticis (ex-neotype CBS 122030). A. Colony on MEA. B. Colony on OA. C. Conidiomata on inoculated apple. D. Conidial mass on apple peel. E. Conidiogenous cells from sporodochium on OA. F. Conidiogenous cells giving rise to macroconidia. G. Microconidia on OA. H. Macroconidia from OA. I. Macroconidia from inoculated apple. J, K. Intermediate conidia between macro- and microconidia. Scale bars: 10 μm, I applies to H, I. Pictures taken from Chen et al. (2016).
Classification: Leotiomycetes, Leotiomycetidae, Helotiales, Dermateaceae.
Type species: Neofabraea malicorticis H.S. Jacks. Neotype and ex-neotype culture: CBS H-22219, CBS 122030 = OSC 100036.
DNA barcodes (genus): LSU.
DNA barcodes (species): ITS, tub2, rpb2. Table 11. Fig. 38.
Table 11.
DNA barcodes of accepted Neofabraea spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | ||
|---|---|---|---|---|---|
| ITS | rpb2 | tub2 | |||
| Neofabraea actinidiae | CBS 121403T | KR859079 | KR859319 | KR859285 | Chen et al. (2016) |
| Na. brasiliensis | CNPUV499T | KR107002 | – | KR107011 | Crous et al. (2015e) |
| Na. inaequalis | CBS 326.75T | KR859081 | KR859321 | KR859287 | Chen et al. (2016) |
| Na. kienholzii | CBS 126461T | KR859082 | KR859322 | KR859288 | Chen et al. (2016) |
| Na. krawtzewii | CBS 102867 | KR859084 | KR859324 | AF281459 | De Jong et al., 2001, Chen et al., 2016 |
| Na. malicorticis | CBS 122030NT | KR859086 | KR859326 | KR859291 | Chen et al. (2016) |
| Na. perennans | CBS 102869 | KR859087 | KR859327 | AF281473 | De Jong et al., 2001, Chen et al., 2016 |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CNPUV: Centro Nacional de Pesquisa de Uva e Vinho, Bento Gonçalves, RS, Brazil. T and NT indicate ex-type and ex-neotype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; rpb2: partial RNA polymerase II second largest subunit gene; tub2: partial β-tubulin gene.
Fig. 38.
RAxML phylogram obtained from the ITS (564 bp) sequences of Neofabraea spp. and related genera. Maximum parsimony and RAxML bootstrap support (BS) values above 70 % are shown in the nodes. The new genus introduced in this study is shown in bold. The tree was rooted to Sclerotinia sclerotiorum CBS 499.50. Numbers between parentheses correspond to GenBank accession numbers. T indicates ex-type strain. TreeBASE: S20877.
Ascomata apothecial, erumpent from bark, sessile to short-stalked, solitary or in clusters on a basal stroma; disc often not well-delimited, circular, elliptical, or irregular and merged, greyish, flesh-coloured to pale reddish or brownish, drying darker, 0.5–2.0 mm diam. Paraphyses numerous, filiform, septate, obtuse, simple or branched, hyaline, smooth-walled, apical cells mostly slightly swollen. Asci inoperculate, cylindrical-clavate, apex rounded or truncate-rounded, attenuated into a stalk of variable length, crozier present, 8-spored; apical apparatus with a well-developed apical thickening, Lugol's + or −, blue in herbarium material, Melzer's + or −. Ascospores inequilateral, elongated ellipsoid, ends rounded, straight or curved, aseptate, thin-walled, smooth, hyaline, with granular contents or small oil droplets; later septate, sometimes germinating or forming conidia from minute openings or phialides. Conidiomata erumpent from bark, stromatic, acervular, plane to pulvinate. Conidiophores simple or branched, smooth, hyaline, acrogenous or acropleurogenous. Conidiogenous cells discrete or integrated, determinate, phialidic, cylindrical to narrowly ampulliform, giving rise to macro- and/or microconidia. Macroconidia cylindrical-fusiform, allantoid to ellipsoid, straight or curved, rounded or somewhat pointed at apex, rounded or attenuated and with an indistinct, barely or non-protruding scar at base, smooth, aseptate, hyaline, and thin-walled when liberated, mostly filled with numerous oil droplets; later becoming septate and brown. Microconidia present or absent, cylindrical, rounded at apex, narrowly truncate at base, aseptate, hyaline, thin- and smooth-walled, with minute granular contents (adapted from Chen et al. 2016).
Culture characteristics: Colonies on OA white, cottony. Colonies on PDA orange or red, slimy.
Optimal media and cultivation conditions: OA at 25 °C under near-ultraviolet light (12 h light, 12 h dark); OA supplemented with sterile nettle stems (Anthriscus sylvestris) or direct inoculation into apple fruit (Malus domestica) can be used to induce sporulation.
Distribution: Worldwide.
Hosts: Pathogens or harmless saprobes of apples and pears, but also of several other hosts such as species of Prunus and Populus.
Disease symptoms: Anthracnose canker, perennial canker and bull's-eye rot.
Notes: Neofabraea was introduced by Jackson (1913) to accommodate the sexual morph Gloeosporium malicorticis. Subsequently, Nannfeldt (1932) synonymised this genus with Pezicula. However, Verkley (1999) revalidated it and observed that species of Neofabraea are more explicitly pathogenic than those of Pezicula. Neofabraea further differs from Pezicula in that Neofabraea produces ascomata with excipular tissues less differentiated and macroconidia more strongly curved with the basal scar less distinct than in Pezicula. Moreover, Pezicula comprises species that have two types of conidiogenesis: conidiogenous cells are determinate and phialidic, or indeterminate and proliferating percurrently, while Neofabraea spp. only produces phialidic conidiogenous cells (Chen et al. 2016). Recently, Chen et al. (2016) carried out a revision of the genus by performing a phylogenetic study based on LSU, ITS, tub2 and rpb2 sequences of Neofabraea, Pezicula and related genera. Consequently, the genus Phlyctema was re-established to accommodate Neofabraea alba, which is the main pathogen causing bull's eye rot in continental Europe. Moreover, the new genera Parafabraea and Pseudofabraea were introduced in order to accommodate Neofabraea eucalypti and Neofabraea citricarpa, respectively (Chen et al. 2016).
References: Verkley 1999 (morphology and pathogenicity), Wang et al. 2015 (morphology and key of Neofabraea spp.), Chen et al. 2016 (phylogeny).
Verkleyomyces Y. Marín & Crous, gen. nov. MycoBank MB820818.
Etymology: Named after Gerard J.M. Verkley, in recognition for his contributions to the understanding of Neofabraea and related genera.
Mycelium hyaline to pale brown, branched, septate. Ascomata apothecial, partly immersed, erumpent, sessile, solitary, sometimes gregarious; medullary excipulum weakly developed, composed of hyaline textura prismatica; ectal excipulum composed of brown to olivaceous brown textura prismatica at the base, and pale brown textura intricata towards the margin; subhymenium hyaline, composed of interwoven hyphae. Paraphyses cylindrical, slender, septate, apex rounded, hyaline, flexuous, numerous. Asci unitunicate, clavate to cylindrical-clavate, base truncate, short pedicellate, with an apical apparatus stained blue or purplish blue in Melzer's reagent, 8-spored, ascospores discharging through apical pore. Ascospores fusoid to ellipsoid, hyaline, ends rounded or somewhat pointed, straight or slightly curved, thin-walled, guttulate or eguttulate, initially aseptate, or later becoming 1-septate. Conidiomata acervular or cupulate, semi-immersed, dark, separate, formed of olivaceous brown textura intricata, dehiscence by irregular fissures, sometimes by a central ostiole. Conidiophores simple, hyaline, smooth, thin-walled, septate at the base, unbranched, discrete, or rarely integrated beneath the aged conidiogenous cell. Conidiogenous cells enteroblastic, phialidic, cylindrical, hyaline, smooth, thin-walled, sometimes with proliferation, periclinal thickening present. Conidia cylindrical, straight, apex obtuse, base abruptly tapered to a distinct scar, hyaline, smooth, thin-walled, aseptate, eguttulate to biguttulate.
Culture characteristics: Colonies on PDA glaucous to sky-grey, with irregular white margin; reverse olivaceous black.
Type species: Verkleyomyces illicii (X. Sun et al.) Y. Marín & Crous. Holotype and ex-type culture: HMAS244704, ASH-3-6-2-5b.
Notes: Verkleyomyces is introduced to accommodate Neofabraea illicii, the most recently published species of Neofabraea (Wang et al. 2015). In the phylogenetic analysis based on ITS sequences (Fig. 38), this species was located in a clade separate from the rest of the species belonging to Neofabraea. Verkleyomyces is mainly differentiated by its endophytic habit. Morphologically both genera are comparable, but Verkleyomyces produces 1-septate ascospores and aseptate conidia, while Neofabraea is characterised by aseptate ascospores and predominately septate conidia. Parafabraea, which is more closely related, also produces aseptate conidia, but this can be differentiated from Verkleyomyces by the production of aseptate ascospores. Other similar genera are Pezicula and Dermea, but these can easily be distinguished by the production of ascospores that are initially hyaline, and then become coloured or contain coloured oil droplets.
Verkleyomyces illicii (X. Sun et al.) Y. Marín & Crous, comb. nov. MycoBank MB820819.
Basionym: Neofabraea illicii X. Sun et al., Mycoscience 56: 334. 2015.
Description and illustration: Wang et al. (2015).
Note: Verkleyomyces illicii is an endophytic fungus isolated from Illicium verum, cultivated in a plantation in southern China.
Authors: Y. Marin-Felix & P.W. Crous
Neofusicoccum Crous et al., Stud. Mycol. 55: 247. 2006. Fig. 39.
Fig. 39.
Neofusicoccum spp. A–D. Disease symptoms. A. Leaf blight on Protea sp. B. Canker on Vitis vinifera. C, D. Cankers on Eucalyptus sp. E–J. Sexual morphs. E, F. Ascomata. E.Neofusicoccum parvum (ex-type ATCC 58191). F.Neofusicoccum luteum (ex-type ATCC 58193). G, H. Asci. G.Neofusicoccum luteum (ex-type ATCC 58193). H.Neofusicoccum australe (ex-type CMW 6837). I. Detail of ascus apex of Neofusicoccum parvum (ex-type ATCC 58191). J. Ascospores of Neofusicoccum parvum (ex-type ATCC 58191). K–S. Asexual morph. K. Conidiomata on pine needles in culture of Neofusicoccum australe (CMW 6837). L, M. Conidiogenous cells. L.Neofusicoccum mediterraneum (ex-type CBS 121718). M.Neofusicoccum parvum (ex-type ATCC 58191). N–P. Conidia. N.Neofusicoccum arbuti (ex-type CBS 116131). O.Neofusicoccum australe (ex-type CMW 6837). P.Neofusicoccum vitifusiforme (ex-type CBS 110887). Q. Coloured, 1- and 2-septate conidia of Neofusicoccum parvum (ex-type ATCC 58191). R. Spermatogenous cells of Neofusicoccum mediterraneum (ex-type CBS 121718). S. Spermatia of Neofusicoccum mediterraneum (ex-type CBS 121718). Scale bar: E–G = 50 μm; H, J, L–P, R = 10 μm; I, Q, S = 5 μm; K = 1 mm. Pictures taken from Phillips et al. (2013).
Classification: Dothideomycetes, Incertae sedis, Botryosphaeriales, Botryosphaeriaceae.
Type species: Neofusicoccum parvum (Pennycook & Samuels) Crous et al. Holotype and ex-type culture: PDD 45438 (Herbarium of Plant Diseases Division), ATCC 58191 = CBS 138823 = PDDCC 8003 = ICMP 8003 = CMW 9081.
DNA barcodes (genus): LSU, rpb2.
DNA barcodes (species): ITS, tef1, tub2, rpb2. Table 12. Fig. 40.
Table 12.
DNA barcodes of accepted Neofusicoccum spp.
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CERC: China Eucalypt Research Centre (CERC), Chinese Academy of Forestry (CAF), China; CMM: Culture collection of Phytopathogenic Fungi “Prof. Maria Menezes”, Universidade Federal Rural de Pernambuco, Recife, Brazil; CMW: Tree Pathology Co-operative Program, Forestry and Agricultural Biotechnology Institute, University of Pretoria, South Africa; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Ria, Thailand; MUCC: Murdoch University, Perth, Western Australia. T, IsoT and PT indicate ex-type, ex-isotype and ex-paratype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; rpb2: partial RNA polymerase II second largest subunit gene; tef1: partial translation elongation factor 1-alpha gene; tub2: partial β-tubulin gene.
Fig. 40.
RAxML phylogram obtained from the combined ITS (541 bp), tef1 (302 bp), rpb2 (594 bp) and tub2 (463 bp) sequences of Neofusicoccum spp. The tree was rooted to Botryosphaeria dothidea CBS 100564. The novel species described in this study are shown in bold. RAxML bootstrap support (BS) values above 70 % and Bayesian posterior probability scores ≥ 0.95 are shown at the nodes. GenBank accession numbers were listed in Berraf-Tebbal et al., 2014, Chen et al., 2015b, and Yang et al. (2017). T and NT indicate ex-type and ex-neotype strains, respectively. TreeBASE: S20877.
Ascomata forming botryose clusters, each comprising many ascomata, erumpent through the bark, globose, with a short, conical papilla, dark brown to black, smooth, thick-walled; ascomatal wall composed of dark brown thick-walled cells of textura angularis, lined with thin-walled hyaline cells of textura angularis. Asci clavate, 8-spored, bitunicate. Ascospores broadly ellipsoidal to fusoid, hyaline, smooth, aseptate, occasionally becoming 1-septate. Conidiomata globose and non-papillate, entire locule lined with conidiogenous cells. Conidiogenous cells holoblastic, hyaline, subcylindrical, proliferating percurrently to form 1–2 annellations, or proliferating at the same level to form periclinal thickenings. Conidia ellipsoidal with apex round and base flat, unicellular, hyaline, old conidia becoming 1–2-septate hyaline, or light brown with the middle cell darker than the terminal cells. Dichomera synasexual morph: Conidia subglobose to obpyriform, brown, apex obtuse, base truncate, 1–3 transverse septa, 1–2 longitudinal septa, and 1–2 oblique septa.
Culture characteristics: Colonies initially white to buff turning olivaceous grey becoming black with age, moderately dense, appressed mycelial mat with irregular very dense aerial aggregations, some conidioma covered by mycelium, immersed-erumpent, conidia and spermatia present. Reverse white to olivaceous black. Reaching 90 mm diam on half strength MEA in 3–4 d.
Optimal media and cultivation conditions: Half strength MEA at 25–30 °C.
Distribution: Worldwide.
Hosts: Plurivorous, mainly pathogenic on Anacardiaceae, Cupressaceae, Ebenaceae, Fagaceae, Juglandaceae, Lauraceae, Moraceae, Myrtaceae, Oleaceae, Pinaceae, Proteaceae, Rosaceae, Rutaceae, Vitaceae, families belonging to Lamiales and various other host plants.
Disease symptoms: Fruit rot, wood canker, leaf spots.
Notes: Neofusicoccum was introduced by Crous et al. (2006b) to accommodate species morphologically similar to, but phylogenetically divergent from Botryosphaeria (= Fusicoccum). To separate Neofusicoccum from Botryosphaeria based solely on morphology can be difficult due to similar morphological characteristics. Therefore, molecular data are required to achieve accurate identification. One morphological difference between both genera is the presence of a Dichomera synasexual morph in Neofusicoccum. However, this synasexual morph is not produced by all Neofusicoccum species, nor even all isolates of any given species. Moreover, dichomera-like conidia were reported in some isolates of Bot. dothidea (Barber et al., 2005, Phillips et al., 2005). Other morphological differences are the absence of paraphyses in the conidiomata of Neofusicoccum spp., while these have been seen in most of the currently accepted Botryosphaeria species, and the conidial L/W ratios being less than 4 in Neofusicoccum. Furthermore, the conidia of Neofusicoccum are more ellipsoidal than the fusiform ones of Fusicoccum s. str.
Species in Neofusicoccum are morphologically similar and hard to differentiate from one another. Neofusicoccum species are currently defined on the basis of conidial dimensions and pigmentation, pigment production in culture media and ITS sequence data. Taxa in some of the species complexes are defined exclusively on DNA sequence data (ITS, often together with tef1, tub2 and rpb2. In some cases, multigene sequence data are essential for species identification.
References: Crous et al., 2006b, Berraf-Tebbal et al., 2014, Yang et al., 2017 (morphology and phylogeny); Pavlic et al. 2009a (phylogeny); Pavlic et al. 2009b (morphology, pathogenicity and phylogeny), Phillips et al. 2013 (morphology, phylogeny and dichotomous key).
Neofusicoccum italicum Dissanayake & K.D. Hyde, sp. nov. MycoBank MB820799, Facesoffungi number FOF02963. Fig. 41.
Fig. 41.
Neofusicoccum italicum (ex-type MFLUCC 15-0900). A. Conidiomata on host substrate. B, C. Cross section of conidiomata. D, E. Immature and mature conidia attached to conidiogenous cells. F. Mature conidia. Scale bars: B, C = 100 μm. D–F = 20 μm.
Etymology: Based on the country where the type specimen was collected, Italy.
Sexual morph not observed. Conidiomata 0.5–1.5 × 1.5–2 mm, black, scattered, uniloculate, globose; conidiomatal wall composed of dark brown textura angularis, becoming hyaline towards conidiogenous region. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 9–16.5 × 2.5–3.5 μm, lining inner wall of pycnidium, holoblastic, cylindrical to subobpyriform, hyaline, discrete, determinate, occasionally indeterminate and proliferating percurrently with indistinct annellations. Conidia 13–18.5 × 3.5–6 μm, obovoid, fusiform, base truncate, apex obtuse to subobtuse, hyaline, guttulate, non-septate, older conidia may become brownish and septate before germination. Dichomera synasexual morph not reported.
Culture characteristics: Colonies white with abundant aerial mycelium reaching 90 mm diam within 1 wk on PDA at 28 °C. Aerial mycelium becoming smoke-grey to olivaceous grey at the surface and dull green to brown-vinaceous at the reverse after 2 wk in the dark at 28 °C.
Materials examined: Italy, on a dead branch of Vitis vinifera, 22 Nov. 2014, E. Camporesi (holotype MFLU 16-2872, culture ex-type MFLUCC 15-0900). New Zealand, on Malus × domestica, unknown date, H.J. Boesewinkel, CBS 719.85.
Notes: According to the phylogenetic analysis Neofusicoccum italicum clustered close to Nm. algeriense, which has larger conidia (17.6 × 5.6 μm) than those of Nm. italicum (15.8 × 5.2 μm). Both species are pathogens of Vitis vinifera, but Nm. algeriense is restricted to this host while Nm. italicum was also isolated from Malus × domestica. Other species of Neofusicoccum associated with Vitis vinifera are Nm. australe, Nm. luteum, Nm. mediterraneum, Nm. parvum, Nm. stellenboschiana, Nm. viticlavatum and Nm. vitifusiforme (Van Niekerk et al., 2004a, Phillips et al., 2013, Yang et al., 2017). Neofusicoccum and other related genera belonging to Botryosphaeriaceae are widely distributed pathogens of grapevines that cause bud mortality, dieback, brown streaking inside the wood, internal necrotic lesions and in some cases bunch rot (Phillips et al. 2013).
Neofusicoccum pistaciicola Crous, sp. nov. MycoBank MB820820. Fig. 42.
Fig. 42.
Neofusicoccum pistaciicola (ex-type CBS 113089). A. Conidiomata forming on PNA. B, C. Conidiomata cells giving rise to conidia. D. Conidia. Scale bars: 10 μm.
Etymology: Named after the host genus from which it was collected, Pistacia.
Sexual morph not observed. Conidiomata stromatic, solitary, globose, up to 300 μm diam; conidiomatal wall 6–8 cell layers thick, of brown textura angularis, becoming hyaline toward inner region. Conidiophores 0–2-septate, branched, hyaline, smooth, subcylindrical, 15–25 × 4–5 μm. Conidiogenous cells holoblastic, hyaline, smooth, subcylindrical, proliferating percurrently, 12–17 × 2.5–3.5 μm. Conidia hyaline, smooth, thin-walled, granular, aseptate, subcylindrical to fusoid-ellipsoid, apex subobtuse, base truncate, 1.5–2.5 μm, straight to irregularly curved, (15–)18–24(–27) × (4–)4.5(–5) μm. Spermatia or Dichomera synasexual morph not observed.
Culture characteristics: Colonies on MEA reaching 90 mm diam within 1 wk, with fluffy moderate aerial mycelium; surface pale mouse-grey, reverse mouse-grey to dark mouse-grey.
Material examined: USA, California, Glenn County, on Pistacia vera, 12 Apr. 2002, T.J. Michailides (holotype CBS H-23108, culture ex-type CBS 113089).
Notes: Neofusicoccum pistaciicola is morphologically similar to Nm. hellenicum, which Chen et al. (2015b) recently described from Pistacia vera in the USA. However, compared with Nm. hellenicum, Nm. pistaciicola possesses smaller conidiomata and narrower conidia. The same features are used to distinguish it from Nm. pistaciarum, which is the closest phylogenetic species, and also a pathogen of Pistacia vera. Other species of Neofusicoccum associated to this host are Nm. australe, Nm. mediterraneum, Nm. nonquaesitum, Nm. parvum and Nm. pistaciae (Inderbitzin et al., 2010, Phillips et al., 2013, Yang et al., 2017).
Neofusicoccum pruni Crous, sp. nov. MycoBank MB820821. Fig. 43.
Fig. 43.
Neofusicoccum prunii (ex-type CBS 121112). A. Conidiomata forming on PNA. B, C. Conidiomata cells giving rise to conidia. D. Conidia. Scale bars: 10 μm.
Etymology: Named after the host genus from which it was collected, Prunus.
Sexual morph not observed. Conidiomata stromatic, solitary, globose to obpyriform, up to 300 μm diam; conidiomata wall 6–10 cell layers thick, of brown textura angularis, becoming hyaline toward inner region. Conidiophores 0–1-septate, hyaline, subcylindrical, 10–20 × 2.5–4 μm. Conidiogenous cells holoblastic, hyaline, subcylindrical, 10–15 × 2.5–3.5 μm, proliferating percurrently with numerous proliferations, or proliferating at the same level (phialidic) with minute periclinal thickening. Conidia hyaline, granular, aseptate, fusoid to ellipsoid, widest in the middle or upper third with an obtuse apex and flattened, subtruncate base, (18–)20–23(–25) × (6.5–)7–7.5(–8.5) μm. Spermatia or Dichomera synasexual morph not observed.
Culture characteristics: Colonies on MEA reaching 90 mm diam within 1 wk, with fluffy moderate aerial mycelium; surface and reverse greenish black.
Material examined: South Africa, Limpopo, Mookgopong, from branches of Prunus salicina, Aug. 2004, U. Damm (holotype CBS H-23109, culture ex-type CBS 121112 = CPC 5912).
Notes: Neofusicoccum vitifusiforme was initially described from Vitis vinifera in South Africa by van Niekerk et al. (2004a). Damm et al. (2007) was the first to report this fungus as a pathogen from Prunus salicina in South Africa, although their phylogenetic tree showed this isolate (CBS 121112) to cluster basal to the grapevine strains based on ITS and tef1 sequence data. A recent study by Yang et al. (2017), which incorporated all Neofusicoccum isolates available in the CBS culture collection, as well as additional genes (rpb2 and tub2) showed the Prunus isolate to represent a distinct species. This isolate (formerly sterile) has subsequently been induced to sporulate, and is therefore named in the present study.
Authors: Y. Marin-Felix, E. Camporesi, A. Dissanayake, K.D. Hyde & P.W. Crous
Pilidium Kunze, Mykol. Hefte 2: 92. 1823. Fig. 44.
Fig. 44.
Pilidium species. A, E, G.Pilidium eucalyptorum (CBS 140662). B, F, H.Pilidium pseudoconcavum (CBS 136433). C, D, I.Pilidium leucospermi (holotype PREM 59602). A, B. Conidiomata on OA and SNA, respectively. C. Vertical section of conidioma. D. Peridium. E, F. Conidiogenous cells. G–I. Conidia. Scale bars: C = 50 μm; D–I = 10 μm. Pictures C, D, I modified from Marincowitz et al. (2008a).
Synonyms: Sclerotiopsis Speg., Anal. Soc. Cient. Argent. 13: 14. 1882.
Hainesia Ellis & Sacc., in Saccardo, Syll. fung. (Abellini) 3: 698. 1884.
Discohainesia Nannf., Nova Acta Regiae Soc. Sci. Upsal., Ser. 4 8: 88. 1932.
Classification: Leotiomycetes, Leotiomycetidae, Helotiales, Chaetomellaceae.
Type species: Pilidium acerinum (Alb. & Schwein.) Kunze. Iconotype in Kunze & Schmidt (1817), table 2, fig. 5. Epitype and ex-epitype culture: BPI 843555, CBS 736.68.
DNA barcode (genus): LSU.
DNA barcode (species): ITS. Table 13. Fig. 45.
Table 13.
DNA barcodes of accepted Pilidium spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | |
|---|---|---|---|---|
| ITS | LSU | |||
| Pilidiumacerinum | CBS 736.68ET | AY487091 | AY487092 | Rossman et al. (2004) |
| Pi. lythri | CBS 114293 | AY487094 | AY487095 | Rossman et al. (2004) |
| Pi. eucalyptorum | CBS 140662T | KT950854 | KT950868 | Crous et al. (2015e) |
| Pi. pseudoconcavum | CBS 136433T | KF777184 | KF777236 | Crous et al. (2013b) |
| Pi. septatum | BCC 79016T | KY922832 | KY922833 | Present study |
BCC: BIOTEC Culture Collection, National Center for Genetic Engineering and Biotechnology (BIOTEC), Khlong Luang, Pathumthani, Thailand; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. T and ET indicate ex-type and ex-epitype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: partial 28S large subunit RNA gene.
Fig. 45.
