Abstract
The Drosophila melanogaster cyclin-dependent protein kinase complex CycD/Cdk4 stimulates both cell cycle progression and cell growth (accumulation of mass). CycD/Cdk4 promotes cell cycle progression via the well-characterized RBF/E2F pathway, but our understanding of how growth is stimulated is still limited. To identify growth regulatory targets of CycD/Cdk4, we performed a loss-of-function screen for modifiers of CycD/Cdk4-induced overgrowth of the Drosophila eye. One mutation that suppressed CycD/Cdk4 was in a gene encoding the mitochondrial ribosomal protein, mRpL12. We show here that mRpL12 is required for CycD/Cdk4-induced cell growth. Cells homozygous mutant for mRpL12 have reduced mitochondrial activity, and exhibit growth defects that are very similar to those of cdk4 null cells. CycD/Cdk4 stimulates mitochondrial activity, and this is mRpL12 dependent. Hif-1 prolyl hydroxylase (Hph), another effector of CycD/Cdk4, regulates growth and is required for inhibition of the hypoxia-inducible transcription factor 1 (Hif-1). Both functions depend on mRpL12 dosage, suggesting that CycD/Cdk4, mRpL12 and Hph function together in a common pathway that controls cell growth via affecting mitochondrial activity.
Keywords: Cyclin D/Cdk4, Drosophila, growth, Hif prolyl hydroxylase, mitochondria
Introduction
In recent years several genetic pathways that are required and sufficient to regulate cell growth have been characterized in Drosophila melanogaster and other organisms. One example is the Drosophila cyclin-dependent protein kinase 4 (Cdk4), bound to Cyclin D (CycD) (Finley et al, 1996; Sauer et al, 1996). This complex stimulates cell growth, as well as cell proliferation (Datar et al, 2000; Meyer et al, 2000; Emmerich et al, 2004). Previous studies show that Drosophila CycD/Cdk4 regulates cell proliferation through inhibition of the pocket protein Rbf1 (Du et al, 1996; Datar et al, 2000; Xin et al, 2002), thereby increasing E2F1 activity, and the expression of cell cycle genes including the factors that are rate-limiting for G1/S and G2/M progression, Cyclin E and String/Cdc25 (Dimova et al, 2003). In contrast, the stimulation of growth by CycD/Cdk4 is independent of Rbf1 (Datar et al, 2000; Xin et al, 2002), but requires Hif prolyl hydroxylase (Hph; Frei and Edgar, 2004). Although the mechanism via which CycD/Cdk4 stimulates growth is still elusive, some data suggest that metabolic rates are increased. Compared to control cells, mitotic cells lacking Cdk4 grow and divide more slowly, and appear to have reduced metabolic activity (Meyer et al, 2000). Ectopic expression of CycD/Cdk4 leads to a stimulation of growth, seen as increased clonal growth in mitotic tissues, and larger cells in endoreplicative tissues (Datar et al, 2000). Interestingly, changes in CycD/Cdk4 activity also lead to significant alterations in organ and body size in adult flies (Datar et al, 2000; Meyer et al, 2000; Emmerich et al, 2004).
In addition to cell-autonomous genetic controls, extrinsic factors also affect growth. In Drosophila, growth is regulated in response to temperature, oxygen concentration and nutrition, but our knowledge of how these factors regulate cellular growth is still limited. At high temperatures, metabolic rates are increased, yet surprisingly, flies reared at high temperatures are smaller. Although flies develop faster at high temperatures, the net increase in mass is reduced, suggesting that high temperatures lead to reduced growth rates (Frazier et al, 2001). Flies reared under hypoxic conditions, 10% O2 instead of the normal 21%, are smaller (Supplementary Figure 1A; Frazier et al, 2001), and thus have reduced growth rates. This phenotype is even more pronounced at high temperatures, suggesting that oxygen could become rate limiting for growth. Accordingly, oxygen solubility does not increase with increasing temperatures (Frazier et al, 2001).
It is still unclear how lack of oxygen is sensed, or how this translates into slower growth. Several models have been proposed for an oxygen sensor, and mitochondria might play an important function (Haddad, 2004). The transcription factor Hif-1α/β, which is essential for the cellular response to hypoxia, is regulated in a complex manner, which includes control by prolyl hydroxylases of the HPH/PHD/EGLN family and the FIH-1 asparaginyl hydroxylase. These hydroxylases target Hif-1α, and regulate its degradation and transcriptional activity, respectively. Importantly, they require oxygen for catalytic activity, linking Hif-1α/β activity to the intracellular oxygen concentration (Bruick, 2003). Furthermore, it was proposed that HPHs are regulated in response to mitochondrial activity (Schroedl et al, 2002), and thus that these hydroxylases, in conjunction with mitochondria, could function as oxygen sensors.
Our understanding of how growth and metabolic activity are reduced during hypoxia is still limited. One obvious explanation involves mitochondria, which require oxygen for oxidative phosphorylation, and thus are the biggest consumers of oxygen. Therefore, a lack of oxygen should lead to reduced ATP levels, and thus to reduced growth. However, syncytial Drosophila embryos exposed to 2% oxygen retain normal ATP levels (DiGregorio et al, 2001), presumably through upregulation of glycolysis. This is also true for other tissues in other organisms. Therefore, more direct pathways are likely to exist to link metabolic rates to oxygen concentrations.
Given the importance of mitochondria, surprisingly little is known about how their activity is regulated in response to cellular growth, or vice versa. The mitochondrial DNA encodes several proteins required for oxidative phosphorylation, including subunits of electron transport enzymes. Thus, mitochondrial ribosomes are essential for energy production, and presumably for oxygen homeostasis. Yet, most mitochondrial ribosomal proteins are poorly characterized. In Drosophila, bonsaï mutants, defective for the mitochondrial ribosomal protein S15, show a strong reduction of mitochondrial activity in the gut. Moreover, growth rates are reduced in mutant animals (Galloni, 2003). To date, it is not known how mitochondrial translation is regulated with respect to growth rates, or how it might be coordinated with the metabolic state of the cell.
Here, we present the characterization of mRpL12, the Drosophila ortholog of mammalian MRPL12, a mitochondrial ribosomal protein. mRpL12 was identified in a genetic screen for modifiers of a CycD/Cdk4-stimulated overgrowth phenotype. mRpL12 mutant cells show mitochondrial defects, and have a cell-autonomous growth defect. The data suggest that CycD/Cdk4 mediates a link between cellular growth rates and mitochondrial activity. Hph, the only known effector of CycD/Cdk4-stimulated growth, depends on mRpL12 for activity, and therefore may be regulated in response to the mitochondrial activity.
Results
Drosophila eye imaginal discs have been used extensively to study cell division, cell cycle arrest, differentiation and apoptosis. GMR-Gal4 drives expression of genes under the control of a UAS-linked promoter, specifically in mostly postmitotic cells in the Drosophila eye (Figure 2C). CycD/Cdk4, driven by GMR-Gal4, led to bigger ommatidia and bristles, as well as an enlargement and rough appearance of the eye (Datar et al, 2000; Figure 1A and C). Using a deficiency collection (Bloomington Drosophila stock center), we screened for dominant modifiers of this phenotype. From 162 deficiencies, which covered 60–70% of the genome, four modifiers were isolated. We previously published the characterization of hph mutants, which were identified as suppressors of CycD/Cdk4-mediated overgrowth using one of these deficiencies (Frei, 2004; Frei and Edgar, 2004). Another deficiency, Df(3L)Scf-R6, led to a dominant suppression of the CycD/Cdk4-induced increases in ommatidia and bristle size (Figure 1A). This deficiency also suppressed the increase in eye size and the rough appearance. Df(3L)Scf-R11, which deletes a smaller segment within Df(3L)Scf-R6, showed the same suppression phenotype (Figure 1B). Subsequently, all available mutants within the region defined by Df(3L)Scf-R11 were tested, and one mutant, l(3)10534, showed the same suppression phenotype as the deficiencies (Figure 1C). l(3)10534 is a P-element insertion into the 5′UTR MRPL12, encoding the mitochondrial ribosomal protein mRpL12 (Figure 1D).
