Abstract
Ubiquitin-dependent proteolysis regulates gene expression in many eukaryotic systems. Pof1 is an essential fission yeast F-box protein that is homologous to budding yeast Met30. Temperature-sensitive pof1 mutants display acute growth arrest with small cell size. Extragenic suppressor analysis identified Zip1, a bZIP (basic leucine zipper) transcription factor, as a target for Pof1. We show Zip1 is stabilized in pof1 mutants, Pof1 binds only phosphorylated forms of Zip1, and Zip1 is ubiquitylated in vivo, indicating that Zip1 is a substrate of SCFPof1. Genome-wide DNA microarray assay shows that many cadmium-induced genes are under the control of Zip1, suggesting Zip1 plays a role in cadmium response. Consistently, zip1 mutants are hypersensitive to cadmium and unlike wild type, lose cell viability under this stress. Intriguingly, cadmium exposure results in upregulation of Zip1 levels and leads wild-type cells to growth arrest with reduced cell size, reminiscent of pof1 phenotypes. Our results indicate that Zip1 mediates growth arrest in cadmium response, which is essential to maintain viability. Normally growing cells prevent this response through constitutive ubiquitylation and degradation of Zip1 via SCFPof1.
Keywords: cadmium, DNA microarray, proteolysis, SCF, ubiquitin
Introduction
All cells must respond to environmental changes, such as UV-irradiation or genotoxic compounds, in order to survive and proliferate. Unicellular organisms in particular have evolved to utilize rapid adaptive responses at a single-cell level against various types of adverse stresses. This adaptation commonly occurs at the level of gene expression. Recent gene expression profiling experiments have shown both the global and selective transcriptional changes that occur in response to various cellular stresses (Gasch et al, 2000; Causton et al, 2001; Chen et al, 2003). These responses involve alterations in the activity of specific transcription factors. New transcription factors must be synthesized or existing ones modified and activated in order to respond to external cues. Cellular responses against these stresses are often accompanied with checkpoint-mediated cell cycle or division arrest (Hartwell and Weinert, 1989). Distinct classes of checkpoint pathways are activated in response to different types of damages, thereby preventing progression of the cell cycle to maintain cell viability. Then, when inappropriate stresses are overcome or removed, checkpoint signaling is turned off, leading to inactivation of these transcription factors and resulting recovery of cell proliferation.
Proteolysis often plays a crucial role in the feedback control underlying transcription-dependent stress responses (Pahl and Baeuerle, 1996). The ubiquitin–proteasome pathway is one of the major pathways for targeted proteolysis in the eukaryotic cell (Hochstrasser, 1996; Hershko and Ciechanover, 1998). It involves the covalent attachment of a polyubiquitin chain to a substrate, marking it for recognition and degradation by the 26S proteasome, a large multisubunit protease. The attachment of ubiquitin to a substrate occurs in a series of reactions, with the final step of substrate recognition being dependent upon a ubiquitin ligase, or E3. Different classes of ubiquitin ligase are now known and one of the largest and most versatile classes identified is that of the SCF (Skp1-Cul1/Cdc53-F-bpx) ubiquitin ligases (Feldman et al, 1997; Skowyra et al, 1997). The SCF ubiquitin ligases are multiprotein complexes. Each one contains at least three common components, Skp1, cullin-1/Cdc53 and Rbx1/Roc1/Hrt1. In addition to this core complex, they contain a variable receptor subunit, known as an F-box protein. Multiple F-box proteins can be identified in all eukaryotic organisms (Bai et al, 1996; Patton et al, 1998). They normally consist of an N-terminally proximal F-box motif, which is necessary for the interaction with Skp1, and a C-terminal protein–protein interaction motif. Each F-box protein recognizes and recruits specific substrates to the SCF complex, thus the F-box proteins are ultimately responsible for the timing and substrate specificity of proteolysis.
The fission yeast Schizosaccharomyces pombe genome contains at least 16 F-box proteins identified by genetic analysis and sequence homology. Of these, only two, Pof1 and Pof6, are essential for cell viability (Kominami et al, 1998; Ikebe et al, 2002; Hermand et al, 2003; Lehmann et al, 2004). Identification of the substrates recognized by these F-box proteins will help in our understanding of the essential role of SCF-mediated proteolysis within the cell. Pof1 is a WD40 repeat containing F-box protein with homology to the budding yeast Saccharomyces cerevisiae F-box protein Met30. The SCFMet30 complex is known to downregulate the activity of the transcription factor Met4 through both proteolytic and proteolysis-independent pathways (Kaiser et al, 2000; Patton et al, 2000; Rouillon et al, 2000; Kuras et al, 2002; Flick et al, 2004). Met4 is a bZIP (basic leucine zipper)-type transcription factor that is required for transcription of genes in the methionine biosynthesis pathway; thus, cells deficient in Met4 activity are methionine auxotrophs. In addition, upregulation of Met4, as seen in a met30 mutant, results in a cell cycle arrest, the mechanism of which remains unknown (Patton et al, 2000).
In this study, we identify a bZIP transcription factor, Zip1, as the crucial substrate of SCFPof1. Inactivation of Zip1 rescues the lethality derived from Pof1 deficiency and overexpression of the zip1+ gene results in a growth arrest phenotype, similar to pof1 mutant cells. Unlike Met4, cells deleted for Zip1 are not auxotrophic for methionine or sulfur amino acids, instead showing acute sensitivity to cadmium stress. DNA microarray analysis suggests that Zip1 plays a critical role in transcription of genes specifically required for cadmium stress, thereby preventing cells from losing viability during exposure to this heavy metal. Thus, Pof1-Zip1-mediated responses comprise an essential network for the cadmium-responsive checkpoint pathway in fission yeast. Evolutional significance, in particular, in comparison with budding yeast Met30-Met4 and human systems, is discussed in the light of stress response mechanisms.
Results
Pof1 is a component of the SCF ubiquitin ligase
We previously showed that Pof1 binds Skp1 (Lehmann et al, 2004). In order to clarify whether Pof1 functions via an SCF complex, binding between Pof1 and cullin-1 (Pcu1) (Kominami et al, 1998) was examined. For this purpose, a strain was constructed in which the chromosomal pof1+ and pcu1+ genes were individually tagged with GST and 13Myc epitopes, respectively, at the C-terminus under the native promoter. Tagging did not interfere with protein function, as growth rate of these strains was indistinguishable compared to untagged wild-type cells. Immunoprecipitation was performed using anti-GST antibody. Immunoblotting showed that Pof1-GST co-precipitated with Pcu1-13Myc (Figure 1A). Thus, Pof1 is a component of the fission yeast SCF ubiquitin ligase.
Figure 1.
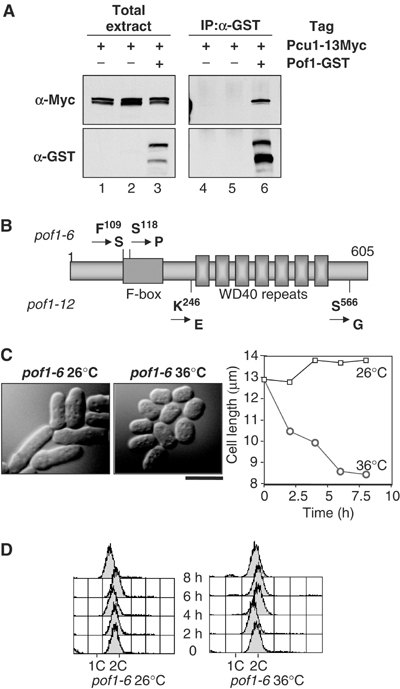
Complex formation between Pof1 and cullin-1 and creation of ts pof1 mutants. (A) Interaction between Pof1 and cullin-1. Protein extracts were prepared from a single tagged (pcu1+-13myc, lanes 1, 2, 4 and 5), or double-tagged strain (pof1+-GST pcu1+-13myc, lanes 3 and 6), and immunoprecipitation was performed with anti-GST antibody (lanes 4–6). After running on SDS–PAGE, immunoblotting was performed with anti-Myc (upper) or anti-GST antibody (lower). (B) Structure of Pof1 and mutation sites in pof1-6 and pof1-12. Pof1-6 contains two amino-acid replacements at P109S and S118P, while Pof1-12 carries K246E and S566G mutations. F-box motif and WD40 repeats are shown with boxes. (C) Cell morphology. A pof1-6 strain was grown at 26°C (left), shifted to 36°C and incubated for 6 h (middle). Phase-contrast microscopy images are shown. In addition, average cell length (n=200) was plotted at each time point. The bar indicates 10 μm. (D) DNA content. A pof1-6 strain was grown at 26°C (left) or 36°C (right) and samples were collected for flow cytometry at 2 h intervals. Note that a small G1 (1C) peak appeared at 6 and 8 h at 36°C.
