Abstract
Modifications of DNA and chromatin are fundamental for the establishment and maintenance of cell type-specific gene expression patterns that constitute cellular identities. To test whether the developmental potential of fetal brain-derived cells that form floating sphere colonies (neurospheres) can be modified by destabilizing their epigenotype, neurosphere cells were treated with chemical compounds that alter the acetylation and methylation patterns of chromatin and DNA. Intravenous infusion of bulk or clonally derived neurosphere cells treated with a combination of trichostatin A (TSA) plus 5-aza-2′-deoxycytidine (AzaC) (TSA/AzaC neurosphere cells) yielded long-term, multilineage and transplantable neurosphere-derived haematopoietic repopulation. Untreated neurosphere cells exhibited no haematopoietic repopulation activity. The neurosphere-derived haematopoietic cells showed a diploid karyotype, indicating that they are unlikely to be products of cell fusion events, a conclusion strengthened by multicolour fluorescence in situ hybridization. Our results indicate that altering the epigenotype of neurosphere cells followed by transplantation enables the generation of neurosphere-derived haematopoietic cells.
Keywords: chromatin-modifying agents, developmental potential of neurosphere cells, haematopoietic activity of neurospheres, trichostatin A, 5-Aza-2′-dC
Introduction
The establishment and maintenance of cellular identity of somatic cells including stem cells require a tight control of gene expression. Common as well as distinct gene expression patterns have been observed for different stem cell types (Terskikh et al, 2001; Ivanova et al, 2002; Ramalho-Santos et al, 2002; D'Amour and Gage, 2003). Chromatin modifications play important roles in determining gene expression (Grewal and Moazed, 2003; Jaenisch and Bird, 2003). In particular, each gene has an epigenotype defined in terms of its cell type- and developmental stage-specific chromatin organization. Transcription factors binding to their recognition sequences recruit multiprotein complexes, such as chromatin remodelling and histone-modifying factors, which alter the access of regulatory proteins to the DNA template. As a consequence, histones may become acetylated, methylated and phosphorylated. This histone code is read by specific trans-acting factors and defines levels of chromatin organization and gene activity (Jenuwein and Allis, 2001). DNA and histone modifications are somatically heritable and the propagation of epigenetic states of the chromatin to the next cell generation is important for maintaining the identity of cells (Avots et al, 2002).
One type of chromatin modification, the acetylation of histones, occurs reversibly on lysine ɛ-NH3+ groups of core histones. Hyperacetylated histones are associated with active chromatin domains, whereas hypoacetylated histones are enriched in nontranscribed loci (Eberharter and Becker, 2002). The level of histone acetylation depends on the opposing activities of histone acetyltransferases (HATs) and deacetylases (HDACs). Several microbially derived compounds specifically inhibit HDACs (Hassig and Schreiber, 1997). One of these inhibitors, the Streptomyces metabolite trichostatin A (TSA), a hydroxamic acid, exerts its activity by interacting with the catalytic site of HDACs (Yoshida et al, 1990; Jung, 2001). TSA indirectly induces acetylation of histones, cell cycle arrest and apoptosis (Johnstone, 2002).
A second epigenetic modification of the vertebrate genome is the methylation of cytosine, predominantly within CpG dinucleotides, which plays a major role in the stable repression of transcription (Bird, 2002; Dillon and Festenstein, 2002). In adult somatic cells, methylation patterns within the genome are stable. On the other hand, DNA methylation is very dynamic during early development and in certain tumour cells (Schmutte and Jones, 1998). AzaC is often used as a DNA methylation inhibitor. When incorporated into DNA, it covalently binds to and irreversibly blocks the maintenance methyltransferase Dnmt1, allowing passive demethylation to take place as cells divide and replicate their DNA (Pietrobono et al, 2002). Previous work described the reactivation of silenced genes by inhibitors of methylation (Lassar et al, 1986) and deacetylation (McCaffrey et al, 1997) and a synergistic effect of both inhibitors in tumour cells (Cameron et al, 1999; Chiurazzi et al, 1999). Interestingly, in vitro-cultured human CD34+ haematopoietic stem/progenitor cells retain the ability to repopulate immunodeficient mice if pretreated with AzaC and TSA (Milhem et al, 2004).
Several recent studies have challenged the traditional concept that development is unidirectional and that cells isolated from differentiated tissues are restricted in the types of cells they can generate. Data published on the developmental potential of committed cells showed for example that clonogenic common lymphoid progenitors can maintain a latent myeloid differentiation programme and that they can be redirected to the myeloid lineage by stimulation through exogenously expressed growth factor receptors (Kondo et al, 2000). Similarly, extracellular signals were reported to induce oligodendrocyte precursors to revert to multipotent neural stem cells (Kondo and Raff, 2000). Several reports further claimed that stem cells isolated from adult tissues may harbour unexpected developmental plasticity. For example, neurosphere cells were reported to generate haematopoietic cells in vivo, and following injection into blastocysts neurosphere cells gave rise to differentiated cells of all three germ layers (Bjornson et al, 1999; Clarke et al, 2000). However, the general plasticity of somatic stem and progenitor cells has also been questioned, as haematopoietically competent neurosphere cells were found to be rare and competence was suggested to depend on epigenetic alterations (Morshead et al, 2002). Other reports further questioned the developmental plasticity of somatic stem cells, as haematopoietic stem cells (HSCs) failed to differentiate into neural cells and transplanted single HSCs did not engraft nonhaematopoietic tissues (Castro et al, 2002; Wagers et al, 2002). In addition, it was demonstrated that cells can adopt the phenotype of other cells by spontaneous cell fusion, suggesting that the plasticity of somatic stem cells could result from cell fusion (Terada et al, 2002; Ying et al, 2002; Vassilopoulos et al, 2003; Wang et al, 2003). But not all reported transdifferentiation events are based on cell fusion: neurosphere cells after coculture with endothelial cells converted to endothelial cells; HSCs when cocultured with injured liver separated by a barrier became liver cells; and bone marrow cells generated epithelial cells in lung, liver and skin without cell fusion (Harris et al, 2004; Jang et al, 2004; Wurmser et al, 2004). Thus, the possibility that somatic stem cells can switch germ layers remains controversial and the available data suggest that transdifferentiation of stem cells is a rare event (Wagers and Weissman, 2004).
To test whether the developmental potential of neurosphere cells can be modified by destabilizing their epigenetic state, we treated neurosphere cells with the HDAC inhibitor TSA and the nucleotide analogue AzaC. Injection of neurosphere cells treated with a combination of TSA plus AzaC (TSA/AzaC) into adult irradiated recipients generated animals with chimeric haematopoietic systems. We show that TSA/AzaC neurosphere cells give rise to haematopoietic engraftment that is multilineage, long-term and transplantable.
Results
Self-renewal and differentiation of bulk and clonal neurosphere cells
Neural stem and progenitor cells form, in culture, floating colonies (neurospheres), which contain both cells that can initiate new colonies and cells that can differentiate into neural and glial cell types (Reynolds and Weiss, 1992). In our experiments, we utilized neurosphere cells from wild-type (wt) and from bcl2-transgenic embryonic day (E) 14.5 embryos (Figure 1A), with expression of the latter gene as protection against the apoptosis-inducing activities of TSA (Koyama et al, 2000; Johnstone, 2002). Ubiquitous expression of the bcl2 transgene under the control of the mouse H-2Kb promoter did not adversely affect embryonic or adult development (Domen et al, 1998). Under limiting dilution, bcl2-transgenic, as compared to wt, neurospheres showed a limited increase in the numbers of neurosphere-initiating cells (Figure 1B). Likewise, bcl2-transgenic mice show a mild increase in HSC numbers (Domen et al, 1998). Wt and eGFP/bcl2 double transgenic (eGFP/bcl2) neurosphere cells differentiated into GFAP+ (glial fibrilaric acidic protein) astroglial and tubulin beta-III+ neuronal cells (Figure 1C). Neurospheres contain cells that self-renew and differentiate into neural and glial cell types.
Figure 1.
