Abstract
To improve radiation therapy-induced quality of life impairments for prostate cancer patients, the development of radio-protectors is needed. Our previous work has demonstrated that MnTE-2-PyP significantly protects urogenital tissues from radiation-induced damage. So, in order for MnTE-2-PyP to be used clinically as a radio-protector, it is fully necessary to explore the effect of MnTE-2-PyP on human prostate cancer progression. MnTE-2-PyP inhibited prostate cancer growth in the presence and absence of radiation and also inhibited prostate cancer migration and invasion. MnTE-2-PyP altered p300 DNA binding, which resulted in the inhibition of HIF-1β and CREB signaling pathways. Accordingly, we also found that MnTE-2-PyP reduced the expression of three genes regulated by HIF-1β and/or CREB: TGF-β2, FGF-1 and PAI-1. Specifically, MnTE-2-PyP decreased p300 complex binding to a specific HRE motif within the PAI-1 gene promoter region, suppressed H3K9 acetylation, and consequently, repressed PAI-1 expression. Mechanistically, less p300 transcriptional complex binding is not due to the reduction of binding between p300 and HIF-1/CREB transcription factors, but through inhibiting the binding of HIF-1/CREB transcription factors to DNA. Our data provide an in depth mechanism by which MnTE-2-PyP reduces prostate cancer growth and metastasis, which validates the clinical use of MnTE-2-PyP as a radio-protector to enhance treatment outcomes in prostate cancer radiotherapy.
Keywords: MnTE-2-PyP, p300, PAI-1, HIF-1β, CREB
INTRODUCTION
Of the individuals diagnosed with prostate cancer, roughly half will undergo radiotherapy as treatment [1]. Although radiation effectively kills most prostate tumor cells, cancer cells that survive radiation therapy become more aggressive and are difficult to treat. Surviving prostate cancer cells after irradiation are more oxidatively stressed [2]. High levels of reactive oxygen species (ROS) drive both tumor survival and migration. Thus, the redox environment in the prostate tumor plays a crucial role in both the survival and ability of the tumor to metastasize.
Accordingly, emerging evidence indicates that oxidative stress may trigger plasminogen activator inhibitor-1 (PAI-1) activation [3]. PAI-1, a major physiologic inhibitor of urokinase-type and tissue-type plasminogen activators, has been reported to promote cancer progression. High tumor PAI-1 protein expression is associated with poor survival in several forms of cancer and is a strong independent prognostic factor in prostate cancer, with elevated levels forecasting shorter recurrence-free and overall survival [4, 5]. Although the PAI-1 activity can be modified at the protein level, the regulation of PAI-1 is achieved mainly by altering the rate of PAI-1 gene expression [6]. Several transcription factors (e.g. CREB, HIF-1α) and co-activator proteins (e.g. p300) of PAI-1 are also activated by fluctuations of the intracellular ROS level [7–9]. Furthermore, recent evidence suggests that oxidative stress may upregulate gene expression by enhancing histone acetyltransferase (HAT) activity of p300. In fact, ROS can enhance complex formation between p300 and transcription factors to facilitate activator-dependent transcription via interactions with basal transcriptional machinery [9]. Thus, alteration in ROS level may regulate gene expression through modulation of transcription factors and co-activator proteins in a redox sensitive fashion, which ultimately affect cancer progression.
MnTE-2-PyP (chemical name: Manganese (III) Meso-Tetrakis-(N-Ethylpyridinium-2-yl), which has 5 plus charges (this is omitted through the manuscript for simplicity), scavenges ROS, including superoxide [10]. We have previously published that MnTE-2-PyP protects the urogenital region from radiation damage. In particular, MnTE-2-PyP protects from erectile dysfunction and testicular atrophy after radiation exposure to the urogenital region [11]. However, there have been no in depth studies conducted to determine the effect of MnTE-2-PyP on human prostate tumors.
The current study was undertaken to determine the effects of MnTE-2-PyP on human prostate cancer progression. This is the first study to demonstrate that MnTE-2-PyP inhibits human prostate cancer cell progression with radiation. We then focused on the mechanisms by which MnTE-2-PyP inhibits prostate cancer. We determined MnTE-2-PyP alters p300 transcriptional complex binding to DNA and inhibits acetylation of histones. Additionally, MnTE-2-PyP inhibits CREB and HIF-1β signaling and reduces PAI-1 expression. These studies are crucial in determining how MnTE-2-PyP can be better used as a radio-protector and radio-sensitizer for the treatment of prostate cancer radiotherapy.
MATERIALS AND METHODS
Cell lines and growth
Three human prostate cancer cell lines were used: PC3, DU145, and LNCaP. All three were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). PC3 and LNCaP cells were cultured in RPMI-1640 media containing L-glutamine, 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. DU145 cells were cultured in Minimum Essential Medium Eagle containing Earle’s salts, L-Glutamine, 15% FBS and 1% penicillin/streptomycin. All cell lines were maintained at 37°C and in 95% air and 5% CO2.
Clonogenic assay
PC3, LNCaP, and DU145 cells were treated with MnTE-2-PyP (0–30 μM) overnight prior to irradiation. PC3 and DU145 cells were exposed to 5 Gy of radiation and LNCaP cells were exposed to 2 Gy. One hour after irradiation, the cells were harvested and re-plated in triplicate in six well plates. The plates were incubated at 37°C for up to 2 weeks while clones formed. The cells were fixed in 4% formalin and stained with 0.5% crystal violet dissolved in 25% methanol. The plates were rinsed with ddH20, dried and counted. Plating efficiency for control (0 Gy/0 μm MnTE-2-PyP) was calculated as the ratio of clones counted to the total number of cells plated. The control was set to 100 and all experimental groups were normalized to control.
Soft agar assay
All cells were treated with MnTE-2-PyP (0–30 μM) overnight prior to irradiation. PC3, DU145, and LNCaP cells were exposed to 2 Gy of radiation. One hour after irradiation, the cells were counted and then suspended in 1.5% DNA grade agarose + media which were subsequently plated atop a 6.0% agarose + media base in a 6 well plastic plate. The plates were incubated at 37°C while clones formed. The plates were incubated overnight in 0.5% crystal violet dissolved in 25% methanol and clones were counted. Plating efficiency was calculated using the formula stated previously.
Migration assay
Cells were incubated and treated with 0 or 30 μM MnTE-2-PyP overnight. Cells were trypsinized, washed, counted and re-suspended in media without FBS. The serum-starved cells were re-plated on a PET track-edged membrane, pore size 8.0 μm (Fisher Scientific, Pittsburg, PA, USA) set inside the well of a six well plate at a concentration of 1.0 × 106 cells/well. The membrane was situated between a top chamber containing media without serum and a bottom chamber filled with serum media. Cells were allowed to migrate across the membrane for 48 hours. The cells and media were isolated from the top and bottom chambers and enumerated.
Invasion assay
Real-time impedance-based cell invasion assays were carried out using the xCELLigence system (Roche Applied Science, Indianapolis, IN). Cells were serum-starved, trypsinized, washed with PBS, and resuspended in serum-free medium. The top chamber wells of the E 16 plates were coated with Matrigel (1:20; BD Biosciences, San Jose, CA) and cells incubated for 4 h. The bottom chamber wells contained medium with 10% FBS, used as a chemo-attractant, and the wells with no serum were used as the negative controls. Roughly 40,000 cells were subsequently seeded onto the Matrigel, and cell invasion was measured.
Histone acetyltransferase (HAT) activity assay
Nuclei were isolated from PC3 cells 1 hour post-radiation using the NXTRACT CelLytic NuCLEAR Extraction Kit (Sigma, St. Louis, MO). Protein concentrations of the nuclear extracts were determined by the Bradford method. Equal amounts of nuclear extract protein (~5 μg) were analyzed for HAT activity using the EpiQuick Activity/Inhibition Assay Kit (Epigetek, Farmingdale, NY) according to manufacturer’s suggestions.
