Abstract
Rhabdomyosarcoma is the most common soft-tissue sarcoma of childhood, and despite clinical advances, subsets of these patients continue to suffer high levels of morbidity and mortality associated with their disease. Recent genetic and molecular characterization of these tumors using sophisticated genomics techniques, including next-generation sequencing experiments, has revealed multiple areas that can be exploited for new molecularly targeted therapies for this disease.
Keywords: Rhabdomyosarcoma, Alveolar Rhabdomyosarcoma, Embryonal Rhabdomyosarcoma, Targeted Therapy, Genomics, Epigenetics, Development
I. INTRODUCTION
Rhabdomyosarcoma (RMS) is the most common soft-tissue sarcoma of childhood, with an annual incidence of 4.5 cases per 1 million children, making it the third most prevalent extracranial solid tumor of childhood after neuroblastoma and Wilms tumor.1 RMS tumors typically are associated with the skeletal muscle lineage, and approximately 50% of cases are diagnosed in the first decade of life. RMS is currently categorized by histopathology into distinct subtypes, including embryonal, alveolar, pleomorphic, and sclerosing/spindle cell pathology, which have distinct molecular and clinical correlates.2 In the 1970s and 1980s a multimodal chemotherapy backbone of vincristine, actinomycin, and cyclophosphamide was established as an effective treatment for RMS.3 Through a series of collaborative group clinical trials, dose modification of this backbone, coupled with improvements in local control and supportive care, have led to impressive gains in survival over the past decades. Patients with low-risk disease now have a 5-year survival that approaches 90%, and current efforts are focused on dose reduction to avoid long-term effects.4 Relapse-free survival for patients with localized disease has improved to 70–80%, albeit with significant toxicity.4 Despite these impressive and important gains, several clinical trends have emerged. First, there has been a general flattening in the improvement in outcome for all patients. This is likely because, despite several randomized studies to evaluate dosing and schedule, the systemic chemotherapy backbone has remained largely unchanged since the 1970s. This effect is most pronounced for patients with intermediate and high-risk disease who, in a series of trials, experienced escalating dose intensification and compression of cytotoxic therapy with minimal gains.5,6 Second, patients with high-risk disease or recurrent disease continue to suffer a dismal prognosis (5-year survival <30% and 17%, respectively).5,7 Finally, clinical trials that integrate the growing knowledge of the oncogenic mechanisms of these tumors with novel therapies have been slow to emerge. This review summarizes recent advances in the understanding of the genetic and molecular basis of RMS and highlights how investigators and clinicians are using this information in an effort to improve outcomes for patients with RMS.
II. RMS GENETICS
The association of RMS with familial cancer syndromes, most notably Li-Fraumeni syndrome,8 neurofibromatosis,9 Beckwith-Wiedemann syndrome,10 and Costello syndrome,11 has made the genetics of RMS an area of intense study. Decades of targeted sequencing and microarray methods have led to the discovery of loss of heterozygosity at 11p15.512; mutations in TP53,13 NRAS, KRAS, HRAS,14 PIK3CA, CTNNB1,15 and FGFR416; and the characteristic translocations involving the PAX3 or PAX7 genes with FOXO117 that have defined the genomic characteristics frequently associated with histologic and clinical features of this disease.
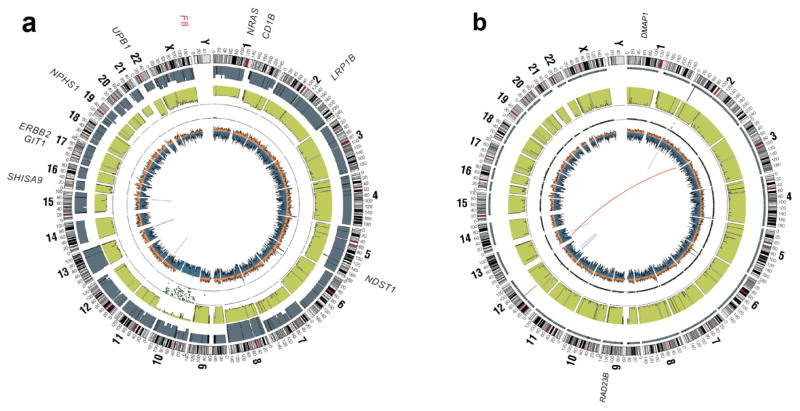
Several more recent large-scale, next-generation sequencing studies of primary RMS tumors have been reported.18–20 These studies revealed in a comprehensive manner the landscape of mutations, copy number changes, and genomic rearrangements that define these tumors. These studies each show that primary RMS has a low overall mutation rate (0.31 protein-coding mutations/Mb) and is characterized by 2 distinct genotypes, which can clearly be defined by the presence or absence of a PAX gene rearrangement (Fig. 1). Next-generation sequencing studies have confirmed that RMS should not be solely diagnosed by histology but by the presence (fusion-positive RMS) or absence (fusion-negative RMS) of a PAX3/7 gene fusion.18
FIG. 1.
Whole-genome sequencing has revealed that rhabdomyosarcoma tumors can be classified into 2 distinct genotypes characterized by the absence (a) or presence (b) of a PAX gene rearrangement. CIRCOS plots from representative tumors are presented (from the outside circle in). Mutated genes: missense mutations (black), non-sense mutations and insertions/deletions (red); genomic location: genome-wide copy number alterations (gray), lesser allele frequency (green), loss of heterozygosity (dotted track), density of heterozygous single nucleotide polymorphisms (SNPs) (orange), homozygous SNPs (blue); intrachromasomal rearrangements (inner circle, gray) and interchromosomal rearrangements (inner circle, red). Adapted from Shern et al.18
A. PAX Fusion–Positive RMS
In RMS, the PAX3 or PAX7 gene fusions were originally found through physical mapping and cloning studies, which revealed the rearrangement of chromosome 2 or 1 in a reciprocal translocation with FOXO1, found on chromosome 13.17,21 Follow-up studies have confirmed that juxtaposition of the N-terminus of the paired box genes with the C-terminus of the forkhead transcription factor characterizes a distinct subset of RMS genotypes. Other infrequent rearrangements of the PAX3 gene also have been observed in tumors with alveolar histology, including the in-frame fusion with the nuclear receptor coactivator NCOA1,22,23 or the chromatin remodeling gene INO80D.18 The tumors that harbor these fusions retain the expression signature characteristic of the canonical fusions PAX3-FOXO1 and PAX7-FOXO1 and define a subset of tumors previously described as fusion-negative alveolar histology.
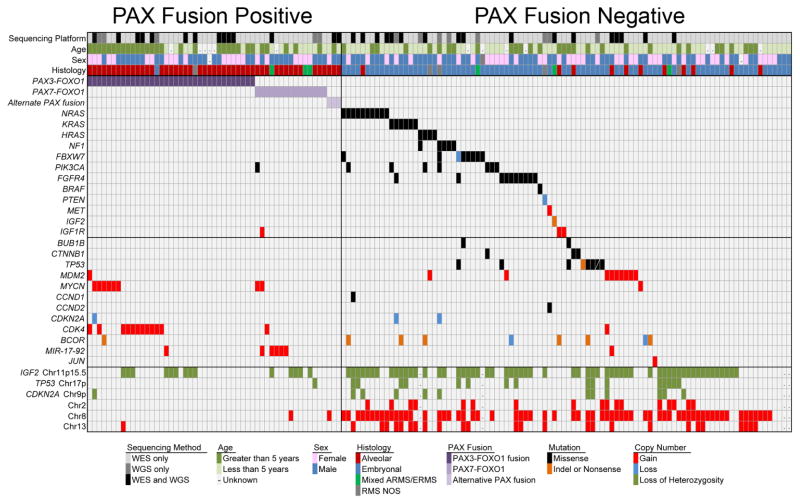
In general, tumors that have a PAX gene translocation have an extremely low overall mutation rate (0.1 protein-coding mutation/Mb) and, interestingly, no recurring genes with single nucleotide mutations18 (Fig. 2). While recurrent collaborating point mutations have not been found in these tumors, regions of focal genomic amplification are frequently observed (Table 1). Multiple genome-wide analyses of copy number alterations in RMS to date have been completed using the single nucleotide polymorphism array technology. The most commonly amplified genomic regions observed in PAX gene fusion–positive tumors are 2p24, containing the MYCN oncogene, and 12q13-q14, which includes CDK4.26 The amplification of MYCN, which occurs in 28% of fusion-positive cases, has been confined to a genomic region less than 1 Mb, including the same region frequently observed in neuroblastoma cases.27 This region includes only 2 genes (MYCN and DDX1) and is observed most commonly in PAX3-FOXO1 fusion–positive RMS. While the number of cases remains small, no correlation between 2p24 amplification and RMS clinical outcome has been shown, in contrast to neuroblastoma.25 Amplifications of 12q13-q14, however, have been associated with significantly worse failure-free and overall survival independent of PAX gene fusion status.25 This amplicon also is observed in multiple other tumor types, including lung cancer, glioblastoma, and osteosarcoma. The observed region has been confined to a common region, 0.55 Mb in length, that contains 27 genes, including CDK4. Expression analysis confirms that the genomic amplification results in overexpression of CDK4. Other amplified regions in fusion-positive tumors include 15q24-26, 1p36, 13q31, 1q21, and 8q13-21.26 The regions of 1p36, which encompasses the PAX7 locus, and 13q31, which includes MIR17HG, are associated specifically with PAX7-FOXO1 tumors.
