Abstract
Elf5 is an epithelial-specific ETS factor. Embryos with a null mutation in the Elf5 gene died before embryonic day 7.5, indicating that Elf5 is essential during mouse embryogenesis. Elf5 is also required for proliferation and differentiation of mouse mammary alveolar epithelial cells during pregnancy and lactation. The loss of one functional allele led to complete developmental arrest of the mammary gland in pregnant Elf5 heterozygous mice. A quantitative mRNA expression study and Western blot analysis revealed that decreased expression of Elf5 correlated with the downregulation of milk proteins in Elf5+/− mammary glands. Mammary gland transplants into Rag−/− mice demonstrated that Elf5+/− mammary alveolar buds failed to develop in an Elf5+/+ mammary fat pad during pregnancy, demonstrating an epithelial cell autonomous defect. Elf5 expression was reduced in Prolactin receptor (Prlr) heterozygous mammary glands, which phenocopy Elf5+/− glands, suggesting that Elf5 and Prlr are in the same pathway. Our data demonstrate that Elf5 is essential for developmental processes in the embryo and in the mammary gland during pregnancy.
Keywords: ETS, mammary gland, milk protein genes, prolactin, prolactin receptor
Introduction
The ETS family of transcription factors now consists of more than 50 members that share a homologous DNA binding domain. These proteins play important roles in development and disease. ETS transcription factors are involved in embryonic development, in hematopoiesis, in immune responses, in development of the skeletal system and differentiation of the small intestine (Bassuk and Leiden, 1997; Ghysdael and Boureux, 1997; Ng et al, 2002; Ristevski et al, 2002, 2004; Wolvetang et al, 2002; Xu et al, 2002). Most ETS genes are expressed in a wide range of cell types, but recently a few that are epithelial-specific have been identified. This group includes ELF3 (ESX/ESE-1/JEN/ERT) (Andreoli et al, 1997; Chang et al, 1997; Oettgen et al, 1997; Tymms et al, 1997; Choi et al, 1998), EHF (ESE-3) (Bochert et al, 1998; Kas et al, 2000), PDEF (PSE) (Oettgen et al, 2000; Yamada et al, 2000) and ELF5 (ESE-2) (Zhou et al, 1998; Oettgen et al, 1999). ELF5, ELF3 and ESE-3 share considerable sequence similarity and are all expressed in a similar subset of epithelial tissues.
Elf3 is the most broadly expressed of the epithelial-specific ETS genes. It is evident in the epithelium of the gastrointestinal tract, the mammary gland, uterus and prostate, and the tongue (Andreoli et al, 1997; Chang et al, 1997; Oettgen et al, 1997; Tymms et al, 1997). Elf5 and Ese-3 have an overlapping but more restricted pattern of expression compared to Elf3. These genes are expressed in a subset of organs containing glandular or secretory epithelium (Oettgen et al, 1999; Kas et al, 2000; Lapinskas et al, 2004). Although there is considerable organ overlap in the expression of Elf5, Elf3 and Ese-3, several differences in their expression patterns have been observed. For instance, Elf3 is expressed strongly in the small intestine and liver, whereas Elf5 and Ese-3 do not appear to be expressed in these organs at all. Both Elf3 and Ese-3 are expressed in the colon and pancreas, Elf5 is not, and Elf3 and Elf5 are present in the stomach, whereas Ese-3 is not.
We have previously shown that Elf5 functions as a transcription factor with similar sequence-specific DNA binding characteristics to other ETS family members (Zhou et al, 1998). ELF5 (ESE-2a) is capable of transactivating a number of epithelial-specific gene promoters, including SPRR2A, PSP and PSA, in vitro (Oettgen et al, 1999). In addition, recombinant ELF5 is capable of transactivating the whey acidic protein (WAP) promoter (Thomas et al, 2000) in mouse mammary epithelial cells. This, together with its expression in the mammary gland (Zhou et al, 1998), implies a function for Elf5 in this organ. In order to establish the biological function of the Elf5 gene in the mouse and to gain an understanding of the ELF5 gene in human biology, we generated and characterized Elf5 null mice.
Results
Elf5 is essential for early mouse embryogenesis
We generated a targeting construct in which part of exon 3 of the Elf5 gene was replaced by an NLS-LacZ-neo cassette (Figure 1A). The targeting construct was designed to interrupt exon 3, which contains the ATG initiation codon of the Elf5 protein, and produce a fusion product containing the first 28 amino acids of the Elf5 protein fused to β-galactosidase. This targeting construct was electroporated into isogenic 129SvJ J1 embryonic stem (ES) cells, and the correctly targeted Elf5+/− ES cell clones #1 and #130 were microinjected into C57Bl/6J murine blastocysts. The resulting male chimeras were then mated with Elf5+/+ C57Bl/6J female mice to generate Elf5+/− mice. Genomic Southern blot analysis and PCR screening were used to confirm the correct targeting event (Figure 1B–D). Southern analysis of tail DNA revealed a polymorphism in the wild-type Elf5 gene locus between 129SvJ and C57Bl/6J genetic backgrounds (Figure 1C).
Figure 1.
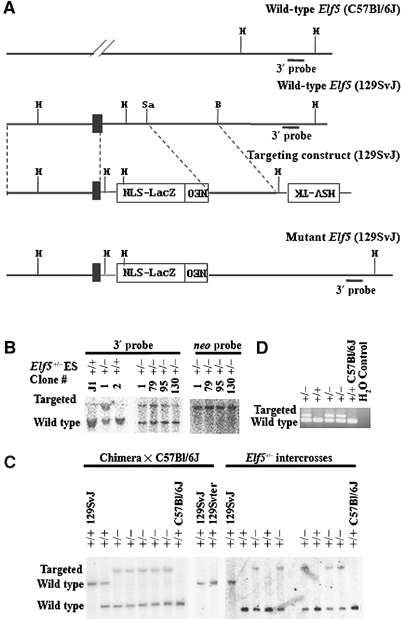
Gene targeting of the murine Elf5 locus. (A) A schematic representation of the targeting strategy. The homologous recombination event replaces a portion of exon 3 and intron 3 with an NLS-LacZ-neo cassette. Black box: exon 3 containing the ATG. The thin line represents plasmid sequences. H: HindIII; Sa: SalI. (B) Representative Southern blot analyses of surviving ES cell clones. A 400 bp 3′-external probe recognizes a 4.6 kb wild-type and an 8.6 kb targeted HindIII DNA fragment, and a neo probe recognizes an 8.6 kb targeted HindIII DNA fragment. (C) Germline transmission of the targeted Elf5 allele. A 3′-external probe recognizes 8.6 kb targeted (129SvJ), 4.6 kb wild-type (129SvJ) and 2.0 kb wild-type (C57Bl/6J) HindIII bands. Left panel: Southern analysis of an entire litter from Chimera × C57Bl/6J; right panel: Southern analysis of an entire litter from Elf5+/− intercrosses. (D) PCR analysis of progeny from Elf5+/− intercrosses detects a 169 bp wild-type and a 237 bp mutant band.