Isolation of pof1 temperature-sensitive (ts) mutants
pof1+ is essential for cell viability (Lehmann et al, 2004), and spores containing deleted pof1 failed to germinate, which hampered the characterization of Pof1 function (C Harrison and T Toda, unpublished results). In order to examine the role of Pof1 further, we created ts pof1 alleles using mutagenic PCR. Briefly, genomic DNA was prepared from a strain in which the GFP gene (linked with the G418-resistance marker gene kanr) was inserted into the C-terminus of Pof1 under its endogenous promoter (pof1+-GFP-kanr). This 4.5 kb pof1+-GFP-kanr cassette was amplified with error-prone PCR, followed by transformation to a wild-type strain. G418-resistant colonies were selected at 27°C and ts transformants then isolated by replica plating at 36°C. This procedure resulted in the isolation of two ts pof1 alleles, pof1-6 and pof1-12. Nucleotide sequencing of the pof1 gene in these ts mutants showed that both contained two mutations (Figure 1B). The pof1-6 allele contained two point mutations in the F-box motif region, F109S and S118P, respectively. These would be predicted to interfere with Pof1 interactions with Skp1. The pof1-12 allele contained two mutations in the C-terminal region following the F-box motif, K246E and S566G. These mutations, situated on either side of the WD40 repeat domain, would thus be predicted to interfere with Pof1-substrate binding.
Initial characterization of these mutants showed that both alleles arrested at 36°C as small cells consisting of a majority of G2 cells and a small G1 population (Figure 1C and D). Average cell length was 8.5 μm after 6 h incubation at 36°C compared to 12.3 μm for wild-type cells under the same conditions (Table I, note that the cell width is indistinguishable between the two strains). This short cell phenotype appeared to be similar to wee mutants, in which mitosis occurs earlier than wild type due to premature activation of the Cdc2 kinase (Nurse, 1990). Unlike authentic wee mutants, however, the pof1 mutants did not show any genetic compensation with the G2 arrest cdc25-22 mutant, nor could any change in the phosphorylation state of Cdc2 at Y15 be observed (C Harrison and T Toda, unpublished results). This suggests that the pof1 phenotype is not due to a ‘wee-like' cell cycle defect, but instead is most likely a direct deficiency in macromolecular synthesis per se, which leads to small cell size and growth arrest.
Table 1.
Cell size comparison between wild type and pof1-6 mutant
| Strains | Cell length (μm) | Cell width (μm) |
|---|---|---|
| Wild type | 12.3±2.1 | 4.3±0.4 |
| pof1-6 | 8.5±1.2 | 4.4±0.4 |
| wee1-50 | 6.6±0.7 | 4.3±0.3 |
| Exponentially growing strains at 26°C were shifted to 36°C and incubated for 6 h. At least 300 cells were measured for each sample. | ||
Identification of zip1 as extragenic suppressors of pof1-6
Pof1 is required for cell viability and ts pof1 mutants displayed growth arrest at the restrictive temperature (Figure 1C). It is likely that uncoordinated accumulation or activation of some substrate(s) in the absence of SCFPof1 function could result in this arrest. If this scenario was the case, the extragenic mutation in a gene encoding this substrate might rescue the ts pof1 mutant. Given this prediction, we next sought to isolate Ts+ suppressors from pof1-6. Lawns of cells (∼108 cells) carrying the pof1-6 mutation were plated onto rich media and these plates left at the restrictive temperature, 36°C. After approximately 10 days, we could observe 23 colonies growing at this temperature. Backcrossing these revertants with a wild-type strain indicated that nine of these colonies contained some intragenic mutation, that is, the pof1-6 allele had reverted to wild-type function, whereas the remaining 14 isolates all contained an extragenic mutation, which suppressed the pof1-6 allele. Further genetic analysis showed that all 14 revertants contain the same, single locus, designated sup9.
Data from DNA microarray experiments (Chen et al, 2003) had identified pof1+ as a gene specifically upregulated in response to cadmium stress. This suggested that SCFPof1 activity is required in response to cadmium. This prompted us to test the sensitivity of cells containing the sup9 mutation to cadmium stress. These cells were indeed hypersensitive to cadmium stress (0.5 mM cadmium sulfate, Figure 2A). To identify this suppressor gene, the suppressor mutant was transformed with a plasmid-based gene library and screened for transformants, that could restore growth on plates containing 1.0 mM cadmium sulfate. Two independent transformants able to grow in the presence of cadmium were isolated and plasmid stability tests showed this phenotype was plasmid dependent. DNA sequencing revealed that both transformants contained identical plasmids, containing the entire ORF encoding a bZIP transcription factor, Zip1 (Ohmiya et al, 1999). Zip1 was originally isolated as a multicopy suppressor gene that rescued the calcium-sensitive phenotype of the mutation in atf1 encoding another bZIP. However, its physiological role remains to be determined.
Figure 2.
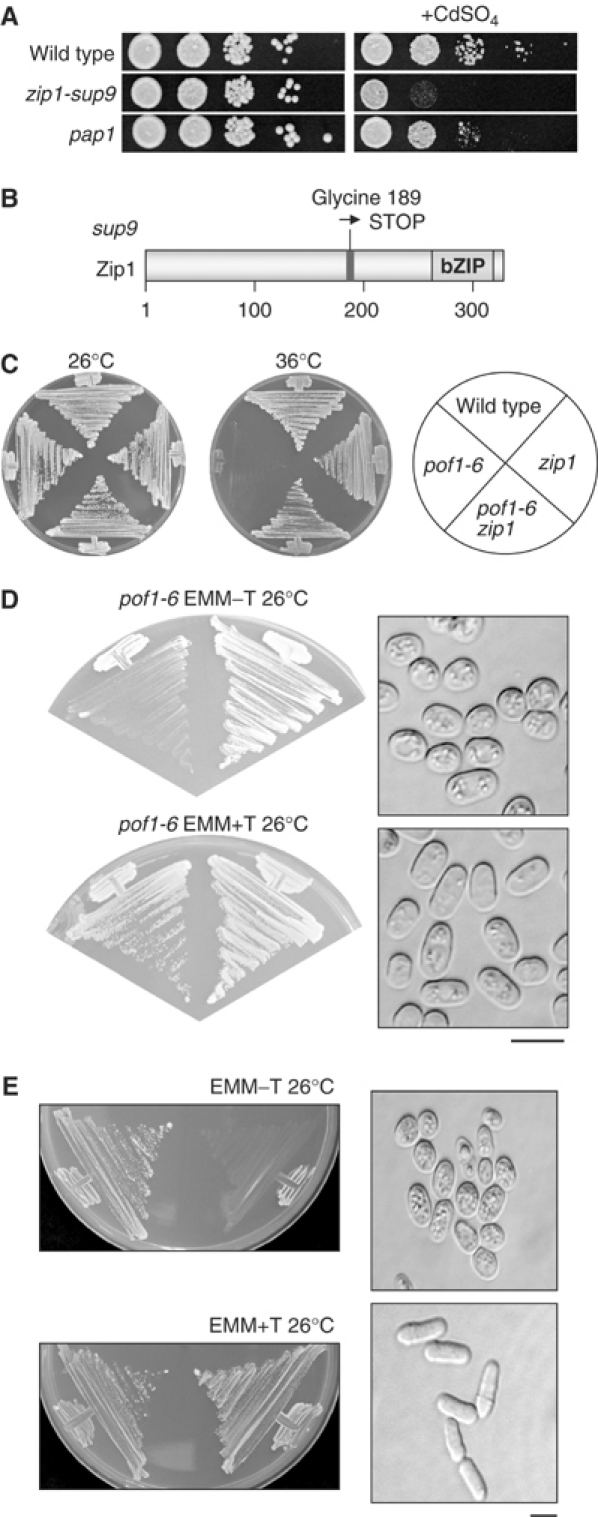
Identification of zip1 as an extragenic suppressor locus of pof1 mutants. (A) Cadmium hypersensitivity of sup9 (zip1). Wild-type (top row), sup9 (=zip1, middle) or pap1-deleted cells (bottom), which are defective in oxidative stress response (Toone and Jones, 1998), were spotted onto rich plates in the absence (left) or presence (right) of 0.5 mM cadmium sulfate (105 cells in the far-left spots for each plate and then diluted 10-fold in each subsequent spot rightwards) at 26°C and incubated for 4 days. (B) Schematic structure of Zip1. The position of the nonsense mutation in sup9 (from glycine to TGA) and the C-terminal ZIP region are shown. (C) Suppression of pof1-6 by zip1 deletion. Four strains (wild type, pof1-6, zip1 deletion and pof1-6 zip1) were streaked on rich medium and incubated at 26 or 36°C. (D, E) Toxicity of overexpressed zip1+. Plasmids containing zip1+ under the thiamine repressible mild nmt41 promoter (D) or strongest nmt1 promoter (E) were transformed into pof1-6 (D) or wild-type cells (E), respectively, streaked on minimal plates in the absence (upper) or presence (lower) of thiamine and incubated at 26°C for 3 days. Cell morphology of pof1-6 or wild-type cells containing zip1+ plasmids under each condition is shown in the right panels together with morphology of cells containing empty plasmids. The bar indicates 10 μm.