Maximum likelihood (ML) tree based on partial sequences of LSU (792 bp) and ITS (477 bp) regions from reference and ex-type strains of Pilidium species. Bootstrap support values and posterior probabilities above 70 % and 0.95, respectively are shown at the nodes. Chaetomella raphigera and Chaetomella oblonga (Chaetomellaceae, Helotiales) were used as outgroup taxa. Numbers within parentheses correspond to GenBank accession numbers of LSU and ITS sequences, respectively.T and ET indicate ex-type and ex-epitype strains, respectively. TreeBASE: S20877.
Ascomata apothecial, flat to funnel-shaped, short stipitate, white, pale brown to amber in the basal portion, wall pseudoparenchymatous (plectenchymatous). Paraphyses narrow, simple or branched, aseptate. Asci unitunicate, cylindrical, clavate, rounded or truncate at the apex, deliquescent. Ascospores ellipsoidal, somewhat enlarged at one side, straight to slightly curved, aseptate, smooth-walled. Conidiomata pycnidial or sporodochial; pycnidia globose, subglobose, obpyriform or oblong, sessile, pale brown when young, dark brown to black at maturity, superficial, solitary or gregarious, uniloculate, smooth; conidiomatal wall with two regions: outer region dark brown, inner region hyaline; opening by a stellate slit, rupturing irregularly, or lacking. Conidiophores hyaline, smooth, branched, cylindrical or filiform. Conidiogenous cells enteroblastic, phialidic, acropleurogenous, hyaline, smooth. Conidia mostly non-septate, hyaline, smooth, fusiform to falcate or cymbiform, with ends slightly pointed, straight to curved. Sporodochia globose becoming cupulate, discoid, with irregularly wavy margin, slimy, pale luteous, superficial, solitary, stalk pale brown near base, becoming dark brown at apex. Conidiophores hyaline, smooth, branched, cylindrical or filiform. Conidiogenous cells enteroblastic, phialidic, acropleurogenous, determinate, integrated, filiform or subcylindrical, hyaline, smooth, with minute collarette. Conidia aseptate, hyaline, smooth, fusiform to falcate or cymbiform to allantoid, with acute ends, straight to curved.
Culture characteristics: Colonies on PDA surface and reverse white to cinnamon, buff, honey, sepia or isabelline, slimy with aerial mycelium absent or sparse, flat, granulose due to production of fruiting bodies; margin smooth and lobate.
Optimal media and cultivation conditions: PDA, OA and MEA incubated at 25 °C for 1 wk at 25 °C under alternating fluorescent (12 h) and near ultraviolet (12 h) light are suitable to determine cultural characteristics and induce sporulation of the asexual morph. The sexual morph is not formed in vitro, and is relatively uncommon and inconspicuous.
Distribution: Worldwide.
Hosts: Species of this genus are mainly found on different hosts of Anacardiaceae, Hippocastanaceae, Myrtaceae and Rosaceae, and also in several other families such as Betulaceae, Ebenaceae, Fabaceae, Geraniaceae, Oleaceae, Paeoniaceae, Pinaceae, Polygonaceae, Salicaceae, Sapindaceae, Saxifragaceae and Vitaceae.
Disease symptoms: Leaf spots, root lesions and tan-brown rot of fruits.
Notes: Species of Pilidium are commonly found as plant-associated fungi or isolated from soil (Sutton 1980), and they are known to produce two kinds of conidiomata. Pilidium lythri (formerly known as Pi. concavum) and Pi. pseudoconcavum form sporodochia in culture. Although, the former species also produces the pycnidial morph, both species can be distinguished based on conidial shape (fusiform vs. cymbiform), sporodochial size (300–1000 μm diam vs. up to 300 μm diam) and DNA sequences (Crous et al. 2013b). Both Pi. acerinum and Pi. eucalyptorum produce brown pycnidia in vitro and they are closely related (Fig. 45). However, they differ in pycnidial size (200–1000 μm diam vs. up to 300 μm diam), conidiophore shape (cylindrical vs. filiform) and in the production of guttulate conidia, which are absent in Pi. acerinum and present in Pi. eucalyptorum (Rossman et al., 2004, Crous et al., 2015e).
Discohainesia oenotherae and Hainesia lythri were considered the sexual and synasexual morphs of Pi. lythri (Rossman et al. 2004). However, after the one fungus = one name initiative the generic name Pilidium was proposed for conservation over Hainesia and Discohainesia (Johnston et al. 2014).
References: Sutton, 1980, Shear and Dodge, 1921, Palm, 1991 (morphology); Sutton & Gibson 1977 (morphology and pathogenicity); Rossman et al. 2004 (morphology, pathogenicity and ecology).
Pilidium septatum Giraldo & Crous, sp. nov. MycoBank MB820871. Fig. 46
Fig. 46.
Pilidium septatum (ex-type BCC 79016). A. Conidiomata on OA. B, C. Longitudinal sections of pycnidia. D. Details of ostiolar region. E. Details of the outer and inner pycnidial walls. F, G. Conidiophores and conidiogenous cells. H. Conidia. Scale bars: B–H = 10 μm.
Etymology: Refers to the presence of septate conidia.
Conidiomata pycnidial, superficial, solitary or gregarious, brown to black, smooth, uniloculate, subglobose to obpyriform, 97–260 × 127–230 μm; outer conidiomatal wall 11–27 μm thick, with textura angularis, formed by thick-walled, brown cells; inner conidiomatal wall 13–20 μm thick, with textura angularis or globulosa, formed by 4–5 layers of thick-walled, hyaline cells. Conidiophores branched, cylindrical, septate, hyaline, smooth, up to 24 μm long, 1.5–2 μm diam. Conidiogenous cells acropleurogenous, monophialidic, cylindrical, slightly curved, smooth, hyaline, delineating the inner part from the pycnidium, 7–11 × 1.5–2 μm. Conidia 1-septate, hyaline, falcate with ends slightly pointed, thin- and smooth-walled, (8.1–)9–11(–12.5) × (1–)1.5(–2) μm.
Culture characteristics: Colonies on OA and PDA reaching 30–40 mm in 2 wk. Colonies flat, granulose due to production of pycnidia, with scarce aerial mycelium, surface honey to isabelline.
Materials examined: Thailand, Nakhon Nayok province, Mueang Nakhon Nayok district, Wang Takhrai waterfall, N14.330023° E101.307168°, 64 m above sea level, from soil, 22 Jul. 2015, A. Giraldo (holotype metabolically inactive, culture ex-type BCC 79016); Nan province, Bo Kluea district, N19.14833333° E101.1566667, from soil, 8 Aug. 2015, A. Giraldo (BCC 79037).
Notes: Presently the genus includes only species with aseptate conidia, and thus Pi. septatum, with septate conidia, expands the generic concept of Pilidium. In addition to the phylogenetic relationship revealed through the analysis of LSU and ITS regions (Fig. 45), morphological characteristics such as the morphology of the pycnidia, the production of acropleurogenous conidiogenous cells and conidial shape, support the inclusion of this species within the genus.
Authors: A. Giraldo, J. Luangsa-ard & P.W. Crous
Pleiochaeta (Sacc.) S. Hughes, Mycol. Pap. 36: 39. 1951. Fig. 47, Fig. 48.
Fig. 47.
Pleiochaeta carotae (ex-type CBS 142644) A. Conidiophores with conidia. B. Conidiophores and conidiogenous cells. C–P. Conidia. Scale bars: 10 μm
Fig. 48.
Pleiochaeta setosa (ex-epitype CBS 496.63, CBS 502.80). A–D. Conidiophores with conidia (ex-epitype CBS 496.63). E, F. Conidiogenous cells (ex-epitype CBS 496.63). G–L. Conidia (ex-epitype CBS 496.63). M–O. Chlamydospores (CBS 502.80). Scale bars: 10 μm.
Synonym: Ceratophorum subgen. Pleiochaeta Sacc., Syll. fung. (Abellini) 11: 622. 1895.
Classification: Dothideomycetes, Pleosporomycetidae, Pleosporales, Dothidotthiaceae.
Type species: Pleiochaeta setosa (Kirchner) S. Hughes. Epitype and ex-epitype culture designated here: CBS H-23058, CBS 496.63 = MUCL 8091).
DNA barcode (genus): LSU.
DNA barcode (species): ITS. Table 14. Fig. 49, Fig. 50.
Table 14.
DNA barcodes of accepted Pleiochaeta spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | |
|---|---|---|---|---|
| ITS | LSU | |||
| Pleiochaetacarotae | CBS 142644T | KY905669 | KY905663 | Present study |
| Plei. ghindensis | CBS 552.92 | EU167561 | EU167561 | Simon et al. (2009) |
| Plei. setosa | CBS 496.63ET | EU167563 | EU167563 | Simon et al. (2009) |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. T and ET indicate ex-type and ex-epitype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: partial 28S large subunit RNA gene.
Fig. 49.
RAxML phylogram obtained from LSU (883 bp) sequences of Dothideomycetes. RAxML bootstrap support (BS) values above 70 % and Bayesian posterior probability scores ≥ 0.95 are shown at the nodes. The tree was rooted to Botryosphaeria dothidea CBS 115476, Neofusicoccum parvum CBS 124491 and Saccharata proteae CBS 115206. Numbers between parentheses correspond to GenBank accession numbers. T, ET, NT and PT indicate ex-type, ex-epitype, ex-neotype and ex-paratype, respectively. TreeBASE: S20877.
Fig. 50.
Phylogenetic tree resulting from a Bayesian analysis of the combined LSU and ITS sequences alignment of Pleiochaeta species. Bayesian posterior probabilities >0.95 are indicated at the nodes. The tree was rooted to Thyrostroma compactum and Thyrostroma cornicola. Numbers between parentheses correspond to GenBank accession numbers of ITS and LSU, respectively. T and ET indicate ex-type and ex-epitype strains, respectively. *, ITS and LSU sequences. TreeBASE: S20877.
Sexual morph unknown. Conidiophores macronematous, mononematous or grouped in fascicles, simple, erect, straight to flexuous, or geniculate, hyaline to pale olivaceous, smooth. Conidiogenous cells mono- and polyblastic, integrated, terminal and intercalary, cylindrical. Conidia solitary, dry, subcylindrical to fusoid, mostly curved, narrowed to obtuse at the apex, truncate at the base, pale to dark brown, smooth, multiseptate; apical cell bears several long, hyaline, subulate appendages which are sometimes branched. Chlamydospores present or absent, brown to dark brown in chains or in groups.
Culture characteristics: Colonies on PDA grey to olivaceous black with aerial mycelium white, cottony, margin fimbriate, effuse; reverse black.
Optimal media and cultivation conditions: MEA, OA, PDA or SNA with sterilised twigs, incubated at 25 °C. Not all strains sporulate well in culture.
Distribution: Worldwide.
Hosts: Mainly pathogens of legumes, with one species reported from carrots.
Disease symptoms: Brown leaf spots, lesions are circular and zonate. It also can attack stems, pods and roots, and destroy whole plants.
Notes: Pleiochaeta was established by Hughes (1951) to accommodate two species previously included in Ceratophorum, namely Plei. setosa and Plei. albizziae. Currently this genus comprises six species, including pathogens and saprobes. Pleiochaeta setosa, the generic type, is the most important species from a phytopathogenic point of view, causing serious damage in Lupinus spp. and other legumes members of Fabaceae. Sequences available to date for the genus are scant. After the analysis of LSU and ITS sequences of the isolates studied with members of Pleosporales (Dothideomycetes), we support the phylogenetic position of Plei. setosa and Plei. ghindensis in the Dothidotthiaceae. Furthermore, our results allow us to describe a new species from South Africa, Plei. carotae, causing a disease on carrot leaves. Cultures of Plei. albiziae, Plei. amazonensis, Plei. cassiae and Plei. stellaris were not available for this study, and their phylogenetic position remains unknown. Further studies with additional molecular data of isolates from different origins and substrates, as well as pathogenicity tests, need to be conducted.
References: Hughes 1951 (taxonomy and morphology); Pirozynski 1974 (morphology and distribution); Bateman 1997 (pathogenicity); Yang & Sweetingham 2002 (morphology and pathogenicity).
Pleiochaeta carotae Hern.-Restr., van der Linde & Crous, sp. nov. MycoBank MB820795. Fig. 47.
Etymology: Named after the host genus from which it was isolated, Daucus carota.
Mycelium partly immersed, partly superficial, composed of branched, septate, hyaline to dark brown, smooth, 3–9 μm wide, hyphae. Conidiophores macronematous, mononematous, usually unbranched, flexuous, frequently geniculate, hyaline or pale olivaceous, smooth. Conidiogenous cells mono- and polyblastic, integrated, terminal and intercalary, sympodial, cylindrical to geniculate. Conidia solitary, dry, subcylindrical ellipsoid to fusoid, mostly curved, narrowed at apex, truncate at the base, at first colourless becoming orange-brown to olivaceous brown, smooth, 92–137 × 16–22 μm, 6–10-septate, usually constricted at the septa; basal cell conical, truncate, subhyaline to pale brown, 8–11 μm wide; apical cell obtuse, arising 2–3 hyaline appendages, with one appendage arising apically which are usually branched 2–4 times, and another two laterally on the sides which are usually branched 1–2(–3) times, appendages 70–114 μm long, 4.5–7 μm wide at the point of origin and pointed at their apices. Chlamydospores not observed.
Culture characteristics: Colonies reaching 40–55 mm diam after 1 wk at 25 °C on OA, PDA and MEA olivaceous black, cottony, with white aerial mycelium in the centre, exudate hyaline, margin fimbriate, effuse, colourless; reverse black.
Material examined: South Africa, Gauteng, Pretoria, on carrot leaf, Mar. 2015, M. Truter (holotype CBS H-23057, culture ex-type CPC 27452 = CBS 142644).
Notes: This is the first species of Pleiochaeta described from carrots, a non-legume host plant. Conidia of this species resemble those of Plei. ghindensis, having branched apical appendages, usually more than twice branched. In Plei. ghindensis conidiogenous cells are monoblastic, terminal and cylindrical with percurrent proliferations. However, in Plei. carotae, conidiogenous cells are mono- and polyblastic, terminal and intercalary and geniculate with sympodial proliferations. Furthermore, conidia in Plei. carotae are larger (92–137 μm vs. 85–115 μm in Plei. ghindensis) and with a larger number of septa (6–10 vs. 6–7 in Plei. ghindensis). Finally, the basal conidial cells are usually paler than the other cells (in Plei. ghindensis conidia are concolourous).
Pleiochaeta setosa (Kirchn.) S. Hughes. Mycol. Pap. 36: 34. 1951. Fig. 48, Fig. 51.
Fig. 51.
Reproduction of the original drawings by Kirchner (1892) illustrating Ceratophorum setosum (original numbers are maintained to indicate the different structures). A. fig. 1. Symptoms in Cytisus capitatus. B. fig. 2. Young conidia. C. fig. 3. Conidiophores and conidia. D, E. figs 4, 5. Conidia. F. fig. 6. Germinating conidia.
Basionym: Ceratophorum setosum Kirchn. Z. Pflanzenkrankh. Pflanzenschutz 2: 324. 1892.
Synonyms: Pestalotia lupini Sorauer, Z. Pflanzenkrankh. Pflanzenschutz 8: 269. 1898.
Mastigosporium lupini (Sorauer) Cavara, Riv. Patol. Veg. 14: 13. 1924.
Mycelium partly immersed, partly superficial, composed of branched, septate, hyaline to brown, smooth, 4–7.5 μm wide, hyphae. Conidiophores macronematous, mononematous, usually unbranched, flexuous, frequently geniculate, hyaline or pale olivaceous, smooth, 34–138 × 5–11 μm. Conidiogenous cells mono-, usually polyblastic, integrated, terminal and intercalary, sympodial, cylindrical, geniculate, hyaline to pale olivaceous, 25–68 × 8–11.5 μm. Conidia solitary, dry, subcylindrical to fusoid, mostly curved, narrowed at the apex, truncate at the base, colourless or with the cell at each end hyaline or subhyaline and intermediate cells straw-coloured to golden brown, smooth, 68–88.5 × 11–25 μm, 8.5–11 μm wide at the base, 4–7-septate; apical cell bears 3–4 hyaline, subulate appendages, 89–150 × 2.5–5.5 μm, apical appendage at first simple later becoming branched, lateral appendages simple. Chlamydospores pale brown to dark brown, terminal and intercalary, in chains or in groups (observed in CBS 142.51 and 502.80, but not in the epitype).
Culture characteristics: Colonies reaching 25–50 mm diam after 1 wk at 25 °C on OA, PDA and MEA, cottony to glabrous, smoke-grey to olivaceous black, with aerial mycelium in the centre white, margin effuse, fimbriate; reverse black. On OA sometimes with hyaline exudate and apricot diffusible pigment.
Material examined: Lectotype designated here: figs 1–6 in Kirchner O. 1892. Über das Absterben junger Cytisus-Pflanzen. Z. Pflanzenkrankh. Pflanzenschutz 2: 324–327, MBT376013. Austria, Wallersberg, near Völkermarkt, on living stem and leaf of Genista sagittale, Aug. 1980, W. Gams, CBS 502.80. Germany, Berlin, on leaf of Cytisus racemosus, unknown date, R. Schneider (epitype designated here CBS H-23058, MBT376012, culture ex-epitype = CBS 496.63 = MUCL 8091). The Netherlands, Boskoop, on spot on stem of Cytisus sp., unknown date, I. de Boer, CBS 142.51. Unknown country, on leaf of Laburnum sp., unknown date, dep. C.M. Doyer, CBS 118.25.
Notes: Pleiochaeta setosa was introduced by Kirchner (1892) as Ceratophorum setosum for a fungus that infects Cytisus in Germany and later was transferred to Pleiochaeta by Hughes (1951). Since type material for Plei. setosa is inexistent, the illustrations included in the protologue reproduced here (Fig. 51) serve as lectotype. In addition, to fix the use of the name the strain CBS 496.63 is designated here as ex-epitype. This isolate was collected, from the same locality and host genus where it was found the first time (Kirchner 1892) and fits well with the description of the protologue. This species has a worldwide distribution and it is frequently reported as pathogen of Lupinus (Hughes, 1951, Pirozynski, 1974). Nevertheless, Crotalaria, Genista, Laburnum and Ornithopus can also be hosts of this species (Pirozynski, 1974, Yang and Sweetingham, 2002). Unfortunately, host specificity studies are not available for this species, even though Yang & Sweetingham (2002) reported morphological and pathogenicity differences among isolates from Lupinus spp. and Ornithopus spp.
Authors: M. Hernández-Restrepo, E.J. van der Linde & P.W. Crous
Plenodomus Preuss, Linnaea 24: 145. 1851. Fig. 52.
Fig. 52.
Plenodomus spp. A. Symptoms of stem canker of Plenodomus biglobosa. B, C. Section through ascomata. B. Plenodomus guttulatus (holotype MFLU 15-1876). C.Plenodomus salviae (holotype MFLU 15-0515). D–G. Asci. D, E.Plenodomus guttulatus (holotype MFLU 15-1876); F, G.Plenodomus salviae (holotype MFLU 15-0515). H–K. Ascospores. H, I.Plenodomus guttulatus (holotype MFLU 15-1876). J, K.Plenodomus salviae (holotype MFLU 15-0515). Scale bars: B = 75 μm; C = 25 μm; D–G, J, K = 10 μm; H, I = 5 μm. Picture A taken from Fitt et al. (2008); B–K from Ariyawansa et al. (2015b).
Synonyms: Phoma sect. Plenodomus (Preuss) Boerema, Kesteren & Loer., Trans. Brit. Mycol. Soc. 77: 61. 1981.
Diploplenodomus Diedicke, Ann. Mycol. 10: 140. 1912.
Plectophomella Moesz, Magyar Bot. Lapok 21: 13. 1922.
Apocytospora Höhn., Mitt. Bot. Lab. TH Wien 1: 43. 1924.
Deuterophoma Petri, Boll. R. Staz. Patalog. Veget. Roma 9: 396. 1929.
For Additional synonyms of the asexual morph and sexual morph genera listed below see Boerema et al. (1994) and Khashnobish et al. (1995), respectively.
Classification: Dothideomycetes, Pleosporomycetidae, Pleosporales, Leptosphaeriaceae.
Type species: Plenodomus lingam (Tode: Fr.) Höhn. Representative strains: CBS 532.66 and CBS 475.81.
DNA barcodes (genus): LSU, ITS.
DNA barcodes (species): tub2, rpb2. Table 15. Fig. 53.
Table 15.
DNA barcodes of accepted Plenodomus spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | ||
|---|---|---|---|---|---|
| ITS | rpb2 | tub2 | |||
| Plenodomusagnitus | CBS 121.89 | JF740194 | KY064036 | KY064053 | de Gruyter et al. (2013), present study |
| Plen. biglobosus | CBS 119951 | JF740198 | KY064037 | KY064054 | de Gruyter et al. (2013), present study |
| Plen. chrysanthemi | CBS 539.63T | JF740253 | KY064038 | KY064055 | de Gruyter et al. (2013), present study |
| Plen. collinsoniae | CBS 120227 | JF740200 | KY064039 | KY064056 | de Gruyter et al. (2013), present study |
| Plen. confertus | CBS 375.64 | AF439459 | KY064040 | KY064057 | Câmara et al. (2002), present study |
| Plen. congestus | CBS 244.64 | AF439460 | KY064041 | KY064058 | Câmara et al. (2002), present study |
| Plen. deqinensis | CGMCC 3.18221 | KY064027 | KY064034 | KY064052 | Present study |
| Plen. enteroleucus | CBS 142.84ET | JF740214 | KY064042 | KT266266 | de Gruyter et al. (2013), present study |
| Plen. fallaciosus | CBS 414.62 | JF740222 | KY064043 | KT266271 | de Gruyter et al. (2013), present study |
| Plen. guttulatus | MFLUCC 151876 | KT454721 | – | – | Ariyawansa et al. (2015b) |
| Plen. hendersoniae | CBS 113702 | JF740225 | KY064044 | KT266271 | de Gruyter et al. (2013), present study |
| Plen. influorescens | CBS 143.84T | JF740228 | KY064045 | KT266267 | de Gruyter et al. (2013), present study |
| Plen. libanotidis | CBS 113795 | JF740231 | KY064046 | KY064059 | de Gruyter et al. (2013), present study |
| Plen. lindquistii | CBS 381.67 | JF740233 | – | AY749028 | Voigt et al., 2005, De Gruyter et al., 2013 |
| Plen. lingam | CBS 260.94 | JF740235 | KY064047 | KY064060 | de Gruyter et al. (2013), present study |
| Plen. lupini | CBS 248.92 | JF740236 | KY064048 | KY064061 | de Gruyter et al. (2013), present study |
| Plen. pimpinellae | CBS 101637T | JF740240 | – | KY064062 | de Gruyter et al. (2013), present study |
| Plen. salviae | MFLUCC 130219 | KT454725 | – | – | Ariyawansa et al. (2015b) |
| Plen. tracheiphilus | CBS 551.93 | JF740249 | KY064049 | KT266269 | de Gruyter et al. (2013), present study |
| Plen. visci | CBS 122783ET | JF740256 | KY064050 | KY064063 | de Gruyter et al. (2013), present study |
| Plen. wasabiae | CBS 120119 | JF740257 | – | KT266272 | de Gruyter et al. (2013), present study |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CGMCC: Chinese General Microbiological Culture Collection Center, Beijing, China; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Ria, Thailand. T and ET indicate ex-type and ex-epitype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; rpb2: partial RNA polymerase II second largest subunit gene; tub2: partial β-tubulin gene.
Fig. 53.
Phylogenetic tree generated from a maximum parsimony analysis based on the combined LSU, ITS, tub2 and rpb2 sequences. Values above the branches represent parsimony bootstrap support values (> 50 %). Novel species are shown in bold. The tree is rooted with Leptosphaeria doliolum CBS 505.75. GenBank accession numbers are listed in Table 15. T and ET indicate ex-type and ex-epitype strains, respectively. TreeBASE: S21048.
Ascomata solitary, scattered or in small groups, erumpent to superficial, subglobose, broadly or narrowly conical, small- to medium sized, dark brown to black, smooth, ostiolate; ostiole apex with a conical, well developed papilla; ascomatal wall composed of two to several layers of scleroplectenchymatous cells. Hamathecium comprising long, septate, pseudoparaphyses. Asci 8-spored, bitunicate, fissitunicate, cylindrical, rounded at the apex, with an ocular chamber, short pedicel. Ascospores cylindrical to ellipsoidal, yellowish brown, septate, not or slightly constricted at septa, guttulate and lacking a mucilaginous sheath, cell above central septum slightly wider. Conidiomata. Type 1: solitary, scattered or in small groups, erumpent to superficial, subglobose or flask shaped with a broad base, mostly black, ostiolate; ostiole with a long neck and well developed poroid papilla in the apex. Type 2: solitary, scattered or in small groups, erumpent to superficial, mostly subglobose, ostiolate; ostiole slightly papillate with a narrow pore or opening via a rupture. Conidiomatal wall composed of several layers with thick-walled cells of textura angularis, surface heavily pigmented. Conidiophores reduced to conidiogenous cells. Conidiogenous cells phialidic, hyaline, smooth, ampulliform. Conidia hyaline, aseptate, ellipsoidal to subcylindrical (adapted from Ariyawansa et al. 2015b).
Culture characteristics: Colonies on OA yellow/green to olivaceous grey, dull green, or translucent, aerial mycelium tenuous, margin irregular and whitish, compact, floccose.
Optimal media and cultivation conditions: OA or PNA near-ultraviolet light (12 h light, 12 h dark) to promote sporulation at 25 °C.
Distribution: Worldwide.
Hosts: As pathogens of herbaceous plants in different families, most records refer to Asteraceae, and on leaves, branches, bark, wood and dead stems of various trees and shrubs of Brassicaceae, Lamiaceae, Rutaceae, Salicaceae and Vitaceae. In addition, some Plenodomus species are found as opportunistic or pathogenic fungi on Apiaceae, Bignoniaceae, Caprifoliaceae, Fabaceae, Rosaceae, Ulmaceae and Umbelliferae.
Disease symptoms: Leaf spots, stem lesions, slow wilt, bark canker, root rot, shoot dieback.
