Abstract
Mitochondria perform a central function in the biogenesis of cellular iron–sulphur (Fe/S) proteins. It is unknown to date why this biosynthetic pathway is indispensable for life, the more so as no essential mitochondrial Fe/S proteins are known. Here, we show that the soluble ATP-binding cassette (ABC) protein Rli1p carries N-terminal Fe/S clusters that require the mitochondrial and cytosolic Fe/S protein biogenesis machineries for assembly. Mutations in critical cysteine residues of Rli1p abolish association with Fe/S clusters and lead to loss of cell viability. Hence, the essential character of Fe/S clusters in Rli1p explains the indispensable character of mitochondria in eukaryotes. We further report that Rli1p is associated with ribosomes and with Hcr1p, a protein involved in rRNA processing and translation initiation. Depletion of Rli1p causes a nuclear export defect of the small and large ribosomal subunits and subsequently a translational arrest. Thus, ribosome biogenesis and function are intimately linked to the crucial role of mitochondria in the maturation of the essential Fe/S protein Rli1p.
Keywords: ABC transporter, iron homeostasis, iron–sulphur cluster, nuclear export, protein translation
Introduction
Mitochondria are essential organelles that perform a variety of functions in metabolism and energy conversion (Attardi and Schatz, 1988; Neupert, 1997; Scheffler, 1999; Pfanner and Geissler, 2001). In the yeast Saccharomyces cerevisiae, none of these processes appear to determine the essential character of these organelles. For instance, the citric acid cycle or oxidative phosphorylation can be inactivated by targeted gene deletions or by the loss of mitochondrial DNA, without affecting the viability of yeast cells in the presence of fermentable carbon sources. Similarly, central processes in the mitochondria of higher eukaryotes such as respiration seem to be dispensable until a rather late stage in embryonic development (Larsson et al, 1998; Li et al, 2000). To date, only one mitochondrial biosynthetic process, the assembly of iron–sulphur (Fe/S) proteins, is known to render mitochondria essential (Lill and Kispal, 2000; Craig and Marszalek, 2002; Balk and Lill, 2004; Lill and Mühlenhoff, 2005). Deletion of several components involved in Fe/S protein biogenesis is lethal in yeast under any growth condition tested. These proteins include the cysteine desulphurase Nfs1p, providing elemental sulphur for biogenesis, the electron transfer chain consisting of the ferredoxin reductase Arh1p and the [2Fe–2S] ferredoxin Yah1p, and the highly conserved protein pair Isu1p/Isu2p serving as a scaffold for the assembly of the Fe/S clusters (Mühlenhoff et al (2003) and references therein). Surprisingly, with the exception of Yah1p, which appears to be involved in its own maturation (Lange et al, 2000), none of the known mitochondrial Fe/S proteins is essential for cell viability. It is therefore a long-standing question which biological process involving an essential Fe/S protein may render mitochondria, and thus Fe/S protein biogenesis, so important for life.
Recently, we provided evidence that the above-mentioned mitochondrial ISC assembly machinery is not only involved in the biogenesis of mitochondrial, but also of cytosolic, Fe/S proteins (Kispal et al, 1999). Several additional components specifically required for cytosolic Fe/S protein maturation have been identified, namely the mitochondrial inner membrane ATP-binding cassette (ABC) transporter Atm1p, the sulphhydryl oxidase Erv1p of the mitochondrial intermembrane space, glutathione, the cytosolic P-loop NTPases Cfd1p/Nbp35p, and the cytosolic iron-only hydrogenase-like Nar1p (Kispal et al, 1999; Lange et al, 2001; Sipos et al, 2002; Roy et al, 2003; Balk et al, 2004; Hausmann et al, 2005). With the exception of Atm1p, all these components are essential for cell viability, suggesting that extra-mitochondrial Fe/S proteins that perform indispensable functions might exist. Indeed, we recently showed that both Nar1p and Nbp35p represent cytosolic Fe/S proteins which, like Yah1p, appear to be involved in their own maturation (Balk et al, 2004; Hausmann et al, 2005). However, this ‘chicken and egg' situation does not explain the essential character of Fe/S protein biogenesis in general. Another candidate protein is the 68 kDa Rli1p, which is encoded by the S. cerevisiae open reading frame YDR091c. At its N-terminus, Rli1p harbours two cysteine-rich motifs predicted to carry two [4Fe–4S] clusters (Figure 1A). The C-terminus of Rli1p contains two domains which share high sequence homology with members of the ABC protein family (Schüller et al, 2003). Unlike ABC transporters, Rli1p does not contain segments predicted to be membrane-spanning. Rli1p is highly conserved in evolution and homologues are present in virtually all eukaryotes and archaebacteria, but are not found in eubacteria.
Figure 1.
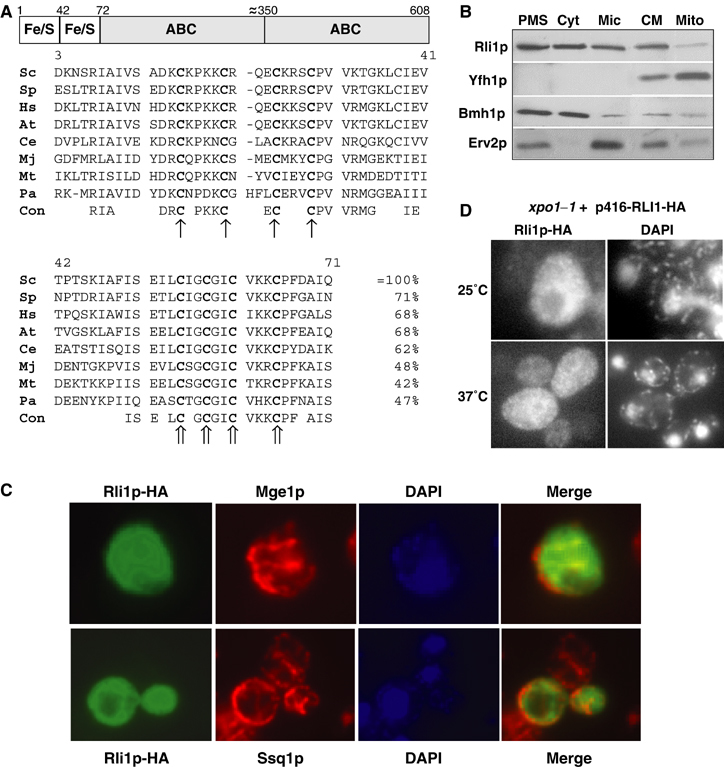
The domain structure and the subcellular localisation of Rli1p. (A) Rli1p contains two domains predicted to carry Fe/S clusters at the N-terminus and two C-terminal ABC domains. The multi-sequence alignment of the cysteine-rich regions at the N-terminus of Rli1p-like proteins was created by the Multalin program (Corpet, 1988). The conserved cysteine residues of the two ferredoxin-like motifs are labelled with different arrows. Sc, S. cerevisiae; Sp, Schizosaccharomyces pombe; Hs, Homo sapiens; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Mj, Methanococcus jannaschii; Mt, Methanobacterium thermoautotrophicum; Pa, Pyrococcus abyssi; Con, consensus sequence. (B) Yeast cells expressing a chromosomal version of Rli1p-HA were subfractionated by differential centrifugation. A post-mitochondrial supernatant (PMS) and a crude mitochondrial (CM) fraction were obtained by centrifugation of the cell lysate for 10 min at 10 000 r.p.m. The PMS fraction was used to separate soluble cytosolic proteins (Cyt) from a microsomal membrane fraction (Mic) by centrifugation at 100 000 r.p.m. for 1 h. Mitochondria (Mito) were purified from the CM fraction by Nycodenz density gradient centrifugation. Equal amounts of protein were analysed by SDS–PAGE and immunostaining for Rli1-HA and the indicated marker proteins (Yfh1p, mitochondrial matrix; Bmh1p, cytosol; Erv2p, endoplasmic reticulum). (C) In situ immunofluorescence labelling of Rli1p-HA and mitochondrial proteins Mge1p and Ssq1p. Wild-type cells (strain PSY581) were transformed with the high-copy expression vector p426 containing RLI1-HA, labelled with antibodies against the HA-tag, Ssq1p or Mge1p, and stained with fluorophore-conjugated secondary antibodies. DNA was counterstained with the fluorescent dye DAPI. Merged images show no co-localisation of Rli1p-HA with Mge1p or Ssq1p. (D) A fraction of Rli1p shuttles between cytosol and nucleus. xpo1–1 cells expressing Rli1p-HA (plasmid p416) were grown in minimal medium at 25°C to OD600 of 0.4. Cells were shifted to 37°C for 15 min and examined by fluorescence microscopy.
