Abstract
Estrogen-related receptor alpha (ERRα), the first orphan nuclear receptor discovered, is crucial for the control of cellular energy metabolism. ERRα and its coactivator, peroxisome proliferator-activated receptor gamma coactivator I-alpha (PGC-1α), are required for rapid energy production in response to environmental challenges. They have been implicated in the etiology of metabolic disorders such as type 2 diabetes and metabolic syndrome. ERRα also plays a role in the pathogenesis of breast cancer. Identification of compounds that modulate ERRα signaling may elucidate environmental factors associated with these diseases. Therefore, we developed stable cell lines containing an intact ERRα signaling pathway, with and without the coactivator PGC-1α, to use as high-throughput screening tools to detect ERRα modulators. The lentiviral PGC-1α expression constructs and ERRα multiple hormone response element (MHRE) reporters were introduced into HEK293T cells that express endogenous ERRα. A cell line expressing the reporter alone was designated “ERR.” A second cell line expressing both reporter and PGC-1α was named “PGC/ERR.” Initial screenings of the Library of Pharmacologically Active Compounds (LOPAC) identified 33 ERR and 22 PGC/ERR agonists, and 54 ERR and 15 PGC/ERR antagonists. Several potent ERRα agonists were dietary plant compounds (e.g., genistein). In conclusion, these cell lines are suitable for high-throughput screens to identify environmental chemicals affecting metabolic pathways and breast cancer progression.
Keywords: ERRα signaling, PGC-1α, stable cell lines, LOPAC library, quantitative high-throughput screening, metabolic syndrome
Introduction
Estrogen-related receptor alpha (ERRα) was the first nuclear receptor found without a known endogenous ligand, and this status remained until recently.1 Questions about the physiological role of this receptor and what regulates its activity have intrigued the scientific community for several decades. It has now been established that ERRα is involved in maintaining energy homeostasis in response to environmental cues. It does so by regulating a broad range of genes important in metabolism, such as those that encode enzymes that function in the glycolytic pathway, the tricarboxylic cycle, oxidative phosphorylation, lipid metabolism, mitochondrial functions, and biogenesis.2,3 Chronic dysregulation of these critical pathways can lead to development of metabolic syndrome, obesity, and diabetes. Recently, ERRα was also shown to be involved in breast cancer progression.4 Because adequate functioning of ERRα is critical to maintaining energy homeostasis, it is crucial to identify the factors involved in regulating or modulating its activity. Although ERRα is expressed in every cell, the expression level varies with cell type and tissue. Levels of ERRα are high in metabolically active tissues such as brown fat, heart, kidney, and skeletal muscle, and expression levels can further increase in response to cold, stress, fasting, and exercise—situations that impose higher energy demands. In contrast to most nuclear receptors, the endogenous activity of the orphan receptor ERRα is not regulated by a small-molecule ligand; rather, its activity directly correlates with the availability of intracellular coregulators. Although ubiquitously expressed coregulators such as steroid receptor coactivator (SRC) or glucocorticoid receptor–interacting protein (GRIP)5 can activate ERRα, the inducible proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) stimulates robust expression of ERRα and has been considered the principal protein ligand for ERRα.6–8 Furthermore, PGC-1α is by itself a key regulator in energy balance.9,10 Therefore, the ERRα receptor and its coregulator PGC-1α are critical molecular partners in managing cellular energy output on demand. Like other nuclear receptors, ERRα is subject to posttranslational modification, through phosphorylation, acetylation, or sumoylation, which regulate its functional activity.11,12 Compounds that modulate these posttranslational signaling pathways may influence ERRα function and its downstream targets.
Humans are constantly exposed from conception through death to a diversity of environmental chemicals. These exposures could potentially impact normal development, as well as result in chronic diseases later in life. Therefore, the ability to identify hazardous chemicals that can perturb normal physiology would be an important advance for public health,13 and the effect of chemical exposures on the ERRα pathway deserves exploration. Due to the large number of compounds without sufficient toxicological characterization, new and innovative methods are needed to evaluate the safety of existing and future chemicals. Since the development of quantitative high-throughput screening (qHTS) approaches in the Tox21 program, a panel of 16 nuclear receptor assays has been screened and many endocrine-disrupting chemicals were identified.14–16 Among these nuclear receptor assays, none were designed to measure effects on the ERRα signaling pathway. To fill this gap, we developed a stable cell line that expresses human PGC-1α (hPGC-1α) in HEK293T cells as a first step to test the usefulness of screening environmental compounds in a cell line that contains an intact PGC-1α/ERRα axis.17 We selected the HEK293T cell line because its endogenous ERRα level is high and ERRγ or ERRβ is low. Using this system in transient transfection experiments, we demonstrated that ERRα is activated by genistein and inhibited by the known ERRα inhibitor, XCT790,18,19 in a dose-dependent manner. In the present study, we developed two cell lines containing a stably integrated ERRα reporter, with or without the presence of the coactivator PGC-1α. The cell lines were optimized for use in a 1536-well plate format on a robotic platform and then further assessed for performance in a high-throughput screen using the Library of Pharmacologically Active Compounds (LOPAC). Here, we present data identifying dietary plant products and phorbol esters as strong agonists, whereas aminopterin and rotenone are antagonists of the ERRα signaling pathway.
Materials and Methods
Design of Viral Constructs
Design of the lentiviral expression construct of hPGC-1 was previously described.17 The sequence of natural ERRα pleiotropic nuclear receptor enhancer multiple hormone response element (MHRE)20,21 designated as AAB was synthesized and cloned into pGreenFire lenti-reporter vector (System Biosciences, Mountain View, CA) that contains both green fluorescent protein (GFP) and Luc marker reporters under contract by GeneScript (Piscataway, NJ) and designated as AAB-GFP-Luc.
Production of Lentivirus
All lentiviruses were packaged in HEK293T/17 cells (ATCC no. CRL-11268) according to published procedures in Current Protocols in Neuroscience by Barde et al.22 A multiplicity of infection (MOI) of 14 was used to create a HEK293T cell line that expresses hPGC. An MOI of 3 was used for the transduction of the aforementioned cell line to create a HEK293T strain that stably expresses hPGC-1 and AAB-GFP-Luc.