Heterozygotes were intercrossed to produce 513 offspring derived from the ES cell clone #130. Genotyping revealed that 34.9% were Elf5+/+ and 65.1% were Elf5+/−. No Elf5−/− mice were detected (Figure 2A). Similar results were obtained for 43 adult mice derived from the ES cell clone #1 (Figure 2A). These data indicated that the Elf5−/− genotype was embryonic lethal.
Figure 2.

Elf5−/− is embryonic lethal. (A) Genotype distribution of Elf5+/− intercrosses. (B) Genotyping blastocysts. PCR analysis detects a 169 bp wild-type (WT) band and a 237 bp mutant band (KO).
Since Elf5−/− animals were not detected among 3-week-old offspring from heterozygote intercrosses, we investigated the genotypes of neonates and embryos. Elf5−/− animals were not detected among neonates nor among embryos at E13.5, E14.5 and E18.5 by Southern blot analysis. PCR analysis of postimplantation embryos dissected from the decidua at E7.5–11.5 (with the morning of vaginal plug detection corresponding to E0.5) showed that none of these embryos were Elf5−/− (Figure 2A). These results suggested that the lethality of Elf5−/− embryos was occurring before E7.5. Genomic PCR analysis of 58 blastocysts at E3.5, collected from intercrossed Elf5+/− females, detected Elf5−/− blastocysts in a ratio expected for a Mendelian distribution (Figure 2A and B). These results indicated that Elf5−/− embryos die between E3.5 and E7.5, during the period of implantation.
Elf5 is expressed in the pregnant and lactating mammary gland
Our earlier studies (Zhou et al, 1998) had established that the Elf5 transcript was expressed in the mammary gland, but we had not examined Elf5 expression in this tissue during late pregnancy and lactation. We have now investigated the temporal expression pattern of Elf5 mRNA in pregnant and day 1 postpartum mammary glands from both Elf5+/+ and Elf5+/− mice. Northern blots of poly(A)+ mRNA were probed with the mouse Elf5 cDNA and the two Elf5 transcripts, Elf5-a (2.5 kb) and Elf5-b (1.5 kb), were observed in both pregnant and day 1 postpartum mammary glands from wild-type and heterozygous mice (Figure 3A and B). It should be noted that we did not detect any spurious Elf5 transcripts on our Northern blots when we compared Elf5+/+ and Elf5+/− mammary gland RNA, indicating that the null allele did not produce detectable alternatively spliced Elf5 messages. Previously, we showed that Elf5 mRNA levels sharply increased between days 2 and 10 of pregnancy (Zhou et al, 1998). Here we show that this surge of Elf5 mRNA occurs after day 8, peaks at day 12 and remains at a high level throughout pregnancy and early lactation (Figure 3A). Elf5 mRNA levels in the heterozygous pregnant and day 1 postpartum mammary gland were reduced compared to that in the corresponding Elf5+/+ organs, as expected (Figure 3B). We also measured Elf5 protein levels and found that Elf5 protein was present but barely detectable before day 10.5 of pregnancy in the wild-type mammary gland (Figure 3C). At day 12.5, there was a dramatic increase in the amount of Elf5 protein, consistent with the increase in Elf5 message at day 12. Surprisingly, Elf5 protein levels in the heterozygous mammary gland were more dramatically reduced than the message, with no protein detectable (Figure 3C).
Figure 3.

Elf5 expression in Elf5+/− and Elf5+/+ mammary glands. (A) Northern blot of total RNA from wild-type mammary glands probed with a murine Elf5 cDNA (top panel) and 18s cDNA as a loading control (lower panel). (B) Northern blot of poly(A)+ mRNA probed with a murine Elf5 cDNA (top panel) and Gapdh cDNA (lower panel). Relative expression levels of Elf5-b/Gapdh are indicated (right panel). Lane 1: day 18.5 pregnant Elf5+/+ mammary gland; lane 2: day 18.5 pregnant Elf5+/− mammary gland; lane 3: day 1 postpartum Elf5+/+ mammary gland; lane 4: day 1 postpartum Elf5+/− mammary gland. Band intensities were quantified on a phosphoimager. (C) Western blot of Elf5 protein in the wild-type and heterozygous glands at days 6.5–14.5 of pregnancy. Elf5 protein is not visible in the 10.5-day wild-type sample on the right-hand panel because a shorter exposure time is shown.
Elf5 is essential for pregnancy-associated mammary gland development
Elf5+/− females derived from ES cell clone #130 were fertile and gave birth to litters of morphologically normal pups whose numbers were comparable to those of Elf5+/+ females. However, despite normal nursing and mothering characteristics displayed by Elf5+/− females, most of their first litter pups died within 24 h of birth, and virtually the entire litter had perished by 48 h. This phenomenon was never observed in our wild-type colonies where only the occasional loss of a maximum of one to two pups per litter was observed. All pups were observed to attach to the nipple and suck, but newborns of Elf5+/− females died of starvation and dehydration. Lethality of the newborns was independent of their genotype (Elf5+/+ or Elf5+/−). Examination of 1-day-old pups revealed that they lacked milk in their stomachs. These results suggested that Elf5+/− females failed to lactate.