In this work, we provide in vivo evidence for the presence of Fe/S clusters in cytosolic Rli1p and for the indispensable character of these Fe/S clusters. Assembly of the Fe/S clusters was strictly dependent on several components of the mitochondrial and cytosolic Fe/S protein assembly machineries. Since Rli1p was found not to be involved in Fe/S protein biogenesis, the maturation of the essential Fe/S protein Rli1p provides the first known process that explains the indispensable character of mitochondria for yeast cell viability.
The function of yeast Rli1p is poorly defined to date. The protein shares 68% amino-acid identity with human ‘RNase L inhibitor' (RLI; Figure 1A). This protein has originally been identified as a binding partner of human RNase L, which cleaves double-stranded RNA in the interferon response to viral infection (Bisbal et al, 1995). Hence, RLI was thought to represent a new component of the interferon-regulated 2′–5′ oligo A pathway. In another study, RLI, together with RNase L, has been reported to regulate the stability of mitochondrial mRNAs in α-interferon-treated cells and a fraction of RLI was claimed to be located in mitochondria (Le Roy et al, 2001). Recently, RLI was shown to perform a crucial function in the post-translational assembly of HIV-1 capsids in vitro by direct binding to several virus proteins including gag (Zimmerman et al, 2002). All these proposed functions do not explain why yeast Rli1p is essential. Further, a homologue of RNase L neither exists in lower eukaryotes nor in archaebacteria. Therefore, Rli1p/RLI must play a more general cellular role that differs from those functions suggested for virus-infected cells.
Here, we report the tight association of Rli1p with cytosolic ribosomes and Hcr1p, a protein involved in rRNA processing and translation initiation (Valasek et al, 2001). Depletion of Rli1p caused a nuclear export defect of both the small and large ribosomal subunits (see also Yarunin et al, 2005), and subsequently a translational arrest (see also Dong et al, 2004). These data demonstrate a close link between the essential role of mitochondria in the assembly of the cytosolic Fe/S protein Rli1p and cytosolic ribosome biogenesis and function.
Results
Rli1p is a cytosolic Fe/S protein and requires both mitochondrial and cytosolic components for Fe/S cluster assembly
To determine the subcellular localisation of yeast Rli1p, a hemagglutinin epitope-tagged version of Rli1p (termed Rli1p-HA) was synthesised in wild-type yeast cells, and cells were fractionated by differential centrifugation. The majority of Rli1p was detected in the post-mitochondrial supernatant (Figure 1B). A smaller amount of Rli1p was present in the crude mitochondria fraction, but this part was largely depleted, as mitochondria were further purified by density gradient centrifugation. Therefore, in contrast to what has been reported for human RLI (Le Roy et al, 2001), yeast Rli1p does not appear to be located in mitochondria, consistent with the lack of a mitochondrial presequence. Instead, Rli1p behaved like typical proteins of the microsomal fraction which can be separated from mitochondria (e.g., Erv2p; Gerber et al, 2001). When the post-mitochondrial supernatant was further fractionated by centrifugation, about half of Rli1p behaved as a soluble protein, whereas the remainder was recovered in the microsomal pellet fraction (Figure 1B).
We further used in situ immunofluorescence microscopy to localise Rli1p. Cells were permeabilised and labelled with antibodies against the HA tag, followed by secondary antibodies conjugated to the fluorophore fluoresceine-isothiocyanate. Rli1-HA was found predominantly in the cytosol (Figure 1C). There was no apparent co-localisation of Rli1p-HA with two mitochondrial proteins. A weak fluorescence in the nucleus indicated that a small amount of Rli1p may reside in this compartment. Accumulation of a fraction of Rli1p-HA in the nucleus was tested using the temperature-sensitive nuclear export mutant xpo1–1 (Stade et al, 1997). The amount of Rli1p-HA in the nucleus was significantly increased at the non-permissive temperature (Figure 1D), whereas no effect was seen with wild-type cells (not shown). In summary, our data show that the majority of Rli1p resides in the cytosol, but a fraction shuttles between the nucleus and cytosol (see also Dong et al, 2004; Yarunin et al, 2005).
Recently, we have shown that Rli1p can bind iron in vivo (Lange et al, 2001). This iron association was dependent on the function of a component of the ISC export machinery, Erv1p, suggesting that Rli1p may be an Fe/S protein. To support this hypothesis, we examined the dependence of iron binding to Rli1p on the function of three mitochondrial proteins which have previously been shown to be central to maturation of cytosolic Fe/S proteins, namely Nfs1p, Yah1p and Atm1p (Lill and Kispal, 2000). For our analysis, we took advantage of mutant cells in which the NFS1, YAH1 or ATM1 genes were under control of a galactose-regulatable promoter, and which were transformed with a plasmid coding for Rli1p-HA. The concentrations of the three mitochondrial proteins were decreased by growth of the cells in glucose-containing medium for various times (not shown; Kispal et al, 1999; Lange et al, 2000). Cells were radiolabelled with 55Fe, a cell extract was prepared, and the binding of 55Fe to overproduced Rli1p-HA was measured by scintillation counting after immunoprecipitation of Rli1p-HA. A high amount of radioactive 55Fe could be precipitated with anti-HA antibodies, whereas hardly any radioactivity was detected using nonspecific antiserum or cells that did not produce Rli1p-HA (Figure 2A). This showed that the co-immunoprecipitated 55Fe was specifically associated with Rli1p-HA. Upon depletion of Nfs1p, Yah1p, or Atm1p, a strong reduction in the amounts of 55Fe precipitated with anti-HA antibodies was observed (Figure 2B). In contrast, the levels of 55Fe co-immunoprecipitated from extracts derived from wild-type cells expressing Rli1p-HA remained virtually unchanged. The content of 55Fe present in the various cell extracts was hardly altered, indicating that depletion of the three mitochondrial proteins did not impair the iron uptake into the mutant cells (Figure 2B). Further, the amounts of Rli1p did not significantly change upon depletion of Nfs1p, Yah1p, or Atm1p (not shown). Taken together, these results demonstrate that the assembly of 55Fe with Rli1p requires the activity of the mitochondrial ISC assembly and export machineries. This strongly suggests that Rli1p is an Fe/S protein.