FIG. 2.
A summary of the genomic alterations frequently occurring in primary rhabdomyosarcoma shows 2 distinct genotypes defined by the presence or absence of a PAX gene fusion and recurrent mutation of RAS pathway genes in fusion-negative tumors. Adapted from Shern et al.18
TABLE 1.
Genetic Alterations Commonly Observed in PAX Gene Fusion–Positive Rhabdomyosarcoma
| Alteration | Type | Chromosomal Locus | Estimated Frequency | References |
|---|---|---|---|---|
| PAX3-FOXO1 | Translocation | (2;13)(q35;q14) | 59% of ARMS | 17, 24 |
| PAX7-FOXO1 | Translocation with amplification | (1;13)(p36;q14) | 19% of ARMS | 21, 24 |
| PAX3-NCOA1 | Translocation | Alveolar | Rare | 23 |
| PAX3-INO80D | Translocation | Alveolar | Rare | 18 |
| CDK4 | Amplification | 12q13-14 | 12% | 25 |
| MYCN | Amplification | 2p24 | 20% | 25 |
| miR-17-92 | Amplification | 13q31-32 | 26 | |
| IGF2 locus | Uniparental disomy | 11p15.5 | 29% | 18 |
ARMS, alveolar rhabdomyosarcoma.
B. Fusion–Negative RMS
In contrast to PAX fusion–positive samples, tumors that do not harbor the fusion are characterized by a more heterogeneous histology, complex karyotype, regions of loss of heterozygosity, and an increased presence of single nucleotide point mutations. These tumors display a wide range of causative mutations. The mutation most frequently occurs within one of the Ras genes (NRAS > KRAS > HRAS), a receptor tyrosine kinase (FGFR4 ≫ ERBB2), or the catalytic component (PIK3CA) of the phosphoinositide-3 kinase (PI3K) complex. Other genes that are recurrently mutated include the ubiquitin ligase FBXW7, as well as NF1, TP53, CTNNB1, and the transcriptional repressor BCOR28 (Table 2). In addition, 2 recent studies have identified that point mutation of the myogenic muscle differentiation transcription factor MYOD1 defines an aggressive subset of embryonal RMS20 and adult spindle cell RMS.31
TABLE 2.
Genetic Alterations Commonly Observed in PAX Gene Fusion–Negative Rhabdomyosarcoma
| Alteration | Type | Chromosome Locus | Estimated Frequency | References |
|---|---|---|---|---|
| IGF2 | Loss of heterozygosity | 11p15.5 | 65% | 18 |
| NRAS | Point mutation | Chr1:115256530 Chr1:115258745 |
7.5% | 18 |
| KRAS | Point mutation | Chr12:25398284 Chr12:25398281 |
4% | 18 |
| HRAS | Point mutation | Chr11:534289 | 3% | 18 |
| NF1 | Point mutation | Multiple | 3.4% | 18 |
| PIK3CA | Point mutation | Chr3:178952084-178952085 Chr3:178936094-178936096 |
5.4% | 18, 29 |
| FBXW7 | Point mutation | Chr4:153247287 Chr4:153249456 Chr4:153251905 |
4.8% | 18 |
| FGFR4 | Point mutation | Chr5:176522551 | 9.3% | 30 |
| BCOR | Point mutation/indel | Multiple | 5.4% | 18 |
| CTNNB1 | Point mutation | Chr3:41266101 Chr3:41266124 |
2% | 18, 29 |
| MYOD1 | Point mutation | p.Leu122Arg | 10% of ERMS | 20, 31 |
| MDM2 | Amplification | 12q15 | 10% | 32 |
| Aneuploidy | Chromosome gain | Typically 2, 7, 8, 11, 13 | Common | 18 |
| Chromosome loss | Typically 1p, 9, 16 | Common | 18 |
ERMS, embryonal rhabdomyosarcoma; indel, insertion/deletion.
Perhaps the most unifying feature of fusion-negative tumors is the loss of heterozygosity at the 11p15.5 locus,12 which occurs in a majority of fusion-negative tumors and also is observed in the Beckwith-Wiedemann syndrome,33 hepatoblastoma,34 and Wilms tumor.35 The observed allelic loss of the maternal allele is frequently associated with duplication of the remaining allele, resulting in paternal isodisomy.36 While many of the genes involved in the region have been implicated in oncogenesis, H19, IGF2, and CDKN1C have been the most extensively studied. Loss of imprinting at the IGF2 gene locus is associated with massive overexpression of IGF2, which is a nearly universal finding in RMS.
Chromosome- and chromosome arm-level gains and losses are frequent events in fusion-negative tumors. Multiple array studies have reported recurrent gains of chromosomes 2, 7, 8, 12, and 13.37–39 In addition, focal losses of 9q32-34, which includes CDKN2A, and 17p, which includes the TP53 and NF1 loci, are observed. One recurrent focal amplification event that occurs in fusion-negative tumors is the high copy gain of the 12q14-15 locus, containing the MDM2 gene. Alteration of the MDM2 locus is a common event in soft-tissue sarcomas,32 and the gene product is known to bind and inactivate TP53. In RMS, the MDM2 amplicon can overlap with the CDK4 amplicon, but more frequently the 2 alterations seem to be mutually exclusive.
III. RMS EPIGENETICS
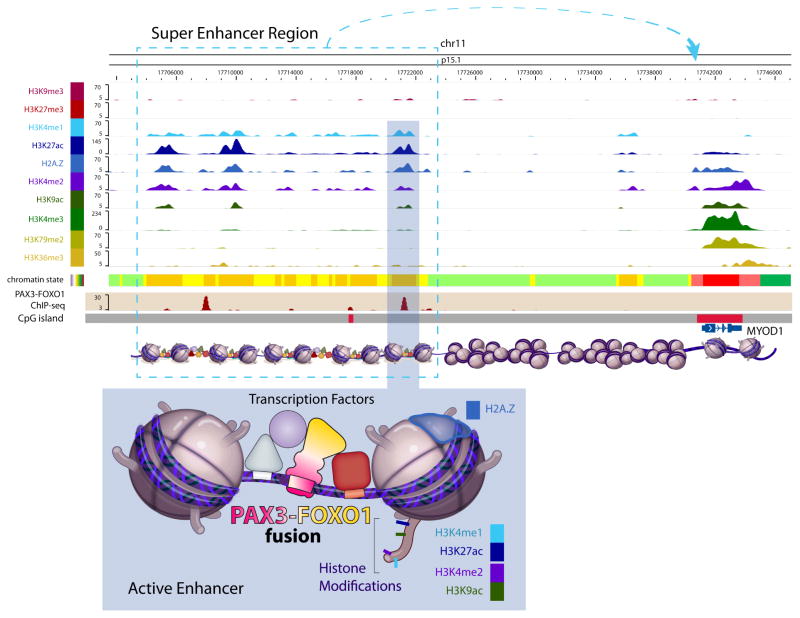
With the emergence of novel techniques to interrogate the epigenome, there have been efforts to define the DNA modifications that affect transcription within these tumors. Hypermethylation of 5′ regulatory regions of cancer genomes results in transcriptional repression of tumor suppressors, and treatment of RMS cell lines with the DNA demethylating agent 5-azacytadine results in a differentiation phenotype.40 Several groups have used a candidate gene approach in RMS tumors to identify methylation changes at the promoters of FGFR1, MYOD1, and PAX3.41–43 A more global approach using genome-wide DNA methylation arrays was recently reported. The authors reported that both fusion-positive and fusion-negative tumors have distinct methylation profiles, with the fusion-positive subtype showing enriched methylation in genes targeted by the polycomb repressive complex.44 In addition, they reported promoter methylation of IRX1, DNAJA4, and P4HTM in cell lines and primary tumors that results in the silencing of these genes when compared with normal skeletal muscle. The gene product of EZH2 is a critical component of the polycomb repressive complex 2 (PRC2), which catalyzes trimethylation of histone H3 lysine 27 and recruits polycomb complexes, DNA methyltransferases, and histone deacetylases (HDACs), resulting in transcriptional repression. Aberrant EZH2 activity, either as a result of mutation or overexpression, is a common feature of cancer.45 In addition, EZH2 expression decreases and the PRC2 dissociates as myogenesis progresses toward mature muscle development.46 While the critical genes that EZH2 modulates remain to be elucidated, certainly EZH2 is highly upregulated in RMS cell lines and tumors.47 One intriguing study found that the PRC2 in RMS may play a role in preventing the binding of MYOD1 at muscle-specific genes.48 PAX3-FOXO1 itself is a fusion of 2 transcription factors; as such, chromatin immunoprecipitation sequencing analysis using an antibody specific to the fusion junction has been used to interrogate how the oncogene interacts with the genome.49 Interestingly, the vast majority of PAX3-FOXO1 binding sites are located more than 4 kb from a transcriptional start site and only infrequently (0.4%) within 1 kb upstream of the transcription initiation site, making it much more likely that the fusion gene exerts its effects through enhancer regions rather than at promoters. Enriched peaks included PAX3 binding motifs and regions distal to the muscle development genes MYOD1 (Fig. 3) and MYF5. In addition, Cao et al. demonstrated a strong association between chromatin immunoprecipitation sequencing peaks and genes that are overexpressed in alveolar RMS and confirmed that ALK, FGFR4, IGF1R, and MYCN are direct targets of PAX3-FOXO1.