P300 activity assay
The Histone Acetyltransferase Assay Kit (Active Motif, Carlsbad, CA) was used according to manufacturer’s suggestions.
CREB activity assay
Nuclei were isolated from PC3 cells 1–24 hours post-radiation using the NXTRACT CelLytic NuCLEAR Extraction Kit (Sigma). Protein concentrations of the nuclear extracts were determined by the Bradford method. Equal amounts of nuclear extract protein (~5 μg) were analyzed for CREB activity using the TransAM CREB Transcription Factor Assay Kits (Active Motif) according to manufacturer’s suggestions.
Apoptosis assay
PC3 cells were treated with MnTE-2-PyP overnight prior to irradiation of 20 Gy. Cells were harvested 48 hours after irradiation, fixed with 4% paraformaldehyde for 30 minutes. The cells were TUNEL stained using the In Situ Cell Death Detection Kit, Fluorescein (Roche, Mannheim, Germany) and then analyzed by fluorescence-activated cell sorter (FACS) (FACScalibur; Becton Dickinson Immunometry Systems, San Jose, CA). Fluorescence data, which indicated TUNEL positive cells, were collected on a log scale, with green fluorescence measured at 530 nm. Data from 10,000 events (cells) were collected and analyzed with CellQuest software (Becton Dickinson Immunometry Systems).
p300 Chromatin Immuno-precipitation (ChIP) assay followed by Next-gen sequencing (ChIP-Seq) and analysis
PC3 cells were treated with or without 30μm MnTE-2-PyP overnight, and then irradiated with 20 Gy. One hour post-irradiation, cells were fixed, lysed (Active Motif ChIP-IT Express #53008) and sheared for Ion Proton ChIP-seq. DNA-protein complexes were immunoprecipitated with an anti-p300 ChIP-grade antibody (Abcam #54984), and the genomic DNA purified (Invitrogen Magnify ChIP Kit #49-2024). The isolated genomic DNA was processed for next-gen library construction using the Ion Plus Fragment Library Kit #4471252. Briefly, libraries were constructed and analyzed for size and quantity using the High Sensitivity Bioanalyzer Kit (Agilent Technologies #5067-4626). The libraries were then sequenced with the Life Technologies Ion Proton, at the NJH Genomics Facility. Total input control libraries were used to normalize ChIP-seq signals, enabling peak comparison in the ChIPed samples. ChIP-sequencing reads were aligned to the human reference genome (hg109) using Ion Torrent Suite software. Peak-calling from aligned reads was performed using the Model-based Analysis for ChIP-Sequencing (MACS2) software [12], comparing p300 IPs to their respective total input controls. Peak significance was determined with a false discovery rate cutoff of 0.001. Peaks found by ChIP-seq were associated with the nearest gene by genomic location, using both the RefSeq, refGene, and UCSC known gene models. The gene list was narrowed by sorting for genes with peak Q-value greater than 10. Gene lists were converted to Ensembl annotation using Biomart and analyzed with GATHER Duke Bioinformatics system [13]. All data is representative of at least two biologically independent ChIP experiments.
ChIP assay
ChIP assays were performed as described previously [14]. Briefly, after pretreatment with MnTE-2-PyP (30 μM) overnight, PC3 cells were exposed to 20 Gy of radiation. One hour later, 6.0 × 106 cells were fixed with 1% of formaldehyde. Genomic DNA was sheared to lengths ranging from 200 to 1000 bp with a Sonic Dismembrator (Fisher Scientific): Ampl 80%, 3 seconds on, 10 seconds off, for 10 cycles. One percent of the cell extract was taken as “input”, and the rest of the extract was incubated with either anti-p300 (Santa Cruz), anti-H3K9ac (Cell Signaling), or control IgG overnight at 4°C, followed by precipitation with protein A agarose beads. The immunoprecipitates were sequentially washed with a low salt buffer, a high salt buffer, a LiCl buffer, and with TE buffer. The DNA-protein complex was eluted and proteins were then digested with proteinase K. The DNA was detected by real-time quantitative PCR analysis and the data obtained by real-time PCR for each specific antibody were normalized to IgG control and plotted as percent input. ChIP primers for five putative HREs in the human PAI-1 gene promoter were: site 1 (−158 to −151 bp) (forward, 5′-GCACACACACACACACACACAT-3′ and reverse, 5′-TGTGGGCAGGAAATAGATGAAC-3′); site 2 (−194 to −187 bp) (forward, 5′-CAGAAAGGTCAAGGGAGGTTC-3′ and reverse, 5′-CTGCTCTGTGTGTGTACGTGTG-3′); site3 (−453 to −446 bp) (forward, 5′-TCTTTCCTGGAGGTGGTCC-3′ and reverse, 5′-TTTGGCGAACCAGGTCAC-3′); site 4 (−566 to −559 bp) (forward, 5′-GGGATGAGGGAAAGACCAAG-3′ and reverse, 5′-CAGCCACGTGATTGTCTAGGT-3′); site 5 (−681 to −674 bp) (forward, 5′-GTTGACACAAGAGAGCCCTCA-3′ and reverse, 5′-TTGGTCTTTCCCTCATCCCT-3′). ChIP primers for human PAI-1 gene promoter acetylation testing were: (proximal, −0.1 to −0.3 kb) (forward, 5′-GGCAGAGGGCAGAAAGGTCA-3′ and reverse, 5′-TGAACAGCCAGCGGGTCC-3′); (distal, −3.6 to −3.8 kb) (forward, 5′-TGCCCAAGGATGGAATGTC-3′ and reverse, 5′-TAGGAGGATCGCTTGAGCCT-3′).
Real-time Quantitative PCR
RNA was isolated from PC3 cells with the ZR RNA MicroPrep RNA isolation kit (Zymo Research) according to the manufacturer’s protocol. For quantitative analysis of mRNA expression, comparative real-time PCR was performed with the use of Power SYBR Green RNA-to-CT 1 step kit (Applied Biosystems). Quantitative PCR was performed under the following conditions: 48 °C for 30 min, 95 °C for 10 min, then 40 cycles with 95 °C for 15 sec and 60°C for 1 min. Following the final cycle, the melt curves of the PCR products were determined to verify the integrity of the PCR products. The sequences for the amplification of human PAI-1 were: 5′-ACCGCAACGTGGTTTTCTCA-3′ (forward) and 5′-TTGAATCCCATAGCTGCTTGAAT-3′ (reverse). The sequences for the amplification of human TGFβ2 were: 5′-CAGCACACTCGATATGGACCA-3′ (forward) and 5′-CCTCGGGCTCAGGATAGTCT-3′ (reverse). The sequences for the amplification of human FGF1 were: 5′-ACACCGACGGGCTTTTATACG-3′ (forward) and 5′-CCCATTCTTCTTGAGGCCAAC-3′ (reverse). 18S rRNA was used as an external endogenous standard. The forward, 5′-CGGCTACATCCAAGGAA-3′, and reverse, 5′-GCTGGAATTACCGCGGCT-3′.
Co-immunoprecipitation (Co-IP) assay
Co-immunoprecipitation was performed as previously described [15]. Briefly, the nuclear fraction was isolated from PC3 cells with the CelLytic NuCLEAR Extraction Kit (Sigma-Aldrich), and 50 μg of nuclear protein extracts were then precleared by incubation with 30 μl of a 50% slurry of protein A/G PLUS-Agarose beads (Santa Cruz) in a total volume of 250 μl, reconstituted with co-precipitation buffer (0.1% Triton X-100, 100 mM NaCl, 15 mM EGTA, PMSF, and a proteinase inhibitor cocktail) at 4°C for 1 h with rotation. After centrifugation the supernatants were removed and incubated with 2 μg p300 Ab (Santa Cruz) at 4°C overnight with rotation. Protein A/G PLUS-Agarose beads (20 μl) were added and incubated at room temperature for 1 h with rotation. The beads were washed with wash buffer (0.1% Triton X-100, 50 mM Tris-Cl (pH 7.4), 300 mM NaCl, 5 mM EDTA) three times. SDS-PAGE sample loading buffer was added to the beads, and then boiled. Proteins were separated by SDS-PAGE, followed by western blotting with anti-p300 (Santa Cruz), anti-CREB (Millipore), anti-HIF-1α (BD Biosciences), or anti-HIF-1β/ARNT (Cell Signaling), and the protein bands were visualized by chemiluminescence (Thermo Scientific).