Selection of Clones
Cells expressing GFP were sorted by an LSRII flow cytometer (Becton Dickenson, Franklin Lakes, NJ) and grown individually in each well of a 96-well plate, and healthy stable clonal cell lines were harvested after expansion. The cell lines were functionally characterized by their response to genistein (agonist; increase in signal) and response to XCT790 (antagonist; decrease in signal). The cell line that expressed GFP/Luc and was also sensitive to puromycin was named ERR; the puromycin-resistant cell line containing the marker for PGC expression was named PGC/ERR (Suppl. Fig. 1). Although different GFP expression levels in the expanded single-cell clone were found, this is not unusual. Cells that express GFP are at a competitive disadvantage. So, as clonal cells divide, any cells that might acquire a mutation that silences GFP are at an evolutionary advantage over GFP-expressing cells.23 Generally, the loss of GFP expression is small (occurring over many passages); HEK239 cell clones in our study demonstrated stable fluorescence over >5 passages.
Miniaturization and Optimization of the Assays
The assays were miniaturized into a 1536-well plate format. Technical factors such as signal-to-background (S/B) ratio, cell growth and viability, reproducibility of response, and EC50 and IC50 values were among the parameters that were optimized to meet the requirements for Tox 21 qHTS assay performance (http://www.ncats.nih.gov/preclinical/drug-dev/assay#criteria).
Quantitative High-Throughput Screening against the LOPAC Collection
Using the optimized ERR or PGC/ERR luciferase reporter gene assays multiplexed with a cell viability assay, the LOPAC1280 (Sigma-Aldrich, St. Louis, MO) and the Tox21 88 duplicate compounds (present on every plate in every assay to monitor plate-to-plate variation) were screened in three independent experiments at eight concentrations, ranging from 0.6 nM to 46 μM (fivefold dilution).
Data Analysis
All assays were run in three independent experiments using eight concentrations per compound to provide three dose—response curves per compound, cell line, or mode. The primary data analysis was performed as previously described.24 Briefly, raw plate reads for each titration point were first normalized relative to the positive control (23 μM genistein for ERR and 46 μM genistein for PGC/ERR, set at 100% for agonist mode; 18 μM XCT790, set at 100% for antagonist mode) and DMSO-only wells (basal, set at 0%), and then corrected by applying a pattern correction algorithm using compound-free control plates (DMSO plates).25 Concentration—response titration points for each compound were fitted to the Hill equation yielding concentrations for half-maximal activity for agonists (EC50) or half-maximal inhibition for antagonists (IC50) and maximal response (Emax) values. The concentration—response curves of the compounds were grouped into four broad classes according to previously published criteria.24
Comparison with Results from Other Assays Screened against the LOPAC
Actives identified from the primary readouts of 11 previously conducted assays (https://tripod.nih.gov/tox/apps/assays/assays.html) targeting nuclear receptor pathways and stress response pathways were compared against the actives from the agonist mode of ERR and PGC/ERR identified in this study. The 11 assays included those measuring signaling pathways for estrogen receptor alpha (ERα), androgen receptor (AR), thyroid receptor alpha (TRα), retinoic acid receptor alpha (RARα), aryl hydrocarbon receptor (AhR), and thyroid-stimulating hormone receptor (TSHR), as well as assays measuring changes in mitochondrial membrane potential (mitotox), activator protein 1 (AP1) activity, activation of the antioxidant response element (ARE), upregulation of luciferase-tagged ATAD5 (a DNA translesion synthesis protein), and activation of the p53 response element (p53RE). Similarly, the actives from four nuclear receptor assays (ERα, AR, TR, and RAR) and a G protein—coupled receptor (GPCR [TSHR]) run in antagonist mode were also compared with the actives from the antagonist mode of ERR and PGC/ERR. Data analysis for all these assays was performed as described above.
Results
Assay Optimization and Miniaturization of ERR and PGC/ERR Luciferase Reporter Gene Assays
The assay was miniaturized into a 1536-well plate format. To optimize the cell density in the well, either 2000 or 3000 cells/well were incubated with various concentrations of genistein, an ERRα agonist, and XCT790, an ERRα antagonist, for 18 h. Genistein increased ERRα induction in a concentration-dependent manner, generating EC50 values of 3.33 μM (2000 cells/well) and 3.23 μM (3000 cells/well), respectively (Fig. 1A). XCT790 (Fig. 1B) significantly inhibited ERRα induction, generating IC50 values of 5.10 μM (2000 cells/well) and 4.16 μM (3000 cells/well). The S/B ratio in the ERR assay run in agonist mode was 2.1-fold for 2000 cells/well and 2.2-fold for 3000 cells/well. In antagonist mode, the S/B ratio was 3.54-fold for 2000 cells/ well and 3.41-fold for 3000 cells/well. In the PGC/ERR assay run with genistein in agonist mode, both cell densities showed very similar EC50 values (5.28 μM for 2000 cells/ well and 5.79 μM for 3000 cells/well) (Fig. 1C). The S/B ratios were also similar (3.16-fold for 2000 cells/well and 3.29-fold for 3000 cells/well). In antagonist mode, the IC50 values for the PGC/ERR assay using XCT790 as the reference antagonist were 3.44 μM for 2000 cells/well and 3.09 μM for 3000 cells/well, and the S/B ratios were 3.43-fold for 2000 cells/well and 2.77-fold for 3000 cells/well (Fig. 1D).
Figure 1.