Figure 2.
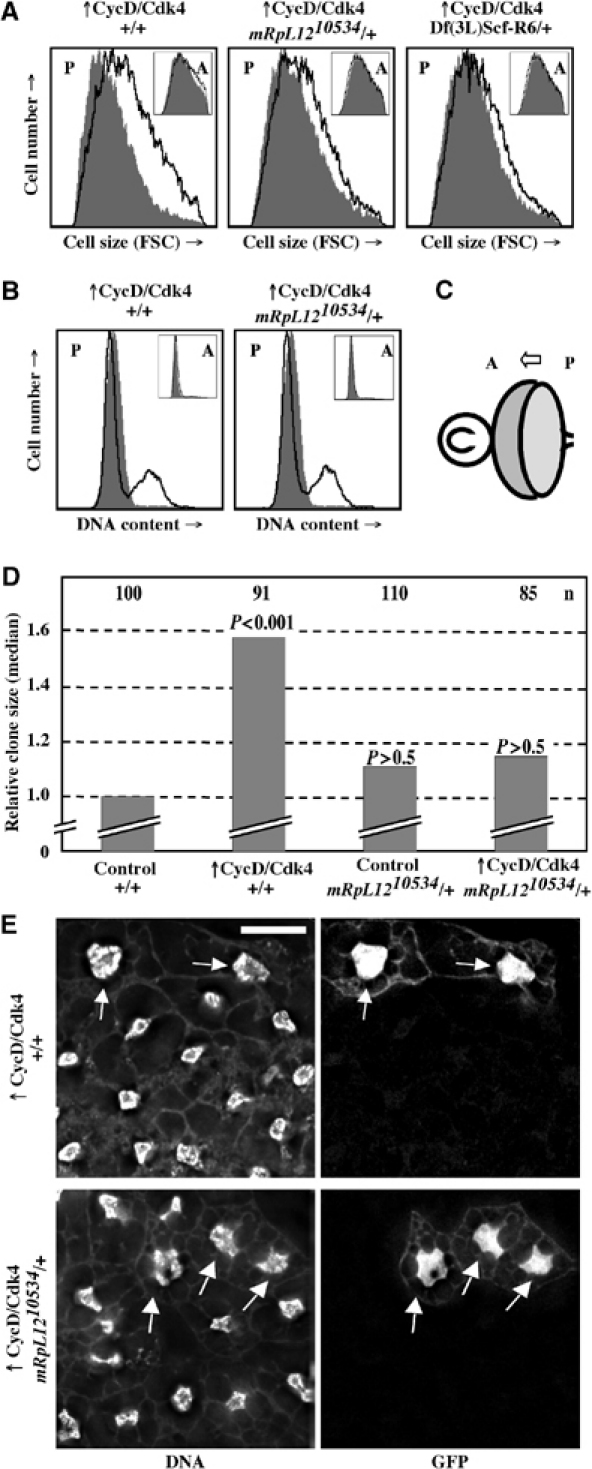
mRpL1210534 suppresses CycD/Cdk4-driven growth in the eye, wing and fat body. (A) FACS of eye imaginal discs from third instar larvae. The forward scatter (FSC; x-axis), indicative of the cell size, is blotted against the number of cells (y-axis). The black line represents posterior cells expressing CycD/Cdk4 together with GFP, compared to posterior cells expressing GFP only in the same background (filled gray). Insets are anterior cells. (B) FACS of pupal eye imaginal discs 48 h after white prepupae formation. DNA content (x-axis) is blotted against cell number (y-axis). GFP-negative cells are non-eye disc cells. (C) Drawing of third instar eye and antenna imaginal discs showing the morphogenetic furrow (arrow) moving from posterior to anterior. GMR-Gal4 leads to the expression of UAS transgenes in the posterior compartment. (D) Random clones in wing imaginal discs expressing GFP or CycD/Cdk4 plus GFP in mRpL1210534 or wild-type backgrounds were induced and the median clone areas were measured as described in Materials and methods. (E) Fat body clones expressing CycD/Cdk4 plus GFP were induced during embryogenesis in the fat body in wild-type (top) or an mRpL1210534/+ background (bottom). Larvae were fed until 72 h AED and starved for 5 days in 20% sucrose in 1 × PBS. Fat bodies were stained with DAPI, mounted and imaged as described in Materials and methods. CycD/Cdk4 leads to a 3.2-fold increase in DNA content in a wild-type background (n=33, P<0.01 compared to GFP alone). In the mRpL1210534/+ background, the increase in DNA is 1.6-fold (n=18, P>0.5). Scale bar, 20 μm.
Figure 1.
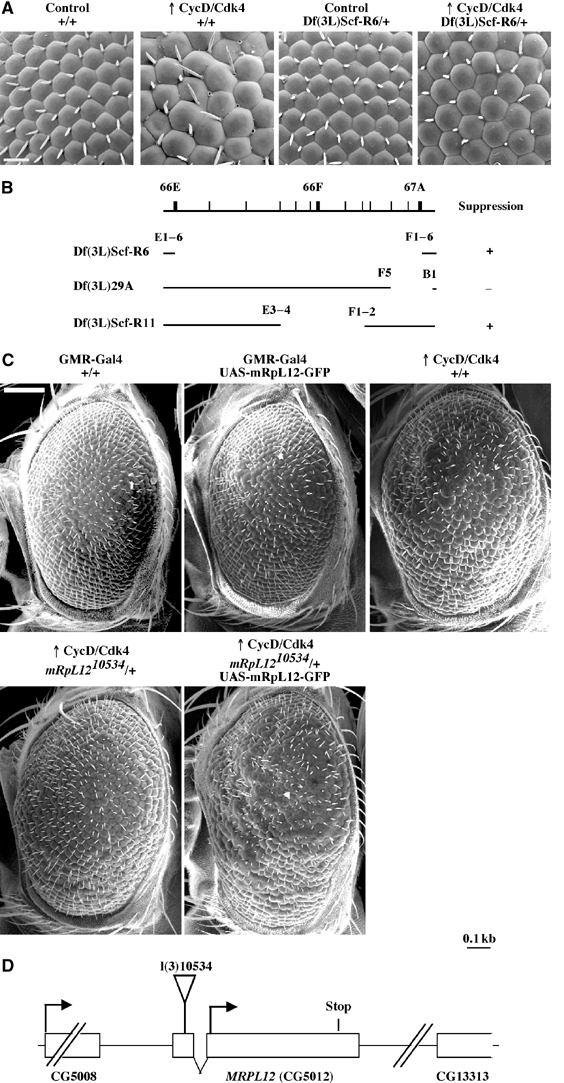
CycD/Cdk4 requires mRpL12 to drive growth in the Drosophila eye. (A) SEM images at × 500 magnification. Genotypes: GMR-Gal4/+; +/+ (left), GMR-Gal4 UAS-CycD UAS-Cdk4/+;+/+ (second from left), GMR-Gal4/+; Df(3L)Scf-R6/+ (third from left) and GMR-Gal4 UAS-CycD UAS-Cdk4/+; Df(3L)Scf-R6/+ (right). Scale bar, 20 μm. (B) Break points of the deficiencies used. ‘+' indicates suppression of the overgrowth phenotype whereas ‘−' indicates no change as compared to a wild-type background. (C) SEM at × 120 magnification. CycD/Cdk4 is driven from the GMR-Gal4 driver. ‘+/+' indicates precise excisions of P{PZ}10534. Scale bar: 100 μm. All flies in A and C are females, reared at 22.5°C. (D) Genomic locus of MRPL12. The lethal P-element (P{PZ}10534) insertion into the 5′UTR of MRPL12 is indicated. The 3′UTR of CG5008 ends 294 bp upstream of MRPL12, and the 5′UTR of CG13313 starts 600 bp downstream of MRPL12.