To quantify this effect, we studied 49 Elf5+/− mothers. Of these, 41 (83.7%) failed to support any pups, three (6.1%) were able to support a reduced number of pups after some had died, and in five cases (10.2%) all the pups born were supported. Two Elf5+/− females derived from ES cell clone #1 were also observed for their lactating abilities. Both females failed to lactate after their first pregnancy. A small number of heterozygous females were able to suckle pups successfully. These mice were on a 129SvJ-C57Bl/6J mixed genetic background and this variable phenotype was most probably due to a genetic modifier present in one of the genetic backgrounds. Elf5+/− mice were subsequently generated on a pure 129Svter genetic background, and six Elf5+/− females were mated with either Elf5+/+ or Elf5+/− males. All six females were incapable of lactation after multiple pregnancies (up to four pregnancies), suggesting that the mammary alveolar developmental defect was 100% penetrant in the 129Svter strain.
Examination of whole mount mammary glands from age-matched virgin Elf5+/+ and Elf5+/− females demonstrated that branching morphogenesis (Hovey et al, 2002) was not affected, as elongation and extension of the mammary ductal tree as well as ductal side branching were comparable between genotypes. Alveolar buds formed at the ductal termini with age in both genotypes (Figure 4A–F). Observations made at day 18.5 of pregnancy and 1 day postpartum showed that Elf5+/− glands entered the proliferation phase of alveolar morphogenesis (Neville et al, 2002), but then development stalled. The degree of retarded alveolar genesis was variable among individuals. In some cases, development stalled at a very early stage of alveolar proliferation and only very few rudimentary alveoli are formed (Figure 4J), whereas in others, development stalled later and alveoli are seen (Figure 4H). However, differentiation and expansion of the alveolar buds into mature lobuloalveolar mammary tissues was severely impaired in Elf5+/− females (Figure 4H and J).
Figure 4.
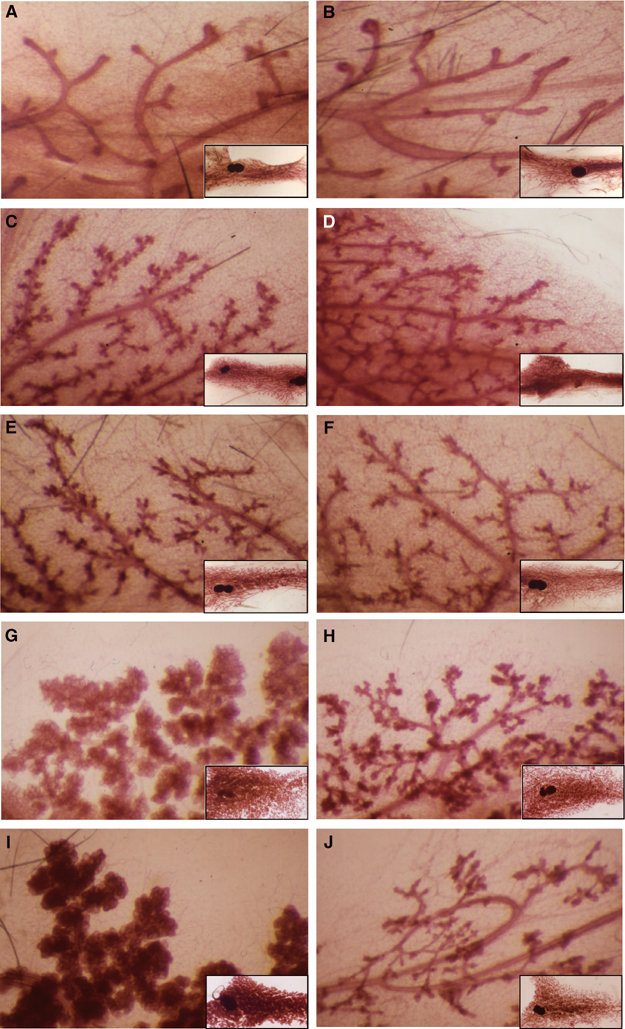
Mammary gland development is defective in pregnant and postpartum Elf5+/− females. Whole-mount analyses of inguinal mammary glands from wild-type (left) and Elf5+/− (right) mice. Shown are virgin mice at 48 days (A, B), 131 days (C, D) and 176 days (E, F), 18.5-day pregnant glands (G, H) and day 1 postpartum glands (I, J).
We examined the proliferation status of the mammary glands from virgin, pregnant and lactating mice by PCNA staining. The virgin mammary glands of Elf5+/+ and Elf5+/− mice showed no difference in the proliferation of the mammary epithelial cells (data not shown). Likewise, no difference was observed between day 6.5 pregnant mammary glands (Figure 5A, B and Q). However, a lower percentage of proliferating mammary epithelial cells was found consistently in day 8.5 (Figure 5C, D and Q) and 10.5 (Figure 5E, F and Q) pregnant Elf5+/− mammary glands compared to wild type. The difference in the percentage of the proliferating mammary epithelial cells between the Elf5+/+ and Elf5+/− mammary glands becomes less evident from day 12.5 to day 18.5 of pregnancy (Figure 5G–N and Q). However, there were fewer proliferating cells in day 1 lactating Elf5+/− mammary glands than in the wild-type control (Figure 5O–Q). From the images shown in Figure 5 at lower magnification, it is obvious that the epithelial content of the heterozygous mammary gland is reduced. We also examined the levels of apoptosis in the mammary glands at all the developmental stages mentioned above. The number of apoptotic cells was negligible in all sections tested and no difference was observed between wild-type and heterozygous mice (data not shown).
Figure 5.