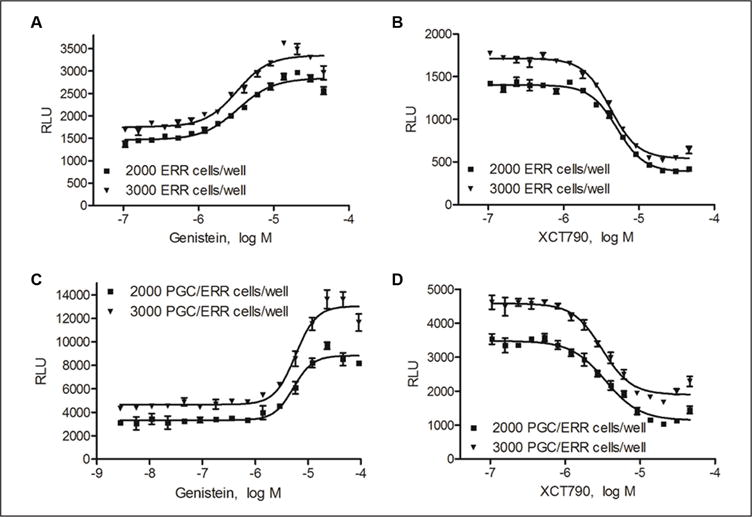
Concentration-response curves for the reference agonist genistein and the reference antagonist XCT790 in the ERR and PGC/ERR cell lines. Data are reported as mean ± SEM from three independent experiments.
Assay Reproducibility in qHTS and LOPAC Library Screening
We screened a LOPAC library three times in both agonist and antagonist modes at eight concentrations ranging from 2.9 nM to 46 μM. Genistein was used as a positive control for the agonist mode screen and XCT790 was used for the antagonist mode screen.17 The concentration—response curves of genistein replicated well across the entire agonist screening with EC50 values of 4.09 ± 0.48 μM (mean ± SD) in the ERR-luc assay and 9.19 ± 2.31 μM in the PCG/ERR-luc assay. IC50 values for XCT790 in the antagonist mode screens were 4.50 ± 0.47 μM in the ERR-luc assay and 3.97 ± 0.29 μM in the PGC/ERR-luc assay. ERR-luc and PGC/ERR-luc assays performed well in both agonist and antagonist modes (Suppl. Table 1).
To evaluate assay performance, response reproducibility over the three replicate screens using the LOPAC library was calculated for the ERR-luc, PGC/ERR-luc, and cell viability assays. The reproducibility was based on active match, inactive match, inconclusives, mismatch rates, and potency differences among the three runs (Suppl. Table 2). All values met requirements for HTS performance, and the assays were approved for online screening of the Tox21 10K chemical library.
Data Analyses
In total, there were 33 and 22 compounds identified as active in the ERR and PGC/ERR agonist mode assays, respectively; 11 compounds were active in both assays (Fig.2A). Chemical names and potency values of the actives are shown in Table 1. For the ERR and PGC/ERR antagonist mode assays, 54 and 15 compounds were identified as active, respectively; 12 compounds were active in both assays (Fig. 2B). The names and potencies of these actives are shown in Table 2. The concentration-response curves of the overlapping actives from both assays in either agonist or antagonist mode are presented in Figure 3. Agonists active in both assays (Fig. 3A,B) show efficacies of >100% at low micromolar concentrations; phorbol 12-myristate 13-acetate (PMA) was active at nanomolar levels. For some of the agonists with outfitted points (e.g., PMA and BIO), cytotoxicity was seen at the concentration accompanying the outfitted points. However, for others (e.g., genistein and kenpaullone), cytotoxicity did not appear to be the direct cause for the outfitted points. The antagonists that were active in both assays (Fig. 3C,D) had activities at least fivefold more potent than their cytotoxicity. Interestingly, rotenone and aminopterin were markedly more potent in inhibiting the ERR and PGC/ERR signaling pathways than the known inhibitor, XCT790.
Figure 2.

Venn diagram illustrating the overlap between agonists (A) and antagonists (B) in the ERR and PGC/ERR assays.
Table 1.
Compounds from the LOPAC That Were Identified as Agonists in Either the ERR or PGC/ERR Assays.a
| Sample Name | CASRNb | ERRαc (Agonist) |
PGC-1α/ERRαc (Agonist) |
|---|---|---|---|
| Phorbol 12-myristate 13-acetate | 16561-29-8 | 0.01 | 0.01 |
| PD173952 | 305820-75-1 | — | 0.05 |
| 6-Bromoindirubin-3-oxime | 667463-62-9 | 1.68 | 1.19 |
| Kenpaullone | 142273-20-9 | 2.98 | 1.33 |
| Forskolin | 66575-29-9 | — | 1.33 |
| BF-170 hydrochloride | 22191-97-5 | 2.66 | 1.58 |
| 2-Phenylaminoadenosine | 53296-10-9 | — | 1.68 |
| Nocodazole | 31430-18-9 | — | 1.88 |
| Apigenin | 520-36-5 | 3.35 | 2.37 |
| N6-Cyclopentyladenosine | 41552-82-3 | — | 2.98 |
| Genistein | 446-72-0 | 2.82 | 3.35 |
| Resveratrol | 501-36-0 | 3.35 | 11.88 |
| Caffeic acid phenethyl ester | 104594-70-9 | — | 11.88 |
| N6-2-(4-Aminophenyl)ethyladenosine | 89705-21-5 | — | 11.88 |
| Rutaecarpine | 84-26-4 | 2.37 | 13.33 |
| SB 206553 hydrochloride | 158942-04-2 (free base) | 13.33 | 13.33 |
| P1,P4-Di(adenosine-5′)tetraphosphate triammonium | 102783-36-8 | — | 13.33 |
| Piceatannol | 10083-24-6 | 8.41 | 18.83 |
| Thio-NADP sodium | 19254-05-8 | — | 19.95 |
| Daidzein | 486-66-8 | 13.33 | 21.13 |
| CGP 57380 | 522629-08-9 | — | 23.71 |
| SU 6656 | 330161-87-0 | — | 23.71 |
| 1,2-Phenylenediamine | 95-54-5 | 21.69 | — |
| 4-Aminoazobenzene | 60-09-3 | 10.87 | — |
| 4-Chloro-1,2-diaminobenzene | 95-83-0 | 6.11 | — |
| 6-Methyl-2-(phenylethynyl)pyridine HCl | 219911-35-0 | 13.33 | — |
| AS 604850 | 648449-76-7 | 3.16 | — |
| Bay 11-7082 | 19542-67-7 | 16.79 | — |
| 3,3 ′-Difluorobenzaldazine | 15332-10-2 | 13.33 | — |
| Diphenyl isophthalate | 744-45-6 | 10.87 | — |
| Flavone | 525-82-6 | 9.69 | — |
| Hydralazine hydrochloride | 304-20-1 | 0.75 | — |
| IMS2186 | 1031206-36-6 | 1.06 | — |
| Indirubin-3′-oxime | 160807-49-8 | 2.66 | — |
| Mevastatin | 73573-88-3 | 2.98 | — |
| Niclosamide | 50-65-7 | 0.04 | — |
| PD 98,059 | 167869-21-8 | 2.11 | — |
| Retinoic acid | 302-79-4 | 7.50 | — |
| Rhodblock 6 | 886625-06-5 | 13.33 | — |
| Ribavirin | 36791-04-5 | 0.94 | — |
| SIB 1757 | 31993-01-8 | 8.41 | — |
| SIB 1893 | 7370-21-0 | 13.33 | — |
| SU 4312 | 5812-07-7 | 3.35 | — |
| Tranilast | 53902-12-8 | 1.68 | — |
Names in bold are plant-derived compounds; a dash indicates inconclusive or no activity.