Notes: The genus Plenodomus was first established by Preuss (1851), and recently re-introduced and placed in the family Leptosphaeriaceae by de Gruyter et al. (2013). The genus mainly consists of species that formerly belonged to Phoma section Plenodomus and the sexual morph Leptosphaeria. Plenodomus includes several well-known important plant pathogens, such as Plen. biglobosus, Plen. lindquistii, Plen. tracheiphilus, and Plen. wasabiae.
References: Boerema et al. 2004 (morphology and pathogenicity); De Gruyter et al., 2013, Ariyawansa et al., 2015b (morphology and phylogeny).
Plenodomus deqinensis Q. Chen & L. Cai, sp. nov. MycoBank MB818821. Fig. 54.
Fig. 54.
Plenodomus deqinensis (ex-type CGMCC 3.18221). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E, F. Colony on PDA (front and reverse). G, H. Pycnidia. I. Section of pycnidium. J. Section of pycnidial wall. K–M. Conidiogenous cells. N. Conidia. Scale bars: G, H = 50 μm; I = 10 μm; J–M = 5 μm; N = 10 μm.
Etymology: Named after the location where the holotype was collected, Deqin, Yunnan Province in China.
Conidiomata pycnidial, solitary, globose to subglobose, glabrous, superficial, (150–)165–355 × (105–)125–305 μm; ostiole single, slightly papillate with a narrow pore or opening via a rupture; conidiomatal wall pseudoparenchymatous, 3–6-layered, 16–28 μm thick, composed of isodiametric to oblong cells, outer layer brown. Conidiophores reduced to conidiogenous cells. Conidiogenous cells phialidic, hyaline, smooth, ampulliform, 5–7 × 4–6.5 μm. Conidia ellipsoidal-cylindrical, smooth- and thin-walled, aseptate, 3.5–5.5 × 1.5–2.5 μm, with 2 minute polar guttules. Conidial exudates not recorded. Sexual morph not observed.
Culture characteristics: Colonies on OA 35 mm diam after 1 wk, margin regular, floccose, white, pale grey near the centre; reverse white to slightly pale olivaceous. Colonies on MEA 17–23 mm diam after 1 wk, margin irregular, aerial mycelia sparse, pale green; reverse concolourous. Colonies on PDA 25–27 mm diam after 1 wk, margin regular, floccose, white, greyish brown near the centre; reverse buff to amber. NaOH test negative.
Material examined: China, Yunnan, Dequin, isolated from soil, Apr. 2011, M.M. Wang (holotype HMAS 247058, culture ex-type CGMCC 3.18221 = LC 5189).
Notes: Plenodomus deqinensis was collected from soil on a snow mountain in China, and proved able to grow at a low temperature (15 °C). This species clustered with Plen. agnitus, Plen. fallaciosus and Plen. lupini in the phylogenetic tree (Fig. 53). The NaOH test of Plen. deqinensis proved negative, while in Plen. agnitus it was positive (Boerema et al. 1994). Morphologically, Plen. deqinensis differs from Plen. lupini in the slightly wider conidiogenous cells (5–7 × 4–6.5 μm vs. 3–8 × 3–6 μm), and being conspicuously biguttulate (de Gruyter et al. 1993). Plenodomus fallaciosus has hitherto only been observed as a sexual morph.
Authors: Q. Chen & L. Cai
Protostegia Cooke, Grevillea 9: 19. 1880. Fig. 55.
Fig. 55.
A–E.Protostegia eucleadicola (ex-type CBS 142615). A, B. Conidiomata on leaf and OA, respectively. C. Conidiogenous cells. D, E. Conidia. F–N.Protostegia eucleae (ex-epitype CBS 137232). F. Leaf symptoms. G. Close-up of conidiomata in vivo. H. Vertical section through conidioma. I–K. Conidia. L. Colony on MEA. M. Conidiogenous cells. N. Conidia in vitro. Scale bars: A, B, G = 250 μm, H = 60 μm, L = 5 mm, all others = 10 μm. Pictures G–N taken from Crous et al. (2015a).
Classification: Dothideomycetes, Dothideomycetidae, Capnodiales, Mycosphaerellaceae.
Type species: Protostegia eucleae Kalchbr. & Cooke. Slide holotype: IMI 230771. Epitype and ex-epitype cultures: PREM 60879, CPC 23549 = CBS 137232.
DNA barcode (genus): LSU.
DNA barcode (species): ITS. Table 16.
Table 16.
DNA barcodes of accepted Protostegia spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | |
|---|---|---|---|---|
| ITS | LSU | |||
| Protostegiaeucleae | CBS 137232ET | KR873252 | KR873280 | Crous et al. (2015a) |
| Pr. eucleicola | CBS 142615T | KY905668 | KY905662 | Present study |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. T and ET indicate ex-type and ex-epitype strains, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: partial 28S large subunit RNA gene.
Sexual morph unknown. Conidiomata pycnidial, immersed, becoming somewhat erumpent, solitary, exuding a mucoid conidial cirrhus, pale brown, splitting the leaf surface, with central ostiole; conidiomatal wall brown, textura intricata. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, lining the inner cavity, lageniform to subcylindrical, proliferating percurrently at apex. Conidia hyaline, smooth, scolecosporous, euseptate (adapted from Crous et al. 2015a).
Culture characteristics: Colonies erumpent, slow growing, with uneven or lobate, feathery margins and sparse to moderate aerial mycelium. On MEA surface and reverse greyish sepia or surface pale olivaceous grey and reverse olivaceous grey; on OA surface mouse-grey or pale olivaceous grey, reverse olivaceous grey; on PDA surface greyish sepia or pale olivaceous grey, reverse mouse-grey or olivaceous grey.
Optimal media and cultivation conditions: PNA incubated at 25 °C under continuous near-ultraviolet light to promote sporulation.
Distribution: South Africa.
Hosts: Euclea divinorum, E. lanceolata, E. natalensis, E. racemosa and E. undulata.
Disease symptoms: Leaf spots.
Notes: The genus Protostegia is thus far only known from South Africa, where it has been reported from leaves of various Euclea spp. However, Euclea is widespread throughout Africa, and therefore Protostegia may be more widespread than currently known. Protostegia was introduced by Kalchbrenner & Cooke (1880) in order to accommodate Stegia magnoliae and the new species Pr. eucleae, and then four more species were allocated in this genus. However, Dyko et al. (1979) transferred three of these species to other genera and another two species were rejected as doubtful. Therefore, only the type species Pr. eucleae was retained and until now this genus has remained monotypic. Protostegia is characterised by immersed conidiomata with walls of textura intricata, splitting the epidermis and appearing acervular, but having a well-developed ostiole (Dyko et al. 1979). Recently Pr. eucleae was placed in the Mycosphaerellaceae together with Cytostagonospora martiniana and Phaeophleospora spp. on the basis of phylogenetic analysis of ITS and LSU sequences (Crous et al. 2015a). Cytostagonospora martiniana can be distinguished from Protostegia by having percurrent and polyphialidic conidiogenous cells, and solitary to aggregated conidiomata embedded in stromatic tissue (Quaedvlieg et al. 2013). Phaeophleospora differs by the production of pigmented conidiogenous cells and conidia (Crous et al. 2009b).
References: Dyko et al. 1979 (morphology); Crous et al. 2015a (morphology and phylogeny).
Protostegia eucleicola Crous, sp. nov. MycoBank MB820822. Fig. 55.
Etymology: Name refers to the host genus it was isolated from, Euclea.
Conidiomata epiphyllous on living leaves, erumpent, solitary, not associated with leaf spots, exuding a mucoid conidial cirrhus that dries to a hard, dark brown crystalline droplet on the leaf surface, up to 250 μm diam, immersed, pale brown, splitting the leaf surface, with central ostiole, 10–30 μm diam; conidiomatal wall brown, textura intricata. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, lining the inner cavity, lageniform to subcylindrical, 8–10 × 4–5 μm, proliferating percurrently at apex. Conidia hyaline, smooth, curved, guttulate, apices subacutely rounded, basal cell tapering to a truncate hilum, 1.5–2 μm diam, 3–7-septate, (40–)50–70(–75) × (2.5–)3–4 μm.
Culture characteristics: Colonies erumpent, slow growing, with lobate, feathery margins and sparse aerial mycelium; on MEA surface and reverse greyish sepia; on OA surface mouse-grey; on PDA surface greyish sepia, reverse mouse-grey.
Material examined: South Africa, Western Cape Province, Porcupine Hills wine farm, between Botrivier and Villiersdorp, on Euclea racemosa, 29 Dec. 2014, A.R. Wood (holotype CBS H-23110, culture ex-type CPC 27224 = CBS 142615).
Notes: With the description of Pr. eucleicola, the genus is presently known from only two species. Protostegia eucleae [conidia (40–)50–75(–80) × (2–)2.5–3 μm] is morphologically similar to Pr. eucleicola [conidia (40–)50–70(–75) × (2.5–)3–4 μm], although the conidia are slightly wider in the latter. The two species are best distinguished based on their DNA data. It is possible that many collections originally reported as Pr. eucleae, actually represent Pr. eucleicola.
Authors: Y. Marin-Felix, A.R. Wood & P.W. Crous
Pseudopyricularia Klaubauf et al., Stud. Mycol. 79: 109. 2014. Fig. 56.
Fig. 56.
Pseudopyricularia spp. A. Sporulation of Pseudopyricularia kyllingae (ex-type CBS 133597) on sterile barley seed on SNA. B. Sporulation of Pseudopyricularia bothriochloae (ex-type CBS 136427) on PNA. C–G. Conidiophores. C.Pseudopyricularia hagahagae (ex-type CPC 25635). D.Pseudopyricularia cyperi (ex-type CBS 133595). E.Pseudopyricularia bothriochloae (ex-type CBS 136427). F.Pseudopyricularia hagahagae (ex-type CPC 25635). G.Pseudopyricularia kyllingae (ex-type CBS 133597). H–K. Conidia. H.Pseudopyricularia cyperi (ex-type CBS 133595). I.Pseudopyricularia bothriochloae (ex-type CBS 136427). J.Pseudopyricularia hagahagae (ex-type CPC 25635). K.Pseudopyricularia kyllingae (ex-type CBS 133597). Scale bars = 10 μm. Pictures A, D, G, H, K taken from Klaubauf et al. (2014); B, E, I taken from Crous et al. (2013b), C, F, J taken from Crous et al. (2015e).
Classification: Sordariomycetes, Sordariomycetidae, Magnaporthales, Pyriculariaceae.
Type species: Pseudopyricularia kyllingae Klaubauf et al. Holotype and ex-type culture: CBS H-21841, CBS 133597.
DNA barcodes (genus): LSU, rpb1.
DNA barcodes (species): ITS, rpb1, act, cal. Table 17. Fig. 57.
Table 17.
DNA barcodes of accepted Pseudopyricularia spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | |||
|---|---|---|---|---|---|---|
| ITS | rpb1 | act | cal | |||
| Pseudopyriculariabothriochloae | CBS 136427T | KF777186 | KY905701 | KY905700 | – | Crous et al. (2013b), present study |
| Py. cyperi | CBS 133595T | KM484872 | AB818013 | AB274453 | AB274485 | Klaubauf et al., 2014, Murata et al., 2014, Hirata et al., 2014 |
| Py. hagahagae | CPC 25635T | KT950851 | KT950877 | KT950873 | – | Crous et al. (2015e) |
| Py. higginsii | CBS 121934 | KM484875 | KM485095 | KM485180 | KM485250 | Klaubauf et al. (2014) |
| Py. kyllingae | CBS 133597T | KM484876 | KM485096 | AB274451 | AB274484 | Klaubauf et al., 2014, Hirata et al., 2014 |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CPC: Culture collection of Pedro Crous, housed at Westerdijk Fungal Biodiversity Institute. T indicates ex-type strains.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; rpb1: partial RNA polymerase II largest subunit gene; act: partial actin gene; cal: partial calmodulin gene.
Fig. 57.
RAxML phylogram obtained from the combined ITS (546 bp) and LSU (750 bp) sequences of members of Pyriculariaceae. The tree was rooted to Bussabanomyces longisporus CBS 125232. The new combination proposed is indicated in bold. RAxML bootstrap support (BS) values above 70 % and Bayesian posterior probability scores ≥ 0.95 are shown at the nodes. Numbers between parentheses correspond to GenBank accession numbers of ITS and LSU sequences, respectively. T and NT indicate ex-type and ex-neotype strains, respectively. TreeBASE: S20877.
Sexual morph unknown. Conidiophores solitary, erect, straight or curved, branched or not, medium brown, smooth or finely roughened, septate. Conidiogenous cells integrated, terminal, rarely intercalary, medium brown, smooth or finely roughened, forming a rachis with several protruding denticles usually flat-tipped. Conidia solitary, obclavate, pale to medium brown, smooth or/to finely roughened, guttulate, 1–2-septate; hila truncate, slightly protruding, unthickened, not darkened (adapted from Klaubauf et al. 2014).
Culture characteristics: Colonies smooth with sparse to moderate aerial mycelium. On MEA transparent, buff, honey to isabelline or white with patches of greyish sepia. On OA transparent sometimes with patches of olivaceous grey or greyish sepia. On PDA transparent, white, greyish sepia or olivaceous black.
Optimal media and cultivation conditions: Sterile barley seed on SNA at 25 °C under continuous near-ultraviolet light to promote sporulation.
Distribution: Mainly found in Asia, but also in North America, Africa and New Zealand.
Hosts: Pathogens of Cyperaceae, but also found on Bothriochloa bladhii (Poaceae) and Typha orientalis (Typhaceae).
Disease symptoms: Leaf spots.
Notes: Pseudopyricularia was one of the genera introduced recently in order to resolve the polyphyletic nature of Pyricularia (Klaubauf et al. 2014). Pseudopyricularia is mainly distinguished from Pyricularia by having short, determinate, brown conidiophores with an apical rachis with flat-tipped denticles.
Reference: Klaubauf et al. 2014 (morphology and phylogeny).
Pseudopyricularia bothriochloae (Crous & Cheew.) Y. Marín & Crous, comb. nov. MycoBank MB819002.
Basionym: Pyricularia bothriochloae Crous & Cheew., Persoonia 31: 229. 2013.
Notes: This fungus was initially described as a new species of Pyricularia (Crous et al. 2013b) before Klaubauf et al. (2014) introduced the new genus Pseudopyricularia. In the latter study, this species was incorporated in the phylogenetic analysis based on LSU sequence data, but not in the combined analysis, since only ITS and LSU sequences were available. Although the ex-type strain of Py. bothriochloae grouped in the Pseudopyricularia clade, a new combination was not proposed, as it could not be incorporated in the combined analysis. However, in the phylogenetic tree based on ITS and LSU sequences (Fig. 57), Py. bothriochloae was located in the Pseudopyricularia clade (100 % bootstrap support / 1 Posterior Probability) and accordingly the new combination, Py. bothriochloae, is made here. This species produces conidiophores with apical rachis with flat-tipped denticles with periclinal thickening, which characterises Pseudopyricularia spp.
Pseudopyricularia spp. are mainly pathogens of Cyperaceae. However, this species was found on Bothriochloa bladhii (Poaceae), producing angular leaf spots. Morphologically, Py. bothriochloae can be easily distinguished by the 1-septate conidia (2-septate in all the other species).
Authors: Y. Marin-Felix & P.W. Crous
Puccinia Pers., Neues Mag. Bot. 1: 118. 1794. Fig. 58.
Fig 58.
Puccinia spp. A, D. Aecia and aeciospores of Puccinia paederiae (BRIP 58338). B, E. Aecia and aeciospores of Puccinia loranthicola (BRIP 59685). C, F. Uredinia and urediniospores of Puccinia oxalidis (BRIP 58379). G, J. Uredinia and urediniospores of Puccinia philippinensis (BRIP 57418). H, K. Telia and teliospores of Puccinia malvacearum (BRIP 60128). I, L. Telia and teliospores of Puccinia thwaitesii (BRIP 58354). Scale bars = 10 μm.
For synonyms see Cunningham (1931).
Classification: Basidiomycota, Pucciniomycotina, Pucciniomycetes, Pucciniales, Pucciniaceae.
Type species: Puccinia graminis Pers. Designated as type species of Puccinia by Cunningham (1931) on cultivated Triticum; lectotypified by Jørstad (1958).
DNA barcodes (genus): ITS, LSU.
DNA barcode (species): ITS (evidence for intraspecific and intra-isolate diversity), LSU. Table 18. Fig. 59.
Table 18.
DNA barcodes of accepted Puccinia spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | |
|---|---|---|---|---|
| ITS | LSU | |||
| Pucciniaabrupta var. partheniicola | BRIP 59295 | – | KX999864 | Present study |
| Pu. acroptili | BPI 863523 | JN204187 | JN204187 | Bruckart et al. (2012) |
| Pu. arthrocnemi | BRIP 57772 | – | KX999865 | Present study |
| Pu. aucta | BRIP 60028 | – | KX999866 | Present study |
| Pu. bassiae | BRIP 57788 | – | KX999867 | Present study |
| Pu. brachypodii | BRIP 59466 | – | KX999868 | Present study |
| Pu. caricina | BRIP 57951 | – | KX999870 | Present study |
| Pu. carissae | BRIP 53242 | – | KX999871 | Present study |
| Pu. chrysanthemi | NA | EU816926 | EU816926 | Pedley (2009) |
| Pu. convolvuli | BPI 871465 | – | DQ354512 | Aime (2006) |
| Pu. coronata var. avenae f. sp. avenae | PUR 22125LT | HM131256 | – | Liu & Hambleton (2013) |
| Pu. coronata var. avenae f. sp. graminicola | PRM 155608 | HM131309 | – | Liu & Hambleton (2013) |
| Pu. coronati-agrostis | PUR N114T | HM131319 | – | Liu & Hambleton (2013) |
| Pu. coronati-brevispora | PUR N652T | HM131235 | – | Liu & Hambleton (2013) |
| Pu. coronati-calamagrostidis | PUR 22155LT | HM131304 | – | Liu & Hambleton (2013) |
| Pu. coronati-hordei | PUR 89857T | HM131225 | – | Liu & Hambleton (2013) |
| Pu. coronati-japonica | PUR F16131T | HM131317 | – | Liu & Hambleton (2013) |
| Pu. coronati-longispora | PRC 196T | HM131232 | – | Liu & Hambleton (2013) |
| Pu. cygnorum | NA | EF490601 | – | Langrell et al. (2008) |
| Pu. cynodontis | BRIP 57556 | – | KX999873 | Present study |
| Pu. dianellae | BRIP 57433 | – | KM249859# | McTaggart et al. (2016a) |
| Pu. dichondrae | BRIP 60027 | – | KX999874 | Present study |
| Pu. dioicae | BPI 879279 | – | GU058019# | Dixon et al. (2010) |
| Pu. duthiei | BRIP 61025 | – | KX999875 | Present study |
| Pu. flavenscentis | BRIP 57992 | – | KX999876 | Present study |
| Pu. gastrolobii | BRIP 57735 | – | KX999877 | Present study |
| Pu. geitonoplesii | BRIP 55679 | KM249860 | KM249860 | McTaggart et al. (2016a) |
| Pu. gilgiana | BRIP 57723 | KF690673 | KF690690 | McTaggart et al. (2014) |
| Pu. graminis f. sp. tritici | CDL 75-36-700-3 | NW_003526581.1* | Duplessis et al. (2011) | |
| Pu. grevilleae | BRIP 55600 | – | KX999878 | Present study |
| Pu. haemodori | BRIP 57777 | KF690676 | KF690694 | McTaggart et al. (2014) |
| Pu. hemerocallidis | BRIP 53476 | KM249855 | KM249855 | McTaggart et al. (2016a) |
| Pu. horiana | NA | HQ201326 | HQ201326 | Alaei et al. (2009) |
| Pu. hypochoeridis | BRIP 57771 | – | KX999879 | Present study |
| Pu. kuehnii | BPI 879137 | GQ283007 | – | Flores et al. (2009) |
| Pu. lagenophorae | BRIP 57563 | KF690677 | KF690696 | McTaggart et al. (2014) |
| Pu. levis var. tricholaenae | BRIP 56867 | – | KX999880 | Present study |
| Pu. liberta | BRIP 59686 | – | KX999881 | Present study |
| Pu. loranthicola | BRIP 59685 | – | KX999882 | Present study |
| Pu. ludwigii | BRIP 60129 | – | KX999883 | Present study |
| Pu. magnusiana | BPI 879281 | – | GU058000# | Dixon et al. (2010) |
| Pu. malvacearum | PBM 2572 | – | EF561641# | Matheny & Hibbett (unpubl. data) |
| Pu. melanocephala | BPI 878929 | – | GU058001# | Dixon et al. (2010) |
| Pu. menthae | BPI 871110 | – | DQ354513# | Aime (2006) |
| Pu. mixta | BRIP 61576 | KU296893 | KU296893 | McTaggart et al. (2016a) |
| Pu. muehlenbeckiae | BRIP 57718 | – | KX999884 | Present study |
| Pu. myrsiphylli | BRIP 57782 | – | KM249854# | McTaggart et al. (2016a) |
| Pu. nakanishikii | BPI 879283 | – | GU058002# | Dixon et al. (2010) |
| Pu. merrilliana | BRIP 56913 | – | KX999885 | Present study |
| Pu. paullula | BRIP 60018 | – | KX999886 | Present study |
| Pu. pelargonii-zonalis | BRIP 57414 | – | KX999887 | Present study |
| Pu. polysora | HSZ1879 | HQ189433 | HQ189433 | Crouch & Szabo (2011) |
| Pu. porri | BRIP 61579 | KU296902 | KU296902 | McTaggart et al. (2016a) |
| Pu. pritzeliana | BRIP 57798 | – | KX999888 | Present study |
| Pu. purpurea | BRIP 57994 | – | KX999889 | Present study |
| Pu. rhagodiae | BRIP 60078 | – | KX999890 | Present study |
| Pu. rhaphidophorae | BRIP 56840 | – | KX999891 | Present study |
| Pu. scirpi | BRIP 61027 | – | KX999892# | Present study |
| Pu. scleriae | BRIP 56911 | – | KX999893 | Present study |
| Pu. smilacis | BPI 871784 | DQ354533 | DQ354533 | Aime (2006) |
| Pu. sparganioidis | BPI 879285A | – | GU058027# | Dixon et al. (2010) |
| Pu. striiformis | HSZ1834 | GQ457306 | GQ457306 | Jin et al. (2010) |
| Pu. stylidii | BRIP 60107 | KJ622216 | KJ622215 | McTaggart et al. (2014) |
| Pu. tetragoniae | BRIP 59703 | – | KX999894 | Present study |
| Pu. triticina | NA | ADAS02000001.1* | Kiran et al. (2016) | |
| Pu. unica | BRIP 56930 | – | KX999895 | Present study |
| Pu. ursiniae | BRIP 57993 | KF690684 | KF690705 | McTaggart et al. (2014) |
| Pu. xanthii | BRIP 56946 | – | KX999896 | Present study |
CDL: US Department of Agriculture, Agricultural Research Service, Cereal Disease Laboratory; BPI: US National Fungus Collections, Beltsville, Maryland, USA; BRIP: Queensland Plant Pathology Herbarium, Brisbane, Australia; HSZ: Cereal Disease Laboratory collection, St. Paul, Minnesota, USA; PBM: P. Brandon Matheny (personal collection); PRC: Charles University in Prague, Prague, Czech Republic; PRM: National Museum, Prague, Czech Republic; PUR: Purdue University, west Lafayette, Indiana, USA. T and LT indicate ex-type and ex-lectotype, respectively.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: partial 28S large subunit RNA gene. *Whole genome sequence. #ITS2–LSU sequence.
Fig 59.
Phylogram obtained from a maximum likelihood search of LSU and cytochrome c oxidase subunit 3 of mitochondrial DNA (co3), partitioned as two separate genes in RAxML. Bootstrap values (≥ 70 %) from 1 000 replicates in a maximum likelihood search shown above nodes. Posterior probabilities (≥0.95) summarised from 30 000 trees obtained by Bayesian inference in MrBayes are shown below nodes. General time-reversible (GTR) with GAMMA distribution was used as a model of evolution for both phylogenetic criteria. Major clades of Puccinia obtained in previous studies are shaded. New combinations made in the present study are in bold. Two species of Sphaerophragmiaceae were selected as outgroup to the Pucciniaceae. Numbers between parentheses correspond to GenBank accession numbers for LSU and co3 sequences, respectively. The locus co3 was used as a second, independent gene in the phylogenetic analyses; however it is not regarded as a molecular barcode for species of Puccinia as there are limited nucleotide differences between closely related species. TreeBASE: S21061.
Spermogonia dark brown to black, often on adaxial leaf surface, subepidermal, concave hymenia with well-developed periphyses at ostiole [Group V, type 4 sensu Hiratsuka & Hiratsuka (1980)]. Spermatia exuded in droplets, small, aseptate, hyaline. Aecia erumpent, usually abaxial, cup-shaped, with well-developed peridium; peridial cells irregular and verrucose. Aeciospores catenulate, globose to subglobose, verruculose. Uredinia subepidermal or erumpent, on both leaf surfaces and stems, without peridium, pale yellow to brown. Paraphyses either absent, peripheral or within the sorus. Urediniospores borne singly on pedicels, mostly echinulate, usually globose, subglobose, ellipsoid to obovoid, germ pores absent or conspicuous. Telia subepidermal or erumpent, mostly dark brown to black, on both leaf surfaces and stems. Teliospores typically 2-celled by transverse or oblique septa (but may have variations of 1–4 cells in some species), borne singly on pedicels, mostly pale to dark brown, cell walls smooth or ornamented. Basidia transversely septate (phragmobasidia), 2–4 celled, external. Basidiospores formed singly from each basidial cell on a sterigma, sometimes ballistosporic.
Distribution: Worldwide.