Figure 2.
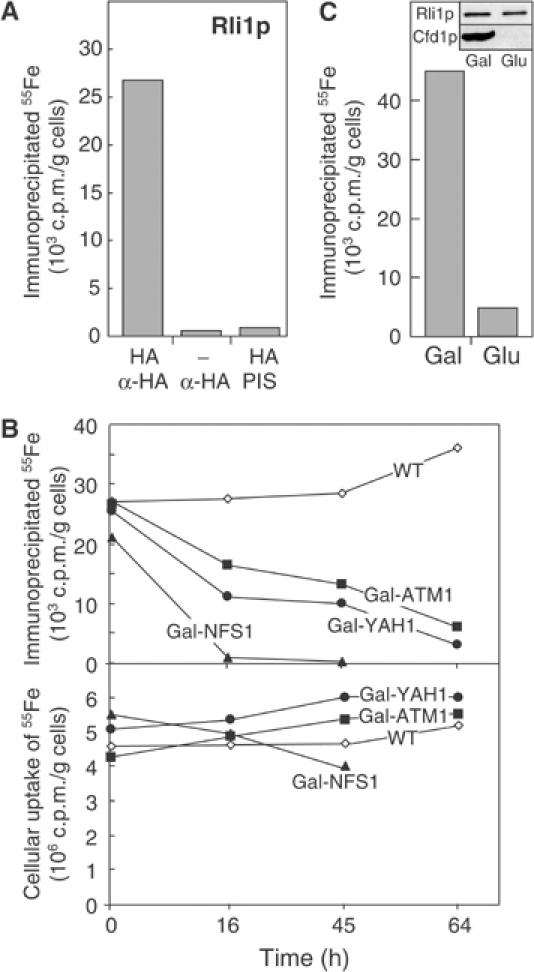
Maturation of the cytosolic Fe/S protein Rli1p requires the mitochondrial and cytosolic Fe/S protein assembly machineries. (A) Wild-type cells (−) or cells expressing Rli1p-HA (HA) were radiolabelled with 55Fe, cell extracts were prepared and an immunoprecipitation was performed using antibodies against the HA-tag or from pre-immune serum (PIS). The binding of 55Fe to the immunobeads was estimated by scintillation counting. (B) The assay for maturation of Rli1p-HA was performed as described in (A) using wild-type (WT), Gal-NFS1, Gal-YAH1, and Gal-ATM1 cells that were transformed with a plasmid encoding Rli1p-HA. Cells were incubated in glucose-containing, iron-poor media for the indicated time periods and an immunoprecipitation was performed using antibodies against the HA tag (upper panel). The lower panel shows the iron uptake into the cells measured by scintillation counting of cell extracts. (C) Gal-CFD1 cells transformed with a plasmid encoding Rli1p-HA were used for 55Fe incorporation into Rli1-HA after growth of cells in the presence of galactose (Gal) or glucose (Glu) as described in part B. The inset shows an immunostain of Rli1p and Cfd1p using cell extracts.
To further substantiate the character of Rli1p as an Fe/S protein, we analysed the role of Cfd1p in the insertion of Fe/S clusters into Rli1p. The cytosolic P-loop ATPase Cfd1p has recently been demonstrated to be essential for maturation of the Fe/S protein Leu1p (Roy et al, 2003). We used Gal-CFD1 cells which carried the CFD1 gene under the control of the galactose-regulatable GAL1–10 promoter and which were transformed with a plasmid coding for Rli1p-HA. Cfd1p was depleted by growth in the presence of glucose and the radiolabelling assay described above was performed to analyse 55Fe binding to Rli1p-HA. Cfd1p-deficient cells were strongly impaired in the incorporation of 55Fe into Rli1p (Figure 2C), supporting the notion that the iron associated with Rli1p is part of an Fe/S cluster.
The Fe/S clusters of Rli1p are essential for yeast cell viability
Systematic gene inactivation studies have revealed that deletion of the RLI1 gene is lethal. This was confirmed by tetrad analysis of a diploid RLI1/Δrli1 yeast strain in which one RLI1 allele was deleted by gene replacement through homologous recombination (Figure 3A). To examine whether the Fe/S clusters represent an essential part of Rli1p, two cysteine residues of the first and second ferredoxin-like motifs (positions 25 and 61; Figure 1A) were mutated either alone or simultaneously to serine residues. Plasmids encoding Rli1p-HA with or without these mutations were transformed into diploid RLI1/Δrli1 cells. After sporulation, the resulting spores carrying the plasmids were analysed for viability by tetrad dissection. Mutation of a single cysteine residue at position 25 or 61 of Rli1p resulted in a 2:2 segregation of the spores, while cells harbouring wild-type RLI1-encoding plasmids gave rise to four viable spores (Figure 3A). Apparently, Δrli1 cells harbouring plasmids encoding the mutated variants of Rli1p were inviable, even in glucose-containing rich medium. The same result was obtained, when both cysteine residues were simultaneously exchanged to serines. These experiments demonstrate the crucial character of cysteine residues 25 and 61 of Rli1p for viability of yeast cells.
Figure 3.
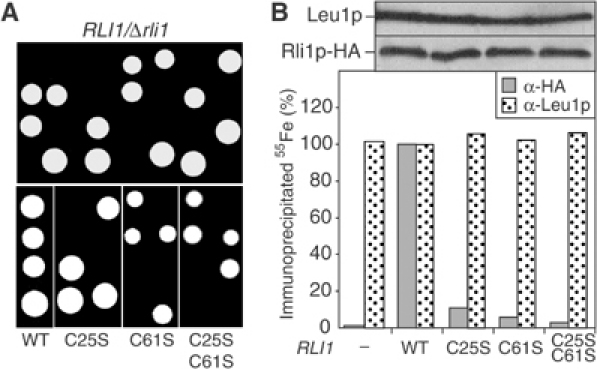
The Fe/S clusters of Rli1p are essential for cell viability. (A) The RLI1/Δrli1 diploid cells were transformed with the multicopy plasmid p426 harbouring wild-type (WT) or the indicated cysteine to serine mutants of Rli1p. In this plasmid, the expression of RLI1 was driven by part of its own promoter (300 bp upstream of RLI1). Cells with (bottom) and without (top) plasmids were subjected to sporulation and tetrad dissection. Colonies arising from individual spores after growth on glucose-containing rich medium are shown. (B) Fe/S cluster incorporation into mutant Rli1p. Wild-type cells were transformed with plasmid p426 (−) or the vectors containing the RLI1 genes described in (A). Analysis of Fe/S cluster assembly in Rli1p-HA (α-HA) or Leu1p was performed as described in Figure 2. The upper panel shows immunostaining of cell extracts for Leu1p and Rli1p-HA. Data were normalised to the levels of 55Fe that was immunoprecipitated in cells carrying plasmid p426 with the WT RLI1 gene.