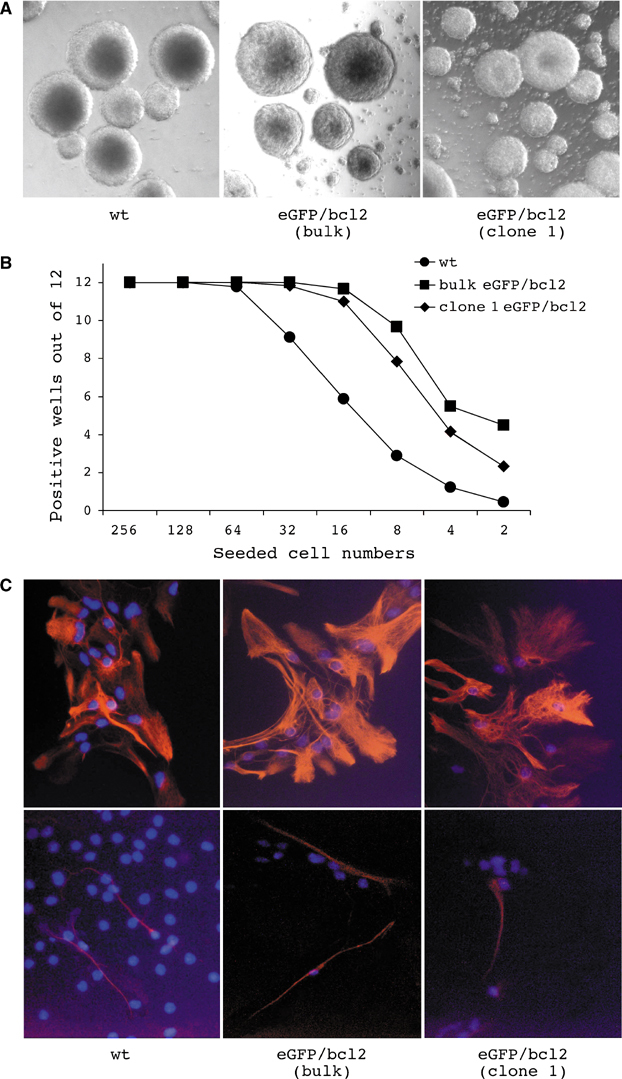
Self-renewal and multilineage differentiation of neurosphere cells. (A) Brain-derived cells from wt and eGFP/bcl2 double transgenic E 14.5 embryos form floating spheres (neurospheres) in culture. (B) To assess the ability of wt and eGFP/bcl2 neurosphere cells to self-renew, graded numbers of cells from single-cell suspensions of neurospheres were seeded into 96-well plates in NSC medium (n⩾6). Numbers of wells that had developed neurospheres 2 weeks postseeding were plotted versus the numbers of cells seeded. (C) wt as well as bulk and cloned eGFP/bcl2 neurosphere cells produced under differentiation conditions GFAP+ (glial fibrilaric acidic protein) astroglial cells (top) and tubulin beta-III+ neuronal cells (bottom). Cells were subjected to nuclear counterstaining (DAPI).
Combined TSA and AzaC treatment confers in vivo haematopoietic activity to neurosphere cells
To ask whether the incubation of neurosphere cells with substances that modify the epigenotype of cells influences their developmental potential, we transiently treated neurosphere cells with a combination of TSA and AzaC prior to transplantation. Treatment increases the level of histone H4 acetylation and decreases the amount of methyl-CpG-binding protein 2 (MeCP2) in nuclei of neurosphere cells, indicating that treatment increases histone acetylation and reduces cytidine methylation (Figure 2A). In addition, treated bcl2 and wt neurospheres show reduced proliferation and neurosphere initiation frequencies (Figure 2B and C) but retain glial and neural differentiation potential (data not shown). Bulk neurospheres were established from male embryos, and then treated and untreated neurospheres were dissociated and intravenously injected into irradiated CD45 congenic female recipients. For animals transplanted with untreated wt or eGFP/bcl2 neurosphere cells, FACS analysis revealed no detectable engraftment of the peripheral blood (Figure 3). In contrast, 2/48 or 5/25 recipients transplanted with the same wt or eGFP/bcl2 neurosphere cell cultures but pretreated with TSA/AzaC had generated donor-derived cells in the peripheral blood that stained with the pan-haematopoietic marker CD45. Donor cells were first detected in the peripheral blood of recipients analysed at 4 weeks post-transplantation. Male-specific PCRs on genomic DNA isolated from peripheral blood further confirmed the donor origin of the cells (data not shown). Repeated sampling of the peripheral blood of one eGFP/bcl2 neurosphere cell transplant recipient with about 80% blood chimaerism showed that the engraftment level was stable over a period of 12 months. In addition to CD45.2+ and eGFP+ donor cells, we also detected in engrafted animals a proportion of CD45.2+ donor cells that do not express eGFP. These cells may have lost eGFP expression, for example, due to silencing of the transgene, since eGFP transgenic mice also carry haematopoietic cells that are eGFP− (data not shown).
Figure 2.
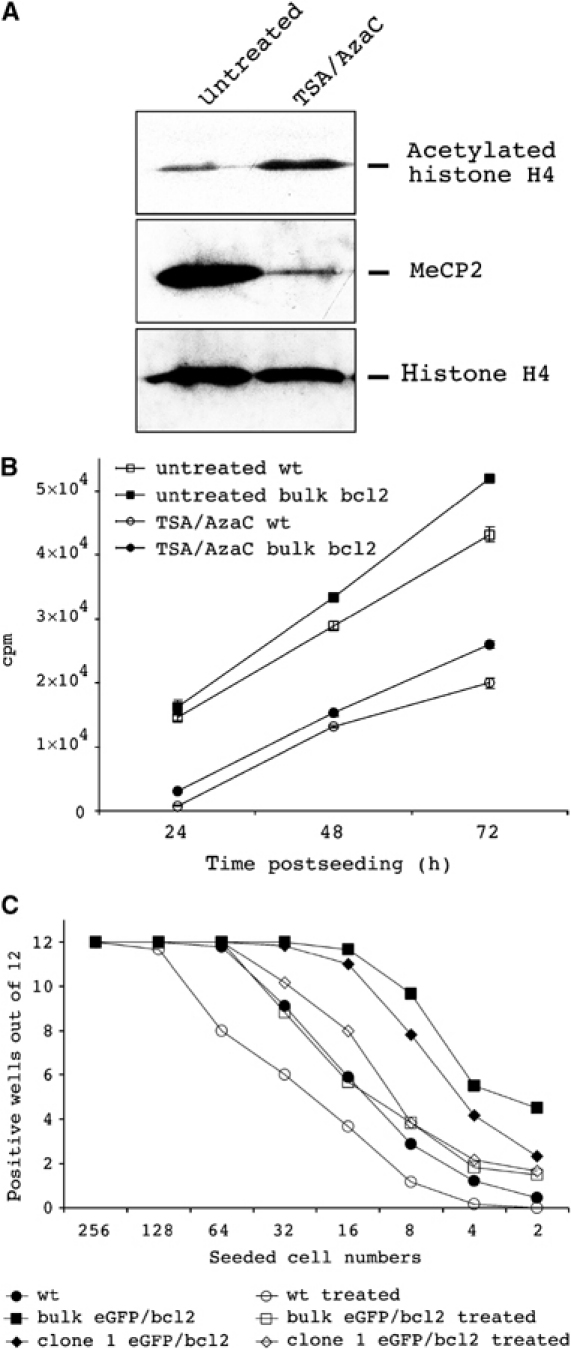
Consequences of TSA/AzaC treatment on epigenetic status, proliferation and neurosphere-initiating frequency of wt and bcl2-transgenic neurosphere cells. (A) Western blot analysis of untreated and treated bcl2-transgenic neurosphere cells with acetylated histone H4- and MeCP2-specific antibodies. Equal loading was verified by histone H4-specific antibody. (B) [3H]-thymidine incorporation into untreated and TSA/AzaC-treated wt neurosphere cells and TSA/AzaC-treated eGFP/bcl2 neurosphere cells. Means and standard deviations are indicated (n=3). (C) Neurosphere-initiating frequencies of untreated, treated wt and eGFP/bcl2 neurosphere cells. To analyse the effect of TSA/AzaC treatment on neurosphere-initiating frequencies, wt and eGFP/bcl2 neurosphere cells were left untreated or were treated for 48 h with TSA/AzaC. Single-cell suspensions were then prepared and graded numbers of cells were seeded into 96-well plates in NSC medium (n⩾6). Numbers of wells that developed neurospheres 2 weeks postseeding were plotted against the numbers of cells seeded. Data for untreated neurosphere cells are from Figure 1B.
Figure 3.