Hosts: Species of Puccinia are obligate plant pathogens that occur on host species in many families, especially Asteraceae, Cyperaceae, Fabaceae, Lamiaceae, Liliaceae s. lat., Malvaceae and Poaceae. Heteroecious species of Puccinia, e.g. Pu. graminis, require two host plant species to complete their life cycle. The spermogonia and aecia of heteroecious species occur on one host species, while the uredinia and telia occur on another, often unrelated, host species. Autoecious species complete their life cycle on one host species. There are many variations in the life cycles of species of Puccinia. For example, some species, e.g. Pu. lagenophorae, do not form spermogonia or uredinia. Other species are known only from their telia, or telia and spermogonia, e.g. Pu. malvacearum and Pu. grevilleae. Frequent host jumps in the evolution of Puccinia and related genera have resulted in closely related species of Puccinia across wide host ranges, as well as distantly related species that occur on the same host plant species (Maier et al., 2007, Van der Merwe et al., 2008, Dixon et al., 2010, McTaggart et al., 2016a).
Disease symptoms: Spermogonia, aecia (Fig. 58A, B), uredinia (Fig. 58C, G) and telia (Fig. 58H, I) occur on leaves and stems, often associated with chlorotic lesions, sometimes on bullate swellings, solitary or scattered or aggregated in groups, arranged linearly or concentrically or irregularly, often erumpent, in cases of severe infection leaves prematurely wilt and senesce.
Notes: The starting publication for names of all rust fungi for purposes of priority as provided by Art. 13 of the International Code of Nomenclature for algae, fungi, and plants (ICN) (McNeill et al. 2012) is the Synopsis Methodica Fungorum by Persoon (1801), who listed 11 species of Puccinia, 19 species of Aecidium and 30 species of Uredo. The genera Aecidium, Uredo and Puccinia were established for rust fungi with aecia, uredinia and telia, respectively. Many species described in these three genera are conspecific, e.g. the lectotype of Aecidium berberidis designated by Clements & Shear (1931) is the aecial stage of Pu. graminis. There is little possibility that Aecidium and Uredo (asexual genera) will displace Puccinia (sexual genus) under Art. 57.2 of the ICN (McNeill et al. 2012). Whether Uredo is a synonym of Puccinia depends on the phylogenetic placement of Uromyces beticola, the lectotype of Uredo (Laundon 1970). A taxonomic working group on the Basidiomycota in 2011 recommended the use of Uredo for uredinial species that could not be assigned to a monophyletic sexual genus (available at: http://www.imafungus.org/Issue/31/05.pdf). Many species of Aecidium and Uredo will need to be transferred to Puccinia, or other monophyletic genera, in order to preserve the one name equals one fungus principle (Hawksworth et al. 2011).
There are about 4 000 described species of Puccinia (Kirk et al. 2008), which have mostly been delimited by host taxon. Many of these species have diversified in the last 50 million years as a result of host jumps (McTaggart et al. 2016b), with the aecial host serving as a pathway for further speciation (van der Merwe et al. 2008). The morphology of teliospores and urediniospores is often sufficient to distinguish species of Puccinia that occur on the same host. Molecular approaches have uncovered cryptic diversity in some species of Puccinia (Liu & Hambleton 2013) as well as linking aecial to telial morphs in the life cycles of heteroecious rusts (Jin et al. 2010). Other studies have shown there is less species biodiversity in some rusts than previously thought, e.g. Pu. lagenophorae and closely related species (Scholler et al., 2011, McTaggart et al., 2014). Intraspecific and intra-isolate diversity of the ITS region was found in Pu. horiana and Pu. kuehnii (Virtudazo et al., 2001, Alaei et al., 2009). Multiple haplotypes and paralogous copies of the ITS region within species of rust must be considered for phylogenetic and molecular barcode studies.
Phylogenetic studies have identified several sexual genera as potentially congeneric with Puccinia. Puccinia is either paraphyletic or polyphyletic with respect to Ceratocoma (McTaggart et al. 2016b), Cumminsiella (Maier et al. 2003), Dietelia (Wingfield et al. 2004), Diorchidium (Beenken & Wood 2015), Endophyllum (Maier et al. 2003), Macruropyxis (Beenken & Wood 2015), Miyagia (Wingfield et al. 2004), Sphenospora (Aime 2006) and Uromyces (Maier et al. 2003). Three major clades that contained Puccinia and related genera were identified in molecular phylogenetic studies (Van der Merwe et al., 2008, Dixon et al., 2010). One clade diversified on Cyperaceae, Juncaceae and orders of plants in the asterids and rosids (The Angiosperm Phylogeny 2016), and the another on Poaceae and Ranunculaceae (van der Merwe et al. 2008). A third clade included species of Puccinia on Poaceae (Dixon et al. 2010). The relationships between the major clades in Puccinia can be observed in our phylogenetic analysis (Fig. 59).
Uromyces requires particular consideration as it has long been thought an aseptate variant of Puccinia (Sydow and Sydow, 1904, Savile, 1978). Morphology alone does not reliably separate Puccinia and Uromyces, because puccinioid (2-celled) and 1-celled spores and characteristics of the pedicel are homoplasious in the Pucciniales (Maier et al., 2007, Minnis et al., 2012, Beenken and Wood, 2015). Several studies have shown that Puccinia and Uromyces are polyphyletic, and furthermore that Puccinia is paraphyletic with respect to the type of Uromyces (U. appendiculatus) and other species of Uromyces on Fabaceae (Maier et al., 2007, Van der Merwe et al., 2008). Consequently, either a taxonomy that accepts Puccinia as a paraphyletic group must be adopted or Uromyces must be synonymised under Puccinia. In the latter case, many important species of Uromyces will require name changes. The traditional use of Uromyces for species with aseptate teliospores has been replaced by a phylogenetic approach; for example, Demers et al. (2017) used a phylogenetic approach to describe two species of Puccinia with aseptate teliospores, which would have been described as Uromyces based on morphology.
The future of Puccinia depends on whether it can be divided into monophyletic genera or sub-genera that reflect synapomorphies or ecological relationships on which a natural classification can be based. A broad concept of Puccinia that accepts species with puccinioid spores that are recovered in closely related clades as defined by van der Merwe et al. (2008) and Dixon et al. (2010) is adopted here. Based on this molecular phylogenetic taxonomic concept, we have transferred four species of Uredo from the Australasian region to Puccinia. Further examples of taxa recovered in Puccinia, include Aecidium kalanchoe (Hernádez et al. 2004) and Uredo guerichiani (Maier et al. 2007). We have chosen not to make new combinations of these species without examination of a specimen. Molecular phylogenetic support must be an essential requirement for the description of new species or new combinations in Puccinia because several species known from an anamorphic stage have an affinity with other genera of rust fungi, e.g. Uredo rolliniae (now Phakopsora rolliniae) (Beenken 2014).
ITS and LSU sequences are available for approximately 200 species of Puccinia on GenBank (accessed 5 Sep. 2016). These two gene regions are generally reliable as a molecular barcode for identification of species of Puccinia. GenBank numbers for some of the most important species of Puccinia that are associated with a herbarium specimen, reference genome sequence, or peer reviewed study, are provided in Table 18.
References: Sydow & Sydow 1904 (morphology); Cummins & Hiratsuka 2003 (biology, morphology and taxonomy).
Puccinia dianellae (Dietel) McTaggart & R.G. Shivas, comb. nov. MycoBank MB819750.
Basionym: Uredo dianellae Dietel, Hedwigia 37: 213. 1898.
Material examined: Philippines, Benguet, Tuba, Mount Santo Tomas, on Dianella javanica, 26 Jun. 2012, K.L. Lancetta, V.A. Felices, T.U. Dalisay, A.I. Llano, A.R. McTaggart, R.G. Shivas & M.D.E. Shivas (BRIP 57433).
Notes: Puccinia dianellae was recovered in a monophyletic group with species of Puccinia on Hemerocallidaceae (McTaggart et al. 2016a). Telia have not been reported.
Puccinia geitonoplesii (McAlpine) McTaggart & R.G. Shivas, comb. nov. MycoBank MB819751.
Basionym: Uredo geitonoplesii McAlpine, The Rusts of Australia, their Structure, Nature and Classification: 203. 1906.
Material examined: Australia, Queensland, Coochiemudlo Island, Victoria Parade, on leaf of Geitonoplesium cymosum, 25 Aug. 2012, C. Doungsa-ard & A.R. McTaggart (BRIP 57603).
Notes: Puccinia geitonoplesii was recovered in a monophyletic group with species of Puccinia on Hemerocallidaceae (McTaggart et al. 2016a). Telia have not been reported. Morphological identification of P. geitonoplesii can be assisted by the Rust Fungi of Australia Lucid Key (Shivas et al. 2014) (http://collections.daff.qld.gov.au/web/key/rustfungi/).
Puccinia merrilliana (Syd. & P. Syd.) McTaggart & R.G. Shivas, nom. nov. MycoBank MB819752.
Basionym: Uredo operculinae Syd. & P. Syd., Philipp. J. Sci., C, Bot. 8: 476. 1913.
Material examined: Australia, Western Australia, Kununurra, Ivanhoe Crossing turnoff, on leaf of Operculina aequisepala, 16 Apr. 2012, M. Butt, C. Doungsa-ard, A.R. McTaggart, R. Berndt, V. Faust-Berndt, M.D.E. Shivas & R.G. Shivas (BRIP 56913).
Notes: Uredo operculinae was first described on Operculina turpethum from the Philippines (Sydow & Sydow 1913). The transfer of U. operculinae to Puccinia requires a new name, Pu. merrilliana, as Pu. operculiniae is already occupied for a rust on O. turpethum in the Malabar region in southern India (Ramakrishnan & Sundaram 1953). The new name honours Elmer Drew Merrill (1876‒1956), an American botanist, who collected this fungus while living in the Philippines, where he became an expert on the flora of the Asia-Pacific region. Puccinia merrilliana has fewer (1‒2) germ pores than Pu. operculinae, which has 3‒4 germ pores. Telia have not been reported for Pu. merrilliana. The specimens examined from Australia are morphologically identical to the type description by Sydow & Sydow (p. 425, 1924). Morphological identification of Pu. merrilliana can be assisted by the Rust Fungi of Australia Lucid Key (Shivas et al. 2014) (http://collections.daff.qld.gov.au/web/key/rustfungi/). Puccinia merrilliana was recovered in Puccinia in Group I sensu van der Merwe et al. (2008).
Puccinia rhagodiae (Cooke & Massee) McTaggart & R.G. Shivas, comb. nov. MycoBank MB819756.
Basionym: Uredo rhagodiae Cooke & Massee, Grevillea 15 (no. 76): 99. 1887.
Material examined: Australia, Tasmania, Lilico Beach, on leaf of Chenopodium candolleanum, 15 Dec. 2013, A.R. McTaggart, L.S. Shuey, M.D.E. Shivas & R.G. Shivas (BRIP 60078).
Notes: Puccinia rhagodiae was recovered in Puccinia group I sensu van der Merwe et al. (2008). Telia have not been reported. Several other species of Puccinia on Amaranthaceae were shown to be closely related, including Pu. arthrocnemi, Pu. bassiae and Pu. tetragoniae, although they did not form a monophyletic group. Morphological identification of Pu. rhagodiae can be assisted by the Rust Fungi of Australia Lucid Key (Shivas et al. 2014) (http://collections.daff.qld.gov.au/web/key/rustfungi/).
Authors: A.R. McTaggart & R.G. Shivas
Saccharata Denman & Crous, CBS Biodiversity Ser. (Utrecht) 2: 104. 2004. Fig. 60.
Fig. 60.
Saccharata spp. A. On Banksia sp. B. Symptomatic leaves of Saccharata proteae (CBS 121406). C. Close-up of subepidermal conidiomata of Saccharata proteae.D–E. Sexual morph of Saccharata proteae (CBS 121406). D, E. Asci, paraphyses and ascospores. F–N. Asexual morphs. F. Colony sporulating on OA of Saccharata capensis (ex-type CBS 122693). G. Pycnidial conidioma of Saccharata capensis (ex-type CBS 122693). H–J. Conidiogenous cells and conidia. H. Saccharata proteae (CBS 121406). I, J.Saccharata capensis (ex-type CBS 122693). K–M. Conidia. K. Saccharata intermedia (ex-type CBS 125546). L.Saccharata kirstenboschensis (ex-type CBS 123537). M.Saccharata proteae (CBS 121406). N. Spermatia of Saccharata capensis (ex-type CBS 122693). Scale bars: G = 100 μm, others = 10 μm; I applies to I, J. Pictures B–K, J–N taken from Crous et al. (2013a); L from Crous et al. (2008).
Classification: Dothideomycetes, Incertae sedis, Botryosphaeriales, Saccharataceae.
Type species: Saccharata proteae (Wakef.) Denman & Crous. Holotype and ex-type culture: PREM 32915, STE-U 1694.
DNA barcode (genus): LSU.
DNA barcodes (species): ITS, rpb2, tef1, tub2. Table 19.
Table 19.
DNA barcodes of accepted Saccharata spp.
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. T indicates ex-type strain.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; rpb2: partial RNA polymerase II second largest subunit gene; tef1: partial translation elongation factor 1-alpha gene; tub2: partial β-tubulin gene.
Ascomata epiphyllous, separate, becoming aggregated, unilocular, immersed, substomatal, with a central, flattened ostiole, surrounded by a continuous, clypeus-like apical thickening of the wall, obovoid, slightly depressed; ascomata wall consisting of 8–11 layers of brown pseudoparenchymatous textura angularis. Pseudoparaphyses hyaline, septate, branched, frequently attached to the top and base of the pseudothecial cavity. Asci clavate to cylindrical, stipitate, bitunicate, fissitunicate; apical chamber visible as a notch-like indentation at the apex. Ascospores uni- to biseriate, hyaline, guttulate, smooth, ellipsoidal, clavate to fusiform, frequently widest in the upper third of the ascospore, tapering to obtuse ends. Conidiomata pycnidial, black, opening by a single, central ostiole, infrequently embedded in stromatic tissue with thickened, darkened upper layer; conidiomatal wall consisting of 2–3 layers of brown textura angularis. Conidiophores hyaline, smooth, subcylindrical, branched, or reduced to conidiogenous cells, lining the inner layer of the cavity, 1–3-septate. Paraphyses intermingled among conidiophores, hyaline, smooth, subcylindrical, unbranched or branched above, with obtuse ends, 0–3-septate, extending above conidiophores or slightly above the conidia. Conidiogenous cells hyaline, smooth, phialidic, proliferating via periclinal thickening or percurrent proliferation, with or without collarettes. Conidia hyaline, smooth, thin-walled, aseptate, granular, fusiform to narrowly ellipsoid or fusoid-ellipsoid, apex subobtuse, base subtruncate or truncate with minute marginal frill, widest in the middle of the conidium. Synasexual morph formed in separate conidiomata, or in same conidiomata with asexual morph. Synasexual conidia pigmented, thick-walled, finely verruculose, ellipsoid or oval, aseptate. Spermatogonia similar to conidiomata in anatomy. Spermatogenous cells ampulliform to lageniform or subcylindrical, hyaline smooth, phialidic. Spermatia developing in conidiomata or spermatogonia, hyaline, smooth, granular, subcylindrical or dumbbell-shaped, with rounded ends (adapted from Crous et al. 2004a and Slippers et al. 2013).
Culture characteristics: Colonies on PDA, OA and MEA spreading, with moderate aerial mycelium, usually erumpent, less frequent flat, margins irregular; surface and reverse show different shades of grey.
Optimal media and cultivation conditions: On OA or PNA at 25 °C under continuous near-ultraviolet light to promote sporulation.
Distribution: Commonly found in South Africa, but also Australia, North America (incl. Hawaii) and Europe.
Hosts: Members of Proteaceae, especially in species of Banksia, Hakea, Isopogon, Lambertia, Leucospermum, Petrophile and Protea. Also found on Daviesia (Fabaceae), Encephalartos (Zamiaceae), and Eucalyptus (Myrtaceae) (see Crous et al. 2016b).
Disease symptoms: Leaf tip die-back and leaf spots.
Notes: Saccharata is the only genus located in the family Saccharataceae, which was recently introduced by Slippers et al. (2013). This genus was described by Crous et al. (2004a) in order to accommodate “Botryosphaeria” proteae, and subsequently several additional species were added to the genus from South Africa. All the species were found on Proteaceae expect Saccharata kirstenboschensis, which was isolated from Encephalartos princeps (Crous et al. 2008). South African Saccharata spp. that occur on Proteaceae can be distinguished from other members of Botryosphaeriales by their asexual morphology, which includes a hyaline, fusicoccum-like and a pigmented diplodia-like asexual morph (Crous et al. 2013a). However, Crous et al. (2016b) introduced eight species from a range of hosts (Myrtaceae and Proteaceae) in Australia, and also widened the generic concept to include the genus Neoseptorioides (3-septate, cylindrical conidia; Crous et al. 2015e). In spite of the range of variation observed in the asexual morphs, morphology of the sexual morphs of Australian and South African species appear remarkably conserved.
References: Crous et al., 2008, Crous et al., 2013a, Crous et al., 2016b (morphology and phylogeny).
Saccharata leucospermi Crous, sp. nov. MycoBank MB820823. Fig. 61.
Fig. 61.
Saccharata leucospermi (ex-type CBS 122694). A. Conidiomata forming on OA. B, C. Conidiogenous cells giving rise to conidia. D. Conidia. Scale bars: A = 300 μm, others = 10 μm.
Etymology: Named for the host genus from which it was collected, Leucospermum.
Conidiomata on PDA pycnidial, black, up to 300 μm diam, with a single, central ostiole; conidiomatal wall consisting of 2–3 layers of brown textura angularis. Conidiophores subcylindrical, hyaline, smooth, frequently reduced to conidiogenous cells or branched in apical part, 1–2-septate, 7–20 × 2–3.5 μm. Paraphyses rarely observed, intermingled among conidiophores, unbranched hyaline, smooth, 0–1-septate, 2–3 μm wide, extending above conidiophores. Conidiogenous cells terminal, subcylindrical, hyaline, 7–10 × 2–3 μm, with periclinal thickening, rarely with percurrent proliferations. Conidia hyaline, smooth, fusiform to narrowly ellipsoid, apex subobtuse, base truncate with minute marginal frill, minutely guttulate, thin-walled, (13–)14–16(–19) × (4–)4.5(–5) μm.
Culture characteristics: Colonies on MEA at 25 °C in the dark after 1 wk: spreading, erumpent, surface crumpled, irregular, with uneven, feathery margin and moderate aerial mycelium; surface pale mouse-grey, reverse mouse-grey.
Material examined: South Africa, Western Cape Province, Kogelberg Nature Reserve, on leaf litter of Leucospermum conocarpodendron subsp. viridum, 11 Jul. 2000, S. Marincowitz (holotype CBS H-20078, culture ex-type CBS 122694 = CPC 13698 = CMW 22197).
Notes: In the treatment of microfungi occurring on leaf litter of Proteceae, Marincowitz et al. (2008a) listed CBS 122694 as a Saccharata sp., acknowledging the fact that it appeared to be different. Three other species are known from Protea leaves in South Africa, namely S. proteae (conidia 20–30 × 4.5–6 μm; Denman et al. 1999), S. intermedia [conidia (17–)18–20(–22) × (3.5–)5–6 μm; Crous et al. 2009a], and S. hawaiiensis [conidia (17–)24–30(–38) × (4–)5–7(–8) μm; Yang et al. 2017]. Saccharata leucospermi can readily be distinguished from these three species by having smaller conidia.
Saccharata protearum Crous, sp. nov. MycoBank MB820824. Fig. 62.
Fig. 62.
Saccharata protearum (ex-type CBS 114569). A. Conidiomata forming on OA. B, C. Conidiogenous cells giving rise to conidia. D. Conidia. Scale bars: A = 400 μm, others = 10 μm.
Etymology: Named after the host genus from which it was collected, Protea.
Conidiomata pycnidial, eustromatic, to 400 μm diam, immersed, subepidermal, erumpent in culture, separate, or aggregated, linked by a stroma, dark brown, uni- to multi-locular, walls consisting of dark brown textura angularis, ostiolate. Fusicoccum-like asexual morph: Conidiophores hyaline, smooth, branched, subcylindrical, 1–3-septate, formed from the inner layer of the locule, 10–30 × 2.5–3.5 μm, intermingled with hyaline, septate paraphyses. Conidiogenous cells phialidic, discrete or integrated, hyaline, smooth, cylindrical, enteroblastic, proliferating percurrently with 1–2 annellations, 9–15 × 2.5–3.5 μm. Conidia hyaline, thin-walled, aseptate, smooth, fusoid, widest in the middle or upper third of the conidium, with a subobtuse apex, and a truncate base, (17–)20–25(–27) × (4–)4.5–5(–6) μm. Microconidial morph occurring in separate or the same conidiomata as the fusicoccum-like asexual morph. Microconidiophores hyaline, smooth, branched, cylindrical, 1–3-septate, formed from the inner layers of the locule, 20–30 × 2.5–3 μm. Microconidiogenous cells phialidic, discrete or integrated, hyaline, smooth, cylindrical, determinate, with prominent periclinal thickening, 5–11 × 2–2.5 μm. Microconidia medium brown, thin-walled, finely verruculose, guttulate, aseptate, subcylindrical to narrowly ellipsoid with rounded ends, (7–)10–15(–17) × (2.5–)3(–4) μm.
Culture characteristics: Colonies on MEA at 25 °C in the dark after 1 wk: flat, spreading, with moderate aerial mycelium; surface pale mouse-grey with patches of dirty white, reverse mouse-grey.
Material examined: USA, Hawaii, Maui, on leaf of Protea sp., 16 Dec. 1998, P.W. Crous & M.E. Palm, (holotype CBS H-23111, culture ex-type CPC 2169 = CBS 114569).
Notes: In the reassessment of Botryosphaeriaceae and allied taxa published by Marincowitz et al. (2008b), the ITS DNA data could not distinguish CBS 114569 from isolates of S. proteae. However, in the recent study of Yang et al. (2017), the combined sequence dataset (ITS, rpb2 and tef1), showed CBS 114569 to cluster basal to S. hawaiiensis. Morphologically, conidia of isolates of CBS 114569 [(17–)20–25(–27) × (4–)4.5–5(–6) μm] are also smaller than those of S. hawaiiensis [(17–)24–30(–38) × (4–)5–7(–8) μm; Yang et al. 2017], and thus this isolate is herewith introduced as a new species, S. protearum.
Authors: Y. Marin-Felix, S. Marincowitz & P.W. Crous
Thyrostroma Höhn., Sitzungsber. Kaiserl. Akad. Wiss., Math.-Naturwiss. Cl., Abt. 1 120: 472 (94 repr.). 1911. Fig. 63.
Fig. 63.
A–H. Thyrostroma cornicola (ex-type CBS 141280). A, B. Symptomatic leaves of Cornus officinalis. C. Sporodochia on PNA. D. Sporulation on PNA. E–H. Conidiogenous cells giving rise to conidia. I–O.Thyrostroma franseriae (ex-type CBS 487.71). I. Sporulation on PNA. J–O. Conidiogenous cells giving rise to conidia. P. Conidia. Scale bars: 20 μm. Pictures B–F, H taken from Crous et al. (2016c).
Classification: Dothideomycetes, Pleosporomycetidae, Pleosporales, Dothidotthiaceae.
Type species: Thyrostroma compactum (Sacc.) Höhn. Holotype could not be located, and a neotype from Europe is required.
DNA barcode (genus): LSU.
DNA barcodes (species): ITS, tef1. Table 20. Fig. 64.
Table 20.
DNA barcodes of accepted Thyrostroma spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | |
|---|---|---|---|---|
| ITS | tef1 | |||
| Thyrostromacompactum | CBS 335.37 | KY905670 | KY905681 | Present study |
| T. cornicola | CBS 141280T | KX228248 | KX228372 | Crous et al. (2016c) |
| T. franseriae | CBS 487.71T | KX228249 | KY905680 | Crous et al. (2016c), present study |
| CBS 700.70 | KX228250 | KY905682 | Crous et al. (2016c), present study | |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. T indicates ex-type strains.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; tef1: partial translation elongation factor 1-alpha gene.
Fig. 64.
RAxML phylogram obtained from the combined ITS (531 bp) and tef1 (389 bp) sequences of members of Dothidotthiaceae. The new species proposed is indicated in bold. RAxML bootstrap support (BS) values above 70 % and Bayesian posterior probability scores ≥ 0.95 are shown in the nodes. Numbers between parentheses correspond to GenBank accession numbers of ITS and tef1 sequences, respectively. T and ET indicate ex-type and ex-epitype strains, respectively. TreeBASE: S20877.
Sexual morph unknown. Conidiomata sporodochial, punctiform, dark brown or black. Stroma immersed to superficial, brown. Conidiophores brown, finely roughened, cylindrical to subcylindrical, 1–3-septate. Conidiogenous cells brown, subcylindrical, finely roughened, proliferating percurrently at apex. Conidia cylindrical, clavate, or ellipsoid to fusoid, pale to medium brown, smooth-walled, with (1–)4 transverse septa, and 0–3 oblique or longitudinal septa, rounded at the apex, base truncate.
Culture characteristics: Colonies reaching 90 mm diam after 2 wk, with sparse or fluffy aerial mycelium. Colonies on MEA, PDA and OA showing different shades of grey or chestnut to umber.
Optimal media and cultivation conditions: MEA, PDA and OA at 25 °C.
Distribution: Asia, Europe and North America.
Hosts: Pathogens of Ulmus spp., Sambucus caerulea, Styphnolobium japonicum, Tilia spp., and Cornus officinalis.
Disease symptoms: Thyrostroma canker, dieback and leaf spots.
Notes: Thyrostroma was introduced in 1911 in order to accommodate T. compactum (von Höhnel 1911). Despite being described more than 100 years ago, the phylogenetic position of Thyrostroma remains unresolved. Thyrostroma was considered the asexual morph of Dothidotthia by Phillips et al. (2008). Subsequently, Slippers et al. (2013) placed Thyrostroma in the Botryosphaeriaceae based on morphology, since molecular data of Thyrostroma spp. were lacking. In the phylogenetic trees based on LSU sequences (Fig. 49), the type species of Thyrostroma, T. compactum, does not cluster with Dothidotthia (Dothidotthiaceae), demonstrating that these genera are not congeneric, as was recently mentioned by Crous et al. (2016c). However, Thyrostroma did cluster in the Dothidotthiaceae clade, as originally proposed by Phillips et al. (2008).
References: Ellis, 1959, Ellis, 1971, Crous et al. 2016c (morphology).