Western blot
PC3 cells were harvested and nuclear extracts were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membranes (Life Technologies). After incubation with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% Tween 20) for 1 hour, the membrane was washed once with TBST and incubated with antibodies against CREB (Millipore) (1:10000), HIF-1α (BD Biosciences) (1:500), or HIF-1β/ARNT (Cell Signaling) (1:1000) at room temperature for 1 h, or p300 (Santa Cruz) (1:200) at 4°C overnight. Membranes were washed and incubated with a 1:10000 dilution of HRP-conjugated goat anti-rabbit antibody (Life Technologies) for 1 hour. Blots were washed with TBST and developed with an ECL detection system (Thermo Scientific).
Electrophoretic mobility shift assay (EMSA)
PC3 cells were pretreated with 30 μM MnTE-2-PyP overnight, and then exposed to 20 Gy of radiation. The nuclear proteins were isolated with the CelLytic NuCLEAR Extraction Kit (Sigma-Aldrich), and its concentration was quantified with using Bradford reagent (Amresco). The following oligonucleotide corresponding the sequence harboring the HRE of human PAI-1 gene was synthesized: 5′-CTGACACTGCACGTCAGAAGGACA-3′ (the element present in the promoter region is underlined). The oligonucleotides were end-labelled using the Biotin 3′ End DNA Labelling kit (Thermo Scientific). The EMSAs were performed using the Light Shift Chemiluminescent EMSA kit (Thermo Scientific). A biotin-labelled oligonucleotide (20 fmol) was incubated with 10 μg of nuclear protein extract for 20 minutes at room temperature in binding buffer. The binding complexes were resolved using electrophoresis with a 6% DNA Retardation Gel (Invitrogen) and transferred to nylon membranes (Thermo Scientific), UV cross-linked and visualized using the Chemiluminescent Nucleic Acid Detection System (Thermo Scientific).
Statistical analysis
All experiments were conducted independently three or more times. Data are expressed as the mean ± standard deviation (SD). The statistical significance between different groups was evaluated with ANOVA followed by Student’s t-test, and a p < 0.05 was considered statistically significant.
RESULTS
MnTE-2-PyP significantly inhibits prostate cancer cells growth, migration and invasion-with or without irradiation
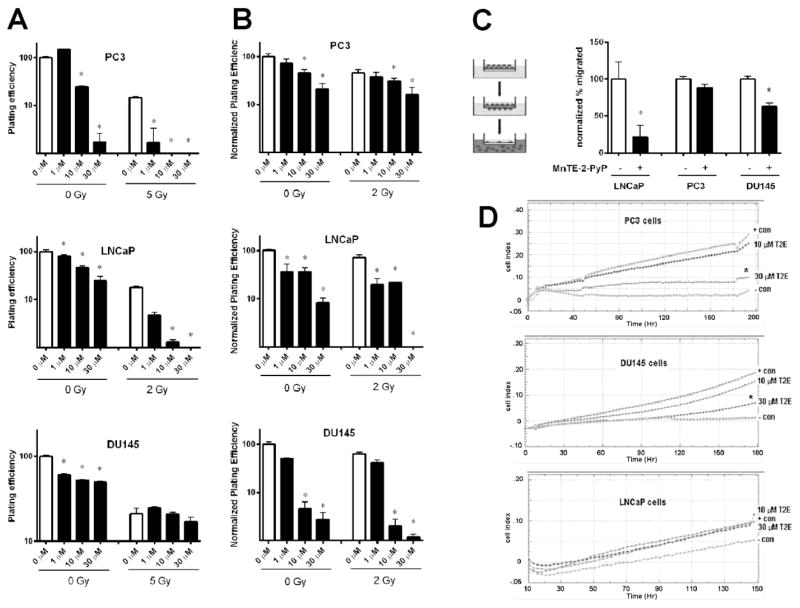
To identify the effect of MnTE-2-PyP on prostate cancer cell growth with or without irradiation, we performed clonogenic assays on three different human prostate cancer cell lines, PC3, LNCaP, and DU145 cells. These cells were treated with 0,1,10, or 30 μM MnTE-2-PyP overnight prior to irradiation. PC3 and DU145 cells were exposed to 0 or 5 Gy of radiation and LNCaP cells were exposed to 0 or 2 Gy. As shown in Fig. 1A, MnTE-2-PyP significantly inhibited the cancer cells growth with or without irradiation. Complementarily, soft agar assays were carried out as well to further confirm this effect. Consistent with the results of the clonogenic assay, MnTE-2-PyP significantly inhibited the growth of these prostate cancer cells in soft agar (Fig. 1B). To identify the effect of MnTE-2-PyP on prostate cancer cell migration, we performed a migration assay on serum starved PC3, LNCaP, and DU145 cells, which were incubated and pre-treated with 0 or 30 μM MnTE-2-PyP overnight. Cells were allowed to migrate across the membrane for 48 hours. As shown in Fig. 1C, a significant decrease in cancer cell migration was detected in LNCaP and DU145 cells treated with MnTE-2-PyP. In order for a tumor to metastasize, the cancer cells must migrate and invade tissues [16]. Thus, we then tested whether MnTE-2-PyP suppressed invasion of the prostate cancer cells. Cells were tracked in real time crossing matrigel using an xCELLigence machine. The positive control wells contained medium with 10% FBS and the wells with no serum were used as the negative controls. In Fig. 1D, a significant decrease in PC3 and DU145 cancer cell invasion activity was detected by MnTE-2-PyP in a dose-dependent fashion. LNCaP cells did not invade well through the matrigel, so the effect of MnTE-2-PyP on inhibition of invasion was inconclusive. Taken together, our data indicate that MnTE-2-PyP inhibits tumor growth and the malignant behavior of prostate cancer cells with or without irradiation.
Figure 1. The inhibition of MnTE-2-PyP on prostate cancer cells growth, migration and invasion with or without irradiation.
(A) PC3, LNCaP, and DU145 cells were treated with 0,1,10, or 30 μm MnTE-2-PyP overnight prior to irradiation. PC3 and DU145 cells were exposed to 0 or 5 Gy of radiation and LNCaP cells were exposed to 0 or 2 Gy. One hour after irradiation, the cells were harvested and re-plated in triplicate in six well plates. After the clones formed, the colonies were enumerated. (B) PC3, LNCaP, and DU145 cells were treated with 0,1,10, or 30 μm MnTE-2-PyP overnight prior to irradiation. All cells were exposed to 0 or 2 Gy of radiation. One hour after irradiation, the cells were counted and then suspended in 1.5% DNA grade agarose + media which was subsequently plated atop a 6.0% agarose + media base. After the clones formed, the soft agar colonies were enumerated. (C) Serum starved PC3, LNCaP, and DU145 cells were incubated and treated with 0 or 30 μM MnTE-2-PyP overnight, then plated on a filter to explore the effects of MnTE-2-PyP on prostate cancer cells migration (right). Cells were allowed to migrate across the membrane for 48 hours. The cells and media were isolated from the top and bottom chambers and then enumerated (left). (D) PC3, LNCaP, and DU145 cells were serum-starved for 24 hours, then real-time impedance-based cell invasion assays were carried out to explore the effects of MnTE-2-PyP on prostate cancer cells invasion. The top chamber wells of the E 16 plates were coated with matrigel and the cells were incubated for 4 hours. The bottom chamber wells contained medium as a chemoattractant. Data represent mean ± SD from 3 independent experiments. * p < 0.05, ** p < 0.01 compared to control, as analyzed with ANOVA followed by two-tailed Student’s t-test.