Do the mutations introduced into Rli1p affect the assembly of its Fe/S clusters? The plasmids encoding the wild-type and mutant forms of Rli1p-HA were transformed into haploid wild-type yeast cells and the formation of 55Fe/S clusters on Rli1p-HA was measured by the immunoprecipitation assay described in Figure 2. Mutation of the cysteine residues 25 and/or 61 to serines almost completely abolished the maturation of Rli1p-HA holoprotein (Figure 3B). On the contrary, the incorporation of 55Fe/S clusters into Leu1p, another cytosolic Fe/S protein (Kispal et al, 1999), was unaffected under these conditions. This result shows that synthesis of mutant Rli1p had no general negative influence on the biogenesis of cytosolic Fe/S proteins. Strikingly, each point mutation in Rli1p resulted in an almost quantitative loss of the ability to incorporate Fe/S clusters. It becomes clear from these data that both clusters have to be inserted simultaneously to allow stable association with the apoprotein. This ‘all or none' behaviour is typical for interacting clusters in complex Fe/S proteins (Smith et al, 1980). We conclude from these findings that the Fe/S clusters of Rli1p are essential parts of the protein. Further, Rli1p requires both Fe/S centres to perform its indispensable function in yeast.
Rli1p is not involved in Fe/S protein biogenesis in the cytosol
Two cytosolic Fe/S proteins have recently been demonstrated to perform a function in Fe/S protein biogenesis, namely Nar1p and Nbp35p (Balk et al, 2004; Hausmann et al, 2005). We therefore tested whether Rli1p may also be required in this pathway. To deplete Rli1p, we constructed a yeast strain (termed Tet-RLI1) in which RLI1 was under the control of a tetracycline-repressible promoter. Addition of the tetracycline-related compound doxycycline to these cells led to a strong depletion of the RLI1 mRNA, Rli1p protein levels, and to growth arrest on rich medium (Figure 4A, data not shown). This demonstrated that the levels of Rli1p were diminished strongly enough under these conditions to detect phenotypic consequences. This made Tet-RLI1 cells suitable for functional studies.
Figure 4.
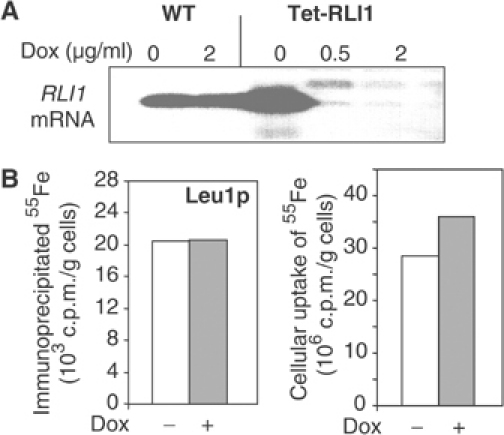
Rli1p is not involved in Fe/S protein biogenesis in the cytosol. (A) Wild-type (WT) and Tet-RLI1 cells in glucose-containing rich media were treated with the indicated amounts of doxycycline (Dox) for 16 h. Total RNA was isolated, separated in a 1% agarose gel under denaturing conditions and blotted onto Hybond membranes. The probe for Northern hybridisation was 32P-labelled RLI1 cDNA. (B) Tet-RLI1 cells were grown in iron-poor minimal media in the presence or absence of 2 μg/ml doxycycline for 16 h. A cell lysate was prepared and analysed for maturation of the Fe/S protein Leu1p by immunoprecipitation (left) or for cellular 55Fe uptake (right) followed by scintillation counting.
The function of Rli1p in the biogenesis of the cytosolic Fe/S protein Leu1p was evaluated by radiolabelling Tet-RLI1 cells with 55Fe after treatment with and without doxycycline for 16 h. No significant influence on the maturation of Leu1p holoprotein was detectable upon depletion of Rli1p (Figure 4B), even after doxycycline treatment for 45 h (not shown). Likewise, the uptake of iron into the cells was hardly changed by depleting Rli1p levels. In conclusion, Rli1p appears to be the first essential cytosolic target of Fe/S protein biogenesis without performing a function in this process.
Rli1p is associated with Hcr1p and cytosolic ribosomes
To gain insight into the cellular function of Rli1p, we attempted to identify cellular interaction partners. Yeast two-hybrid analyses identified an interaction between Rli1p and Hcr1p (Table I), a protein involved in rRNA processing and translation initiation (Ito et al, 2001; Valasek et al, 2001; Dong et al, 2004; Krogan et al, 2004). We were further interested whether the Fe/S clusters of Rli1p were needed to establish this interaction. The mutant proteins Rli1pC25S and Rli1pC61S defective in Fe/S cluster incorporation showed strong interaction with Hcr1p, demonstrating that the presence of the Fe/S clusters in Rli1p is not required for binding to this protein (Table I). By contrast, site-specific mutants in the Walker A motifs of the two nucleotide-binding domains of Rli1p were unable to interact with Hcr1p, indicating that the ABC domains are crucial for binding.
Table 1.
Interaction between Hcr1p and Rli1p mutants studied by yeast two-hybrid analysis
| Insert in pGAD424 | Insert in pGBT9 | Growth on MMD −Leu, Trp, His |
|---|---|---|
| — | — | − |
| HCR1 | — | − |
| — | RLI1 | − |
| HCR1 | RLI1 | +++ |
| — | RLI1C25S | − |
| HCR1 | RLI1C25S | +++ |
| — | RLI1C61S | − |
| HCR1 | RLI1C61S | +++ |
| — | RLI1C25S, C61S | − |
| HCR1 | RLI1C25S, C61S | − |
| HCR1 | RLI1K116L | − |
| HCR1 | RLI1K391L | − |
| Yeast two-hybrid analyses were performed as described (Clontech) using various RLI1 mutants. +++Indicates a strong interaction between the two proteins under study. MMD, minimal medium with 2% dextrose. | ||
We created a yeast strain encoding a TAP-affinity-tagged version of Rli1p under control of its endogenous promoter. A cell extract was prepared and subjected to tandem affinity purification (TAP) steps on IgG- and calmodulin-affinity columns (Puig et al, 2001). The samples from the purification procedure were analysed by SDS–PAGE and revealed a complex pattern of proteins of diverse molecular mass (Figure 5A). The identity of the TAP-purified proteins was resolved by MALDI-TOF analysis. In addition to Rli1p, numerous proteins of the large (60S) and small (40S) subunits of cytosolic ribosomes were identified (Table II). These data suggest that Rli1p was associated with 80S cytosolic ribosomes. To verify the specificity of ribosome binding to Rli1p-TAP, we analysed in parallel a sample purified from a strain expressing Nbp35p-TAP. Only low amounts of Rps3p (protein of the small ribosomal subunit) and Rpl10p (large subunit) were present in the Nbp35p-TAP-purified sample, showing that the interaction of Rli1p with ribosomes was specific (Figure 5B).
Figure 5.