Thyrostroma franseriae Crous, sp. nov. MycoBank MB820825. Fig. 63.
Etymology: Named after the host genus from which it was collected, Franseria.
Sporodochia dark brown, punctiform, up to 250 μm diam. Stromata brown, superficial, 100–150 μm diam. Conidiophores brown, finely roughened, subcylindrical, 0–1-septate, 10–18 × 6–11 μm. Conidiogenous cells brown, subcylindrical, finely roughened, proliferating percurrently at apex, 5–10 × 6–11 μm. Conidia brown, ellipsoid to fusoid, with 2–4 oblique or longitudinal septa, 1–3 transverse septa, apex broadly obtuse, base truncate, 8–9 μm diam, (25–)28–33(–35) × (18–)20–25 μm.
Culture characteristics: Colonies flat, spreading, with sparse aerial mycelium and feathery margins, reaching 60 mm diam after 2 wk on MEA, PDA and OA; surface and reverse iron-grey.
Material examined: USA, Nevada, Death Valley, on dead leaf of Franseria sp., 7 Jul. 1970, F.W. Went (holotype CBS H-23112, culture ex-type CBS 487.71); Nevada, north end of Death Valley, on green, living leaf of Franseria sp., Jul. 1970, F.W. Went, CBS H-18568, culture CBS 700.70.
Notes: Thyrostroma franseriae is known from two isolates, both of which were collected from leaves of Franseria sp. in Death Valley, Nevada (USA) in 1970. Morphologically, isolate CBS 700.70 differs from CBS 487.71 in having larger conidia that are more cylindrical, clavate to ellipsoid, with 2–4 transverse septa, 2–8 oblique or longitudinal septa, 40–65 × 18–25 μm. However, the two isolates are phylogenetically indistinguishable (Fig. 64).
Thyrostroma compactum is a European species originally described from Ulmus in Italy. One such isolate was available for study, namely CBS 335.37, collected by J.C. Carter (a US-based researcher), but the origin of this strain remains unknown, and it proved to be sterile in culture. Phylogenetically, however, CBS 335.37 is distinct from T. franseriae (Fig. 64), although we could not confirm that CBS 335.37 is authentic for the name it was deposited under by J.C. Carter.
Authors: Y. Marin-Felix & P.W. Crous
Venturia Sacc., Syll. fung. (Abellini) 1: 586. 1882. Fig. 65.
Fig. 65.
Venturia spp. A–C. Disease symptoms. A. Symptoms caused by Venturia martianoffiana (HMAS 247008). B. Symptoms caused by Venturia catenospora (HMAS 247006). C. Symptoms caused by Venturia fuliginosa (HMAS 247007). D–O. Sexual morphs. D–F. Ascomata on the host. D.Venturia chinensis (HMAS 246485). E.Venturia canadensis (NY 00914436). F.Venturia atriseda (K 189232). G–J. Asci. G.Venturia cephalariae (K 189236). H.Venturia chinensis (HMAS 246485). I.Venturia inaequalis (NY 00914442). J.Venturia asperata (PDD 31846). K–O. Ascospores. K.Venturia atriseda (K 189232). L.Venturia cephalariae (K 189236). M.Venturia carpophila (K 189234). N.Venturia inaequalis (NY 00914442). O.Venturia helvética (ZT 49111). P–T. Asexual morphs. P. Conidial chains of Venturia phaeosepta (ex-type CGMCC 3.18368). Q. Conidiophores sporulation of Venturia inaequalis (CGMCC 3.18372). R. Conidia of Venturia inaequalis (CGMCC 3.18372). S. Fasciculate conidiophores of Venturia pyrina (HMAS 03923). T. Conidium of Venturia pyrina (HMAS 03923). Scale bars: D = 300 μm; E, F = 0.2 mm; G, H, J–M, Q, T = 10 μm; I, N, O, S = 5 μm; P, R = 20 μm.
Synonyms: Fusicladium Bonord., Handb. Mykol.: 80. 1851.
Apiosporina Höhn., Sitzungsber. Kaiserl. Akad. Wiss., Math.-Naturw. Cl., Abt. 1, 119: 439. 1910.
Metacoleroa Petr., Ann. Mycol. 25: 332. 1927.
Caproventuria U. Braun, A Monograph of Cercosporella, Ramularia and Allied Genera (Phytopathogenic Hyphomycetes) 2: 396. 1998.
Pseudocladosporium U. Braun, A Monograph of Cercosporella, Ramularia and Allied Genera (Phytopathogenic Hyphomycetes) 2: 392. 1998.
Classification: Dothideomycetes, Pleosporomycetidae, Venturiales, Venturiaceae.
Type species: Venturia inaequalis (Cooke) G. Winter. Type material in Kew: IMI 47413.
DNA barcode (genus): LSU.
DNA barcodes (species): ITS, tef1, tub2. Table 21. Fig. 66.
Table 21.
DNA barcodes of accepted Venturia spp.
| Species | Isolates1 | GenBank accession numbers2 |
References | ||
|---|---|---|---|---|---|
| ITS | tef1 | tub2 | |||
| Venturiaanemones | CBS 370.55 | EU035447 | KF853965 | KF808264 | Crous et al. (2007d), Hamelin et al. (unpubl. data) |
| V. aucupariae | CBS 365.35 | EU035450 | – | – | Crous et al. (2007d) |
| V. catenospora | CBS 447.91T | EU035427 | KF853957 | KF808256 | Crous et al. (2007d), Hamelin et al. (unpubl. data) |
| V. chinensis | CGMCC 3.17685T | KP689596 | – | – | Zhang et al. (2016) |
| V. fraxini | CBS 140930 | KT823548 | KT823582 | KT823514 | Ibrahim et al. (2016) |
| V. fuliginosa | CGMCC 3.18370T | KU220965 | – | – | Shen et al. (2017) |
| V. helvetica | CBS 474.61 | EU035458 | KF853974 | KF808274 | Crous et al. (2007d), Hamelin et al. (unpubl. data) |
| V. hystrioides | CBS 117727 | EU035459 | KF853975 | Crous et al. (2007d) | |
| V. inaequalis | CBS 476.61 | EU282478 | GU456288 | – | Sanchez-Torres et al., 2009, Zhang et al., 2011 |
| V. inopina | MYA 2852T | AY177406 | – | – | Newcombe (2003) |
| V. macularis | CBS 477.61 | EU035462 | KF853977 | KF808277 | Crous et al. (2007d), Hamelin et al. (unpubl. data) |
| V. martianoffiana | CGMCC 3.18376 | KU985131 | – | – | Present study |
| V. nashicola | OYO-1 | HQ434393 | HQ434349 | HQ434437 | Zhao et al. (2012) |
| V. orni | CBS 140924T | KT823564 | KT823598 | KT823530 | Ibrahim et al. (2016) |
| V. phaeosepta | CGMCC 3.18368T | KU985133 | – | – | Present study |
| V. polygoni-vivipari | CBS 114207 | EU035466 | KF853984 | KF808284 | Crous et al. (2007d) |
| V. pyrina | 38995 | HQ434425 | HQ434381 | HQ434469 | Zhao et al. (2012) |
| V. saliciperda | CBS 480.61 | EU035471 | – | – | Crous et al. (2007d) |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CGMCC: Chinese General Microbiological Culture Collection Center, Beijing, China; MYA: the American Type Culture Collection; OYO: Private collection. T indicates ex-type strain.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; tef1: partial translation elongation factor 1-alpha gene; tub2: partial beta-tubulin gene.
Fig. 66.
Maximum likelihood tree generated from a sequence analysis of the ITS rDNA dataset. The outgroup is Fusicladium africanum CPC 12829. Maximum likelihood bootstrap support values above 50 % are shown at the nodes and based on 1 000 replicates. Bayesian posterior probability values above 0.70 are shown at the nodes. The species from poplar are in bold. Numbers between parentheses correspond to GenBank accession numbers. T indicates ex-type strain. TreeBASE: S21068.
Ascomata pseudothecial, globose, subglobose, black, initially immersed, becoming erumpent, solitary, scattered or gregarious, covered with setae; ostiole central, papillate; ascomatal wall composed of a few layers of pigmented cells of textura angularis, which is of equal thickness or slightly thickened at apex. Hamathecium comprising septate, filiform pseudoparaphyses, evanescent in mature ascomata. Asci bitunicate, oblong to obclavate, fissitunicate dehiscence unknown, with or without a short, thick pedicel, rounded at the apex with an inconspicuous ocular chamber. Ascospores obliquely uniseriate, partially overlapping to biseriate, especially at the base, ellipsoidal, with broadly rounded ends, pale brown, 1-septate, slightly constricted at the septum, the upper cell shorter than the lower one, smooth-walled. Conidiophores single, sometimes arranged in small groups, straight to flexuous, pale olivaceous to dark brown, unbranched or occasionally branched, thin- to slightly thick-walled, conidiophores often reduced to conidiogenous cells or composed of several cells. Conidiogenous nodes smooth to verruculose. Conidia in simple or branched acropetal chains, ellipsoid-ovoid, obovoid, fusoid, obclavate-subcylindrical, canoe-shaped, straight to curved, subhyaline to medium brown, but mostly olivaceous, thin- to thick-walled, smooth to verruculose, 0–3(–4)-euseptate, germinating by production of germination tubes from middle or polar cells; hila often denticle-like, somewhat protuberant, unthickened or almost so, occasionally somewhat darkened-refractive; septum ontogeny: first septum median to sub-median.
Culture characteristics: Colonies on PDA fuscous black, and reverse dark fuscous-black, with moderate aerial mycelium and regular, but feathery margins. Colonies normally reaching not more than 15 mm diam after 1 mo on PDA at 25 °C in the dark.
Optimal media and cultivation conditions: PDA, MEA and CMA. Optimal growing temperature is 24–28 °C. Sometimes grows faster after cold-shock under 10 °C for 1 wk.
Distribution: Worldwide.
Hosts: Mainly on woody dicotyledonous plants. Twenty-four families of plants have been reported hosting venturiaceous fungi, i.e. Aceraceae, Amaryllidaceae, Asteraceae, Betulaceae, Caprifoliaceae, Cornaceae, Dipsacaceae, Ericaceae, Fagaceae, Gentianaceae, Geraniaceae, Iridaceae, Juncaginaceae, Liliaceae, Onagraceae, Oleaceae, Polygonaceae, Ranunculaceae, Rhamnaceae, Rosaceae, Rubiaceae, Salicaceae, Sapindaceae and Ulmaceae (Barr, 1968, Barr, 1989, Sivanesan, 1977). After studying a large number of type materials of Venturia species, many have been found to be representative of other genera (Shen et al. in prep.).
Disease symptoms: Leaf spots, flower and fruit canker.
Notes: Species of Venturia are widely distributed in the northern temperate region of the world, and are saprobic or parasitic on a large variety of dicotyledonous plants. Venturia comprises 198 species according to Index Fungorum. Based on the morphology of type specimens studied, the diagnostic characteristics of Venturia are as follows: Ascomata immersed, semi-immersed or superficial, scattered or gregarious, often papillate and ostiolate with setae. Hamathecium narrowly cellular, hyaline, evanescent in mature ascomata. Asci 8-spored, bitunicate, fissitunicate, broadly cylindrical to obclavate, usually lacking a pedicel. Ascospores pale olivaceous to brown, 1-septate, usually asymmetrical. Morphological discrimination of the sexual morph is limited, and the asexual morph is more informative (Sivanesan 1977). The genus is morphologically comparable to the Mycosphaerella morph of Ramularia in having bitunicate, oblong to obclavate asci with a short, thick pedicel or pedicel lacking, ellipsoidal, 1-septate ascospores which are slightly constricted at the septum. However, Venturia can be distinguished from the sexual morph of Ramularia by its setose ascomata, pale olivaceous to brown and asymmetrical ascospores. In addition, pseudoparaphyses are lacking in the sexual morph of Ramularia. Although several studies have been conducted on the phylogeny of Venturia, they mostly relied on rDNA sequences of the ITS and LSU, which proved insufficient in distinguishing some species (Crous et al., 2007d, Zhang et al., 2011). More genes, especially protein coding genes are required to provide a better resolution at the species level.
References: Menon, 1956, Nüesch, 1960, Barr, 1968, Sivanesan, 1977 (morphology); Schubert et al. 2003 (morphology of asexual stage); Crous et al., 2007d, Zhang et al., 2011, Zhang et al., 2016 (morphology and phylogeny).
Venturia martianoffiana (Thüm.) Y. Zhang ter & J.Q. Zhang, comb. nov. Mycobank MB821418.
Basionym: Cladosporium martianoffianum Thüm., Byull. Moskovsk. Obshch. Isp. Prir., Otd. Biol. 55: 74. 1880.
Venturia phaeosepta Y. Zhang ter & J.Q. Zhang, sp. nov. MycoBank MB817355. Fig. 67.
Fig. 67.
Venturia phaeosepta (ex-type CGMCC3.18368). A–F. On MEA. A. Colony on MEA. B. Conidial chains. C–D. Ramoconidia and conidia. E. Germinating conidium. F. Conidium. G–M. On leaves. G. Leaves infected by Venturia phaeosepta. H. Conidiophores and conidia. I. Conidiogenous cells giving rise conidia. J. Conidiogenous cell. K. Conidial chains. L. Conidia. M. Germinating conidium. Scale bars: B–D = 20 μm; E, F = 10 μm; G = 0.5 cm; H–M = 10 μm.
Etymology: Latin “phaeo-”, in reference to “dark” septum.
On Populus: Leaf spots amphigenous, subcircular to angular, 1.5–13 mm wide, often confluent, diffuse, mostly spread along leaf veins, dark brown to black, with an irregular margin. Colonies amphigenous, caespitose, greenish brown to blackish. Mycelium mainly subcuticular. Stromata variable in size, composed of pale olivaceous to brown, angular to rounded, thick-walled, pseudoparenchymatous cells, 4–8 μm diam. Conidiophores solitary or loosely fasciculate, arising mostly from stromata or from hyphae, erect, straight, sometimes flexuous at the apex, unbranched or apically branched, 12–29 × 5–8 μm, 0–1-septate, pale to medium brown, smooth, with somewhat thickened walls, sometimes conidiophores reduced to conidiogenous cells. Conidiogenous cells integrated, terminal, 15–27 × 5–8 μm, with a 1–2(–3) denticle-like conidiogenous loci, proliferation sympodial, loci unthickened, not or only somewhat darkened-refractive, 2–3 μm wide. Conidia in simple or branched chains, clavate, subcylindrical, ellipsoid or rarely fusiform, (12–)16–29 × 4–7 μm, pale olivaceous brown, 0–1(–3)-septate, smooth, tapering towards both ends, apex mostly truncate, occasionally rounded or pointed, base truncate; hila often somewhat thickened and darkened-refractive, 1.5–3 μm wide. Sexual morph not observed. On MEA: Mycelium consisting of pale olivaceous, smooth, branched, 1.5–3 μm wide hyphae. Conidiophores integrated, produced in the middle of the mycelium, 3–6-septate, visible as small, protruding, denticle-like loci, up to 92 μm long, 5–6 μm wide. Conidiogenous cells subcylindrical, 15–25 × 5–7 μm, pale to medium olivaceous, smooth, tapering to 1–2 apical truncate loci, 2–4 μm wide. Conidia pale olivaceous, smooth, subcylindrical to narrowly ellipsoid, occurring in simple or branched chains, 0–1(–2)-septate, tapering towards subtruncate ends, ends 2–4 μm wide, aseptate conidia 12–21.5 × 5–7 μm, septate conidia up to 28 μm long and 5–7 μm wide; basal hila usually thickened and darkened-refractive; microcyclic conidiation common in older cultures. Sexual morph not observed.
Culture characteristics: Colonies reaching 43 mm diam after 1 mo on PDA at 25 °C in the dark. Colonies sporulated, erumpent, spreading, with abundant aerial mycelium and feathery to smooth margins; grey olivaceous (surface), reverse dark olivaceous.
Habitat and distribution: China (Henan, Shannxi), on leaves of Populus spp.
Material examined: China, Henan, Puyang City Academy Experimental Farm, on leaves of Populus × euramericana cv. 74/76 (sect. Aigeiros), 20 May 2015, W. He (holotype, HMAS 246998, culture ex-type CGMCC3.18368); on leaves of Populus × euramericana cv. 74/76 (sects. Aigeiros), Y.F. Zhang, 20 Jun. 2015 (paratype, HMAS 246999, CGMCC3.18371); 6 August 2015 (paratype, HMAS 247000, CGMCC3.18373); 7 Aug. 2015 (paratype, HMAS 247002, CGMCC3.18374); 8 Aug. 2015 (paratype, HMAS 247001, CGMCC3.18375); Shanxi, Yangling, on leaves of Populus sp. (sects. Aigeiros), 4 Sep. 2015, Y.F. Zhang (paratype, HMAS 247004, CGMCC3.18378); ibid. (paratype, HMAS 247005, CGMCC3.18379).
Notes: Among the reported venturiaceous species occurring on Populus, the asexual morph of Venturia phaeosepta is more comparable with Venturia martianoffiana and F. romellianum in the morphology of the conidiophore and mode of conidia production (Schubert et al. 2003). Venturia phaeosepta, however, can readily be distinguished from V. martianoffiana by its 1–2(–3) apical denticle-like conidiogenous loci (vs. a single or several (>3) conidiogenous loci of V. martianoffiana). Venturia phaeosepta differs from F. romellianum by its septate (vs. chiefly aseptate) conidia (Schubert et al. 2003).
Authors: Y. Zhang, M. Shen & J.Q. Zhang
Wilsonomyces Adask. et al., Mycotaxon 37: 283. 1990. Fig. 68.
Fig. 68.
Wilsonomyces carpophilus (ex-epitype CBS 231.89). A. Conidiomata. B–G. Conidiogenous cells giving rise to conidia. E. Conidia. Scale bars: 10 μm.
Classification: Dothideomycetes, Pleosporomycetidae, Pleosporales, Dothidotthiaceae.
Type species: Wilsonomyces carpophilus (Lév.) Adask. et al. Lectotype: plate 7, fig. 5 in Léveillé JH. 1843. Ann. Sci. Nat., Bot., sér. 2 19: 215. Epitype and ex-epitype culture designated here: CBS H-23113, CBS 231.89.
DNA barcode (genus): LSU.
DNA barcodes (species): ITS, tef1. Table 22. Fig. 64.
Table 22.
DNA barcodes of accepted Wilsonomyces sp.
| Species | Isolates1 | GenBank accession numbers2 |
References | |
|---|---|---|---|---|
| ITS | tef1 | |||
| Wilsonomycescarpophilus | CBS 231.89ET | KY905672 | KY905684 | Present study |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. ET indicates ex-epitype strain.
ITS: internal transcribed spacers and intervening 5.8S nrDNA; tef1: partial translation elongation factor 1-alpha gene.
Sexual morph unknown. Conidiomata sporodochial, usually punctiform, tan to olivaceous brown, finally becoming black. Stroma present in host tissue, compact, immersed, erumpent, discoid on leaves, fusoid on twigs. Conidiophores macronematous, mononematous, densely compacted, straight or flexuous, branched or not, subcylindrical, geniculate, smooth, subhyaline to pale brown, cicatrised, 1–4-septate. Conidiogenous cells terminal and intercalary, subcylindrical, subhyaline, smooth, proliferating sympodially at apex, scars unthickened. Conidia holoblastic, solitary, dry, acrogenous, simple, thick-walled, cylindrical, clavate, ellipsoidal or fusiform, occasionally forked, rounded to acute at the apex, truncate at the base, generally (2–)3–5(–10) transverse with occasionally 1–2 oblique septa, rarely with 1 longitudinal septum, subhyaline, becoming medium brown to golden-brown, dark olivaceous to black in mass, conidia in vivo are larger (adapted from Adaskaveg et al. 1990).
Culture characteristics: Growth moderate, PDA plates covered in 4 wk, mostly consisting of submerged hyphae with sparse aerial mycelium. In cultures grown in the dark, aerial mycelium sub-felty to felty, initially white becoming pale olive-grey to greyish olive, occasionally olive-ochre. In cultures grown exposed to light, submerged light brownish olive to olive-brown hyphae radiate outward from mycelial mat; aerial hyphae buffy-brown; olivaceous black to black conidia produced in mass (more details in Adaskaveg et al. 1990).
Optimal media and cultivation conditions: PDA at 20 °C on the laboratory bench.
Distribution: Worldwide.
Hosts: Pathogens mainly of Prunus spp., particularly peaches and apricots, but also of other members of the family Rosaceae, i.e. Pyrus communis, Malus domestica, Sorbus aucuparia and Cydonia oblonga. Also reported in Cleome sp. (Cleomaceae) and Quercus ilex (Fagaceae).
Disease symptoms: Wilsonomyces causes a disease known as shot-hole disease because of the symptoms on the host leaves: small circular purple lesions with pale centres that gradually enlarge and become necrotic in the centre until the centre falls out. This genus also produces necrotic spots on the twigs and necrotic lesions on fruit.
Notes: Wilsonomyces is a monotypic genus. Wilsonomyces carpophilus was initially described as a new species of Helminthosporium, and was subsequently transferred to different genera until Adaskaveg et al. (1990) introduced Wilsonomyces to accommodate it. The taxonomy of the genus was controversial, and Sutton (1997) regarded it as synonym of Thyrostroma. However, all the strains of Wilsonomyces carpophilus included in the phylogenetic analysis based on LSU, ITS and tef1 (Fig. 49, Fig. 64) sequences were located in a clade separate from the rest of the taxa incorporated in the tree including the type species of Thyrostroma, T. compactum. Therefore, it is herewith supported that Wilsonomyces represents a distinct genus. Finally, its location in the Dothidotthiaceae is also supported.
References: Ellis, 1959, Adaskaveg et al., 1990 (morphology); Ahmadpour et al. 2012a (morphology and pathogenicity).
Wilsonomyces carpophilus (Lév.) Adask. et al., Mycotaxon 37: 283. 1990. Fig. 68.
Basionym: Helminthosporium carpophilum Lév., Annls Sci. Nat., Bot., sér. 2 19: 215. 1843.
Synonyms: Clasterosporium carpophilum (Lév.) Aderh., Landw. Jahrb. 30: 815. 1901.
Coryneum carpophilum (Lév.) Jauch, Int. Bull. Pl. Protect. 14: 99. 1940.
Stigmina carpophila (Lév.) M.B. Ellis, Mycol. Pap. 72: 56. 1959.
Sciniatosporium carpophilum (Lév.) Morgan-Jones, Canad. J. Bot. 49: 995. 1971.
Sporocadus carpophilus (Lév.) Arx, Gen. Fungi Sporul. Cult., Edn 3 (Vaduz): 224. 1981.
Thyrostroma carpophilum (Lév.) B. Sutton, Arnoldia 14: 34. 1997.
For additional synonyms see Adaskaveg et al. (1990).
Conidiomata sporodochial, brown, with immersed to erumpent stromata, 30–200 μm diam. Conidiophores subcylindrical, branched or not, geniculate, 10–70 × 5–7 μm, subhyaline to pale brown, smooth, 1–4-septate. Conidiogenous cells terminal and intercalary, subcylindrical, subhyaline, smooth, 10–30 × 5–7 μm, proliferating sympodially, scars unthickened, 3.5–5 μm diam. Conidia narrowly ellipsoid to subcylindrical or fusoid, subhyaline, becoming medium brown to golden-brown, smooth, with (2–)3–7(–11) dark, transverse septa, rarely with any oblique septum, (27–)32–45(–55) × (12–)13–14(–16) μm, base truncate, 4–6 μm diam in vitro. Conidia in vivo are larger, namely 20–90 × 7–16 μm (adapted from Adaskaveg et al. 1990).
Culture characteristics: Colonies flat, spreading, with moderate aerial mycelium, and feathery margins, reaching 40 mm diam after 2 wk. On MEA, PDA and OA surface umber, reverse isabelline.
Material examined: Lectotype: plate 7, fig. 5 in Léveillé JH. 1843. Ann. Sci. Nat., Bot., sér. 2 19: 215. Unknown country, on petiole of Prunus subhirtella, 1989, J.W. Veenbaas-Rijks (epitype designated here CBS H-23113, MBT376057, culture ex-epitype CBS 231.89).
Notes: The holotype of W. carpophilus was not located by Adaskaveg et al. (1990) when they introduced the genus Wilsonomyces to accommodate Helminthosporium carpophilum, although they searched in several herbaria in Europe. The holotype was probably lost when Léveillé's collection was destroyed in the Franco-Prussian War in 1870–1871. Therefore, Adaskaveg et al. (1990) selected the drawings of Léveillé present in the original description of this taxon as lectotype (Ann. Sci. Nat., Bot., sér. 2 19: 215, plate 7, fig. 5). To fix the application of the generic name, an epitype for this species is therefore designated here.
Authors: Y. Marin-Felix & P.W. Crous
Acknowledgements
Yasmina Marin-Felix is grateful for the financial support received from the Vice-Chancellor's postdoctoral fellowship programme from University of Pretoria, South Africa. Min Shen, Jiaqi Zhang and Ying Zhang are supported by the National Natural Science Foundation of China (General Program) (31370063). Sincere thanks are due to the curators Rossella Marcucci (PAD) and Lisa A. Castlebury (BPI).
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Contributor Information
Y. Marin-Felix, Email: y.marin@westerdijkinstitute.nl.
P.W. Crous, Email: p.crous@westerdijkinstitute.nl.