The zip1 gene in the sup9 suppressor strain was sequenced and found to contain a GGA to TGA nonsense mutation corresponding to amino-acid residue glycine 189. This resulted in the insertion of a STOP opal codon in the middle of the zip1 ORF prior to the C-terminal bZIP region (Figure 2B, sup9 is referred to as zip1 hereafter). This nonsense zip1 allele was also able to rescue growth of ts pof1-12 at 36°C (data not shown). To directly confirm that inactivation of Zip1 suppresses ts pof1-6, a complete deletion of the zip1+ ORF was constructed and crossed with a pof1-6 strain. The pof1-6 zip1 double mutant could grow at 36°C, verifying that loss of Zip1 function rescues ts pof1-6 (Figure 2C).
If Zip1 accumulation or activation was a direct cause for the phenotypic appearance of a pof1-6 strain at the restrictive temperature, further overexpression of the zip1+ gene in this mutant might result in inhibition of cell division even at the permissive temperature. To address this, the zip1+ gene was subcloned into a multicopy plasmid under the control of a thiamine-repressible mild promoter (nmt41-zip1+) (Basi et al, 1993). This plasmid was toxic to only pof1-6 cells, but not wild type, at 26°C and arrested cells mimicked the phenotype normally only seen at 36°C, with small cell size (Figure 2D). Moreover, when the zip1+ gene was expressed under the strongest promoter (nmt1-zip1+), growth was inhibited even in wild-type cells with reduced cell size (Figure 2E). Taken together, these data suggest that the small-size phenotype and temperature sensitivity of pof1 mutants can be explained by an excess or hyperactivation of the zip1+ gene product.
Zip1 is ubiquitylated in vivo
We addressed ubiquitylation of Zip1. In many cases, ubiquitylation of a protein leads to rapid proteasome-dependent degradation, which makes detection of ubiquitylated intermediates difficult. To circumvent this situation, ts mts3-1, a proteasome mutant (Seeger et al, 1996), was used and Zip1 was C-terminally tagged with an HA epitope (Zip1-HA) in this mutant. In order to visualize conjugated ubiquitin, this strain was transformed with a multicopy plasmid containing ubiquitin tagged with an HIS epitope (His-Ubi). After incubation for 1 h at the restrictive temperature to inactivate mts3, Ni2+ chelate resin was used to pull down all ubiquitylated conjugates in the cell and the resulting extract blotted with anti-HA antibody to detect the presence of Zip1-HA. A characteristic smear pattern was seen in the His-Ubi lane (Figure 3A, lanes 1 and 3), which was missing in the extract from cells transformed with empty vector (lanes 2 and 4). This indicates that Zip1 is ubiquitylated in vivo.
Figure 3.
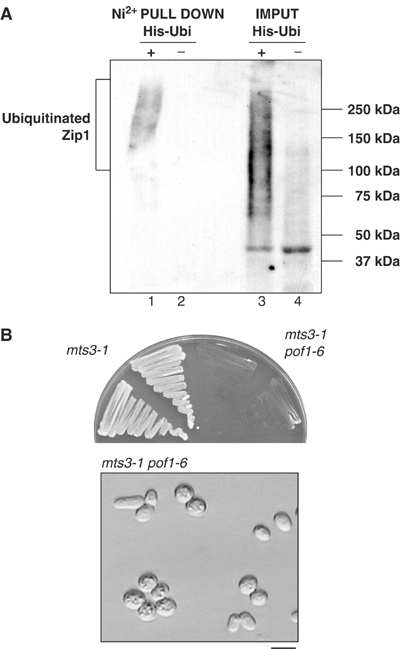
Ubiquitylation of Zip1. (A) Zip1 is polyubiquitylated in vivo. Plasmids containing 6His-ubiquitin (His-Ubi, lanes 1 and 3) or empty vector (lanes 2 and 4) were introduced into the mts3-1 mutant containing Zip1-HA. After 1 h incubation at 36°C, ubiquitylated proteins were purified with Ni2+-NTA beads (lanes 1 and 2). Total extracts (60 μg) are also run (lanes 3 and 4). Immunoblotting was performed with anti-HA antibody. (B) Synthetic phenotype between pof1-6 and mts3-1. mts3-1 single (left) or mts3-1 pof1-6 double mutants (right) containing Zip1-HA were streaked on rich medium and incubated at 26°C for 3 days (upper). Cell morphology for mts3-1 pof1-6 is also shown (lower). The bar indicates 10 μm.
The next obvious question to ask was whether ubiquitylation of Zip1 was dependent upon Pof1 activity. In order to answer this question, we created pof1-6mts3-1 double mutants carrying Zip1-HA. This strain was, however, extremely slow growing even at 26°C and practically impossible to propagate (Figure 3B, upper). Examination of these cells from plates showed that they displayed small-sized cell phenotype, identical to pof1-6 at the restrictive temperature (Figure 3B, lower). Although this meant dependency of Zip1 ubiquitylation upon Pof1 could not be directly addressed, these data in itself strongly imply that Pof1 and the proteasome are acting additively to inactivate the same substrate, Zip1.
Pof1 is required for Zip1 instability
We next asked whether the stability of the Zip1 protein is dependent upon Pof1 by examining the half-life of Zip1-HA in wild-type and pof1-6 cells. Exponentially growing cultures of both strains were treated with protein synthesis inhibitor cycloheximide (CHX) to shut off de novo protein synthesis and Zip1 levels were examined at regular intervals afterwards. It was clear from the initial time point that pof1-6 cells have increased total Zip1 levels when compared to wild-type cells (Figure 4A, lane 2). In addition, Zip1 in this mutant accumulated in slower migrating forms, while only the fastest band was seen in wild-type cells. Zip1-HA disappeared quickly in wild-type cells upon addition of CHX; after 15 min, very little signal was detected (upper panel, lane 3). In clear contrast, in the pof1-6 mutant, the faster migrating Zip1-HA band showed an increased half-life compared to wild type (lower panel and Figure 4B for quantification). Slower migrating bands, however, seemed to be still degraded relatively efficiently even in pof1-6. It is of note that not only Zip1 but also the Pof1 protein was upregulated in pof1-6, although in this case Pof1-GFP was as unstable as in wild-type cells (Figure 4A, lower panel). These increased levels of Pof1 in pof1-6 are probably attributable to transcriptional activation of the pof1+ gene via accumulation of Zip1, as pof1+ gene expression is dependent upon Zip1 activity (see below).
Figure 4.