We next asked whether l(3)10534 is an allele of MRPL12. A precise excision of the P element rescued the lethality associated with l(3)10534 (data not shown), and restored the CycD/Cdk4-driven overgrowth phenotype (Figure 1C). Furthermore, a GFP-tagged transgene under the control of a UAS promoter (UAS-mRpL12-GFP; Amikura et al, 2001) was recombined on the l(3)10534 chromosome. Whereas expression of mRpL12-GFP alone was not sufficient to drive growth in the eye, this chromosome restored the CycD/Cdk4-specific suppression phenotype of l(3)10534 (Figure 1C). Finally, l(3)10534/Df(3L)Scf-R6 animals die during larval development, 70–80 h after egg deposition (AED). UAS-mRpL12-GFP, driven from the hs-Gal4 driver by three 1 h heat shocks per day (37°C), suppressed the larval lethality and the small size (Figure 3) of l(3)10534/Df(3L)Scf-R6 animals. These larvae were alive for up to 10 days AED, but most failed to enter pupal stages. Taken together, these data indicate that l(3)10534 is an allele of MRPL12, which we refer to as mRpL1210534.
Figure 3.
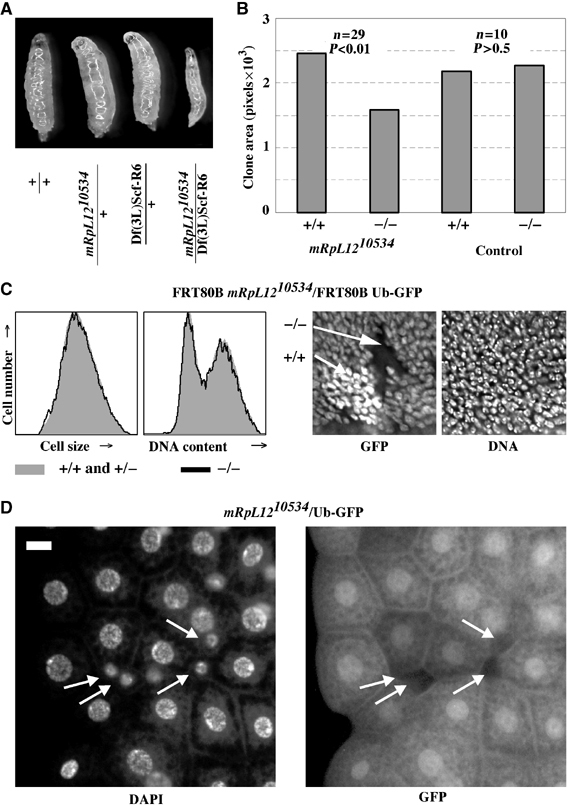
Homozygous mRpL1210534 cells have a cell-autonomous growth defect. (A) Images of larvae at 70 h AED. (B) Homozygous mRpL1210534 or control clones were induced at 66 h and dissected at 114 h AED, and wing discs were stained with DAPI and imaged. The area without GFP (−/−) and two copies of GFP (+/+) was measured in Photoshop. Genotypes: hs-Flp122; FRT80B mRpL1210534 or FRT80B/FRT80B Ub-GFP13A. (C) Wing discs from (B) were analyzed by FACS and GFP-negative cells (black line) were separated from GFP-positive cells (one or two copies; filled gray histogram. Shown are the forward scatters and DNA contents blotted against cell numbers (left). DAPI and GFP staining of a representative FRT80B mRpL1210534/FRT80B Ub-GFP13A twinspot (right). (D) Homozygous mRpL1210534 cells were induced in the fat body during embryogenesis by ionizing radiation, and third instar larvae were dissected and their fat body mounted. Homozygous mutant cells are marked by the absence of GFP (white arrows). Scale bar, 10 μm.
CycD/Cdk4 requires mRpL12 to drive growth in the eye imaginal disc
When CycD/Cdk4 is overexpressed in postmitotic cells in the larval eye, it augments cell growth, which is evidenced as an increase in cell size. Overexpressed CycD/Cdk4 also promotes ectopic cell proliferation posterior to the morphogenetic furrow (MF), and this can be assayed as an increase in cells in S, G2 and M phase of the cell cycle (Datar et al, 2000; Frei and Edgar, 2004). To test whether either or both of these effects might be suppressed by mRpL1210534, we expressed CycD/Cdk4 together with GFP and analyzed eye imaginal discs by fluorescence activated cell sorting (FACS). In a wild-type background, as shown previously (Datar et al, 2000; Frei and Edgar, 2004), CycD/Cdk4 led to an increase in forward scatter (FSC), indicating increased cell size (Figure 2A, left). In heterozygous mRpL1210534 or Df(3L)Scf-R6 backgrounds, this increase in FSC was reduced by about 50% (Figure 2A, middle and right). When cellular DNA content was assessed, we found that overexpressed CycD/Cdk4 was sufficient to drive cell cycle progression in eye imaginal disc cells. This was seen in third instar larvae (data not shown) or in pupae 48 h after prepupae formation (Figure 2B). Importantly, the increase of cells in S and G2/M phases was not reduced in mRpL1210534 heterozygotes. Therefore, as shown previously (Frei and Edgar, 2004), the growth and proliferation functions of CycD/Cdk4 can be separated. mRpL12 mutations specifically suppressed growth, but not proliferation induced by CycD/Cdk4. This also suggests that mRpL12 functions downstream of CycD/Cdk4.
mRpL1210534 suppresses CycD/Cdk4-driven growth in wings and fat bodies
Since CycD/Cdk4 stimulates growth in many tissues (all tested so far), we asked whether mRpL1210534 could also suppress CycD/Cdk4-driven growth in the wing imaginal disc and the fat body. During larval growth, wing imaginal disc cells grow exponentially and divide every 8–14 h. Expression of CycD/Cdk4 was induced in random clones, and clone areas were measured after a 48 h growth period. As shown previously (Datar et al, 2000; Frei and Edgar, 2004), CycD/Cdk4 stimulated growth, seen as a 60% increase in clone areas (Figure 2D). Since these cells also proliferate at an increased rate, they retain their normal size (Datar et al, 2000). When CycD/Cdk4 was expressed in a heterozygous mRpL1210534 background, the clone area was not statistically different from control clones (Figure 2D). Therefore, as seen in eye imaginal disc, reducing the dose of mRpL1210534 also suppresses growth stimulated by CycD/Cdk4 in the wing. Importantly, in either background, we did not observe any differences in cell size or cell cycle phasing (data not shown).
Endoreplicative tissues react very rapidly to changes in nutrient availability, and their DNA content correlates well with cell size. Upon starvation, fat body cells, which normally reach a C value of ∼256, stop endoreduplication and become autophagic (Britton and Edgar, 1998; Rusten et al, 2004; Scott et al, 2004). CycD/Cdk4 stimulates additional rounds of endoreduplication in normally fed animals (Datar et al, 2000) and also, to a greater extent, in starved animals. To test whether CycD/Cdk4 requires mRpL12 to stimulate growth in the fat body, we expressed CycD/Cdk4 in wild-type and mRpL1210534 heterozygous mutant backgrounds, and starved the animals for 5 days. The amount of DNA in CycD/Cdk4-expressing cells, marked with coexpressed GFP (Figure 2E, arrows), was compared to the DNA content of surrounding, GFP-negative cells (see Materials and methods). Under these conditions, CycD/Cdk4 induced a 3.2-fold increase in DNA content in a wild-type background, and a 1.6-fold increase in a heterozygous mRpL1210534 background. Thus, mRpL1210534 suppressed CycD/Cdk4-stimulated growth in eye and wing imaginal discs, as well as in the fat body.