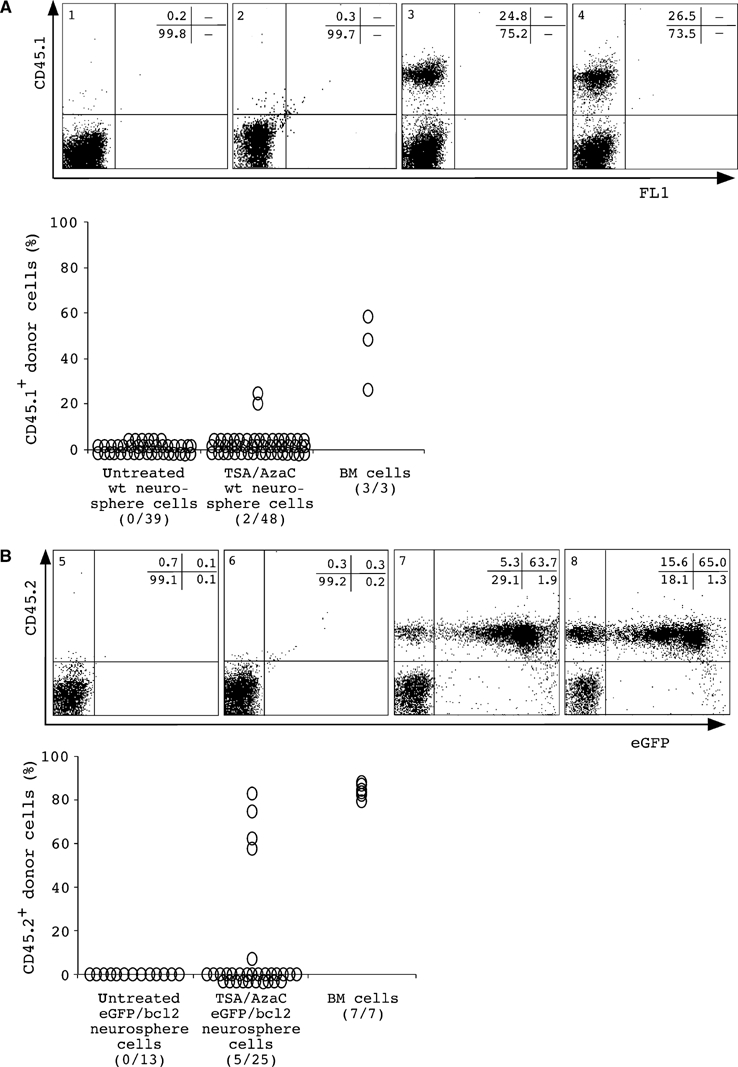
Donor cells in recipients after transplantation of bulk TSA/AzaC-treated wt (A) or eGFP/bcl2 (B) neurosphere cells. Donor-specific FACS analysis of peripheral blood cells of nontransplanted CD45.2 and CD45.1 animals (panels 1 and 5) and of recipients receiving untreated (panels 2 and 6) or treated CD45.1 congenic wt or CD45.2 congenic eGFP/bcl2 neurosphere cells (panels 3 and 7). Animals receiving CD45.1 congenic or eGFP transgenic, CD45.2 congenic bone marrow cells are shown (panels 4 and 8). Recipient #154 is shown in panel 7. Percentages of cells positive for donor or lineage markers are shown. Also shown are summaries of the engraftment (%) in the peripheral blood by wt or eGFP/bcl2 neurosphere-derived cells. Numbers of chimerics per total number of analysed animals are given. Animals were analysed 1.5-2 months after transplantation. The results from wt and bulk eGFP/bcl2 neurosphere cell transplantations were from two and six independent experiments; 2/2 and 3/6 transplantations produced haematopoietic chimaeras.
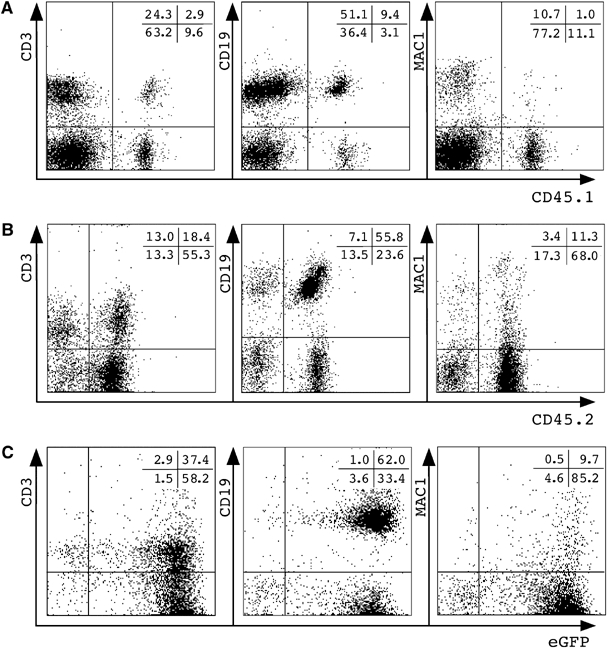
Further analysis of splenocytes (Figure 4) and bone marrow cells (data not shown) showed in all engrafted animals the presence of donor-derived cells that stained with monoclonal antibodies against T cells (CD3), B cells (CD19) or macrophages/granulocytes (MAC1). Control transplantations of bone marrow cells resulted in a similar repopulation pattern. Importantly, transplantation of bone marrow cells from primary into secondary recipients demonstrated that the donor-derived haematopoietic activity is serially transplantable (Figure 4C). Taken together, the transplantation of bulk TSA/AzaC neurosphere cells from wt and eGFP/bcl2 origin resulted in long-term and multilineage engraftment of the haematopoietic system of irradiated recipients. As wt and eGFP/bcl2 neurosphere cells differ in TSA/AzaC sensitivity and eGFP/bcl2 neurosphere cells show higher engraftment frequencies, only eGFP/bcl2 neurosphere cells were further investigated.
Figure 4.
Lymphoid/myeloid donor cells in recipients after transplantation of bulk TSA/AzaC-treated neurosphere cells. Shown are splenocytes of wt (A) or eGFP/bcl2 (B) neurosphere transplant recipients stained with CD45 donor-type and lineage marker antibodies. (C) FACS analysis of the peripheral blood of a representative secondary CD45.1 recipient (n=3) receiving bone marrow cells of animal #154. Gated CD45.1− cells were plotted against lineage markers and eGFP.
Cloned neurosphere cells generate multilineage haematopoietic reconstitution with normal haematopoietic cells
To test whether the progeny of a single neurosphere cell have the potential to generate haematopoietic cells in vivo, we established clonal cultures. Three clonal eGFP/bcl2 neurosphere lines were established and transplanted into irradiated recipients. Two of the three clonal lines engrafted and generated CD45.2+ and eGFP+ donor cells (Figure 5A and B). As seen with bulk neurosphere cells, only TSA/AzaC-treated but not untreated cloned eGFP/bcl2 neurosphere cells successfully engrafted. Phenotypic analysis of sorted donor cells from bone marrow revealed normal haematopoietic morphology (Figure 5C). The haematopoietic differentiation potential of eGFP/bcl2 neurosphere-derived cells was studied by in vitro clonogenic assays of sorted CD45.2− host and CD45.2+ donor cells from the bone marrow of chimeric recipients. The frequencies of erythro/myeloid colony-forming cells in the donor and host cell populations were essentially identical (Figure 5D).
Figure 5.