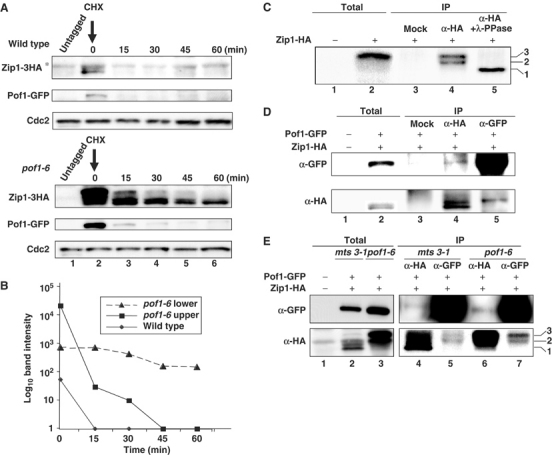
Stabilization of Zip1 in pof1 mutants and binding between Pof1 and phosphorylated Zip1. (A) Accumulation and partial stabilization of Zip1 in pof1-6. Exponentially growing wild-type pof1+-GFP (upper gel) or pof1-6-GFP (lower) cells containing Zip1-HA were shifted to 36°C for 2 h and CHX (100 μg/ml) was then added (time 0). Protein samples were prepared at indicated time points. Immunoblotting was performed with anti-HA (top), anti-GFP (middle) or anti-Cdc2 antibody (bottom). Samples from untagged strains were also run as a negative control (lane 1). Nonspecific band is marked with asterisk. (B) Quantification of Zip1 levels. Zip1-HA amount shown in (A) was quantified. Intensities of upper (squares), or lower bands (triangles) of Zip1-HA in pof1-6 and a lower band (diamonds) in wild type were measured. (C) Phosphorylation of Zip1. pof1-6 mutant cells containing Zip1-HA were shifted to 36°C for 4 h and immunoprecipitation was performed with mock (lane 3) or anti-HA antibody (lanes 4 and 5). Immunoprecipitates were treated with λ-protein phosphatase for 30 min at 30°C (lane 5). Total extract (30 μg) was also run (lane 2). As a negative control, protein samples prepared from untagged pof1-6 were also run (lane 1). Immunoblotting was performed with anti-HA antibody. Three Zip1-HA bands are marked. (D, E) Binding between Zip1 and Pof1. Immunoprecipitation was performed with anti-HA (lanes 4 and 6), anti-GFP antibody (lanes 5 and 7) or mock (lane 3 in D), using protein extracts prepared from mts3-1 (D and lanes 2, 4 and 5 in E) or pof1-6 mutants containing Pof1-GFP Zip1-HA (lanes 3, 6 and 7 in E), which were incubated at 36°C for 1 h. Total extracts from a tagged (lane 2) or untagged strain (lane 1) were also run. Immunoblotting was performed with anti-GFP (upper) or anti-HA antibody (lower).
Pof1 binds phosphorylated forms of Zip1
Phosphorylation is known to be a requirement of many F-box proteins for substrate recognition (Patton et al, 1998; Nash et al, 2001). Slower migrating bands of Zip1-HA existing in pof1-6 cells prompted us to examine whether these bands are phosphorylated forms. We tested this possibility by immunopurifying Zip1-HA from pof1-6 cells, then treating with λ-phosphatase. This confirmed that Zip1 accumulates in apparently two modified forms in pof1-6 cells, both of which are phosphorylated (Figure 4C, lanes 4 and 5, two phosphorylated bands are marked with 2 and 3, while the fastest migrating unphosphorylated band is shown as 1).
If Zip1 is a substrate for the SCFPof1, Pof1 is expected to bind Zip1. Immunoprecipitation was performed in a strain containing Pof1-GFP and Zip1-HA. To prevent degradation of Zip1, experiments were again carried out in an mts3-1 background. Under this condition, Zip1 was detected as a doublet, the lower band of which corresponded to the band seen in wild-type cells (band 1 in Figure 4C) and the upper of which appeared to correlate to the faster migrating form (band 2 in Figure 4C) of the phosphorylated Zip1 bands (Figure 4D, lanes 2 and 4). Immunoprecipitation using anti-GFP antibody showed that Pof1 co-immunoprecipitated with Zip1, specifically the upper band of the doublet (Figure 4D, lane 5). Reciprocal immunoprecipitation with anti-HA antibody also co-precipitated Pof1-GFP, albeit very faintly (lane 4). This is probably due to the existence of an excess amount of free Zip1-HA dissociated from Pof1-GFP in the mts3-1 mutant.
In order to examine which of Zip1 forms binds Pof1 in the pof1-6 background, immunoprecipitation was performed using samples prepared from the pof1-6 mutant used in Figure 4A (doubly tagged with Pof1-6-GFP and Zip1-HA). For comparison, parallel immunoprecipitation was also performed in extracts from mts3-1 cells and precipitates were run side-by-side. As shown in Figure 4E, it is evident that two phosphorylated Zip1 forms interacted with Pof1-6 (lane 7, marked as bands 2 and 3). Taken together, these findings indicate that Zip1 is phosphorylated and this phosphorylation leads to an interaction with Pof1 and subsequent degradation.
Zip1 regulates expression of cadmium stress specific genes
Having identified Zip1 as a critical substrate of SCFPof1, we next explored the physiological role of Zip1. The hypersensitivity of zip1 mutant cells to cadmium suggested that Zip1 plays some role in the cellular response against cadmium stress. In order to study this further, we utilized DNA microarrays to compare genome-wide transcriptional profiles of wild-type and zip1 cells upon cadmium stress. Both strains were exposed to cadmium sulfate and at 0, 15 and 60 min time points, total RNA was prepared and microarray data were collected as performed previously (Chen et al, 2003). This analysis showed that a set of 27 genes appeared to be specific targets of Zip1, having less than 50% reduction in expression in a zip1 strain compared to wild type at either 15 or 60 min after addition of cadmium (Table II). Noticeably, 20 out of these 27 genes were induced more than two-fold in wild-type cells in response to cadmium exposure (Figure 5A and see genes shown with bold letters in Table II).
Table 2.
Zip1 target genes
| Gene name | Function | Budding yeast | Induction by cadmium | zip1 versus wild type |
|---|---|---|---|---|
| SPAC869.05c | Sulfate transporter | Sul1 | 16.4 | 0.1 |
| SPCC1739.06c | Uroporphyrin methyltransferase | Met1 | 7.8 | 0.1 |
| SPCPB1C11.03 | Membrane transporter | 4.1 | 0.1 | |
| SPBPB10D8.02c | Arylsulfatase | 7.3 | 0.1 | |
| SPBPB10D8.01 | Membrane transporter | 2.7 | 0.1 | |
| pof1+ | F-box protein | Met30 | 5.7 | 0.2 |
| SPBC106.17c | Homoserine O-acetyltransferase | Met6 | 3.5 | 0.1 |
| SPBPB2B2.08 | Unknown | 25 | 0.1 | |
| SPBP16F5.08c | Flavin-dependent monooxygenase | Fmo1 | 4.4 | 0.1 |
| SPAC10F6.01c | Sulfite reductase | Ecm17 | 1.3 | 0.3 |
| frp1+ | Ferric-chelate reductase | Fre1–4 | 3.8 | 0.4 |
| SPBP8B7.05c | Carbonic anhydrase | Nce103 | 1.7 | 0.3 |
| cad1+/hmt2+ | Sulfide-quinone oxidoreductase | 2.2 | 0.4 | |
| SPBC725.04 | Oxalyl-CoA decarboxylase | 3.0 | 0.4 | |
| SPAC23A1.14c | Cystathionine γ-synthase | 5.8 | 0.4 | |
| SPCC584.01c | Sulfite reductase | Met10 | 1.3 | 0.4 |
| SPCC965.06 | Potassium channel subunit | 21.8 | 0.5 | |
| SPBPB10D8.04c | Malate permease | Ssu1 | 1.2 | 0.5 |
| SPCC1827.03c | Long-chain fatty acid transporter | Fat2 | 1.2 | 0.3 |
| SPBC27.04 | Coiled-coil protein | Rad50 | 2.6 | 0.2 |
| fip1+ | Iron permease | Fth1, Ftr1 | 1.3 | 0.2 |
| SPBC1348.06c | Unknown | Fun19 | 21.0 | 0.3 |
| Similarity to SPAC977.05c and SPBPB2B2.15 | ||||
| SPAC977.05c | Unknown | Fun19 | 20.0 | 0.3 |
| Similarity to SPBC1348.06c and SPBPB2B2.15 | ||||
| cdc15+ | SH3 and FCH containing | Hof1 | 1.8 | 0.3 |
| SPBPB2B2.05 | GMP synthase | 5.6 | 0.4 | |
| cdc22+ | Ribonucleoside-diphosphate | 2.2 | 0.4 | |
| reductase (large subunit) | Rnr1, Rnr3 | |||
| SPCC622.12c | NADP glutamate dehydrogenase | Gdh1, Gdh3 | 2.6 | 0.5 |
| A total of 27 genes, in which the induction rate between zip1 (sup9) and wild-type strains (zip1 versus wild type) was less than 50%, are listed. Particularly, those (20 genes) that show more than two-fold induction at either 15 or 60 min upon cadmium stress in wild-type cells are emphasized by bold letters. Furthermore, among 20 of these genes, the induction of 15 genes is specific to cadmium stress (Chen et al, 2003) and these genes are shown with underlined bold letters (see Figure 5A). | ||||
Figure 5.