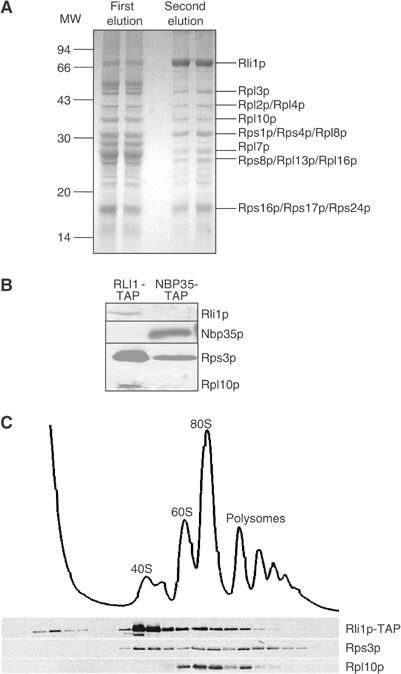
Rli1p is associated with ribosomes. (A) RLI1-TAP cells were grown in glucose-containing rich medium and used to perform a TAP of Rli1p-TAP as described using 100 mM NaCl and 5 mM CaCl2 in the buffers (Puig et al, 2001). Rli1p-associated proteins were resolved by SDS–PAGE. The identity of major bands was determined by MALDI-TOF mass spectroscopy (see Table II). (B) RLI1-TAP and NBP35-TAP cells were subjected to TAP and aliquots were analysed by immunostaining for the indicated proteins. (C) RLI1-TAP cells were grown in glucose-containing rich medium, and a cell lysate was subjected to sucrose density gradient centrifugation (10–50% sucrose). Fractions were collected and analysed by SDS–PAGE and by immunostaining for the indicated proteins. The OD 260 nm reading (top) shows the positions of the 40S, 60S, and 80S ribosomes and polysomes.
Table 2.
Proteins associated with Rli1p-TAP identified by MALDI-TOF
| Protein | Calc. MW | App. MW |
|---|---|---|
| Rpl3p | 43.8 | 45 |
| Rpl4ap | 39.1 | 39 |
| Rpl4bp | 39.1 | 39 |
| Rpl2bp | 39.1 | 39 |
| Rpl10p | 33.7 | 34 |
| Rps1bp | 28.8 | 30 |
| Rps1ap | 28.7 | 30 |
| Rps4bp | 29.4 | 30 |
| Rps4ap | 29.4 | 30 |
| Rpl8bp | 28.1 | 30 |
| Rpl7ap | 27.6 | 26 |
| Rpl7bp | 27.7 | 26 |
| Rpl16ap | 22.2 | 24 |
| Rps8ap | 22.5 | 24 |
| Rps8bp | 22.5 | 24 |
| Rpl13bp | 22.5 | 24 |
| Rpl13ap | 22.6 | 24 |
| Rps17ap | 15.8 | 16 |
| Rps16bp | 15.8 | 16 |
| Rps24ap | 15.3 | 16 |
| RLI1-TAP cells were subjected to TAP of Rli1p-TAP as described in Figure 5A (Puig et al, 2001). Rli1p-TAP-bound, lower molecular mass proteins were identified by MALDI-TOF mass spectroscopy. | ||
To obtain independent support for an interaction between Rli1p and cytosolic ribosomes, we fractionated cell extracts by sucrose density gradient centrifugation. To facilitate detection of Rli1p by immunostaining, we used cells synthesising the TAP-tagged (Figure 5C) or HA-tagged (not shown) versions of Rli1p. The individual fractions of the sucrose gradient were separated by SDS–PAGE and analysed for Rli1p, Rps3p and Rpl10p by immunostaining. The position of ribosomes was estimated by reading the absorption at 260 nm. Most of Rli1p migrated in fractions containing the small 40S ribosomal subunit indicated by the presence of Rps3p, but not Rpl10p (Figure 5C). This suggested that the majority of Rli1p was bound to the 40S ribosomal subunit under these conditions. In addition, minor amounts of Rli1p co-migrated with 60S and 80S ribosomes. We conclude from these analyses that Rli1p was associated with cytosolic ribosomes, thus corroborating the affinity purification data (see above). The amount of proteins associated with Rli1p strongly depends on the ionic strength. At lower salt concentrations, Rli1p was found associated with both ribosomes and the initiation factor complex eIF3, while higher salt concentrations lead to dissociation of these complexes (Yarunin et al, 2005; Dong et al, 2004). Thus, Rli1p binding to ribosomes and eIF3 complex is rather sensitive to increases in the ionic strength, implying a dynamic nature of the interactions.
Rli1p is required for both protein translation and ribosome export from the nucleus
What may be the functional implication for the association of Rli1p with ribosomes? We tested the importance of Rli1p for cytosolic protein translation and for biogenesis of ribosomes. To address the potential role of Rli1p in protein synthesis, wild-type and Tet-RLI1 cells were first treated with doxycycline for 24 h in order to deplete Rli1p. Cells were then briefly radiolabelled with 35S-methionine, and equal amounts of cell extracts were analysed by SDS–PAGE for the radioactive protein pattern. Depletion of Rli1p strongly impaired protein translation, when cells were grown in rich medium, whereas no effects were observed after growth in minimal (SD) medium (Figure 6A). The strong effect in rich medium may readily be explained by the more efficient depletion of Rli1p, as under these conditions cells undergo more divisions than in minimal medium. The amounts of ribosomes, in particular the levels of the ribosomal proteins Rps3p and Rpl10p, were only slightly (rich medium) or not at all (minimal medium) decreased upon depletion of Rli1p (Figure 6B; data not shown), indicating that the translation defect cannot fully be explained by ribosome depletion. In summary, Rli1p depletion in rich medium causes a defect in cytosolic protein translation. This effect might be due to the requirement of Rli1p in the assembly of the translation initiation factor eIF3 (Dong et al, 2004).
Figure 6.
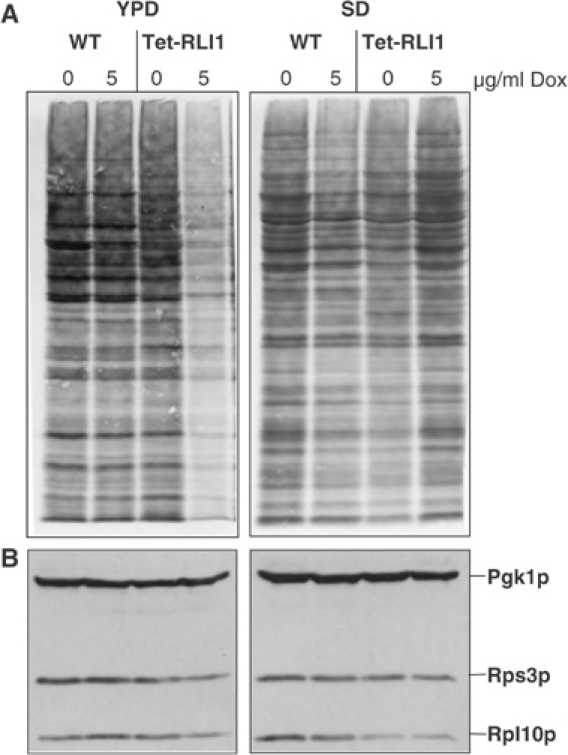
Depletion of Rli1p impairs protein synthesis in cells grown in glucose-containing rich medium. Wild-type (WT) and Tet-RLI1 cells were grown in glucose-containing rich (YPD) and minimal (SD) media and treated with doxycycline (Dox) for 24 h. (A) Cells were radiolabelled with 35S-methionine for 10 min at 30°C. A cell extract was prepared and analysed by SDS–PAGE and autoradiography. (B) Post-mitochondrial supernatants were analysed by immunostaining for the indicated proteins. Pgk1p, cytosolic phosphoglycerate kinase.