FIG. 3.
The interaction of PAX3-FOXO1 with genomic locations. As determined by chromatin precipitation, the PAX3-FOXO1 fusion gene is typically associated with enhancer regions of the genome. Presented here are the fusion gene binding sites in the MYOD1 enhancer region (top panel) and its relationship to other epigenetic and transcriptional marks (bottom panel).
IV. PATHOGENESIS AND BIOLOGY OF RMS
A. Fusion-Positive RMS
PAX3-FOXO1 clearly defines a distinct genotype, and given its role as the key prognostic marker in RMS,50,51 multiple groups have attempted to characterize the effects of the fusion gene on the cell. The translocations create a break within intron 7 of the PAX gene and intron 1 of the FOXO1 gene, resulting in a chimeric transcript that encodes the N-terminal DNA-binding domain of the PAX gene with the C-terminal activation domain of the FOXO1 gene.52,53 Expression of the fusion gene causes transformation and anchorage-independent growth of fibroblasts.54 Knockdown of PAX3-FOXO1 decreased the proliferation rate in the RMS cell line RH30 and the metastatic phenotype observed in the corresponding xenograft model.55 In addition, conditional simultaneous biallelic PAX3-FOXO1 expression from the PAX3 locus and homozygous deletion of either TP53 or CDKN2A in Myf6-expressing maturing mouse myofibers leads to alveolar RMS development with 100% penetrance.56,57
Expression analysis has been the tool of choice to globally dissect the downstream effects of the presence of the fusion gene. These studies revealed that the oncogene alters the myogenic program of the cell, inducing or repressing a large set of muscle development genes, including MYOD1, MYOG, and SIX1.59,59 Expression of the fusion protein also massively upregulates several receptor tyrosine kinase molecules important for cell growth, including FGFR4,60 ALK,61 and MET,62 which may provide a feed-forward loop driving proliferation. Universally, these studies have demonstrated a common signature that is associated with a tumor’s fusion status and that can be used for diagnosis and prognosis.51,63
B. Fusion-Negative RMS as an “RAS-opathy”
As discussed above, the most common causative single nucleotide variant in fusion-negative RMS is an oncogenic mutation in one of the Ras isoforms, namely, NRAS, HRAS, or KRAS. The role of Ras mutation as a prognostic marker in fusion-negative RMS is emerging. In one recent study 75% of high-risk fusion-negative RMS tumors harbored a Ras isoform mutation, in contrast to 45% of intermediate-risk and 0% of low-risk fusion-negative RMS tumors with a Ras isoform mutation.19 A subset of fusion-negative RMS tumors harbors mutations in the Ras GTPase-activating protein NF1, which also potentially leads to aberrant Ras signaling in these tumors. Finally, activating mutations in known Ras effectors such as BRAF and PIK3CA also have been identified in fusion-negative RMS tumors; however, the signaling pathways downstream of activated Ras that are necessary for tumorigenesis in fusion-negative RMS have yet to be fully characterized. Fusion-negative RMS is unique in that mutations in all 3 of the major Ras isoforms may function as genetic driver mutations.64 The functional consequences of individual Ras isoform mutations in fusion-negative RMS tumors are not yet known.
The available in vitro and in vivo models of fusion-negative RMS confirm a central role for aberrant Ras activity in fusion-negative RMS tumorigenesis. A knock-in model in which oncogenic KRAS is conditionally expressed in the skeletal muscle of adult mice deficient in TP53 leads to the development of pleomorphic RMS.65 In addition, human postnatal skeletal muscle myoblasts engineered to overexpress large T oncoprotein, small t oncoprotein, human telomerase reverse transcriptase, and oncogenic HRAS show anchorage-independent growth and form tumors capable of local invasion and distant metastasis when implanted in immunodeficient mice.66 Most of the established human fusion-negative RMS cell lines currently in use harbor oncogenic mutations in one of the Ras isoforms.67 Finally, zebrafish embryos injected with oncogenic KRAS at the one cell stage develop tumors that histologically resemble fusion-negative RMS.68 In sum, the available next-generation sequencing and model system data indicate that the major driver of fusion-negative RMS tumorigenesis is oncogenic Ras.
Mutation of FGFR4 has been identified in approximately 7% of fusion-negative RMSs.30 Interestingly, FGFR4 is expressed in myoblasts during normal development and in regenerating muscle following injury, but not in mature skeletal muscle. Based on microarray-based gene expression analysis, this gene has been reported to be the most differentially expressed gene in RMS tumors.59,63,69 These observations led to the hypothesis that overexpression or mutational activation of this gene may be involved in the tumorigenesis of RMS. Taylor et al. found that suppression of the wild-type FGFR4 in RMS led to reduced growth and lung metastasis.16 Mutations in the tyrosine kinase domain were predicted and confirmed to be activating and resulted in increased growth and reduced RMS cell death, and enhanced the ability of RMS cells to metastasize. The investigators found that the mutations lead to the activation of an oncogenic pathway involving STAT3.30 Subsequent studies have shown that stimulation of wild-type FGFR4 in fusion-positive RMS cell lines leads to activation of the RAS-RAF-MEK-ERK mitogen-activated protein kinase pathway, indicating that RMS tumors expressing activating mutations in FGFR4 may phenocopy tumors with activating mutations in Ras isoforms.70
Insulin-like growth factor (IGF) 2 promotes myoblast proliferation and nutrient uptake. Loss of imprinting at the IGF2 locus is associated with massive overexpression of IGF2 at the messenger RNA and protein levels in fusion-negative RMS cell lines, and expression of IGF2 is also induced by PAX3-FOXO1.60 The cellular effects of IGF2 are mediated by the type I IGF receptor (IGF1R), which also is expressed on RMS cells. IGF2 is secreted by RMS cell lines and is able to act as a mitogen in an autocrine manner in this model system.71 Mouse C2C12 myoblasts expressing IGF2 in combination with PAX3-FOXO1 are transformed in vitro and form undifferentiated tumors in immunodeficient mice.72
C. Evidence for a Commonly Disrupted Pathway in Fusion-Positive and Fusion-Negative Tumors
Our genomic characterization of RMS tumors identified some interesting overlap between the genes mutated in fusion-negative tumors and the genes that are under the control of the PAX3-FOXO1 fusion gene. The results demonstrate that both alveolar RMS and embryonal RMS tumors hijack the common receptor tyrosine kinase/RAS/PIK3CA axis through alternative mechanisms: either mutation or translocation. Up to 93% of RMS tumors have genetic evidence indicating alteration of this axis.18 With the proliferation of approved clinical agents that target these pathways, these findings provide a molecular basis for the rapid movement of these agents into clinical trials of RMS.
D. Alteration of Developmental Programs in RMS
Anatomic location of RMS tumors within skeletal muscle and the characteristic expression of muscle markers such as myogenin73 and MYOD174 provide evidence that RMS tumors reflect a disordered or corrupted muscle development program. The process of normal myogenic differentiation is driven by sequential expression of myogenic regulatory factors that include the basic helix-loop-helix transcription factors MYOD1, MYF5, MYOG, and MYF6 (reviewed in refs. 75 and 76). The paired box transcription factors PAX3 and PAX7 in turn regulate MYOD1 and MYF5.77 Forced expression of the PAX3-FOXO1 fusion alters a myogenic program including upregulation of MYOD1, MYOG, SIX1, and IGF2 and induces myoblast-like cells.60 Collectively, these findings indicate that in RMS the oncogenic damage to a muscle precursor cell leads to a persistent or static developmental state that drives both the survival and proliferation signals in the tumor.