Homozygous mRpL1210534 cells have reduced growth rates
Homozygous mRpL12 mutants die during larval stages 3–4 days AED. At 70 h AED, mutant mRpL1210534/Df(3L)Scf-R6 larvae were thinner than controls (Figure 3A) but did not have a developmental delay as they entered the second instar on time (data not shown). After 70 h AED, these animals died within 24 h, without reaching the third larval instar. To test whether this growth defect was cell autonomous, we recombined mRpL1210534 onto an FRT80B chromosome, and induced homozygous mutant cells in imaginal wing discs. Homozygous mutant cells were recognized by the absence of GFP, whereas heterozygous cells contained one copy and wild-type cells contained two copies of GFP. As shown in Figure 3B, mRpL12 mutant cells survived, but had a growth disadvantage that led to smaller clones compared to paired control clones. The reduced clone size did not appear to be due to an increase in apoptosis (data not shown). When assayed for their cell size and cell cycle phasing by FACS and microscopy, we did not detect any differences between homozygous mutant and control cells (Figure 3C). Therefore, mRpL1210534 mutant cells must grow and divide at a slower rate than normal cells. This phenotype is indistinguishable from cells lacking Cdk4 or Cyclin D (Meyer et al, 2000; Emmerich et al, 2004).
To test a cell-autonomous effect in the fat body, we induced homozygous mutant cells by irradiation of flies heterozygous for mRpL1210534 over a GFP marked chromosome during embryogenesis. Homozygous mutant cells were recognized by the absence of GFP. When analyzed in third instar larvae, mutant cells were very small and contained less DNA than control cells (Figure 3D, arrows). Hence, at least in wing imaginal discs and the fat body, cells lacking mRpL12 have a cell-autonomous growth defect.
mRpL12 mutants affect mitochondria
MRPL12 encodes a protein predicted to localize to mitochondria, and expression of a functional GFP-tagged protein colocalized with ribosomes in the germ plasm of Drosophila embryos (Amikura et al, 2001). To test whether mRpL12-GFP would localize to mitochondria in the fat body, we expressed the protein in random clones. To mark mitochondria, we used MitoTracker red, a dye specific for mitochondria. In fat body cells, we detected an almost perfect colocalization of mRpL12-GFP and MitoTracker red (Figure 4A). Furthermore, in wing imaginal disc cells, mRpL12-GFP also showed a localization pattern typical for mitochondria (data not shown), suggesting that mRpL12 is a mitochondrial protein. Next, cells homozygous mutant for mRpL12 were induced in the fat body. These cells, recognized by the absence of GFP, showed a strong reduction in MitoTracker red staining (Figure 4B, arrows).
Figure 4.
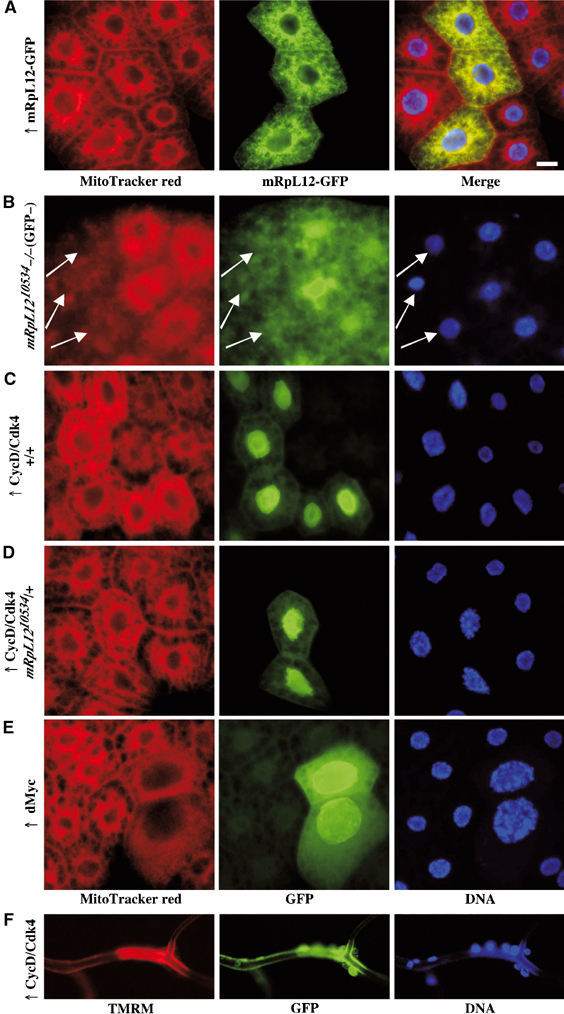
CycD/Cdk4 regulates mitochondrial function. (A–E) Fat body from third instar larvae were stained using MitoTracker red (left; see Materials and methods). (A) Ectopic expression of UAS-mRpL12-GFP in the fat body using the hs-Flp Act>CD2>Gal4 system. (B) Cells homozygous mutant for mRpL1210534 were induced by ionizing radiation. Mutant cells lack GFP (arrows). (C–F) Ectopic expression of CycD/Cdk4 or dMyc using the hs-Flp Act>CD2>Gal4 UAS-GFP system in a wild-type (C, E and F) or mRpL1210534/+ background (D). Expression was induced during embryogenesis without any heat shock. CycD/Cdk4- or dMyc-expressing cells are GFP positive. (E) Trachea were stained with TMRM. Scale bar, 20 μm.
CycD/Cdk4 affects mitochondrial activity. CycD/Cdk4 is a potent growth driver, suggesting that cells expressing CycD/Cdk4 require increased ATP levels. To test whether CycD/Cdk4 would increase mitochondrial activity, we used TMRM, as well as MitoTracker red. Both dyes require the proton gradient across the inner-mitochondrial membrane for binding, and thus correlate with mitochondrial activity. When trachea were stained with TMRM, CycD/Cdk4-expressing cells stained much brighter than control cells (Figure 4F). Similarly in the fat body, expression of CycD/Cdk4 led to a more intense MitoTracker red staining. These data suggest that CycD/Cdk4 stimulates the mitochondrial activity and/or mitochondrial numbers. Importantly, the increase in MitoTracker red was seen in a wild-type background (Figure 4C), but not in a heterozygous mRpL1210534 background (Figure 4D), suggesting that mRpl12 is required for CycD/Cdk4 to stimulate mitochondria. When expressed in a heterozygous hph background, another mutant that suppresses CycD/Cdk4-driven growth in the fat body (Frei and Edgar, 2004), CycD/Cdk4 still stimulated mitochondrial activity (data not shown). This indicates that increased MitoTracker red staining is not caused indirectly by increased cell size. To test whether other growth drivers would induce MitoTracker red staining, we expressed dMyc, a transcription factor known to stimulate growth in fat body cells (Saucedo and Edgar, 2002; Pierce et al, 2004). Although dMyc-expressing cells had several additional cycles of endoreplication, we did not detect an increase in MitoTracker red staining (Figure 4E), demonstrating that the increase seen for CycD/Cdk4 is not just a response to the higher ATP requirements.
Cytochrome c oxidase and succinate dehydrogenase activities are induced by CycD/Cdk4
To assay mitochondrial oxidative phosphorylation directly, we measured the activity of cytochrome c oxidase (COX), the complex IV of the electron transport chain. COX is a multiprotein enzyme, with subunits encoded by nuclear as well as mitochondrial genes. CycD/Cdk4, expressed in random clones in the fat body, led to increased COX stainings (Figure 5A, arrows). When stained for succinate dehydrogenase (SDH), complex II of the electron transport chain, we also detected an increase upon CycD/Cdk4 overexpression, however to a lesser extent than for COX (Figure 5A, arrows). We conclude that CycD/Cdk4 stimulates mitochondrial activity and/or abundance of mitochondria, most likely leading to increased ATP synthesis.
Figure 5.