We next analysed whether Rli1p may be of importance for the biogenesis of ribosomes. Wild-type and Tet-RLI1 cells were transformed with plasmids encoding fusion proteins of the ribosomal proteins Rpl25p or Rps2p and green fluorescent protein (GFP; Gadal et al, 2001). Cells were then cultivated in minimal medium with doxycycline to deplete Rli1p. After various time periods, cells were analysed by immunofluorescence microscopy for the subcellular localisation of the GFP fusion proteins. The position of the nucleus was visualised by DAPI staining. Upon depletion of Rli1p, both ribosomal fusion proteins accumulated in the cell nucleus, whereas normal cytosolic localisation was seen in doxycycline-treated wild-type cells or in untreated Tet-RLI1 cells (Figures 7A and B). This clearly shows that Rli1p is necessary for nuclear export of both ribosomal subunits to the cytosol, suggesting a crucial role of Rli1p for ribosome biogenesis (see also Yarunin et al, 2005). Strong accumulation of the small and large ribosomal subunits was observed after depletion of Rli1p for 24 and 15 h, respectively (not shown). As noted above, no protein translation defect was evident after this period in this growth medium. Apparently, nuclear export of ribosomal subunits is impaired before effects on translation become detectable.
Figure 7.
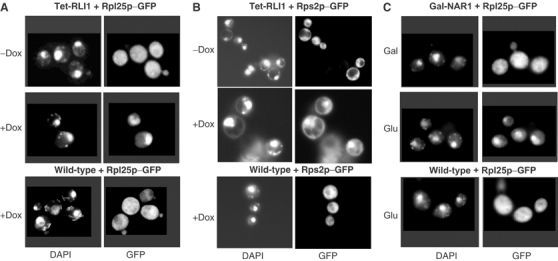
Depletion of Rli1p and the cytosolic Fe/S protein assembly component Nar1p results in nuclear accumulation of ribosomal subunits. (A, B) Wild-type and Tet-RLI1 cells were transformed with plasmids carrying the fusion proteins Rpl25p-GFP (A) or Rps2p-GFP (B). Cells were grown in minimal medium and treated with 5 μg/ml doxycycline (Dox). Cells were analysed after 19 h (Rpl25p-GFP) or 24 h (Rps2p-GFP) by immunofluorescence microscopy. Nuclear accumulation of Rpl25p and Rps2p was seen in at least 80 and 50%, respectively, of the cells. (C) Gal-NAR1 cells were transformed with a plasmid coding for Rpl25p-GFP, and were grown in glucose-containing rich medium for depletion of Nar1p (Balk et al, 2004). After 24 h, cells were analysed by immunofluorescence. Wild-type and Nar1p-containing Gal-NAR1 cells are shown as a control.
Since Rli1p is an Fe/S protein, we finally examined whether the nuclear export defect of ribosomes can be observed in mutants defective in Fe/S protein biogenesis. Nar1p, a component of the cytosolic Fe/S protein assembly machinery, was depleted using Gal-NAR1 cells by growth in the presence of glucose (Balk et al, 2004). The subcellular localisation of Rpl25p-GFP was measured by immunofluorescence. A strong nucleus accumulation was seen upon depletion of Nar1p (Figure 7C). Since Rli1p apoprotein is present at wild-type levels under these conditions (Balk et al, 2004), our data demonstrate that the presence of Fe/S clusters on Rli1p is crucial for its function in ribosome export from the nucleus. No translational defect was detected upon depletion of Nar1p (not shown), supporting the notion that the nuclear export defect occurred earlier than the translational arrest.
Discussion
In the present work, we report a surprising functional connection between two central cellular processes, namely Fe/S protein maturation involving mitochondria and the biogenesis of cytosolic ribosomes. Mitochondria were shown to be required for the assembly of the essential Fe/S clusters in Rli1p, a novel cytosolic Fe/S protein with two C-terminal ABC domains. Mutation of two conserved cysteine residues predicted to co-ordinate the Fe/S clusters rendered yeast cells inviable and fully impaired the ability of Rli1p to assemble its Fe/S clusters. Further, this protein was demonstrated to be required for ribosome export from the nucleus to the cytosol and, under some conditions, for cytosolic protein translation (see also Yarunin et al, 2005). The latter function of Rli1p fits nicely to and extends a recent observation that Rli1p is involved in the assembly of translation pre-initiation complexes (Dong et al, 2004). All these data suggest that Rli1p may perform multiple roles within the cell. Importantly, Rli1p closely links two central and ancient biosynthetic pathways, Fe/S protein maturation and ribosome biogenesis. Since Fe/S protein biogenesis is the only known essential biochemical process performed by mitochondria, these data provide the first explanation why these organelles are indispensable in all eukaryotes studied to date, a long-standing question in mitochondriology. Recently, even those organisms which were thought to lack classical mitochondria altogether, such as microsporidia and diplomonads (so-called amitochondriate protists), have been shown to harbour remnants of these organelles (Williams et al, 2002; Tovar et al, 2003). These so-called mitosomes have lost most functions of classical mitochondria, yet retained homologues of central components of the ISC assembly machinery, such as Nfs1 and Isu1. Since Rli1 is present in virtually all eukaryotes including microsporidia and diplomonads, the involvement of mitochondria/mitosomes in the maturation of cytosolic Fe/S proteins such as Rli1, and in turn in ribosome biogenesis, appears to represent an essential and minimal function of these organelles.
Biogenesis of the essential Fe/S clusters of Rli1p is dependent on the function of the mitochondrial ISC assembly and export machineries. In addition, components of the emerging cytosolic Fe/S protein assembly apparatus are required for Rli1p maturation. These include the P-loop NTPases Cfd1p and Nbp35p (this work; Vitale et al, 1996; Roy et al, 2003; Hausmann et al, 2005) and the iron-only hydrogenase-like protein Nar1p (Balk et al, 2004). Many of the mitochondrial ISC components (such as Nfs1p, Yah1p and Erv1p) and all the three cytosolic assembly proteins are encoded by essential genes, suggesting that they perform a critical function in the assembly of the Fe/S clusters in Rli1p. Interestingly, Nbp35p and Nar1p contain Fe/S clusters, and therefore they appear to be involved in their own maturation. Thus, these proteins cannot be regarded as true targets of the Fe/S cluster biogenesis pathway, unlike Rli1p, which is not needed for efficient Fe/S protein biogenesis in the cytosol. With the identification of Rli1p, the list of known extra-mitochondrial Fe/S proteins is further growing (Lill et al, 1999; Balk and Lill, 2004; Lill and Mühlenhoff, 2005). All these proteins have in common that their maturation depends on mitochondrial ISC assembly components (Lill and Kispal, 2000). According to a current working hypothesis, the mitochondrial ISC assembly machinery synthesises a component which is exported through the ABC transporter Atm1p to the cytosol, where it can be further used for assembly of cytosolic Fe/S proteins (Balk and Lill, 2004; Lill and Mühlenhoff, 2005).