V. POTENTIAL NEW THERAPEUTIC OPTIONS
A. Signaling Inhibitors, Including Combination Strategies
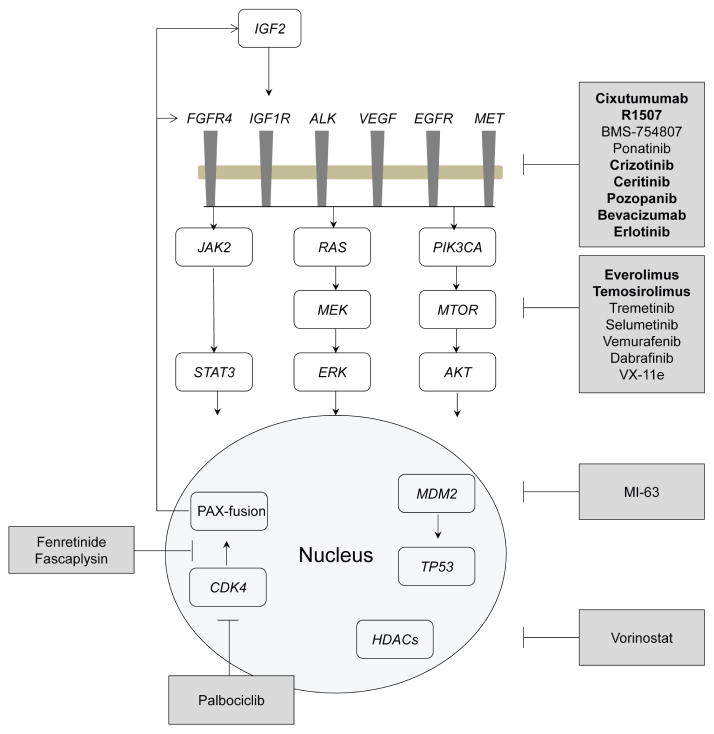
Perhaps the most immediate translational clinical opportunity that can be derived from the molecular and genomic study of RMS is that these tumors result from alteration of signaling and growth pathways, many of which can be targeted with small-molecule inhibitors or biological agents (Fig. 4).
FIG. 4.
Summary of the potential therapeutic options outlined in this review. Adapted from Shern et al.18
The nearly universal alteration of the insulin signaling pathway in RMS tumors has made it an intense focus for novel therapies. Cixutumumab, a human immunoglobulin G monoclonal antibody directed against IGF1R, the receptor for IGF2, showed promising single-agent activity in xenograft models of RMS.78 Based on these results, 20 pediatric patients with RMS were treated in a phase II clinical trial of cixutumumab as a single agent. In that study, 1 patient had a partial response to anti-IGF1R therapy, 1 patient had prolonged stable disease, and 2 patients had short-term stable disease.79 The efficacy of cixutumumab in combination with intensive multiagent, interval, compressed cytotoxic chemotherapy was studied in metastatic RMS (www.clinicaltrials.gov identifier NCT01055314). An additional anti-IGF1R antibody, R1507, also showed promising results as a single-agent therapy in patients with RMS: Of 36 patients with RMS treated in the study, 1 patient had a partial response and 6 patients had stable disease.80 BMS-754807, a small-molecule, ATP-competitive inhibitor of IGF1R, has shown promising activity as a single agent in RMS cell lines in vitro and is currently under investigation in phase I clinical trials for adults with solid tumors.78
FGFR4 signaling is altered in both fusion-positive (by overexpression) and fusion-negative (by mutation) RMS, making FGFR4 an attractive candidate for targeted therapy. Recent work demonstrated that the tyrosine kinase inhibitor ponatinib potently inhibited FGFR4 signaling in cell lines expressing either wild-type or mutant FGFR4. This drug also inhibited growth of RMS cell lines in vitro, induced apoptosis in RMS cell lines, and inhibited tumor growth in xenografts of cell lines expressing mutant FGFR4.81
The ALK receptor, a member of the insulin receptor family of receptor tyrosine kinases, activates the signal transducer and activator of transcription 3, PI3K/AKT, and Ras/ERK pathways, and is highly expressed in both fusion-positive and fusion-negative RMS. This high level of protein expression can be the result of ALK gene copy number gain. In addition, ALK gene expression is enhanced by PAX3-FOXO1.61 Genomic gains or amplifications of ALK also occur in pediatric tumor neuroblastoma, as well as non-small-cell lung cancer. Small-molecule inhibitors of ALK (crizotinib, ceritinib) are currently being studied in these tumor types (www.clinicaltrials.gov identifier NCT01742286) and may prove to be beneficial in the treatment of RMS. Additional receptor tyrosine kinases, such as MET, epidermal growth factor receptor, and vascular endothelial growth factor receptor also are overexpressed in RMS compared with normal muscle, and small-molecule inhibitors or antibody-based therapies directed against these receptor tyrosine kinases could also be developed as effective RMS therapies.82
RMS likely represents a malignant tumor of myoblast-like cells failing to exit the cell cycle and differentiate. The cell cycle exit and differentiation of myoblasts into myotubes are mediated, in part, by downregulation of cyclin D1 and a decrease in CDK4/CDK6 activity. Genetic amplification of CDK4 and genetic loss of the CDK4/6-specific inhibitor p16Ink4a (CDKN2A locus) frequently occur in RMS, making CDK4 an attractive target in RMS. Treatment of RMS cell lines with the CDK4/6 inhibitor palbociclib (PD-0332991) leads to G1 arrest and induces the expression of muscle-specific markers. This inhibitor is currently in early phase clinical trials in adults and may represent a therapeutic option for patients with RMS.83 In addition, as discussed above, the MDM2 locus also is frequently amplified in RMS. An inhibitor of MDM2–TP53 interaction, MI-63, decreases proliferation and increases apoptosis in RMS cell lines expressing wild-type TP53.84 This compound is currently awaiting phase I testing.
Because Ras isoforms also are commonly mutated in fusion-negative RMS, targeting Ras effectors such as PI3K and BRAF is a logical strategy. Several BRAF (veumurafenib, dabrafenib) and MEK (trametinib) inhibitors have gained US Food and Drug Administration (FDA) approval for the treatment of other Ras-driven cancers, such as metastatic melanoma. Recent work has shown that the small-molecule inhibitors of MEK and PI3K—U0126 and PI103, respectively—have a synthetic lethal interaction in RD, a fusion-negative RMS cell line harboring an activating mutation in NRAS.85 Further studies have shown that growth of RD is dependent on the RAS-RAF-MEK-ERK signaling pathway because an MEK inhibitor, AZD244 (selumetinib), and an ERK2 inhibitor, VX-11e, inhibit proliferation of these cells.86 Finally, treatment with the MEK inhibitor PD-0352901 leads to differentiation of RD.87
Ultimately, single-agent targeted therapy for RMS seems unlikely to provide durable responses. The mechanisms of resistance to therapy are relatively unstudied in RMS; however, some evidence exists that the mechanism likely includes genetic selection of a minor clone that is present at the time of diagnosis.19 Given the evidence for altered signaling in RMS through the RAS signaling pathway and the insulin growth signaling pathway, one therapeutic strategy might be to target both the mitogen-activated protein kinase as well as the AKT pathways. This strategy is bolstered by a variety of evidence from adult tumor models showing extensive cross-talk between the 2 pathways. One exciting preclinical study using AZD8055, a mammalian target of rapamycin inhibitor, and AZD6244, an MEK inhibitor, demonstrated significant synergy in both fusion-positive and fusion-negative cell lines both in vitro and in vivo.88 With the explosion of clinical trials examining similar combinations in adult solid tumors, it seems possible that these results could rapidly inform novel therapies for RMS. Combinations of signaling inhibitors similar to those described above might also be efficacious.
B. Modulating the Epigenome
The convergence point for abnormal signaling in cancer is transcriptional regulation. Therefore, novel mechanisms to target the epigenome might provide new therapeutic modalities for the treatment of RMS. HDACs are critical regulators of gene expression and linked to key oncogenic events in a variety of tumor types, including colon, breast, and lung tumors and leukemia.89 In preclinical xenograft models of both fusion-positive and fusion-negative RMS, HDAC inhibitors inhibited growth and induced apoptosis.90 Also, in preclinical models, HDAC inhibition radiosensitized RMS tumors.91 Finally, in the therapeutically challenging fusion-positive subtype, the combination of HDAC inhibition with the multikinase inhibitor PKC412 released the transcriptional repression of p21, resulting in a synergistic therapeutic combination.92
Inhibiting the epigenetic silencing of myogenic genes involved in normal skeletal differentiation by pharmacologic inhibition of EZH2 has restored myogenic differentiation of the embryonal cell line RD in both culture as well as xenograft. Ciarapica et al. 93 used the catalytic inhibitor MC1945 and demonstrated antiproliferative effects as well as increased expression of markers of terminal muscle differentiation compared with placebo. Given the pursuit of EZH2 inhibitors for the treatment of diffuse large B-cell lymphoma, these results provide an exciting new therapeutic avenue in fusion-negative RMS.