References
- Adaskaveg J.E., Ogawa J.M., Butler E.E. Morphology and ontogeny of conidia in Wilsonomyces carpophilus, gen. nov. and comb. nov., causal pathogen of shot hole disease of Prunus species. Mycotaxon. 1990;37:275–290. [Google Scholar]
- Ahmadpour A., Ghosta Y., Javan-Nikkhah M. Study on morphology, pathogenicity and genetic diversity of Wilsonomyces carpophilus isolates, the causal agent of shot hole of stone fruit trees based on RAPD-PCR in Iran. Archives of Phytopathology and Plant Protection. 2012;45:1–11. [Google Scholar]
- Ahmadpour A., Heidarian Z., Donyadoost-Chelan M. A new species of Bipolaris from Iran. Mycotaxon. 2012;120:301–307. [Google Scholar]
- Aime M.C. Toward resolving family-level relationships in rust fungi (Uredinales) Mycoscience. 2006;47:112–122. [Google Scholar]
- Al Adawi A.O., Barnes I., Khan I.A. Ceratocystis manginecans associated with a serious wilt disease of two native legume trees in Oman and Pakistan. Australasian Plant Pathology. 2013;42:179–193. [Google Scholar]
- Alaei H., de Backer M., Nuytinck J. Phylogenetic relationships of Puccinia horiana and other rust pathogens of Chrysanthemum x morifolium based on rDNA ITS sequence analysis. Mycological Research. 2009;113:668–683. doi: 10.1016/j.mycres.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Alfenas R.F., Lombard L., Pereira O.L. Diversity and potential impact of Calonectria species in Eucalyptus plantations in Brazil. Studies in Mycology. 2015;80:89–130. doi: 10.1016/j.simyco.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfenas R.F., Pereira O.L., Ferreira M.A. Calonectria metrosideri, a highly aggressive pathogen causing leaf blight, root rot, and wilt of Metrosideros spp. in Brazil. Forest Pathology. 2013;43:257–265. [Google Scholar]
- Alvarez L.V., Groenewald J.Z., Crous P.W. Revising the Schizoparmaceae: Coniella, Pilidiella and Schizoparme. Studies in Mycology. 2016;85:1–34. doi: 10.1016/j.simyco.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrie R.M., Schoch C.L., Hedges R. Homologs of ToxB, a host-selective toxin gene from Pyrenophora tritici-repentis, are present in the genome of sister-species Pyrenophora bromi and other members of the Ascomycota. Fungal Genetics and Biology. 2008;45:363–377. doi: 10.1016/j.fgb.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Ariyawansa H.A., Hyde K.D., Jayasiri S.C. Fungal diversity notes 111–252-taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity. 2015;75:27–274. [Google Scholar]
- Ariyawansa H.A., Phukhamsakda C., Thambugala K.M. Revision and phylogeny of Leptosphaeriaceae. Fungal Diversity. 2015;74:19–51. [Google Scholar]
- Arzanlou M., Bakhshi M., Karimi K. Multigene phylogeny reveals three new records of Colletotrichum spp. and several new host records for the mycobiota of Iran. Journal of Plant Protection Research. 2015;55:198–211. [Google Scholar]
- Aveskamp M.M., de Gruyter J., Woudenberg J.H.C. Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in Mycology. 2010;65:1–60. doi: 10.3114/sim.2010.65.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aveskamp M.M., Verkley G.J., de Gruyter J. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia. 2009;101:363–382. doi: 10.3852/08-199. [DOI] [PubMed] [Google Scholar]
- Aveskamp M.M., Woudenberg J.H., de Gruyter J. Development of taxon-specific sequence characterized amplified region (SCAR) markers based on actin sequences and DNA amplification fingerprinting (DAF): a case study in the Phoma exigua species complex. Molecular Plant Pathology. 2009;10:403–414. doi: 10.1111/j.1364-3703.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.J., Harrington T.C., Krauss U. Genetic variability and host specialization in the Latin American clade of Ceratocystis fimbriata. Phytopathology. 2003;93:1274–1284. doi: 10.1094/PHYTO.2003.93.10.1274. [DOI] [PubMed] [Google Scholar]
- Barber P.A., Burgess T.J., Hardy GEStJ. Botryosphaeria species from Eucalyptus in Australia are pleoanamorphic, producing Dichomera synanamorphs in culture. Mycological Research. 2005;109:1347–1363. doi: 10.1017/s0953756205003989. [DOI] [PubMed] [Google Scholar]
- Barimani M., Pethybridge S.J., Vaghefi N. A new anthracnose disease of pyrethrum caused by Colletotrichum tanaceti sp. nov. Plant Pathology. 2013;62:1248–1257. [Google Scholar]
- Barnes I., Roux J., Wingfield B.D. Ceratocystis pirilliformis, a new species from Eucalyptus nitens in Australia. Mycologia. 2003;95:865–871. [PubMed] [Google Scholar]
- Barr M.E. The Venturiaceae in North America. Canadian Journal of Botany. 1968;46:799–864. [Google Scholar]
- Barr M.E. The Venturiaceae in North America: revisions and additions. Sydowia. 1989;41:25–40. [Google Scholar]
- Bateman G.L. Pathogenicity of fungi associated with winter loss and injury in white lupin. Plant Pathologist. 1997;46:157–167. [Google Scholar]
- Batra L.R. Monilinia gaylussaciae, a new species pathogenic on huckleberries (Gaylussacia) in North America. Mycologia. 1988;80:653–659. [Google Scholar]
- Batra L.R. World species of Monilinia (Fungi): their ecology, biosystematics and control. Mycologia Memoir. 1991;16:1–246. [Google Scholar]
- Beenken L. Pucciniales on Annona (Annonaceae) with special focus on the genus Phakopsora. Mycological Progress. 2014;13:791–809. [Google Scholar]
- Beenken L., Wood A. Puccorchidium and Sphenorchidium, two new genera of Pucciniales on Annonaceae related to Puccinia psidii and the genus Dasyspora. Mycological Progress. 2015;14:1–13. [Google Scholar]
- Begoude B.A.D., Slippers B., Wingfield M.J. Botryosphaeriaceae associated with Terminalia catappa in Cameroon, South Africa and Madagascar. Mycological Progress. 2010;9:101–123. [Google Scholar]
- Benduhn B., Krauthausen H.J., Schult T. [Regulation of necrosis in organic elderberry growing]. Dienstleistungszentrum Ländlicher Raum-Rheinpfalz, Kompetenzzentrum Gartenbau; D-Rheinbach: 2011. Regulierung der Doldenwelke im ökologischen Holunderanbau.http://orgprints.org/20875/ [Google Scholar]
- Bensch K., Braun U., Groenewald J.Z. The genus Cladosporium. Studies in Mycology. 2012;72:1–401. doi: 10.3114/sim0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch K., Groenewald J.Z., Braun U. Common but different: the expanding realm of Cladosporium. Studies in Mycology. 2015;82:23–74. doi: 10.1016/j.simyco.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch K., Groenewald J.Z., Dijksterhuis J. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales) Studies in Mycology. 2010;67:1–94. doi: 10.3114/sim.2010.67.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee M.L., Pirseyedi M., Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977. [Google Scholar]
- Berner D., Cavin C., Woudenberg J.H.C. Assessment of Boeremia exigua var. rhapontica, as a biological control agent of Russian knapweed (Rhaponticum repens) Biological Control. 2015;81:65–75. [Google Scholar]
- Berraf-Tebbal A., Guerreiro M.A., Phillips A.J.L. Phylogeny of Neofusicoccum species associated with grapevine trunk diseases in Algeria, with description of Neofusicoccum algeriense sp. nov. Phytopathologia Mediterranea. 2014;53:416–427. [Google Scholar]
- Boerema G.H., de Gruyter J., van Kesteren H.A. Contributions towards a monograph of Phoma (Coelomycetes) – III. 1. Section Plenodomus: taxa often with a Leptosphaeria teleomorph. Persoonia. 1994;15:431–487. [Google Scholar]
- Boerema G.H., de Gruyter J., Noordeloos M.E. CABI Publishing; Wallingford, UK: 2004. Phoma identification manual. Differentiation of specific and infra-specific taxa in culture. [Google Scholar]
- Bragança C.A., Damm U., Baroncelli R. Species of the Colletotrichum acutatum complex associated with anthracnose diseases of fruit in Brazil. Fungal Biology. 2016;120:547–561. doi: 10.1016/j.funbio.2016.01.011. [DOI] [PubMed] [Google Scholar]
- Braun U., Crous P.W., Dugan F.M. Phylogeny and taxonomy of cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s. str. Mycological Progress. 2003;2:3–18. [Google Scholar]
- Bruckart W.L., Eskandari F.M., Berner D.K. Comparison of Puccinia acroptili from Eurasia and the USA. Botany. 2012;90:465–471. [Google Scholar]
- Burgess T.I., Barber P.A., Hardy GEStJ. Botryosphaeria spp. associated with eucalypts in Western Australia, including the description of Fusicoccum macroclavatum sp. nov. Australasian Plant Pathology. 2005;34:557–567. [Google Scholar]
- Cai L., Hyde K.D., Taylor P.W.J. A polyphasic approach for studying Colletotrichum. Fungal Diversity. 2009;39:183–204. [Google Scholar]
- Câmara M.P., Palm M.E., van Berkum P. Molecular phylogeny of Leptosphaeria and Phaeosphaeria. Mycologia. 2002;94:630–640. doi: 10.1080/15572536.2003.11833191. [DOI] [PubMed] [Google Scholar]
- Cannon P.F., Damm U., Johnston P.R. Colletotrichum current status and future directions. Studies in Mycology. 2012;73:181–213. doi: 10.3114/sim0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter E., Boudreaux C. Fatal cerebral phaeohyphomycosis due to Curvularia lunata in an immunocompetent patient. Journal of Clinical Microbiology. 2004;42:5419–5423. doi: 10.1128/JCM.42.11.5419-5423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castlebury L.A., Rossman A.Y., Jaklitsch W.J. A preliminary overview of the Diaporthales based on large subunit nuclear ribosomal DNA sequences. Mycologia. 2002;94:1017–1031. [PubMed] [Google Scholar]
- Chen C., Verkley G.J.M., Sun G. Redefining common endophytes and plant pathogens in Neofabraea, Pezicula, and related genera. Fungal Biology. 2016;120:1291–1322. doi: 10.1016/j.funbio.2015.09.013. [DOI] [PubMed] [Google Scholar]
- Chen Q., Jiang J.R., Zhang G.Z. Resolving the Phoma enigma. Studies in Mycology. 2015;82:137–217. doi: 10.1016/j.simyco.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Li G., Liu F. Novel species of Botryosphaeriaceae associated with shoot blight of pistachio. Mycologia. 2015;107:780–792. doi: 10.3852/14-242. [DOI] [PubMed] [Google Scholar]
- Chen S.F., Lombard L., Roux J. Novel species of Calonectria associated with Eucalyptus leaf blight in Southeast China. Persoonia. 2011;26:1–12. doi: 10.3767/003158511X555236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.J., Kim W.G., Kim H.G. Morphology, molecular phylogeny and pathogenicity of Colletotrichum panacicola causing anthracnose of Korean ginseng. The Plant Pathology Journal. 2011;27:1–7. [Google Scholar]
- Clements F.E., Shear C.L. The H.W. Wilson Company; New York, USA: 1931. The genera of fungi. [Google Scholar]
- Cote M.J., Tardif M.C., Meldrum A.J. Identification of Monilinia fructigena, M. fructicola, M. laxa, and Monilia polystroma on inoculated and naturally infected fruit using multiplex PCR. Plant Disease. 2004;88:1219–1225. doi: 10.1094/PDIS.2004.88.11.1219. [DOI] [PubMed] [Google Scholar]
- Crouch J.A. Colletotrichum caudatum s. l. is a species complex. IMA Fungus. 2014;5:1–30. doi: 10.5598/imafungus.2014.05.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch J.A., Beirn L.A., Cortese L.M. Anthracnose disease of switchgrass caused by the novel fungal species Colletotrichum navitas. Mycological Research. 2009;113:1411–1421. doi: 10.1016/j.mycres.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Crouch J.A., Clarke B.B., Hillman B.I. Unraveling evolutionary relationships among the divergent lineages of Colletotrichum causing anthracnose disease in turfgrass and corn. Phytopathology. 2006;96:46–60. doi: 10.1094/PHYTO-96-0046. [DOI] [PubMed] [Google Scholar]
- Crouch J.A., Clarke B.B., White J.F., Jr. Systematic analysis of the falcate-spored graminicolous Colletotrichum and a description of six new species from warm-season grasses. Mycologia. 2009;101:717–732. doi: 10.3852/08-230. [DOI] [PubMed] [Google Scholar]
- Crouch J.A., Szabo L.J. Real-time PCR detection and discrimination of the southern and common corn rust pathogens Puccinia polysora and Puccinia sorghi. Plant Disease. 2011;95:624–632. doi: 10.1094/PDIS-10-10-0745. [DOI] [PubMed] [Google Scholar]
- Crouch J.A., Tomaso-Peterson M. Anthracnose disease of centipedegrass turf caused by Colletotrichum eremochloae, a new fungal species closely related to Colletotrichum sublineola. Mycologia. 2012;104:1085–1096. doi: 10.3852/11-317. [DOI] [PubMed] [Google Scholar]
- Crouch J.A., Tredway L.P., Clarke B.B. Phylogenetic and population genetic divergence correspond with habitat for the pathogen Colletotrichum cereale and allied taxa across diverse grass communities. Molecular Ecology. 2009;18:123–135. doi: 10.1111/j.1365-294X.2008.04008.x. [DOI] [PubMed] [Google Scholar]
- Crous P.W. APS Press; St. Paul, Minnesota, USA: 2002. Taxonomy and pathology of Cylindrocladium (Calonectria) and allied genera. [Google Scholar]
- The genus Cladosporium and similar dematiaceous hyphomycetes. Crous P.W., Braun U., Schubert K., editors. Studies in Mycology. 2007;58:1–253. doi: 10.3114/sim.2007.58.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Braun U., Schubert K. Delimiting Cladosporium from morphologically similar genera. Studies in Mycology. 2007;58:33–56. doi: 10.3114/sim.2007.58.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Carris L.M., Giraldo A. The Genera of Fungi – fixing the application of the type species of generic names – G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus. 2015;6:163–198. doi: 10.5598/imafungus.2015.06.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Denman S., Taylor J.E. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2004. Cultivation and diseases of Proteaceae: Leucadendron, Leucospermum and Protea. CBS Biodiversity Series 2. [Google Scholar]
- Crous P.W., Denman S., Taylor J.E. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2013. Cultivation and diseases of Proteaceae: Leucadendron, Leucospermum and Protea. CBS Biodiversity Series 13. [Google Scholar]
- Crous P.W., Gams W., Stalpers J.A. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology. 2004;50:19–22. [Google Scholar]
- Crous P.W., Giraldo A., Hawksworth D.L. The Genera of Fungi: fixing the application of the type species of generic names. IMA Fungus. 2014;5:141–160. doi: 10.5598/imafungus.2014.05.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Groenewald J.Z. Why everlastings don't last. Persoonia. 2011;26:70–84. doi: 10.3767/003158511X574532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Groenewald J.Z., Risède J.-M. Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology. 2004;50:415–430. doi: 10.3114/sim.55.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Groenewald J.Z., Risède J.-M. Calonectria species and their Cylindrocladium anamorphs: species with clavate vesicles. Studies in Mycology. 2006;55:213–226. doi: 10.3114/sim.55.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Groenewald J.Z., Shivas R.G. Fungal Planet description sheets: 69–91. Persoonia. 2011;26:108–156. doi: 10.3767/003158511X581723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Groenewald J.Z., Slippers B. Global food and fibre security threatened by current inefficiencies in fungal identification. Philosophical Transactions of the Royal Society B: Biological Sciences. 2016;371:1709. doi: 10.1098/rstb.2016.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Groenewald J.Z., Taylor J.E. Saccharata intermedia. Fungal Planet 43. Persoonia. 2009;23:198–199. [Google Scholar]
- Crous P.W., Groenewald J.Z., Wingfield M.J. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2007. Neofusicoccum mediterraneum. Fungal Planet 19. [Google Scholar]
- Crous P.W., Hawksworth D.L., Wingfield M.J. Identifying and naming plant-pathogenic fungi: past, present, and future. Annual Review of Phytopathology. 2015;53:247–267. doi: 10.1146/annurev-phyto-080614-120245. [DOI] [PubMed] [Google Scholar]
- Crous P.W., Kang J.-C. Phylogenetic confirmation of Calonectria spathulata and Cylindrocladium leucothoes based on morphology, β-tubulin and ITS rDNA sequence data. Mycoscience. 2001;42:51–57. [Google Scholar]
- Crous P.W., Kang J.-C., Schoch C.L. Phylogenetic relationships of Cylindrocladium pseudogracile and Cylindrocladium rumohrae with morphologically similar taxa, based on morphology and DNA sequences of internal transcribed spacers and β-tubulin. Canadian Journal of Botany. 1999;77:1813–1820. [Google Scholar]
- Crous P.W., Schubert K., Braun U. Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Studies in Mycology. 2007;58:185–217. doi: 10.3114/sim.2007.58.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Schumacher R.K., Wingfield M.J. Fungal Systematics and Evolution: FUSE 1. Sydowia. 2015;67:81–118. [Google Scholar]
- Crous P.W., Shivas R.G., Quaedvlieg W. Fungal Planet description sheets 214–280. Persoonia. 2014;32:184–306. doi: 10.3767/003158514X682395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Shivas R.G., Wingfield M.J. Fungal Planet description sheets: 128–153. Persoonia. 2012;29:146–201. doi: 10.3767/003158512X661589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Slippers B., Wingfield M.J. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology. 2006;55:235–254. doi: 10.3114/sim.55.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Summerell B.A., Carnegie A.J. Unravelling Mycosphaerella: do you believe in genera? Persoonia. 2009;23:99–118. doi: 10.3767/003158509X479487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Summerell B.A., Shivas R.G. Fungal Planet description sheets: 107–127. Persoonia. 2012;28:138–182. doi: 10.3767/003158512X652633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Tanaka K., Summerell B.A. Additions to the Mycosphaerella complex. IMA Fungus. 2011;2:49–64. doi: 10.5598/imafungus.2011.02.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Verkley G.J.M., Groenewald J.Z. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2009. Fungal biodiversity. CBS Laboratory Manual Series 1. [Google Scholar]
- Crous P.W., Wingfield M.J., Burgess T.I. Fungal Planet description sheets: 469–557. Persoonia. 2016;37:218–403. doi: 10.3767/003158516X694499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wingfield M.J., Guarro J. Fungal Planet description sheets: 154–213. Persoonia. 2013;31:188–296. doi: 10.3767/003158513X675925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wingfield M.J., Guarro J. Fungal Planet description sheets: 320–370. Persoonia. 2015;34:167–266. doi: 10.3767/003158515X688433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wingfield M.J., le Roux J.J. Fungal Planet description sheets: 371–399. Persoonia. 2015;35:264–327. doi: 10.3767/003158515X690269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wingfield M.J., Park R.F. Mycosphaerella nubilosa a synonym of M. molleriana. Mycological Research. 1991;95:628–632. [Google Scholar]
- Crous P.W., Wingfield M.J., Richardson D.M. Fungal Planet description sheets: 400–468. Persoonia. 2016;36:316–458. doi: 10.3767/003158516X692185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wood A.R., Okada G. Foliicolous microfungi occurring on Encephalartos. Persoonia. 2008;21:135–146. doi: 10.3767/003158508X380612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins G.B., Hiratsuka Y. American Phytopathological Society; St. Paul, Minnesota, USA: 2003. Illustrated genera of rust fungi. [Google Scholar]
- Cunningham G.H. John McIndoe; Dunedin, New Zealand: 1931. The rust fungi of New Zealand: together with the biology, cytology and therapeutics of the Uredinales. [Google Scholar]
- da Cunha K.C., Sutton D.A., Fothergill A.W. Diversity of Bipolaris species in clinical samples in the United States and their antifungal susceptibility profiles. Journal of Clinical Microbiology. 2012;50:4061–4066. doi: 10.1128/JCM.01965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha K.C., Sutton D.A., Fothergill A.W. In vitro antifungal susceptibility and molecular identity of 99 clinical isolates of the opportunistic fungal genus Curvularia. Diagnostic Microbiology and Infectious Disease. 2013;76:168–174. doi: 10.1016/j.diagmicrobio.2013.02.034. [DOI] [PubMed] [Google Scholar]
- Damm U., Cannon P.F., Liu F. The Colletotrichum orbiculare species complex: important pathogens of field and weeds. Fungal Diversity. 2013;61:29–59. [Google Scholar]
- Damm U., Cannon P.F., Woudenberg J.H.C. The Colletotrichum boninense species complex. Studies in Mycology. 2012;73:1–36. doi: 10.3114/sim0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U., Cannon P.F., Woudenberg J.H.C. The Colletotrichum acutatum species complex. Studies in Mycology. 2012;73:37–113. doi: 10.3114/sim0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U., Crous P.W., Fourie P.H. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia. 2007;99:664–680. doi: 10.3852/mycologia.99.5.664. [DOI] [PubMed] [Google Scholar]
- Damm U., O'Connell R.J., Groenewald J.Z. The Colletotrichum destructivum species complex – hemibiotrophic pathogens of forage and field crops. Studies in Mycology. 2014;79:49–84. doi: 10.1016/j.simyco.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U., Woudenberg J.H.C., Cannon P.F. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity. 2009;39:45–87. [Google Scholar]
- David J.C. A contribution to the systematics of Cladosporium. Revision of the fungi previously referred to Heterosporium. Mycological papers. 1997;172:1–157. [Google Scholar]
- De Beer Z.W., Duong T.A., Barnes I. Redefining Ceratocystis and allied genera. Studies in Mycology. 2014;79:187–219. doi: 10.1016/j.simyco.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer Z.W., Seifert K.A., Wingfield M.J. The ophiostomatoid fungi: their dual position in the Sordariomycetes. In: Seifert K.A., De Beer Z.W., Wingfield M.J., editors. The ophiostomatoid fungi: expanding frontiers. CBS Biodiversity Series 12. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2013. pp. 1–19. [Google Scholar]
- De Beer Z.W., Seifert K.A., Wingfield M.J. A nomenclator for ophiostomatoid genera and species in the Ophiostomatales and Microascales. In: Seifert K.A., De Beer Z.W., Wingfield M.J., editors. The ophiostomatoid fungi: expanding frontiers. CBS Biodiversity Series 12. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2013. pp. 245–322. [Google Scholar]
- De Gruyter J., Noordeloos M.E., Boerema G.H. Contributions towards a monograph of Phoma (Coelomycetes) – I. 2. Section Phoma: additional taxa with very small conidia and taxa with conidia up to 7 μm long. Persoonia. 1993;15:369–400. [Google Scholar]
- De Gruyter J., Woudenberg J.H.C., Aveskamp M.M. Redisposition of phoma-like anamorphs in Pleosporales. Studies in Mycology. 2013;75:1–36. doi: 10.3114/sim0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong S.N., Levesque C.A., Verkley G.J.M. Phylogenetic relationships among Neofabraea species causing tree cankers and bull's-eye rot of apple based on DNA sequencing of ITS nuclear rDNA, mitochondrial rDNA, and the β-tubulin gene. Mycological Research. 2001;105:658–669. [Google Scholar]
- Demers J.E., Liu M., Hambleton S. Rust fungi on Panicum. Mycologia. 2017;109:1–17. doi: 10.1080/00275514.2016.1262656. [DOI] [PubMed] [Google Scholar]
- Deng H., Tan Y.P., Shivas R.G. Curvularia tsudae comb. nov. et nom. nov., formerly Pseudocochliobolus australiensis, and a revised synonymy for Curvularia australiensis. Mycoscience. 2014;56:24–28. [Google Scholar]
- Denman S., Crous P.W., Groenewald J.Z. Circumscription of Botryosphaeria species associated with Proteaceae based on morphology and DNA sequence data. Mycologia. 2003;95:294–307. [PubMed] [Google Scholar]
- Denman S., Crous P.W., Wingfield M.J. A taxonomic reassessment of Phyllachora proteae, a leaf pathogen of Proteaceae. Mycologia. 1999;91:510–516. [Google Scholar]
- De Silva D.D., Ades P.K., Crous P.W. Colletotrichum species associated with chili anthracnose in Australia. Plant Pathology. 2017;66:254–267. [Google Scholar]
- Diao Y.Z., Zhang C., Liu F. Colletotrichum species causing anthracnose disease of chili in China. Persoonia. 2017;38:20–37. doi: 10.3767/003158517X692788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.J., Castlebury L.A., Aime M.C. Phylogenetic relationships of sugarcane rust fungi. Mycological Progress. 2010;9:459–468. [Google Scholar]
- Doyle V.P., Oudemans P.V., Rehner S.A. Habitat and host indicate lineage identity in Colletotrichum gloeosporioides sl. from wild and agricultural landscapes in North America. PLoS One. 2013;8:e62394. doi: 10.1371/journal.pone.0062394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., Schardl C.L., Vaillancourt L.J. Using mating-type gene sequences for improved phylogenetic resolution of Colletotrichum species complexes. Mycologia. 2005;97:641–658. doi: 10.3852/mycologia.97.3.641. [DOI] [PubMed] [Google Scholar]