Chemical Abstracts Service (CAS) Registry Number.
EC50 values (μM).
Table 2.
Compounds from the LOPAC Collection That Were Identified as Antagonists in Either the ERR or PGC/ERR Assays.a
| Chemical Name | CASRNb | ERRαc (Antagonist) | ERRαc (Viability) | PGC-1α/ERRαc (Antagonist) | PGC-1α/ERRαc (Viability) |
|---|---|---|---|---|---|
| Methotrexate hydrate | 133073-73-1 | 0.04 | — | — | — |
| Aminopterin | 54-62-6 | 0.05 | — | 0.06 | — |
| Rotenone | 83-79-4 | 0.07 | 1.33 | 0.04 | 6.68 |
| (S)-(+)-Camptothecin | 7689-03-4 | 0.12 | — | — | — |
| Thapsigargin | 67526-95-8 | 0.24 | 10.59 | — | — |
| Papaverine hydrochloride | 61-25-6 | 0.25 | — | — | — |
| AC-93253 iodide | 108527-83-9 | 0.30 | 8.41 | 0.42 | 3.76 |
| Gemcitabine hydrochloride | 122111-03-9 | 0.33 | — | — | — |
| Artemether | 71963-77-4 | 0.53 | — | 0.75 | — |
| S-(p-Azidophenacyl)glutathione | 73322-71-1 | 0.53 | — | — | — |
| SB 205384 | 160296-13-9 | 0.53 | — | — | — |
| Amsacrine hydrochloride | 54301-15-4 | 0.75 | 23.71 | — | — |
| Topotecan hydrochloride hydrate | 123948-87-8 (free base) | 0.75 | — | — | — |
| RepSox | 446859-33-2 | 1.33 | — | 1.19 | — |
| Brefeldin A from Penicillium brefeldianum | 20350-15-6 | 1.50 | — | — | — |
| AGK2 | 304896-28-4 | 1.68 | — | — | — |
| Auranofin | 34031-32-8 | 1.68 | 9.44 | — | — |
| Tyrphostin AG 879 | 148741-30-4 | 2.37 | — | — | — |
| AMG 9810 | 545395-94-6 | 2.66 | — | — | — |
| CP-471474 | 210755-45-6 | 2.66 | — | — | — |
| Etoposide | 33419-42-0 | 2.66 | — | — | — |
| Staurosporine aglycone | 85753-43-1 | 2.66 | — | — | — |
| IRAK-1/4 inhibitor I | 509093-47-4 | 2.82 | — | — | — |
| BAY 61-3606 hydrochloride hydrate | 732983-37-8 | 2.98 | — | 2.37 | — |
| PD-184161 | 212631-67-9 | 2.98 | — | — | — |
| PD-161570 | 192705-80-9 | 3.35 | 21.13 | — | — |
| PAC-1d | 315183-21-2 | 3.76 | — | — | — |
| SB 242084 dihydrochloride hydrate | 181632-25-7 | 4.73 | 26.60 | — | — |
| Azoxystrobin | 131860-33-8 | 4.86 | 30.64 | — | — |
| 4-Chloroaniline | 106-47-8 | 6.11 | — | — | — |
| XCT790 | 725247-18-7 | 6.68 | — | 3.35 | — |
| PF-4708671 | 1255517-76-0 | 6.68 | — | — | — |
| MG 624 | 77257-42-2 | 9.44 | — | 12.59 | — |
| SMER28 | 307538-42-7 | 9.44 | — | 10.59 | — |
| NSC 95397 | 93718-83-3 | 9.44 | — | — | — |
| SB 202190 | 152121-30-7 | 10.59 | — | — | — |
| WIN 62,577 | 138091-43-7 | 10.59 | — | — | — |
| 3-(1H-Imidazol-4-yl)propyl | 182069-10-9 | 11.22 | — | — | — |
| di(p-fluorophenyl)methyl ether | |||||
| hydrochloride | |||||
| R(+)-Butylindazone | 81166-47-4 | 11.22 | — | — | — |
| U0126 | 109511-58-2 | 11.22 | — | — | — |
| TG003 | 719277-26-6 | 11.88 | — | 4.73 | — |
| Clotrimazole | 23593-75-1 | 11.88 | — | — | — |
| Dipyridamole | 58-32-2 | 11.88 | — | — | — |
| Progesterone | 57-83-0 | 11.88 | — | — | — |
| AA-861 | 80809-81-0 | 12.59 | — | — | — |
| Danazol | 17230-88-5 | 14.96 | — | — | — |
| Ritanserin | 87051-43-2 | 16.79 | — | — | — |
| 4,5,6,7-Tetrabromobenz-imidazole | 577779-57-8 | 16.79 | — | — | — |
| Trequinsin hydrochloride | 78416-81-6 | 16.79 | — | — | — |
| SKF 96365 | 130495-35-1 | 17.78 | — | — | — |
| 4,5,6,7-Tetrabromobenzo-triazole | 17374-26-4 | 18.83 | — | 21.13 | — |
| Spironolactone | 52-01-7 | 18.83 | — | 9.44 | — |
| H-8 dihydrochloride | 113276-94-1 | 21.13 | — | — | — |
| Mifepristone | 84371-65-3 | 23.71 | — | — | — |
| MK-886 | 118414-82-7 | — | — | 17.78 | — |
| 3-Deazaadenosine | 6736-58-9 | — | — | 16.79 | — |
| Actinonin | 13434-13-4 | — | — | 8.91 | — |
IC50 values in μM; dash indicates inactive or inconclusive response.