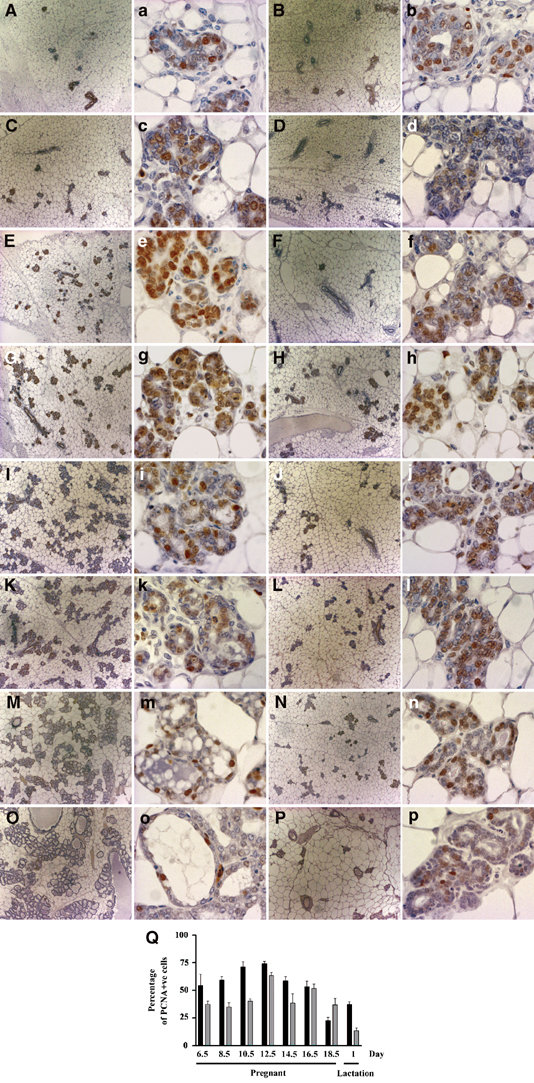
Histological analyses and PCNA staining of wild-type and Elf5 heterozygous pregnant mammary glands. Left two panels: wild type; right two panels: Elf5+/−. Low-magnification images (A–P) show under development of Elf5+/− glands. Day 6.5 (A, B); day 8.5 (C, D); day 10.5 (E, F); day 12.5 (G, H); day 14.5 (I, J); day 16.5 (K, L); day 18.5 (M, N); day 1 lactation (O, P). PCNA staining is shown as a brown nuclear precipitate in epithelial cells (A–P). The percentage of PCNA-positive cells throughout pregnancy-associated mammary gland development is shown (Q). Results are the average of four sections per mouse, n=2/3, error bars represent ±s.e.m.
Taken together, the results in Figures 4 and 5 indicate that the Elf5+/− mammary gland defect occurred during pregnancy at the proliferative phase of alveolar morphogenesis and that this is a direct result of a partially impaired proliferation of mammary epithelial cells, and not due to increased apoptosis. Notably in the wild type, histology shows that the alveoli subunits expanded and filled with milk, indicating the functional secretory state of the mammary gland (Figure 5M and O). In contrast, there was no expansion of the alveolar lumens in Elf5+/− mammary glands in which alveoli had formed. These observations indicate that in individuals where alveolar development proceeded past the proliferative phase, it was blocked during the secretory initiation phase (Figure 5N and P).
The Elf5+/− mammary defect is epithelial cell autonomous
In order to determine if the defect in mammary gland development observed in Elf5 heterozygous females was due to the lack of Elf5 in the mammary epithelium per se or whether the defect was secondary due to defects in other endocrine systems, we transplanted wild-type and Elf5+/− mammary epithelium (Naylor et al, 2003) into Rag1−/− recipient females (Mombaerts et al, 1992), which cannot reject transplants due to their compromised immune systems. These females were then mated. The wild-type transplants displayed ductal branching and alveolar proliferation comparable to unmanipulated wild-type glands (Figure 6A and B). The Elf5 heterozygous glands showed side branching but no lobuloalveolar development (Figure 6C and D), demonstrating that the Elf5+/− mammary defect was epithelial cell autonomous and not dependent on other factors.
Figure 6.
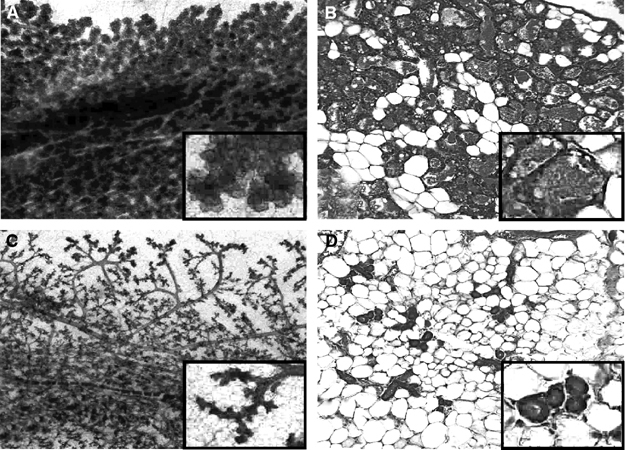
Whole-mount analysis of wild-type and Elf5 heterozygous mammary epithelium transplanted to a normal host mammary fat pad. Wild-type (A, B) and Elf5+/− (C, D) glands are shown. (A, C) Whole mount; (B, D) histology sections.
Gene expression profiles in Elf5 and Prlr heterozygous mammary glands
The observation that a similar mammary gland developmental defect also occurs in Prlr+/− mice (Ormandy et al, 1997) prompted us to examine the levels of Elf5 in the mammary glands of Prlr+/− females. A quantitative PCR analysis demonstrated that Elf5 mRNA was significantly reduced in the Prlr+/− day 1 postpartum mammary glands compared to wild-type controls (t-test, P=0.0157) (Figure 7A). On the contrary, Prlr expression was unchanged in Elf5+/− glands compared to wild types, indicating that Elf5 is downstream in the Prl signalling pathway (Figure 7B). We also used quantitative PCR to examine the expression levels of a panel of genes encoding milk proteins in the Elf5+/− glands. WDNM1/EXP1, WAP, CEL and β-casein were all reduced in the heterozygous glands compared to wild-type controls (Figure 7C). Furthermore, Western blot analysis of the milk proteins α-casein, β-casein and WAP in Elf5+/− mammary gland extracts revealed that these milk proteins were not synthesized in the heterozygous gland (Figure 7D).
Figure 7.

Gene expression analysis in Elf5 and Prlr heterozygous mammary glands. (A) Elf5 mRNA expression (isoforms a and b) in Prlr+/− versus Prlr+/+ glands. Results are shown as the average number of Elf5 transcripts (Prlr+/+: n=4, Prlr+/−: n=3). A statistical t-test was performed between the two groups. (B) Prlr mRNA expression in Elf5+/− versus Elf5+/+ glands. There is no significant difference between wild types and heterozygotes. (C) Milk protein gene expression in Elf5+/− versus Elf5+/+ glands. The genes tested are as indicated (CSNβ is β-casein). (D) Milk protein expression in Elf5+/− versus Elf5+/+ glands.