C. Directly Targeting the Fusion
A fusion protein that occurs specifically in tumor tissue while being absent in normal tissue theoretically presents an ideal opportunity for a “precision” therapy. In fusion-positive RMS this strategy is further enhanced by the observation that targeted knockdown of PAX394 or the PAX3-FOXO1 gene product95 induces apoptosis and inhibition of proliferation. Unfortunately, unlike the BCR-ABL or EML4-ALK fusion oncogenes that occur in adult cancers, which involve a targetable tyrosine kinase, the translocations observed in pediatric solid tumors almost uniformly involve transcription factors. While it is generally thought that specific inhibition of the transcriptional machinery is difficult,96 there has been recent success inhibiting the EWS-FLI1 translocation in Ewing sarcoma97 and the SS18-SSX translocation in synovial sarcoma.98 Similar efforts have been completed in PAX3-FOXO1-driven RMS, with interesting results. Using a luciferase reporter placed downstream of the promoter of a PAX3-FOXO1-induced gene (TFAP2B), Martin and colleagues99 screened a small-molecule library and identified the synthetic retinoid fenretinide as a specific repressor of PAX3-FOXO1 at both the messenger RNA and protein levels, which induced apoptosis of RMS cell lines both in vitro and in vivo. A second drug screen using a similar approach identified that fascaplysin, an inhibitor of CDK4, worked by abrogating the phosphorylation and subsequent subcellular localization of PAX3-FOXO1.100
D. Immunotherapy
Antibodies or immune cells engineered against specific antigens expressed by cancer cells have been increasingly used as therapeutic agents either alone or in combination with chemotherapeutic drugs.101,102 Following the revolutionary success of rituximab targeting CD20 on B cells as an FDA-approved drug for non-Hodgkin lymphoma, a number of monoclonal antibodies have been developed and are being used as approved drugs in the clinic or are currently in various phases of clinical trials for the treatment of leukemias,103–106 as well as solid tumors including mesothelioma107 and neuroblastoma.108,109 Most recently, chimeric antigen receptor–based therapies have shown stunning successes in refractory pediatric cancers.110 Given their unique expression pattern, including FGFR4, RMS tumors present a unique opportunity for immunotherapy.111 A consortium funded by a StandUp2Cancer and a St. Baldrick’s grant is actively pursuing this novel therapeutic modality.
VI. FUTURE PERSPECTIVES
Clear progress has been made in the understanding of the molecular and genetic causes that are the basis of RMS oncogenesis. This groundwork is ushering in a new era in which molecular knowledge will inform risk stratification and clinical decisions in real time. While this goal will be challenging in cases of a relatively rare pediatric tumor, the growing knowledge of the oncogenic and survival mechanisms underlying RMS give clinicians remarkable insights into their patients’ disease. Given the mortality associated with high risk and relapsed disease, priority should be given to innovative treatment modalities for these patients. While determining a statistically significant clinical improvement in such a small patient population is daunting, many of the genetic and molecular features of RMS overlap with alterations that occur in more common tumors. Previously established, FDA-approved drugs might provide significant benefit to these patients. In addition, these patients can provide crucial preliminary information for candidate therapies before expanding their use in the intermediate-risk patient cohort. Proper design of these trials should include a tumor biopsy before, during, and upon relapse so that the genomic and proteomic informations can be used to properly link the patient with the appropriate therapy and evaluate how the tumor responds to the treatment. During the same procedure, adequate tissue should be collected so that banking cell lines and in vivo tumor grafts can be generated, allowing the physician and researcher to fully evaluate the tumor’s response to the drug. These generated model systems would serve as critical tools to guide the development of novel therapies.
Acknowledgments
The authors thank Berkley Gryder for his help in the preparation of this manuscript. The reviewers are supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government. J.F.S. receives additional grant support from the Sarcoma Research Alliance for Collaboration and the St. Baldrick’s Foundation.
ABBREVIATIONS
- FDA
US Food and Drug Administration
- HDAC
histone deacetylase
- IGF
insulin-like growth factor
- IGF1R
type I insulin-like growth factor receptor
- PI3K
phosphoinositide-3 kinase
- PRC2
polycomb repressive complex 2
- RMS
rhabdomyosarcoma
References
- 1.Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975–2005. Cancer. 2009;115:4218–26. doi: 10.1002/cncr.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parham DM, Barr FG. Classification of Rhabdomyosarcoma and its molecular basis. Adv Anat Pathol. 2013;20:387–97. doi: 10.1097/PAP.0b013e3182a92d0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappo AS, Shapiro DN, Crist WM, Maurer HM. Biology and therapy of pediatric rhabdomyosarcoma. J Clin Oncol. 1995;13:2123–39. doi: 10.1200/JCO.1995.13.8.2123. [DOI] [PubMed] [Google Scholar]
- 4.Malempati S, Hawkins DS. Rhabdomyosarcoma: review of the Children’s Oncology Group (COG) Soft-Tissue Sarcoma Committee experience and rationale for current COG studies. Pediatr Blood Cancer. 2012;59:5–10. doi: 10.1002/pbc.24118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappo AS, Lyden E, Breitfeld P, Donaldson SS, Wiener E, Parham D, Crews KR, Houghton P, Meyer WH Children’s Oncology Group. Two consecutive phase II window trials of irinotecan alone or in combination with vincristine for the treatment of metastatic rhabdomyosarcoma: the Children’s Oncology Group. J Clin Oncol. 2007;25:362–9. doi: 10.1200/JCO.2006.07.1720. [DOI] [PubMed] [Google Scholar]
- 6.Arndt CA, Stoner JA, Hawkins DS, Rodeberg DA, Hayes-Jordan AA, Paidas CN, Parham DM, Teot LA, Wharam MD, Breneman JC, Donaldson SS, Anderson JR, Meyer WH. Vincristine, actinomycin, and cyclophosphamide compared with vincristine, actinomycin, and cyclophosphamide alternating with vincristine, topotecan, and cyclophosphamide for intermediate-risk rhabdomyosarcoma: Children’s Oncology Group Study D9803. J Clin Oncol. 2009;27:5182–8. doi: 10.1200/JCO.2009.22.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pappo AS, Anderson JR, Crist WM, Wharam MD, Breitfeld PP, Hawkins D, Raney RB, Womer RB, Parham DM, Qualman SJ, Grier HE. Survival after relapse in children and adolescents with rhabdomyosarcoma: areport from the intergroup rhabdomyosarcoma study group. J Clin Oncol. 1999;17:3487–93. doi: 10.1200/JCO.1999.17.11.3487. [DOI] [PubMed] [Google Scholar]
- 8.Li FP, Fraumeni JF. Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst. 1969;43:1365–73. [PubMed] [Google Scholar]
- 9.Sung L, Anderson JR, Arndt C, Raney B, Meyer WH, Pappo AS. Neurofibromatosis in children with rhabdomyosarcoma: a report from the intergroup Rhabdomyosarcoma Study IV. J Pediatr. 2004;144:666–8. doi: 10.1016/j.jpeds.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Steenman M, Westerveld A, Mannens M. Genetics of Beckwith-Wiedemann syndrome-associated tumors: common genetic pathways. Gene Chromosome Cancer. 2000;28:1–13. doi: 10.1002/(sici)1098-2264(200005)28:1<1::aid-gcc1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 11.Philip NM, Sigaudy S, Lacombe D, Vittu G, David A, Vigneron J, Moncla A, Flori E. Costello syndrome: report of eight patients including one with a rhabdomyosarcoma. Am J Hum Genet. 1999;65:A338-A. doi: 10.1007/s004310050037. [DOI] [PubMed] [Google Scholar]
- 12.Scrable H, Cavenee W, Ghavimi F, Lovell M, Morgan K, Sapienza C. A model for embryonal rhabdomyosarcoma tumorigenesis that involves genome imprinting. Proc Natl Acad Sci U S A. 1989;86:7480–4. doi: 10.1073/pnas.86.19.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor AC, Shu L, Danks MK, Poquette CA, Shetty S, Thayer MJ, Houghton PJ, Harris LC. P53 mutation and MDM2 amplification frequency in pediatric rhabdomyosarcoma tumors and cell lines. Med Pediatr Oncol. 2000;35:96–103. doi: 10.1002/1096-911x(200008)35:2<96::aid-mpo2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 14.Stratton MR, Fisher C, Gusterson BA, Cooper CS. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Res. 1989;49:6324–7. [PubMed] [Google Scholar]