MnTE-2-PyP does not activate the apoptosis pathway, but inhibits histone acetyltransferase (HAT) p300 activity
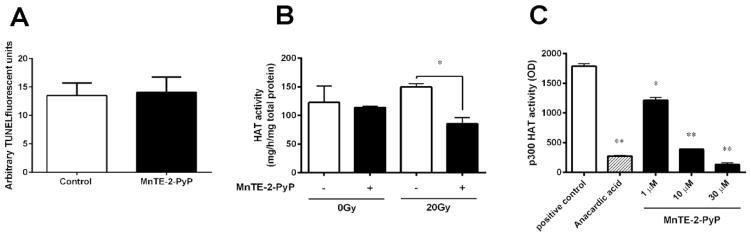
To better understand how MnTE-2-PyP is inhibiting prostate cancer growth, we chose to use PC3 cells as our model because they are the most aggressive human prostate cancer cell tested and use the dose of 30 μM of MnTE-2-PyP because it produced the largest anti-tumor effect. Defects in apoptosis inducing pathways contribute to neoplastic transformation, progression and metastasis [17]. Furthermore, there have been other studies indicating that MnTE-2-PyP works through apoptotic pathways [18, 19]. To explore the underlying mechanisms by which MnTE-2-PyP inhibits prostate cancer cell malignant potential, we performed a TUNEL assay to detect apoptotic PC3 cancer cells. As shown in Fig. 2A, MnTE-2-PyP did not significantly enhance apoptosis; therefore, the drug must be working through another mechanism(s).
Figure 2. MnTE-2-PyP does not activate the apoptosis pathway, but inhibits histone acetyltransferase (HAT) p300 activity.
(A) PC3 cells were treated with or without MnTE-2-PyP overnight prior to irradiation of 20 Gy. Cells were harvested 48 hours after radiation. The cells were TUNEL stained and analyzed by fluorescence-activated cell sorting (FACS). (B) The nuclei were isolated from PC3 cells 1 hour post-radiation, and nuclear proteins (5 μg) were analyzed for HAT activity with or without MnTE-2-PyP. (C) The p300 HAT activity assay was performed with various concentrations of MnTE-2-PyP (0–30 μM). The p300 activity inhibitor, anacardic acid (15 μM), was used as a positive control for p300 HAT inhibition in this experiment. Data represent mean ± SD from 3 independent experiments. * p < 0.05, ** p < 0.01 compared to control, as analyzed with ANOVA followed by two-tailed Student’s t-test.
It was observed that 96% of the genes in a pathway gene array were down-regulated by MnTE-2-PyP in irradiated tumors (data not shown). This indicated that MnTE-2-PyP may globally inhibit transcription. One protein family shown to regulate gene transcription is the histone acetyltransferase (HAT) enzymes [20]. Cells were treated with MnTE-2-PyP or PBS then radiated with 20 Gy. As shown in Fig. 2B, MnTE-2-PyP treated cells exhibited significantly less HAT activity as compared to control cells following irradiation. p300 is the only HAT enzyme shown to be redox sensitive and it has been reported that p300 expression is increased in tumor cells and enhanced following radiation treatment [21]. In Fig. 2C, a significant decrease of p300 HAT activity was detected in the presence of MnTE-2-PyP in a dose-dependent fashion. MnTE-2-PyP (10 and 30 μM) significantly inhibited p300 HAT activity as well or better than the known p300 HAT inhibitor, anacardic acid (15 μM). These data suggested that MnTE-2-PyP does not activate apoptosis, but significantly inhibits the p300 HAT activity.
MnTE-2-PyP alters gene expression of p300 associated transcription factors with or without irradiation
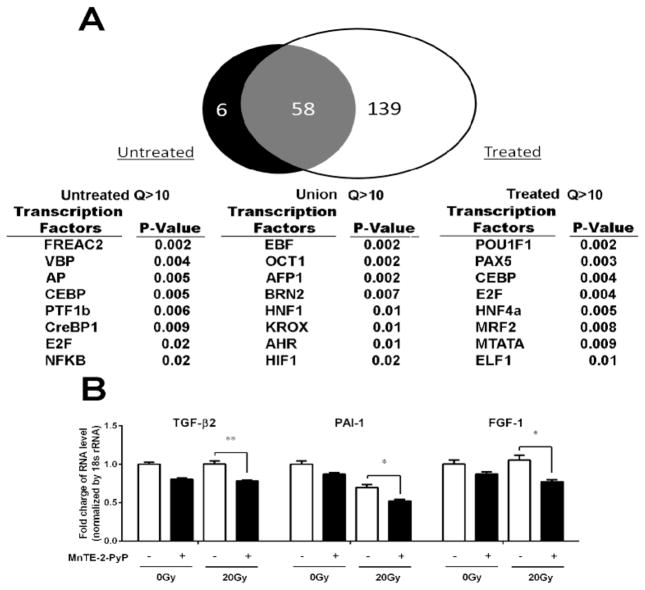
In order to identify how MnTE-2-PyP potentially affects the genome-wide transcriptional activity of p300 via altered chromatin association, we performed a p300 ChIP sequence assay on irradiated PC3 cells treated with or without MnTE-2-PyP (Figure 3A). From the Venn diagram, we found 6 genes associated with p300 without MnTE-2-PyP, 139 genes associated with p300 with MnTE-2-PyP, and 58 genes associated with p300 that were not affected by MnTE-2-PyP. Thus, MnTE-2-PyP significantly affected p300 associating with 150 gene promoters. Based on the analysis of the list of statistically-called ChIP-seq peaks proximal to specific gene loci, we predicted that treatment with MnTE-2-PyP may reduce the genome-wide DNA association of the HIF-1β and CREB transcription factors resulting in reduced transcription of the associated gene loci. To further confirm the result of the ChIP-seq, we conducted real-time PCR for genes regulated by both CREB and HIF-1β: TGF-β2, PAI-1 or FGF1. PC3 cells treated with 30 μm MnTE-2-PyP, followed by 20 Gy of radiation were harvested for RNA 24 hours after irradiation. Decreased expression of TGF-β2, PAI-1 or FGF1 was observed in cells treated with MnTE-2-PyP with or without irradiation, compared with control cells with or without irradiation (Fig. 3B). In addition, we also repeated this real-time PCR assay using 5 Gy instead of 20 Gy. We found that 5 Gy also caused these changes and that the addition of MnTE-2-PyP inhibited these changes (data not shown). Thus, these data indicate that MnTE-2-PyP inhibits the expression of several genes that are regulated by the p300/CREB/HIF-1β complex.
Figure 3. MnTE-2-PyP alters p300 association and specific gene expression with or without irradiation.
(A) PC3 cells were treated with or without MnTE-2-PyP (30 μm) overnight prior to irradiation of 20 Gy. One hour post-irradiation, the cells were fixed. Cells were ChIPed, DNA was prepared for Next-Gen sequencing and then sequenced on a Life Technologies Ion Proton. Global p300 association in both cells with and without MnTE-2-PyP was analyzed by ChIP-Seq using MACS2. A Venn diagram of all the peak associated Ensembl genes that passed multiple statistical tests for peak-gene analysis for the two conditions was generated (up). The list of genes proximal to peaks enriched with a Q-value greater than 10 was analyzed using GATHER Duke Bioinformatics System. The most significant transcription factors and GO Terms are reported (down) in the cells without MnTE-2-PyP treatment. (B) PC3 cells were treated with 30 μM MnTE-2-PyP overnight, and then exposed to 20 Gy of radiation. 24 hours after irradiation, the cells were harvested and RNA was isolated from PC3 cells. The gene expression levels of TGF-β2, PAI-1 and FGF1 were measured by real-time PCR using the ΔΔCT method. 18S was used as the housekeeping gene. The primer sets produced parallel curves with similar slopes. Data represent mean ± SD from 3 independent experiments. * p < 0.05, ** p < 0.01 compared to control, as analyzed with ANOVA followed by two-tailed Student’s t-test.
