Abstract
In eukaryotes, nuclear export of the large (60S) ribosomal subunit requires the adapter protein Nmd3p to provide the nuclear export signal. Here, we show that in yeast release of Nmd3p from 60S subunits in the cytoplasm requires the ribosomal protein Rpl10p and the G-protein, Lsg1p. Mutations in LSG1 or RPL10 blocked Nmd3-GFP shuttling into the nucleus and export of pre-60S subunits from the nucleus. Overexpression of NMD3 alleviated the export defect, indicating that the block in 60S export in lsg1 and rpl10 mutants results indirectly from failing to recycle Nmd3p. The defect in Nmd3p recycling and the block in 60S export in both lsg1 and rpl10 mutants was also suppressed by mutant Nmd3 proteins that showed reduced binding to 60S subunits in vitro. We propose that the correct loading of Rpl10p into 60S subunits is required for the release of Nmd3p from subunits by Lsg1p. These results suggest a coupling between recycling the 60S export adapter and activation of 60S subunits for translation.
Keywords: LSG1, NMD3, nuclear export, ribosome, RPL10
Introduction
In eukaryotic cells, manufacturing the large (60S) and small (40S) subunits of the ribosome is a highly dynamic and regulated process (Fatica and Tollervey, 2002; Fromont-Racine et al, 2003; Johnson, 2003; Schafer et al, 2003; Tschochner and Hurt, 2003). The subunits are assembled in the nucleolus, a nuclear subcompartment, where the primary 35S and 5S rRNA transcripts are synthesized by RNA polymerases I and III, respectively. In yeast, more than 170 different trans-acting factors are required for the complex series of endo- and exoribonucleolytic cleavages, rRNA base modifications, and RNA folding and assembly events that resolve the primary rRNA transcripts into 40S and 60S subunits (Venema and Tollervey, 1995; Kressler et al, 1999; Fromont-Racine et al, 2003). Most of these trans-acting factors are shed as the pre-60S and pre-40S subunits are released from the nucleolus into the nucleoplasm (Fatica and Tollervey, 2002; Nissan et al, 2002; Fromont-Racine et al, 2003), allowing for the reutilization of these factors within the nucleolus. Additional trans-acting factors are removed in the nucleoplasm, greatly simplifying the complement of nonribosomal proteins that remain associated with the large subunit as it is exported to the cytoplasm.
Nuclear export of the large subunit also requires recruitment of export factors including the adapter protein Nmd3p, which provides the nuclear export signal (NES) for the subunit (Ho et al, 2000b; Gadal et al, 2001b; Johnson et al, 2002; Thomas and Kutay, 2003; Trotta et al, 2003). The leucine-rich NES of Nmd3p is recognized by the export receptor Crm1p, allowing for the formation of an export complex with RanGTP and unidirectional trafficking of nascent 60S subunits to the cytoplasm. This export pathway is conserved in humans (Johnson et al, 2002; Thomas and Kutay, 2003; Trotta et al, 2003).
The recruitment of Nmd3p to the 60S subunit has been thought to depend on Rpl10p (Gadal et al, 2001b). In yeast, Rpl10p is one of the last proteins to assemble into the nascent 60S subunit (Kruiswijk et al, 1978), and is one of three proteins that exchange on 60S subunits (Zinker and Warner, 1976). In the cytoplasm, release of Nmd3p from the subunit occurs prior to subunit joining during translation initiation, as it is not found on translating polysomes (Ho and Johnson, 1999). Conversely, Rpl10p remains associated with the subunit and is required for subunit joining (Dick et al, 1997a; Eisinger et al, 1997). How Nmd3p is released from the subunit for recycling to the nucleus has not been described previously.
GTPases have been implicated in 60S subunit biogenesis in the nucleus (Fromont-Racine et al, 2003) and in the cytoplasm. Deletion of the cytoplasmic GTPase Ria1p/Efl1p leads to cytoplasmic accumulation of Tif6p, a 60S binding protein that inhibits subunit joining in vitro (Russell and Spremulli, 1979). The slow growth phenotype of an efl1 mutant can be suppressed by tif6 mutants that restore its normal cellular localization (Becam et al, 2001; Senger et al, 2001), leading to the conclusion that Efl1p/Ria1p is needed for the release of Tif6p from the subunit in the cytoplasm (Senger et al, 2001). Work in our lab has demonstrated the importance of the cytoplasmic GTPase Lsg1p in the biogenesis of the large subunit. Lsg1p associates with free 60S subunits in the cytoplasm but does not shuttle to the nucleus (Kallstrom et al, 2003). However, lsg1 mutants accumulate pre-60S subunits in the nucleolus. We proposed previously that Lsg1p, like Efl1p, is necessary for the release of a nucle(ol)ar biogenesis factor(s) from cytoplasmic subunits prior to translation initiation, conferring an indirect effect on nuclear events in 60S assembly. Here, we show that Nmd3p is the target of Lsg1p and that release of Nmd3p from 60S subunits requires both Rpl10p and Lsg1p.
Results
Dominant-negative mutations in LSG1
We previously characterized LSG1 as a cytoplasmic protein that is essential for biogenesis and nucleolar release of 60S ribosomal subunits in Saccharomyces cerevisiae (Kallstrom et al, 2003). LSG1 belongs to the YawG/YlqF family of circularly permuted GTPases (Leipe et al, 2002), including the bacterial protein YjeQ, which has been shown to have GTPase activity (Daigle et al, 2002). Although a cellular function has not been ascribed to this family of GTPases, we suggested that, in yeast, Lsg1p recycles an exported factor back to the nucleus (Kallstrom et al, 2003).
To further study Lsg1p, we made dominant-negative mutants under control of a galactose-inducible promoter. Out of 35 000 transformants, 10 dominant-negative mutants were identified that exhibited a strong galactose-dependent growth arrest (Figure 1A). These mutants were noncomplementing when expressed at wild-type levels as the sole source of Lsg1p (data not shown).
Figure 1.
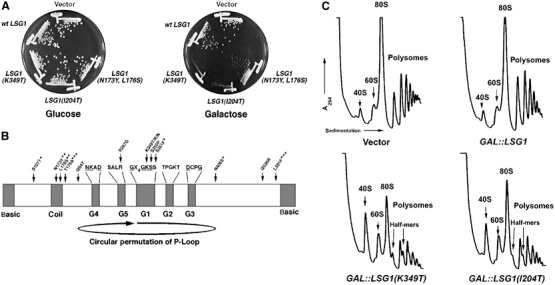
LSG1 dominant-negative mutants. (A) Growth of LSG1 dominant-negative mutants. CH1305 (wild-type) transformants containing empty vector (pRS425), pAJ879 (GAL10∷LSG1) or LSG1 dominant mutants (LSG1(K349T) (pAJ1109), LSG1(I204T) (pAJ1132) or LSG1(N173Y, L176S) (pAJ1131)) under control of the GAL10 promoter were streaked onto selective plates containing either glucose (repressing) or galactose (inducing) as the carbon source. Plates were incubated at 30°C for 5 days. (B) Diagram depicting positions of dominant-negative mutations within LSG1. Predicted functional motifs are demarcated. The designation of the putative G5 motif is tentative due to the degeneracy of G5 residues among G-proteins and varies from that published previously (Kallstrom et al, 2003). Arrowheads indicate positions of point mutations in dominant-negative mutants. Dominant-negative alleles are as follows: LSG1-20(Q536R), LSG1-5(T179A,L591F), LSG1-7(S107T,S351F,N406S), LSG1-10 (K349T), LSG1-14(S350P), LSG1-30(I204T), LSG1-41(K349N), LSG1-51(R267G), LSG1-52(N173Y,L176S) and LSG1-53(K349R). *, **, *** denote multiple mutations in LSG1-7, LSG1-52 and LSG1-5, respectively. For polysomes in (C), corresponding cultures in (A) were grown to saturation, diluted to OD600∼0.1 in fresh raffinose-containing medium and incubated to OD600∼0.3. All cultures were then induced with galactose for 4 h, treated with cycloheximide and extracts run on 7–47% linear sucrose gradients as described in Materials and methods.
The majority of the dominant-negative mutations clustered within the G1 (Walker A) motif (GX4GKS/T) (Figure 1B) that is required for coordination and catalysis of GTP (Saraste et al, 1990). Within this motif, mutations in the invariant lysine and adjacent serine residues result in dominant-negative phenotypes in heterologous GTPases (Fujimura et al, 1993; Damke et al, 2001; Park et al, 2001; Daigle et al, 2002). We identified three independent mutants, each containing different substitutions of this lysine (amino acid 349 in Lsg1p). These G1 mutations may trap the mutant LSG1 proteins in dead-end complexes. Other mutations (e.g. LSG1(N173Y,L176S)) mapping to a predicted coiled-coil region did not inhibit growth as completely as mutations in the G1 motif (Figure 1A). Several additional mutations mapped to regions of Lsg1p outside the recognized motifs and exhibited varying degrees of growth inhibition, with one mutant, LSG1(I204T), being particularly strong (Figure 1A).
LSG1 dominant-negative mutants recapitulate the 60S export defect of temperature-sensitive (ts) lsg1 mutants
We previously showed that lsg1ts mutants have reduced levels of free 60S subunits and accumulate half-mer polysomes (Kallstrom et al, 2003). Half-mers, representing mRNAs with 40S ribosomes not joined to 60S, result from either a lack of available 60S subunits or from a defect in subunit joining itself. As with the lsg1ts mutants, LSG1(K349T) and LSG1(I204T) showed reduced 60S levels and half-mer polysomes (Figure 1C).
We also monitored 60S subunit export in the LSG1 mutants using the ribosomal reporter Rpl25-eGFP (Gadal et al, 2001b). Rpl25-eGFP showed marked accumulation in the nucleus after 3 h of induction of LSG1(K349T) or LSG1(I204T) (Figure 2A), indicating that 60S export was blocked. Since both wild-type and the dominant-negative Lsg1p mutant proteins are cytoplasmic ((Kallstrom et al, 2003) and data not shown), we conclude that the dominant-negative Lsg1 proteins, like the conditional mutants, block the nuclear recycling of a biogenesis factor needed for subunit export, resulting in entrapment of Rpl25-eGFP-containing subunits in the nucleus.
Figure 2.
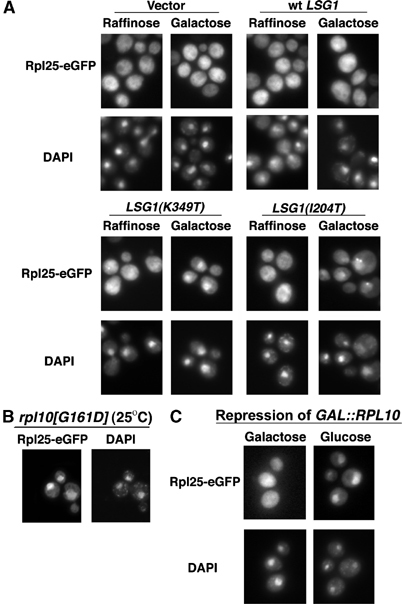
Nuclear accumulation of Rpl25-eGFP in lsg1 and rpl10 mutants. (A) Rpl25-eGFP (pAJ908) was continuously expressed in wild-type cells (CH1305) carrying empty vector (pRS425), pAJ879 (GAL10∷LSG1), pAJ1109 (GAL10∷LSG1[K349T]) or pAJ1132 (GAL10∷LSG1[I204T]). Localization after growth in raffinose was compared to that after a 3 h induction with galactose. Rpl25-eGFP (pAJ907) was visualized in the rpl10[G161D] mutant (AJY1657) at semipermissive temperature (B) and in GAL∷RPL10 cells (DEH221+) with or without 4 h of repression by glucose (C). Cells were prepared for visualization as described in Materials and methods.
Nuclear export of 60S subunits is also blocked in the conditional rpl10(G161D) mutant and by repression of RPL10
RPL10 was identified in a screen for conditional mutants that blocked nuclear export of 60S subunits (Gadal et al, 2001b). The rpl10(G161D)ts mutant also accumulates the 60S reporter Rpl25-eGFP in the nucleus at semipermissive temperature (Figure 2B). Similarly, repression of RPL10 transcription led to nuclear accumulation of Rpl25-eGFP (Figure 2C). Although the effects of rpl10 and lsg1 mutants on 60S export are common to a number of 60S biogenesis mutants and do not by themselves indicate related functions of Rpl10p and Lsg1p, results below show a close functional relationship between these two proteins.
Dominant-negative Lsg1p, rpl10(G161D)p or repression of RPL10 blocks nuclear recycling of the 60S export adapter, Nmd3p, from the cytoplasm
Ts nmd3 mutants accumulate Rpl25-eGFP in the nucleolus at restrictive temperature (Kallstrom et al, 2003). In addition, mutant human NMD3 that is defective for nuclear export accumulates in the nucleolus in human cells (Trotta et al, 2003). These results suggest that Nmd3p is required for the release of subunits from the nucleolus and could explain the nucleolar accumulation of subunits in lsg1 mutants if Nmd3p were the factor that failed to shuttle in these mutants. The steady-state distribution of Nmd3p is predominantly cytoplasmic (Ho and Johnson, 1999). As it is exported from the nucleus in a Crm1p-dependent manner, Nmd3p that is shuttling can be trapped in the nucleus in the presence of the Crm1 inhibitor leptomycin B (LMB) (Ho et al, 2000b; Gadal et al, 2001b; Kallstrom et al, 2003).
To test if dominant-negative LSG1 mutants block Nmd3p recycling to the nucleus, we first tried expressing Nmd3-GFP ectopically from a plasmid and assayed for shuttling by monitoring the nuclear accumulation of Nmd3-GFP in the presence of LMB. Under these conditions, dominant-negative LSG1 did not prevent Nmd3-GFP shuttling to the nucleus (data not shown and see below). We then considered the possibility that the regulation of Nmd3p shuttling depends on its stoichiometry with other factors. To address this concern, we introduced GFP at the 3′-end of the genomic NMD3 locus through homologous recombination. We also integrated the CRM1(T539C) allele for a more uniform response to LMB throughout the cell population. Genomically expressed Nmd3-GFP showed the expected nuclear accumulation when treated with LMB in the absence of expression of mutant LSG1 (Figure 3A). However, in cells expressing either Lsg1(K349T)p or Lsg1(I204T)p, genomically expressed Nmd3-GFP exhibited a dramatic cytoplasmic retention (Figure 3A) and was virtually absent from the nucleus (see arrows, Figure 3A) in the presence of LMB. Thus, when Nmd3-GFP was expressed at wild-type stoichiometry with other cellular factors, its ability to shuttle was blocked by dominant-negative Lsg1p mutants.
Figure 3.
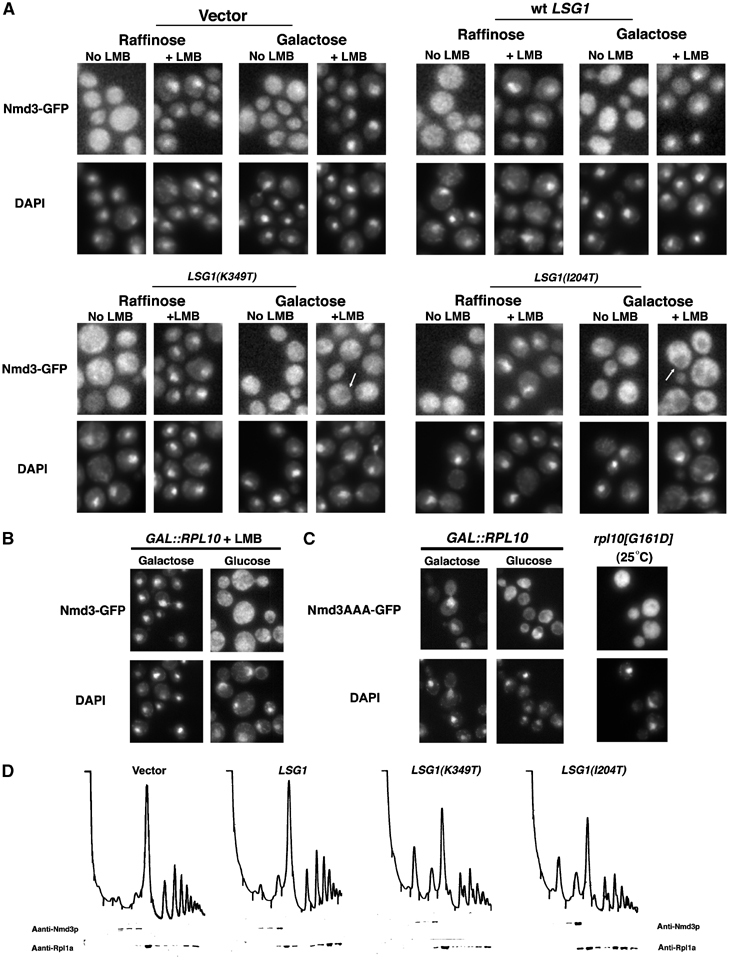
lsg1 and rpl10 mutants trap Nmd3-GFP on cytoplasmic 60S. Localization of chromosomally expressed Nmd3-GFP in the presence of LMB. (A) Nmd3-GFP was visualized in strain AJY1705 (NMD3-GFP∷KanMX6 CRM1[T539C]) carrying empty vector (pRS425), pAJ879 or pAJ1109. Cells were induced with galactose or maintained in glucose for 3 h before the addition of LMB. (B) RPL10 expression was maintained in AJY1836 (GAL1-10∷RPL10 NMD3-GFP CRM1[T539C]) in galactose or repressed in glucose before the addition of LMB and visualization of Nmd3-GFP. In (C), Nmd3AAA-GFP (pAJ754) was visualized in strain DEH221+ (GAL1-10∷RPL10) after RPL10 repression or in the rpl10[G161D] mutant AJY1657 cultured at 25°C. (D) Cells were cultured as in Figure 1C, extracts prepared and fractionated by sucrose gradient sedimentation. Western blotting of proteins in each fraction was carried out using α-Nmd3p or α-Rpl1ap.
Considering that repression of RPL10 or expression of mutant Rpl10p also led to nuclear accumulation of 60S subunits, we asked if Nmd3p was able to shuttle under these conditions as well. As with LSG1 mutants, genomically expressed Nmd3-GFP was retained in the cytoplasm when transcription of RPL10 was repressed (Figure 3B). As a second means of assaying Nmd3p shuttling in rpl10 mutants, we used an NMD3 reporter, referred to as NMD3AAA, that contains three point mutations (I493A, L497A and L500A) within its NES. This mutant is mildly dominant negative when expressed at low levels (data not shown), and the corresponding mutant in human NMD3 shows impaired nuclear export (Trotta et al, 2003). Nmd3AAA-GFP was predominantly nuclear in wild-type cells because nuclear export was rate limiting. However, Nmd3AAA-GFP was redistributed to the cytoplasm upon repression of RPL10 and in the rpl10(G161D) mutant (Figure 3C). The cytoplasmic accumulation of this reporter suggests that import has become the rate-limiting step in these mutants. The similar results obtained with Rpl10p and Lsg1p suggest that both of these proteins are needed for Nmd3p recycling to the nucleus.
A failure to recycle Nmd3p could be due to a failure in the release of Nmd3p from the subunit or due to a subsequent failure to import Nmd3p after release from the subunit. However, Western blotting of sucrose gradient fractions showed that Nmd3p cosedimented with free 60S subunits in the presence of dominant-negative Lsg1p or when RPL10 was repressed (Figure 3D and data not shown). A free pool of Nmd3p was not observed at the top of the gradients. Thus, Nmd3p that is unable to recycle to the nucleus remains associated with 60S subunits in the cytoplasm. It should be noted that, in extracts prepared from wild-type cells, no free pool of Nmd3p is observed, even though the protein must be free transiently as it shuttles into the nucleus. However, a free Nmd3p population can be observed if the protein is ectopically expressed.
Synthetic lethality between lsg1 and nmd3 mutants
Functionally related genes often show negative synergy, or synthetic lethality, when mutant alleles are combined. To test this, we crossed nmd3ts mutants to an lsg1Δ mutant containing LSG1 on an URA3 plasmid. Single and double mutants were obtained after sporulating the diploids. We then asked if wild-type LSG1 could be replaced by a conditional allele by counter selection against the wild-type allele on 5-FOA-containing media. While strains bearing a single conditional allele of either NMD3 or LSG1 grew well at permissive temperature in the presence of 5-FOA, double mutants exhibited either a pronounced growth defect (nmd3-3 lsg1-2 and nmd3-4 lsg1-2, Figure 4A) or synthetic lethality (nmd3-3 lsg1-3 and nmd3-4 lsg1-3, Figure 4A). These genetic interactions are consistent with a close functional relationship between NMD3 and LSG1.
Figure 4.
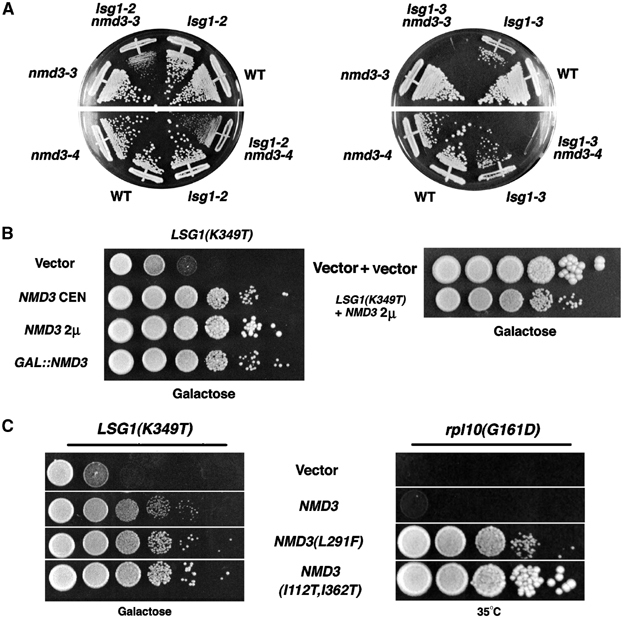
Genetic interactions among LSG1, NMD3 and RPL10. (A) lsg1 and nmd3 mutants exhibit synthetic lethality. AJY1513 (nmd3–3), AJY1518 (nmd3–4), AJY1511 (lsg1∷KanMX4/pAJ626[LSG1 URA3]), AJY1512 (lsg1∷KanMX4nmd3–3/pAJ626[LSG1 URA3]), AJY1521 (lsg1∷KanMX4nmd3–4/pAJ626[LSG1 URA3]) and a congenic wild-type strain were transformed with lsg1-2 and lsg1-3 mutant alleles on LEU2 plasmids. Transformants were restreaked onto 5-FOA plates to exclude the wild-type LSG1∷URA3 plasmids. Plates were incubated at 25°C for 10 days. (B) Growth suppression of a dominant-negative LSG1 allele by high-copy NMD3. In all, 10-fold serial dilutions of stationary-phase cultures were spotted onto selective plates containing galactose and incubated for 5 days at 30°C. The strains tested were: wild-type (CH1305) transformed with pAJ1109 (GAL10∷LSG1[K349T]) in combination with empty vector (pRS416), pAJ409 (NMD3, CEN), pAJ363 (NMD3, 2μ) or pAJ1143 (GAL1∷NMD3,CEN). The growth of wild-type (CH1305), containing empty vectors, and LSG1(K349T), suppressed with high-copy NMD3, is shown for comparison. (C) Growth suppression of dominant-negative LSG1 and rpl10ts mutants by NMD3 suppressor alleles. Cultures of either CH1305 containing pAJ1278 (GAL10∷LSG1[K349T]) or AJY1657 (rpl10[G161D]), each also containing empty vector (pRS415), pAJ538 (NMD3-myc), pAJ415 (NMD3[L291F]-myc) or pAJ1315 (NMD3[I112T, I362T]-myc), were diluted and plated as for (B). The CH1305 plate was incubated 5 days at 30°C and the AJY1657 plate for 3 days at 35°C.
Common suppression of LSG1(K349T) and rpl10(G161D) by NMD3
NMD3 is a high-copy suppressor of the conditional rpl10 mutant (rpl10[F85S]) (Zuk et al, 1999). Independently, we found that high-copy NMD3 suppressed LSG1(K349T) (Figure 4B). The degree of suppression increased with copy number up to a point (Figure 4B, compare NMD3 on CEN versus 2μ vectors) but was reduced at very high levels of Nmd3p expression (galactose induction). Note that suppression by high-copy NMD3 was partial and did not fully restore growth to wild-type levels (Figure 4B). High-copy NMD3 also suppressed lsg1ts mutants but not mutants of NOG1, a nuclear GTPase required for 60S biogenesis (Kallstrom et al, 2003), indicating that suppression was specific (data not shown).
The ability of high-copy NMD3 to suppress both rpl10 and lsg1 mutants hinted at the possibility that Rpl10p and Lsg1p have a common effect on Nmd3p function. To explore this idea in more detail, we looked for allele specificity in these interactions. Three dominant alleles of NMD3, including NMD3(L291F), have been described previously as extragenic suppressors of rpl10(G161D) (Karl et al, 1999). We found that, in comparison to wild-type NMD3, low-copy NMD3(L291F) also modestly suppressed LSG1(K349T) (Figure 4C). We then mutagenized NMD3 to find additional mutants that could suppress rpl10(G161D) and tested these for suppression of the lsg1 mutants. One mutant in particular, NMD3(I112T, I362T), showed robust suppression of both rpl10G161D and LSG1(K349T) (Figure 4C). Thus, both rpl10 and lsg1 mutants can be suppressed by the same NMD3 mutations.
High-copy NMD3 or mutant NMD3 bypasses the block in shuttling in lsg1 and rpl10 mutants
In the work above, we showed that genomically expressed Nmd3-GFP was retained on 60S subunits in the cytoplasm by lsg1 or rpl10 mutants. However, high-copy NMD3 suppressed the growth defects of these mutants. If high-copy NMD3 bypasses the block in nuclear recycling in these mutants, we should be able to observe its accumulation in the nucleus in an LMB-dependent manner. Indeed, the addition of LMB led to the nuclear accumulation of genomically expressed Nmd3-GFP when untagged Nmd3p was expressed ectopically (Figure 5A). Similar results were obtained in RPL10-repressed cells (data not shown). Thus, high-copy NMD3 over-rides defects in both RPL10 and LSG1, indicating that free Nmd3p can recycle to the nucleus. In previous work, we reported that Nmd3p appeared not to be the target of Lsg1p because we did not observe cytoplasmic retention of Nmd3p in lsg1 mutants (Kallstrom et al, 2003). We can now ascribe our initial failure to identify Nmd3p as the target of Lsg1p to the fact that Nmd3p shuttling is highly dependent on its relative abundance.
Figure 5.
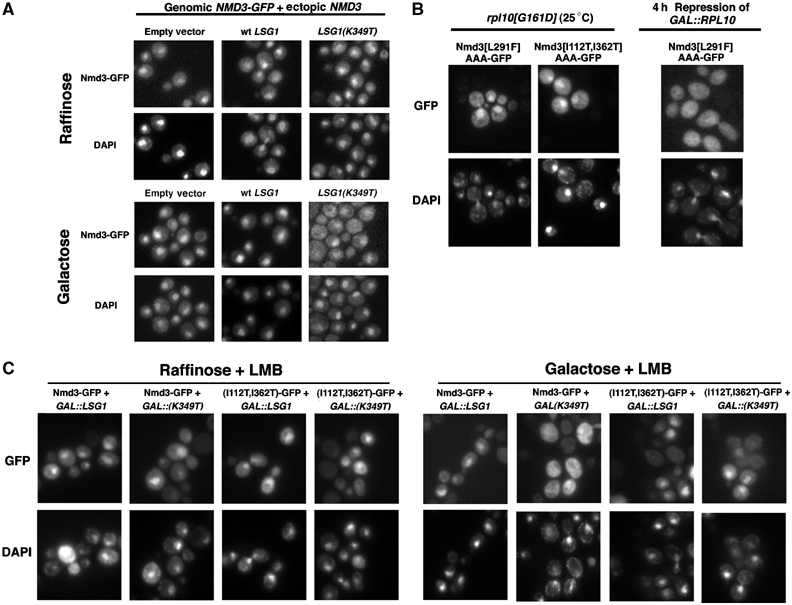
Ectopic expression of wild-type NMD3 and introduction of suppressor mutations allows Nmd3-GFP to recycle in the presence of lsg1 and rpl10 mutants. (A) Cultures of strain AJY1705 (NMD3-GFP CRM1[T539C]) containing pAJ363 (NMD3, 2μ) and either empty vector (pRS425), pAJ879 (GAL10∷LSG1) or pAJ1109 (GAL10∷LSG1[K349T]) were maintained in raffinose or galactose was added to induce the LSG1 alleles. Cells were then treated with LMB, fixed, DAPI stained and visualized as described in Materials and methods. For (B), AJY1657 (rpl10[G161D]) containing either pAJ1069 (NMD3[L291F]AAA-GFP) or pAJ1288 (NMD3[I112T, I362T]AAA-GFP) was grown constitutively at 25°C and treated as in (A). DEH221+ containing pAJ1069 (NMD3[L291F]AAA-GFP) was cultured and prepared for microscopy as described for Figure 3C. (C) Overnight cultures of strain AJY1896 (nmd3∷TRP1 CRM1[T539C]) containing pAJ582 (NMD3-GFP) or pAJ1287 (NMD3[I112T, I362T]-GFP) as the sole copies of NMD3 and either empty vector (pRS426), pAJ1312 (GAL10∷LSG1) or pAJ1278 (GAL10∷LSG1[K349T]) in medium containing raffinose were diluted two-fold in fresh medium in the presence of either raffinose or galactose to induce LSG1 expression. After 3 h, cells were fixed and treated for visualization as in (A).
Considering that high-copy NMD3 bypassed the block in shuttling, we reasoned that the NMD3 mutants might also show enhanced recycling to the nucleus. To examine this in an rpl10(G161D) mutant, we introduced the NMD3(L291F) or NMD3(I112T, I362T) mutations into the Nmd3AAA-GFP. In contrast to Nmd3AAA-GFP, which was cytoplasmic in rpl10(G161D), the introduction of the L291F or I112T, I362T mutations allowed the protein to be relocalized to the nucleus (Figure 5B). The NMD3(L291F) and NMD3(I112T, I362T) mutations on their own had no obvious detrimental effects when expressed in wild-type cells and did not block nuclear export of 60S subunits (data not shown). Additionally, these mutant proteins were expressed at levels identical to wild-type Nmd3p, eliminating the possibility that they act as high-copy suppressors (data not shown). Thus, the redistribution of these proteins to the nucleus appears to be due to increased nuclear re-entry rather than an increased block in export (see Discussion).
As these mutations also suppressed dominant LSG1 mutants, we assayed their effects in LSG1(K349T)-expressing cells as well. For this, we introduced plasmid-borne Nmd3-GFP or Nmd3(I112T, I362T)-GFP as the sole copy of NMD3 in a LMB-sensitive strain deleted for NMD3. These strains exhibited the expected nuclear accumulation of both Nmd3-GFP alleles upon treatment of cells with LMB in the absence of LSG1(K349T) expression (Figure 5C, raffinose). However, the expression of LSG1(K349T) but not wild-type LSG1 resulted in a marked absence of nuclear accumulation for wild-type Nmd3-GFP following LMB treatment (Figure 5C, galactose) similar to that observed for the strain possessing genomically integrated NMD3-GFP (Figure 3A). In contrast, cells possessing the Nmd3(I112T, I362T)-GFP suppressor allele were not nearly as sensitive to the expression of LSG1(K349T), exhibiting a persistent nuclear accumulation following LMB treatment. These results support the idea that the suppressor mutations enhance recycling to the nucleus.
The Nmd3(L291F) and Nmd3(I112T, I362T) mutant proteins have weakened affinity for 60S subunits
The fact that the mutant proteins but not wild-type Nmd3p were able to accumulate in the nucleus in rpl10 and lsg1 mutants suggested that the mechanism of suppression was by facilitating release of Nmd3p from cytoplasmic 60S subunits. Enhanced release could be due to bypass of the dependence of Nmd3p release on Lsg1p, possibly by lower affinity of mutant Nmd3p for the 60S subunit. To test this idea, we adapted a native gel assay (Dahlberg and Grabowski, 1990) to measure qualitatively the affinity of Nmd3p for 60S subunits in vitro. Wild-type and mutant Nmd3 proteins were purified as GST fusions from yeast, and wild-type 60S subunits were prepared by dissociating 80S ribosomes. 60S subunits were then incubated with the different Nmd3 proteins under conditions to promote binding (Ho et al, 2000a), and complexes were electrophoresed on native agarose/polyacrylamide gels. The migration of 60S subunits was determined by ethidium bromide staining rRNA, and the position of Nmd3p was monitored by Western blotting. In this gel system, 60S subunits migrate as a discrete species (Figure 6A), whereas free Nmd3p migrates as a diffuse band with lower relative mobility. However, when preincubated with 60S subunits, wild-type Nmd3p comigrated with 60S subunits (Figure 6A, lanes 4 and 5). On the other hand, when increasing amounts of Nmd3(I112T, I362T)p were incubated with wild-type 60S subunits, little binding was observed (Figure 6A, lanes 8 and 9). Nmd3(L291F)p, which was a weaker suppressor of both rpl10 and lsg1 mutants, also bound free 60S subunits, but approximately two to three times more mutant protein was required to achieve levels of binding similar to that of wild-type Nmd3p (Figure 6A, lanes 13 and 14). As a negative control, Nmd3(V340D)p showed no detectable binding to 60S subunits (Figure 6A, lanes 17 and 18). This mutant, identified in a screen for nmd3 null mutants (Chen and Johnson, unpublished), does not support growth and binds only weakly to 60S subunits as measured by co-immunoprecipitation (data not shown and Figure 6B).
Figure 6.
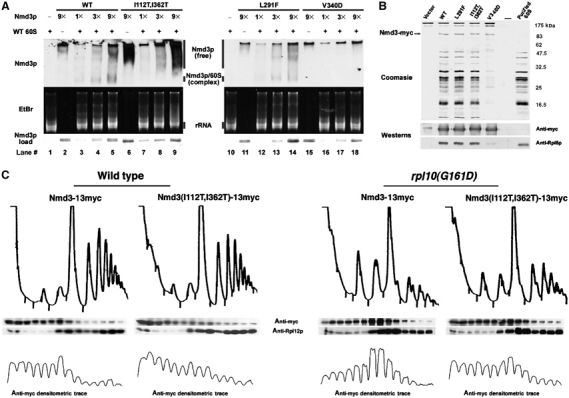
Nmd3 suppressor proteins have lower affinity for 60S subunits. (A) Three-fold increasing amounts (1 × ∼30 ng) of affinity-purified GST-Nmd3p wild-type, [I112T, I362T] suppressor, [L291F] suppressor or [V340D] loss of function mutant proteins were incubated alone or with purified 60S subunits and run on 2.5% acrylamide/0.5% agarose composite gels as described in Materials and methods. The position of Nmd3p and 60S subunits was determined by Western blotting using anti-GST and ethidium bromide staining, respectively. (B) Cultures of strain W303 containing empty vector (pRS425), pAJ538 (NMD3-myc), pAJ1315 (NMD3[I112T, I362T]-myc), pAJ1070 (NMD3[L291F]-myc) or pAJ1299 (nmd3[V340D]-myc) were collected and α-myc immunoprecipitations performed as described in Materials and methods. Samples and a purified 60S control were run on a 12% SDS–PAGE gel and analyzed by Western blotting using α-myc (Nmd3p) or α-Rpl8p. The gel was stained with Coomassie Blue. (C) Cultures of strains W303 or AJY1657 transformed with either pAJ538 (NMD3-myc) or pAJ1315 (NMD3[I112T, I362T]-myc) were grown constitutively at 30°C (W303) or shifted to 37°C for 3 h (AJY1657) before harvesting. Extracts were prepared in the presence of cycloheximide followed by analysis on sucrose gradients as described in Materials and methods. Sucrose gradient fractions were analyzed by Western blotting using α-myc (Nmd3p) or α-Rpl12p. Densitometric traces of α-myc blots were conducted using NIH Image V.1.62.
As Nmd3(I112T, I362T)p did not appreciably bind purified subunits in this reconstituted system, we tested if this mutant could bind subunits under more in vivo-like conditions. Wild-type and cmyc-tagged mutant Nmd3p proteins were expressed in vivo, immunoprecipitated and assayed for copurification of 60S subunits (Figure 6B). Indeed, Nmd3(I112T, I362T)p co-immunoprecipitated 60S subunits, but not as efficiently as did Nmd3(L291F)p and wild-type Nmd3p. Only trace amounts of 60S were associated with Nmd3(V340D)p (Figure 6B). We also analyzed the association of Nmd3(I112T, I362T)p with wild-type and rpl10(G161D) mutant subunits by sucrose gradient sedimentation. Nmd3(I112T, I362T)p showed a bias in distribution to the top of the gradient, indicating a larger pool of free protein in both wild-type and rpl10(G161D) extracts (Figure 6C). However, in rpl10(G161D) extracts, a greater proportion of wild-type Nmd3p cosedimented with free 60S subunits, consistent with our fluorescence analysis that this rpl10 mutant traps Nmd3p on cytoplasmic subunits. Interestingly, the distribution of Nmd3(I112T, I362T)p in the rpl10(G161D) mutant was similar to that of wild-type Nmd3p in wild-type cells, suggesting that this mutant restores the function of Nmd3p in the context of rpl10(G161D). These results show that Nmd3p suppressors have reduced affinity for 60S subunits and provide physical evidence that enhancing the release of Nmd3p from subunits can bypass the defect of rpl10 and lsg1 mutants.
High-copy Nmd3p alleviates the nuclear accumulation of Rpl25-eGFP caused by dominant-negative Lsg1p mutants or by repression of RPL10
If high-copy Nmd3p suppresses dominant-negative LSG1 mutants by restoring export of 60S subunits, the steady-state distribution of Rpl25-eGFP should be shifted back to the cytoplasm in the presence of elevated Nmd3p levels. Rpl25-eGFP localization was monitored in cells ectopically expressing NMD3 from a 2μ (high-copy) vector following induction of LSG1(K349T). Indeed, the localization of Rpl25-eGFP in these cells showed a pronounced redistribution to the cytoplasm dependent on the increased copy number of Nmd3p (Figure 7A, compare K349T/NMD3 to K349T/vector). The bulk shift of Rpl25-eGFP from the nucle(ol)us to the cytoplasm in cells coexpressing dominant Lsg1p alleles and high-copy NMD3 suggests that Nmd3p was rate limiting for 60S export and is consistent with the idea that Nmd3p is the primary target of Lsg1p.
Figure 7.
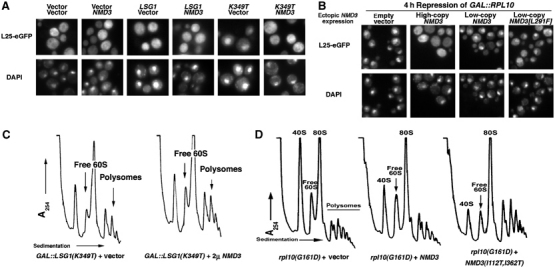
NMD3 suppresses ribosome biogenesis/export defects in lsg1 and rpl10 mutants. (A) Wild-type strain CH1305 was cotransformed with the various combinations of vectors: two empty vectors (pRS425) and (pRS426); pRS425 and pAJ363 (NMD3); pAJ879 (GAL10∷LSG1) and pRS426; pAJ879 and pAJ363; pAJ1109 (GAL10∷LSG1[K349T]) and pRS426; or pAJ1109 and pAJ363. Cells were grown in the presence of galactose for 3 h and prepared for microscopy as described in Materials and methods. (B) DEH221+ (GAL1-10∷RPL10) cells containing pASZ11-RPL25-eGFP (RPL25-eGFP) with either pRS315 (empty vector), pAJ410 (NMD3 2μ), pAJ123 (NMD3 CEN) or pAJ415 (NMD3[L291F] CEN) were grown in galactose-containing medium and diluted four-fold into glucose-containing medium to repress RPL10 expression. After 4 h, cells were prepared for visualization as above. Similar results were obtained with NMD3(I112T, I362T) (data not shown). (C) Cultures of AJY1657 (rpl10[G161D]) cells carrying empty vector (pRS425), pAJ538 (NMD3-myc) or pAJ1315 (NMD3[I112T, I362T]-myc) were shifted to 37°C for 3 h before harvesting. Extracts were prepared in the presence of cycloheximide followed by analysis on sucrose gradients as described in Materials and methods. (D) CH1305 cells carrying pAJ1109 (GAL10∷LSG1[K349T]) with either empty vector (pRS426) or high-copy pAJ363 (NMD3) were cultured in raffinose media followed by induction with galactose. After 4 h, cells were collected, extracts prepared and gradients run as in (C). Arrows indicate increased 60S levels and polysomes in cells coexpressing Lsg1p dominant-negative alleles and high-copy Nmd3p.
Although high-copy NMD3 cannot rescue the inviability due to loss of Rpl10p, it was possible that overexpression of Nmd3p could suppress specific defects, such as the block in nuclear export of 60S subunits, that result from loss of Rpl10p. We therefore determined if the overexpression of Nmd3p could bypass the nuclear localization of Rpl25-eGFP in RPL10-repressed cells as observed in lsg1 mutant cells. Rpl25-eGFP localization was monitored 4 h after repression of transcription of RPL10. Remarkably, high-copy expression of Nmd3p led to relocalization of Rpl25-eGFP to the cytoplasm (Figure 7B), indicating that Nmd3p can indeed bypass the requirement for Rpl10p in nuclear export of 60S. On the other hand, low-copy expression of wild-type NMD3 or NMD3(L291F), which strongly suppresses rpl10(G161D), did not support redistribution of Rpl25-eGFP to the cytoplasm (Figure 7B). The lack of suppression from low-copy NMD3 suggests that, in the absence of Rpl10p, there is a more severe block in recycling Nmd3p than in the rpl10(G161D) mutant or in cells expressing dominant-negative LSG1. These results are consistent with our suggestion that NMD3(L291F) facilitates release of Nmd3p from 60S subunits in the cytoplasm but does not directly enhance 60S subunit export.
Suppression by NMD3 partially restores 60S levels in rpl10 and lsg1 mutants
In order to assess whether suppression by Nmd3p correlated with restored 60S biogenesis, we analyzed polysome profiles in an rpl10(G161D) mutant suppressed by NMD3(I112T, I362T) or in strains coexpressing high-copy NMD3 and LSG1(K349T). In the case of rpl10(G161D), expression of NMD3(I112T, I362T) led to a dramatic improvement in the ratio of free 60S to 40S subunits along with a significant restoration of polysomes (Figure 7C) and restoration of the 60S export defect of rpl10(G161D) cells (data not shown). High-copy NMD3 also led to an improved subunit ratio in LSG1(K349T)p-expressing cells (Figure 7D) and a modest increase in polysomes (see arrows, Figure 7D). Similar results were seen with NMD3(I112T, I362T) (data not shown). Thus, high-copy Nmd3p enhances translation in lsg1 and rpl10 mutants by increasing the export of 60S subunits out of the nucleus (Figure 7A). Although the block in 60S export due to dominant-negative Lsg1p was efficiently alleviated by increasing Nmd3p expression, polysome profiles were only modestly restored, and the free subunit pools remained relatively high. This is likely due to the persistence of dominant Lsg1p and/or Nmd3p on 60S subunits, thereby blocking subunit joining.
Discussion
In eukaryotic cells, nascent ribosomal subunits emerging from the nucleus are not immediately incorporated into translating ribosomes (Warner, 1971). This lag in the utilization of subunits has been taken as evidence for cytoplasmic maturation of the subunit. The nascent 60S subunit is accompanied to the cytoplasm by several trans-acting proteins that recycle to the nucleus (Kressler et al, 1999; Nissan et al, 2002; Fromont-Racine et al, 2003; Tschochner and Hurt, 2003). Among these is the export adapter Nmd3p that provides the NES for the subunit (Ho et al, 2000b; Gadal et al, 2001b). The function of Nmd3p in subunit export is conserved from yeast to humans (Thomas and Kutay, 2003; Trotta et al, 2003); however, Nmd3p orthologs are found in archaeal organisms as well. As Archaeons lack nuclei, Nmd3p must have a function that predates the evolution of the nuclear envelope. It is intriguing that in many archaea, Nmd3p is fused to an eIF5A-like domain (Aravind and Koonin, 2000). Although the function of eIF5A is not well understood, in vitro eIF5A stimulates the formation of the first peptide bond during translation (Benne and Hershey, 1978). Thus, the physical association of Nmd3p with an eIF5A-like domain in archaea suggests that Nmd3p function is coupled to translation.
The molecular events that regulate Nmd3p association with the 60S subunit have been poorly understood. Here, we have shown that release of Nmd3p from subunits in the cytoplasm is blocked by mutations in Lsg1p, a cytoplasmic 60S-associated G-protein, as well as by mutations in the 60S subunit protein Rpl10p. Although Lsg1p and Rpl10p have not previously been shown to be functionally related, mutations in both are suppressed by high-copy NMD3 and by the same dominant NMD3 mutants, suggesting that the effects of lsg1 and rpl10 mutants on NMD3 are related. These dominant Nmd3 proteins display weaker binding to 60S subunits in vitro and recycle to the nucleus more efficiently in vivo, indicating that suppression of rpl10 and lsg1 mutants is the result of enhancing release of Nmd3p that is trapped in the cytoplasm on free 60S subunits. Although we have not yet been successful in showing GTPase activity for Lsg1p in vitro, mutations predicted to be necessary for GTP binding and hydrolysis are lethal and inhibit Nmd3p cycling, suggesting that Lsg1p GTPase activity is necessary for Nmd3p release.
Figure 8A depicts our model for the function of Rpl10p and Lsg1p in recycling Nmd3p. Nmd3p loads onto pre-60S particles in the nucleolus and provides the NES to direct their export to the cytoplasm. Initial binding in the nucleolus is based on analysis of human NMD3 (Trotta et al, 2003) and the defect in pre-60S release from the nucleolus seen in nmd3 and lsg1 mutants. Although it has been suggested that Rpl10p recruits Nmd3p to nuclear pre-60S subunits, our results suggest that Rpl10p is not directly required for 60S export (see below). Nevertheless, once in the cytoplasm, release of Nmd3p requires the presence of functional Rpl10p in the 60S subunit. Lsg1p, which is restricted to the cytoplasm, binds nascent subunits after they emerge from the nucleus. By analogy to other GTPases, Lsg1p likely binds to the 60S subunit initially in a GTP-bound form. In response to some molecular cue, possibly the correct loading of Rpl10p into the subunit, the GTPase activity of Lsg1p is triggered, leading to a structural rearrangement of the subunit and the dissociation of Nmd3p and Lsg1p prior to translation initiation. In this way, Lsg1p could serve a proofreading function in assessing the structural integrity of the subunit, allowing only properly assembled subunits to enter the translationally active pool.
Figure 8.
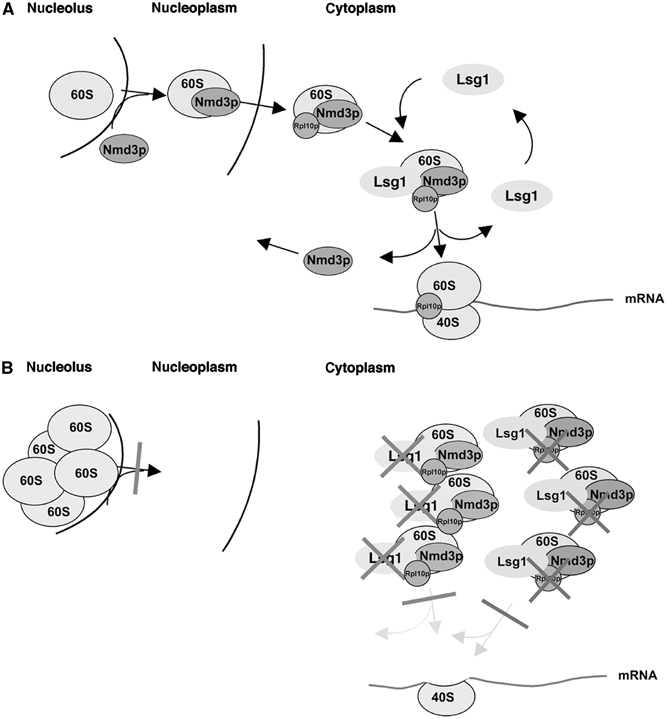
Model explaining the effects of Lsg1p and Rpl10p on Nmd3p shuttling and 60S export. (A) The interactions between Rpl10p, Nmd3p and Lsg1p are shown under normal cellular conditions. (B) The consequence of failing to recycle Nmd3p due to defects in Lsg1p or Rpl10p. Both models are described in the Discussion.
Disruption of Rpl10p or Lsg1p function prevents Nmd3p release from subunits. This may result in a steric block to subunit joining (neither Nmd3p nor Lsg1p are observed on 80S couples) and the accumulation of unjoined subunits in the cytoplasm (Figure 8B). Under these conditions, the free pool of Nmd3p would be rapidly depleted due to its retention on cytoplasmic subunits. The depletion of free Nmd3p available to recycle to the nucleus for subunit export would in turn lead to accumulation of nascent subunits in the nucle(ol)us. Hence, both 60S subunit export and activation of cytoplasmic subunits would be affected simultaneously.
Does Nmd3p bind before Rpl10p?
The suppression of rpl10(G161D) by dominant mutations in NMD3 has lent credence to the idea that Rpl10p provides part of the binding site for Nmd3p, thereby recruiting Nmd3p to the subunit for export from the nucleus. However, the bypass of the nuclear export defect that we observed in RPL10-repressed cells by high-copy NMD3 cannot easily be explained if Nmd3p binding requires Rpl10p. We suggest instead that Rpl10p is loaded onto the subunit after Nmd3p. This could occur in the cytoplasm or nucleus. However, Rpl10p is not retained in the nucleus by LMB or by expression of mutant Nmd3p lacking an NES (unpublished), conditions that do trap Nmd3p and 60S subunits (Ho et al, 2000b), suggesting that Rpl10p loads into the subunit after export to the cytoplasm. In this scenario, Lsg1p could couple the loading of Rpl10p, as the final step in subunit assembly, with the release of the export factor Nmd3p.
Could Lsg1p be involved in loading Rpl10p?
Rpl10p is one of three large subunit proteins that exchanges on cytoplasmic subunits (Zinker and Warner, 1976). Based on the crystal structure of the 50S subunit of Haloarcula marismortui (Ban et al, 2000) along with the cryo-EM structure of the large subunit from yeast (Spahn et al, 2001), Rpl10p fits into a cleft between the GTPase stalk and the central protuberance of the large subunit where it binds both 25 and 5S rRNAs. Considering its extensive interactions with the subunit, it is difficult to imagine insertion of Rpl10p into the subunit without significant conformational changes in the surrounding structure. Attempts to load Rpl10p into the subunit in vitro have also not been successful (Eisinger et al, 1997). It is possible that the GTPase activity of Lsg1p drives a conformational change that allows Rpl10p to exchange and/or that locks Rpl10p into place on subunits.
Coordination with other 60S maturation events
Like Lsg1p, another cytoplasmic GTPase, Efl1p/Ria1p, was shown to be required for a late step in 60S biogenesis. Disruption of Efl1p activity leads to a 60S biogenesis defect (Becam et al, 2001; Senger et al, 2001), coinciding with the cytoplasmic accumulation of the nucle(ol)ar biogenesis factor, Tif6p (Senger et al, 2001).The authors proposed that Efl1p induces a conformational change in nascent 60S subunits in the cytoplasm, thereby releasing Tif6p and contributing to translational competence (Senger et al, 2001). As Lsg1p appears to play a similar role in facilitating the release of Nmd3p, it is possible that Lsg1p and Efl1p act on subunits in a concerted manner. Although lsg1 mutants do not accumulate Tif6p on cytoplasmic subunits (West and Johnson, unpublished), Lsg1p and Efl1p may act in tandem or sequentially to induce structural rearrangements important for final ribosomal protein assembly and release of persistent biogenesis factors, priming subunits for translation. These structural changes may be rate limiting and account for the lag that has been observed for incorporation of the nascent large subunit into polysomes.
Materials and methods
Strains and plasmids used in this study are listed in Tables I and II, respectively, and described in Supplementary Material.
Table 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303 (wt) | MATa leu2-3,112 his3-11 ura3-1 trp1-1 ade2-1 can1-100 SSD1-d | J Warner |
| DEH221+ | MATα qsr1Δ1::HIS3 ade2-1 trp1-1 leu2-3,112 ura3-1 can1-100 (pDEGQ2) | Eisinger et al (1997) |
| AJY729 | MATa ade2 ade3 leu2 lys2-801 ura3-52 trp1 nmd3-3 | Ho and Johnson (1999) |
| AJY736 | MATa ade2 ade3 leu2 lys2-801 ura3-52 trp1 his3 nmd3-4 | Ho and Johnson (1999) |
| AJY1171 | MATa his3 leu2 met15Δ0 ura lsg1Δ::KanMX4 | Kallstrom et al (2003) |
| AJY1433 | MATα sqt1::KanMX4 met15Δ10 leu2Δ0 ura3Δ0 his3Δ1 (pAJ336) | This study |
| AJY1511 | MATα leu2 ura3 lsg1Δ::KanMX4 | This study |
| AJY1512 | MATa leu2 ura3 lsg1Δ::KanMX4 nmd3-3 | This study |
| AJY1513 | MATa leu2 ura3 nmd3-3 | This study |
| AJY1518 | MATa leu2 ura3 nmd3-4 | This study |
| AJY1521 | MATα leu2 ura3 lsg1Δ::KanMX4 nmd3-4 | This study |
| AJY1548 | MATα leu2 ura3 his3 met15 crm1(T539C) | This study |
| AJY1605 | MATα sqt1::KanMX4 met15Δ10 leu2Δ0 ura3Δ0 his3Δ1 (pAJ1062) | This study |
| AJY1640 | MATα sqt1::KanMX4 met15Δ10 leu2Δ0 ura3Δ0 his3Δ1 (pAJ1065) | This study |
| AJY1657 | MATa ura3 leu2 rpl10[G161D] | This study |
| AJY1705 | MATa leu2 ura3 NMD3-GFP::KanMX6 CRM1[T539C] | This study |
| AJY1708 | MATa ade2 ade3 leu2 lys2-801 ura3-52 NMD3:GFP::KanMX6 | This study |
| AJY1836 | MATa NMD3-GFP::KanMX6 CRM1[T539C] rpl10::KanMX4 ura3 leu2 (pDEGQ2) | This study |
| AJY1896 | MATa ura3 his3 leu2 nmd3::TRP1 CRM1[T539C] (pAJ112) | This study |
| CH1305 | MATa ade2 ade3 leu2 lys2-801 ura3-52 | Kranz and Holm (1990) |
Table 2.
Plasmids used in this study
| Plasmid | Relevant markers | Source or reference |
|---|---|---|
| pDC-CRM1 | LEU2-CEN (CRM1-HA) | Neville and Rosbash (1999) |
| pDC-CRM1[T539C] | LEU2-CEN (CRM1[T539C]-HA) | Neville and Rosbash (1999) |
| pASZ11-Rpl25eGFP | ADE2-CEN (RPL25-eGFP) | Gadal et al (2001) |
| pAJ60 | ADE3 URA3-CEN | This study |
| pAJ78 | LEU2-2μ (NMD3) | Ho and Johnson (1999) |
| pAJ118 | (LEU2-2μ (GAL10::NMD3) | Ho and Johnson (1999) |
| pAJ123 | LEU2-CEN (NMD3) | Ho and Johnson (1999) |
| pAJ289 | LEU2-CEN (myc-LSG1) | Kallstrom et al (2003) |
| pAJ363 | URA3 ADE3-2μ (NMD3) | This study |
| pAJ369 | URA3-CEN (GAL10-RPL25-GFP) | Ho et al (2000b) |
| pAJ409 | URA3-CEN (NMD3) | Kallstrom et al (2003) |
| pAJ410 | LEU2-2μ (NMD3) | This study |
| pAJ415 | LEU2-CEN (NMD3[L291F]) | This study |
| pAJ535 | LEU2-CEN (NMD3Δ100-myc) | This study |
| pAJ582 | LEU2-CEN (NMD3-GFP) | This study |
| pAJ689 | URA3-2μ (GAL10::GST) | This study |
| pAJ690 | URA3-2μ (GAL10::GST-NMD3) | This study |
| pAJ698 | URA3-2μ (GAL10::GST-nmd3Δ194) | This study |
| pAJ741 | LEU2-CEN (lsg1-2) | Kallstrom et al (2003) |
| pAJ742 | LEU2-CEN (lsg1-3) | This study |
| pAJ752 | LEU2-CEN (NMD3AAA-myc) | This study |
| pAJ754 | LEU2-CEN (NMD3AAA-GFP) | This study |
| pAJ755 | URA3-CEN (NMD3-GFP) | This study |
| pAJ879 | URA3-CEN (GAL10::myc-LSG1) | This study |
| pAJ907 | LEU2-CEN (RPL25-eGFP) | This study |
| pAJ908 | URA3-CEN (RPL25-eGFP) | This study |
| pAJ1002 | URA3-CEN (NMD3-myc) | This study |
| pAJ1069 | LEU2-CEN (NMD3[L291F]AAA-GFP) | This study |
| pAJ1070 | LEU2-CEN (NMD3[L291F]-myc) | This study |
| pAJ1109 | LEU2-CEN (GAL10::myc-LSG1[K349T]) | This study |
| pAJ1131 | LEU2-CEN (GAL10::myc-LSG1[N173Y,L176S]) | This study |
| pAJ1132 | LEU2-CEN (GAL10::myc-LSG1[I204T]) | This study |
| pAJ1143 | URA3-CEN (GAL1::NMD3) | This study |
| pAJ1278 | URA3-CEN (GAL10::myc-LSG1[K349T]) | This study |
| pAJ1287 | LEU2-CEN (NMD3[I112T, I362T]-GFP) | This study |
| pAJ1291 | URA3-2μ (GAL10::GST-NMD3[L291F]) | This study |
| pAJ1292 | URA3-2μ (GAL10::GST-NMD3[I112 T, I362T]) | This study |
| pAJ1293 | URA3-2μ (GAL10::GST-NMD3[V340D]) | This study |
| pAJ1299 | LEU2-CEN (NMD3[V340D]-myc) | This study |
| pAJ1312 | URA3-CEN (GAL10::myc-LSG1) | This study |
LSG1 mutagenesis
LSG1 was mutagenized by PCR with Taq polymerase using pAJ879 as template and the primers 5′-CATGCCATGGAACAAAAG TTGATTTCTGAAGAAGACTTGAGCTCTATGCCACC AAAAGAAGCT and 5′-CGTGA CGTCTAATTATTTTCAATGCT. The PCR product was cotransformed with BclI and BglII linearized pAJ879 into wild-type CH1305 cells. Cells were plated onto dropout media supplemented with glucose and mutants were identified by their inability to grow after replica plating to galactose-containing plates.
Composite gels
GST-fusion proteins expressed from pAJ690, pAJ1291, pAJ1292 or pAJ1293 were purified from yeast as described previously (Ho et al, 2000a). Protein concentrations were measured using the Bradford assay. Composite gels were as described previously (Dahlberg and Grabowski, 1990). For in vitro reconstitutions, increasing amounts of each GST-Nmd3p (see Figure 6A, 1 × ∼30 ng) were mixed with 0.018 OD260U of purified free 60S subunits (prepared as in Ho et al, 2000a) in 10 μl of low TKM buffer (25 mM Tris–OAc (pH 7.6), 60 mM KOAc and 1 mM MgOAc2) plus protease inhibitors. After 30 min incubation at 25°C, samples were placed on ice and sucrose loading dye was added to each reaction. Gels were prerun at 60 V for 1 h in TKM buffer with continual cooling at 4°C. After a fresh buffer change, samples were then loaded and run with cooling for 4 h at 60 V with a second fresh buffer change after 2 h, Proteins were transferred to nitrocellulose in 25 mM Tris–OAc (pH 7.6) after presoaking gel and membrane in same buffer containing 0.1% SDS for 10 min. Western blotting was carried out as in Figure 6A and the gel was stained post-transfer with ethidium bromide rRNA.
Detailed descriptions of additional methods are provided in the Supplementary Material.
Supplementary Material
Supplementary Material
Acknowledgments
We thank M Yoshida for generously providing LMB, E Hurt for providing the Rpl25-eGFP vector, M Rosbash for providing the pDC-CRM1(T539C) vector, A Bretscher for providing the GAL1-regulated yeast cDNA library, J Ballesta for the anti-Rpl12p antibody and F Lacroute for the anti-Rpl1ap antibody. We thank B Trumpower for strain DEH221+ and plasmid pDEGQ2. We also thank A Chen for making NMD3 suppressor mutants and G Chen for making NMD3 loss of function mutants. This work was supported by NIH Grant RO1-GM53655 to A Johnson.
References
- Aravind L, Koonin EV (2000) Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res 10: 1172–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Becam AM, Nasr F, Racki WJ, Zagulski M, Herbert CJ (2001) Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol Genet Genom 266: 454–462 [DOI] [PubMed] [Google Scholar]
- Benne R, Hershey JW (1978) The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem 253: 3078–3087 [PubMed] [Google Scholar]
- Dahlberg AE, Grabowski PJ (1990) Gel electrophoresis of ribonucleoproteins. In Gel Electrophoresis of Nucleic Acids: A Practical Approach, D Rickwood, BD Hames (eds) pp 275–289. Oxford: Oxford University Press [Google Scholar]
- Daigle DM, Rossi L, Berghuis AM, Aravind L, Koonin EV, Brown ED (2002) YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry 41: 11109–11117 [DOI] [PubMed] [Google Scholar]
- Damke H, Binns DD, Ueda H, Schmid SL, Baba T (2001) Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages. Mol Cell Biol 12: 2578–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Eisinger DP, Trumpower BL (1997a) Exchangeability of Qsr1p, a large ribosomal subunit protein required for subunit joining, suggests a novel translational regulatory mechanism. FEBS Lett 419: 1–3 [DOI] [PubMed] [Google Scholar]
- Eisinger DP, Dick FA, Trumpower BL (1997) Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40 and 60S subunits. Mol Cell Biol 17: 5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D (2002) Making ribosomes. Curr Opin Cell Biol 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F (2003) Ribosome assembly in eukaryotes. Gene 313: 17–42 [DOI] [PubMed] [Google Scholar]
- Fujimura K, Tanaka K, Toh-e A (1993) A dominant interfering mutation in RAS1 of Saccharomyces cerevisiae. Mol Gen Genet 241: 280–286 [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E (2001b) Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Johnson AW (1999) NMD3 encodes an essential cytoplasmic protein required for stable ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol 19: 2389–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW (2000a) Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA 6: 1625–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW (2000b) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AW (2003) Nuclear export of ribosomal subunits. In Nucleolus, M Olson (ed). pp 286–301. New York: Kluwer Academic/Plenum Publishers [Google Scholar]
- Johnson AW, Lund E, Dahlberg J (2002) Nuclear export of ribosomal subunits. Trends Biochem Sci 27: 580–585 [DOI] [PubMed] [Google Scholar]
- Kallstrom G, Hedges J, Johnson A (2003) The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol Cell Biol 23: 4344–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl T, Onder K, Kodzius R, Pichova A, Wimmer H, Th r A, Hundsberger H, Loffler M, Klade T, Beyer A, Breitenbach M, Koller L (1999) GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr Genet 34: 419–429 [DOI] [PubMed] [Google Scholar]
- Kressler D, Linder P, de La Cruz J (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 19: 7897–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T, Planta RJ, Krop JM (1978) The course of the assembly of ribosomal subunits in yeast. Biochim Biophys Acta 517: 378–389 [DOI] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L (2002) Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317: 41–72 [DOI] [PubMed] [Google Scholar]
- Neville M, Rosbash M (1999) The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J 18: 3746–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E (2002) 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J 21: 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Jensen BC, Kifer CT, Parsons M (2001) A novel nucleolar G-protein conserved in eukaryotes. J Cell Sci 114: 173–185 [DOI] [PubMed] [Google Scholar]
- Russell DW, Spremulli LL (1979) Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J Biol Chem 254: 8796–8800 [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci 15: 430–434 [DOI] [PubMed] [Google Scholar]
- Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E (2003) The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J 22: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F (2001) The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Thomas F, Kutay U (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE (2003) Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J 22: 2841–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D (1995) Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11: 1629–1650 [DOI] [PubMed] [Google Scholar]
- Warner JR (1971) The assembly of ribosomes in yeast. J Biol Chem 246: 447–454 [PubMed] [Google Scholar]
- Zinker S, Warner JR (1976) The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J Biol Chem 251: 1799–1807 [PubMed] [Google Scholar]
- Zuk D, Belk JP, Jacobson A (1999) Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics 153: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