Chemical Abstracts Service (CAS) Registry Number.
Antagonists must have an IC50 value at least five-fold lower than the IC50 value in the viability assay (antagonism must be demonstrated independently of decreased viability).
Precaspase-activating compound 1.
Figure 3.
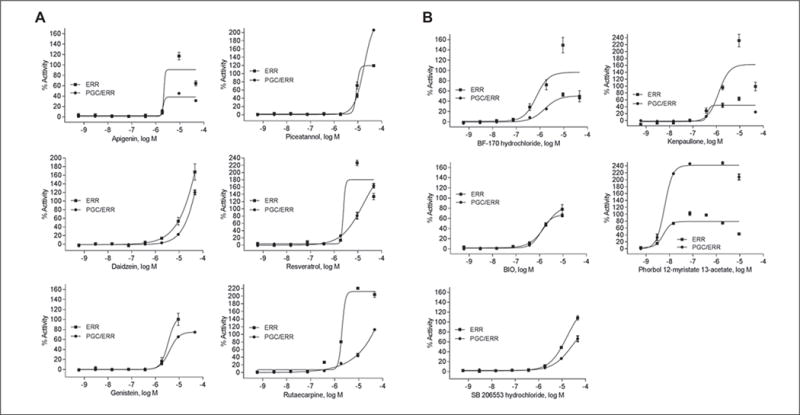
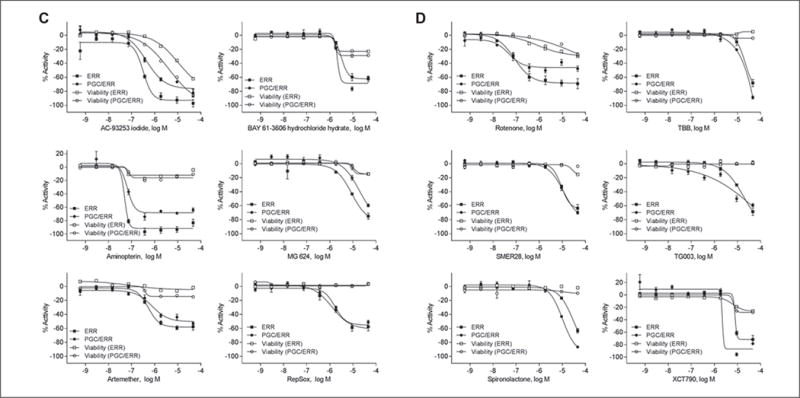
Dose-response curves for compounds identified as actives in both ERR and PGC/ERR. Cell lines, agonists (A, B) and antagonists (C, D); black squares, ERR line; black circles, PGC/ERR line; open squares, cytotoxicity for ERR line; open circles, cytotoxicity for PGC/ERR line; X axis, concentration of compounds; Y axis, response, as a percentage of the positive control. All data points are shown as mean ± SEM from three independent experiments.
Cross-Checking Results with Other LOPAC Library qHTS Screens
Concern has been expressed regarding different approaches used for interpretation of the actives in HTS screens such as those reported here.26 To verify whether the actives in the ERR and PGC/ERR screens were true actives (i.e., activity was specific for these cell lines), we cross-checked the overlapping compounds from both cell lines in agonist and antagonist mode with results from other LOPAC library screens conducted as part of the Tox21 project. For the agonist mode, we selected LOPAC screening results from five nuclear receptor assays, one GPCR assay, and five stress response pathway assays (Table 3A). It is clear that many, but not all, of the actives in the ERR and PGC/ERR pathways also activate functionally related pathways, such as ERα, RARα, AhR, mitochondrial function, ATAD5, and nrf2/ARE, while the AR, TRα, TSHR, AP1, and p53 signaling pathways were not affected. Six of the 11 top actives in the ERR and PGC/ERR assays are dietary plant compounds: apigenine, daidzin, genistein, piceatannol, resveratrol, and rutaecarpine.
Table 3A.
Activities (AC50) of ERR and PGC/ERR Agonists in Other qHTS Agonist Screens with the LOPAC Collection.
| Chemical Name | ERR ↑ | PGC-ERR ↑ | ER ↑ | AR ↑ | TR ↑ | RAR ↑ |
AhR ↑ |
TSHR ↑ | AP1 ↑ | Nrf2 ↑ | MitoTox ↑ |
ATAD5 ↑ |
TP53 ↑ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Apigenin | 5.48 | 5.63 | 4.64 | 0.00 | 0.00 | 5.78 | 5.28 | 0.00 | 0.00 | 0.00 | 5.24 | 0.00 | 0.00 |
| BF-170 hydrochloride | 5.58 | 5.78 | 6.54 | 0.00 | 0.00 | 6.53 | 4.63 | 0.00 | 0.00 | 0.00 | 4.89 | 5.74 | 0.00 |
| BIO | 5.78 | 5.93 | 0.00 | 0.00 | 0.00 | 0.00 | 5.48 | 0.00 | 0.00 | 0.00 | 7.44 | 6.14 | 4.89 |
| Daidzein | 4.88 | 4.68 | 5.74 | 0.00 | 0.00 | 5.68 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 4.69 | 0.00 |
| Genistein | 5.53 | 5.48 | 5.34 | 0.00 | 0.00 | 5.63 | 0.00 | 0.00 | 0.00 | 0.00 | 4.74 | 4.79 | 0.00 |
| Kenpaullone | 5.53 | 5.88 | 4.74 | 0.00 | 0.00 | 6.93 | 5.53 | 0.00 | 0.00 | 5.49 | 5.59 | 6.24 | 0.00 |
| Phorbol 12-myristate 13-acetate |
8.08 | 8.18 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 8.09 | 8.04 | 0.00 |
| Piceatannol | 5.08 | 4.73 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 4.74 | 5.24 | 0.00 | 0.00 |
| Resveratrol | 5.48 | 4.93 | 4.74 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 5.14 | 5.24 | 0.00 |
| Rutaecarpine | 5.63 | 4.88 | 5.24 | 0.00 | 0.00 | 6.28 | 5.53 | 0.00 | 4.92 | 5.09 | 5.19 | 5.24 | 0.00 |
| SB 206553 hydrochloride | 4.88 | 4.88 | NA | NA | NA | 5.18 | 6.38 | 0.00 | 0.00 | NA | NA | NA | NA |
Gray shades, active; NA, not tested; 0, inactive or inconclusive; AC50, log10(M)*–1 transformed; ER, estrogen receptor; AR, androgen receptor; TR, thyroid receptor; RAR, retinoic acid receptor; AhR, aryl hydrocarbon receptor; TSHR, thyroid-stimulating hormone receptor; AP1, activator protein 1; MitoTox, mitochondrial toxicity.