- 15.Shukla N, Ameur N, Yilmaz I, Nafa K, Lau CY, Marchetti A, Borsu L, Barr FG, Ladanyi M. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–57. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor JG, 6th, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, Yu Y, Chen QR, Shah K, Youngblood V, Fang J, Kim SY, Yeung C, Helman LJ, Mendoza A, Ngo V, Staudt LM, Wei JS, Khanna C, Catchpoole D, Qualman SJ, Hewitt SM, Merlino G, Chanock SJ, Khan J. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the Pax3 paired box gene in the pediatric solid tumor alveolar rhabdomyosarcoma. Nat Genet. 1993;3:113–7. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- 18.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, Tolman C, Hurd L, Liao H, Zhang S, Bogen D, Brohl AS, Sindiri S, Catchpoole D, Badgett T, Getz G, Mora J, Anderson JR, Skapek SX, Barr FG, Meyerson M, Hawkins DS, Khan J. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–31. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, Spunt S, Valentine MB, Valentine V, Krafcik F, Lang WH, Wierdl M, Tsurkan L, Tolleman V, Federico SM, Morton C, Lu C, Ding L, Easton J, Rusch M, Nagahawatte P, Wang J, Parker M, Wei L, Hedlund E, Finkelstein D, Edmonson M, Shurtleff S, Boggs K, Mulder H, Yergeau D, Shapek S, Hawkins DS, Ramierez N, Potter PM, Sandoval JA, Davidoff AM, Mardis ER, Wilson RK, Zhang J, Downing JR, Dyer MA St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24:710–24. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohsaka S, Shukla N, Ameur N, Ito T, Ng CK, Wang L, Lim D, Marchetti A, Viale A, Pirun M, Socci ND5, Qin LX, Sciot R, Bridge J, Singer S, Meyers P, Wexler LH, Barr FG, Dogan S, Fletcher JA, Reis-Filho JS, Ladanyi M. A Recurrent point mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma. Lab Invest. 2014;94:20a–1a. doi: 10.1038/ng.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis RJ, Dcruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of Pax7 to Fkhr by the variant T(1,13)(P36,Q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–72. [PubMed] [Google Scholar]
- 22.Sumegi J, Streblow R, Frayer RW, Dal Cin P, Rosenberg A, Meloni-Ehrig A, Bridge JA. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer. 2010;49:224–36. doi: 10.1002/gcc.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wachtel M, Dettling M, Koscielniak E, Stegmaier S, Treuner J, Simon-Klingenstein K, Bühlmann P, Niggli FK, Schäfer BW. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–45. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 24.Barr FG, Smith LM, Lynch JC, Strzelecki D, Parham DM, Qualman SJ, Breitfeld PP. Examination of gene fusion status in archival samples of alveolar rhabdomyosarcoma entered on the intergroup rhabdomyosarcoma study-III trial: a report from the Children’s Oncology Group. J Mol Diagn. 2006;8:202–8. doi: 10.2353/jmoldx.2006.050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barr FG, Duan FH, Smith LM, Gustafson D, Pitts M, Hammond S, Gastier-Foster JM. Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. Genes Chromosomes Cancer. 2009;48:661–72. doi: 10.1002/gcc.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon AT, Brinkschmidt C, Anderson J, Coleman N, Dockhorn-Dworniczak B, Pritchard-Jones K, Shipley J. A novel and consistent amplicon at 13q31 associated with alveolar rhabdomyosarcoma. Gene Chromosome Canc. 2000;28:220–6. [PubMed] [Google Scholar]
- 27.Schwab M. MYCN in neuronal tumours. Cancer Lett. 2004;204:179–87. doi: 10.1016/S0304-3835(03)00454-3. [DOI] [PubMed] [Google Scholar]
- 28.Paulson V, Chandler G, Rakheja D, Galindo RL, Wilson K, Amatruda JF, Cameron S. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes Chromosomes Cancer. 2011;50:397–408. doi: 10.1002/gcc.20864. [DOI] [PubMed] [Google Scholar]
- 29.Shukla N, Ameur N, Yilmaz I, Nafa K, Lau CY, Marchetti A, Borsu L, Barr FG, Ladanyi M. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–57. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li SQ, Cheuk AT, Shern JF, Song YK, Hurd L, Liao H, Wei JS, Khan J. Targeting wild-type and mutationally activated FGFR4 in rhabdomyosarcoma with the inhibitor ponatinib (AP24534) PLoS One. 2013;8:e76551. doi: 10.1371/journal.pone.0076551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szuhai K, de Jong D, Leung WY, Fletcher CDM, Hogendoorn PCW. Transactivating mutation of the MYOD1 gene is a frequent event in adult spindle cell rhabdomyosarcoma. J Pathol. 2014;232:300–7. doi: 10.1002/path.4307. [DOI] [PubMed] [Google Scholar]
- 32.Leach FS, Tokino T, Meltzer P, Burrell M, Oliner JD, Smith S, Hill DE, Sidransky D, Kinzler KW, Vogelstein B. P53 mutation and Mdm2 amplification in human soft-tissue sarcomas. Cancer Res. 1993;53:2231–4. [PubMed] [Google Scholar]
- 33.Ping AJ, Reeve AE, Law DJ, Young MR, Boehnke M, Feinberg AP. Genetic-linkage of Beckwith-Wiedemann syndrome to 11p15. Am J Hum Genet. 1989;44:720–3. [PMC free article] [PubMed] [Google Scholar]
- 34.Rainier S, Dobry CJ, Feinberg AP. Loss of imprinting in hepatoblastoma. Cancer Res. 1995;55:1836–8. [PubMed] [Google Scholar]
- 35.Besnard-Guérin C, Newsham I, Winqvist R, Cavenee WK. A common region of loss of heterozygosity in Wilms’ tumor and embryonal rhabdomyosarcoma distal to the D11S988 locus on chromosome 11p15.5. Hum Genet. 1996;97:163–70. doi: 10.1007/BF02265259. [DOI] [PubMed] [Google Scholar]
- 36.Zhan SL, Shapiro DN, Helman LJ. Activation of an imprinted allele of the insulin-like growth-factor-II gene implicated in rhabdomyosarcoma. J Clin Invest. 1994;94:445–8. doi: 10.1172/JCI117344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber-Hall S, Anderson J, McManus A, Abe S, Nojima T, Pinkerton R, Pritchard-Jones K, Shipley J. Gains, losses, and amplification of genomic material in rhabdomyosarcoma analyzed by comparative genomic hybridization. Cancer Res. 1996;56:3220–4. [PubMed] [Google Scholar]
- 38.Bridge JA, Liu J, Qualman S, Suijkerbuijk R, Wenger G, Zhang J, Wan X, Baker KS, Sorensen P, Barr FG. Genomic gains and losses are similar in genetic and histologic subsets of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer. 2002;33:310–21. doi: 10.1002/gcc.10026. [DOI] [PubMed] [Google Scholar]
- 39.Bridge JA, Liu J, Weibolt V, Baker KS, Perry D, Kruger R, Qualman S, Barr F, Sorensen P, Triche T, Suijkerbuijk R. Novel genomic imbalances in embryonal rhabdomyosarcoma revealed by comparative genomic hybridization and fluorescence in situ hybridization: an intergroup rhabdomyosarcoma study. Genes Chromosomes Cancer. 2000;27:337–44. doi: 10.1002/(sici)1098-2264(200004)27:4<337::aid-gcc1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 40.Lollini PL, De Giovanni C, Del Re B, Landuzzi L, Nicoletti G, Prodi G, Scotlandi K, Nanni P. Myogenic differentiation of human rhabdomyosarcoma cells induced invitro by antineoplastic drugs. Cancer Res. 1989;49:3631–6. [PubMed] [Google Scholar]
- 41.Goldstein M, Meller I, Orr-Urtreger A. FGFR1 over-expression in primary rhabdomyosarcoma tumors is associated with hypomethylation of a 5′ CpG wand and abnormal expression of the AKT1, NOG, and BMP4 genes. Genes Chromosomes Cancer. 2007;46:1028–38. doi: 10.1002/gcc.20489. [DOI] [PubMed] [Google Scholar]
- 42.Gastaldi T, Bonvini P, Sartori F, Marrone A, Iolascon A, Rosolen A. Plakoglobin is differentially expressed in alveolar and embryonal rhabdomyosarcoma and is regulated by DNA methylation and histone acetylation. Carcinogenesis. 2006;27:1758–67. doi: 10.1093/carcin/bgl008. [DOI] [PubMed] [Google Scholar]
- 43.Chen B, Dias P, Jenkins JJ, Savell V, Parham DM. Methylation alterations of the MyoD1 upstream region predict rhabdomyosarcoma histology. Modern Pathol. 1998;11:167a-a. [PMC free article] [PubMed] [Google Scholar]