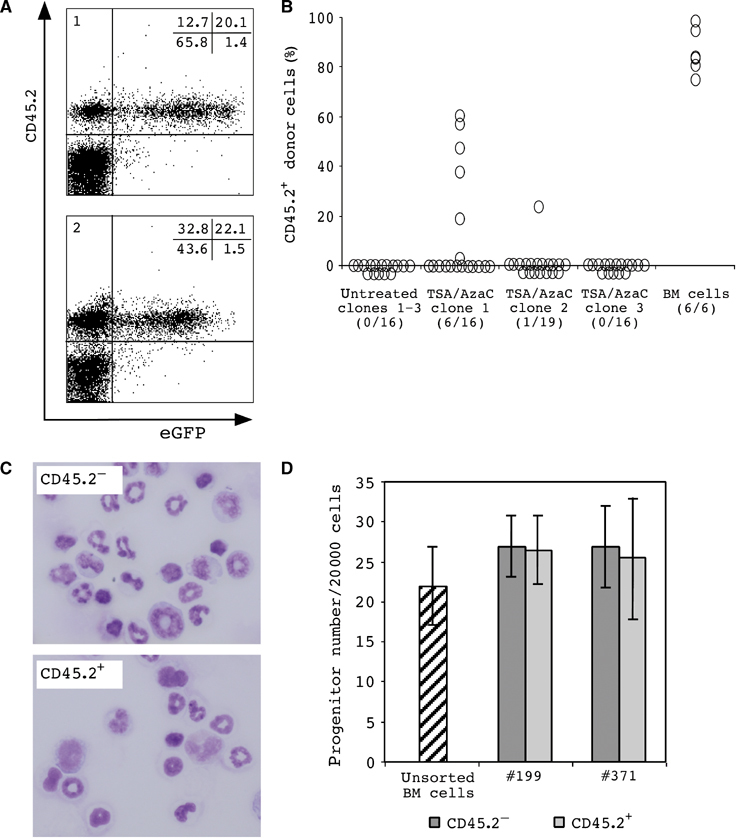
Donor cells in transplant recipients of cloned TSA/AzaC eGFP/bcl2 neurosphere cells. (A) FACS analysis of peripheral blood cells of two representative TSA/AzaC clone 1 recipients 3 months post-transplantation (panel 1: recipient #197; panel 2: recipient #199). (B) Percentage CD45.2+ donor cells in the peripheral blood 5–8 weeks post-transplant. Numbers of chimeric animals per total number of analysed animals are indicated. (C) Morphology of sorted recipient (CD45.2−) and neurosphere-derived (CD45.2+) cells from chimeric bone marrow of a clone 2 recipient. (D) Erythro/myeloid colony formation of sorted recipient (CD45.2−) and clone 1-derived (CD45.2+) bone marrow cells (transplant recipients #199 and #371).
To further characterize the neurosphere-derived cells, we performed immunophenotyping of donor T- and B-lymphoid cells in chimeric animals (Figure 6A and B). We detected CD4+ and CD8+ donor cell populations in thymus and spleen and immature (IgM+ IgD+/−) and mature (IgM+ IgD+) donor B cells in spleen and bone marrow. Additionally, bone marrow and spleen of transplant recipients contained neurosphere-derived MAC1+ myeloid cells (Figure 6C). Using four-colour flow cytometry, we assessed the presence of normal combinations of myeloid and lymphoid differentiation markers on donor cells in chimeric bone marrow and spleen. Like their HSC-derived counterparts, neurosphere-derived myeloid eGFP+ MAC1+ Gr1+ cells did not express the B-lymphoid marker CD19, nor did they show T-lymphoid-specific CD3 staining. Similarly, donor B-lymphoid eGFP+ B220+ CD19+ cells did not stain with Gr1 (Figure 7). After transplantation of bone marrow cells from two primary TSA/AzaC neurosphere cell recipients into six secondary recipients each, all secondary recipients were engrafted (data not shown). Our data indicate that transplantation of cloned TSA/AzaC eGFP/bcl2 neurosphere cells into recipient mice results in haematopoietic chimaerism. Donor cells showed normal morphology, produced erythro/myeloid colony-forming cells in clonogenic in vitro cultures and exhibited normal expression of lineage-specific myeloid/lymphoid differentiation markers. Importantly, none of the primary or secondary recipients displayed signs of haematological malignancies.
Figure 6.
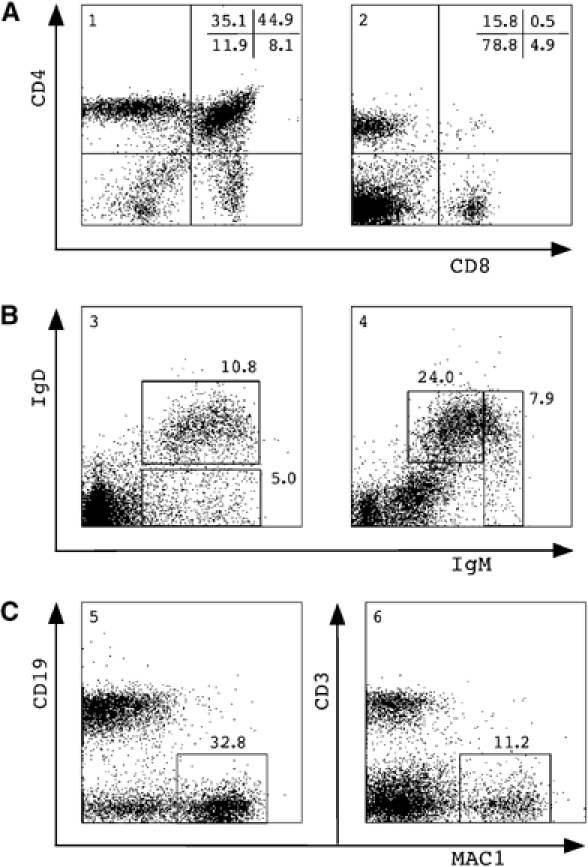
Lymphoid/myeloid repopulation by cloned TSA/AzaC eGFP/bcl2 neurosphere cells. (A) Thymocytes (panel 1) and splenocytes (panel 2) and (B) bone marrow cells (panel 3) and splenocytes (panel 4) were isolated and stained; gated eGFP+ donor-derived cells were stained with antibodies against CD4 and CD8 (A) or IgD and IgM (B). (C) Gated eGFP+ donor cells in bone marrow were stained with antibodies against CD19 and MAC1 (panel 5), and donor splenocytes were stained for CD3 and MAC1 (panel 6). Percentages of gated eGFP+ cells plotted against lineage-specific staining are shown. Analysis of a representative clone 1 recipient 3 months post-transplant is shown.
Figure 7.
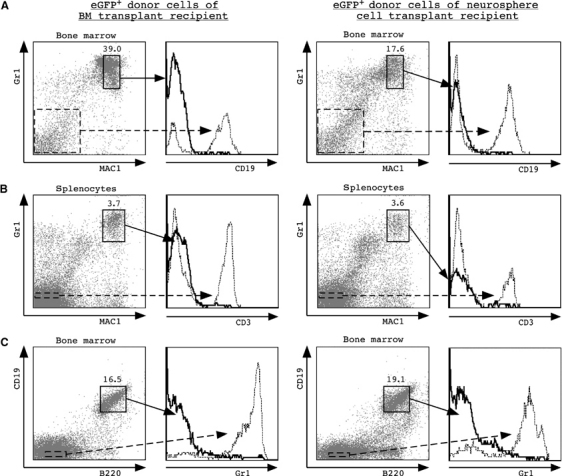
Myeloid/lymphoid-specific marker expression of neurosphere-derived bone marrow cells and splenocytes. Analyses of representative animals 3 months post-transplant are shown (bone marrow transplant recipient (left); TSA/AzaC clone 1 recipient (right); two transplanted animals were analysed with similar results). Percentages of gated eGFP+ donor cells are plotted against lineage-specific staining. (A) eGFP+ donor cells in bone marrow were gated and plotted versus MAC1 and Gr1 staining; eGFP+ MAC1+ Gr1+ cells were gated and plotted versus CD19 staining. As a control, eGFP+ MAC1− Gr1− non-myeloid cells were plotted versus CD19 immunoreactivity (hatched line). (B) eGFP+ splenocytes were stained with anti-MAC1 and anti-Gr1 antibodies and counterstained with anti-CD3 antibodies. Gated eGFP+ MAC1− Gr1− cells were plotted versus CD3 staining (hatched line). (D) eGFP+ bone marrow cells were gated and plotted versus B220 and CD19 immunoreactivity; eGFP+ B220+ CD19+ cells were counterstained with anti-Gr1 antibodies. Gated eGFP+ B220− CD19− cells were plotted versus Gr1 staining (hatched line). Analyses of a representative animal 3 months post-transplant are shown.
To test whether neurosphere cells have in vitro haematopoietic activity, we plated untreated (1 × 106) and treated (2 × 106 bulk and 1 × 106 cloned) eGFP/bcl2 neurosphere cells into methylcellulose under haematopoietic growth conditions. While control cultures seeded with bone marrow cells generated the expected numbers of colonies (26±2.2 colonies per 20 000 bone marrow cells), cultures seeded with neurosphere cells did not generate haematopoietic colonies.