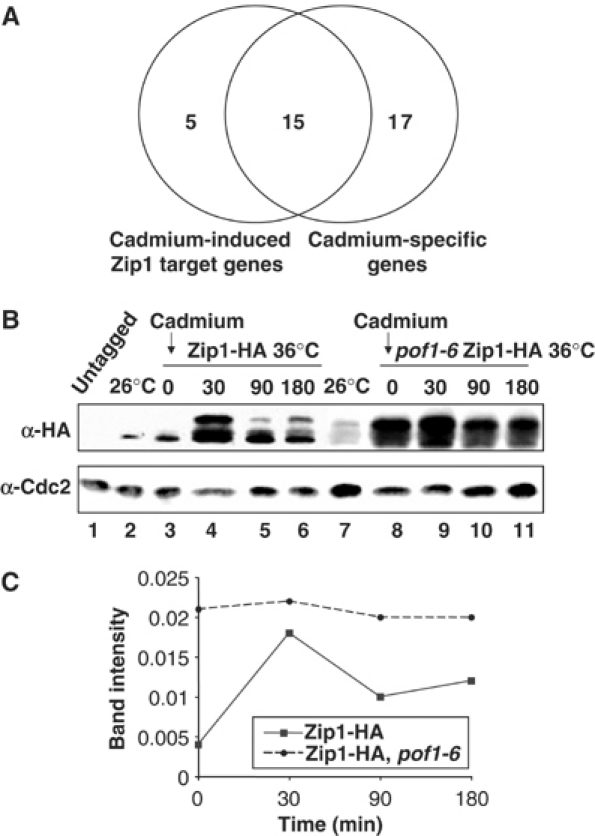
Comparison of Zip1-target and cadmium-specific genes and accumulation of Zip1 upon cadmium exposure. (A) Summary of DNA microarray analysis. Genes induced by cadmium stress in a Zip1-dependent manner and those induced in a cadmium stress-specific manner (Chen et al, 2003) are compared. (B) Increase of Zip1 upon cadmium exposure. Exponentially growing wild-type (lanes 2) or pof1-6 cells containing Zip1-HA (lane 7) were shifted from 26 to 36°C and incubated for 1 h (lanes 3 and 8). Then, 0.5 mM cadmium sulfate was added, followed by protein extracts at 0, 30, 90 and 180 min time points (lanes 3–6 and 8–11). Immunoblotting was performed with anti-HA (upper) or anti-Cdc2 antibody (lower). Untagged wild type was used as a negative control (lane 1). (C) Quantification. Zip1-HA levels shown in (B) are quantified using Cdc2 as a control.
Previous global transcriptional analysis in response to various types of stresses has shown that responsive genes fall into two categories, core environmental stress response (CESR) that is common to most stresses and stress-specific responses (Chen et al, 2003). To explore which of these categories Zip1-dependent genes fell into, we compared our data with the previous stress data (Chen et al, 2003). Out of 20 Zip1-target genes, 15 belong to a subgroup consisting of the 32 cadmium-stress-specific genes (Figure 5A, these 15 genes are shown in underlined bold letters in Table II). It is worth noting that at least 10 of these 15 Zip1- and cadmium stress-specific target genes are independent of the Sty1-Atf1 stress-activated mitogen-activated protein kinase pathway, as shown previously (Chen et al, 2003). This substantiates the notion that Sty1-Atf1 primarily acts on CESR genes, while Pof1-Zip1 plays a specific role in cadmium stress response. Interestingly, pof1+ itself is also seen to be induced in a cadmium-specific, Zip1-dependent manner, suggesting a negative feedback mechanism exists to ensure Zip1 degradation at some later time point following the cadmium response (Tables II and III). Overall, this microarray analysis shows that Zip1 is the major, if not sole, factor required for transcriptional induction upon cadmium stress.
Table 3.
Cadmium induction of pof1+ and zip1+ genes
| Strains | Wild type |
zip1 (=sup9) |
||||
|---|---|---|---|---|---|---|
| Cd2+ (min) | 0 | 15 | 60 | 0 | 15 | 60 |
| Gene expression | ||||||
| pof1 | 1.0 | 5.7 | 2.1 | 1.0 | 1.0 | 0.8 |
|
zip1 |
ND |
1.1 |
1.6 |
1.0 |
1.3 |
0.9 |
| Comparative transcription levels of the pof1 and zip1 genes in response to cadmium stress (0, 15 and 60 min) are taken from microarray data in wild-type and zip1-sup9 mutant cells. Note that, as zip1-sup9 contains a nonsense mutation, zip1 mRNA levels could be measured. | ||||||
| ND stands for not determined. | ||||||
One of the most adverse consequences by the presence of cadmium in the cell is the absorption of sulfur molecules. Glutathione (GSH) functions to protect cells from cadmium stress by direct binding to this metal, leading to sequestration into the vacuole (Clemens and Simm, 2003). In budding yeast, GSH is synthesized, along with sulfur-containing amino acids, methionine and cysteine, through the sulfate assimilation pathway (Marzluf, 1997). Most of the enzymes in this pathway are induced under cadmium stress in this yeast (Fauchon et al, 2002). In contrast, in fission yeast only a subset of these sulfate assimilation genes appeared to be induced (Table II) (Chen et al, 2003), suggesting that the sulfate assimilation pathways are regulated differently between the two organisms. Nonetheless, the sulfate assimilation genes induced by cadmium in fission yeast were Zip1 dependent, suggesting that the primary role of Zip1 in response to cadmium is to upregulate uptake and assimilation of inorganic sulfur from the environment, presumably for the increased production of GSH.
Exogenous cadmium upregulates Zip1 protein levels
Given the critical role for Zip1 in cadmium response and tolerance, zip1+ gene expression might have been expected to be upregulated upon cadmium stress. None of the wild-type microarray data, however, indicated the increase of zip1+ gene expression upon exposure to cadmium (Table III). In order to examine Zip1 protein levels in response to cadmium stress, immunoblotting was performed in a Zip1-HA strain. As shown in Figure 5B, after 30 min in the presence of cadmium, cells had increased Zip1 levels (lane 4). Intriguingly, a slower migrating form of Zip1, apparently corresponding to the phosphorylated form observed in pof1-6 mutants, as well as unphosphorylated form, appeared after cadmium exposure. As zip1+ gene expression does not change after cadmium stress (Table III), inhibition of Zip1 protein degradation is likely to be responsible for this increase in Zip1 levels upon cadmium addition. Prolonged exposure with cadmium appeared to result in a slight decrease of Zip1 levels (90 and 180 min, lanes 5 and 6 and Figure 5C for quantification). This may be ascribable to induction of the pof1+ gene upon cadmium stress as a part of negative feedback loop (Table III), although other possibilities can not be excluded.
Cadmium was also added to pof1-6 mutants preincubated at 36°C. In this mutant, as shown earlier (Figure 4A), 1 h incubation at 36°C prior to cadmium exposure resulted in the increase of Zip1-HA levels together with the appearance of slower migrating bands (lane 8). In clear contrast to wild-type cells, pof1-6 mutants did not show any further increase of Zip1-HA levels upon cadmium addition (lanes 9–11 and Figure 5C for quantification). These results indicate that one of the cellular responses to cadmium includes Zip1 stabilization and in pof1-6 mutants, Zip1 levels are maximally upregulated. This also suggests that upregulation of Zip1 levels under cadmium stress is mediated by inhibition of Pof1-dependent ubiquitylation and degradation of Zip1.