Discussion
We demonstrate that Elf5 is an ETS factor required for early mouse development. A number of ETS transcription factors play essential roles in mouse embryogenesis. Of the 10 ETS family members that have been disrupted in mice, Pu.1−/−, Tel−/−, Fli1−/−, Elf3−/− and Gabpα−/− were embryonic lethal around E16.5, E10.5–11.5, E11–12.5, E11.5 and prior to implantation, respectively (Scott et al, 1994; Wang et al, 1997; Hart et al, 2000; Spyropoulos et al, 2000; Ng et al, 2002, Ristevski et al, 2004). In contrast, Ets2−/− embryos died before E8.5 due to growth arrest of the ectoplacental cone and apoptosis of the embryonic ectoderm (Yamamoto et al, 1998). Breakdown in the vascular interactions between the embryo and the maternal circulation also contributed to the Ets2−/− embryonic lethality (Yamamoto et al, 1998). In this regard, it is important to note that while the ETS proteins all share a similar DNA binding domain structure, have similar cognate DNA binding elements and in many cases overlapping expression, there is no functional redundancy during development.
Although the status of Elf5 expression in the early mouse embryo and its corresponding extraembryonic tissues (before E9.5) is unknown, Elf5 was found to be expressed in the placenta throughout E9.5–18, indicating a potential function for Elf5 in this tissue (Zhou et al, 1998). The placenta is derived from the trophoblast (Kaufman and Bard, 1999), and the trophectoderm of the blastocyst is important for the exchange of metabolites between the mother and the embryo. Therefore, the early lethality of Elf5−/− embryos may be due to the loss of Elf5 function in the embryonic trophoblast. However, we cannot rule out possible defects in the inner cell mass of the developing embryo.
Although it may be argued that the phenotype observed is due to the dominant behavior of a truncated Elf5 protein generated from the targeted allele, we believe that this is extremely unlikely. The disruption in the Elf5 gene occurs 28 amino acids after the initiating methionine and if splicing occurred from the truncated exon 3 (skipping the LacZ-neo cassette) through to exons 4, 5 or 6, there would be a frameshift such that no functional domains of the Elf5 protein would be produced. Also, we do not see any unexpected Elf5 transcripts in Northern blots performed on our heterozygous mice. Unfortunately, we were unable to extract sufficient quantities of high-quality RNA from the Elf5−/− blastocysts for Northern blot analysis.
A number of epithelial-specific ETS transcription factors (ELF3, ELF5 and ESE-3) are expressed in the mammary gland, suggesting potential functions for these proteins there (Chang et al, 1997; Neve et al, 1998; Oettgen et al, 1999; Kas et al, 2000). We have shown that both ELF3 and ELF5 can transactivate a pregnancy- and lactation-associated milk protein (WAP) gene promoter in vitro (Thomas et al, 2000). However, neither Elf3 nor Ese-3 is able to compensate for the lack of Elf5 in the mouse mammary gland. In this study, we have demonstrated that Elf5 has a specific, nonredundant role in mammary gland development during pregnancy and lactation. Furthermore, the phenotype is observed in the heterozygous state but intriguingly, even though we can detect Elf5 message in the heterozygous glands, the levels of Elf5 protein are reduced more than expected. Indeed, the protein is undetectable on our Western blots. With the exceptions of Prlr+/−, Plg+/− (plasminogen) and Ptc1+/− (patched-1) females (Ormandy et al, 1997; Lewis et al, 1999; Lund et al, 2000), no other gene identified as being involved in mammary gland development and function displayed a phenotype in the heterozygous state and this phenomenon has never been observed with any other ETS factor.
The mammary ductal tree can elongate and branch quickly through the entire mammary fat pad under the stimulation of ovarian hormones following puberty (reviewed in Hovey et al, 2002). Pregnancy hormones then promote alveolar proliferation, and the alveolar epithelial cells from the resulting lobuloalveolar structures eventually differentiate into secretory epithelial cells at parturition (reviewed in Neville et al, 2002). Our results demonstrate that mammary epithelial cell proliferation and differentiation during pregnancy and the postpartum period depends on the expression of Elf5, and that one functional Elf5 allele is not sufficient for function. In contrast, the earlier developmental events of ductal morphogenesis do not show this sensitivity to reduced Elf5. Death of the pups born to Elf5+/− females was due to a failure of mammary lobuloalveolar development and the consequent failure in maternal milk production. Thus, Elf5 has a dual function in the mammary gland. It is required for proliferation in early pregnancy, then for differentiation later on. Notably, a significant increase in Elf5 protein was observed during mid-pregnancy (at day 12.5) when the mammary alveolar epithelial cells start to undergo secretory initiation, suggesting that Elf5 expression may trigger the onset of lactogenesis of the mammary alveolar epithelial cells during pregnancy. The expression of WDNM1/EXP1 and β-casein begins around day 9 of pregnancy and that of WAP at day 14 (Robinson et al, 1995; Kannius-Janson et al, 1998), coinciding with the upsurge in Elf5 expression. Thus, an apparent correlation exists between the onset of milk protein gene-associated differentiation of the wild-type mammary alveolar epithelial cells and the expression levels of Elf5. Our data suggest that in the Elf5 heterozygous mammary gland, the level of Elf5 is insufficient to trigger the expression of genes required for differentiation of the epithelial cells and subsequent lactogenesis. In effect, what we observe at day 18.5 of pregnancy is cells blocked at an earlier stage of differentiation.
The mammary defect in Elf5+/− females closely resembled that in Prlr+/− females where an impaired differentiation of lobuloalveolar units resulted in an inability to lactate (Ormandy et al, 1997). Since disruptions in either Elf5 or Prlr produce a similar phenotype, it is likely that Elf5 and Prlr participate in the same pathway. This study and the one by Naylor et al (2003) support that a possible downstream link for Elf5 is in the prolactin signalling pathway.