Neurosphere-derived haematopoietic cells carry a diploid donor karyotype
It was previously observed that bone marrow and neurosphere cells are both able to fuse with unrelated cell types (Terada et al, 2002; Ying et al, 2002; Vassilopoulos et al, 2003; Wang et al, 2003). Consequently, cell fate changes of somatic stem cells could be the result of fusion with recipient cells. The high-level chimaerism in haematopoietic tissues described above allows flow cytometric analysis of the DNA content of donor cells. The total DNA content of cells as a marker revealing polyploidy can be determined spectofluorometrically after staining cells with the DNA dye 7-aminoactinomycin D (7-AAD; Schmid et al, 2000). We injected bulk and clonally derived TSA/AzaC eGFP/bcl2 neurosphere cells isolated from CD45.2 embryos into irradiated CD45.1 congenic recipients. At 3–4 months post-transplantation, splenocytes were isolated from recipients that had received either neurosphere or bone marrow cells from a control mouse. CD45.2 expression, as a marker for transplanted donor cells, was plotted against DNA content (Figure 8A). Most splenocytes of control mice were noncycling and had a 2n DNA content, and only a minor population had more than 2n DNA content, indicating cycling. After transplantation of CD45.2 bone marrow cells or treated CD45.2 eGFP/bcl2 neurosphere cells, CD45.2+ cells were evident in the periphery, and 7-AAD analysis of both donor and recipient splenocytes revealed 2n DNA for the great majority of cells. The minor fraction of cells displaying a 4n DNA is expected due to cell cycling. Importantly, no cells with higher DNA content were detectable. We obtained similar results with peripheral blood cells (data not shown). The distribution of cycling and noncycling cells, revealed by 7-AAD DNA content analysis, is shown on stimulated splenocytes. To determine the karyotype of neurosphere-derived splenocytes, multicolour fluorescence in situ hybridization (M-FISH) analysis was performed (Figure 8B). The results indicate in all 20 evaluated metaphase spreads a normal diploid male karyotype (2n). In addition, we determined whether the CD45.1 allele, a marker of recipient cells, and the donor alleles CD45.2 (both chromosome 1) and eGFP (chromosome 15) were present in distinct cells (Figure 8C). The CD45.1 and CD45.2 alleles are biallelically expressed (Ema and Nakauchi, 2000). Analysis by flow cytometry of the peripheral blood cells or splenocytes revealed that most CD45.1+ recipient cells are eGFP−, while CD45.2+ donor cells are eGFP+. Thus, CD45.1+ and CD45.2+ cells comprise distinct cell populations. Together, these results indicate that the majority of the eGFP/bcl2 neurosphere-derived cells are diploid and are thus unlikely to be products of cell fusion.
Figure 8.
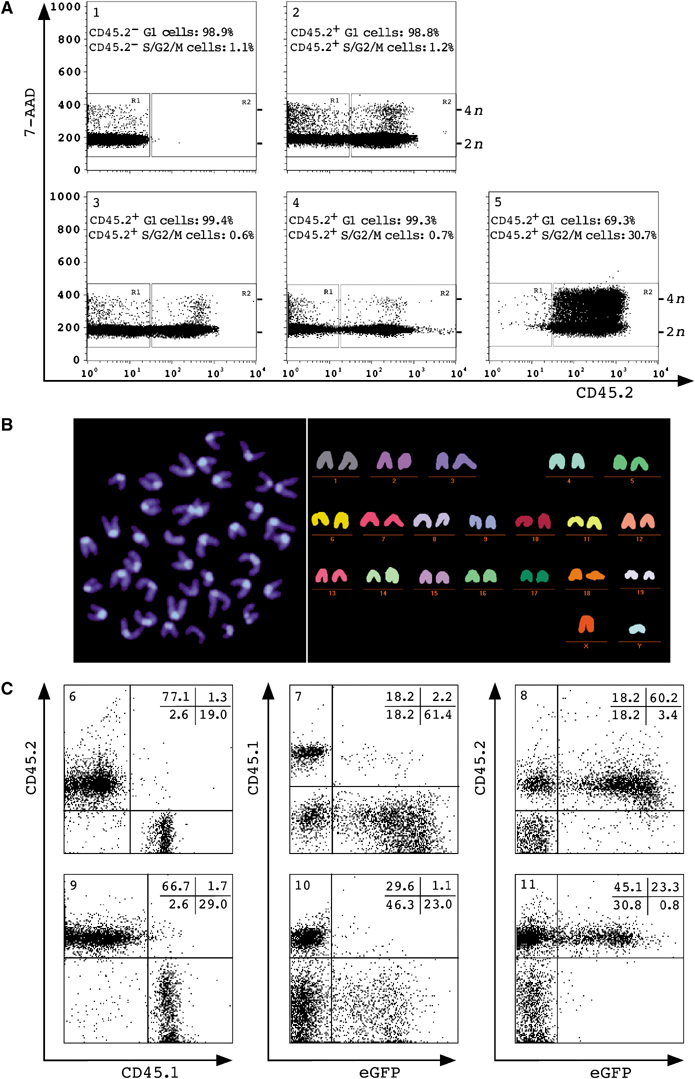
DNA content and M-FISH analysis of TSA/AzaC eGFP/bcl2 neurosphere-derived haematopoietic cells. (A) Splenocytes of a nontransplanted CD45.1 animal (panel 1), of CD45.1 recipients of CD45.2 bone marrow cells (panel 2), of treated bulk (panel 3) or clone 1 CD45.2 neurosphere cells (panel 4) and splenocytes of a nontransplanted CD45.2 control animal grown in vitro (panel 5) were stained with anti-CD45.2 antibody and 7-AAD. Panels 1–4 show the analysis of freshly isolated splenocytes; cells in panel 5 were ConA-stimulated. Position of recipient (R1), donor cells (R2), 2n, 4n cells and % of cells with 2n (G1) or >2n (S/G2/M) are shown. (B) DAPI image (left) and multicolour karyogram of a representative donor splenocyte (right) (n=20). Sorted CD45.2+ splenocytes of a clone 2 recipient (6 months post-transplant) were grown in vitro and analysed. (C) Allele-specific FACS analysis of peripheral blood cells of a transplant recipient of bulk TSA/AzaC neurosphere cells (panels 6–8) or splenocytes of a TSA/AzaC clone 1 recipient (panels 9–11). Shown are cells stained with anti-CD45.1 and anti-CD45.2 antibody (panels 6 and 9) and with either anti-CD45.1 (panels 7 and 10) or anti-CD45.2 (panels 8 and 11) antibody and plotted against eGFP. Animals shown were analysed 3–5 months after transplantation.
Discussion
Our studies show that neurosphere cells treated with agents that induce epigenetic modifications generate haematopoietic cells after transplantation into irradiated adult recipients. The neurosphere-derived haematopoietic repopulation of adult recipients is long-term, multilineage and transplantable. Furthermore, neurosphere-derived haematopoietic cells have a 2n donor karyotype, so the induced neural-to-haematopoietic fate change is unlikely to be the result of cell fusion. Our data imply that altering the epigenetic state of neurosphere cells followed by transplantation enables neural-to-haematopoietic fate change.
How fixed is the differentiation state of somatic cells?
Developmental processes are generally regarded as being unidirectional, but several lines of evidence indicate that the commitment and differentiation states of somatic cells are not invariantly fixed. Reprogramming of developmental stage-specific transcription can be achieved by transplanting cells to heterochronic microenvironments. We previously observed the reactivation of embryonic-type gene expression upon transplanting HSCs from adult donors into embryonic microenvironments, and, conversely, that embryonic haematopoietic progenitors express adult-type genes when transplanted to adult environments (Geiger et al, 1998). Gene expression of differentiated cells can also be reprogrammed by heterokaryon formation. Following muscle cell fusion, a range of muscle functions was induced in fibroblasts, while non-muscle genes were repressed (Blau et al, 1985). In addition, functional reprogramming of mature fibroblasts into cells with lymphoid character was observed upon incubating them with nuclear or cytoplasmic extracts of T cells (Hakelien et al, 2002). Reprogramming was to some extent heritable and is believed to depend on loss of nuclear regulatory proteins from recipient nuclei and their replacement by factors from heterologous cellular environments. Finally, the successful cloning of mammals by nuclear transplantation proves that nuclei of differentiated cells can be reprogrammed to a zygotic state when transplanted into an oocyte (Wilmut et al, 1997). Studies on the frequency of successful cloning using embryonic stem (ES) cells and terminally differentiated cells indicate that cloning is more effective using stem cells rather than differentiated cells as nuclear donors, suggesting that the nucleus of a stem cell is easier to reprogramme (Liu, 2001). Thus, the differentiation state of a somatic cell is not inalterably fixed and can be modified by experimental manipulations.