For the antagonists in both the ERR and PGC/ERR assays, we selected results from the ERα, AR, TR, RARα, and TSHR assays for cross-checking (Table 3B). Few compounds inhibited these nuclear receptor pathways with potency comparable to the potency seen in the ERR and PGC/ERR assays. Thus, it appears that the actives identified in the ERR and PGC/ERR assays with the LOPAC library are not promiscuous.
Table 3B.
Activities (AC50) of ERR and PGC/ERR Antagonists in Other qHTS Antagonist Screens with the LOPAC Collection.
| Chemical Name | ERR↓ | PGC/ERR ↓ | ER ↓ | AR ↓ | TR ↓ | RAR ↓ | TSHR ↓ |
|---|---|---|---|---|---|---|---|
| Aminopterin | 7.33 | 7.23 | 0.00 | 7.24 | 7.04 | 0.00 | 0.00 |
| Artemether | 6.28 | 5.98 | NA | NA | NA | 0.00 | 0.00 |
| AC-93253 iodide | 6.53 | 6.38 | 6.29 | 6.44 | 6.24 | 0.00 | 4.68 |
| RepSox | 5.88 | 5.93 | NA | NA | NA | 0.00 | 0.00 |
| MG 624 | 4.98 | 4.93 | NA | NA | NA | 0.00 | 0.00 |
| TG003 | 4.93 | 5.23 | 4.74 | 0.00 | 4.94 | 0.00 | 0.00 |
| TBB | 4.68 | 4.68 | 0.00 | 0.00 | 4.74 | 0.00 | 0.00 |
| SMER28 | 5.03 | 4.98 | NA | NA | 0.00 | 0.00 | |
| Spironolactone | 4.78 | 5.03 | 0.00 | 5.19 | 0.00 | 0.00 | 0.00 |
| Rotenone | 7.18 | 7.13 | NA | NA | NA | 0.00 | 0.00 |
| BAY 61-3606 hydrochloride hydrate | 5.53 | 5.63 | 0.00 | 0.00 | 5.64 | 0.00 | 0.00 |
| XCT790 | 5.18 | 5.48 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
Gray shades, active; NA, not tested; 0, inactive or inconclusive; AC50, log10(M)*–1 transformed; ER, estrogen receptor; AR, androgen receptor; TR, thyroid receptor; RAR, retinoic acid receptor; TSHR, thyroid-stimulating hormone receptor.
Discussion
We previously developed a stable cell line that expresses PGC-1α in HEK293T cells.17 The two current cell lines that are the subject of the work presented here, ERR and PGC/ ERR, have several advantages over the previous one. First, the assay conditions for both cell lines were optimized to fit the 1536-well plate format utilized for qHTS. In addition, the two cell lines contain a reporter that carries the natural hormone response element of the ERRα gene such that test compounds not only activate the reporter, but also activate the transcription of the endogenous ERRα gene and enhance its expression.17 Furthermore, compounds that modulate the ERRα signaling pathway in either agonist or antagonist mode can be detected in a single run of the assay, thus creating significant savings over the cost of screening independently for both modes, which is the approach that must typically be taken in assays with dual modes of signaling. These advantages, along with the easy technical performance features of the assays, make the present cell lines well suited for use as a screening tool for the Tox21 10K library.
In the studies reported here, we identified 33 compounds from the LOPAC collection as agonists in the ERR assay (23 were strongly active, with EC50 values < 10 μM), 22 compounds as agonists in the PGC/ERR assay, and 11 compounds as agonists in both assays (Table 1). Eleven compounds were identified as unique agonists in the PGC/ERR assay (i.e., no activity in ERR assay). A greater number of compounds (54) were identified as antagonists in the ERR assay (35 with IC50 values < 10 μM). An additional 15 compounds were identified as antagonists in the PGC/ERR assay, and 12 were antagonists in both assays (Table 2). Only three compounds were uniquely identified as antagonists in the PGC/ERR assay. Additional studies are needed to better understand the mechanisms whereby some compounds can influence signaling in both assays and others only in the presence or absence of the PGC cofactor.
The search for ligands and modulators of the ERRα and PGC/ERR signaling pathways, which are metabolic-sensing nuclear receptor and homeostatic switches, has been a decades-long effort by academic scientists and pharmaceutical companies. Recently, several dietary compounds and signal transduction pathway modulators were identified as ERRα agonists or antagonists.17,27–31 We and others have found that genistein, an isoflavoid, stimulates ERRα reporter activity in transient transfection experiments17,32 and enhances its expression.33 In the present study, several more dietary compounds were demonstrated to stimulate ERR and PGC/ERR signaling pathways (Table 1, Fig. 3A,B). Whether these dietary plant compounds can bind ERRα and act as agonists is still debatable, as alternative mechanisms of influencing the transcriptional activity and function of the ERRα by these dietary compounds are also likely.32,34 In fact, modulation of ERRα transcriptional activity via phosphorylation by mitogen-activated protein kinases (MAPK and AKTS) of the ErbB2/HER2 signaling pathway has been documented.35 By using AMPKα2 knockout mice, as well as by manipulating AMPKα2 expression in isolated neonatal cardiomyocytes, the importance of this pathway was demonstrated in regulation of ERRα expression and its role in heart failure.27 Similarly, PKC epsilon affects mitochondrial function through the ERRα pathway,28 suggesting that compounds that interfere with the PKC epsilon signaling pathway could also affect ERRα function via modulating the ERRα signaling pathway. Recently, regulation of nuclear receptors by botanical compounds used in some traditional Chinese medicines has generated attention.36 Since members of the nuclear receptor family, such as steroid receptors (ER, GR, and AR) and metabolic receptors (PPAR, LXR, FXR, NR1H, PXR, CAR, and RXR), are regulated by lipophilic molecules derived from endogenous hormones or the diet and the environment, they are obvious targets for some of the compounds in traditional Chinese medicines.37 Interestingly, in our studies, none of the compounds identified as antagonists in either of the two assays were plant-derived dietary compounds.