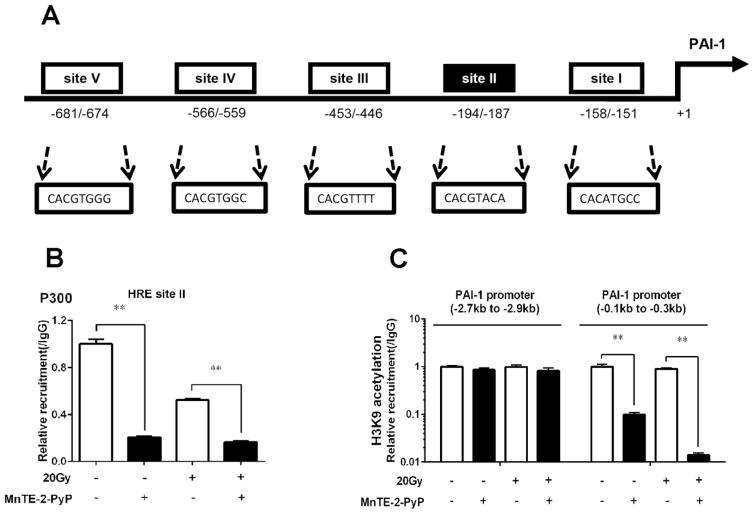
MnTE-2-PyP inhibits p300 association to the promoter region and reduces H3K9 acetylation of human PAI-1 gene locus
We then focused on the mechanism of the inhibitory effects of MnTE-2-PyP on human PAI-1 gene transcription, as increased PAI-1 expression correlates with poor prognosis in prostate cancer [22]. Based on TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and MOTIF (http://motif.genome.jp/) database searches, there are five putative hypoxia-response elements (HREs 1–5) identified in the promoter region of human PAI-1 gene (Fig. 4A). Using an antibody against p300 and PCR primers covering each HRE site, we performed a p300 ChIP-qPCR assay to test the binding of the p300 complex to this PAI-1 gene promoter region. As shown in Fig. 4B, a significant decrease in association of p300 with the HRE-II motif of PAI-1 gene promoter region was detected in cells following MnTE-2-PyP treatment with or without irradiation. No changes of p300 association to the other four HRE sites were observed after MnTE-2-PyP treatment in PC3 cells (data not shown). p300 enhances transcription by acting as a bridge between transcription factors and transcriptional machinery and by acetylating histones to make DNA more accessible for transcription. We then determined whether the inhibition of gene transcription by MnTE-2-PyP is also due to reduced acetylation of histones in the PAI-1 promoter. In a previous study, H3K9 was shown to be regulated by p300 within the proximal promoter region of PAI-1 [23]. Consistent with these results, we detected a decrease of H3K9 acetylation within the PAI-1 proximal promoter region (0.1 to 0.3 kb region) in cells following MnTE-2-PyP stimulation by H3K9Ac ChIP analysis. However, no change of H3K9 acetylation within the PAI-1 distal promoter region (2.7 to 2.9 kb region) was observed with or without MnTE-2-PyP treatment (Fig. 4C). Thus, MnTE-2-PyP can inhibit p300 binding to PAI-1 gene promoter region and, at the same time, reduce H3K9 acetylation of human PAI-1 gene locus.
Figure 4. MnTE-2-PyP inhibits p300 association to the promoter region and reduces H3K9 acetylation of PAI-1 gene locus.
(A) The sequences of five potential HIF-1 binding sites on human PAI-1 gene promoter region were shown. The canonical hypoxia-response element (HRE) sequence is BACGTSSK: B = G/C/T, S = C/G, and K = G/T. (B) P300 binding to site II on human PAI-1 gene promoter by MnTE-2-PyP with or without irradiation. PC3 cells were treated with 30 μM MnTE-2-PyP overnight, and then exposed to 20 Gy of radiation. 1 hour after irradiation, the cells were harvested, followed by ChIP assay using anti-p300 and PCR primer covering binding site II region of the PAI-1 gene promoter. (C) PC3 cells were treated with 30 μM MnTE-2-PyP overnight, and then exposed to 20 Gy of radiation. 1 hour after irradiation, the cells were collected and applied with ChIP analysis for H3K9 acetylation associated with human PAI-1 promoter region. PCR primers covered different promoter regions relative to transcription start site. Data represent mean ± SD from 3 independent experiments. * p < 0.05, ** p < 0.01 compared to control, as analyzed with ANOVA followed by two-tailed Student’s t-test.
MnTE-2-PyP inhibits p300 transcriptional complex binding to the PAI-1 promoter region, not due to interference of binding between p300 and CREB/HIF-1 transcription factors, but through inhibition of these transcription factors to bind DNA
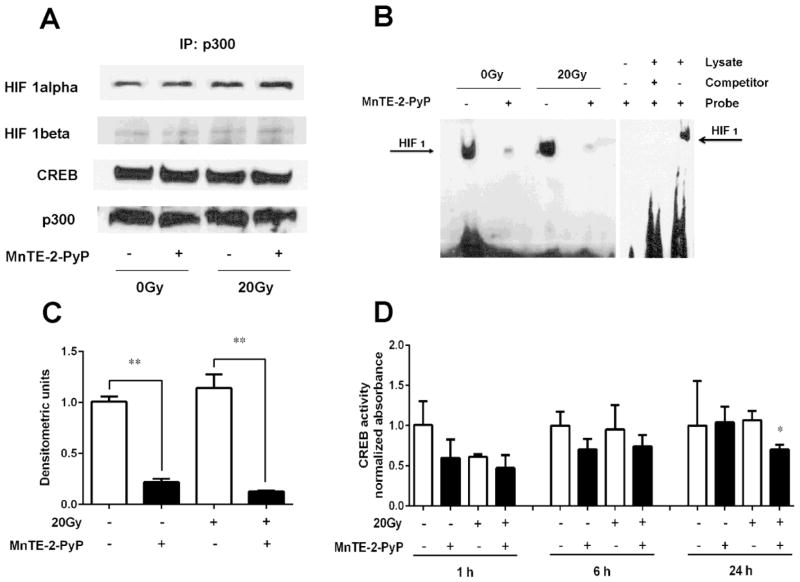
From the p300 ChIP data above, it is clear that MnTE-2-PyP inhibits p300 binding to the promoter region of human PAI-1 gene. To determine the underlying mechanisms by which MnTE-2-PyP inhibits p300 association to human PAI-1 promoter region, a co-IP assay was performed using a p300 antibody to test whether less p300 transcriptional complex binding to DNA was due to decreased binding of p300 to other transcription factors. After the p300 protein complex was pulled down, western blotting for CREB, HIF-1α, or HIF-1β/ARNT was performed. No changes of p300 binding to the transcription factors (CREB, HIF-1α and HIF-1β) were observed after MnTE-2-PyP treatment in PC3 cells with or without irradiation (Fig. 5A). We then carried out a HIF-1α/β EMSA assay with biotin-labelled oligonucleotide corresponding to a HRE sequence of HIF-1. As shown in Fig. 5B and Fig. 5C, a significant decrease of binding to the biotin-labelled target DNA was detected in cells following MnTE-2-PyP stimulation with or without irradiation. Moreover, CREB activity analysis was also performed in PC3 cells with or without MnTE-2-PyP treatment. As evident by CREB activity assay in Fig. 5D, a significant decrease of CREB activity was detected after MnTE-2-PyP post-treatment overnight in the presence of irradiation. Taken together, the above data suggest that MnTE-2-PyP inhibits p300 transcriptional complex association to PAI-1 promoter region, not due to the reduction of binding between p300 and CREB/HIF-1 transcription factors, but through inhibition of these transcription factors to bind DNA.