We have provided evidence that Rli1p performs a crucial role in the biogenesis of ribosomes. Rli1p was found to be associated with ribosomal particles by using co-precipitation and co-sedimentation methods. Depletion of Rli1p resulted in a strong defect in the nuclear export of the small and large ribosomal subunits to the cytosol and in an impairment of late steps of rRNA processing (see Yarunin et al, 2005). In light of these results and the proposed function of human RLI as an RNase inhibitor (Bisbal et al, 1995), it is tempting to speculate that Rli1p may regulate the activity of an RNase, thereby controlling the rate and/or efficiency of ribosome assembly and nuclear export. Taken together, these findings strongly suggest that Rli1p performs a central function in a partial reaction of the complex pathway of ribosome biogenesis (reviewed by Tschochner and Hurt, 2003). Our work now opens the way to define this step.
Depletion of Rli1p in glucose-containing, rich medium diminished the protein synthesis activity of cytosolic ribosomes, suggesting a role of Rli1p also in protein translation. Strikingly, this defect developed well after the impairment of nuclear ribosome export. Since the effect on translation was stronger than the depletion of ribosomes, the late occurrence of translational defects likely does not represent a secondary effect. Rather, the physical interaction of Rli1p with the translation initiation complex eIF3 including the bifunctional Hcr1p may suggest an additional role of Rli1p in early steps of protein synthesis (this work; Ito et al, 2001; Valasek et al, 2001; Krogan et al, 2004; Yarunin et al, 2005). This contention is further supported by recent work showing an involvement of Rli1p in assembly of the eIF3 complex (Dong et al, 2004).
Rli1p belongs to the most strongly conserved proteins in evolution and homologues are encoded in the genomes of Eukarya and Archaea. No eubacterial Rli1p homologues have been identified hitherto, indicating that eukaryotic Rli1 proteins were inherited from archaebacteria. The evolutionary origin of Rli1 is of considerable interest with regard to both the biogenesis of its Fe/S clusters and the cellular function of the protein. It is currently unknown how archaebacteria assemble their Fe/S proteins. The genomes of archaebacteria generally do not contain homologues of the bacterial or mitochondrial ISC assembly machinery (Takahashi and Tokumoto, 2002). It is therefore unlikely that the assembly of the Fe/S clusters in archaebacterial Rli1 is facilitated by a process involving ISC assembly components. Instead, archaebacterial genomes contain members of the SUF machinery which assembles Fe/S proteins in eubacteria, particularly under oxidative stress and iron-deficient conditions (Zheng et al, 2001). However, most Archaea contain only a limited set of the suf genes with sufB and sufC being present in virtually all archaebacterial genomes (Takahashi and Tokumoto, 2002). Despite the poor conservation of the Suf proteins, it may now be tested whether this machinery may be responsible for the maturation of archaebacterial Rli1.
The conspicuous evolutionary conservation of Rli1 in Archaea and Eukarya suggests that the protein carries out an ancient and central cellular task that is common to members of both kingdoms. The functions of yeast Rli1p in cytosolic protein translation and rRNA processing defined herein and by others (Dong et al, 2004; Yarunin et al, 2005) are likely to be fulfilled also by its archaebacterial homologues. In contrast, the requirement of Rli1p in ribosome export from the nucleus in eukaryotes may either represent an additional function not present in archaebacteria, or, more likely to us, represent an indirect consequence of the defect in rRNA processing in the absence of functional Rli1p.
Work described in this and the accompanying manuscript has identified a critical role of Rli1p in ribosome biogenesis and function. The studies have opened the door to unravel the detailed molecular function(s) of Rli1p in distinct steps of the complex pathway of ribosome biogenesis and protein translation. It will also be a future challenge to address the precise role of the Fe/S clusters in Rli1p and define at which steps the input of ATP is needed.
Materials and methods
Yeast strains and cell growth
The following strains of S. cerevisiae were used: Strain W303 (MATα, ura3–1, ade2–1, trp1–1, his3–11,15, leu2–3,112) in diploid and haploid form served as wild type. Disruption of the RLI1 gene was achieved by a PCR-based gene replacement technique to yield the diploid RLI1/Δrli1 cells (Wach et al, 1994). Exchange of the endogenous promoter of the RLI1 gene for a tetracycline-regulatable promoter (to yield strain Tet-RLI1) was performed by integrating the promoter substitution cassette of plasmid pCM225 in front of the RLI1 gene (Belli et al, 1998). The cassette containing the KANR tetO7 promoter and the tTA activator was amplified by PCR and was transformed into W303α haploid cells for homologous recombination. Addition of a C-terminal TAP tag was achieved by adding the coding sequence of the TAP tag to RLI1 (Puig et al, 2001). The resulting fusion gene was integrated into the yeast genome to yield strain RLI1-TAP. NBP35-TAP cells carry the NBP35 gene fused to a TAP-tag coding sequence (Euroscarf accession number SC0234). All cells were analysed for correct insertion by PCR.
Cells were grown as reported previously using rich (YP) or minimal media (Sherman, 1991; Kispal et al, 1997) and lactate medium (Diekert et al, 2001) containing the required carbon sources. For experiments involving cell labelling with radioactive 55Fe, no iron salt was added to the synthetic minimal medium.
Expression and mutagenesis of RLI1
Chromosomal expression of the gene encoding a hemagglutinin (HA)-tagged version of Rli1p (termed Rli1p-HA) was achieved by amplifying the RLI1 gene with the primers AAA GAG CTC GAC TAT ACG CTG CTT CAG TCA and AAA GTC GAC TAC CGG TGT TAT CCA AGA AA. The resulting DNA piece was inserted into the SacI and SalI restriction sites of plasmid YIplac211/HA integrative plasmid. The recombinant plasmid was linearised by digestion with BglII restriction enzyme, and the DNA was transformed into wild-type (W303α) cells. For use in complementation and radioactive 55Fe incorporation studies, the coding sequence of Rli1p-HA was amplified by PCR and inserted into the SacI and XhoI restriction sites of plasmid p426GPD (Mumberg et al, 1994).
The exchange of the two conserved N-terminal cysteine residues 25, 61 of Rli1p to serines and the lysine residues 116, 391 to leucines was accomplished by PCR-based mutagenesis of the RLI1 gene using appropriate degenerate primers. Single (C25S, C61S, K116L, K391L) and double mutations (C25S, C61S) were created, and PCR fragments encompassing the mutated RLI1 genes were cloned into the SacI and XhoI restriction sites of vector pRS426 or into pGBT9 (Clontech). DNA sequencing was used to verify the desired sequences of all mutated RLI1 genes.
Miscellaneous methods
The following published methods were used: manipulation of DNA and PCR (Sambrook and Russell, 2001); transformation of yeast cells (Gietz et al, 1992); isolation of plasmids from yeast (Sambrook and Russell, 2001); isolation and purification of yeast mitochondria by Nycodenz density gradient centrifugation (Diekert et al, 2001); raising antibodies using purified proteins or peptides coupled to maleimide-activated ovalbumine (Pierce Co., Harlow and Lane, 1998); TAP-tag affinity purification (Puig et al, 2001); sucrose density gradient centrifugation (Bassler et al, 2001); interaction analysis by Matchmaker GAL4 Two-Hybrid system (Clontech).