The current search for modulators of ERRα using the LOPAC library has yielded valuable information on dietary compounds that can indeed activate or disrupt this signaling pathway, suggesting a need for additional, broader searches for dietary compounds with this capability. In the studies reported here, we found that certain signal transduction regulators (e.g., PMA) and herbal compounds (e.g., apigenin, resveratrol, and rutaecarpine) included in the LOPAC library are among the most potent ERRα agonists (Fig. 3A,B, Table 1). Additional investigations are needed to establish the mechanisms of action of these dietary components (6 of the 11 most potent agonists) and their potential long-term health benefits. It is also important to investigate some of the adverse effects on ERRα signaling seen with some of the pharmaceuticals that were tested in the LOPAC, as these may be associated with long-term side effects that are currently poorly understood.
Now that the ERR and PGC/ERR assays have been validated for use in qHTS, we intend to follow up by screening the full Tox2110K library in both cell lines to identify compounds with the ability to modulate these important signaling pathways. Our intention is to identify environmental chemicals that may be important stimulators of metabolic diseases, as well as certain cancers, in order to provide evidence in support of reducing or eliminating exposures to these compounds. Identification of ERRα signaling modulators may also assist in the development of effective therapeutic interventions for the diseases that arise from such disruptions.
Supplementary Material
Acknowledgments
We thank Drs. Sifre and Bortner of the NIEHS Signal Transduction laboratory for cell sorting, Dr. Romero of the Viral Core laboratory for lentiviral infection, and Drs. Jetten and Liao for maintaining the cell stock. We also thank Carleen Klumpp-Thomas and Sam Michael for assisting with screening. We also are grateful to Drs. Sipes and Smith-Roe for critically reading the manuscript. Lastly, we are indebted to Drs. Bucher, DeVito, and Paules for strong support of this project.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This project was supported in part by Interagency Agreement NTR12003-001-0100 from the National Institute of Environmental Health Sciences/Division of the National Toxicology Program to the NIH Center for Advancing Translational Sciences, National Institutes of Health.
Footnotes
Supplementary material is available online with this article.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Giguere V, Yang N, Segui P, et al. Identification of a New Class of Steroid Hormone Receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 2.Audet-Walsh E, Giguere V. The Multiple Universes of Estrogen-Related Receptor Alpha and Gamma in Metabolic Control and Related Diseases. Acta Pharmacol Sin. 2015;36(1):51–61. doi: 10.1038/aps.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huss JM, Garbacz WG, Xie W. Constitutive Activities of Estrogen-Related Receptors: Transcriptional Regulation of Metabolism by the ERR Pathways in Health and Disease. Biochim Biophys Acta. 2015;1852(9):1912–1927. doi: 10.1016/j.bbadis.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 4.May FE. Novel Drugs That Target the Estrogen-Related Receptor Alpha: Their Therapeutic Potential in Breast Cancer. Cancer Manag Res. 2014;6:225–252. doi: 10.2147/CMAR.S35024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z, Teng CT. Estrogen Receptor-Related Receptor Alpha 1 Interacts with Coactivator and Constitutively Activates the Estrogen Response Elements of the Human Lactoferrin Gene. J Biol Chem. 2000;275(27):20837–20846. doi: 10.1074/jbc.M001880200. [DOI] [PubMed] [Google Scholar]
- 6.Kamei Y, Ohizumi H, Fujitani Y, et al. PPARgamma Coactivator 1beta/ERR Ligand 1 Is an ERR Protein Ligand, Whose Expression Induces a High-Energy Expenditure and Antagonizes Obesity. Proc Natl Acad Sci USA. 2003;100(21):12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mootha VK, Handschin C, Arlow D, et al. Err{Alpha} and Gabpa/b Specify PGC-1{Alpha}-Dependent Oxidative Phosphorylation Gene Expression That Is Altered in Diabetic Muscle. Proc Natl Acad Sci USA. 2004;101(17):6570–6775. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber SN, Knutti D, Brogli K, et al. The Transcriptional Coactivator PGC-1 Regulates the Expression and Activity of the Orphan Nuclear Receptor ERRalpha. J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 9.Lehman JJ, Kelly DP. Transcriptional Activation of Energy Metabolic Switches in the Developing and Hypertrophied Heart. Clin Exp Pharmacol Physiol. 2002;29(4):339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 10.Puigserver P, Spiegelman BM. Peroxisome Proliferator-Activated Receptor-Gamma Coactivator 1 Alpha (PGC-1 Alpha): Transcriptional Coactivator and Metabolic Regulator. Endocr Rev. 2003;24(1):78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 11.Matsuyama R, Takada I, Yokoyama A, et al. Double PHD Fingers Protein DPF2 Recognizes Acetylated Histones and Suppresses the Function of Estrogen-Related Receptor Alpha through Histone Deacetylase 1. J Biol Chem. 2010;285(24):18166–18176. doi: 10.1074/jbc.M109.077024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Tremblay AM, Wilson BJ, Yang XJ, et al. Phosphorylation-Dependent Sumoylation Regulates Estrogen-Related Receptor-Alpha and -Gamma Transcriptional Activity through a Synergy Control Motif. Mol Endocrinol. 2008;22(3):570–584. doi: 10.1210/me.2007-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birnbaum LS. When Environmental Chemicals Act Like Uncontrolled Medicine. Trends Endocrinol Metab. 2013;24(7):321–323. doi: 10.1016/j.tem.