Zip1 is not necessary for functioning of the sulfur amino-acid biosynthesis genes
Pof1 is most homologous to budding yeast Met30, the F-box protein required for inactivation of bZIP transcription factor Met4 (Patton et al, 2000). Met4 is known to be responsible for the transcription of many of the genes in the sulfate assimilation pathway, specifically for the production of methionine and cysteine when these are missing from the environment. Thus, met4-deleted cells cannot grow without an external source of methionine. As our microarray data suggested that Zip1 is able to activate transcription of only some, if not many, of the genes in this pathway, we tested if Zip1 is similarly essential for growth on media lacking sulfur amino acids. In contrast to cells with mutations in genes in the sulfur amino-acid pathway, such as met5-1, zip1-deleted mutant cells were able to grow on minimal media containing only the inorganic sulfur source, Na2SO4 (Figure 6). This suggests that while Zip1 is capable of upregulating some of the genes in the sulfur assimilation pathway in response to cadmium stress, it is not essential for the cells to synthesize sulfur amino acids under conditions where these amino acids are lacking.
Figure 6.
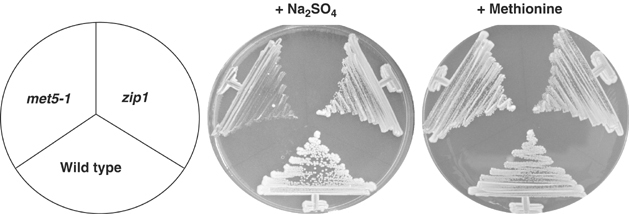
The absence of Zip1 does not result in sulfur amino-acid auxotrophy. Wild-type, zip1+-deleted and met5-1 mutant cells were streaked on minimal medium whose sulfur sources are provided from only Na2SO4 (middle) or methionine (right) and incubated at 26°C for 5 days. Some growth of met5-1 in the presence of Na2SO4 is most likely ascribable to the existence of residual methionine pool inside met5-1 cells.
Zip1 is required for maintenance of viability in cells exposed to cadmium
Previous results led us to the possibility that Zip1 is specifically required to initiate cadmium stress response and that phenotypic consequences of pof1 mutants, in which Zip1 accumulates, are in fact mimicking those of cells exposed to cadmium. In line with this notion, as shown earlier, cadmium exposure also results in elevated Zip1 levels (see Figure 5B and C). If this was the case, wild-type cells exposed to cadmium should show some of the defective phenotypes seen in a pof1 mutant, such as small cell size and growth arrest. We investigated this by exposing wild-type, pof1-6- and zip1-deleted cells growing in culture to cadmium stress and observing their response. Cadmium sulfate (0.5 mM) stopped growth of all cells at 26°C in liquid medium. However, if samples of these liquid cultures were plated onto cadmium-free plates, even after 6 h exposure to cadmium, wild-type and pof1-6 cells retained high viability (Figure 7A). This implies that growth arrest observed in these two strains is reversible. In sharp contrast, zip1 cells lost viability, with only 15% of cells surviving after 6 h in the presence of cadmium. This indicates that apparent division arrest in wild-type and pof1-6 cells upon cadmium stress is not due to cell death; instead, that cells arrest growth but remain viable and, importantly, that this response is Zip1 dependent.
Figure 7.
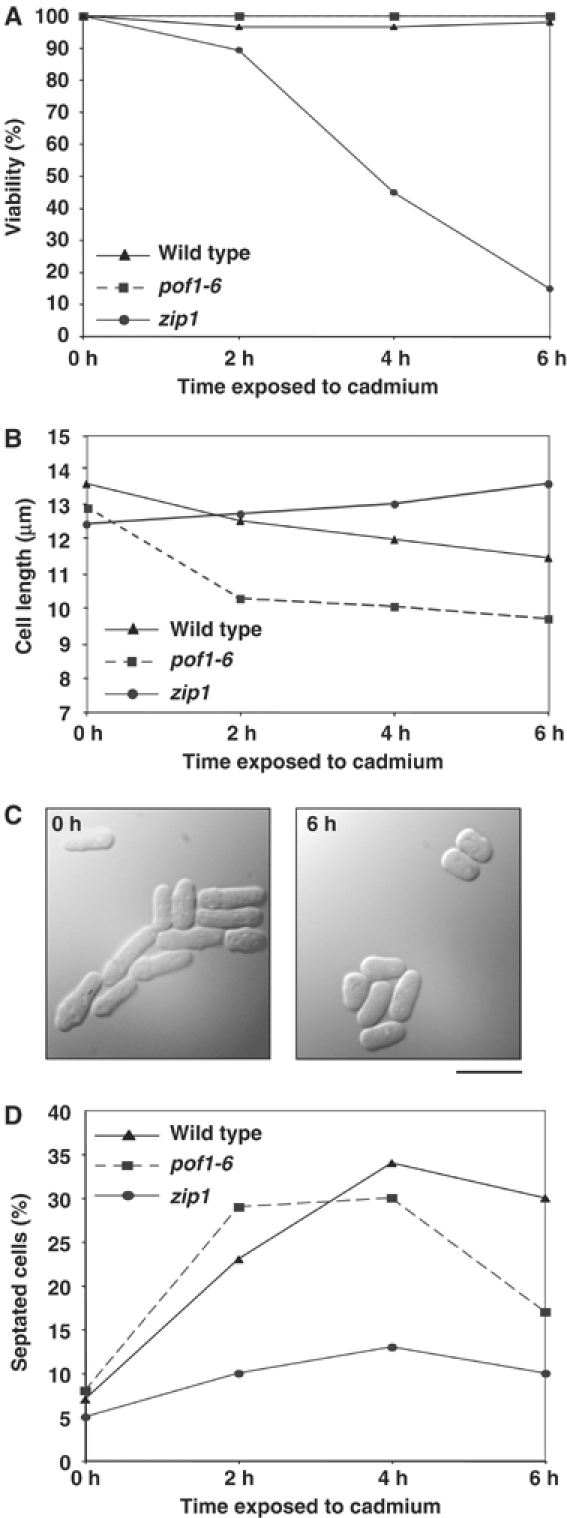
Zip1 is required for maintenance of viability during cadmium exposure. (A) Viability under cadmium stress. Wild-type (triangles), pof1-6 (squares) or zip1-deleted cells (circles) were treated with 0.5 mM cadmium sulfate at 26°C and cell viability was examined at every 2 h interval. At each time point, 2 × 200 cells were plated onto rich medium (lacking cadmium) and incubated at 26°C for 5 days. The number of viable colonies was counted and viability was calculated by dividing this number by 200 and calculating an average. (B) Cell size change upon cadmium exposure. The cell size was measured and plotted (n=100 per time point). (C) Cell morphology under cadmium stress. Phase-contrast micrographs of wild-type cells before (left) or after cadmium treatment (6 h, right) are shown. The bar indicates 10 μm. (D) Percentage of septated cells.
We next examined whether cadmium exposure could induce small cells as seen in the pof1 mutant at the restrictive temperature. In agreement with our prediction, even at the permissive temperature, pof1-6 cells became smaller in response to cadmium as did wild-type cells (Figure 7B and C). On the contrary, the cell size of zip1 mutants did not show any reduction, rather it was increased (Figure 7B).
Finally, while observing the cadmium-exposed cells, we noticed that a larger than average number of them were septating. We tested whether this response was Zip1 dependent. Again, consistent with our notion, this was indeed the case. Wild-type and pof1-6 cells showed a peak of septation 2–4 h after cadmium exposure, while the percentage of septated cells in zip1 mutants did not display a clear increase (Figure 7D). The biological significance of this remains unclear, but it is obviously a Zip1-dependent response. In summary, this analysis has unveiled the physiological role for Zip1 in cadmium stress, namely that Zip1 plays a key role in survival under adverse cadmium exposure.
Discussion
This report provides evidence that the F-box protein Pof1 is essential because of its ability to inactivate Zip1. This conclusion is drawn from the following results. Firstly, genetic data show that nonsense mutation or deletion of zip1+ rescues the growth defects of pof1 mutants. Secondly, zip1+ overexpression is toxic in wild-type cells, which mimic the phenotypes of pof1 mutants at the restrictive temperature. Thirdly, Zip1 is stabilized in a pof1 mutant and Zip1 itself is ubiquitylated in vivo. Finally, Pof1 binds to phosphorylated forms of Zip1 in the cell.