Figure 5. MnTE-2-PyP inhibits p300 associaiton to PAI-1 promoter region, not due to the disruption of binding between p300 and its associated transcription factors, but through inhibiting the binding of transcription factors to DNA.
(A) A co-IP for p300 binding to transcription factors, HIF-1α, HIF-1β and CREB, ± MnTE-2-PyP with or without irradiation. PC3 cells were treated with 30 μm MnTE-2-PyP overnight, and exposed to 20 Gy of radiation. 1 hour after irradiation, the nuclear protein was isolated, followed by co-IP assay using a p300 antibody. (B) Representative HIF-1 EMSA. PC3 cells were pretreated with 30 μM MnTE-2-PyP overnight, and then exposed to 20 Gy of radiation. After the nuclei were isolated from PC3 cells, EMSA assay was performed with biotin-labelled oligonucleotide corresponding the sequence harboring the HRE of HIF-1α/β. Controls for the HIF-1 EMSA assay are also shown. The blank control contained only biotin-labeled oligonucleotide and the negative control contained cold HIF-1α/β oligonucleotide added to the EMSA reaction. (C) Densitometric analysis of HIF-1 EMSA blots treated by MnTE-2-PyP with or without irradiation. The mean OD of the reactive bands in the control group was normalized to 1.0. (D) The nuclei were isolated from PC3 cells 1, 6 and 24 hours post-radiation, CREB DNA binding was then analyzed with or without MnTE-2-PyP using 5 μg nuclear protein. Data represent mean ± SD from 3 independent experiments. * p < 0.05, ** p < 0.01 compared to control group, as analyzed with ANOVA followed by two-tailed Student’s t-test.
DISCUSSION
After irradiation, the surviving tumor cells have increased levels of ROS, which is thought to drive cancer progression. In particular, superoxide drives HIF-1α expression, which promotes tumor angiogenesis and ultimately tumor survival [24]. Radiation exposure also leads to free radical-mediated oxidative damage to normal tissues leading to fibrosis. Thus, scavenging superoxide in both normal and tumor cells following irradiation would protect from radiation induced fibrosis and reduce tumor regrowth following radiation exposure. Redox-active manganese porphyrins, such as MnTE-2-PyP, have been developed to be potent scavengers of superoxide [25]. We have previously published that MnTE-2-PyP protects the urogenital region from radiation damage. In particular, MnTE-2-PyP protects from erectile dysfunction and testicular atrophy after radiation exposure to the urogenital region [11].
In order for MnTE-2-PyP to be considered a good radio-protector for prostate cancer, the drug cannot protect prostate tumor cells from radiation induced killing. In this study, we found that MnTE-2-PyP inhibited human prostate cancer growth, migration and invasion in vitro. These findings are in accordance with previous work demonstrating that manganese porphyrins do not protect prostate cancer cells from radiation injury [26, 27]. To our knowledge, this is the first study to show that MnTE-2-PyP also inhibits migration and invasion of prostate cancer cells.
We next wanted to determine how MnTE-2-PyP was inhibiting prostate cancer progression. Based on preliminary gene array data, MnTE-2-PyP appeared to down regulate many genes. Accordingly, we investigated whether MnTE-2-PyP reduced gene transcription by inhibiting HAT activity. We determined that MnTE-2-PyP inhibited HAT activity and specifically inhibited p300 HAT activity. To further investigate whether MnTE-2-PyP inhibited p300 activity, we performed a p300 ChIP-seq experiment. We found that MnTE-2-PyP significantly alters p300 association with DNA. Based on the genes that were enriched with p300 in the absence of MnTE-2-PyP but were no longer associated with p300 in the presence of MnTE-2-PyP, it was predicted that MnTE-2-PyP was inhibiting HIF-1β and CREB signaling pathways.
To confirm the p300 ChIP-seq analysis, we carried out a qRT-PCR assay for genes that are involved in cancer progression and regulated by HIF-1 and/or CREB. We found that TGF-β2, FGF-1 and PAI-1 were all inhibited in the presence of MnTE-2-PyP with or without irradiation. We decided to focus on the human PAI-1 gene, as it is not only critically important to cancer growth and metastasis through promoting angiogenesis, enhancing breakdown of extracellular matrix and inhibiting apoptosis of cancer cells, but also has a simple promoter region [22].
The highly conserved cofactor p300 is an important component of the transcriptional complex that participates in regulation both at the level of chromatin organization and transcription initiation [28]. p300 mediates recruitment of basal transcription machinery to the promoter of many genes, but unlike DNA-binding transcription factors, p300 does not directly bind to DNA. To find the DNA-binding transcription factors associated with p300, we performed a p300 ChIP assay for the promoter region of the human PAI-1 gene and found that in the presence of MnTE-2-PyP, a significant reduction in association of p300 to the hypoxia-response element (HRE) site II was detected compared with the other four HRE sites with or without irradiation (data not shown). CREB binds to certain DNA sequences, thereby increasing or decreasing the transcription of the downstream genes. Just like CREB, HIF-1 is also a key regulator, which is responsible for the induction of genes that facilitate adaptation and survival of cells and the whole organism [29]. HIF-1, is a heterodimeric complex consisting of a hypoxically inducible subunit HIF-1α and a constitutively expressed subunit HIF-1β that binds to the HRE sequence. HIF-1β is also known as the aryl hydrocarbon nuclear translocator (ARNT), which was originally identified as a binding partner of the aryl hydrocarbon receptor, whereas HIF-1α expression and activity are tightly regulated [29]. These proteins belong to the basic helix-loop-helix-Per-ARNT-Sim (bHLH-PAS) protein family. The bHLH and PAS motifs are required for heterodimer formation between the HIF-1α and HIF-1β subunits, and the downstream basic region affords specific binding to the HRE DNA sequence [30]. Both HIF-1 and CREB can bind to the HRE of the PAI-1 promoter [31, 32]; thus, it is not clear which transcription factor MnTE-2-PyP is affecting in this model.
Changes in the acetylation state of chromatin-associated histones directly affect gene expression [33, 34]. Acetylation is generally associated with activation of gene expression, while deacetylation is associated with repression. These two processes are normally in a dynamic equilibrium. p300 contains a catalytic HAT domain, which remodels chromatin to “relax” its superstructure and makes DNA more accessible for transcription by acetylating histones [28]. In this study, we detected a decrease of H3K9 acetylation within the PAI-1 proximal promoter region following MnTE-2-PyP stimulation by H3K9ac ChIP analysis. However, no change of H3K9 acetylation within the PAI-1 distal promoter region was observed. Thus, MnTE-2-PyP can both inhibit the p300 protein complex from binding to the PAI-1 gene promoter region, and at the same time reduce H3K9 acetylation of human PAI-1 gene locus.
There are two possible mechanisms by which MnTE-2-PyP inhibits H3K9 acetylation on human PAI-1 gene promoter. First, as shown in the p300 HAT activity assay, MnTE-2-PyP may directly inhibit p300 HAT activity. MnTE-2-PyP at low concentrations directly inhibited p300 HAT activity as well or better than a known p300 inhibitor, anacardic acid. At the present time, it is unclear how MnTE-2-PyP is inhibiting p300 HAT activity directly. We explored the possibility that MnTE-2-PyP was structurally interfering with p300 binding to acetyl CoA. However, excess acetyl CoA did not change the ability of MnTE-2-PyP to inhibit p300 HAT activity (data not shown). Conversely, we also added excess amount of histones to determine if MnTE-2-PyP may interfere with the acetylation of the histone by binding directly to the histone acetylation site. Again, we found no change in histone acetylation in the presence of MnTE-2-PyP when there was an excess of histone substrate. p300 has been reported to contain cysteine residues in its functional domain [35], thus, we postulate that MnTE-2-PyP may be oxidizing p300 and causing a conformational change resulting in reduced HAT activity. This has been speculated for curcumin, another antioxidant enzyme that has been shown to significantly reduce p300 HAT activity [36, 37]. Second, since MnTE-2-PyP interferes with the DNA binding of p300 associated transcription factors, this could result in the inability of p300 to interact with histones and, therefore, acetylation of histones in the PAI-1 gene promoter region is reduced.