Studies on the influence of the epigenetic state on reprogramming demonstrated that modification of the methylation status can induce cell fate changes. Brief exposure of embryonic fibroblasts to AzaC converts fibroblasts to myoblasts and to a lesser extent to adipocytes and chondrocytes (Lassar et al, 1986). Further studies into the molecular mechanism of the AzaC-induced myogenic differentiation of fibroblasts led to the identification of the transcription factor MyoD1 as a key regulator of myogenic determination (Thayer et al, 1989). These results showed that epigenetic modification leads to the conversion of fibroblasts into myoblasts. Our present results support the notion that fate changes of somatic cells can be induced by epigenetic modification.
Neurosphere-derived haematopoietic activity
Our observation that untreated neurosphere cells are unable to generate haematopoietic cells agrees with some previous results but is inconsistent with others. Bjornson et al (1999) reported that in vitro-cultured neurosphere cells are capable of generating haematopoietic cells upon injection into irradiated recipient animals. However, we were unable to detect in transplant recipients of untreated neurospheres any haematopoietic potential that can engraft adult recipients. And similar to our results, two other reports failed to identify neurosphere-derived haematopoietic reconstitution (Morshead et al, 2002; Magrassi et al, 2003). We do observe however that injection of untreated neurosphere cells into murine blastocysts generates embryos with transient expression in donor cells of globin transgenes and the differentiation of cells that express the erythroid cell surface marker TER119 in chimeric fetal livers and peripheral blood. This indicates that neurosphere-derived donor cells in chimeric tissues display haematopoietic markers when exposed to embryonic microenvironments (Harder et al, 2004).
It should be emphasized that we employed cultured neurospheres in our analyses. Evidence exists that the potentials of cells such as primordial germ cells and oligodendrocyte precursor cells were increased by in vitro culture (Matsui et al, 1992; Kondo et al, 2000). Interestingly, cultured neurosphere cells and cultured mesenchymal stem cells (multipotent adult progenitor cells, MAPCs) were also reported to generate unrelated cell types when injected into blastocysts (Clarke et al, 2000; Jiang et al, 2002). These cells display the ability to change tissue-specific cell characteristics and they can cross germ layer borders. The multipotentiality of neurosphere cells and MAPCs is associated with in vitro culture, which was shown to result in epigenetic changes (Jost et al, 2001). It is therefore possible that freshly isolated stem cells are committed to one stem cell system and that they acquire broader lineage potentials only after extended time in culture. Thus, while our experiments give some insights into the developmental potentials of cultured neurospheres, we did not investigate whether freshly isolated neurosphere cells can contribute to the haematopoietic lineage after treatment and transplantation.
We established neurospheres from bcl2-transgenic mice because it has been shown that bcl2 expression confers protection against apoptosis-inducing challenges (Domen et al, 1998). Since both wt and bcl2-transgenic neurosphere cells engraft only when treated, it is unlikely that bcl2 transgene expression by itself confers haematopoietic activity to neurospheres. Bcl2 transgene expression increases the in vivo haematopoietic potential of treated neurosphere cells. This may be due to a reduction of apoptosis and a consequent increase of progenitor cell numbers in the injected samples.
But not all adult transplant recipients receiving neurosphere cells were engrafted. The relatively moderate engraftment frequency and the variability of the engraftment levels could indicate that engraftment was due to there being a few repopulating cells in the transplanted samples. The reason for the different engrafting activities of the three clones is unclear, but it might be due to heterogeneity between individual clones or could reflect low fate change frequencies. Altogether, we transplanted in eight independent experiments 25 animals with 0.5 × 106 bulk neurosphere cells each and 51 animals with 0.5 × 106 clonal cells each, respectively resulting in five and seven engrafted recipients. Since a single HSC is able to engraft the haematopoietic system of a transplant recipient, our available data indicate that the frequency of the haematopoietic system engrafting cells in the injected TSA/AzaC neurosphere cells is 1 per 2.5 × 106 in bulk cells and 1 per 3.6 × 106 clonal cells. In comparison, fresh bone marrow contains 1–10 engrafting cells per 105 cells. Thus, treated neurosphere cultures contain a considerably lower proportion of engrafting cells than fresh bone marrow.
Analysis of treated neurosphere cells in vitro did not generate haematopoietic colonies in methylcellulose cultures supplemented with haematopoietic growth factors. This may indicate that, in order to transdifferentiate, neurosphere cells require the destabilization of the epigenotype and the in vivo haematopoietic microenvironment. TSA/AzaC treatment or transplantation alone is not sufficient for neural-to-haematopoietic transdifferentiation of cultured neurosphere cells.
A series of recent papers reported that somatic stem cells can fuse with ES cells and hepatocytes and generate tetraploid cells (Terada et al, 2002; Ying et al, 2002; Vassilopoulos et al, 2003; Wang et al, 2003). This led us to test whether after transplantation TSA/AzaC neurosphere cells fuse with recipient haematopoietic cells. We found that most donor-derived haematopoietic cells originating both from bulk and clonal cultures have a 2n DNA content and that neurosphere-derived splenocytes have a 2n karyotype. Furthermore, in chimeric spleen and peripheral blood, most donor cells show normal expression of eGFP and CD45.2 (chromosomes 1 and 15) but not CD45.1 (chromosome 1). However, a small proportion of donor cells express markers both of donor and recipient cells. Thus, although we cannot rule out cell fusion completely, our data indicate that fusion is not an explanation for the haematopoietic reconstitution.
Molecular prerequisites for neurosphere-derived haematopoiesis
Interestingly, it has been demonstrated that different somatic stem cell types express distinct as well as overlapping transcriptional repertoires (Terskikh et al, 2001; Ivanova et al, 2002; Ramalho-Santos et al, 2002; D'Amour and Gage, 2003). In addition to the subsets of genes commonly expressed in all somatic stem cells, the TSA/AzaC-induced fate change of neurosphere cells might be caused by either an increase in the expression of genes already expressed or the activation of genes involved in survival and differentiation in haematopoietic microenvironments. After TSA plus AzaC treatment, some cells may show, in addition to a neural transcript repertoire, transcription of further loci. This, together with existing signalling components, could further enhance and stabilize expression of loci appropriate for the new haematopoietic microenvironment. Distinct molecular interactions provided by the new environment could then trigger the differentiation of haematopoietic cell types from treated cells.
In conclusion, our data indicate that transient incubation of cultured neurosphere cells with chromatin-modifying agents enables the generation of neurosphere-derived haematopoietic cells in vivo.
Materials and methods
Establishment of bulk and clonal neurospheres and isolation of bone marrow cells
Bulk neurospheres were established from individually isolated E 14.5 forebrains of male CD45.1 wt embryos or male CD45.2 embryos that carry eGFP and human bcl2 transgenes (Okabe et al, 1997; Domen et al, 1998) as described (Reynolds and Weiss, 1992; Kirchhof et al, 2002). To eliminate any differences between mouse strains, transgenic mice were backcrossed with C57BL/6 mice (Charles River). Cells were cultured in NSC medium consisting of neural basal medium (Life Technologies) supplemented with B27 (2% v/v) (Life Technologies), penicillin/ampicillin (100 U/ml) (Gibco), streptomycin (100 μg/ml) (Gibco), L-glutamine (2 mM) (Gibco), bFGF (20 ng/ml) (Cell Concepts) and EGF (20 ng/ml) (Cell Concepts). wt neurospheres were passaged about once per week while bcl2-transgenic neurospheres, which grow faster, were passaged every 2–3 days. Clonal cultures were established by plating dissociated neurosphere cells of a bulk eGFP/bcl2 culture individually into wells of 96-well plates by limiting dilution. A cell concentration was prepared that seeded in about 30% of the wells a single cell while the rest of the wells remained empty. The presence of a single cell per well was confirmed by microscopy. Then, the single cells were expanded. wt neurosphere cells were of passages 10–15 and bulk eGFP/bcl2 neurosphere cells were of passages 15–30; however, cloned eGFP/bcl2 neurosphere cells were established from bulk cultures after 20 passages and further expanded for 12–18 passages. For isolation of bone marrow cells, mice were killed, tibiae and femura were flushed with PBS and erythrocytes were lysed by incubation in Gey's solution.
Differentiation of neurosphere cells
Differentiation was carried out by plating neurosphere cells on slides coated with poly-ornithine. Differentiation media consisted of neural basal medium (Life Technologies) supplemented with B27 (2% v/v) (Life Technologies), penicillin (100 U/ml) (Gibco), streptomycin (100 μg/ml) (Gibco), L-glutamine (2 mM) (Gibco) and 10% murine neural stem cell differentiation supplement (Stem Cell Tech.). Differentiation proceeded for 6–8 days.