Pof1 is homologous to the essential budding yeast F-box protein Met30, known to regulate the Met4 transcription factor (Kaiser et al, 2000; Patton et al, 2000; Rouillon et al, 2000; Kuras et al, 2002; Flick et al, 2004). Met4 transcribes, among others, genes of the MET network required for the biosynthesis of methionine (Patton et al, 2000; Rouillon et al, 2000). It has been shown that Met30 downregulates Met4 activity in response to extracellular methionine or intracellular S-adenosylmethionine levels (Kuras and Thomas, 1995; Thomas et al, 1995; Kaiser et al, 2000; Rouillon et al, 2000) and this is achieved through both proteolytic and nonproteolytic mechanisms (Kaiser et al, 2000; Rouillon et al, 2000; Kuras et al, 2002). Our observations, however, suggest that only proteolytic mechanism may be shared in fission yeast. Firstly, pof1-6 mutants clearly have increased levels of Zip1. Secondly, Zip1 is a protein of short half-life (<15 min) in wild-type cells and displays an increased stability in pof1 mutants. Thirdly, unlike Met4 (Flick et al, 2004), no monoubiquitylated form of Zip1 has been found. Finally, pof1 mutants are lethal when combined with a proteasome mutation. All of these data suggest that Pof1 is responsible for the degradation of Zip1.
The differences in regulation of Met4 and Zip1 may be attributable to their different functions. As described above, Met4 is crucial for the production of sulfur amino acids when none are available in the environment, thus the met4 mutant cannot grow without the addition of methionine exogenously to the growth media. However, in addition to the MET genes, Met4 is responsible for the regulation of many other genes; most importantly, Met4 has been shown, like Zip1, to be involved in the upregulation of GSH in response to cadmium stress (Dormer et al, 2000) and this appears to be regulated independently of the sulfate assimilation pathway (Wheeler et al, 2002). Zip1, on the other hand, plays no role in the sulfate assimilation pathway in terms of normal growth, as the zip1 mutant can grow on media containing only the inorganic sulfur source. This would suggest that although both transcription factors are able to induce transcription of genes in the sulfate assimilation pathway, S. cerevisiae has evolved to rely upon Met4 for all sulfur amino-acid biosynthesis, whereas S. pombe uses Zip1 only to upregulate this pathway to make more glutathione in response to cadmium stress. Thus, S. pombe could be envisaged to use a simple system as described in Figure 8, where Zip1 is constantly made and degraded via SCFPof1 and only stabilized Zip1 is capable of transcribing downstream genes in the presence of cadmium stress. This system allows Zip1 to be readily available – presumably cadmium was a commonly encountered stress for fission yeast – yet protects cells from the dangerous effects of Zip1 transcription, noticeably complete cell growth arrest. S. cerevisiae, on the other hand, cannot just switch Met4 completely on and off as it relies on Met4 for sulfur amino-acid biosynthesis, and so has evolved a more complex level of regulation involving ubiquitylation without degradation.
Figure 8.
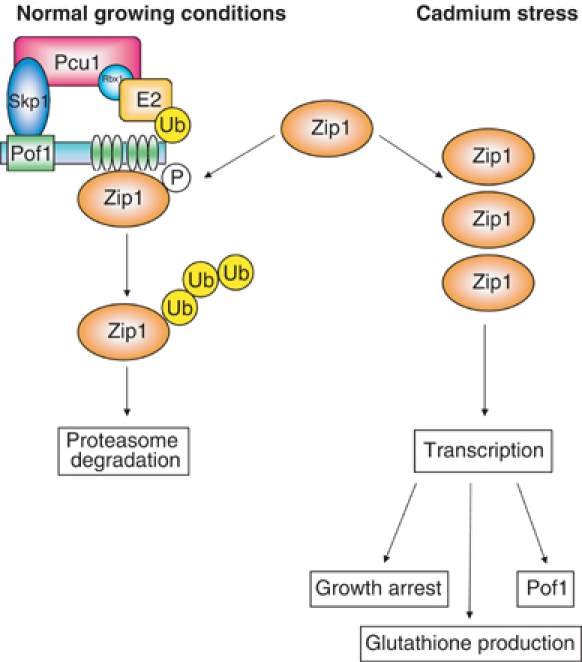
A model for the function and regulation of Zip1. Zip1 is constitutively produced, but its cellular levels are kept low under normal, noncadmium, growing conditions (left). This is achieved by constitutive SCFPof1-dependent ubiquitylation and proteasome-mediated degradation. Binding between Pof1 and Zip1 is dependent upon Zip1 phosphorylation. Under cadmium stress, some switch triggers an increase in Zip1 protein levels and its transcriptional activity. This results in an increased production of glutathione coupled with an arrest of cell growth, thereby ensuring high cell viability during cadmium exposure. The mechanism of this switch is unknown, but the accumulation of phosphorylated Zip1 species during cadmium exposure suggests that the phosphorylation of Zip1 could play some role in transcriptional activity.
How the cell regulates Zip1 activity in response to cadmium remains unknown. We have shown that Zip1 protein levels increase under cadmium stress without transcriptional stimulation and it is, therefore, tempting to speculate that Pof1-dependent degradation of Zip1 is switched off under this condition. Pof1 interacts with a phosphorylated form of Zip1, so the kinase responsible for this phosphorylation would be a candidate effector of this molecular switch. However, the data suggest the mechanism of Zip1 regulation is more complex, as a phosphorylated form of Zip1 appears to accumulate in cadmium-exposed cells. It may be that this form is capable of, or even required for, Zip1 transcriptional activity.
At the moment, it remains to be determined whether slower migrating phosphorylated forms of Zip1 in pof1-6 are identical to those seen in mts3-1 or wild-type cells exposed to cadmium. What is clear, on the other hand, is that the slowest form in pof1-6 (band 3) is still unstable. This phosphorylated form might either be degraded by some pathway other than SCFPof1 or be easily dephosphorylated. It is also possible that Zip1 phosphorylation under cadmium stress might be performed by a kinase different from that phosphorylating Zip1 under normally growing conditions or pof1-6 mutant background. One such candidate kinase includes the Sty1/Spc1 stress-activated protein kinase (Toone and Jones, 1998). In any case, Pof1 is required for Zip1 ubiquitylation and degradation under normal growing conditions, but it should be noted that in the presence of cadmium stress, Zip1 levels might be regulated in a different manner.
The most interesting aspect of the Zip1 transcription factor is its ability to induce growth arrest. This is seen in pof1 mutants, which arrest as small cells in a Zip1-dependent manner, and also when zip1+ is overexpressed. This growth arrest does inhibit cell division, but is not lethal to the cells, as pof1 mutants grown at the restrictive temperature still retain high viability even after 8 h (C Harrison and T Toda, unpublished observations). The growth arrest seen in both wild-type cells treated with cadmium stress and pof1 mutants at the restrictive temperature is likely to be caused by the same mechanism, as in either case, Zip1 accumulates and the arrest is Zip1 dependent. We envisage two possibilities for the mechanisms underlying Zip1-dependent growth arrest. One is that genes, which encode growth inhibitor-type molecules, are induced in a Zip1-dependent manner. The other possibility is that growth arrest is derived from intracellular sulfur deficiency. In particular, the pof1-6 small cell phenotype is remarkably similar to that of spermidine-deprived S. pombe cells (Chattopadhyay et al, 2002). Spermidine is synthesized via the sulfur assimilation pathway, and absolutely required for eukaryotic cell growth (Tabor and Tabor, 1984). These two possibilities are currently under investigation.
Cadmium has a biological half-life of 10–30 years and one of the most serious environmental pollutants (Hengstler et al, 2003). Given that many cells in the body will be exposed to cadmium, it is likely that higher organisms also have some conserved aspects of cadmium stress response seen in lower eukaryotes. Consistent with this notion, the mammalian bZIP Nrf2 protein has been shown to be an important transcription factor for cadmium response and its overexpression leads to upregulation of genes involved in glutathione production in certain cell types (Hayes et al, 2000). Interestingly, Nrf2 is an extremely labile protein and is degraded by the ubiquitin–proteasome system, while in the presence of cadmium it becomes stabilized (Stewart et al, 2003). This is an identical mechanism to that which seems to exist to control Zip1 activity in fission yeast. Recent reports indicate that Nrf2 is degraded via the SCF-related Cul3 pathway (Cullinan et al, 2004; Kobayashi et al, 2004), suggesting that Zip1 and Nrf2 systems are evolutionarily interconnected.
Oncoprotein transcription factors such as c-Jun and c-Myc have recently been shown to be degraded via SCF (Cardozo and Pagano, 2004). Here, we describe how a growth arrest mechanism can be coupled to cadmium stress response through a bZIP transcription factor. This mechanism should be conserved through evolution and its elucidation is of great significance for our understanding of eukaryotic growth control upon environmental stresses in general.