We have also determined that MnTE-2-PyP inhibits the p300 transcriptional complex binding to the PAI-1 promoter region not due to interfering with the binding between p300 and CREB/-HIF-1 transcription factors, but through inhibiting the binding of these transcription factors to DNA. The mechanism by which MnTE-2-PyP inhibits CREB/-HIF-1 binding to PAI-1 promoter region is not clear. We speculate that the transcription factors, CREB and HIF-1, are both redox-sensitive primarily through cysteine residues present in DNA binding domain [7, 8]. As a metalloporphyrin-based catalytic antioxidant, by scavenging superoxide, MnTE-2-PyP could enhance local levels of hydrogen peroxide, which would oxidize cysteine residues on the transcription factors and inhibit DNA binding [38, 39]. In addition to making H2O2 (via different pathways see details in refs [38, 39]), MnTE-2-PyP could also use H2O2 produced by irradiation to oxidize thiols. In agreement with this rationale, it has previously been demonstrated that MnTE-2-PyP oxidizes the p50 [40] and p65 subunits [18] of NF-κB, which results in reduced DNA binding. Alternatively, other metalloporphyrins have been reported to bind directly to specific DNA sequences and potentially mask the transcription factor binding site [41]. Another manganese porphyrin, MnTM-4-PyP, has been shown to bind to RNA and DNA [42]. MnTE-2-PyP consists of a porphyrin ring with four positively charged pyridyl groups, which could bind to the negatively charged phosphate groups in DNA. Experiments are currently underway to further explore these possible mechanisms.
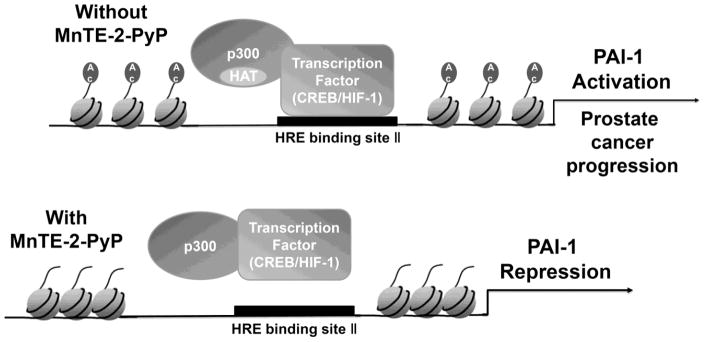
In summary, our data demonstrate a novel mechanism by which MnTE-2-PyP represses pro-cancerous genes expression and inhibit prostate cancer growth, migration and invasion. As is shown in Fig. 6, without MnTE-2-PyP, the p300 binding complex can directly bind to the promoter region of human PAI-1 gene, acetylate the chromatin and promote PAI-1 gene transcription. In the presence of MnTE-2-PyP, the p300 transcriptional complex no longer binds to the promoter region of human PAI-1 gene, histone acetylation is reduced and thus PAI-1 gene expression is down-regulated. This study provides a more in depth mechanistic understanding of the mechanisms by which MnTE-2-PyP reduces prostate cancer growth during prostate cancer radiotherapy. Thus, MnTE-2-PyP works as a unique radio-protector of normal tissues that simultaneously enhances tumor killing. Using MnTE-2-PyP in conjunction with radiation therapy could enhance treatment outcomes in patients undergoing prostate cancer radiotherapy.
Figure 6. Proposed model of how MnTE-2-PyP effects PAI-1 expression.
Without MnTE-2-PyP, the p300 binding complex can directly bind to the promoter region of human PAI-1 gene, acetylate the chromatin and promote PAI-1 gene transcription. In the presence of MnTE-2-PyP, with or without radiation, the p300 transcriptional complex no longer binds to the promoter region of human PAI-1 gene, histone acetylation is reduced and, thus, PAI-1 gene expression is down-regulated.
Highlights.
MnTE-2-PyP combined with radiation further inhibits growth and metastasis of human prostate cancer cells.
MnTE-2-PyP inhibits p300 HAT activity and alters p300 transcriptional complex binding to DNA.
Specifically, MnTE-2-PyP reduces histone acetylation of the promoter of PAI-1, which is regulated by HIF-1, CREB and p300.
This is the first study to show that MnTE-2-PyP can epigenetically regulate gene expression.
Acknowledgments
This work was supported by National Institutes of Health Grants 1R01CA178888 (ROD), by the National Natural Science Foundation of China No. 81172186 (QT) and by the Fred and Pamela Buffet Cancer Center Support Grant P30CA036727 (ROD).
Footnotes
Conflicts of interest statement: Dr. Oberley-Deegan is a consultant with BioMimetix Pharmaceutical, Inc. and holds equities in BioMimetix Pharmaceutical, Inc. There are no conflicts of interest for the other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cooperberg MR, Broering JM, Carroll PR. Time trends and local variation in primary treatment of localized prostate cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:1117–1123. doi: 10.1200/JCO.2009.26.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao L, Holley AK, Zhao Y, St Clair WH, St Clair DK. Redox-mediated and ionizing-radiation-induced inflammatory mediators in prostate cancer development and treatment. Antioxidants & redox signaling. 2014;20:1481–1500. doi: 10.1089/ars.2013.5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimova EY, Samoylenko A, Kietzmann T. Oxidative stress and hypoxia: implications for plasminogen activator inhibitor-1 expression. Antioxidants & redox signaling. 2004;6:777–791. doi: 10.1089/1523086041361596. [DOI] [PubMed] [Google Scholar]
- 4.Dong Z, Saliganan AD, Meng H, Nabha SM, Sabbota AL, Sheng S, Bonfil RD, Cher ML. Prostate cancer cell-derived urokinase-type plasminogen activator contributes to intraosseous tumor growth and bone turnover. Neoplasia. 2008;10:439–449. doi: 10.1593/neo.08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harbeck N, Kruger A, Sinz S, Kates RE, Thomssen C, Schmitt M, Janicke F. Clinical relevance of the plasminogen activator inhibitor type 1--a multifaceted proteolytic factor. Onkologie. 2001;24:238–244. doi: 10.1159/000055086. [DOI] [PubMed] [Google Scholar]
- 6.Liu RM. Oxidative stress, plasminogen activator inhibitor 1, and lung fibrosis. Antioxidants & redox signaling. 2008;10:303–319. doi: 10.1089/ars.2007.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jornot L, Petersen H, Junod AF. Modulation of the DNA binding activity of transcription factors CREP, NFkappaB and HSF by H2O2 and TNF alpha. Differences between in vivo and in vitro effects. FEBS letters. 1997;416:381–386. doi: 10.1016/s0014-5793(97)01244-1. [DOI] [PubMed] [Google Scholar]
- 8.Nikinmaa M, Pursiheimo S, Soitamo AJ. Redox state regulates HIF-1alpha and its DNA binding and phosphorylation in salmonid cells. Journal of cell science. 2004;117:3201–3206. doi: 10.1242/jcs.01192. [DOI] [PubMed] [Google Scholar]
- 9.Rahman I, Marwick J, Kirkham P. Redox modulation of chromatin remodeling: impact on histone acetylation and deacetylation, NF-kappaB and pro-inflammatory gene expression. Biochemical pharmacology. 2004;68:1255–1267. doi: 10.1016/j.bcp.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 10.Batinic-Haberle I, SI, Hambright P, Benov L, Crumbliss A, Fridovich I. Relationship among redox potentials, proton dissociation constants of pyrrolic nitrogens, and in vivo and in vitro superoxide dismutating activities of manganese (III) and iron (III) water-soluble porphyrins. Inorganic Chemistry. 1999;38:4011–4022. [Google Scholar]
- 11.Oberley-Deegan RE, Steffan JJ, Rove KO, Pate KM, Weaver MW, Spasojevic I, Frederick B, Raben D, Meacham RB, Crapo JD, Koul HK. The antioxidant, MnTE-2-PyP, prevents side-effects incurred by prostate cancer irradiation. PloS one. 2012;7:e44178. doi: 10.1371/journal.pone.0044178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 14.Das PM, Ramachandran K, van Wert J, Singal R. Chromatin immunoprecipitation assay. BioTechniques. 2004;37:961–969. doi: 10.2144/04376RV01. [DOI] [PubMed] [Google Scholar]