- 44.Mahoney SE, Yao Z, Keyes CC, Tapscott SJ, Diede SJ. Genome-wide DNA methylation studies suggest distinct DNA methylation patterns in pediatric embryonal and alveolar rhabdomyosarcomas. Epigenetics. 2012;7:400–8. doi: 10.4161/epi.19463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chase A, Cross NCP. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–8. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 46.Marchesi I, Giordano A, Bagella L. Roles of enhancer of zeste homolog 2: from skeletal muscle differentiation to rhabdomyosarcoma carcinogenesis. Cell Cycle. 2014;13:516–27. doi: 10.4161/cc.27921. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–81. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchesi I, Fiorentino FP, Rizzolio F, Giordano A, Bagella L. The ablation of EZH2 uncovers its crucial role in rhabdomyosarcoma formation. Cell Cycle. 2012;11:3828–36. doi: 10.4161/cc.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao L, Yu Y, Bilke S, Walker RL, Mayeenuddin LH, Azorsa DO, Yang F, Pineda M, Helman LJ, Meltzer PS. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70:6497–508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Missiaglia E, Williamson D, Chisholm J, Wirapati P, Pierron G, Petel F, Concordet JP, Thway K, Oberlin O, Pritchard-Jones K, Delattre O, Delorenzi M, Shipley J. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol. 2012;30:1670–7. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 51.Skapek SX, Anderson J, Barr FG, Bridge JA, Gastier-Foster JM, Parham DM, Rudzinski ER, Triche T, Hawkins DS. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a Children’s Oncology Group report. Pediatr Blood Cancer. 2013;60:1411–7. doi: 10.1002/pbc.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis RJ, Bennicelli JL, Macina RA, Nycum LM, Biegel JA, Barr FG. Structural characterization of the Fkhr gene and its rearrangement in alveolar rhabdomyosarcoma. Hum Mol Genet. 1995;4:2355–62. doi: 10.1093/hmg/4.12.2355. [DOI] [PubMed] [Google Scholar]
- 53.Fitzgerald JC, Scherr AM, Barr FG. Structural analysis of PAX7 rearrangements in alveolar rhabdomyosarcoma. Cancer Genet Cytogenet. 2000;117:37–40. doi: 10.1016/s0165-4608(99)00130-2. [DOI] [PubMed] [Google Scholar]
- 54.Scheidler S, Fredericks WJ, Rauscher FJ, Barr FG, Vogt PK. The hybrid PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc Natl Acad Sci U S A. 1996;93:9805–9. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kikuchi K, Tsuchiya K, Otabe O, Gotoh T, Tamura S, Katsumi Y, Yagyu S, Tsubai-Shimizu S, Miyachi M, Iehara T, Hosoi H. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365:568–74. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 56.Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, De-Pinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Devel. 2004;18:2614–26. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishijo K, Chen QR, Zhang L, McCleish AT, Rodriguez A, Cho MJ, Prajapati SI, Gelfond JA, Chisholm GB, Michalek JE, Aronow BJ, Barr FG, Randall RL, Ladanyi M, Qualman SJ, Rubin BP, LeGallo RD, Wang C, Khan J, Keller C. Credentialing a preclinical mouse model of alveolar rhabdomyosarcoma. Cancer Res. 2009;69:2902–11. doi: 10.1158/0008-5472.CAN-08-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khan J, Bittner ML, Chen YD, Faller AJ, Saal LH, Azorsa DA, Teichman U, Pavan W, Trent JM, Meltzer PS. Elucidation of the downstream targets of the PAX3-FKHR fusion oncogene found in aveolar rhabdomyosarcoma using cDNA microarrays [abstract] Pediatr Res. 1999;45:148A. [Google Scholar]
- 59.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–46. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 60.Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, Pavan WJ, Trent JM, Meltzer PS. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci U S A. 1999;96:13264–9. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Gaal JC, Flucke UE, Roeffen MH, de Bont ES, Sleijfer S, Mavinkurve-Groothuis AM, Suurmeijer AJ, van der Graaf WT, Versleijen-Jonkers YM. Anaplastic lymphoma kinase aberrations in rhabdomyosarcoma: clinical and prognostic implications. J Clin Oncol. 2012;30:308–15. doi: 10.1200/JCO.2011.37.8588. [DOI] [PubMed] [Google Scholar]
- 62.Taulli R, Scuoppo C, Bersani F, Accornero P, Forni PE, Miretti S, Grinza A, Allegra P, Schmitt-Ney M, Crepaldi T, Ponzetto C. Validation of met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006;66:4742–9. doi: 10.1158/0008-5472.CAN-05-4292. [DOI] [PubMed] [Google Scholar]
- 63.Khan J, Wei JS, Ringnér M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–9. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsumura H, Yoshida T, Saito H, Imanaka-Yoshida K, Suzuki N. Cooperation of oncogenic K-ras and p53 deficiency in pleomorphic rhabdomyosarcoma development in adult mice. Oncogene. 2006;25:7673–9. doi: 10.1038/sj.onc.1209749. [DOI] [PubMed] [Google Scholar]
- 66.Linardic CM, Downie DL, Qualman S, Bentley RC, Counter CM. Genetic modeling of human rhabdomyosarcoma. Cancer Res. 2005;65:4490–5. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- 67.Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol. 2013;3:183. doi: 10.3389/fonc.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Storer NY, White RM, Uong A, Price E, Nielsen GP, Langenau DM, Zon LI. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis. Development. 2013;140:3040–50. doi: 10.1242/dev.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res. 2005;65:9226–35. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 70.Wachtel M, Rakic J, Okoniewski M, Bode P, Niggli F, Schafer BW. FGFR4 signaling couples to Bim and not Bmf to discriminate subsets of alveolar rhabdomyosarcoma cells. Int J Cancer. 2014;135:1543–52. doi: 10.1002/ijc.28800. [DOI] [PubMed] [Google Scholar]
- 71.Elbadry OM, Minniti C, Kohn EC, Houghton PJ, Daughaday WH, Helman LJ. Insulin-like growth factor-II acts as an autocrine growth and motility factor in human rhabdomyosarcoma tumors. Cell Growth Differ. 1990;1:325–31. [PubMed] [Google Scholar]
- 72.Wang W, Kumar P, Wang W, Epstein J, Helman L, Moore JV, Kumar S. Insulin-like growth factor II and PAX3-FKHR cooperate in the oncogenesis of rhabdomyosarcoma. Cancer Res. 1998;58:4426–33. [PubMed] [Google Scholar]
- 73.Kumar S, Perlman E, Harris CA, Raffeld M, Tsokos M. Myogenin is a specific marker for rhabdomyosarcoma: an immunohistochemical study in paraffin-embedded tissues. Mod Pathol. 2000;13:988–93. doi: 10.1038/modpathol.3880179. [DOI] [PubMed] [Google Scholar]
- 74.Sebire NJ, Malone M. Myogenin and MyoD1 expression in paediatric rhabdomyosarcomas. J Clin Pathol. 2003;56:412–6. doi: 10.1136/jcp.56.6.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keller C, Guttridge DC. Mechanisms of impaired differentiation in rhabdomyosarcoma. FEBS J. 2013;280:4323–34. doi: 10.1111/febs.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saab R, Spunt SL, Skapek SX. Myogenesis and rhabdomyosarcoma: the Jekyll and Hyde of skeletal muscle. Curr Top Dev Biol. 2011;94:197–234. doi: 10.1016/B978-0-12-380916-2.00007-3. [DOI] [PubMed] [Google Scholar]
- 77.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–73. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 78.Kolb EA, Gorlick R, Lock R, Carol H, Morton CL, Keir ST, Reynolds CP, Kang MH, Maris JM, Billups C, Smith MA, Houghton PJ. Initial testing (stage 1) of the IGF-1 receptor inhibitor BMS-754807 by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer. 2011;56:595–603. doi: 10.1002/pbc.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weigel B, Malempati S, Reid JM, Voss SD, Cho SY, Chen HX, Krailo M, Villaluna D, Adamson PC, Blaney SM. Phase 2 trial of cixutumumab in children, adolescents, and young adults with refractory solid tumors: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2014;61:452–6. doi: 10.1002/pbc.24605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pappo AS, Vassal G, Crowley JJ, Bolejack V, Hogendoorn PC, Chugh R, Ladanyi M, Grippo JF, Dall G, Staddon AP, Chawla SP, Maki RG, Araujo DM, Geoerger B, Ganjoo K, Marina N, Blay JY, Schuetze SM, Chow WA, Helman LJ. A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: results of a Sarcoma Alliance for Research Through Collaboration study. Cancer. 2014;120:2448–56. doi: 10.1002/cncr.28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li SQ, Cheuk AT, Shern JF, Song YK, Hurd L, Liao H, Wei JS, Khan J. Targeting wild-type and mutationally activated FGFR4 in rhabdomyosarcoma with the inhibitor ponatinib (AP24534) PloS One. 2013;8:e76551. doi: 10.1371/journal.pone.0076551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ferguson M, Hingorani P, Gupta AA. Emerging molecular-targeted therapies in early-phase clinical trials and preclinical models. Am Soc Clin Oncol Educ Book. 2013:420–4. doi: 10.14694/EdBook_AM.2013.33.420. [DOI] [PubMed] [Google Scholar]