- Dugan F.M., Braun U., Groenewald J.Z. Morphological plasticity in Cladosporium sphaerospermum. Persoonia. 2008;21:9–16. doi: 10.3767/003158508X334389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan F.M., Schubert K., Braun U. Check-list of Cladosporium names. Schlechtendalia. 2004;11:1–103. [Google Scholar]
- Duplessis S., Cuomo C.A., Lin Y.-C. Obligate biotrophy features unravelled by the genomic analysis of rust fungi. Proceedings of the National Academy of Sciences (USA) 2011;108:9166–9171. doi: 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyko B.J., Sutton B.C., Roquebert M.F. The genus Protostegia. Mycologia. 1979;71:918–934. [Google Scholar]
- Ellis J.B., Dearness J. New species of Canadian fungi. Proceedings of the Royal Canadian Institute. 1897;1:89–93. [Google Scholar]
- Ellis M.B. Clasterosporium and some allied Dematiaceae – Phragmosporae. II. Mycological Papers. 1959;72:1–72. [Google Scholar]
- Ellis M.B. Commonwealth Mycological Institute; Kew, UK: 1971. Dematiaceous Hyphomycetes. [Google Scholar]
- Farr D.F., Aime M.C., Rossman A.Y. Species of Colletotrichum on Agavaceae. Mycological Research. 2006;110:1395–1408. doi: 10.1016/j.mycres.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Farr D.F., Elliott M., Rossman A.Y. Fusicoccum arbuti sp. nov. causing cankers on Pacific madrone in western North America with notes on Fusicoccum dimidiatum, the correct name for Scytalidium dimidiatum and Natrassia mangiferae. Mycologia. 2005;97:730–741. doi: 10.3852/mycologia.97.3.730. [DOI] [PubMed] [Google Scholar]
- Farr D.F., Rossman A.Y. U.S. National Fungus Collections, ARS, USDA; 2017. Fungal databases.https://nt.ars-grin.gov/fungaldatabases [Retrieved April 4, 2017] [Google Scholar]
- Fitt B.D.L., Hu B.C., Li Z.Q. Strategies to prevent spread of Leptosphaeria maculans (phoma stem canker) onto oilseed rape crops in China; costs and benefits. Plant Pathology. 2008;57:652–664. [Google Scholar]
- Flores R.C., Loyo J.R., Ojeda R.A. First report of orange rust of sugarcane caused by Puccinia kuehnii in Mexico, El Salvador, and Panama. Plant Disease. 2009;93:1347. doi: 10.1094/PDIS-93-12-1347B. [DOI] [PubMed] [Google Scholar]
- Fourie A., Wingfield M.J., Wingfield B.D. Molecular markers delimit cryptic species in Ceratocystis sensu stricto. Mycological Progress. 2015;14:1–18. [Google Scholar]
- Fulton C.E., Brown A.E. Use of SSU rDNA group-I intron to distinguish Monilinia fructicola from M. laxa and M. fructigena. FEMS Microbiology Letters. 1997;157:307–312. doi: 10.1111/j.1574-6968.1997.tb12790.x. [DOI] [PubMed] [Google Scholar]
- Gehesquière B., Crouch J.A., Marra R.E. Characterization and taxonomic reassessment of the box blight pathogen Calonectria pseudonaviculata, introducing Calonectria henricotiae sp. nov. Plant Pathology. 2016;65:37–52. [Google Scholar]
- Gell I., Cubero J., Melgarejo P. Two different PCR approaches for universal diagnosis of brown rot and identification of Monilinia spp. in stone fruit trees. Journal of Applied Microbiology. 2007;103:2629–2637. doi: 10.1111/j.1365-2672.2007.03495.x. [DOI] [PubMed] [Google Scholar]
- Giraldo A., Crous P.W., Schumacher R.K. The Genera of Fungi – G3: Aleurocystis, Blastacervulus, Clypeophysalospora, Licrostroma, Neohendersonia and Spumatoria. Mycological Progress. 2017;16:325–348. [Google Scholar]
- Gril T., Celar F., Javornik B. Fluorescent AFLP fingerprinting of Monilinia fructicola. Journal of Plant Diseases and Protection. 2010;117:168–172. [Google Scholar]
- Hansford C.G. Contribution towards the fungus flora of Uganda. V. Fungi Imperfecti. Proceedings of the Linnean Society London. 1943;155:34–67. [Google Scholar]
- Harada Y., Nakao S., Sasaki M. Monilia mumecola, a new brown rot fungus on Prunus mume in Japan. Journal of General Plant Pathology. 2004;70:297–307. [Google Scholar]
- Harada Y., Sasaki M., Sasaki Y. Monilinia ssiori sp. nov. in the Sclerotiniaceae, causing leaf blight and young fruit rot of Prunus ssiori in Japan. Mycoscience. 2005;46:376–380. [Google Scholar]
- Harrington T.C., Kazmi M.R., Al-Sadi A.M. Intraspecific and intragenomic variability of ITS rDNA sequences reveals taxonomic problems in Ceratocystis fimbriata sensu stricto. Mycologia. 2014;106:224–242. doi: 10.3852/106.2.224. [DOI] [PubMed] [Google Scholar]
- Hawksworth D.L., Crous P.W., Redhead S.A. The Amsterdam Declaration on fungal nomenclature. IMA Fungus. 2011;2:105–112. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R.N., Wingfield M.J., Wingfield B.D. Ceratocystis species on Acacia mearnsii and Eucalyptus spp. in eastern and southern Africa including six new species. Fungal Diversity. 2009;34:41–68. [Google Scholar]
- Hernádez J.R., Aime M.C., Newbry B. Aecidium kalanchoe sp. nov., a new rust on Kalanchoe blossfeldiana (Crassulaceae) Mycological Research. 2004;108:846–848. doi: 10.1017/s0953756204000681. [DOI] [PubMed] [Google Scholar]
- Hernández-Restrepo M., Groenewald J.Z., Elliott M.L. Take-all or nothing. Studies in Mycology. 2016;83:19–48. doi: 10.1016/j.simyco.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuchert B., Braun U., Schubert K. Morphotaxonomic revision of fungicolous Cladosporium species (hyphomycetes) Schlechtendalia. 2005;13:1–78. [Google Scholar]
- Hirata K., Kusaba M., Chuma I. Speciation in Pyricularia inferred from multilocus phylogenetic analysis. Mycological Research. 2014;111:799–808. doi: 10.1016/j.mycres.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Hiratsuka Y., Hiratsuka N. Morphology of spermogonia and taxonomy of rust fungi. Reports of the Tottori Mycological Institute. 1980;18:257–268. [Google Scholar]
- Holst-Jensen A., Kohn L.M., Jakobsen K.S. Molecular phylogeny and evolution of Monilinia (Sclerotiniaceae) based on coding and noncoding rDNA sequences. American Journal of Botany. 1997;84:686–701. [PubMed] [Google Scholar]
- Honey E.E. The monilioid species of Sclerotinia. Mycologia. 1928;20:127–157. [Google Scholar]
- Honey E.E. North American species of Monilinia. I. Occurrence, grouping, and life-histories. American Journal of Botany. 1936;23:100–106. [Google Scholar]
- Hou L.W., Liu F., Duan W.J. Colletotrichum aracearum and C. camelliae-japonicae, two holomorphic new species from China and Japan. Mycosphere. 2016;7:1111–1123. [Google Scholar]
- Hu M.J., Cox K.D., Schnabel G. Monilinia species causing brown rot of peach in China. PloS ONE. 2011;6:1–14. doi: 10.1371/journal.pone.0024990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Chen G.Q., Hou X. Colletotrichum species associated with cultivated citrus in China. Fungal Diversity. 2013;61:61–74. [Google Scholar]
- Hughes S. Studies on microfungi III. Mastigosporium, Camposporium and Ceratophorum. Mycological Papers. 1951;36:1–45. [Google Scholar]
- Hunt J. Taxonomy of the genus Ceratocystis. Lloydia. 1956;19:1–58. [Google Scholar]
- Hyde K.D., Hongsanan S., Jeewon R. Fungal diversity notes 367–491: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity. 2016;80:1–270. [Google Scholar]
- Hyde K.D., Nilsson R.H., Alias S.A. One stop shop: backbones trees for important phytopathogenic genera: I. Fungal Diversity. 2014;67:21–125. [Google Scholar]
- Ibrahim M., Schlegel M., Sieber T.N. Venturia orni sp. nov., a species distinct from Venturia fraxini, living in the leaves of Fraxinus ornus. Mycological Progress. 2016;15:29. [Google Scholar]
- Inderbitzin P., Trouillas F.P., Bostock R.M. A six-locus phylogeny reveals high levels of species diversity in Botryosphaeriaceae from California almond. Mycologia. 2010;102:1350–1368. doi: 10.3852/10-006. [DOI] [PubMed] [Google Scholar]
- Jackson H.S. Apple tree anthracnose. Oregon Agricultural Experiment Station Biennial Crop, Pest and Horticulture Report. 1913;1911–1912:178–197. [Google Scholar]
- Jayawardena R.S., Huang J., Jin B. An updated account of Colletotrichum species associated with strawberry anthracnose in China based on molecular data. Mycosphere. 2016;7:1147–1163. [Google Scholar]
- Jayawardena R.S., Hyde K.D., Damm U. Notes on currently accepted species of Colletotrichum. Mycosphere. 2016;7:1192–1260. [Google Scholar]
- Jin Y., Szabo L.J., Carson M. Century-old mystery of Puccinia striiformis life history solved with the identification of Berberis as an alternate host. Phytopathology. 2010;100:432–435. doi: 10.1094/PHYTO-100-5-0432. [DOI] [PubMed] [Google Scholar]
- Johnson J.A., Harrington T.C., Engelbrecht C.J.B. Phylogeny and taxonomy of the North American clade of the Ceratocystis fimbriata complex. Mycologia. 2005;97:1067–1092. doi: 10.3852/mycologia.97.5.1067. [DOI] [PubMed] [Google Scholar]
- Johnston P.R., Seifert K.A., Stone J.K. Recommendations on generic names competing for use in Leotiomycetes (Ascomycota) IMA Fungus. 2014;5:91–120. doi: 10.5598/imafungus.2014.05.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørstad I. The genera Aecidium, Uredo and Puccinia of Persoon. Blumea – Biodiversity, Evolution and Biogeography of Plants. 1958;9:1–20. [Google Scholar]
- Kajitani Y., Masuya H. Ceratocystis ficicola sp. nov., a causal fungus of fig canker in Japan. Mycoscience. 2011;52:349–353. [Google Scholar]
- Kalchbrenner K., Cooke M.C. South African fungi. Grevillea. 1880;9:17–34. [Google Scholar]
- Kamgan N.G., Jacobs K., De Beer Z.W. Ceratocystis and Ophiostoma species including three new taxa, associated with wounds on native South African trees. Fungal Diversity. 2008;29:37–59. [Google Scholar]
- Kamgan Nkuekam G., Wingfield M.J., Mohammed C. Ceratocystis species, including two new species associated with nitidulid beetles, on eucalypts in Australia. Antonie Van Leeuwenhoek. 2012;101:217–241. doi: 10.1007/s10482-011-9625-7. [DOI] [PubMed] [Google Scholar]
- Kang J.-C., Crous P.W., Schoch C.L. Species concepts in the Cylindrocladium floridanum and Cy. spathiphylli complexes (Hypocreaceae) based on multi-allelic sequence data, sexual compatibility and morphology. Systematic and Applied Microbiology. 2001;24:206–217. doi: 10.1078/0723-2020-00026. [DOI] [PubMed] [Google Scholar]
- Khashnobish A., Shearer C.A., Crane J.L. Reexamination of species of Leptosphaeria on asteraceous hosts. Mycotaxon. 1995;54:91–106. [Google Scholar]
- Khemmuk W., Shivas R.G., Henry R.J. Fungi associated with foliar diseases of wild and cultivated rice (Oryza spp.) in northern Queensland. Australasian Plant Pathology. 2016;45:297–308. [Google Scholar]
- Kile G.A. Plant diseases caused by species of Ceratocystis sensu stricto and Chalara. In: Wingfield M.J., Seifert K.A., Webber J., editors. Ceratocystis and Ophiostoma: taxonomy, ecology and pathogenicity. APS Press; St. Paul, Minnesota, USA: 1993. pp. 173–183. [Google Scholar]
- Kiran K., Rawal H.C., Dubey H. Draft genome of the wheat rust pathogen (Puccinia triticina) unravels genome-wide structural variations during evolution. Genome Biology and Evolution. 2016;8:2702–2721. doi: 10.1093/gbe/evw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner O. Über das Absterben junger Cytisus-Pflanzen. Zeitschrift für Pflanzenkrankheiten. 1892;2:324–327. [Google Scholar]
- Kirk P.M., Cannon P.F., Minter D.W. 10th edn. CABI; Wallingford, UK: 2008. Dictionary of the fungi. [Google Scholar]
- Kirk P.M., Stalpers J.A., Braun U. A without-prejudice list of generic names of fungi for protection under the International Code of Nomenclature for algae, fungi and plants. IMA Fungus. 2013;4:381–443. doi: 10.5598/imafungus.2013.04.02.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaubauf S., Tharreau D., Fournier E. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae) Studies in Mycology. 2014;79:85–120. doi: 10.1016/j.simyco.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G., Schmidt J.K. Vol. 2. 1817. (Mykologische Hefte). Leipzig, Germany. [Google Scholar]
- Langrell S.R.H., Glen M., Alfenas A.C. Molecular diagnosis of Puccinia psidii (guava rust) – a quarantine threat to Australian eucalypt and Myrtaceae biodiversity. Plant Pathology. 2008;57:687–701. [Google Scholar]
- Laundon G.F. The lectotype for Uredo. Taxon. 1970;19:947. [Google Scholar]
- Léveillé J.H. Observations sur quelques champignons de la flore des environs de Paris. Annales des Sciences Naturelles Botanique. 1843;19:213–231. [Google Scholar]
- Li G.J., Hyde K.D., Zhao R.L. Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity. 2016;78:1–237. [Google Scholar]
- Liu F., Cai L., Crous P.W. Circumscription of the anthracnose pathogens Colletotrichum lindemuthianum and C. nigrum. Mycologia. 2013;105:844–860. doi: 10.3852/12-315. [DOI] [PubMed] [Google Scholar]
- Liu F., Cai L., Crous P.W. The Colletotrichum gigasporum species complex. Persoonia. 2014;33:83–97. doi: 10.3767/003158514X684447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Damm U., Cai L. Species of the Colletotrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Diversity. 2013;61:89–105. [Google Scholar]
- Liu F., Hyde K.D., Cai L. Neotypification of Colletotrichum coccodes, the causal agent of potato black dot disease and tomato anthracnose. Mycology. 2011;2:248–254. [Google Scholar]
- Liu F., Mbenoun M., Barnes I. New Ceratocystis species from Eucalyptus and Cunninghamia in South China. Antonie Van Leeuwenhoek. 2015;107:1451–1473. doi: 10.1007/s10482-015-0441-3. [DOI] [PubMed] [Google Scholar]
- Liu F., Wang M., Damm U. Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evolutionary Biology. 2016;16:81. doi: 10.1186/s12862-016-0649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Weir B.S., Damm U. Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia. 2015;35:63–86. doi: 10.3767/003158515X687597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.K., Hyde K.D., Jones E.B.G. Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity. 2015;72:1–197. [Google Scholar]
- Liu M., Hambleton S. Laying the foundation for a taxonomic review of Puccinia coronata s. l. in a phylogenetic context. Mycological Progress. 2013;12:63–89. [Google Scholar]
- Liu X., Xie X., Duan J. Colletotrichum yunnanense sp. nov., a new endophytic species from Buxus sp. Mycotaxon. 2007;100:137–144. [Google Scholar]
- Lombard L., Chen S.F., Mou X. New species, hyper-diversity and potential importance of Calonectria spp. from Eucalyptus in South China. Studies in Mycology. 2015;80:151–188. doi: 10.1016/j.simyco.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L., Crous P.W., Wingfield B.D. Phylogeny and systematics of the genus Calonectria. Studies in Mycology. 2010;66:31–69. doi: 10.3114/sim.2010.66.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L., Crous P.W., Wingfield B.D. Species concepts in Calonectria (Cylindrocladium) Studies in Mycology. 2010;66:1–14. doi: 10.3114/sim.2010.66.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L., Crous P.W., Wingfield B.D. Multigene phylogeny and mating tests reveal three cryptic species related to Calonectria pauciramosa. Studies in Mycology. 2010;66:15–30. doi: 10.3114/sim.2010.66.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L., Polizzi G., Guarnaccia V. Calonectria spp. causing leaf spot, crown and root rot of ornamental plants in Tunisia. Persoonia. 2011;27:73–79. doi: 10.3767/003158511X615086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L., Rodas C.A., Crous P.W. Calonectria (Cylindrocladium) species associated with dying Pinus cuttings. Persoonia. 2009;23:41–47. doi: 10.3767/003158509X471052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L., Wingfield M.J., Alfenas A.C. The forgotten Calonectria collection: pouring old wine into new bags. Studies in Mycology. 2016;85:159–198. doi: 10.1016/j.simyco.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L., Zhou X.D., Crous P.W. Calonectria species associated with cutting rot of Eucalyptus. Persoonia. 2010;24:1–11. doi: 10.3767/003158510X486568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchi N., Ghelardini L., Belbahri L. Rapid detection of Ceratocystis platani inoculum by quantitative real-time PCR assay. Applied and Environmental Microbiology. 2013;79:5394–5404. doi: 10.1128/AEM.01484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo D.M., Barreto R.W. Colletotrichum dracaenophilum causes anthracnose on Dracaena braunii in Brazil. Australasian Plant Disease Notes. 2016;11:5. [Google Scholar]
- Macedo D.M., Pereira O.L., Hora Júnior B.T. Mycobiota of the weed Tradescantia fluminensis in its native range in Brazil with particular reference to classical biological control. Australasian Plant Pathology. 2016;45:45–56. [Google Scholar]
- Madrid H., da Cunha K.C., Gene J. Novel Curvularia species from clinical specimens. Persoonia. 2014;33:48–60. doi: 10.3767/003158514X683538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W., Begerow D., Weiss M. Phylogeny of the rust fungi: an approach using nuclear large subunit ribosomal DNA sequences. Canadian Journal of Botany. 2003;81:12–23. [Google Scholar]
- Maier W., Wingfield B.D., Mennicken M. Polyphyly and two emerging lineages in the rust genera Puccinia and Uromyces. Mycological Research. 2007;111:176–185. doi: 10.1016/j.mycres.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Manamgoda D.S., Cai L., Bahkali A.H. Cochliobolus: an overview and current status of species. Fungal Diversity. 2011;51:3–42. [Google Scholar]
- Manamgoda D.S., Cai L., Hyde K.D. Two new species of Curvularia from nothern Thailand. Sydowia. 2012;64:255–266. [Google Scholar]
- Manamgoda D.S., Cai L., Hyde K.D. A taxonomic evaluation of the holomorphic species complex: Cochliobolus, Bipolaris and Curvularia through multilocus phylogeny. Fungal Diversity. 2012;56:131–144. [Google Scholar]
- Manamgoda D.S., Cai L., McKenzie E.H.C. A phylogenetic and taxonomic re-evaluation of the Bipolaris – Cochliobolus – Curvularia complex. Fungal Diversity. 2012;56:131–144. [Google Scholar]
- Manamgoda D.S., Rossman A.Y., Castlebury L.A. The genus Bipolaris. Studies in Mycology. 2014;79:221–288. doi: 10.1016/j.simyco.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manamgoda D.S., Rossman A.Y., Castlebury L.A. A taxonomic and phylogenetic re-appraisal of the genus Curvularia (Pleosporaceae): human and plant pathogens. Phytotaxa. 2015;212:175–198. [Google Scholar]
- Manamgoda D.S., Udayanga D., Cai L. Endophytic Colletotrichum from tropical grasses with a new species C. endophytica. Fungal Diversity. 2013;61:107–115. [Google Scholar]
- Marin M., Castro B., Gaitan A. Relationships of Ceratocystis fimbriata isolates from Colombian coffee-growing regions based on molecular data and pathogenicity. Journal of Phytopathology. 2003;151:395–405. [Google Scholar]
- Marincowitz S., Crous P.W., Groenewald J.Z. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2008. Microfungi occurring on Proteaceae in the fynbos. CBS Biodiversity Series 7. [Google Scholar]
- Marincowitz S., Groenewald J.Z., Wingfield M.J. Species of Botryosphaeriaceae occurring on Proteaceae. Persoonia. 2008;21:111–118. doi: 10.3767/003158508X372387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques M.W., Lima N.B., de Morais M.A., Jr. Botryosphaeria, Neofusicoccum, Neoscytalidium and Pseudofusicoccum species associated with mango in Brazil. Fungal Diversity. 2013;61:195–208. [Google Scholar]
- Martini C., Mari M. Monilinia fructicola, Monilinia laxa (Monilinia Rot, Brown Rot) In: Bautista-Banos S., editor. Postharvest decay, control strategies. Academic Press; USA: 2014. pp. 233–265. [Google Scholar]
- Mayers C.G., Mcnew D.L., Harrington T.C. Three genera in the Ceratocystidaceae are the respective symbionts of three independent lineages of ambrosia beetles with large, complex mycangia. Fungal Biology. 2015;119:1075–1092. doi: 10.1016/j.funbio.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Mbenoun M., Wingfield M.J., Begoude Boyogueno A.D. Molecular phylogenetic analyses reveal three new Ceratocystis species and provide evidence for geographic differentiation of the genus in Africa. Mycological Progress. 2014;13:219–240. [Google Scholar]
- McNeill J., Barrie F.R., Buck W.R. Koeltz Scientific Books; Germany: 2012. International Code of Nomenclature for algae, fungi and plants (Melbourne Code) adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. [Google Scholar]
- McTaggart A.R., Geering A.D.W., Shivas R.G. The rusts on Goodeniaceae and Stylidiaceae. Mycological Progress. 2014;13:1017–1025. [Google Scholar]
- McTaggart A.R., Shivas R.G., Doungsa-ard C. Identification of rust fungi (Pucciniales) on species of Allium in Australia. Australasian Plant Pathology. 2016;45:581–592. [Google Scholar]
- McTaggart A.R., Shivas R.G., van der Nest M.A. Host jumps shaped the diversity of extant rust fungi (Pucciniales) New Phytologist. 2016;209:1149–1158. doi: 10.1111/nph.13686. [DOI] [PubMed] [Google Scholar]
- Menon R. Studies on Venturiaceae on rosaceous plants. Phytopathologische Zeitschrift. 1956;27:117–146. [Google Scholar]
- Michel V.V., Hollenstein R., Stensvand A. Colletotrichum acutatum, agent of anthracnose on the new host black elderberry (Sambucus nigra) in Switzerland. Plant Disease. 2013;97:1246. doi: 10.1094/PDIS-08-12-0751-PDN. [DOI] [PubMed] [Google Scholar]
- Minnis D., McTaggart A.R., Rossman A. Taxonomy of mayapple rust: the genus Allodus resurrected. Mycologia. 2012;104:942–950. doi: 10.3852/11-350. [DOI] [PubMed] [Google Scholar]
- Miranda B.E.C., Barreto R.W., Crous P.W. Pilidiella tibouchinae sp. nov. associated with foliage blight of Tibouchina granulosa (quaresmeira) in Brazil. IMA Fungus. 2012;3:1–7. doi: 10.5598/imafungus.2012.03.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohali S., Slippers B., Wingfield M.J. Two new Fusicoccum species from Acacia and Eucalyptus in Venezuela, based on morphology and DNA sequence data. Mycological Research. 2006;110:405–413. doi: 10.1016/j.mycres.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Moriwaki J., Tsukiboshi T. Colletotrichum echinochloae, a new species on Japanese barnyard millet (Echinochloa utilis) Mycoscience. 2009;50:273–280. [Google Scholar]
- Muchovej J.J., Carvalho A.O. A new combination for Helminthosporium euphorbiae. Mycotaxon. 1989;35:159–162. [Google Scholar]
- Murata N., Aoki T., Kusaba M. Various species of Pyricularia constitute a robust clade distinct from Magnaporthe salvinii and its relatives in Magnaporthaceae. Journal of General Plant Pathology. 2014;80:66–72. [Google Scholar]
- Nag Raj T.R. Mycologue Publications; Waterloo, Canada: 1993. Coelomycetous anamorphs with appendage bearing conidia. [Google Scholar]
- Nag Raj T.R., Kendrick B. Wilfrid Laurier University Press; Waterloo, Canada: 1975. A monograph of Chalara and allied genera. [Google Scholar]
- Naidoo K., Steenkamp E., Coetzee M.P.A. Concerted evolution in the ribosomal RNA cistron. PLoS ONE. 2013;8:e59355. doi: 10.1371/journal.pone.0059355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Ohzono M., Iwai H. Anthracnose of Sansevieria trifasciata caused by Colletotrichum sansevieriae sp. nov. Journal of General Plant Pathology. 2006;72:253–256. [Google Scholar]
- Nannfeldt J.A. Studien uber die morphologie und systematik der nicht-lichenisierten inoperculaten discomyceten. Nova Acta Regiae Societatis Scientiarum Upsaliensis, ser. 4. 1932;8:1–368. [Google Scholar]
- Newcombe G. Native Venturia inopina sp. nov., specific to Populus trichocarpa and its hybrids. Mycological Research. 2003;107:108–116. doi: 10.1017/s0953756202007086. [DOI] [PubMed] [Google Scholar]
- Nirenberg H.I., Feiler U., Hagedorn G. Description of Colletotrichum lupini comb. nov. in modern terms. Mycologia. 2002;94:307–320. [PubMed] [Google Scholar]
- Noireung P., Phoulivong S., Liu F. Novel species of Colletotrichum revealed by morphology and molecular analysis. Cryptogamie Mycologie. 2012;33:347–362. [Google Scholar]
- Nüesch J. Beitrag zur Kenntnis der weidenbewohnenden Venturiaceae. Phytopathologische Zeitschrift. 1960;39:329–360. [Google Scholar]
- O'Connell R.J., Thon M.R., Hacquard S. Life-style transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Genetics. 2012;44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OEPP/EPPO Monilinia fructicola PM 7/18(2) Bulletin OEPP/EPPO Bulletin. 2009;39:337–343. [Google Scholar]
- Palm M.E. Taxonomy and morphology of the synanamorphs Pilidium concavum and Hainesia lythri (coelomycetes) Mycologia. 1991;83:787–796. [Google Scholar]
- Paulin-Mahady A.E., Harrington T.C., McNew D. Phylogenetic and taxonomic evaluation of Chalara, Chalaropsis, and Thielaviopsis anamorphs associated with Ceratocystis. Mycologia. 2002;94:62–72. [PubMed] [Google Scholar]
- Pavlic D., Slippers B., Coutinho T.A. Multiple gene genealogies and phenotypic data reveal cryptic species of the Botryosphaeriaceae: a case study on the Neofusicoccum parvum/N. ribis complex. Molecular Phylogenetics and Evolution. 2009;51:259–268. doi: 10.1016/j.ympev.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Pavlic D., Slippers B., Coutinho T.A. Molecular and phenotypic characterization of three phylogenetic species discovered within the Neofusicoccum parvum/N. ribis complex. Mycologia. 2009;101:636–647. doi: 10.3852/08-193. [DOI] [PubMed] [Google Scholar]