TSA and AzaC treatment of neurosphere cells
Before treatment, single-cell suspensions were prepared by enzymatic dissociation of neurospheres with Accumax (PAA Laboratories). Cells were counted and seeded at a concentration of 1 × 105 cells/ml. For treatment, 150 nM TSA (Sigma) plus 500 nM AzaC (Sigma) were added to the culture medium for 2 days, while control flasks were left untreated.
Western blot analyses
For Western blot analysis, nuclear lysates were prepared with histone isolation buffer (10 mM Tris–HCl, pH 6.5, 1% Triton X-100, 10 mM MgCl2, 250 mM sucrose, 20 mM NaHSO3) followed by protein extraction in 28.5 mM Tris–HCl, pH 7.4, 47 mM EDTA and 1% H2SO4 conc. The protein content was quantified using the quantiPro BCA assay kit (Sigma-Aldrich). Equal protein amounts were separated electrophoretically on a 12% SDS–PAA gel and transferred to nitrocellulose membranes. After blocking with 5% skim milk, membranes were probed with anti-acetyl histone H4 rabbit antiserum (recognizing acetylation on K5, 8, 12 and 16) (Upstate Biotechnology) and anti-MeCP2 polyclonal rabbit antibody (Upstate Biotechnology), washed and incubated with horseradish peroxidase-conjugated bovine anti-rabbit antibody (Santa Cruz). Membranes were developed using ECL reagents (Amersham Biosciences). After stripping of membranes, equal loading of the lanes was verified using anti-histone H4 polyclonal rabbit antibody (Upstate Biotechnology).
[3H]-thymidine incorporation into neurosphere cells
To evaluate wt and eGFP/bcl2 neurosphere cell proliferation upon TSA/AzaC treatment, 2.5 × 104 dissociated neurosphere cells were plated in 200 μl NSC medium supplemented with [3H]-thymidine (2 μCi/ml) for 16 h. At 24, 48 and 72 h postseeding, cells were harvested onto glass fibre filters, washed, dried and counted by liquid scintillation.
Transplantation of neurosphere cells into recipient mice
Female CD45.1 or CD45.2 C57Bl/6 transplant recipients (8–14 weeks old; Charles River) received sublethal gamma-irradiation with 9.5 Gy (split dose, 137Cs-Gammatron, 0.511 MeV, dose rate=0.322 Gy/min). Single-cell suspensions of untreated or treated neurosphere cells were prepared by enzymatic dissociation, then per primary recipient 5 × 105 CD45 congenic neurosphere cells or 1 × 106 CD45 congenic control bone marrow cells were injected into the tail vein. Secondary recipients received lethal irradiation (11.5 Gy, split dose) and 1 × 105 to 8 × 106 bone marrow cells from primary transplant recipients. Animals were given an antibiotic (neomycin sulphate; Applichem) in the drinking water for 2 days before and 2 weeks after irradiation. All animal experimentation was carried out according to permitted procedures (Government of Unterfranken, Würzburg, Germany).
Analysis of donor contribution
For FACS analysis, single-cell suspensions were prepared from thymus, spleen and bone marrow in ice-cold PBS/0.3% BSA. Whole blood was isolated in PBS/5 mM EDTA. Where appropriate, erythrocytes were lysed by incubation in Gey's solution. Cells were rinsed with FACS buffer (PBS supplemented with 0.4% BSA and 0.02% NaN3, pH 7.4), and, after blocking of Fc receptors (FcγRIII/II) with 2.4G2 antibody, 5 × 105 cells were added to 50 μl of FACS buffer supplemented with the appropriate primary antibodies and incubated at 4°C for 30 min. After washing, to detect biotinylated primary antibodies streptavidin-R-PE (Gibco), streptavidin-Red670 (Gibco) or streptavidin-APC (BD PharMingen) was added for incubation at 4°C for 30 min. The following antibodies were used: CD3-biotin, CD3-PE (145-2C11, BD PharMingen), CD4-biotin (RM4-5), CD8-PE (53-6.7, BD PharMingen), CD19-biotin, CD19-PE (1D3, BD PharMingen), CD45.1-PE (A20, BD PharMingen), CD45.2-biotin (104, BD PharMingen), IgDd-PE (217-170, BD PharMingen), IgMb-biotin (AF6-78, BD PharMingen), B220-PE (RA3-6B2, BD PharMingen), F4/80 (A3-1), Gr1-PerCPCy5.5 (RB6-8C5, BD PharMingen), MAC1-biotin (M1/70), MAC1-PE (M1/70, BD PharMingen). FACS analysis was performed on a Becton Dickinson four-colour FACS Calibur™. Male-specific PCR was performed on genomic DNA as described (Geiger et al, 1998).
Methylcellulose cultures and morphological analysis
Cells were seeded in 1 ml of methylcellulose (MethoCult M3134, Stem Cell Tech.) containing 30% FCS, 1% BSA, 10−4 M 2-ME and 10 ng/ml rmIL3 (PeproTech). Where indicated, cultures were supplemented with 4 U/ml rmEPO and 2% pokeweed mitogen murine-spleen cell-conditioned medium (PWM-SCCM, Stem Cell Tech.) or 2% PWM-SCCM alone. Cell suspensions were grown for 9–12 days. For morphological analysis of neurosphere-derived bone marrow cells, CD45.2+ donor cells were FACS-sorted (BD FACS Vantage SE system), Cytospin preparations were made and slides were stained with DiffQuik (Baxter).
DNA content and M-FISH analysis
Spleens were isolated, and after erythrocyte lysis with Gey's solution, single-cell suspensions were prepared. Where indicated, splenocytes were stimulated with concanavalin A (5 μg/ml, Sigma) and human IL2 (50 U/ml, Strathman Biotech) in complete RPMI 1640 medium (10% FCS, 2 mM L-glutamine, 10 mM Hepes, 100 U/ml penicillin/streptomycin) for 3–5 days. After blocking Fc receptors (FcγRIII/II) with 2.4G2 antibody, 1 × 106 splenocytes were stained with biotinylated anti-CD45.2 (104, BD PharMingen) and, for biotin detection, with streptavidin-R-PE (Gibco). Subsequently, the cellular DNA was labelled by a combination of 7-AAD (Sigma) and actinomycin D (AD, Sigma) (Schmid et al, 2000). Samples were measured with a Becton Dickinson FACS Calibur™ equipped with a Doublet Discrimination module for exclusion of cell clumps. Gated on the singlet cell population, the specific fluorescence signals were analysed with Becton Dickinson CellQuest™ software. Metaphase spreads were prepared from sorted and concanavalin A/Il2-stimulated splenocytes. For M-FISH analysis, mouse chromosome-specific painting probes were combinatorially labelled using seven different fluorochromes and hybridized as described (Jentsch et al, 2003). Images of 20 metaphase spreads were acquired using a Leica DM RXA epifluorescence microscope (Leica, Bensheim, Germany) equipped with a Sensys CCD camera (Photometrics, Tucson, AZ) and controlled by the Leica Q-FISH software (Leica Microsystems Imaging Solution, Cambridge, UK). Image processing and karyotyping were performed using the Leica MCK software.
Acknowledgments
We thank C Bonifer, R Cassada, S Petrovic, M Porsch, K Pramanik and S Wiese for stimulating discussion and comments. We thank V Hornich for technical assistance, B Schoell and H Holtgreve-Grez for help with M-FISH analysis and C Linden for flow cytometric isolation of donor cells. We are indebted for transgenic mouse lines made available by J Domen and IL Weissman (human bcl2), and M Okabe (eGFP). This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (to AMM and MZ), the Bundesministerium für Bildung und Forschung (BMBF) (to AJ) and the Novartis-Stiftung für Therapeutische Forschung (to AMM).