Quantitative RT–PCR showed that Elf5 expression is downregulated in the Prlr+/− day 1 postpartum mammary gland and that Prlr levels are unchanged in Elf5+/− glands, placing Elf5 downstream in the Prl/Prlr signalling cascade. Significantly, the expression of four genes encoding the milk proteins WDNM1/EXP1, β-casein, CEL and WAP is downregulated. Our data demonstrate that these genes are all regulated, either directly or indirectly, by Elf5; thus, it is likely that the mammary gland developmental defect observed in the Elf5+/− pregnant female is due to a block in milk protein-associated alveolar differentiation.
The action of prolactin on Elf5 gene expression is most likely mediated via the JAK2/STAT or MAP kinase signalling pathways. Stat5 is activated by tyrosine phosphorylation in response to Prl stimulation. Stat5a null mice exhibit defects in lobuloalveolar proliferation and differentiation (Liu et al, 1997); therefore, it is possible that Stat5a regulates expression of Elf5. Interestingly, the Elf5 promoter has a putative Stat binding site but this site is yet to be proven functional. Other ETS factors are known to be targets of MAP kinases (Graves and Petersen, 1998), but the conserved MAP kinase phosphorylation site present in ETS1, ETS2 and pntp2 (Yang et al, 1996) is not present in Elf5 (Zhou et al, 1998). Other ligand/receptor pathways may also involve Elf5. For instance, mouse knockouts of the ligand OPGL and its receptor RANK (Fata et al, 2000) and of the LAR receptor-like protein tyrosine phosphatase (Schaapveld et al, 1997) all produce mammary gland defects characterized by a failure of alveolar terminal differentiation similar to that of Elf5+/− female mice, but the involvement of Elf5 in these pathways has yet to be tested.
Finally, our transplant studies strongly suggested that the mammary gland defect in the Elf5+/− mice is epithelial cell autonomous and not due to other influences such as systemic endocrine alterations since the Elf5+/− mammary epithelium transplanted to a wild-type mammary fat pad failed to undergo pregnancy-associated lobuloalveolar development.
Epithelial cell proliferation and differentiation is tightly controlled during mammalian development. This study has provided in vivo evidence of the involvement of Elf5 in these critical developmental processes and further defined a dual role for Elf5. It is initially required for early embryonic development and again later in life during development of the mammary gland in pregnancy. The role of Elf5 in the mammary gland is extremely interesting since in the absence of one Elf5 allele, the development of the gland during pregnancy is completely shut down. How Elf5 exerts its effects on the processes of proliferation and differentiation in the mammary gland is the subject of an ongoing study and has important implications for both development and diseases such as breast cancer.
Materials and methods
Gene targeting
Murine Elf5 genomic clones were isolated from a 129SvJ λFIXII genomic library (Stratagene) using an mElf5 cDNA probe. The targeting construct contained a 2.2 kb fragment, encoding the first 84 bp of mElf5 cDNA coding sequence as the left arm and a 1.65 kb SalI–BamHI fragment as the right arm. The homologous recombination event replaces a portion of exon 3 and intron 3 with an NLS-LacZ-neo cassette in which the pMC1-neo cassette was in a reverse orientation to that of Elf5 transcription. A pMC1-TK cassette was placed 3′ of the right arm to allow for enrichment of targeted ES cells. HindIII-digested genomic DNA was used to determine the genotype by Southern blot analysis using a 400 bp probe as indicated in Figure 1. PCR genotyping used two mElf5-specific primers, PS (5′-GCACACCCAGAATTGAAGATTCC-3′) and PAS1 (5′-CCTTCACTGCACGTGGACTG-3′), and one neo-specific primer PNEO (5′-ATTCGCCAATGACAAGACGC-3′).
Mammary gland whole mounts and histology
For whole mounts, inguinal mammary glands were fixed in Carnoy's fixative and stained in carmine alum as described (Kordon et al, 1995). For histology, inguinal mammary glands were fixed in Bouin's fixative and paraffin sections (10 μm) were stained with hematoxylin and eosin.
Western blots
Protein was extracted from thoracic mammary glands removed at different stages of pregnancy using Trizol (Invitrogen). The protein extracts were resolved on a 12% SDS–PAGE gel and then transferred to Immobilon PVDF membrane (Millipore). After blocking, membranes were incubated with goat anti-Elf5 (N-20) antibody (Santa Cruz) at a 1:100 dilution. Secondary antibody was a 1:4000 dilution of horseradish peroxidase-conjugated anti-goat immunoglobulin (DAKO) incubated at room temperature for 1 h. SuperSignal West Pico Chemiluminescent Substrate (Pierce) was used to visualize binding after exposure of the membrane to CLX-Posure film (Progen). To control for protein loading and epithelial content, the membrane was washed and then incubated with a mouse anti-cytokeratin 5/8 monoclonal antibody (BD Pharmingen) at a 1:1000 dilution for 1 h at room temperature. Signal was visualized by incubating with a 1:2000 dilution of horseradish peroxidase-conjugated anti-mouse immunoglobulin (DAKO). To determine milk protein expression, the membrane was stripped by washing three times in 50 mM glycine pH 2.0, then washed once in TBST and incubated with a 1:10 000 dilution of polyclonal rabbit antisera for milk-specific proteins (Accurate Chemical and Scientific Corporation). Horseradish peroxidase-conjugated anti-rabbit immunoglobulin (DAKO) was used at a 1:2000 dilution and signal visualized as described above.
PCNA and TUNEL staining
After fixation in Bouin's fixative for 4 h at room temperature, inguinal mammary gland tissue from Elf5+/+ and Elf5+/− females was embedded in paraffin and sectioned at 5 μm. Sections were cleared in histosol and rehydrated. To detect apoptosis, the ApopTag® Peroxidase In situ Apoptosis Detection Kit (Serological Corporation) was used essentially according to the manufacturer's instructions, except no proteinase K pretreatment was required. To detect cell proliferation, antigen retrieval was performed by heat treatment in 10 mM sodium citrate buffer (pH 6.0). Endogenous peroxidase activity was quenched using 6% (v/v) H2O2 in methanol for 30 min at room temperature. Tissue sections were blocked using CAS Block (Zymed Laboratories Inc.) for 40 min at room temperature. Sections were immunoreacted with (neat) mouse monoclonal antiproliferating cell nuclear antigen antibody EPOS anti-PCNA/HRP, clone PC10 (DAKO) or the EPOS Immunoglobulins/HRP Negative Control (DAKO) for 60 min at room temperature. Immunoperoxidase staining was detected using the Liquid DAB Substrate-Chromogen System (DAKO). All sections were counterstained lightly with Mayer's hematoxylin.