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt CW. TOX 21: New Dimensions of Toxicity Testing. Environ Health Perspect. 2009;117(8):A348–A353. doi: 10.1289/ehp.117-a348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R, Xia M, Cho MH, et al. Chemical Genomics Profiling of Environmental Chemical Modulation of Human Nuclear Receptors. Environ Health Perspect. 2011;119(8):1142–1148. doi: 10.1289/ehp.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tice RR, Austin CP, Kavlock RJ, et al. Improving the Human Hazard Characterization of Chemicals: A Tox21 Update. Environ Health Perspect. 2013;121(7):756–765. doi: 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng CT, Beames B, Alex Merrick B, et al. Development of a Stable Cell Line with an Intact PGC-1alpha/ERRalpha Axis for Screening Environmental Chemicals. Biochem Biophys Res Commun. 2014;444(2):177–181. doi: 10.1016/j.bbrc.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busch BB, Stevens WCJr, Martin R, et al. Identification of a Selective Inverse Agonist for the Orphan Nuclear Receptor Estrogen-Related Receptor Alpha. J Med Chem. 2004;47(23):5593–5596. doi: 10.1021/jm049334f. [DOI] [PubMed] [Google Scholar]
- 19.Willy PJ, Murray IR, Qian J, et al. Regulation of PPARgamma Coactivator 1alpha (PGC-1alpha) Signaling by an Estrogen-Related Receptor Alpha (ERRalpha) Ligand. Proc Natl Acad Sci USA. 2004;101(24):8912–8917. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Zhang Z, Gladwell W, et al. Estrogen Stimulates Estrogen-Related Receptor {Alpha} Gene Expression through Conserved Hormone Response Elements. Endocrinology. 2003;144(11):4894–4904. doi: 10.1210/en.2003-0432. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Zhang Z, Teng CT. Estrogen-Related Receptor-Gamma and Peroxisome Proliferator-Activated Receptor-Gamma Coactivator-1alpha Regulate Estrogen-Related Receptor-Alpha Gene Expression via a Conserved Multi-Hormone Response Element. J Mol Endocrinol. 2005;34(2):473–487. doi: 10.1677/jme.1.01586. [DOI] [PubMed] [Google Scholar]
- 22.Barde I, Salmon P, Trono D. Production and Titration of Lentiviral Vectors. Curr Protoc Neurosci. 2010 doi: 10.1002/0471142301.ns0421s53. Chapter 4, Unit 4.21. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan M, Park JM, Cao F, et al. Effects of Epigenetic Modulation on Reporter Gene Expression: Implications for Stem Cell Imaging. FASEB J. 2006;20(1):106–108. doi: 10.1096/fj.05-4551fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inglese J, Auld DS, Jadhav A, et al. Quantitative High-Throughput Screening: A Titration-Based Approach That Efficiently Identifies Biological Activities in Large Chemical Libraries. Proc Natl Acad Sci USA. 2006;103(31):11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Huang R. In: High-Throughput Screening Assays in Toxicology. Zhu H, Xia M, editors. Humana Press; New York: 2016. pp. 123–134. chapter correction of microplate data from high throughput screening. [Google Scholar]
- 26.Baell JB, Holloway GA. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J Med Chem. 2010;53(7):2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 27.Hu X, Xu X, Lu Z, et al. AMP Activated Protein Kinase-Alpha2 Regulates Expression of Estrogen-Related Receptor-Alpha, a Metabolic Transcription Factor Related to Heart Failure Development. Hypertension. 2011;58(4):696–703. doi: 10.1161/HYPERTENSIONAHA.111.174128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu N, Wang W, Liu J, et al. Protein Kinase C Epsilon Affects Mitochondrial Function through Estrogen-Related Receptor Alpha. Cell Signal. 2011;23(9):1473–1478. doi: 10.1016/j.cellsig.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Ranhotra HS. Estrogen-Related Receptor Alpha and Mitochondria: Tale of the Titans. J Recept Signal Transduct Res. 2015;35(5):386–390. doi: 10.3109/10799893.2014.959592. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Fang F, Huang Z, et al. Kaempferol Is an Estrogen-Related Receptor Alpha and Gamma Inverse Agonist. FEBS Lett. 2009;583(4):643–647. doi: 10.1016/j.febslet.2009.01.030. [DOI] [PubMed] [Google Scholar]
- 31.Wei W, Schwaid AG, Wang X, et al. Ligand Activation of ERRalpha by Cholesterol Mediates Statin and Bisphosphonate Effects. Cell Metab. 2016;23(3):479–491. doi: 10.1016/j.cmet.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suetsugi M, Su L, Karlsberg K, et al. Flavone and Isoflavone Phytoestrogens Are Agonists of Estrogen-Related Receptors. Mol Cancer Res. 2003;1(13):981–991. [PubMed] [Google Scholar]
- 33.Ambra R, Rimbach G, de Pascual Teresa S, et al. Genistein Affects the Expression of Genes Involved in Blood Pressure Regulation and Angiogenesis in Primary Human Endothelial Cells. Nutr Metab Cardiovasc Dis. 2006;16(1):35–43. doi: 10.1016/j.numecd.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Huang Z, Fang F, Wang J, et al. Structural Activity Relationship of Flavonoids with Estrogen-Related Receptor Gamma. FEBS Lett. 2010;584(1):22–26. doi: 10.1016/j.febslet.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 35.Ariazi EA, Kraus RJ, Farrell ML, et al. Estrogen-Related Receptor Alpha1 Transcriptional Activities Are Regulated in Part via the ErbB2/HER2 Signaling Pathway. Mol Cancer Res. 2007;5(1):71–85. doi: 10.1158/1541-7786.MCR-06-0227. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Bonneton F, Chen XY, et al. Botanical Compounds and Their Regulation of Nuclear Receptor Action: The Case of Traditional Chinese Medicine. Mol Cell Endocrinol. 2015;401:221–237. doi: 10.1016/j.mce.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 37.Lazar MA. East Meets West: An herbal Tea Finds a Receptor. J Clin Invest. 2004;113(1):23–25. doi: 10.1172/JCI200420661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