- 15.Ganster RW, Guo Z, Shao L, Geller DA. Differential effects of TNF-alpha and IFN-gamma on gene transcription mediated by NF-kappaB-Stat1 interactions. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2005;25:707–719. doi: 10.1089/jir.2005.25.707. [DOI] [PubMed] [Google Scholar]
- 16.Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, Hengstschlager M, Dolznig H. In vitro cell migration and invasion assays. Mutation research. 2013;752:10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Malaguarnera L. Implications of apoptosis regulators in tumorigenesis. Cancer metastasis reviews. 2004;23:367–387. doi: 10.1023/B:CANC.0000031774.32572.df. [DOI] [PubMed] [Google Scholar]
- 18.Jaramillo MC, BM, Crapo JD, Batinic-Haberle I, Tome ME. Manganese porphyrin, MnTE-2-PyP5+, Acts as a pro-oxidant to potentiate glucocorticoid-induced apoptosis in lymphoma cells. Free radical biology & medicine. 2012;52:1272–1284. doi: 10.1016/j.freeradbiomed.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans MK, TA, Batinic-Haberle I, Devi GR. Mn porphyrin in combination with ascorbate acts as a pro-oxidant and mediates caspase-independent cancer cell death. Free radical biology & medicine. 2014;68:302–314. doi: 10.1016/j.freeradbiomed.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan AN, Tomasi TB. Histone deacetylase regulation of immune gene expression in tumor cells. Immunologic research. 2008;40:164–178. doi: 10.1007/s12026-007-0085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heemers HV, Debes JD, Tindall DJ. The role of the transcriptional coactivator p300 in prostate cancer progression. Advances in experimental medicine and biology. 2008;617:535–540. doi: 10.1007/978-0-387-69080-3_54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Current pharmaceutical design. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 23.Yuan H, Reddy MA, Sun G, Lanting L, Wang M, Kato M, Natarajan R. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta1-mediated gene transcription in mesangial cells. American journal of physiology. Renal physiology. 2013;304:F601–613. doi: 10.1152/ajprenal.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasabe E, YZ, Ohno S, Yamamoto T. Reactive oxygen species produced by the knockdown of manganese-superoxide dismutase up-regulate hypoxia-inducible factor-1alpha expression in oral squamous cell carcinoma cells. Free Radic Biol Med. 2010;48:1321–1329. doi: 10.1016/j.freeradbiomed.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Miriyala S, SI, Tovmasyan A, Salvemini D, Vujaskovic Z, St Clair D, Batinic-Haberle I. Manganese superoxide dismutase, MnSOD and its mimics. Biochim Biophys Acta. 2012;1822:794–814. doi: 10.1016/j.bbadis.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gridley DS, Makinde AY, Luo X, Rizvi A, Crapo JD, Dewhirst MW, Moeller BJ, Pearlstein RD, Slater JM. Radiation and a metalloporphyrin radioprotectant in a mouse prostate tumor model. Anticancer research. 2007;27:3101–3109. [PubMed] [Google Scholar]
- 27.Makinde AY, Luo-Owen X, Rizvi A, Crapo JD, Pearlstein RD, Slater JM, Gridley DS. Effect of a metalloporphyrin antioxidant (MnTE-2-PyP) on the response of a mouse prostate cancer model to radiation. Anticancer research. 2009;29:107–118. [PubMed] [Google Scholar]
- 28.Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cellular and molecular life sciences: CMLS. 2013;70:3989–4008. doi: 10.1007/s00018-012-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang GL, JB, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.STC Control of cell lineage-specific development and transcription by bHLH–PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 31.Dimova EY, Jakubowska MM, Kietzmann T. CREB binding to the hypoxia-inducible factor-1 responsive elements in the plasminogen activator inhibitor-1 promoter mediates the glucagon effect. Thrombosis and haemostasis. 2007;98:296–303. [PubMed] [Google Scholar]
- 32.Dimova EY, Samoylenko A, Kietzmann T. FOXO4 induces human plasminogen activator inhibitor-1 gene expression via an indirect mechanism by modulating HIF-1alpha and CREB levels. Antioxidants & redox signaling. 2010;13:413–424. doi: 10.1089/ars.2009.2999. [DOI] [PubMed] [Google Scholar]
- 33.Adcock IM, Lee KY. Abnormal histone acetylase and deacetylase expression and function in lung inflammation. Inflammation research: official journal of the European Histamine Research Society … [et al.] 2006;55:311–321. doi: 10.1007/s00011-006-0081-1. [DOI] [PubMed] [Google Scholar]
- 34.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiology and molecular biology reviews: MMBR. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu J, MJ, Huang LE. Molecular mechanism of hypoxia-inducible factor 1alpha -p300 interaction. A leucine-rich interface regulated by a single cysteine. J Biol Chem. 2001;276:3550–3554. doi: 10.1074/jbc.M009522200. [DOI] [PubMed] [Google Scholar]
- 36.Morimoto T, Sunagawa Y, Kawamura T, Takaya T, Wada H, Nagasawa A, Komeda M, Fujita M, Shimatsu A, Kita T, Hasegawa K. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. The Journal of clinical investigation. 2008;118:868–878. doi: 10.1172/JCI33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuter S, Gupta SC, Park B, Goel A, Aggarwal BB. Epigenetic changes induced by curcumin and other natural compounds. Genes & nutrition. 2011;6:93–108. doi: 10.1007/s12263-011-0222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batinic-Haberle I, TA, Roberts ER, Vujaskovic Z, Leong KW, Spasojevic I. SOD therapeutics: latest insights into their structure-activity relationships and impact on the cellular redox-based signaling pathways. Antioxidants & redox signaling. 2014;20:2372–2415. doi: 10.1089/ars.2012.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batinic-Haberle I, TA, Spasojevic I. An educational overview of the chemistry, biochemistry and therapeutic aspects of Mn porphyrins - From superoxide dismutation to H2O2-driven pathways. Redox Biol. 2015;7:43–65. doi: 10.1016/j.redox.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tse HM, Milton MJ, Piganelli JD. Mechanistic analysis of the immunomodulatory effects of a catalytic antioxidant on antigen-presenting cells: implication for their use in targeting oxidation-reduction reactions in innate immunity. Free radical biology & medicine. 2004;36:233–247. doi: 10.1016/j.freeradbiomed.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 41.Odom DT, Parker CS, Barton JK. Site-specific inhibition of transcription factor binding to DNA by a metallointercalator. Biochemistry. 1999;38:5155–5163. doi: 10.1021/bi9827969. [DOI] [PubMed] [Google Scholar]
- 42.Batinić-Haberle I, BL, Spasojević I, Fridovich I. The ortho effect makes manganese(III) meso-tetrakis(N-methylpyridinium-2-yl)porphyrin a powerful and potentially useful superoxide dismutase mimic. J Biol Chem. 1998;273:24521–24528. doi: 10.1074/jbc.273.38.24521. [DOI] [PubMed] [Google Scholar]