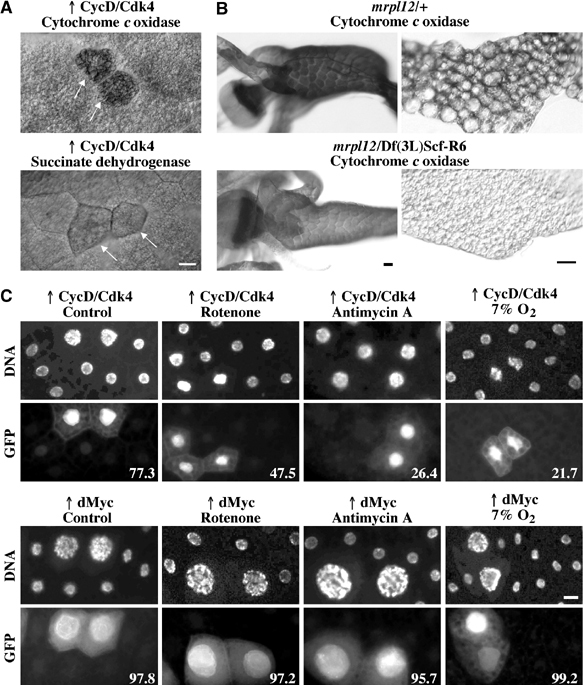
CycD/Cdk4 stimulates mitochondria in fat body cells. (A) COX (top) and SDH (bottom) activity in fat body cells. CycD/Cdk4-expressing cells were detected by coexpression of GFP in random clones, and are marked by arrows. (B) COX staining in second instar larvae, 77 h AED for the midgut and the proventriculus (left) and 96 h AED for the fat body (right). Images of heterozygous (mrpl1210534 or Df(3L)Scf-R6/+) or homozygous mutant (mrpl1210534/Df(3L)Scf-R6) larvae were exposed and treated identically in Photoshop. (C) Random clones expressing CycD/Cdk4 (top) or dMyc (bottom) were induced as described in Figure 2E, and marked by coexpression of GFP. At 24 h AED, larvae were transferred to normal food, or food supplemented with rotenone (5 μg/ml), antimycin A (5 μg/ml), or normal food and larvae were incubated at 7% O2. All were dissected at 116 h AED, fixed and stained with DAPI, and fat bodies were mounted. Numbers indicate the percentages of GFP-positive cells that are increased in ploidity, compared to neighboring cells. Quantifications were carried out in blind (n⩾150). Scale bar, 20 μm.
When mrpl12 homozygous mutant larvae were compared to heterozygous animals, we detected a significant reduction in COX staining in the midgut and the fat body (Figure 5B). This reduced staining was above background levels, possibly due to the fact that mrpl1210534 is a hypomorphic allele, and not a null. Together with the strong reduction in MitoTracker staining in mrpl12−/− clones (Figure 4B), we conclude that mRpL12 is required for mitochondrial activity.
CycD/Cdk4 requires mitochondrial activity to drive growth
To further test whether CycD/Cdk4 requires mitochondrial activity to drive growth, we overexpressed CycD/Cdk4 in random clones, and grew larvae in food supplemented with mitochondrial inhibitors. Rotenone and antimycin A are specific inhibitors of the mitochondrial electron transport enzyme NADH reductase (complex I) or cytochrome reductase (complex III), respectively. Since these drugs needed to be absorbed by the digestive system of the larvae, we first tested several different concentrations. Whereas 10 μg/ml was lethal, 5 μg/ml was tolerated and led only to a minimal delay in development (data not shown). Therefore, we used 5 μg/ml as a standard concentration. To measure CycD/Cdk4-driven growth, we coexpressed GFP, and counted the percentage of GFP-positive cells that showed a clear increase in ploidity upon expression of CycD/Cdk4. Under normal conditions, 77% of CycD/Cdk4 expression cells had increased DNA levels, whereas only 47% and 26% did so in the presence of rotenone or antimycin A, respectively (Figure 5C, top). Since the drugs are absorbed through the digestive tract, we also counted increased DNA levels in the gut, and found that both drugs greatly affected CycD/Cdk4-driven growth (data not shown).
We have shown previously that Hph is critical for CycD/Cdk4-driven growth (Frei and Edgar, 2004). To test further the dependence on Hph, we expressed CycD/Cdk4 in larvae grown under hypoxic conditions (7% O2). As mentioned above, this O2 concentration is expected to allow the retention of normal ATP levels (DiGregorio et al, 2001), but it is unclear whether this is achieved by normal ATP synthesis or by reduced ATP demand. Lavista-Llanos et al (2002) have shown that 7% O2 leads to Hif-1 activation, although to a lesser extent than 3–5% O2. Furthermore, the Km of mammalian HPH enzymes for oxygen is close to 21% (Hirsila et al, 2003), suggesting that 7% O2 might lead to greatly reduced activity of Drosophila Hph. Indeed, we found that hypoxia abolished CycD/Cdk4-driven overgrowth to a similar extent as mitochondrial inhibitors (Figure 5C). In the eye, the growth phenotype stimulated by CycD/Cdk4 is very sensitive and dosage dependent. In agreement with previous data (Frei and Edgar, 2004), we found that hypoxia suppressed CycD/Cdk4-dependent overgrowth of the eye to a similar extent as heterozygosity of mrpl12 or hph (Supplementary Figure 1A). These data further strengthen the importance of Hph for CycD/Cdk4-stimulated growth.
As described above, dMyc expression does not stimulate mitochondrial activity. To test whether mitochondria are required for dMyc function, we expressed dMyc in clones and added rotenone or antimycin A. Under these conditions, we did not see a reduction but rather a slight increase in growth stimulation in fat body cells. Furthermore, we did not see an effect of hypoxia treatment upon dMyc-driven growth (Figure 5C). These findings are surprising, since we would expect that increased growth necessarily requires increased mitochondrial activity. However, this is not the case, suggesting that dMyc does not require excess ATP to drive growth, or that increased ATP is provided by upregulation of glycolysis. In either case, these data further demonstrate that stimulation of mitochondrial activity by CycD/Cdk4 is not a secondary effect upon stimulation of growth.
Hph requires mRpL12 for activity
We recently found that mutations in hph also suppress CycD/Cdk4-driven overgrowth phenotypes (Frei and Edgar, 2004). In all tissues tested, the suppression by hph mutants is qualitatively and quantitatively similar to mRpL12 mutants. Particularly in the eye, both mutants suppressed the CycD/Cdk4-driven increase in cell size, but did not suppress increased proliferation (Figure 2A and B; Frei and Edgar, 2004). Ectopic expression of Hph is sufficient to drive growth in the wing imaginal disc. In contrast, overexpression of mRpL12 is not sufficient (data not shown). To test whether mRpL12 and Hph might function in the same pathway, we expressed Hph in an mRpL12 heterozygous mutant background, and measured clone area as described in Figure 2D. We found that Hph could not drive extra growth in this background (Figure 6A). Therefore, Hph requires mRpL12 for its growth-promoting activity, suggesting that CycD/Cdk4, mRpL12 and Hph function in the same pathway.
Figure 6.
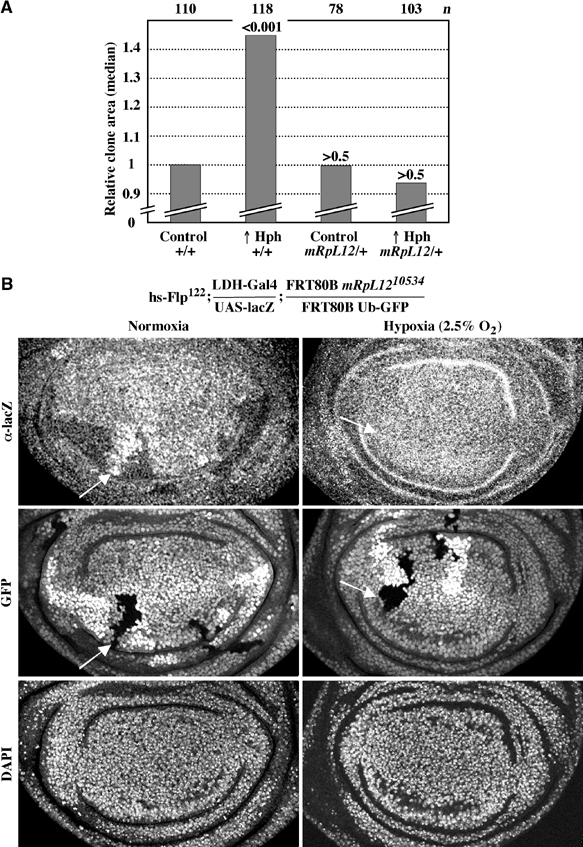
Normal mRpL12 levels are required for Hph function. (A) Hph was overexpressed in random clones in wing imaginal discs and clone area was measured as in Figure 2D. (B) Cells homozygous mutant for mRpL12 were induced by a heat shock 48 h AED. Larvae were either left at normoxia (left) or incubated at 2.5% oxygen for the last 7 h before dissection (right). Wing discs from third instar larvae were stained for lacZ or DNA, or imaged for GFP. Arrows point to a homozygous mutant clone.