- 83.Saab R, Bills JL, Miceli AP, Anderson CM, Khoury JD, Fry DW, Navid F, Houghton PJ, Skapek SX. Pharmacologic inhibition of cyclin-dependent kinase 4/6 activity arrests proliferation in myoblasts and rhabdomyosarcomaderived cells. Mol Cancer Ther. 2006;5:1299–308. doi: 10.1158/1535-7163.MCT-05-0383. [DOI] [PubMed] [Google Scholar]
- 84.Canner JA, Sobo M, Ball S, Hutzen B, DeAngelis S, Willis W, Studebaker AW, Ding K, Wang S, Yang D, Lin J. MI-63: a novel small-molecule inhibitor targets MDM2 and induces apoptosis in embryonal and alveolar rhabdomyosarcoma cells with wild-type p53. Br J Cancer. 2009;101:774–81. doi: 10.1038/sj.bjc.6605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guenther MK, Graab U, Fulda S. Synthetic lethal interaction between PI3K/Akt/mTOR and Ras/MEK/ERK pathway inhibition in rhabdomyosarcoma. Cancer Lett. 2013;337:200–9. doi: 10.1016/j.canlet.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 86.Li Z, Zhang Y, Ramanujan K, Ma Y, Kirsch DG, Glass DJ. Oncogenic NRAS, required for pathogenesis of embryonic rhabdomyosarcoma, relies upon the HMGA2-IGF2BP2 pathway. Cancer Res. 2013;73:3041–50. doi: 10.1158/0008-5472.CAN-12-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen EY, DeRan MT, Ignatius MS, Grandinetti KB, Clagg R, McCarthy KM, Lobbardi RM, Brockmann J, Keller C, Wu X, Langenau DM. Glycogen synthase kinase 3 inhibitors induce the canonical WNT/beta-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc Natl Acad Sci U S A. 2014;111:5349–54. doi: 10.1073/pnas.1317731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Renshaw J, Taylor KR, Bishop R, Valenti M, De Haven Brandon A, Gowan S, Eccles SA, Ruddle RR, Johnson LD, Raynaud FI, Selfe JL, Thway K, Pietsch T, Pearson AD, Shipley J. Dual blockade of the PI3K/AKT/mTOR (AZD8055) and RAS/MEK/ERK (AZD6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clin Cancer Res. 2013;19:5940–51. doi: 10.1158/1078-0432.CCR-13-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–9. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kutko MC, Glick RD, Butler LM, Coffey DC, Rifkind RA, Marks PA, Richon VM, LaQuaglia MP. Histone deacetylase inhibitors induce growth suppression and cell death in human rhabdomyosarcoma in vitro. Clin Cancer Res. 2003;9:5749–55. [PubMed] [Google Scholar]
- 91.Blattmann C, Oertel S, Ehemann V, Thiemann M, Huber PE, Bischof M, Witt O, Deubzer HE, Kulozik AE, Debus J, Weber KJ. Enhancement of radiation response in osteosarcoma and rhabomyosarcoma cell lines by histone deacetylase inhibition. Int J Radiat Oncol. 2010;78:237–45. doi: 10.1016/j.ijrobp.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 92.Hecker RM, Amstutz RA, Wachtel M, Walter D, Niggli FK, Schafer BW. p21 Downregulation is an important component of PAX3/FKHR oncogenicity and its reactivation by HDAC inhibitors enhances combination treatment. Oncogene. 2010;29:3942–52. doi: 10.1038/onc.2010.145. [DOI] [PubMed] [Google Scholar]
- 93.Ciarapica R, Carcarino E, Adesso L, De Salvo M, Bracaglia G, Leoncini PP, Dall’agnese A, Verginelli F, Milano GM, Boldrini R, Inserra A, Stifani S, Screpanti I, Marquez VE, Valente S, Mai A, Puri PL, Locatelli F, Palacios D, Rota R. Pharmacological inhibition of EZH2 as a promising differentiation therapy in embryonal RMS. BMC Cancer. 2014;14:139. doi: 10.1186/1471-2407-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bernasconi M, Remppis A, Fredericks WJ, Rauscher F, Jr, Schafer BW. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc Natl Acad Sci U S A. 1996;93:13164–9. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kikuchi K, Tsuchiya K, Otabe O, Gotoh T, Tamura S, Katsumi Y, Yagyu S, Tsubai-Shimizu S, Miyachi M, Iehara T, Hosoi H. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Bioph Res Commun. 2008;365:568–74. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 96.Gibbs JB. Mechanism-based target identification and drug discovery in cancer research. Science. 2000;287:1969–73. doi: 10.1126/science.287.5460.1969. [DOI] [PubMed] [Google Scholar]
- 97.Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen QR, Yeung C, Currier DG, Davis S, Khanna C, Khan J, McMahon JB, Helman LJ. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst. 2011;103:962–78. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martin DH, Boro A, Schafer BW. Cell-based small-molecule compound screen identifies fenretinide as potential therapeutic for translocation-positive rhabdomyosarcoma. PloS One. 2013;8(1):e55072. doi: 10.1371/journal.pone.0055072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu LL, Wu J, Ong SS, Chen TS. Cyclin-dependent kinase 4 phosphorylates and positively regulates PAX3-FOXO1 in human alveolar rhabdomyosarcoma cells. PloS One. 2013;8:e55072. doi: 10.1371/journal.pone.0058193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheson BD. Ofatumumab, a novel anti-CD20 monoclonal antibody for the treatment of B-cell malignancies. J Clin Oncol. 2010;28:3525–30. doi: 10.1200/JCO.2010.27.9836. [DOI] [PubMed] [Google Scholar]
- 102.Cheson BD, Leonard JP. Monoclonal antibody therapy for B-cell non-Hodgkin’s lymphoma. N Engl J Med. 2008;359:613–26. doi: 10.1056/NEJMra0708875. [DOI] [PubMed] [Google Scholar]
- 103.FitzGerald DJ, Wayne AS, Kreitman RJ, Pastan I. Treatment of hematologic malignancies with immunotoxins and antibody-drug conjugates. Cancer Res. 2011;71:6300–9. doi: 10.1158/0008-5472.CAN-11-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kreitman RJ, Arons E, Stetler-Stevenson M, Fitzgerald DJ, Wilson WH, Pastan I. Recombinant immunotoxins and other therapies for relapsed/refractory hairy cell leukemia. Leuk Lymphoma. 2011;52(Suppl 2):82–6. doi: 10.3109/10428194.2011.565843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kreitman RJ, Tallman MS, Robak T, Coutre S, Wilson WH, Stetler-Stevenson M, Fitzgerald DJ, Lechleider R, Pastan I. Phase I trial of anti-CD22 recombinant immunotoxin moxetumomab pasudotox (CAT-8015 or HA22) in patients with hairy cell leukemia. J Clin Oncol. 2012;30:1822–8. doi: 10.1200/JCO.2011.38.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wayne AS, Kreitman RJ, Findley HW, Lew G, Delbrook C, Steinberg SM, Stetler-Stevenson M, Fitzgerald DJ, Pastan I. Anti-CD22 immunotoxin RFB4(dsFv)-PE38 (BL22) for CD22-positive hematologic malignancies of childhood: preclinical studies and phase I clinical trial. Clin Cancer Res. 2010;16:1894–903. doi: 10.1158/1078-0432.CCR-09-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hassan R, Cohen SJ, Phillips M, Pastan I, Sharon E, Kelly RJ, Schweizer C, Weil S, Laheru D. Phase I clinical trial of the chimeric anti-mesothelin monoclonal antibody MORAb-009 in patients with mesothelin-expressing cancers. Clin Cancer Res. 2010;16:6132–8. doi: 10.1158/1078-0432.CCR-10-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cheung NK, Cheung IY, Kushner BH, Ostrovnaya I, Chamberlain E, Kramer K, Modak S. Murine anti-GD2 monoclonal antibody 3F8 combined with granulocyte-macrophage colony-stimulating factor and 13-cis-retinoic acid in high-risk patients with stage 4 neuroblastoma in first remission. J Clin Oncol. 2012;30:3264–70. doi: 10.1200/JCO.2011.41.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cheung NK, Kushner BH, Yeh SD, Larson SM. 3F8 monoclonal antibody treatment of patients with stage 4 neuroblastoma: a phase II study. Int J Oncol. 1998;12:1299–306. doi: 10.3892/ijo.12.6.1299. [DOI] [PubMed] [Google Scholar]
- 110.Lee DW, Barrett DM, Mackall C, Orentas R, Grupp SA. The future is now: chimeric antigen receptors as new targeted therapies for childhood cancer. Clin Cancer Res. 2012;18:2780–90. doi: 10.1158/1078-0432.CCR-11-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Orentas RJ, Yang JJ, Wen X, Wei JS, Mackall CL, Khan J. Identification of cell surface proteins as potential immunotherapy targets in 12 pediatric cancers. Front Oncol. 2012;2:194. doi: 10.3389/fonc.2012.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]