Quantification of cellular proliferation and apoptosis
Images from PCNA and TUNEL reacted mammary gland consecutive sections were captured with a CoolSNAP-Pro color camera attached to an Olympus BX60 microscope with Image-Pro Plus software. The total number of cells and the number of proliferating or apoptotic cells in four representative fields from each mouse were enumerated.
Mammary epithelium transplants
Transplants were performed as described (Brisken et al, 1999). Mammary gland fragments from 16-week-old Elf5 heterozygotes and wild-type littermates were transplanted into the cleared fat pads of 3-week-old C57Bl6/Rag1−/− mice (Mombaerts et al, 1992). The recipients were mated at 8 weeks post-transplant and the transplants examined by whole-mount microscopy and histology at day 1 postpartum. The whole mounts were stained with carmine alum and the sections stained with hematoxylin and eosin.
Gene expression studies
Poly(A)+ mRNA was extracted from the inguinal mammary glands of Elf5 heterozygotes and wild-type littermates at day 18.5 of pregnancy and at day 1 postpartum. Two samples of each were reverse transcribed using AMV reverse transcriptase (Promega). PCR primers were designed for WDNM1, CEL, β-casein and WAP using Macvector. To ensure that the reaction was specific for cDNA and not genomic DNA, the primers were designed spanning an intron. PCR primers for Elf5 were (1) sense primer 5′-TGGACTCCGTAACCCATAGCACCT-3′ (2) antisense primer 5′-ATTGCTTAAGGGCTGATGGCATCG-3′. This primer pair detects both mRNA isoforms Elf5a and Elf5b (Zhou et al, 1998). The PCR reactions were performed in a LightCycler (Roche) using 1 μl of the cDNA diluted 1:2, 5 pmol of primers and the FastStart DNA master SYBR Green I enzyme mix (Roche) in a 10 μl reaction volume. Relative quantification of the product was performed by comparing the crossing points of different samples above background and assuming that a difference of one cycle in the linear phase of the reaction corresponds to a two-fold difference in transcript levels between samples. The samples were analyzed twice and the results reported as an average of both animal and analysis duplicates. For quantitative analysis of Prlr expression, an ABI TaqMan Gene Expression Assay was used.
Acknowledgments
We thank Caroline Wang and Belinda Duscio for excellent technical assistance.
References
- Andreoli JM, Jang SI, Chung E, Coticchia CM, Steinert PM, Markova NG (1997) The expression of a novel, epithelium-specific ets transcription factor is restricted to the most differentiated layers in the epidermis. Nucleic Acids Res 25: 4287–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, Leiden JM (1997) The role of Ets transcription factors in the development and function of the mammalian immune system. Adv Immunol 64: 65–104 [DOI] [PubMed] [Google Scholar]
- Bochert MA, Kleinbaum LA, Sun LY, Burton FH (1998) Molecular cloning and expression of Ehf, a new member of the ets transcription factor/oncoprotein gene family. Biochem Biophys Res Commun 246: 176–181 [DOI] [PubMed] [Google Scholar]
- Brisken C, Kaur S, Chavarria TE, Binart N, Sutherland RL, Weinberg RA, Kelly PA, Ormandy CJ (1999) Prolactin controls mammary gland development via direct and indirect mechanisms. Dev Biol 210: 96–106 [DOI] [PubMed] [Google Scholar]
- Chang CH, Scott GK, Kuo WL, Xiong X, Suzdaltseva Y, Park JW, Sayre P, Erny K, Collins C, Gray JW, Benz CC (1997) ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene 14: 1617–1622 [DOI] [PubMed] [Google Scholar]
- Choi SG, Yi Y, Kim YS, Kato M, Chang J, Chung HW, Hahm KB, Yang HK, Rhee HH, Bang YJ, Kim SJ (1998) A novel ets-related transcription factor, ERT/ESX/ESE-1, regulates expression of the transforming growth factor-beta type II receptor. J Biol Chem 273: 110–117 [DOI] [PubMed] [Google Scholar]
- Fata JE, Kong YY, Li J, Sasaki T, Irie-Sasaki J, Moorehead RA, Elliott R, Scully S, Voura EB, Lacey DL, Boyle WJ, Khokha R, Penninger JM (2000) The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103: 41–50 [DOI] [PubMed] [Google Scholar]
- Ghysdael J, Boureux A (1997) The Ets family of transcriptional regulators. In Oncogenes as Transcriptional Regulators, Yaniv M, Ghysdael J (eds) Vol. 1, p 22 Basel, Switzerland: Birkhauser Verlag [Google Scholar]
- Graves BJ, Petersen JM (1998) Specificity within the ets family of transcription factors. Adv Cancer Res 75: 1–55 [DOI] [PubMed] [Google Scholar]
- Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, Favier R, Bernstein A (2000) Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity 13: 167–177 [DOI] [PubMed] [Google Scholar]
- Hovey RC, Trott JF, Vonderhaar BK (2002) Establishing a framework for the functional mammary gland: from endocrinology to morphology. J Mammary Gland Biol Neoplasia 7: 17–38 [DOI] [PubMed] [Google Scholar]
- Kannius-Janson M, Lidberg U, Hulten K, Gritli-Linde A, Bjursell G, Nilsson J (1998) Studies of the regulation of the mouse carboxyl ester lipase gene in mammary gland. Biochem J 336: 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas K, Finger E, Grall F, Gu X, Akbarali Y, Boltax J, Weiss A, Oettgen P, Kapeller R, Libermann TA (2000) ESE-3, a novel member of an epithelium-specific ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J Biol Chem 275: 2986–2998 [DOI] [PubMed] [Google Scholar]
- Kaufman MH, Bard JBL (1999) The Anatomical Basis of Mouse Development. San Diego, CA: Academic Press [Google Scholar]
- Kordon EC, McKnight RA, Jhappan C, Hennighausen L, Merlino G, Smith GH (1995) Ectopic TGF beta 1 expression in the secretory mammary epithelium induces early senescence of the epithelial stem cell population. Dev Biol 168: 47–61 [DOI] [PubMed] [Google Scholar]
- Lapinskas EJ, Palmer J, Ricardo S, Hertzog PJ, Hammacher A, Pritchard MA (2004) A major site of expression of the ets transcription factor Elf5 is epithelia of exocrine glands. Histochem Cell Biol (in press) [DOI] [PubMed] [Google Scholar]