In addition to driving growth, Hph is required for the cellular response to hypoxia. Mammalian HPH hydroxylates two proline residues of the hypoxia-induced transcription factor Hif-1α; upon hydroxylation, Hif-1α is recognized by an E3-ubiquitin ligase complex, and targeted for degradation. At hypoxia, hydroxylation activity is abolished, leading to stabilization of Hif-1α and dimerization with Hif-1β. Hif-1α/β then binds to the hypoxia responsive element (HRE) in promoters of target genes, and induces transcription (Bruick, 2003). There is good evidence that Drosophila Hph has a very similar function (Bruick and McKnight, 2001; Lavista-Llanos et al, 2002). To test whether mRpL12 might be involved in the hypoxic response, we used the LDH-gal4 reporter line, which expresses Gal4 under the control of four HREs and two cyclic AMP responsive elements (CREs). This construct is derived from the promoter of mammalian lactate dehydrogenase (Lavista-Llanos et al, 2002). HREs and CREs are often found in close proximity, but the importance of CREs in respect to hypoxia is still poorly understood in mammalian cells (Kvietikova et al, 1997; Braun et al, 2001; Miyazaki et al, 2002), and has not been addressed in Drosophila. Upon binding of Drosophila Hif-1 to the HRE, Gal4 induces synthesis of UAS-lacZ. Since Hif-1α/Sima is unstable at normoxia, lacZ is only detected under hypoxic conditions (Lavista-Llanos et al, 2002).
Cells homozygous mutant for mRpL12 were induced in the wing imaginal disc as described in Figure 3C, and stained for lacZ. We found that cells lacking mRpL12 showed ectopic lacZ staining (Figure 6B, left). Remarkably, wild-type cells had a lower lacZ staining compared to heterozygous mRpL1210534 cells. This suggests that Hif-1 activity is tightly regulated in response to mRpL12 levels. When fat body cells lacking mRpL12 were tested, we also detected ectopic Hif-1 activity (data not shown). Since this experiment was performed at normoxia, we conclude that mRpL12 is required for the repression of Hif-1 activity under normal oxygen concentrations. To test whether Hph is required for this regulation, we abolished Hph's activity by hypoxia, or by the addition of the iron chelator DFO. In either case, we detected increased Hif-1 activity, but did not see an effect in cells lacking mRpL12 (Figure 6B, right, and data not shown). Importantly, when we used an Hph-specific serum, we did not see any change in Hph protein levels in mrpl12−/− cells (Supplementary Figure 1B). This suggests that Hph activity, rather than protein level, is regulated in response to mRpL12. Since mRpL12 is required for mitochondrial activity, we propose that Hph activity may be regulated in response to mitochondrial activity.
Discussion
Mitochondria are required for the synthesis of ATP, as well as for many biological functions. Most mitochondrial proteins are encoded by nuclear genes, and are imported into mitochondria. The mitochondrial DNA contains only a handful of genes, encoding several subunits of enzymes required for oxidative phosphorylation. Therefore, translation of mitochondrial-encoded mRNAs is a prerequisite of mitochondrial function and thus ATP synthesis. However, our understanding of how mitochondrial protein synthesis is regulated and how it is regulated in response to nuclear genes is still limited.
Mammalian MRPL12 was the first mitochondrial ribosomal protein to be characterized, and is encoded by a nuclear gene. The protein forms a homodimer, localizes predominantly to mitochondria and binds to the large mitochondrial ribosomal subunit (Marty and Fort, 1996; Marty et al, 1997). In cultured cells, MRPL12 mRNA levels are induced by the addition of serum, and ectopic expression of a truncated version leads to reduced ATP synthesis and reduced growth. In bacteria, ribosomal proteins L7 and L12 are orthologs of MRPL12. L7 is identical to L12, except for an N-terminal acetyl group. The two proteins form a dimer and localize to a special structure on the large ribosomal subunit. This structure, the ‘ribosomal stalk', is composed of two L7/L12 dimers, as well as the L10 protein, and is required for the recruitment of translation elongation factors Tu and G to the large ribosomal subunit (Wahl and Moller, 2002; Gonzalo and Reboud, 2003). Therefore, L7/L12 are essential for mitochondrial protein synthesis, and thus for the generation of ATP.
The Drosophila genome encodes one mitochondrial L7/L12 homolog: mRpL12. This protein was first identified for its localization to mitochondria-type ribosomes in the germ plasm of Drosophila embryos (Amikura et al, 2001). We present here the identification of mRpL12 as a protein required for CycD/Cdk4 to drive cell growth. Importantly, the mRpL12 mutant showed no dominant suppression of other growth drivers, like dMyc or the insulin signaling pathway, suggesting that the suppression is specific for CycD/Cdk4 (data not shown). Cells lacking mRpL12 had strongly reduced MitoTracker and COX stainings. This suggests that the inner-mitochondrial membrane potential is reduced, presumably due to a decrease in the translation of mitochondrial-encoded subunits of the electron transport chain. Therefore, mRpL12 might have a function similar to bacterial L7/L12.
Our data show that CycD/Cdk4 stimulates mitochondrial activity, and that this increase is required for the stimulation of cellular growth. This induction requires mRpL12, suggesting that mitochondrial protein synthesis might be regulated in response to CycD/Cdk4. We still do not know how this is achieved. Our data do not exclude the possibility that mRpL12 has a function outside mitochondria, and that loss of this function causes suppression of CycD/Cdk4-stimulated growth. However, this seems very unlikely, since we saw an almost perfect colocalization of mRpL12-GFP with mitochondria, and because mitochondrial activity correlates well with CycD/Cdk4-driven growth.
We also tested whether other mutants, defective for mitochondrial ribosomes, would suppress CycD/Cdk4-driven overgrowth phenotypes. We found that the only other characterized mutant, bonsaï (mRpS15), did not suppress CycD/Cdk4-driven overgrowth in the eye when heterozygous (data not shown). Furthermore, we tested several lines predicted to be specific mutants for mRpL4, mRpL15, mRpL17, mRpS32, but none suppressed CycD/Cdk4 in heterozygous conditions (data not shown). Although none of the latter mutants are characterized, these data suggest that mRpL12 might be special among mitochondrial ribosomal proteins in its ability to suppress the action of CycD/Cdk4. Since our tests were performed in heterozygotes, this is not surprising; mRpL12 may be the only one of these components that is dosage-limiting for the activity of mitochondrial ribosomes. Alternatively, mRpL12 might uniquely be targeted by CycD/Cdk4. Nevertheless, the finding that CycD/Cdk4-driven growth is significantly reduced by the mitochondrial inhibitors rotenone and antimycin A supports the requirement of mitochondria.
Mammalian Hif prolyl hydroxylases (HPHs) are required for the cellular response to hypoxia (Bruick and McKnight, 2001; Epstein et al, 2001). These enzymes hydroxylate Hif-1α, leading to its ubiquitin-dependent degradation. Cells lacking Drosophila Hph have increased Hif-1α/Sima protein levels and transcriptional activity, demonstrating that fly Hph is an important regulator of Hif-1α/Sima (Lavista-Llanos et al, 2002). More recently, we showed that Drosophila Hph is also required for cellular growth, a function that is likely to be independent of Hif-1α/Sima (Frei and Edgar, 2004).