References
- Avots A, Harder F, Schmittwolf C, Petrovic S, Müller AM (2002) Plasticity of hematopoietic stem cells and cellular memory. Immunol Rev 187: 9–21 [DOI] [PubMed] [Google Scholar]
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bjornson CR, Rietze RL, Reynolds BA, Magli MC, Vescovi AL (1999) Turning brain into blood: a hematopoietic fate adopted by adult neural stem cells in vivo. Science 283: 534–537 [DOI] [PubMed] [Google Scholar]
- Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC, Webster C (1985) Plasticity of the differentiated state. Science 230: 758–766 [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB (1999) Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21: 103–107 [DOI] [PubMed] [Google Scholar]
- Castro RF, Jackson KA, Goodell MA, Robertson CS, Liu H, Shine HD (2002) Failure of bone marrow cells to transdifferentiate into neural cells in vivo. Science 297: 1299. [DOI] [PubMed] [Google Scholar]
- Chiurazzi P, Pomponi MG, Pietrobono R, Bakker CE, Neri G, Oostra BA (1999) Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum Mol Genet 8: 2317–2323 [DOI] [PubMed] [Google Scholar]
- Clarke DL, Johansson CB, Wilbertz J, Veress B, Nilsson E, Karlstrom H, Lendahl U, Frisen J (2000) Generalized potential of adult neural stem cells. Science 288: 1660–1663 [DOI] [PubMed] [Google Scholar]
- D'Amour KA, Gage FH (2003) Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc Natl Acad Sci USA 100: 11866–11872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N, Festenstein R (2002) Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet 18: 252–258 [DOI] [PubMed] [Google Scholar]
- Domen J, Gandy KL, Weissman IL (1998) Systemic overexpression of BCL-2 in the hematopoietic system protects transgenic mice from the consequences of lethal irradiation. Blood 91: 2272–2282 [PubMed] [Google Scholar]
- Eberharter A, Becker PB (2002) Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 3: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema H, Nakauchi H (2000) Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95: 2284–2288 [PubMed] [Google Scholar]
- Geiger H, Sick S, Bonifer C, Müller AM (1998) Globin gene expression is reprogrammed in chimeras generated by injecting adult hematopoietic stem cells into mouse blastocysts. Cell 93: 1055–1065 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Hakelien AM, Landsverk HB, Robl JM, Skalhegg BS, Collas P (2002) Reprogramming fibroblasts to express T-cell functions using cell extracts. Nat Biotechnol 20: 460–466 [DOI] [PubMed] [Google Scholar]
- Harder F, Kirchhof N, Petrovic S, Wiese S, Müller AM (2004) Erythroid-like cells from neural stem cells injected into blastocysts. Exp Hematol 32: 673–682 [DOI] [PubMed] [Google Scholar]
- Harris RG, Herzog EL, Bruscia EM, Grove JE, Van Arnam JS, Krause DS (2004) Lack of a fusion requirement for development of bone marrow-derived epithelia. Science 305: 90–93 [DOI] [PubMed] [Google Scholar]
- Hassig CA, Schreiber SL (1997) Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol 1: 300–308 [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR (2002) A stem cell molecular signature. Science 298: 601–604 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33: 245–254 [DOI] [PubMed] [Google Scholar]
- Jang YY, Collector MI, Baylin SB, Diehl AM, Sharkis SJ (2004) Hematopoietic stem cells convert into liver cells within days without fusion. Nat Cell Biol 6: 532–539 [DOI] [PubMed] [Google Scholar]
- Jentsch I, Geigl J, Klein CA, Speicher MR (2003) Seven fluorochrome mouse M-FISH for high resolution analysis of interchromosomal rearrangements. Cytogenet Genome Res 103: 84–88 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM (2002) Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418: 41–49 [DOI] [PubMed] [Google Scholar]
- Johnstone RW (2002) Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 1: 287–299 [DOI] [PubMed] [Google Scholar]
- Jost JP, Oakeley EJ, Zhu B, Benjamin D, Thiry S, Siegmann M, Jost YC (2001) 5-Methylcytosine DNA glycosylase participates in the genome-wide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic Acids Res 29: 4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M (2001) Inhibitors of histone deacetylase as new anticancer agents. Curr Med Chem 8: 1505–1511 [DOI] [PubMed] [Google Scholar]
- Kirchhof N, Harder F, Petrovic S, Kreutzfeldt S, Schmittwolf C, Dürr M, Mühl B, Merkel A, Müller AM (2002) Developmental potential of hematopoietic and neural stem cells: unique or all the same? Cells Tissues Organs 171: 77–89 [DOI] [PubMed] [Google Scholar]
- Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL (2000) Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature 407: 383–386 [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M (2000) Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science 289: 1754–1757 [DOI] [PubMed] [Google Scholar]
- Koyama Y, Adachi M, Sekiya M, Takekawa M, Imai K (2000) Histone deacetylase inhibitors suppress IL-2-mediated gene expression prior to induction of apoptosis. Blood 96: 1490–1495 [PubMed] [Google Scholar]
- Lassar AB, Paterson BM, Weintraub H (1986) Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 47: 649–656 [DOI] [PubMed] [Google Scholar]
- Liu L (2001) Cloning efficiency and differentiation. Nat Biotechnol 19: 406. [DOI] [PubMed] [Google Scholar]
- Magrassi L, Castello S, Ciardelli L, Podesta M, Gasparoni A, Conti L, Pezzotta S, Frassoni F, Cattaneo E (2003) Freshly dissociated fetal neural stem/progenitor cells do not turn into blood. Mol Cell Neurosci 22: 179–187 [DOI] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BL (1992) Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70: 841–847 [DOI] [PubMed] [Google Scholar]
- McCaffrey PG, Newsome DA, Fibach E, Yoshida M, Su MS-S (1997) Induction of gamma-globin by histone deacetylase inhibitors. Blood 90: 2075–2083 [PubMed] [Google Scholar]
- Milhem M, Mahmud N, Lavelle D, Araki H, Desimone J, Saunthararajah Y, Hoffman R (2004) Modification of hematopoietic stem cell fate by 5aza 2′deoxycytidine and trichostatin A. Blood 103: 4102–4110 [DOI] [PubMed] [Google Scholar]
- Morshead CM, Benveniste P, Iscove NN, van der Kooy D (2002) Hematopoietic competence is a rare property of neural stem cells that may depend on genetic and epigenetic alterations. Nat Med 8: 268–273 [DOI] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y (1997) ‘Green mice' as a source of ubiquitous green cells. FEBS Lett 407: 313–319 [DOI] [PubMed] [Google Scholar]
- Pietrobono R, Pomponi MG, Tabolacci E, Oostra B, Chiurazzi P, Neri G (2002) Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic Acids Res 30: 3278–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA (2002) ‘Stemness': transcriptional profiling of embryonic and adult stem cells. Science 298: 597–600 [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S (1992) Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255: 1707–1710 [DOI] [PubMed] [Google Scholar]
- Schmid I, Cole SW, Zack JA, Giorgi JV (2000) Measurement of lymphocyte subset proliferation by three-color immunofluorescence and DNA flow cytometry. J Immunol Methods 235: 121–131 [DOI] [PubMed] [Google Scholar]
- Schmutte C, Jones PA (1998) Involvement of DNA methylation in human carcinogenesis. Biol Chem 379: 377–388 [DOI] [PubMed] [Google Scholar]
- Terada N, Hamazaki T, Oka M, Hoki M, Mastalerz DM, Nakano Y, Meyer EM, Morel L, Petersen BE, Scott EW (2002) Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature 416: 542–545 [DOI] [PubMed] [Google Scholar]
- Terskikh AV, Easterday MC, Li L, Hood L, Kornblum HI, Geschwind DH, Weissman IL (2001) From hematopoiesis to neuropoiesis: evidence of overlapping genetic programs. Proc Natl Acad Sci USA 98: 7934–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MJ, Tapscott SJ, Davis RL, Wright WE, Lassar AB, Weintraub H (1989) Positive autoregulation of the myogenic determination gene MyoD1. Cell 58: 241–248 [DOI] [PubMed] [Google Scholar]
- Vassilopoulos G, Wang PR, Russell DW (2003) Transplanted bone marrow regenerates liver by cell fusion. Nature 422: 901–904 [DOI] [PubMed] [Google Scholar]
- Wagers AJ, Sherwood RI, Christensen JL, Weissman IL (2002) Little evidence for developmental plasticity of adult hematopoietic stem cells. Science 297: 2256–2259 [DOI] [PubMed] [Google Scholar]
- Wagers AJ, Weissman IL (2004) Plasticity of adult stem cells. Cell 116: 639–648 [DOI] [PubMed] [Google Scholar]
- Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Olson S, Grompe M (2003) Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature 422: 897–901 [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385: 810–813 [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Nakashima K, Summers RG, Toni N, D'Amour KA, Lie DC, Gage FH (2004) Cell fusion-independent differentiation of neural stem cells to the endothelial lineage. Nature 430: 350–356 [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Evans EP, Smith AG (2002) Changing potency by spontaneous fusion. Nature 416: 545–548 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265: 17174–17179 [PubMed] [Google Scholar]