Materials and methods
Strains used in this study are listed in Table IV. See Supplementary data for Materials and methods.
Table 4.
Strains used in this study
| Strain name | Genotype | Derivation |
|---|---|---|
| 22 | h− leu1 | Our stock |
| TP108-3C | h− leu1 ura4 pap1∷ura4+ | Our stock |
| wee1 | h+ wee1-50 | Paul Nurse |
| met5 | h+ met5-1 | Paul Nurse |
| SKP407 | h− leu1 ura4 pof1+-GST-kanr | This study |
| SKP414-17 | h− leu1 ura4 pcu1+-13myc-kanr | Our stock |
| CLP30 | h− leu1 pof1+-GFP-kanr | This study |
| CLP30-6 | h− leu1 pof1-6-GFP-kanr | This study |
| CLP30-12 | h− leu1 pof1-12-GFP-kanr | This study |
| CLP41 | h− pof1-6-GFP-kanR sup9 | This study |
| CLP32 | h− pof1-6-GFP-kanR | This study |
| CLP52-1 | h− sup9 | This study |
| CLP064-1 | h+ leu1 ura4 his2 zip1∷kanr | This study |
| CLP066-1 | h+ leu1 his2 ura4 pof1-6-GFP-kanr zip1∷kanr | This study |
| CLP057-2A | h− leu1 mts3-1 zip1-3HA-kanr pof1-6-GFP-kanr | This study |
| CLP053-5 | h− leu1 ura4 pof1-6-GFP-kanr zip1+-3HA- kanr | This study |
| CLP054-5 | h+ leu1 ura4 his2 zip1+-3HA- kanr | This study |
| CLP056-5C | h− leu1 mts3-1 zip1+-3HA-kanr | This study |
Supplementary Material
Supplementary Information
Acknowledgments
We thank Drs Paul Nurse, Hiroaki Seino and Minoru Yoshida for providing strains and materials used in this study. We thank Dr Yasmine Mamnun for useful discussion and suggestions, and Dr Axel Behrens for critical reading of the manuscript. DC was supported by the EMF Biological Research Trust. This work was supported by the Cancer Research UK (to JB, NJ and TT) and the HFSP research grant (to TT).
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K (1993) TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcriptional efficiency but not the transcription start point or thiamine repressibility. Gene 123: 131–136 [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 5: 739–751 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H (2002) Absolute requirement of spermidine for growth and cell cycle progression of fission yeast (Schizosaccharomyces pombe). Proc Nat Acad Sci USA 99: 10330–10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J (2003) Global transcriptional responses of fission yeast to environmental stress. Mol Cell Biol 14: 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Simm C (2003) Schizosaccharomyces pombe as a model for metal homeostasis in plant cells: the phytochelatin-dependent pathway is the main cadmium detoxification mechanism. New Phytol 159: 323–330 [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper W, Diehl JA (2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24: 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormer UH, Westwater J, McLaren NF, Kent NA, Mellor J, Jamieson DJ (2000) Cadmium-inducible expression of the yeast GSH1 gene requires a functional sulfur-amino acid regulatory network. J Biol Chem 275: 32611–32616 [DOI] [PubMed] [Google Scholar]
- Fauchon M, Lagniel G, Aude J-C, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J (2002) Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell 9: 713–723 [DOI] [PubMed] [Google Scholar]
- Feldman RMR, Correll CC, Kaplan KB, Deshaies RJ (1997) A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230 [DOI] [PubMed] [Google Scholar]
- Flick K, Ouni I, Wohlschlegel JA, Capati C, McDonald WH, Yates JR III, Kaiser P (2004) Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat Cell Biol 6: 634–641 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Cell Biol 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA (1989) Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634 [DOI] [PubMed] [Google Scholar]
- Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, Wolf CR, Yamamoto M (2000) The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem Soc Trans 28: 33–41 [DOI] [PubMed] [Google Scholar]
- Hengstler JG, Bolm-Audorff U, Faldum A, Janssen K, Reifenrath M, Gotte W, Jung D, Mayer-Popken O, Fuchs J, Gebhard S, Bienfait HG, Schlink K, Dietrich C, Faust D, Epe B, Oesch F (2003) Occupational exposure to heavy metals: DNA damage induction and DNA repair inhibition prove co-exposures to cadmium, cobalt and lead as more dangerous than hitherto expected. Carcinogenesis 24: 63–73 [DOI] [PubMed] [Google Scholar]
- Hermand D, Bamps S, Tafforeau L, Vandenhaute J, Makela TP (2003) Skp1 and the F-box protein Pof6 are essential for cell separation in fission yeast. J Biol Chem 278: 9671–9677 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (1996) Ubiquitin-dependent protein degradation. Annu Rev Genet 30: 405–439 [DOI] [PubMed] [Google Scholar]
- Ikebe C, Kominami K, Toda T, Nakayama K (2002) Isolation and characterization of a novel F-box protein Pof10 in fission yeast. Biochem Biophys Res Commun 290: 1399–1407 [DOI] [PubMed] [Google Scholar]
- Kaiser P, Flick K, Wittenberg C, Reed SI (2000) Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCFMet30-mediated inactivation of the transcription factor Met4. Cell 102: 303–314 [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24: 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K, Ochotorena I, Toda T (1998) Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-F-box) ubiquitin ligase. Genes Cells 3: 721–735 [DOI] [PubMed] [Google Scholar]
- Kuras L, Rouillon A, Lee T, Barbey R, Tyers M, Thomas D (2002) Dual regulation of the Met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol Cell 10: 69–80 [DOI] [PubMed] [Google Scholar]
- Kuras L, Thomas D (1995) Functional analysis of Met4, a yeast transcriptional activator responsive to S-adenosylmethionione. Mol Cell Biol 15: 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A, Katayama S, Harrison C, Dhut S, Kitamura K, McDonald N, Toda T (2004) Molecular interactions of fission yeast Skp1 and its role in the DNA damage checkpoint. Genes Cells 9: 367–382 [DOI] [PubMed] [Google Scholar]
- Marzluf GA (1997) Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol 51: 73–96 [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [DOI] [PubMed] [Google Scholar]
- Nurse P (1990) Universal control mechanism regulating onset of M-phase. Nature (Lond.) 344: 503–508 [DOI] [PubMed] [Google Scholar]
- Ohmiya R, Kato C, Yamada H, Aiba H, Mizuno T (1999) Isolation of multicopy suppressors of the calcium sensitivity of a mutant lacking the bZIP transcription factor Atf1 in fission yeast. Mol Gen Genet 261: 297–306 [DOI] [PubMed] [Google Scholar]
- Pahl HL, Baeuerle PA (1996) Control of gene expression by proteolysis. Curr Opin Cell Biol 8: 340–347 [DOI] [PubMed] [Google Scholar]
- Patton EE, Peyraud C, Rouillon A, Surdin-Kerjan Y, Tyers M, Thomas D (2000) SCFMet30-mediated control of the transcriptional activator Met4 is required for the G1-S transition. EMBO J 19: 1613–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M (1998) Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet 14: 236–243 [DOI] [PubMed] [Google Scholar]
- Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D (2000) Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCFMet30 complex. EMBO J 19: 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Gordon C, Ferrell K, Dubiel W (1996) Characteristics of 26 S proteasomes from fission yeast mutants, which arrest in mitosis. J Mol Biol 263: 423–431 [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell 91: 209–219 [DOI] [PubMed] [Google Scholar]
- Stewart D, Killeen E, Naquin R, Alam S, Alam J (2003) Degradation of transcription factor Nrf2 via the ubiquitin–proteosome pathway and stabilization by cadmium. J Biol Chem 278: 2396–2402 [DOI] [PubMed] [Google Scholar]
- Tabor CW, Tabor H (1984) Polyamines. Annu Rev Biochem 53: 749–790 [DOI] [PubMed] [Google Scholar]
- Thomas D, Kuras L, Barbey R, Cherest H, Blaiseau P-L, Surdin-Kerjan Y (1995) Met30p, a yeast transcriptional inhibitor that responds to S-adenosylmethionine, is an essential protein with WD40 repeats. Mol Cell Biol 15: 6526–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone WM, Jones N (1998) Stress-activated signalling pathways in yeast. Genes Cells 3: 485–498 [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Quinn KA, Perrone G, Dawes IW, Grant CM (2002) Glutathione regulates the expression of γ-glutamycysteine synthetase via the Met4 transcription factor. Mol Micobiol 46: 545–556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