- Lewis MT, Ross S, Strickland PA, Sugnet CW, Jimenez E, Scott MP, Daniel CW (1999) Defects in mouse mammary gland development caused by conditional haploinsufficiency of Patched-1. Development 126: 5181–5193 [DOI] [PubMed] [Google Scholar]
- Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L (1997) Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev 11: 179–186 [DOI] [PubMed] [Google Scholar]
- Lund LR, Bjorn SF, Sternlicht MD, Nielsen BS, Solberg H, Usher PA, Osterby R, Christensen IJ, Stephens RW, Bugge TH, Dano K, Werb Z (2000) Lactational competence and involution of the mouse mammary gland require plasminogen. Development 127: 4481–4492 [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE (1992) RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68: 869–877 [DOI] [PubMed] [Google Scholar]
- Naylor MJ, Ginsburg E, Iismaa TP, Vonderhaar BK, Wynick D, Ormandy CJ (2003) The neuropeptide galanin augments lobuloalveolar development. J Biol Chem 278: 29145–29152 [DOI] [PubMed] [Google Scholar]
- Neve R, Chang CH, Scott GK, Wong A, Friis RR, Hynes NE, Benz CC (1998) The epithelium-specific ets transcription factor ESX is associated with mammary gland development and involution. FASEB J 12: 1541–1550 [DOI] [PubMed] [Google Scholar]
- Neville MC, McFadden TB, Forsyth I (2002) Hormonal regulation of mammary differentiation and milk secretion. J Mammary Gland Biol Neoplasia 7: 49–66 [DOI] [PubMed] [Google Scholar]
- Ng AY, Waring P, Ristevski S, Wang C, Wilson T, Pritchard M, Hertzog P, Kola I (2002) Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology 122: 1455–1466 [DOI] [PubMed] [Google Scholar]
- Oettgen P, Alani RM, Barcinski MA, Brown L, Akbarali Y, Boltax J, Kunsch C, Munger K, Libermann TA (1997) Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol Cell Biol 17: 4419–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettgen P, Finger E, Sun Z, Akbarali Y, Thamrongsak U, Boltax J, Grall F, Dube A, Weiss A, Brown L, Quinn G, Kas K, Endress G, Kunsch C, Libermann TA (2000) PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem 275: 1216–1225 [DOI] [PubMed] [Google Scholar]
- Oettgen P, Kas K, Dube A, Gu X, Grall F, Thamrongsak U, Akbarali Y, Finger E, Boltax J, Endress G, Munger K, Kunsch C, Libermann TA (1999) Characterization of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J Biol Chem 274: 29439–29452 [DOI] [PubMed] [Google Scholar]
- Ormandy CJ, Camus A, Barra J, Damotte D, Lucas B, Buteau H, Edery M, Brousse N, Babinet C, Binart N, Kelly PA (1997) Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev 11: 167–178 [DOI] [PubMed] [Google Scholar]
- Ristevski S, O'Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ (2004) The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol Cell Biol 13: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristevski S, Tam PP, Hertzog PJ, Kola I (2002) Ets2 is expressed during morphogenesis of the somite and limb in the mouse embryo. Mech Dev 116: 165–168 [DOI] [PubMed] [Google Scholar]
- Robinson GW, McKnight RA, Smith GH, Hennighausen L (1995) Mammary epithelial cells undergo secretory differentiation in cycling virgins but require pregnancy for the establishment of terminal differentiation. Development 121: 2079–2090 [DOI] [PubMed] [Google Scholar]
- Schaapveld RQ, Schepens JT, Robinson GW, Attema J, Oerlemans FT, Fransen JA, Streuli M, Wieringa B, Hennighausen L, Hendriks WJ (1997) Impaired mammary gland development and function in mice lacking LAR receptor-like tyrosine phosphatase activity. Dev Biol 188: 134–146 [DOI] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H (1994) Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573–1577 [DOI] [PubMed] [Google Scholar]
- Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK (2000) Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol 20: 5643–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Ng AN, Zhou J, Tymms MJ, Doppler W, Kola I (2000) The Elf group of Ets-related transcription factors, ELF3 and ELF5. Adv Exp Med Biol 480: 123–128 [DOI] [PubMed] [Google Scholar]
- Tymms MJ, Ng AY, Thomas RS, Schutte BC, Zhou J, Eyre HJ, Sutherland GR, Seth A, Rosenberg M, Papas T, Debouck C, Kola I (1997) A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene 15: 2449–2462 [DOI] [PubMed] [Google Scholar]
- Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH (1997) Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J 16: 4374–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolvetang EJ, Wilson TJ, Sanij E, Busciglio J, Hatzistavrou T, Seth A, Hertzog PJ, Kola I (2002) Ets2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet 12: 247–255 [DOI] [PubMed] [Google Scholar]
- Xu D, Wilson TJ, Chan D, De Luca E, Zhou J, Hertzog PJ, Kola I (2002) Ets1 is required for p53 transcriptional activity in UV-induced apoptosis in embryonic stem cells. EMBO J 21: 4081–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N, Tamai Y, Miyamoto H, Nozaki M (2000) Cloning and expression of the mouse Pse gene encoding a novel Ets family member. Gene 241: 267–274 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG (1998) Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev 12: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BS, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, Hume DA, Maki RA, Ostrowski MC (1996) Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol 16: 538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ng AY, Tymms MJ, Jermiin LS, Seth AK, Thomas RS, Kola I (1998) A novel transcription factor, ELF5, belongs to the ELF subfamily of ETS genes and maps to human chromosome 11p13–15, a region subject to LOH and rearrangement in human carcinoma cell lines. Oncogene 17: 2719–2732 [DOI] [PubMed] [Google Scholar]