Mutants of mRpL12 and hph show very similar suppression phenotypes with respect to CycD/Cdk4. Furthermore, Hph requires mRpL12 to drive cell growth, and cells lacking mRpL12 have ectopic activation of Hif-1 (Figure 6). This suggests that CycD/Cdk4, mRpL12 and Hph function in the same pathway. The data presented here suggest that CycD/Cdk4 could have a dual function in growth control: First, CycD/Cdk4 stimulates mitochondrial activity, in an mRpL12-dependent but Hph-independent manner (Figure 4D, and data not shown). Second, Hph protein is regulated post-transcriptionally in response to CycD/Cdk4 (Frei and Edgar, 2004). Moreover, cells lacking mRpL12 have normal Hph protein levels, but increased Hif-1 activity. This suggests that Hph's hydroxylation activity may be regulated in response to the mitochondrial activity (see model in Figure 7).
Figure 7.
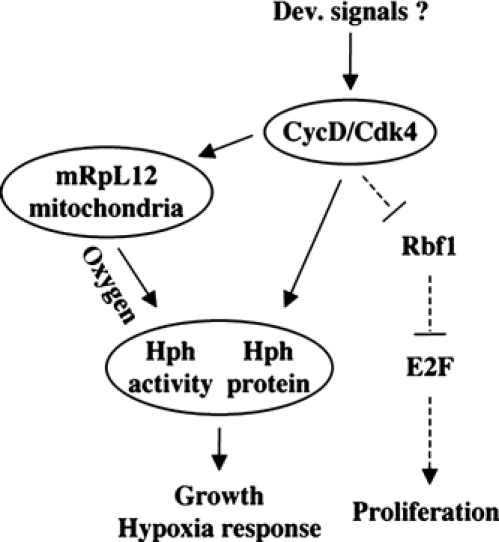
Model for the CycD/Cdk4–mRpL12–Hph pathway. mRpL12 and Hph are required for CycD/Cdk4-stimulated growth, but not proliferation, suggesting that they function downstream of CycD/Cdk4. CycD/Cdk4 has a dual function: post-transcriptional regulation of Hph protein levels (Frei and Edgar, 2004), and induction of mitochondrial activity, which is dependent on mRpL12. Furthermore, Hph activity depends on mRpL12, hence may be regulated in response to mitochondrial activity. Hph hydroxylation activity is required for stimulation of growth and inhibition of the transcription factor Hif-1α/Sima, which is essential for the cellular response to hypoxia.
Several previous studies in mammalian cells have shown that cells lacking functional mitochondria (ρ° cells) did not induce Hif-1 activity under hypoxic conditions (Chandel et al, 1998, 2000; Agani et al, 2000, 2002; Schroedl et al, 2002). In contrast, other studies did not find a mitochondrial role in the regulation of Hif-1 in mammalian cells (Srinivas et al, 2001; Vaux et al, 2001; Enomoto et al, 2002). Nevertheless, several models have been proposed for a mechanism that links mitochondrial activity to HPH and/or Hif-1α activity: First, reactive oxygen species (ROS), a by-product of oxidative phosphorylation, could regulate Hif-1α levels, but a possible mechanism is not known (Schumacker, 2002). Second, HPH hydroxylation activity depends on 2-oxoglutarate (α-ketoglutarate) for activity. 2-Oxoglutarate is an intermediate of the citrate cycle, and shuttles between mitochondria and the cytosol, where it is required for amino-acid synthesis. Third, the glycolysis product pyruvate, which gets imported into mitochondria, and oxaloacetate, a citrate cycle intermediate, can stabilize Hif-1α protein at normoxia (Lu et al, 2002; Dalgard et al, 2004). Therefore, in the latter two models, metabolites are good candidates for a link between mitochondria and Hph and/or Hif-1.
The data presented here are the first genetic evidence for a mitochondrial role in the regulation of Hph activity. We propose that Hph activity is induced by mitochondrial function (Figure 7), which contrasts the model proposed by Schroedl et al (2002), where mitochondria inhibit mammalian HPH hydroxylases. The reason for this discrepancy is unclear. In mammalian cells, the evidence that the effect of mitochondria on Hif-1 depends on HPH is indirect, and only supported by studies where HPH activity was inhibited by the iron chelator DFO (Chandel et al, 2000). Our model posits a link between oxygen sensing and growth rates: Hph's hydroxylation activity depends on oxygen, which is thus required for inhibition of Hif-1, as well as for stimulation of growth (Frei and Edgar, 2004). Therefore, Hph is a prime candidate for a protein that couples growth rates to oxygen concentrations. Since our data link Hph activity to mitochondrial activity, this pathway might also link growth and oxygen to the metabolic activity.
Materials and methods
Fly stocks
The fly stocks used were as follows: UAS-CycD, UAS-Cdk4 (Datar et al, 2000), mRpL1210534, Df(3L)Scf-R6, Df(3L)29A, Df(3L)Scf-R11, GMR-Gal4, FRT80B Ub-GFP13A (Bloomington), hs-Flp122 Act>CD2>Gal4 UAS-GFPS65T, hs-Flp122 Act>CD2>Gal4 (Neufeld et al, 1998), UAS-Hph (Frei and Edgar, 2004), UAS-mRpL12-GFP (Amikura et al, 2001), LDH-Gal4 (Lavista-Llanos et al, 2002) and UAS-dMyc42 (Zaffran et al, 1998).
Microscopy
Fat bodies were imaged on a Deltavision microscope (Applied Precision Inc.). Multiple 0.3 μm sections were deconvolved, and the amount (pixels3 × intensity) of the DAPI staining was calculated using SoftWoRx 2.5 software for GFP-positive cells as well as for 15–20 surrounding, GFP-negative cells of the same image. Pierce et al (2004) have shown that quantitation of DAPI signal by this method gives an accurate measurement of the DNA content. Wing discs were imaged on a Leica SP2 confocal microscope, using a × 40 objective. Rabbit α-lacZ (Cappel) was used at 1/1000.
Mitochondrial stainings
For MitoTracker red (Molecular Probes), five third instar larvae were fixed with paraformaldehyde for 90 min, and washed 30 min in PBS 1% Tween 20. MitoTracker red was used at 300 nM in PBS 1% Tween 20. The tissues were stained with DAPI (0.5 μg/ml), washed 30 min in PBS 1% Tween 20 and fat body was mounted. TMRM (tetramethylrhodamine methyl ester, at 2.5 nM; Molecular Probes) was added to live tissues in PBS, and DNA was stained using Hoechst 33342 (1 μg/ml) and washed in PBS.
Cytochrome c oxidase and succinate dehydrogenase assays
Second or third instar larvae were stained for COX and SDH as described previously (Maier et al, 2001; Galloni, 2003). For COX, four larvae/ml staining solution were incubated for 4 h, and mounted in 80% glycerol. For SDH, larvae were not fixed, dissected in staining solution and mounted in glycerol.
Hypoxia treatment
N2 (100%) was mixed with normal air using a ventilation pump (VENT2 pump, EMKA Technologies, France), and pumped at ∼1 l/min to a plethysmograph. Oxygen concentration was measured leaving the plethysmograph using a specific sensor (Electrovac, Austria).
Clone size measurements
Larvae from 4 h egg collection were transferred to yeasted vials, 50 larvae/vial, 24 h AED. Overexpression clones were induced using the hs-Flp Act>CD2>Gal4/UAS system at 37°C for 10 min at 66 h AED and third instar larvae were dissected at 114 h AED. Wing discs were dissected, mounted, imaged on a Leitz DMRD microscope using a × 20 objective, and the size of clones expressing GFP was measured in Photoshop. Imaging and quantitation were performed in blind and control was set to 1. P-values were calculated using standard T test with unequal variances compared to the control in the wild-type background.
Supplementary Material
Supplementary Figure
Acknowledgments
We thank S Buvelot Frei for critical reading of the manuscript, and members of the Edgar and Hafen laboratories for helpful discussions. We thank the Bloomington stock collection, S Kobayashi, P Wappner and K Basler for flies. Ch Hugentobler is thanked for help with SEM in Figure 1C. ChF was supported by fellowships from the Swiss National Science Foundation, the Swiss Cancer League and the Human Frontier Science Program. This study was supported by GM68105 to BAE.