Determination of enzyme activities was performed according to procedures described earlier (Kispal et al, 1999). The standard error of the determination of enzyme activities varied between 5 and 15%. The labelling of yeast cells with radioactive 55Fe and the measurement of the incorporation of 55Fe into cytosolic Fe/S proteins by immunoprecipitation and liquid scintillation counting was performed as reported earlier (Kispal et al, 1999). The standard error for detection of 55Fe associated with the cytosolic Fe/S proteins in a cell lysate was less than 15%, the cellular uptake of 55Fe varied by 30%.
Acknowledgments
We thank K Neumann for expert technical assistance, Drs E Hurt and M Seedorf for generous gifts of antisera against Rps3p and Rpl10p and for the plasmids encoding eGFP fusion proteins, Drs A Yarunin and E Hurt for sharing unpublished data and discussion. Our work was supported by grants of the Sonderforschungsbereiche 286 and 593, Deutsche Forschungsgemeinschaft (Gottfried Wilhelm Leibniz program), the European Commission (Marie Curie European Fellowship HPMF-CT-2002-01750 to JB), Volkswagen-Stiftung, Fonds der chemischen Industrie, the Hungarian Fund OKTA (T34305, F37187), and the Alexander-von-Humboldt-Stiftung.
References
- Attardi G, Schatz G (1988) The biogenesis of mitochondria. Annu Rev Cell Biol 4: 289–333 [DOI] [PubMed] [Google Scholar]
- Balk J, Lill R (2004) The cell's cookbook for iron–sulfur clusters: recipes for fool's gold? Chem Biol Chem 5: 1044–1049 [DOI] [PubMed] [Google Scholar]
- Balk J, Pierik AJ, Aguilar Netz D, Mühlenhoff U, Lill R (2004) The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron–sulfur proteins. EMBO J 23: 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E (2001) Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529 [DOI] [PubMed] [Google Scholar]
- Belli G, Gari E, Aldea M, Herrero E (1998) Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14: 1127–1138 [DOI] [PubMed] [Google Scholar]
- Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T (1995) Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2–5 A pathway. J Biol Chem 270: 13308–13317 [DOI] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Marszalek J (2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell Mol Life Sci 59: 1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert K, deKroon AIPM, Kispal G, Lill R (2001) Isolation and sub-fractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol 65: 37–51 [DOI] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG (2004) The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168 [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E (2001) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Mühlenhoff U, Hofhaus G, Lill R, Lisowsky T (2001) Yeast Erv2p is the first microsomal FAD-linked sulfhydryl oxidase of the Erv1p/Alrp protein family. J Biol Chem 276: 23486–23491 [DOI] [PubMed] [Google Scholar]
- Gietz D, StJean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucl Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D (1998) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory: Cold Spring Harbor, NY [Google Scholar]
- Hausmann A, Aguilar Netz D, Balk J, Pierik AJ, Mühlenhoff U, Lill R (2005) The eukaryotic P-loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron–sulfur protein assembly machinery. Proc Natl Acad Sci USA 102 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G, Csere P, Guiard B, Lill R (1997) The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett 418: 346–350 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are required for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, Beattie BK, Lalev A, Zhang W, Davierwala AP, Mnaimneh S, Starostine A, Tikuisis AP, Grigull J, Datta N, Bray JE, Hughes TR, Emili A, Greenblatt JF (2004) High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell 13: 225–239 [DOI] [PubMed] [Google Scholar]
- Lange H, Kispal G, Kaut A, Lill R (2000) A mitochondrial ferredoxin is essential for biogenesis of intra- and extra-mitochondrial Fe/S proteins. Proc Natl Acad Sci USA 97: 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Lisowsky T, Gerber J, Mühlenhoff U, Kispal G, Lill R (2001) An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep 2: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18: 231–236 [DOI] [PubMed] [Google Scholar]
- Le Roy F, Bisbal C, Silhol M, Martinand C, Lebleu B, Salehzada T (2001) The 2′–5′A/RNase L/RNase L inhibitor (RLI) pathway regulates mitochondrial mRNAs stability in interferon alpha-treated H9 cells. J Biol Chem 276: 48473–48482 [DOI] [PubMed] [Google Scholar]
- Li K, Li Y, Shelton JM, Richardson JA, Spencer E, Chen ZJ, Wang X, Williams RS (2000) Cytochrome c deficiency causes embryonic lethality and attenuates stress-induced apoptosis. Cell 101: 389–399 [DOI] [PubMed] [Google Scholar]
- Lill R, Diekert K, Kaut A, Lange H, Pelzer W, Prohl C, Kispal G (1999) The essential role of mitochondria in the biogenesis of cellular iron–sulfur proteins. Biol Chem 380: 1157–1166 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G (2000) Maturation of cellular Fe/S proteins: the essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U (2005) Iron–sulfur protein biogenesis in eukaryotes. Trends Biochem Sci 30 (in press) [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Ann Rev Biochem 66: 861–915 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Geissler A (2001) Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol 2: 339–349 [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE (2003) A novel eukaryotic factor for cytosolic Fe–S cluster assembly. EMBO J 22: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- Scheffler IE (1999) Mitochondria. New York: Wiley-Liss [Google Scholar]
- Schüller C, Bauer BE, Kuchler K (2003) Inventory and evolution of fungal ABC protein genes. In ABC Proteins: From Bacteria to Man, Holland BI, Cole SPC, Kuchler K, Higgins CF (eds), pp 279–293. Amsterdam: Academic Press [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Sipos K, Lange H, Fekete Z, Ullmann P, Lill R, Kispal G (2002) Maturation of cytosolic iron–sulfur proteins requires glutathione. J Biol Chem 277: 26944–26949 [DOI] [PubMed] [Google Scholar]
- Smith BE, O'Donnell MJ, Lang G, Spartalian K (1980) A Mössbauer spectroscopic investigation of the redox behaviour of the molybdenum–iron protein from Klebsiella pneumoniae nitrogenase. Biochem J 191: 449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Tokumoto U (2002) A third bacterial system for the assembly of iron–sulfur clusters with homologs in archaea and plastids. J Biol Chem 277: 28380–28383 [DOI] [PubMed] [Google Scholar]
- Tovar J, Leon-Avila G, Sanchez LB, Sutak R, Tachezy J, Van Der Giezen M, Hernandez M, Muller M, Lucocq JM (2003) Mitochondrial remnant organelles of Giardia function in iron–sulphur protein maturation. Nature 426: 172–176 [DOI] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Valasek L, Hasek J, Nielsen KH, Hinnebusch AG (2001) Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J Biol Chem 276: 43351–43360 [DOI] [PubMed] [Google Scholar]
- Vitale G, Fabre E, Hurt EC (1996) NBP35 encodes an essential and evolutionary conserved protein in Saccharomyces cerevisiae with homology to a superfamily of bacterial ATPases. Gene 178: 97–106 [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Poehlmann R, Phillipsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808 [DOI] [PubMed] [Google Scholar]
- Williams BA, Hirt RP, Lucocq JM, Embley TM (2002) A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418: 865–869 [DOI] [PubMed] [Google Scholar]
- Yarunin A, Panse V, Petfalski E, Tollervey D, Hurt E (2005) Functional link between ribosome formation and biogenesis of iron–sulfur proteins. EMBO J 2005, 20 January [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G (2001) DNA microarra-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183: 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman C, Klein KC, Kiser PK, Singh AR, Firestein BL, Riba SC, Lingappa JR (2002) Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415: 88–92 [DOI] [PubMed] [Google Scholar]