- Pedley K.F. PCR-based assays for the detection of Puccinia horiana on Chrysanthemums. Plant Disease. 2009;93:1252–1258. doi: 10.1094/PDIS-93-12-1252. [DOI] [PubMed] [Google Scholar]
- Peng L.J., Sun T., Yang Y.L. Colletotrichum species on grape in Guizhou and Yuannan provinces, China. Mycoscience. 2013;54:29–41. [Google Scholar]
- Persoon C.H. Henricus Dieterich; Göttingen, Germany: 1801. Synopsis Methodica Fungorum. [Google Scholar]
- Phillips A.J., Alves A., Abdollahzadeh J. The Botryosphaeriaceae: genera and species known from culture. Studies in Mycology. 2013;76:51–167. doi: 10.3114/sim0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.J.L., Alves A., Pennycook S.R. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia. 2008;21:29–55. doi: 10.3767/003158508X340742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.J.L., Rumbos I.C., Alves A. Morphology and phylogeny of Botryosphaeria dothidea causing fruit rot of olives. Mycopathologia. 2005;159:433–439. doi: 10.1007/s11046-005-0256-2. [DOI] [PubMed] [Google Scholar]
- Phoulivong S., Cai L., Chen H. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Diversity. 2009;44:33–43. [Google Scholar]
- Pirozynski K.A. 1974. Pleiochaeta setosa. Fungi Canadenses No. 12. [Google Scholar]
- Preuss C.G.T. Übersicht untersuchter Pilze, besonders aus der Umgegend von Hoyerswerda. Linnaea. 1851;24:99–153. [Google Scholar]
- Prihastuti H., Cai L., Chen H. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Diversity. 2009;39:89–109. [Google Scholar]
- Prihastuti H., Cai L., Hyde K.D. Neotypification of Colletotrichum falcatum, the causative agent of red-rot disease in sugarcane. Sydowia. 2010;62:283–293. [Google Scholar]
- Quaedvlieg W., Verkley G.J.M., Shin H.-D. Sizing up Septoria. Studies in Mycology. 2013;75:307–390. doi: 10.3114/sim0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeshkumar K.C., Hepat R.P., Gaikwad S.B. Pilidiella crousii sp. nov. from the northern Western Ghats, India. Mycotaxon. 2011;115:155–162. [Google Scholar]
- Rakotoniriana E.F., Scauflaire J., Rabemanantsoa C. Colletotrichum gigasporum sp. nov., a new species of Colletotrichum producing long straight conidia. Mycological Progress. 2013;12:403–412. [Google Scholar]
- Ramakrishnan T.S., Sundaram N.V. Notes on some fungi from south India. I. Indian Phytopathology. 1953;5:110–115. [Google Scholar]
- Rayner R.W. Commonwealth Mycological Institute; Kew, UK: 1970. A mycological colour chart. [Google Scholar]
- Robert V., Vu D., Amor A.B.H. MycoBank gearing up for new horizons. IMA Fungus. 2013;4:371–379. doi: 10.5598/imafungus.2013.04.02.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodas C.A., Roux J., van Wyk M. Ceratocystis neglecta sp. nov., infecting Eucalyptus trees in Colombia. Fungal Diversity. 2008;28:73–84. [Google Scholar]
- Rojas E.I., Rehner S.A., Samuels G.J. Colletotrichum gloeosporioides s. l. associated with Theobroma cacao and other plants in Panama: multilocus phylogenies distinguish pathogen and endophyte clades. Mycologia. 2010;102:1318–1338. doi: 10.3852/09-244. [DOI] [PubMed] [Google Scholar]
- Rossman A.Y. Calonectria and its type species, C. daldiniana, a later synonym of C. pyrochroa. Mycotaxon. 1979;8:321–328. [Google Scholar]
- Rossman A.Y., Aime M.C., Farr D.F. The coelomycetous genera Chaetomella and Pilidium represent a newly discovered lineage of inoperculate discomycetes. Mycological Progress. 2004;3:275–290. [Google Scholar]
- Rossman A.Y., Seifert K.A., Samuels G.J. Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus. 2013;4:41–51. doi: 10.5598/imafungus.2013.04.01.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo P.A., Sydow P. Supplementum Universale, Pars IV. Sylloge Fungorum. 1899;14:1–1316. [Google Scholar]
- Sakalidis M.L., Hardy GEStJ., Burgess T.I. Use of the Genealogical Sorting Index (GSI) to delineate species boundaries in the Neofusicoccum parvum-Neofusicoccum ribis species complex. Molecular Phylogenetics and Evolution. 2011;60:333–344. doi: 10.1016/j.ympev.2011.04.026. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Houbraken J., Thrane U. CBS-KNAW Fungal Biodiversity Centre; Utrecht, the Netherlands: 2010. Food and indoor fungi. CBS Laboratory Manual Series 2. [Google Scholar]
- Sanchez-Torres P., Hinarejos R., Tuset J.J. Characterization and pathogenicity of Fusicladium eriobotryae, the fungal pathogen responsible for loquat scab. Plant Disease. 2009;93:1151–1157. doi: 10.1094/PDIS-93-11-1151. [DOI] [PubMed] [Google Scholar]
- Sandoval-Denis M., Gené J., Sutton D.A. New species of Cladosporium associated with human and animal infections. Persoonia. 2016;36:281–298. doi: 10.3767/003158516X691951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Moriwaki J., Uzuhashi S. Molecular phylogenetic analyses and morphological re-examination of strains belonging to three rare Colletotrichum species in Japan. Microbiology and Culture Collections. 2012;28:121–134. [Google Scholar]
- Savile D.B.O. Paleoecology and convergent evolution in rust fungi (Uredinales) Biosystems. 1978;10:31–36. [Google Scholar]
- Schoch C.L., Crous P.W., Groenewald J.Z. A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology. 2009;64 doi: 10.3114/sim.2009.64.01. 1–15-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Crous P.W., Wingfield B.D. The Cylindrocladium candelabrum species complex includes four distinct mating populations. Mycologia. 1999;91:286–298. [Google Scholar]
- Schoch C.L., Crous P.W., Wingfield B.D. Phylogeny of Calonectria based on comparisons of β-tubulin DNA sequences. Mycological Research. 2001;105:1045–1052. [Google Scholar]
- Schoch C.L., Robbertse B., Robert V. Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. Database. 2014;2014:bau061. doi: 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Seifert K.A., Huhndorf S. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences (USA) 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler M., Lutz M., Wood A. Taxonomy and phylogeny of Puccinia lagenophorae: a study using rDNA sequence data, morphological and host range features. Mycological Progress. 2011;10:175–187. [Google Scholar]
- Schubert K. Mathematisch-Naturwissenschaftlich-Technischen Fakultät, Martin-Luther-University Halle-Wittenberg; Germany: 2005. Morphotaxonomic revision of foliicolous Cladosporium species (hyphomycetes) Ph.D. dissertation. [Google Scholar]
- Schubert K., Greslebin A., Groenewald J.Z. New foliicolous species of Cladosporium from South America. Persoonia. 2009;22:111–122. doi: 10.3767/003158509X449381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K., Groenewald J.Z., Braun U. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Studies in Mycology. 2007;58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K., Ritschel A., Braun U. A monograph of Fusicladium s. lat. (hyphomycetes) Schlechtendalia. 2003;9:1–132. [Google Scholar]
- Sharma G., Kumar-Pinnaka A., Shenoy B.D. Resolving the Colletotrichum siamense species complex using ApMat marker. Fungal Diversity. 2015;71:247–264. [Google Scholar]
- Shear C.L., Dodge B.O. The life history and identity of Patellina fragariae, Leptothryrium macrothecium, and Peziza oenotherae. Mycologia. 1921;13:135–170. [Google Scholar]
- Shen M., Zhang J.Q., Zhang Y. Venturia species form sooty mold-like colonies on leaves of Salix: introducing Venturia fuliginosa sp. nov. Mycosphere. 2017;7:1292–1300. [Google Scholar]
- Shivas R.G., Bathgate J., Podger F.D. Colletotrichum xanthorrhoeae sp. nov. on Xanthorrhoea in Western Australia. Mycological Research. 1998;102:280–282. [Google Scholar]
- Shivas R.G., Beasley D.R., McTaggart A.R. Online identification guides for Australian smut fungi (Ustilaginomycotina) and rust fungi (Pucciniales) IMA Fungus. 2014;5:195–202. doi: 10.5598/imafungus.2014.05.02.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva D.N., Talhinhas P., Varzea V. Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Mycologia. 2012;104:396–409. doi: 10.3852/11-145. [DOI] [PubMed] [Google Scholar]
- Simon U.K., Groenewald J.Z., Crous P.W. Cymadothea trifolii, an obligate biotrophic leaf parasite of Trifolium, belongs to Mycosphaerellaceae as shown by nuclear ribosomal DNA analyses. Persoonia. 2009;22:49–55. doi: 10.3767/003158509X425350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanesan A. Lubrecht & Cramer Ltd; Vaduz, Liechtenstein: 1977. The taxonomy and pathology of Venturia species. [Google Scholar]
- Sivanesan A. Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. Mycological Papers. 1987;158:1–261. [Google Scholar]
- Slippers B., Boissin E., Phillips A.J. Phylogenetic lineages in the Botryosphaeriales: a systematic and evolutionary framework. Studies in Mycology. 2013;76:31–49. doi: 10.3114/sim0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slippers B., Crous P.W., Denman S. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia. 2004;96:83–101. [PubMed] [Google Scholar]
- Slippers B., Fourie G., Crous P.W. Multiple gene sequences delimit Botryosphaeria australis sp. nov. from B. lutea. Mycologia. 2004;96:1030–1041. [PubMed] [Google Scholar]
- Slippers B., Fourie G., Crous P.W. Speciation and distribution of Botryosphaeria spp. on native and introduced Eucalyptus trees in Australia and South Africa. Studies in Mycology. 2004;50:343–358. [Google Scholar]
- Slippers B., Johnson G.I., Crous P.W. Phylogenetic and morphological re-evaluation of the Botryosphaeria species causing diseases of Mangifera indica. Mycologia. 2005;97:99–110. doi: 10.3852/mycologia.97.1.99. [DOI] [PubMed] [Google Scholar]
- Smith H., Crous P.W., Wingfield M.J. Botryosphaeria eucalyptorum sp. nov., a new species in the B. dothidea-complex on Eucalyptus in South Africa. Mycologia. 2001;93:277–285. [Google Scholar]
- Smith H., Wingfield M.J., Crous P.W. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany. 1996;62:86–88. [Google Scholar]
- Summerell B.A., Groenewald J.Z., Carnegie A.J. Eucalyptus microfungi known from culture. 2. Alysidiella, Fusculina and Phlogicylindrium genera nova, with notes on some other poorly known taxa. Fungal Diversity. 2006;23:323–350. [Google Scholar]
- Sutton B.C. Commonwealth Mycological Institute; Kew, UK: 1980. The Coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. [Google Scholar]
- Sutton B.C. On Stigmina, Wilsonomyces and Thyrostroma (Hyphomycetes) Arnoldia. 1997;14:33–35. [Google Scholar]
- Sutton B.C., Gibson I.A.S. Pezizella oenotherae. CMI Descriptions of Pathogenic Fungi and Bacteria. 1977;535:1–2. [Google Scholar]
- Sydow H. Mycotheca Germanica Fasc. LXIX–LXXII (No. 3401–3600) Annales Mycologici. 1942;40:193–218. [Google Scholar]
- Sydow H., Sydow P. Enumerations of Philippine fungi, with notes and descriptions of new species. Part 1: Micromycetes. Philippine Journal of Science Section C Botany. 1913;8:265–285. [Google Scholar]
- Sydow P., Sydow H. Verlag Von J. Cramer; Lipsiae, Germany: 1904. Monographia Uredinearum seu Specierum Omnium ad hunc usque Diem Descriptio et Adumbratio Systematica. Volume 1. Genus Puccinia. [Google Scholar]
- Sydow P., Sydow H. Bornträger; Leipzig, Germany: 1924. Monographia Uredinearum. Volume 4. [Google Scholar]
- Takahashi Y., Ichihashi Y., Sano T. Monilinia jezoensis sp. nov. in the Sclerotiniaceae, causing leaf blight and mummy fruit disease of Rhododendron kaempferi in Hokkaido, northern Japan. Mycoscience. 2005;46:106–109. [Google Scholar]
- Tan Y.P., Crous P.W., Shivas R.G. Eight novel Bipolaris species identified from John L. Alcorn's collections at the Queensland Plant Pathology Herbarium (BRIP) Mycological Progress. 2016;15:1203–1214. [Google Scholar]
- Tan Y.P., Madrid H., Crous P.W. Johnalcornia gen. et. comb. nov., and nine new combinations in Curvularia based on molecular phylogenetic analysis. Australasian Plant Pathology. 2014;43:589–603. [Google Scholar]
- Tao G., Liu Z.Y., Liu F. Endophytic Colletotrichum species from Bletilla ochracea (Orchidaceae), with description of seven new species. Fungal Diversity. 2013;61:139–164. [Google Scholar]
- Taylor K., Barber P.A., Hardy GEStJ. Botryosphaeriaceae from tuart (Eucalyptus gomphocephala) woodland, including descriptions of four new species. Mycological Research. 2009;113:337–353. doi: 10.1016/j.mycres.2008.11.010. [DOI] [PubMed] [Google Scholar]
- The Angiosperm Phylogeny G An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society. 2016;181:1–20. [Google Scholar]
- Tomaso-Peterson M., Jo Y.-K., Vines P. Curvularia malina sp. nov. incites a new disease of warm-season turfgrasses in the southeastern. Mycologia. 2016;108:915–924. doi: 10.3852/15-238. [DOI] [PubMed] [Google Scholar]
- Udayanga D., Manamgoda D.S., Liu X.-Z. What are the common anthracnose pathogens of tropical fruits? Fungal Diversity. 2013;61:165–179. [Google Scholar]
- Uematsu S., Kageyama K., Moriwaki J. Colletotrichum carthami comb nov., an anthracnose pathogen of safflower, garland chrysanthemum and pot marigold, revived by molecular phylogeny with authentic herbarium specimens. Journal of General Plant Pathology. 2012;78:316–330. [Google Scholar]
- Upadhyay H.P. University of Georgia Press; Athens, Georgia, USA: 1981. A monograph of Ceratocystis and Ceratocystiopsis. [Google Scholar]
- Van der Merwe M.M., Walker J., Ericson L. Coevolution with higher taxonomic host groups within the Puccinia/Uromyces rust lineage obscured by host jumps. Mycological Research. 2008;112:1387–1408. doi: 10.1016/j.mycres.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Van der Nest M.A., Beirn L.A., Crouch J.A. Draft genomes of Amanita jacksonii, Ceratocystis albifundus, Fusarium circinatum, Huntiella omanensis, Leptographium procerum, Rutstroemia sydowiana, and Sclerotinia echinophila. IMA Fungus. 2014;5:473–486. doi: 10.5598/imafungus.2014.05.02.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Nest M.A., Bihon W., De Vos L. Draft genome sequences of Diplodia sapinea, Ceratocystis manginecans, and Ceratocystis moniliformis. IMA Fungus. 2014;5:135–140. doi: 10.5598/imafungus.2014.05.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen G.C.M. Wageningen University; The Netherlands: 2000. The brown rot fungi of fruit crops (Monilinia spp.), with special reference to Monilinia fructigena (Aderh. & Ruhl.) Honey. Ph.D. dissertation. [Google Scholar]
- Van Leeuwen G.C.M., Baayen R.P., Holb I.J. Distinction of the Asiatic brown rot fungus Monilia polystroma sp. nov. from M. fructigena. Mycological Research. 2002;106:444–451. [Google Scholar]
- Van Niekerk J.M., Crous P.W., Groenewald J.Z. DNA phylogeny, morphology and pathogenicity of Botryosphaeria species on grapevines. Mycologia. 2004;96:781–798. [PubMed] [Google Scholar]
- Van Niekerk J.M., Groenewald J.Z., Verkley G.J.M. Systematic reappraisal of Coniella and Pilidiella, with specific reference to species occurring on Eucalyptus and Vitis in South Africa. Mycological Research. 2004;108:283–303. doi: 10.1017/s0953756204009268. [DOI] [PubMed] [Google Scholar]
- Van Wyk M., Al-Adawi A.O., Khan I.A. Ceratocystis manginecans sp. nov., causal agent of a destructive mango wilt disease in Oman and Pakistan. Fungal Diversity. 2007;27:213–230. [Google Scholar]
- Van Wyk M., Al-Adawi A.O., Wingfield B.D. DNA based characterization of Ceratocystis fimbriata isolates associated with mango decline in Oman. Australasian Plant Pathology. 2005;34:587–590. [Google Scholar]
- Van Wyk M., Roux J., Barnes I. Ceratocystis polychroma sp. nov., a new species from Syzygium aromaticum in Sulawesi. Studies in Mycology. 2004;50:273–282. [Google Scholar]
- Van Wyk M., Roux J., Nkuekam G.K. Ceratocystis eucalypticola sp. nov. from Eucalyptus in South Africa and comparison to global isolates from this tree. IMA Fungus. 2012;3:45–58. doi: 10.5598/imafungus.2012.03.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wyk M., Van der Merwe N.A., Roux J. Population genetic analyses suggest that the Eucalyptus fungal pathogen Ceratocystis fimbriata has been introduced into South Africa. South African Journal of Science. 2006;102:259–263. [Google Scholar]
- Van Wyk M., Wingfield B.D., Al-Adawi A.O. Two new Ceratocystis species associated with mango disease in Brazil. Mycotaxon. 2011;117:381–404. [Google Scholar]
- Van Wyk M., Wingfield B.D., Clegg P.A. Ceratocystis larium sp. nov., a new species from Styrax benzoin wounds associated with incense harvesting in Indonesia. Persoonia. 2009;22:75–82. doi: 10.3767/003158509X439076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wyk M., Wingfield B.D., Marin M. New Ceratocystis species infecting coffee, cacao, citrus and native trees in Colombia. Fungal Diversity. 2010;40:103–117. [Google Scholar]
- Van Wyk M., Wingfield B.D., Mohali S. Ceratocystis fimbriatomima, a new species in the C. fimbriata sensu lato complex isolated from Eucalyptus trees in Venezuela. Fungal Diversity. 2009;34:173–183. [Google Scholar]
- Van Wyk M., Wingfield B.D., Wingfield M.J. Four new Ceratocystis spp. associated with wounds on Eucalyptus, Schizolobium and Terminalia trees in Ecuador. Fungal Diversity. 2011;46:111–131. [Google Scholar]
- Van Wyk M., Wingfield B.D., Wingfield M.J. Ceratocystis species in the Ceratocystis fimbriata complex. In: Seifert K.A., De Beer Z.W., Wingfield M.J., editors. The Ophiostomatoid Fungi: expanding frontiers. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2013. pp. 65–73. [Google Scholar]
- Vasić M., Duduk N., Vico I. Comparative study of Monilinia fructigena and Monilia polystroma on morphological features, RFLP analysis, pathogenicity and histopathology. European Journal of Plant Pathology. 2016;144:15–30. [Google Scholar]
- Verkley G.J.M. A monograph of the genus Pezicula and its anamorphs. Studies in Mycology. 1999;44:1–180. [Google Scholar]
- Virtudazo E., Nakamura H., Kakishima M. Ribosomal DNA-ITS sequence polymorphism in the sugarcane rust, Puccinia kuehnii. Mycoscience. 2001;42:447–453. [Google Scholar]
- Voigt K., Cozijnsen A.J., Kroymann J. Phylogenetic relationships between members of the crucifer pathogenic Leptosphaeria maculans species complex as shown by mating type (MAT1-2), actin, and beta-tubulin sequences. Molecular Phylogenetics and Evolution. 2005;37:541–557. doi: 10.1016/j.ympev.2005.07.006. [DOI] [PubMed] [Google Scholar]
- von Arx J.A. 3rd edn. J Cramer; Liechtenstein: 1981. The genera of fungi sporulating in pure culture. [Google Scholar]
- von Höhnel F. Fragmente zur Mykologie. XIII Mitteilung (Nr. 642 bis 718) Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften Math.-naturw. Klasse Abt. I. 1911;120:379–484. [Google Scholar]
- Wang L., Sun X., Wei J.-G. A new endophytic fungus Neofabraea illicii isolated from Illicium verum. Mycoscience. 2015;56:332–339. [Google Scholar]
- Wang Y.C., Hao X.Y., Wang L. Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Kuntze) in China. Scientific Reports. 2016;6:35287. doi: 10.1038/srep35287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B.S., Johnston P.R., Damm U. The Colletotrichum gloeosporioides species complex. Studies in Mycology. 2012;73:115–180. doi: 10.3114/sim0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene N.N., Hyde K.D., Wanasinghe D.N. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Diversity. 2016;77:1–316. [Google Scholar]
- Wilken P.M., Steenkamp E.T., Wingfield M.J. Draft nuclear genome sequence for the plant pathogen, Ceratocystis fimbriata. IMA Fungus. 2013;4:357–358. doi: 10.5598/imafungus.2013.04.02.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield B.D., Ambler J.M., Coetzee M.P.A. Draft genome sequences of Armillaria fuscipes, Ceratocystiopsis minuta, Ceratocystis adiposa, Endoconidiophora laricicola, E. polonica and Penicillium freii DAOMC 242723. IMA Fungus. 2016;7:217–227. doi: 10.5598/imafungus.2016.07.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield B.D., Barnes I., De Beer Z.W. Draft genome sequences of Ceratocystis eucalypticola, Chrysoporthe cubensis, C. deuterocubensis, Davidsoniella virescens, Fusarium temperatum, Graphilbum fragrans, Penicillium nordicum, and Thielaviopsis musarum. IMA Fungus. 2015;6:493–506. doi: 10.5598/imafungus.2015.06.02.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield M.J., De Beer Z.W., Slippers B. One fungus, one name promotes progressive plant pathology. Molecular Plant Pathology. 2012;13:604–613. doi: 10.1111/j.1364-3703.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield B.D., Duong T.A., Hammerbacher A. Draft genome sequences for Ceratocystis fagacearum, C. harringtonii, Grosmannia penicillata, and Huntiella bhutanensis. IMA Fungus. 2016;7:317–323. doi: 10.5598/imafungus.2016.07.02.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield B.D., Ericson L., Szaro T. Phylogenetic patterns in the Uredinales. Australasian Plant Pathology. 2004;33:327–335. [Google Scholar]
- Wingfield B.D., Van Wyk M., Roos H. Ceratocystis: emerging evidence for discrete generic boundaries. In: Seifert K.A., De Beer Z.W., Wingfield M.J., editors. The Ophiostomatoid Fungi: expanding frontiers. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2013. pp. 57–64. [Google Scholar]
- Wingfield M.J., Roux J., Wingfield B.D. Ceratocystis and Ophiostoma: International spread, new associations and plant health. In: Seifert K.A., de Beer Z.W., Wingfield M.J., editors. The Ophiostomatoid Fungi: expanding frontiers. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2013. pp. 191–200. [Google Scholar]
- Xu J.-J., Qin S.-Y., Hao Y.-Y. A new species of Calonectria causing leaf disease of water lily in China. Mycotaxon. 2012;122:177–185. [Google Scholar]
- Yan J.Y., Jayawardena M.M.R.S., Goonasekara I.D. Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal Diversity. 2015;71:233–246. [Google Scholar]
- Yang H.A., Sweetingham M.W. Variation in morphology and pathogenicity of Pleiochaeta setosa isolates from Lupinus spp. and other legumes. Australasian Plant Pathology. 2002;31:273–280. [Google Scholar]
- Yang H.C., Haudenshield J.S., Hartman G.L. Colletotrichum incanum sp. nov., a curved-conidial species causing soybean anthracnose in USA. Mycologia. 2014;106:32–42. doi: 10.3852/13-013. [DOI] [PubMed] [Google Scholar]
- Yang T., Groenewald J.Z., Cheewangkoon R. Families, genera and species of Botryosphaeriales. Fungal Biology. 2017;121:322–346. doi: 10.1016/j.funbio.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Yang Y.L., Lei Z.Y., Cai L. New species and notes of Colletotrichum on daylilies (Hemerocallis spp.) Tropical Plant Pathology. 2012;37:165–174. [Google Scholar]
- Yang Y.L., Liu Z.Y., Cai L. Colletotrichum anthracnose of Amaryllidaceae. Fungal Diversity. 2009;39:123–146. [Google Scholar]
- Yin L.F., Chen S.N., Cai M.L. First report of brown rot of apricot caused by Monilia mumecola. Plant Disease. 2014;98:694. doi: 10.1094/PDIS-09-13-0995-PDN. [DOI] [PubMed] [Google Scholar]
- Yin L.G., Chen S.N., Chen G.K. Identification and characterization of three Monilinia species from plum in China. Plant Disease. 2015;99:1775–1783. doi: 10.1094/PDIS-12-14-1308-RE. [DOI] [PubMed] [Google Scholar]
- Yip H.Y. Coniella duckerae sp. nov. Transactions of the British Mycological Society. 1987;89:587–589. [Google Scholar]
- Zalar P., de Hoog G.S., Schroers H.-J. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Studies in Mycology. 2007;58:157–183. doi: 10.3114/sim.2007.58.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Q., Dou ZhP., Zhou Y.P. Venturia chinensis sp. nov., a new venturialean ascomycete from Khingan Mountains. Saudi Journal of Biological Sciences. 2016;23:592–597. doi: 10.1016/j.sjbs.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Li M.J. A new species of Bipolaris from the halophyte Sesuvium portulacastrum in Guangdong Province, China. Mycotaxon. 2009;109:289–300. [Google Scholar]
- Zhang Y., Crous P.W., Schoch C. A molecular, morphological and ecological re-appraisal of Venturiales – a new order of Dothideomycetes. Fungal Diversity. 2011;51:249–277. doi: 10.1007/s13225-011-0141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Kakishima M., Uzuhashi S. Multigene phylogenetic analysis of inter- and intraspecific relationships in Venturia nashicola and V. pirina. European Journal of Plant Pathology. 2012;132:245–258. [Google Scholar]
- Zhu X.Q., Niu C.W., Chen X.Y. Monilinia species associated with brown rot of cultivated apple and pear fruit in China. Plant Disease. 2016;100:2240–2250. doi: 10.1094/PDIS-03-16-0325-RE. [DOI] [PubMed] [Google Scholar]