References
- Agani FH, Pichiule P, Carlos Chavez J, LaManna JC (2002) Inhibitors of mitochondrial complex I attenuate the accumulation of hypoxia-inducible factor-1 during hypoxia in Hep3B cells. Comp Biochem Physiol A 132: 107–109 [DOI] [PubMed] [Google Scholar]
- Agani FH, Pichiule P, Chavez JC, LaManna JC (2000) The role of mitochondria in the regulation of hypoxia-inducible factor 1 expression during hypoxia. J Biol Chem 275: 35863–35867 [DOI] [PubMed] [Google Scholar]
- Amikura R, Kashikawa M, Nakamura A, Kobayashi S (2001) Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc Natl Acad Sci USA 98: 9133–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun L, Kardon T, Reisz-Porszasz ZS, Banhegyi G, Mandl J (2001) The regulation of the induction of vascular endothelial growth factor at the onset of diabetes in spontaneously diabetic rats. Life Sci 69: 2533–2542 [DOI] [PubMed] [Google Scholar]
- Britton JS, Edgar BA (1998) Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125: 2149–2158 [DOI] [PubMed] [Google Scholar]
- Bruick RK (2003) Oxygen sensing in the hypoxic response pathway: regulation of the hypoxia-inducible transcription factor. Genes Dev 17: 2614–2623 [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT (1998) Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci USA 95: 11715–11720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT (2000) Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem 275: 25130–25138 [DOI] [PubMed] [Google Scholar]
- Dalgard CL, Lu H, Mohyeldin A, Verma A (2004) Endogenous 2-oxoacids differentially regulate expression of oxygen sensors. Biochem J 380: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar SA, Jacobs HW, de la Cruz AF, Lehner CF, Edgar BA (2000) The Drosophila cyclin D–Cdk4 complex promotes cellular growth. EMBO J 19: 4543–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGregorio PJ, Ubersax JA, O'Farrell PH (2001) Hypoxia and nitric oxide induce a rapid, reversible cell cycle arrest of the Drosophila syncytial divisions. J Biol Chem 276: 1930–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ (2003) Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17: 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Vidal M, Xie JE, Dyson N (1996) RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev 10: 1206–1218 [DOI] [PubMed] [Google Scholar]
- Emmerich J, Meyer CA, de la Cruz AF, Edgar BA, Lehner CF (2004) Cyclin D does not provide essential cdk4-independent functions in Drosophila. Genetics 168: 867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto N, Koshikawa N, Gassmann M, Hayashi J, Takenaga K (2002) Hypoxic induction of hypoxia-inducible factor-1alpha and oxygen-regulated gene expression in mitochondrial DNA-depleted HeLa cells. Biochem Biophys Res Commun 297: 346–352 [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Finley RL Jr, Thomas BJ, Zipursky SL, Brent R (1996) Isolation of Drosophila cyclin D, a protein expressed in the morphogenetic furrow before entry into S phase. Proc Natl Acad Sci USA 93: 3011–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier MR, Woods HA, Harrison JF (2001) Interactive effects of rearing temperature and oxygen on the development of Drosophila melanogaster. Physiol Biochem Zool 74: 641–650 [DOI] [PubMed] [Google Scholar]
- Frei C (2004) Cyclin D/Cdk4: new insights from Drosophila. Cell Cycle 3: 558–560 [PubMed] [Google Scholar]
- Frei C, Edgar BA (2004) Drosophila cyclin D/cdk4 requires hif-1 prolyl hydroxylase to drive cell growth. Dev Cell 6: 241–251 [DOI] [PubMed] [Google Scholar]
- Galloni M (2003) Bonsai, a ribosomal protein S15 homolog, involved in gut mitochondrial activity and systemic growth. Dev Biol 264: 482–494 [DOI] [PubMed] [Google Scholar]
- Gonzalo P, Reboud JP (2003) The puzzling lateral flexible stalk of the ribosome. Biol Cell 95: 179–193 [DOI] [PubMed] [Google Scholar]
- Haddad JJ (2004) Oxygen sensing and oxidant/redox-related pathways. Biochem Biophys Res Commun 316: 969–977 [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J (2003) Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem 278: 30772–30780 [DOI] [PubMed] [Google Scholar]
- Kvietikova I, Wenger RH, Marti HH, Gassmann M (1997) The hypoxia-inducible factor-1 DNA recognition site is cAMP-responsive. Kidney Int 51: 564–566 [DOI] [PubMed] [Google Scholar]
- Lavista-Llanos S, Centanin L, Irisarri M, Russo DM, Gleadle JM, Bocca SN, Muzzopappa M, Ratcliffe PJ, Wappner P (2002) Control of the hypoxic response in Drosophila melanogaster by the basic helix–loop–helix PAS protein similar. Mol Cell Biol 22: 6842–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Forbes RA, Verma A (2002) Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J Biol Chem 277: 23111–23115 [DOI] [PubMed] [Google Scholar]
- Maier D, Farr CL, Poeck B, Alahari A, Vogel M, Fischer S, Kaguni LS, Schneuwly S (2001) Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol Biol Cell 12: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty L, Fort P (1996) A delayed-early response nuclear gene encoding MRPL12, the mitochondrial homologue to the bacterial translational regulator L7/L12 protein. J Biol Chem 271: 11468–11476 [DOI] [PubMed] [Google Scholar]
- Marty L, Taviaux S, Fort P (1997) Expression and human chromosomal localization to 17q25 of the growth-regulated gene encoding the mitochondrial ribosomal protein MRPL12. Genomics 41: 453–457 [DOI] [PubMed] [Google Scholar]
- Meyer CA, Jacobs HW, Datar SA, Du W, Edgar BA, Lehner CF (2000) Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J 19: 4533–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Kawamoto T, Tanimoto K, Nishiyama M, Honda H, Kato Y (2002) Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem 277: 47014–47021 [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA (1998) Coordination of growth and cell division in the Drosophila wing. Cell 93: 1183–1193 [DOI] [PubMed] [Google Scholar]
- Pierce SB, Yost C, Britton JS, Loo LW, Flynn EM, Edgar BA, Eisenman RN (2004) dMyc is required for larval growth and endoreplication in Drosophila. Development 131: 2317–2327 [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H (2004) Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell 7: 179–192 [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Edgar BA (2002) Why size matters: altering cell size. Curr Opin Genet Dev 12: 565–571 [DOI] [PubMed] [Google Scholar]
- Sauer K, Weigmann K, Sigrist S, Lehner CF (1996) Novel members of the cdc2-related kinase family in Drosophila: cdk4/6, cdk5, PFTAIRE, and PITSLRE kinase. Mol Biol Cell 7: 1759–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroedl C, McClintock DS, Budinger GR, Chandel NS (2002) Hypoxic but not anoxic stabilization of HIF-1alpha requires mitochondrial reactive oxygen species. Am J Physiol Lung Cell Mol Physiol 283: L922–L931 [DOI] [PubMed] [Google Scholar]
- Schumacker PT (2002) Hypoxia, anoxia, and O2 sensing: the search continues. Am J Physiol Lung Cell Mol Physiol 283: L918–L921 [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP (2004) Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7: 167–178 [DOI] [PubMed] [Google Scholar]
- Srinivas V, Leshchinsky I, Sang N, King MP, Minchenko A, Caro J (2001) Oxygen sensing and HIF-1 activation does not require an active mitochondrial respiratory chain electron-transfer pathway. J Biol Chem 276: 21995–21998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux EC, Metzen E, Yeates KM, Ratcliffe PJ (2001) Regulation of hypoxia-inducible factor is preserved in the absence of a functioning mitochondrial respiratory chain. Blood 98: 296–302 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Moller W (2002) Structure and function of the acidic ribosomal stalk proteins. Curr Protein Pept Sci 3: 93–106 [DOI] [PubMed] [Google Scholar]
- Xin S, Weng L, Xu J, Du W (2002) The role of RBF in developmentally regulated cell proliferation in the eye disc and in Cyclin D/Cdk4 induced cellular growth. Development 129: 1345–1356 [DOI] [PubMed] [Google Scholar]
- Zaffran S, Chartier A, Gallant P, Astier M, Arquier N, Doherty D, Gratecos D, Semeriva M (1998) A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development 125: 3571–3584 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure


