Abstract
E1 enzymes facilitate conjugation of ubiquitin and ubiquitin-like proteins through adenylation, thioester transfer within E1, and thioester transfer from E1 to E2 conjugating proteins. Structures of human heterodimeric Sae1/Sae2-Mg·ATP and Sae1/Sae2-SUMO-1-Mg·ATP complexes were determined at 2.2 and 2.75 Å resolution, respectively. Despite the presence of Mg·ATP, the Sae1/Sae2-SUMO-1-Mg·ATP structure reveals a substrate complex insomuch as the SUMO C-terminus remains unmodified within the adenylation site and 35 Å from the catalytic cysteine, suggesting that additional changes within the adenylation site may be required to facilitate chemistry prior to adenylation and thioester transfer. A mechanism for E2 recruitment to E1 is suggested by biochemical and genetic data, each of which supports a direct role for the E1 C-terminal ubiquitin-like domain for E2 recruitment during conjugation.
Keywords: Aos1, conjugation, Sae1, Sae2, Uba2
Introduction
Ubiquitin (Ub) and ubiquitin-like (Ubl) modifiers are ∼100-amino-acid proteins that regulate differentiation, apoptosis, the cell cycle, and responses to stress through post-translational covalent attachment to lysine residues in targeted proteins (Hochstrasser, 1996; Hershko and Ciechanover, 1998; Laney and Hochstrasser, 1999; Melchior, 2000; Muller et al, 2001; Johnson, 2004). The small ubiquitin-related modifier SUMO-1 is a Ub/Ubl family member, and although SUMO-1 shares structural similarity to Ub, SUMO's cellular functions remain distinct insomuch as SUMO modification alters protein function through changes in activity, cellular localization, or by protecting substrates from ubiquitination (Hershko and Ciechanover, 1998; Melchior, 2000; Melchior et al, 2003; Johnson, 2004).
Saccharomyces cerevisiae Smt3 was the first SUMO ortholog identified and Smt3 conjugation is critical for septin ring formation, chromosomal segregation, and G2–M cell cycle progression (Johnson and Blobel, 1999; Li and Hochstrasser, 1999; Takahashi et al, 1999; Tanaka et al, 1999; Johnson, 2004). In contrast to the single yeast SMT3 gene, four SUMO family members have been identified in human. SUMO-1 shares ∼43% sequence identity with the almost identical SUMO-2 and SUMO-3, which share 96% sequence identity to each other. SUMO-4 shares 87% sequence homology to SUMO-2, and exists in native and disease-specific isoforms (Bohren et al, 2004). Although SUMO isoforms are involved in distinct cellular processes, we will use SUMO to describe the entire system and refer by name to protein components when necessary.
The mechanisms utilized to conjugate Ub and Ubl modifiers share many similarities and some important differences. The Ub pathway utilizes a single E1 to activate Ub and unique combinations of several E2s and numerous E3s to direct conjugation and ensure specificity (Hershko and Ciechanover, 1998). In contrast, SUMO and Nedd8 pathways utilize a single E1 and a single E2 in combination with a few known E3s. Ubc9 is the only known SUMO E2 enzyme (Seufert et al, 1995; Johnson and Blobel, 1997), and while SUMO conjugation can be reconstituted under select conditions in vitro using only E1, Ubc9, SUMO, and ATP, several SUMO E3 factors have been recently identified that facilitate conjugation in vivo and in vitro (Johnson, 2004).
Conjugation ultimately results in isopeptide bond formation between a protein lysine ɛ-amino group and the Ub/Ubl C-terminus (Hershko and Ciechanover, 1998). To catalyze this, full-length Ub/Ubls are proteolytically processed by specific cysteine proteases to produce the conserved C-terminal di-glycine (Gly–Gly) motif that is subsequently adenylated by E1 enzymes in an ATP-dependent process, thus activating the C-terminus for transfer to an E1-thioester adduct via a conserved E1 cysteine. The E1-thioester complex must then recruit an E2 to facilitate transfer of the E1-thioester to a conserved E2 cysteine. The energy stored in the E2-Ub/Ubl thioester is utilized to conjugate Ub/Ubls to target lysine ɛ-amino groups directly or through complexes mediated by E3s.
To determine the molecular basis for E1-dependent SUMO transfer mechanisms, and to elaborate interactions utilized by the Sae1/Sae2 heterodimeric human SUMO E1 for recruitment of the SUMO E2 Ubc9 to the E1-SUMO-thioester adduct, we have structurally characterized complexes between Sae1/Sae2-Mg·ATP and Sae1/Sae2-SUMO-1-Mg·ATP. The presence of an unreacted SUMO-1-Mg·ATP substrate complex suggests that conformational changes may be required to facilitate adenylation, thioester transfer, and subsequent recruitment of E2 to the E1-SUMO complex. Deletion and biochemical analysis revealed an essential and direct role in vitro and in vivo for the Sae2 C-terminal ubiquitin-like domain (UbL) in E1-SUMO-thioester transfer to the SUMO E2 conjugating protein.
Results and discussion
Sae1/Sae2-Mg·ATP and Sae1/Sae2-SUMO-1-Mg·ATP structure determination
Human SAE1 and SAE2 were cloned and coexpressed to isolate the heterodimeric 113 kDa Sae1/Sae2 protein complex previously characterized as the E1 for SUMO activation (Johnson et al, 1997; Desterro et al, 1999; Okuma et al, 1999). Native Sae1/Sae2-ATP crystals were soaked with thimerosal to obtain a mercury derivative suitable for isomorphous replacement. Data were obtained at the mercury LIII absorption edge at NSLS X4A for one native crystal and one mercury derivative crystal. A total of 12 mercury atoms were identified and used to calculate phases with two-fold NCS averaging between the two Sae1/Sae2-ATP complexes observed per asymmetric unit (ASU) (Table I and Materials and methods). The SUMO E1 structure was built at 2.6 Å using these data. Crystals were obtained for native protein, but superior diffraction was observed for crystals containing a single point mutation for the catalytic cysteine (Cys173Ala) of the Sae2 subunit. A crystal that diffracted X-rays to 2.2 Å at the Advanced Photon Source was obtained and used to refine the Sae1/Sae2-Mg·ATP complex at 2.25 Å to an R-factor and Rfree of 20.8 and 24.8, respectively. Structures containing Mg·ATP and ATP differed only in that the Mg·ATP complex contained one magnesium ion per ATP ligand as determined by difference Fourier analysis.
Table 1.
Crystallographic data and refinement statistics
| Native Mg·ATP | Native SUMO Mg·ATP | Native/ATP | Hg/ATP | |
|---|---|---|---|---|
| PDB ID | 1Y8Q | 1Y8R | ||
| Source | APS 31ID | APS 31ID | X4A | X4A |
| Wavelength (Å) | 0.9790 | 0.9790 | 1.008 | 1.008 |
| Resolution limits (Å) | 20–2.2 | 20–2.75 | 20–2.5 | 20–2.8 |
| Space group | P21 | P212121 | P21 | P21 |
| Unit cell (Å) a, b, c, α, β, γ | 101.2, 116.7, 106.1, 90, 112.7, 90 | 117.1, 214.1, 100.7, 90, 90, 90 | 101.2, 116.7, 106.0, 90, 112.6, 90 | 101.8, 116.6, 105.6, 90, 112.6, 90 |
| Number of observations | 351 595 | 727 056 | 310 888 | 236 755 |
| Number of reflections | 108 617 | 67 058 | 74 210 | 104 523a |
| Completeness (%) | 97.3 (92.7) | 93.4 (75.9) | 98.5 (97.0) | 96.3 (92.8) |
| Mean I/σI | 16.4 (2.6) | 9.7 (3.0) | 6.2 (1.5) | 4.3 (1.0) |
| Rmerge on Ib | 7.1 (43.5) | 15.0 (48.4) | 13.2 (57.7) | 17.8 (84.1) |
| Cutoff criteria I/σI | −0.5 | −0.7 | −0.5 | −1.0 |
| Number of sites | 12 | |||
| MFID (%)c | 18.6 | |||
| Mean FOM (SOLVE)d | 0.15 (acentric)/0.21 (centric) | |||
| FOM (RESOLVE with NCS) | 0.50 (acentric)/0.61 (centric) | |||
| Refinement statistics | ||||
| Resolution limits (Å) | 20–2.25 | 20–2.75 | ||
| Number of reflections | 104 390 | 63 543 | ||
| Completeness (%) | 97.0 (93.2) | 94.9 (84.3) | ||
| Cutoff criteria I/σI | 0 | 0 | ||
| Protein/ligand/water atoms | 12 935/66/820 | 14 316/66/317 | ||
| Rcryste | 0.208 (0.350) | 0.220 (0.370) | ||
| Rfree (5% of data) | 0.248 (0.371) | 0.276 (0.391) | ||
| Bonds (Å)f | 0.006 | 0.009 | ||
| Angles (deg)f | 1.1 | 1.2 | ||
| B-factors (mc/sc in Å2)f | 1.50/2.14 | 1.4/1.8 | ||
| aSIRAS data completeness treats Bijvoët mates independently. | ||||
| bRmerge=∑hkl∑i∣I(hkl)i−〈I(hkl)〉∣/∑hkl∑i〈I(hkl)i〉. | ||||
| cMFID (mean fractional isomorphous difference)=∑∣∣Fph∣−∣Fp∣∣/∑∣Fp∣, where Fp is the protein structure factor amplitude and ∣Fph∣ is the heavy-atom derivative structure factor amplitude. | ||||
| dMean FOM=combined figure of merit. | ||||
| eRcryst=∑hkl∣Fo(hkl)−Fc(hkl)∣ /∑hkl∣Fo(hkl)∣, where Fo and Fc are the observed and calculated structure factors, respectively. | ||||
| fValues indicate root-mean-square deviations in bond lengths, bond angles, and B-factors of bonded atoms. | ||||
| Values in parentheses indicate statistics for the high-resolution data bin. | ||||
The Sae1/Sae2-SUMO-ATP complex was obtained by cocrystallization. A 2.6 Å data set was collected that belonged to a different space group, so the structure was solved by molecular replacement using native Sae1/Sae2 coordinates with two complexes observed per ASU. Electron density was observed for SUMO-1 in both complexes and modeled using a SUMO-1 crystal structure obtained from a complex between SUMO-1 and Senp2 (Reverter and Lima, 2004). Sae1/Sae2-SUMO-1-Mg·ATP was obtained by cocrystallization and by incubating crystals of Sae1/Sae2-SUMO-ATP in magnesium sulfate for 4–5 h prior to cryopreservation. Although magnesium and ATP were expected to result in adenylation of the SUMO-1 C-terminal glycine, this complex was nearly identical to Sae1/Sae2-SUMO-ATP except for a single magnesium ion coordinated to each ATP. Similar to Sae1/Sae2-Mg·ATP, we utilized the mutant Sae2 Cys173Ala subunit and a magnesium soak to obtain crystals of Sae1/Sae2-SUMO-1-Mg·ATP that diffracted X-rays to superior resolution. These data were used to refine the complex at 2.75 Å to a final R-factor and Rfree of 22.0 and 27.6, respectively (Table I and Materials and methods).
Structural description of Sae1/Sae2-Mg·ATP and Sae1/Sae2-SUMO-1-Mg·ATP
Full-length human proteins for Sae1 (1–346) and Sae2 (1–640) were used in crystallization. Disordered regions not observed in electron density included Sae1 residues 178–203 and 346, and Sae2 residues 1–3, 219–237, 291–304, and 551–640. The Sae1/Sae2 structure revealed the pseudosymmetric heterodimer between the Sae2 adenylation domain (1–158, 384–438) and Sae1 (1–346) (Figure 1; Supplementary Figure 1). These domains and the adenylation active site are related to those described in the structures of MoeB-MoaD (Lake et al, 2001) and the APPBP1/UBA3 Nedd8 E1 (Walden et al, 2003b; for review, see Huang et al, 2004b). Sae2 includes three domains, the adenylation domain and two additional domains located on either side of the Sae2 adenylation active site (Figure 1). The first will be referred to as the catalytic Cys domain (159–386) since it contains Cys173, the catalytic cysteine responsible for E1-SUMO-thioester bond formation. The second will be termed the UbL or UBiquitin-Like domain (442–549) due to its structural similarity to Ub and other Ubl modifiers.
Figure 1.
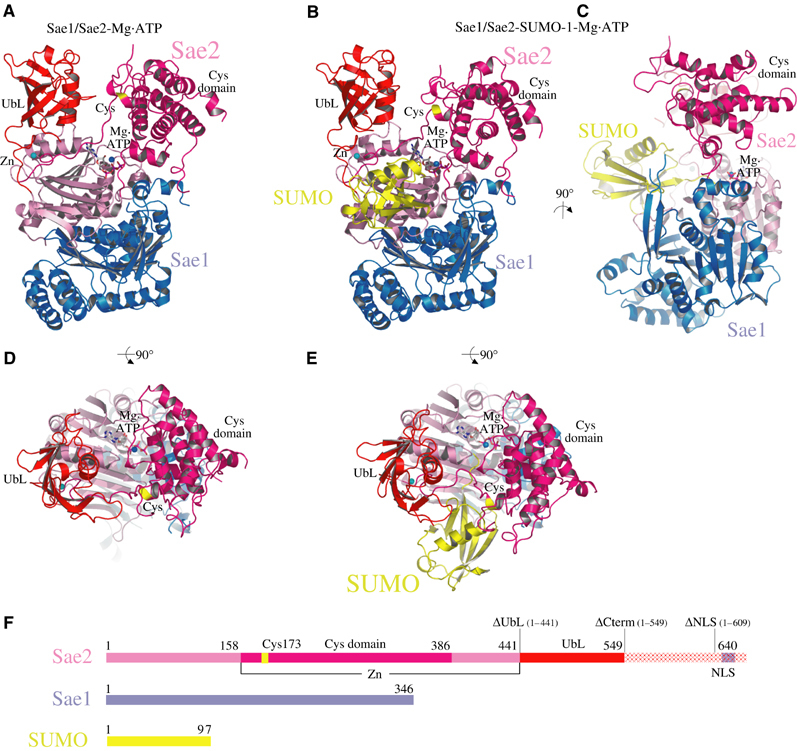
Structure of the SUMO activating enzyme. (A) Ribbon diagram of the Sae1/Sae2 heterodimer Mg·ATP complex. Sae1 is colored blue and Sae2 is colored shades of pink, magenta, and red. (B) Ribbon diagram of the Sae1/Sae2-SUMO-1-Mg·ATP complex. SUMO is colored yellow. Sae1/Sae2 is colored as in (A). Mg·ATP and zinc are labeled and shown in stick and sphere representation. Catalytic Cys and UbL domains are labeled. The active site cysteine is labeled and colored yellow. (C) Orthogonal view of Sae1/Sae2-SUMO-1-Mg·ATP complex as in (B). (D) Orthogonal view of Sae1/Sae2-Mg·ATP complex as in (A). (E) Orthogonal view of Sae1/Sae2-SUMO-1-Mg·ATP complex as in (B). (F) Schematic representation of Sae1/Sae2 and SUMO-1 colored as in (A–E). C-terminal truncation mutants described in the text are indicated above the Sae2 peptide as ΔUbL, ΔCterm, and ΔNLS. Stereo images of panels A and B are provided in Supplementary Figure 1. Images were generated with SETOR or PYMOL unless noted otherwise (Evans, 1993; Delano, 2002).
The Sae2 polypeptide chain emerges from the adenylation domain under the E1 UbL domain at the first of two Cys-X-X-Cys motifs that coordinate a single zinc ion (residues Cys158 and Cys161). A crossover strand of ∼11 amino acids passes over the adenylation active site, leading to Cys173 and the site of thioester bond formation. After completion of the catalytic Cys domain at amino acid 386, the Sae2 polypeptide passes into the adenylation domain before emerging once again at the second Cys-X-X-Cys motif (Cys441 and Cys444) to form the C-terminal E1 UbL domain (Figure 1). As such, the crossover strand links the catalytic Cys and UbL domains via the zinc motif in a more direct manner than is obvious through inspection of the linear polypeptide sequence (Figure 1A and F). The catalytic Cys domain appears perched over the rest of the molecule (Figure 1C), burying a relatively small interface between Sae1/Sae2 and the Cys domain (1800 Å2 of total surface area; GRASP; Nicholls et al, 1991).
The SUMO E1 Sae2 subunit includes a C-terminal extension from amino acids 550–640 that was not apparent in electron density maps, despite its presence in our protein and crystalline preparations. Interestingly, sequence and structural alignment between the SUMO Sae2, Ub Uba1, and Nedd8 Uba3 suggest that the Ub and Nedd8 E1 enzymes have their C-termini located near Sae2 residue 540, indicating that the Sae2 C-terminal extension is not conserved in all E1 enzymes. As will be discussed below, the yeast SUMO E1 Uba2 C-terminal extension was previously shown to contain a nuclear localization signal (NLS) (Dohmen et al, 1995), suggesting that at least a portion of amino acids 550–640 are not directly involved in catalytic activities associated with SUMO activation or transfer to E2.
The structure of Sae1/Sae2-SUMO-1-Mg·ATP complex revealed a similar overall topology for Sae1 and the Sae2 adenylation, catalytic Cys, and UbL domains (Figure 1B, C, and E). Full-length mature SUMO-1 (1–97) was utilized in crystallization trials although only residues 14–97 were apparent in electron density maps. SUMO-1 is bound on the surface of Sae2 between the UbL and catalytic Cys domains in an orientation approximating that observed for MoaD in complex with MoeB, and for Nedd8 in complex with APPBP1/Uba3 (Lake et al, 2001; Walden et al, 2003a).
SUMO-1 recognition
SUMO-1 is recognized exclusively by residues emanating from Sae2, as no direct interactions are observed between SUMO-1 and the Sae1 subunit (Figure 1E). The Sae2-SUMO interface buries 1650 Å2 of total surface area between Sae2 and SUMO-1 (∼20% of the SUMO-1 total surface area). Contacts are observed between Sae2 and SUMO-1 residues Gln29, Ser31, Asn60, Arg70, Glu89, Tyr91, Glu93, Gln94, Thr95, Gly96, and Gly97 (Figures 2A, B, and 3F). SUMO-1 Glu93 contacts Sae2 Arg119 and Tyr159 and is conserved as Glu in SUMO-1 and Smt3 or Gln in SUMO-2 or -3 (Figures 2 and 3F). The analogous position is substituted to Ala or Arg in Nedd8 or Ub, a position previously identified as a critical determinant for Nedd8 E1 discrimination between Nedd8 and Ub (Walden et al, 2003a).
Figure 2.
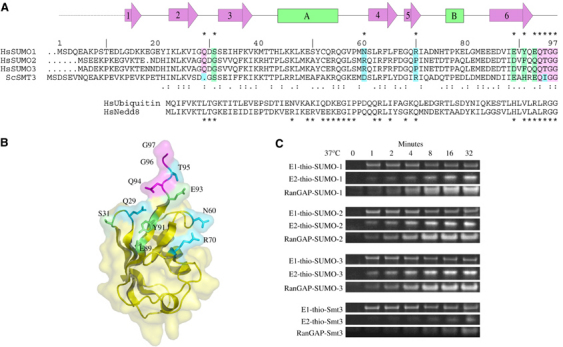
Conserved SUMO-1 residues and E1 activation. (A) Structure-based alignment of human SUMO family members SUMO-1, -2, -3, S. cerevisiae Smt3, Ub, and Nedd8. SUMO-1 amino-acid numbering, secondary structure, and contact residues with Sae2 (*) are shown above the sequence with sequence similarity ( . ) and identity ( : ) shown below the aligned SUMO sequences. SUMO-1 residues in contact with Sae2 are colored pink (identical), green (similar), or blue (dissimilar). Ub and Nedd8 alignment is shown below the SUMO alignment with (*) denoting contacts observed between Nedd8 and the Nedd8 E1. (B) Ribbon and transparent surface for SUMO-1 with surface residues contacting Sae2 shaded pink, green, or blue as in (A). (C) Time course for formation of E1-thioester, E2-thioester, and SUMO conjugation at 37°C using human SUMO-1, -2, -3, and S. cerevisiae Smt3 as substrates (see Materials and methods and Supplementary Figure 2 for assay details). Labels indicate the positions for E1-thio-SUMO, E2-thio-SUMO, and RanGAP1-SUMO.
Figure 3.
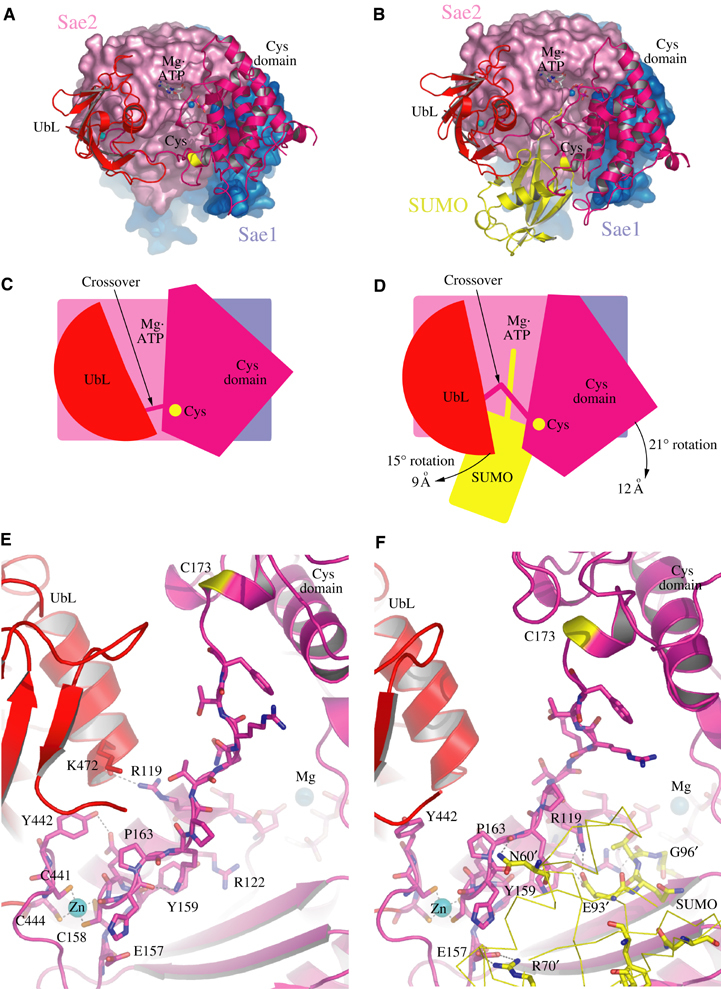
Comparison between Sae1/Sae2-Mg·ATP and Sae1/Sae2-SUMO-1-Mg·ATP complexes. (A) Ribbon and surface representations for Sae1/Sae2-Mg·ATP colored as in Figure 1. (B) Sae1/Sae2-SUMO-1-Mg·ATP complex with SUMO-1 colored yellow. (C) Cartoon representation of (A) with Mg·ATP, catalytic cysteine, crossover loop, with UbL and Cys domains labeled. (D) Cartoon representation of (B) with arrows indicating the direction of observed domain rotations (in degrees) and maximal displacement (in Å). (E) Close-up of the Sae1/Sae2-Mg·ATP complex and crossover loop. Mg·ATP and zinc are labeled with amino acids in stick representation. (F) Close-up of the Sae1/Sae2-SUMO-1-Mg·ATP complex and crossover loop as in (E) but now showing amino-acid residues from SUMO-1 (yellow) interacting with Sae2 residues.
SUMO-1 Gln94 is strictly conserved among SUMO isoforms and contacts Sae2 Ala142 and Leu145. Although Gln94 could serve as specificity determinant for SUMO E1 discrimination between SUMO, Nedd8 (Leu), and Ub (Leu), no specific hydrogen bonding interactions are observed between Sae2 and either Gln Oɛ or Nɛ atoms (Figures 2A, B, and 3F). Main-chain contacts are observed between SUMO Thr95, Gly96, and Gly97 with residues in the adenylation active site and ATP. The C-terminal di-glycine motif is conserved in SUMO, Nedd8, and Ub and cannot serve as a point of discrimination between these molecules.
SUMO-1 Arg70 makes salt-bridging contacts to Glu157 and Asp435, and while not strictly conserved in SUMO isoforms, this position is substituted to Gln or Asp in Nedd8 or Ub, substitutions that could generate unfavorable interactions with negatively charged Sae2 residues (Figures 2A, B, and 3F). Hydrophobic contacts are observed between Sae2 Phe417 and Val440 and SUMO-1 Tyr91. Tyr91 also contacts SUMO-1 Glu89, which in turn contacts Sae2 Asn423. The negatively charged SUMO Glu89 and bulky Tyr91 are substituted to histidine and valine in both Ub and Nedd8.
Of the 11 SUMO-1 side chains that make direct contact to Sae2, five are strictly conserved in SUMO-1, SUMO-2, and SUMO-3, four are conserved at the level of amino-acid property, and two are divergent between SUMO-1 (Asn60 and Arg70) and SUMO-2/3 (substituted to Arg and Pro, respectively) (Figure 2A and B). To assess whether these substitutions give rise to any differences in reaction kinetics for E1-thioester, E2-thioester, and conjugate formation, human Sae1/Sae2 was used to activate and conjugate SUMO-1, -2, and -3 (Figure 2C). Data are presented as individual gel segments because three independent assays were used to assess each activity (see Materials and methods and Supplementary Figure 2). Results show that each SUMO isoform can be activated by E1, transferred to an E2-thioester, and conjugated to RanGAP1 with similar kinetics, suggesting that amino-acid differences observed between human SUMO isoforms are not critical for E1, E2, or substrate interaction in vitro.
Comparison of SUMO sequences to S. cerevisiae Smt3 (Figure 2A) revealed that Smt3 shared many of the conserved SUMO residues observed in contact with human SUMO E1. Consistent with this observation, Smt3 was efficiently activated and transferred to a SUMO E1-thioester adduct (Figure 2C). Smt3 can also be transferred to human Ubc9 and conjugated to RanGAP1, although it is less efficient than the other human SUMO isoforms tested. It is intriguing to speculate that despite efficient transfer to the E1-thioester, Smt3 presents determinants within the E1-Smt3 complex that prevent efficient transfer to human Ubc9, a defect that could not be overcome even when human Ubc9 was replaced with yeast Ubc9 (not shown). As such, it appears that determinants required for efficient E2-thioester transfer are provided by a combination of SUMO and E1 surfaces within the E1-SUMO-thioester complex.
Comparison of Sae1/Sae2-Mg·ATP and Sae1/Sae2-SUMO-1-Mg·ATP
Domain movements were observed for the E1 UbL and catalytic Cys domains when comparing Sae1/Sae2-Mg·ATP and Sae1/Sae2-SUMO-1-Mg·ATP complexes. Although both Sae1/Sae2-SUMO-1-Mg·ATP complexes in the ASU share the same lattice environment and make a similar number of contacts to the same respective domains in the lattice, one complex was observed in which the E1 UbL and catalytic Cys domains were rotated by 15 and 21°, respectively, maximally displacing the domains by as much as ∼9 and ∼12 Å (Figure 3; calculated with Dyndom; Hayward and Berendsen, 1998). Movement of the catalytic Cys domain disrupted interactions in the adenylation active site between Sae2 Asp347, Lys348, and ATP observed in the absence of SUMO-1 (Figure 4; see below). E1 UbL domain movement disrupted interactions between Sae2 Arg119 (adenylation domain) and the main chain of Lys472 in the E1 UbL helix. In addition, a hydrogen bonding interaction is disrupted between Tyr442 in the E1 UbL domain and Glu160 in the crossover strand (Figure 3E). In their place, contacts were observed between Sae2 Arg119 in its alternate conformation, Sae2 Tyr159, and the conserved SUMO-1 residue Glu93 (Figure 3F). The absence of direct contacts between SUMO-1 and either E1 UbL or catalytic Cys domains suggests the possibility that E1 UbL and catalytic Cys domain movements might indirectly change contacts in the adenylation active site that could alter the active site environment between SUMO-1, ATP, the crossover strand, and adenylation domains.
Figure 4.
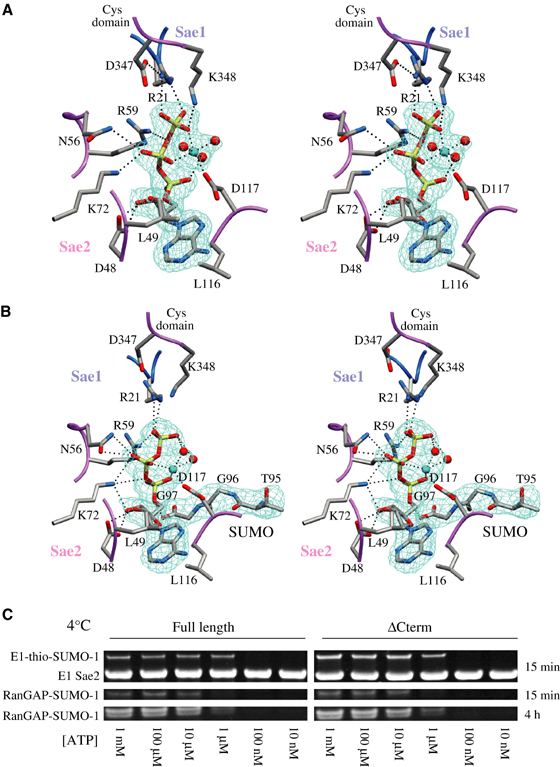
Sae1/Sae2 active sites in complex with Mg·ATP and SUMO-1-Mg·ATP. (A) Stereo representation of the Sae1/Sae2-Mg·ATP complex with a 2.25 Å simulated annealing (SA) omit map contoured at 1.0σ covering the Mg·ATP ligand. (B) Stereo representation of the Sae1/Sae2-SUMO-1-Mg·ATP complex with a 2.75 Å SA omit map contoured at 1σ covering Mg·ATP ligand and SUMO-1 C-terminal amino acids. Residues are numbered in stick representation on a color-coded backbone worm with Sae1 (blue) and Sae2 (pink). The catalytic Cys domain is labeled. Dashed lines represent potential hydrogen bonds; several water molecules were omitted for clarity. (C) ATP dependence of Sae1/Sae2-catalyzed E1-thioester formation and E2-dependent SUMO-1 conjugation to RanGAP1 at 4°C (see Materials and methods). Labels indicate positions for E1-thio-SUMO-1, the E1 Sae2 subunit, and RanGAP1-SUMO-1 after 15 min and 4 h.
Catalytic basis for adenylation and thioester transfer
The adenylation active sites observed in both Sae1/Sae2-SUMO-1-Mg·ATP and Sae1/Sae2-Mg·ATP reveal nearly identical positions for amino-acid residues involved in adenylation when compared to structures of MoeB-MoaD (Figure 4; Lake et al, 2001). These highly conserved residues include Sae1 Arg21 and Sae2 Asp48, Asn56, Arg59, Lys72, and Asp117. While the relative positions of active site residues and ATP were reported in the Nedd8-APPBP1/Uba3-ATP complex, this structure was determined at 3.6 Å without a divalent cation, so direct comparison of atomic details was not warranted (Walden et al, 2003a). As was observed for MoeB-MoaD, we expected that magnesium addition to the crystallization buffer would result in adenylation of the SUMO C-terminus, but inspection of electron density maps revealed a substrate complex composed of Mg·ATP and an unreacted SUMO-1 C-terminal glycine (Figure 4B).
Differences could be observed within the adenylation active sites when comparing Sae1/Sae2-SUMO-1-Mg·ATP and Sae1/Sae2-Mg·ATP complexes. Two residues from the catalytic Cys domain make contacts to ATP in the Sae1/Sae2-Mg·ATP structure (Figure 4A). Lys348 makes a direct contact to the ATP γ-phosphate, while Asp347 makes a salt-bridging interaction with Sae1 Arg21, which in turn contacts the ATP γ-phosphate. These contacts are disrupted as a result of domain movements observed for both UbL and catalytic Cys domains in a Sae1/Sae2-SUMO-1-Mg·ATP complex (Figure 4B; see above). Contacts between the C-terminal carboxylate of SUMO-1 Gly97 and the ATP α-phosphate place the C-terminal SUMO-1 glycine in an optimal orientation for attack at the ATP α-phosphate.
Magnesium is coordinated by the ATP β- and γ-phosphate oxygen atoms in the Sae/Sae2-Mg·ATP complexes and in one of the Sae1/Sae2-SUMO-1-Mg·ATP complexes in the ASU, with nearly ideal octahedral geometry provided by three additional water molecules and the Asp117 side chain (Figure 4A). Mg·ATP alters conformation within the other Sae1/Sae2-SUMO-1-Mg·ATP complex, losing contacts between Mg and the β- and γ-phosphate oxygen atoms in favor of Mg contacts to the α- and β-phosphate oxygen atoms and two water molecules (Figure 4B). Although a definitive analysis of the contacts observed in the Sae1/Sae2-SUMO-1-Mg·ATP complex is precluded due to the resolution limits of the structure determination (2.75 Å), magnesium coordination of the α-phosphate would be consistent with direct stabilization of the proposed transition state during adenylation and inversion of the α-phosphate.
Since adenylated intermediates were not observed in the presence of Mg·ATP, SUMO E1 was assayed for thioester formation and conjugation under temperatures similar to those used for crystallization (4°C). Proteins utilized for these assays included Sae1 with full-length Sae2 (1–640), Sae2ΔNLS (1–609) (the predicted NLS deleted), and Sae2ΔCterm (1–549) (C-terminal residues up to the E1 UbL domain deleted). Results are not shown for Sae2ΔNLS since it behaved identically to Sae1/Sae2ΔCterm in all in vitro assays. Sae1/Sae2 and Sae1/Sae2ΔCterm form E1-thioester SUMO adducts with similar ATP dependencies, and both transfer SUMO to an E2-thioester as evidenced by SUMO conjugation reaction products (Figure 4C; see Materials and methods), suggesting that the C-terminal 89 amino acids are dispensable for in vitro SUMO E1 activity and that SUMO E1 can form E1- and E2-thioester intermediates under similar temperature conditions used to generate the Sae1/Sae2-SUMO-1-Mg·ATP structure.
Two factors might explain the absence of adenylated SUMO intermediates in the crystal lattice. First, reaction products may diffuse more slowly in the lattice, favoring reverse reaction and reconstitution of ATP. We do not favor this model since the active site is exposed to solvent in the lattice and no structural evidence was observed for adenylation intermediates in either simulated annealing omit maps or difference maps (Figure 4B). A second model suggests that additional conformational changes are required for adenylation to occur, and that adenylation might be coupled to changes in the orientation of the catalytic Cys and/or E1 UbL domains. Consistent with our observations, attempts to generate adenylated complexes between the Nedd8 E1 and Nedd8 also failed to produce the desired reaction products (Walden et al, 2003a), suggesting that both Nedd8 and SUMO E1 enzymes may entail a more complex reaction mechanism for adenylation than was observed for MoeB-MoaD (Lake et al, 2001).
In vitro and in vivo requirements for C-terminal Sae2 domains
Human Sae2 and yeast Uba2 share 31% sequence identity throughout the adenylation, catalytic Cys, UbL, and NLS domains (Figure 5). To assess the importance of these domains in vivo and in vitro, we constructed mutant alleles for yeast UBA2 and human SAE2 to generate the C-terminal deletion mutants ΔNLS, ΔCterm, and ΔUbL (Figure 5A). Yeast mutants were placed into a centromeric plasmid under control of the endogenous UBA2 promoter and used to complement a S. cerevisiae Δuba2 null strain (Johnson et al, 1997; see Materials and methods) (Figure 5B). Smt3 conjugation patterns were analyzed to evaluate if C-terminal deletions altered Smt3 conjugate patterns in viable strains (Figure 5C). Expression of a yUBA2ΔNLS mutant under the control of a strong promoter was previously reported (Dohmen et al, 1995). While viable, yUba2ΔNLS mislocalized from the nucleus to the cytoplasm, suggesting the presence of a bona fide C-terminal nuclear targeting sequence. Yeast strains containing yUBA2ΔNLS under control of the natural UBA2 promoter were also viable, but exhibited a slightly altered Smt3 conjugation pattern (Figure 5C). Deletion of the C-terminal domain (1–554) also resulted in viable yeast colonies and slightly altered Smt3 conjugation patterns when compared to wild type.
Figure 5.
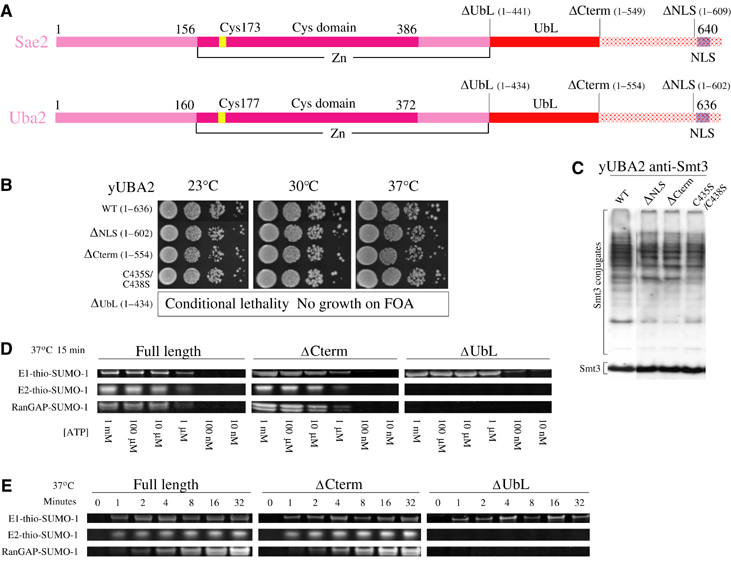
Biochemical and genetic analysis of human Sae2 and yeast Uba2. (A) Schematic representation of human Sae2 and yeast Uba2 subunits indicating domain boundaries between the adenylation (pink), catalytic Cys (magenta), UbL (red), C-terminal (hashed red), and nuclear localization domains (blue hash). Amino-acid numbering is indicated above schematics for yeast and human proteins. (B) Serial dilutions of S. cerevisiae Δuba2 cultures bearing indicated UBA2 alleles were spotted on YPD agar and tested for growth at 23°C (left panel), 30°C (middle panel), and 37°C (right panel). (C) Smt3 conjugation patterns for the strains in panel C as analyzed by SDS–PAGE and Western blotting against Smt3. Conjugates and free Smt3 are indicated by labels. (D) ATP-dependent formation of E1-thioester, E2-thioester, and SUMO conjugation at 37°C after 15 min as catalyzed by Sae1 complexes with full-length Sae2 (1–640), ΔCterm Sae2 (1–549), and ΔUbL Sae2 (1–441). (E) Time course showing formation of E1-thioester, E2-thioester, and SUMO conjugation at 37°C using indicated Sae2 proteins (see Materials and methods). Labels indicate respective positions for E1-thio-SUMO, E2-thio-SUMO, and RanGAP1-SUMO.
Deletion of the E1 UbL domain from UBA2 (1–434) resulted in lethality as measured by the inability to obtain colonies after 8 days at 23, 30, or 37°C using plasmid shuffle and selection on media containing 5-FOA (Figure 5B). Two cysteine residues from the UbL domain and two cysteine residues from the crossover strand link the two elements through a zinc motif. To assess if the zinc motif was essential, Cys435 and Cys438 were substituted to serine. This mutation did not alter cell growth or significantly alter Smt3 conjugate patterns. Taken together, these data suggest that the E1 UbL domain is essential in vivo, and that association of the E1 UbL domain to the rest of Uba2 is not strictly dependent on tight association of zinc between the UbL and crossover strand (Figure 5B and C).
Analogous C-terminal deletions for the respective human Sae2 protein were assayed in vitro for E1-SUMO-thioester formation, E2-SUMO-thioester formation, or E2-dependent conjugation to RanGAP1 (Figure 5D and E). Sae1/Sae2ΔNLS and Sae1/Sae2ΔCterm exhibited no defect in vitro for these activities. Sae1/Sae2ΔUbL was able to form the E1-thioester intermediate with similar rates and ATP dependencies as wild type, indicating that the E1 UbL domain is not required for adenylation or E1-thioester formation. However, Sae1/Sae2ΔUbL was unable to transfer the E1-SUMO-thioester to E2. This defect could not be overcome by 100-fold excess E2 in the reactions (10 μM) or by stoichiometric or 10-fold excess UbL (446–549) supplied in trans (0.1–1.0 μM) (not shown). These data suggest that the E1 UbL domain must be covalently linked to E1 to facilitate SUMO transfer from the E1-SUMO-thioester to E2.
Properties and function of the UbL domain in E2 recruitment to E1
The E1 UbL domain is conserved in all E1 enzymes that transfer Ub or Ubl modifiers to E2 conjugating proteins, and biochemical activities of SUMO E1 show a strict dependency on the E1 UbL domain for thioester transfer between E1 and E2. Although sequence alignments did not reveal conserved functional surfaces between the UbL domains from SUMO, Nedd8, and Ub E1s, a complete structural model for the E1 UbL domain in the SUMO E1 enzyme enabled structure-based sequence alignment to the sequence from the Ub E1 UbL and the partial structure obtained for the Nedd8 E1 UbL domain (Figure 6A; Walden et al, 2003b).
Figure 6.
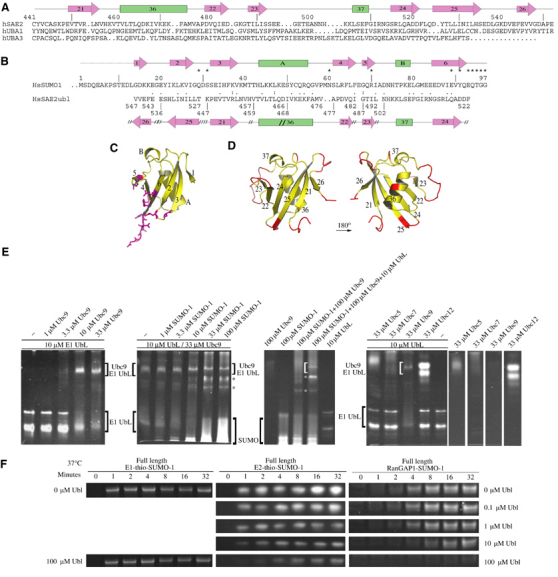
Sae2 UbL domain. (A) Structure-based sequence alignment for UbL domains from human SUMO Sae2, Ub Uba1, and Nedd8 Uba3. Sae2 amino-acid numbering and secondary structure are shown above the sequence. Strands and helices are numbered on arrows and bars, respectively. (B) Structure-based sequence alignment for SUMO-1 and the Sae2 UbL domain. Secondary structure for SUMO-1 and UbL domain is shown above and below the sequence, respectively, with discontinuity between the UbL and SUMO-1 structural alignment indicated by ( // ) and residue numbers below the alignment. Asterisks indicate SUMO-1 residues that contact Sae2 with sequence similarity ( . ) and identity ( : ) between the alignment. (C) SUMO-1 structure aligned to the UbL domain as in (D). SUMO-1 residues contacting Sae2 are colored magenta. Secondary structure labeled as in (B). (D) Opposing orientations of the Sae2 UbL domain. Yellow demarcates regions of structure that superimpose with SUMO-1 and magenta indicates insertions or large deviations from the SUMO-1 structure. Strands and helices are numbered as in (A, B). (E) Gel-shift analysis indicating direct interactions between human Ubc9 and the Sae2 UbL domain. Both Ubc9 and Sae2 UbL domain show multiple bands by native gel electrophoresis, thus complexes between Ubc9 and UbL manifest as multiple bands within the gel. Positions of the respective proteins are indicated by brackets and labels with concentrations of the various components indicated at the top of each panel. Asterisks indicate the positions of additional bands described in the text. The bottom of the respective gels corresponds to the positive electrode. Ubc9 cannot be visualized directly on the same gel as it migrates toward the negative electrode. The first panel suggests a stoichiometric interaction between Ubc9 and the UbL domain. The second panel suggests that the Ubc9-UbL interaction cannot be competed for by addition of exogenous SUMO-1 concentrations. The third panel includes controls for panel 2. The fourth and the remaining panels indicate that gel shift of the SUMO E1 UbL domain is specific to Ubc9. (F) Time course for formation of E1-thioester, E2-thioester, and SUMO conjugation at 37°C using Sae2 (1–640) with increasing concentrations of exogenous Sae2 UbL (446–549) (see Materials and methods). Labels indicate positions for E1-thio-SUMO, E2-thio-SUMO, and RanGAP1-SUMO.
Alignment of SUMO-1 and the Sae2 UbL domain revealed few conserved residues but significant structural overlap that included some interesting differences in the polypeptide topology within the UbL fold (Figure 6B). Several loops and insertions in the UbL domain that are not observed in SUMO-1 create surfaces unique to the UbL domain, and while not conserved at the level of amino-acid identity, insertions occur at similar positions in both Ub and Nedd8 UbL domains (Figure 6A and D). One possible role for the E1 UbL domain might be to mimic or compete with SUMO-1 after adenylation or thioester transfer in order to displace SUMO from its original binding site, although two points argue against this. First, sequence conservation between SUMO and the UbL domain does not identify a conserved surface (Figure 6). Second, Sae2ΔUbL was able to form E1-thioester intermediates with similar rates and ATP dependencies as wild type, indicating that the UbL domain is not required for E1 SUMO activation or for thioester transfer to the E1 catalytic Cys domain.
SUMO's negative charge potential has been noted as a feature distinguishing it from the electrostatic potentials observed for Ub or other Ubl modifiers (Bayer et al, 1998). It has also been proposed that charge complementarity may facilitate interaction between SUMO and the positively charged E2 conjugating protein Ubc9 (Liu et al, 1999; Bencsath et al, 2002; Tatham et al, 2003). The electrostatic properties of SUMO and the E1 UbL domain reveal that both are acidic, although if structurally aligned, each appears to focus negative charge potential onto different surfaces (not shown), suggesting that if SUMO-1 and UbL interact with Ubc9 through charge complementarity, they are likely to do so in different orientations.
To test whether the Sae2 UbL interacts directly with Ubc9, we conducted gel-shift assays with SUMO E1 UbL domains consisting of residues 442–549 or 446–549 (Figure 6E). In both instances, increasing concentrations of Ubc9 shifted the Sae2 UbL domain to a retarded position in a concentration-dependent manner consistent with the formation of a stoichiometric Sae2 UbL-Ubc9 complex (see Materials and methods). The Ubc9-UbL interaction is not disrupted by increasing SUMO-1 concentrations, and although a unique band appears in the SUMO-1 titration (indicated by asterisks in Figure 6E), the Ubc9-UbL complex does not diminish in intensity, even in the presence of 10-fold excess SUMO-1. These data suggest that Ubc9-UbL interactions do not substantially overlap with previously characterized binding surfaces between Ubc9 and SUMO (Liu et al, 1999; Bencsath et al, 2002; Tatham et al, 2003). Ubc9-UbL interactions appear specific insomuch as the SUMO E1 UbL domain is shifted by Ubc9, but not by excess Ubc5, Ubc7, or Ubc12. To confirm that the E1 UbL domain competes specifically for E2 recruitment by E1, we tested whether exogenous UbL domain affected E1-thioester formation, E2-thioester formation, or conjugation. Increasing UbL concentrations adversely affected E2-thioester transfer and E2-mediated conjugation while E1-SUMO-1-thioester formation remained unaffected (Figure 6F).
Comparison of E1 activating enzymes for Ub, SUMO, and Nedd8
Structural comparison of the SUMO and Nedd8 E1 enzymes reveals similarities with respect to the global positions for the adenylation, UbL, and catalytic Cys domains (Figure 7). However, interactions with SUMO-1 are much less extensive than observed for the Nedd8-Nedd8 E1 complex. This is due, in large part, to the absence in SUMO E1 of the 200-amino-acid insertion observed in the Nedd8 APPBP1 subunit that interacts directly with Nedd8 across a large surface (Figure 7D–F). This APPBP1 domain insertion is absent and has no apparent structural or sequence correlate in either SUMO or Ub E1 enzymes, respectively.
Figure 7.
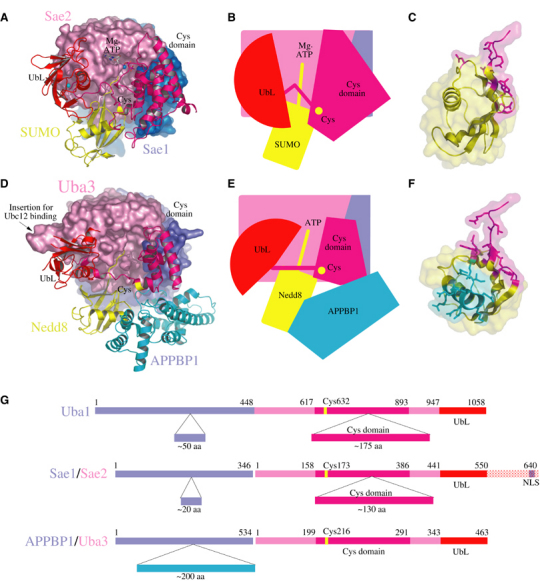
Comparison between SUMO, Nedd8, and Ub E1 enzymes. (A) Ribbon and surface representations for the Sae1/Sae2-SUMO-1-Mg·ATP complex. (B) Cartoon representation of SUMO E1 with Mg·ATP, catalytic cysteine, crossover loop, UbL and Cys domains labeled. (C) Side view of SUMO-1 in ribbon and partial surface representation showing the contact surfaces with Sae2 (magenta). (D) APPBP1/Uba3-Nedd8 complex with Uba3 colored pink and the catalytic Cys and UbL domains in ribbon representation colored magenta and red, respectively. Nedd8 is colored yellow. The APPBP1 domain is colored blue in surface representation with the APPBP1 insertion in ribbon representation (cyan). The Ubc12 binding site is labeled. (E) Cartoon of Nedd8 E1 indicating domain positions and contacts between the APPBP1 insertion and Nedd8. (F) Nedd8 side view in ribbon and surface representation showing contact surfaces with Uba3 (magenta) and APPBP1 (cyan). SUMO-1 in (C) and Nedd8 in (F) are shown as observed in the respective complexes if E1 subunits are aligned. (G) Schematic domain representation observed in human Ub, SUMO, and Nedd8 E1 activating enzymes with domain insertions indicated below each schematic. Numbering and color coding represent domain boundaries.
Previous biochemical and structural characterization of the Nedd8 E1 enzyme revealed structural elements required for efficient recruitment of the Nedd8 E2 (Ubc12) that differentiate it from the mechanisms employed for E2 recruitment by the SUMO E1. Deletion analysis of the Nedd8 E1 UbL domain diminished but did not eliminate the ability of Ubc12 to interact with Nedd8 E1 (Walden et al, 2003b). Subsequent studies reported the importance of a unique Ubc12 N-terminal extension in interactions with an insertion in the Uba3 subunit (Figure 7D; Huang et al, 2004a). Moreover, the Uba3 insertion responsible for direct interaction with the Ubc12 N-terminal element is absent in other E1s (Huang et al, 2004a). While partially disordered, the Nedd8 UbL domain is rotated nearly 90° relative to the SUMO E1 UbL, suggesting that precise orientation of the catalytic Cys and UbL domains might be less important than their global positions over the adenylation active site. Interestingly, conformational changes were also observed for the catalytic Cys and UbL domains in response to Nedd8 interaction when APPBP1/Uba3 and APPBP1/Uba3-Nedd8 were compared (Walden et al, 2003a).
Structure-based sequence and domain alignments between the E1 UbL and catalytic Cys domains can be better accomplished based on both SUMO and Nedd8 E1 structures, also enabling alignment of the Ub E1 sequence to these elements. The overall architecture of the SUMO E1 more closely resembles that of the Ub E1 with respect to the overall length and composition of the adenylation domains (corresponding to Sae1/Sae2), the catalytic Cys domain, and the C-terminal UbL domain (Figure 7G). As such, it appears that the functional significance of individual domains as observed in the SUMO E1 structure might provide additional insights into those elements required for Ub activation and E2 transfer by the Ub E1.
Conclusions
E1s are thought to have evolved from the bacterial enzymes MoeB and ThiF, which catalyze a similar adenylation reaction on respective Ubl proteins. E1s that catalyze protein conjugation have a more complex structure with additional domains that catalyze E1- and E2-thioester transfer following adenylation. The fact that a substrate acyl-adenylate intermediate has only been observed for the MoeB substrate MoaD, and that conformational changes in the catalytic Cys and UbL domains, which are missing in MoeB, are observed in both SUMO E1 and Nedd8 E1 complexes suggest that eukaryotic E1 enzymes may coordinate adenylation and thioester transfer through domain movements that facilitate these reactions. Interestingly, E2-thioester transfer is not required for efficient adenylation and E1-thioester transfer insomuch as the SUMO E1 ΔUbL mutant is competent for E1-SUMO-thioester formation but deficient in transfer of SUMO to E2.
The SUMO E1 UbL domain is essential for E2 recruitment to E1 since the UbL domain is sufficient for E2 interaction, essential in vivo, and essential in vitro for thioester transfer between E1 and E2. This observation is unique to the SUMO E1 insomuch as the Nedd8 E1 UbL domain contributes only partially to Nedd8 E2 recruitment and other elements within the Nedd8 E1 are involved in this interaction (see above). As such, it appears that activation mechanisms, including Ubl modifier interactions, conformational changes, and E2 recruitment, may be specific to each conjugation pathway. Despite these differences, it appears that a common feature of these enzymes will include potential coupling of these reactions through concerted movements of respective domains above the adenylation active site, changes that may also be required for productive interaction and thioester transfer to the cognate E2.
Materials and methods
Cloning, expression, and protein purification
Human SAE1 and SAE2 were amplified by PCR from cDNA obtained from testes (Clontech). SAE1 was cloned into pET-11c to encode a native polypeptide and SAE2 was cloned into pET-28b to encode an N-terminal thrombin-cleavable hexahistidine fusion protein. Plasmids were cotransformed into Escherichia coli BL21(DE3) Codon Plus RIL (Stratagene). Mutant SAE2 alleles were generated by QuikChange mutagenesis (Stratagene). Human SUMO-1 was isolated by PCR as described and full-length human SUMO-1 (1–101) was fused to a C-terminal hexahistidine tag by excluding the native stop codon. Cultures (10 l) were fermented at 37°C to A600 of 3 before 0.75 mM IPTG induction for 4–6 h at 30°C. Cells were harvested and suspended in 50 mM Tris–HCl (pH 8.0), 20% w/v sucrose, 350 mM NaCl, 20 mM imidazole, 0.1% IGEPAL, 1 mM PMSF, 1 mM β-mercaptoethanol (BME), and 10 μg/ml DNAse prior to sonication and removal of insoluble material by centrifugation.
Sae1/Sae2 was purified by metal-affinity chromatography (Qiagen), gel filtration (Superdex200; Pharmacia), and anion exchange chromatography (MonoQ; Pharmacia). SUMO-1 was purified by metal-affinity chromatography, processed with Ulp1(1–403)p to generate mature SUMO-1 (1–97) (Mossessova and Lima, 2000; Reverter and Lima, 2004), and purified by gel filtration (Superdex75) and anion exchange (MonoQ). Sae1/Sae2 was concentrated to 10–15 mg/ml in 87 mM NaCl, 10 mM Tris–HCl pH 8.0, and 1 mM DTT, and SUMO-1 was concentrated to 17 mg/ml in 180 mM NaCl, 20 mM Tris–HCl pH 8.0, and 1 mM DTT before freezing in liquid nitrogen and storage at −80°C.
Crystallographic analysis
Crystals were obtained for wild-type Sae1/Sae2 and Sae1/Sae2(Cys173Ala) at 18 and 4°C, but diffraction quality crystals were only obtained at 4°C by hanging drop vapor diffusion against a well solution containing 0.1 M sodium acetate (pH 5.6), 10 mM ATP, 10–20% polyethylene glycerol 4000, and 0.1–1.0 M ammonium acetate. Crystals were transferred in a stepwise fashion and stabilized for 5–18 h prior to cryopreservation in crystallization solutions that contained 15% glycerol and 2% additional PEG. Magnesium sulfate was added to this solution to a final concentration of 20 mM to obtain Sae1/Sae2-Mg·ATP and Sae1/Sae2-SUMO-1-Mg·ATP crystals. Data were processed with DENZO, SCALEPACK (Otwinowski and Minor, 1997), and CCP4 (Collaborative Computational Project, 1994) (Table I). Phases (2.6 Å) were calculated with SOLVE and RESOLVE (Terwilliger and Berendzen, 1999) using 12 mercury positions and two-fold noncrystallographic averaging. The ASU contained two Sae1/Sae2 heterodimers. A complete atomic model for Sae1/Sae2 was built using O (Jones et al, 1991) and refined using CNS (Brunger et al, 1998). Model details, refinement, and content of the ASU are provided in the text and in Table I.
Mutagenesis, complementation, and biochemical assays
Point mutations in the C-terminal domain of SAE2 were engineered into pET28b-SAE2 and pIS3-UBA2 (Johnson et al, 1997) using PCR-based mutagenesis (QuikChange, Stratagene). Proteins were expressed and purified as described above. E1-thioester formation was assayed in reactions containing 1 μM E1, 5 mM MgCl2, 20 mM Hepes pH 7.5, 50 mM NaCl, and 2 μM mature SUMO (1–97). Reactions were initiated by addition of 1 mM ATP unless otherwise indicated. E2-thioester formation was assayed in reactions containing 100 nM E1, 1 μM E2, 5 mM MgCl2, 20 mM Hepes pH 7.5, 50 mM NaCl, and 2 μM mature SUMO. E2 was used at a final concentration of 100 nM in reactions presented in Figure 6F and detected by Western blot using an antibody against Ubc9 (Boston Biochem). E1- and E2-mediated SUMO-1 conjugation to RanGAP1(420–589)p was assessed using reactions containing 100 nM E1, 100 nM Ubc9 (E2), 5 mM MgCl2, 20 mM Hepes pH 7.5, 50 mM NaCl, 2 μM RanGAP1, 2 μM mature SUMO, and 1 mM ATP. Samples were denatured in nonreducing SDS–PAGE buffer and analyzed by SDS–PAGE and Sypro staining (Bio-Rad). Mutations were confirmed by DNA sequencing. Gel-shift experiments were conducted under native conditions that included 50 mM NaCl and 20 mM Tris–HCl pH 8.0 using a Phast-gel apparatus and related gel supplies (Pharmacia). Ubc5, Ubc7, and Ubc12 were purchased from Boston Biochemical. Protein bands were visualized by staining with Sypro (Bio-Rad).
Supplementary Material
Supplementary Figure
Supplementary Figure
Acknowledgments
We thank the staff of the APS SGX-CAT and NSLS X4A. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38. Use of the SGX Collaborative Access Team (SGX-CAT) beamline facilities at Sector 31 of the Advanced Photon Source was provided by Structural GenomiX Inc., who constructed and operates the facility. We also thank Xin Zhou and Mike Lake for technical help during initial crystallization trials. We would like to thank Erica Johnson for providing us with strains and plasmids used for yeast complementation assays. LML and CDL were supported in part by a grant from the National Institutes of Health (GM65872). CDL acknowledges additional support from the Rita Allen Foundation.
References
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280: 275–286 [DOI] [PubMed] [Google Scholar]
- Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA (2002) Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J Biol Chem 277: 47938–47945 [DOI] [PubMed] [Google Scholar]
- Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D (2004) A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem 279: 27233–27238 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL molecular graphics system, DeLano Scientific, San Carlos, CA, USA http://www.pymol.org [Google Scholar]
- Desterro JM, Rodriguez MS, Kemp GD, Hay RT (1999) Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem 274: 10618–10624 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem 270: 18099–18109 [DOI] [PubMed] [Google Scholar]
- Evans SV (1993) SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph 11: 127–138 [DOI] [PubMed] [Google Scholar]
- Hayward S, Berendsen HJ (1998) Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30: 144–154 [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (1996) Ubiquitin-dependent protein degradation. Annu Rev Genet 30: 405–439 [DOI] [PubMed] [Google Scholar]
- Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA (2004a) A unique E1–E2 interaction required for optimal conjugation of the ubiquitin-like protein Nedd8. Nat Struct Mol Biol 11: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Walden H, Duda D, Schulman BA (2004b) Ubiquitin-like protein activation. Oncogene 23: 1958–1971 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272: 26799–26802 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G (1999) Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol 147: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J 16: 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H (2001) Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 414: 325–329 [DOI] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M (1999) Substrate targeting in the ubiquitin system. Cell 97: 427–430 [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398: 246–251 [DOI] [PubMed] [Google Scholar]
- Liu Q, Jin C, Liao X, Shen Z, Chen DJ, Chen Y (1999) The binding interface between an E2 (UBC9) and a ubiquitin homologue (UBL1). J Biol Chem 274: 16979–16987 [DOI] [PubMed] [Google Scholar]
- Melchior F (2000) SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol 16: 591–626 [DOI] [PubMed] [Google Scholar]
- Melchior F, Schergaut M, Pichler A (2003) SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci 28: 612–618 [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5: 865–876 [DOI] [PubMed] [Google Scholar]
- Muller S, Hoege C, Pyrowolakis G, Jentsch S (2001) SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol 2: 202–210 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H (1999) In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun 254: 693–698 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2004) A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure 12: 1519–1531 [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S (1995) Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373: 78–81 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Iwase M, Konishi M, Tanaka M, Toh-e A, Kikuchi Y (1999) Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem Biophys Res Commun 259: 582–587 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y (1999) Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol 19: 8660–8672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Kim S, Yu B, Jaffray E, Song J, Zheng J, Rodriguez MS, Hay RT, Chen Y (2003) Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry 42: 9959–9969 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL Jr, Holton JM, Schulman BA (2003a) The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell 12: 1427–1437 [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Schulman BA (2003b) Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 422: 330–334 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure
Supplementary Figure


