Abstract
The nematode Caenorhabditis elegans uses simple building blocks from primary metabolism and a strategy of modular assembly to build a great diversity of signaling molecules, the ascarosides, which function as a chemical language in this model organism. In the ascarosides, the dideoxysugar ascarylose serves as a scaffold to which diverse moieties from lipid, amino acid, neurotransmitter, and nucleoside metabolism are attached. However, the mechanisms that underlie the highly specific assembly of ascarosides are not understood. We show that the acyl-CoA synthetase ACS-7, which localizes to lysosome-related organelles, is specifically required for the attachment of different building blocks to the 4′-position of ascr#9. We further show that mutants lacking lysosome-related organelles are defective in the production of all 4′-modified ascarosides, thus identifying the waste disposal system of the cell as a hotspot for ascaroside biosynthesis.
Keywords: biosynthesis, chemical signaling, metabolites, natural products, pheromones
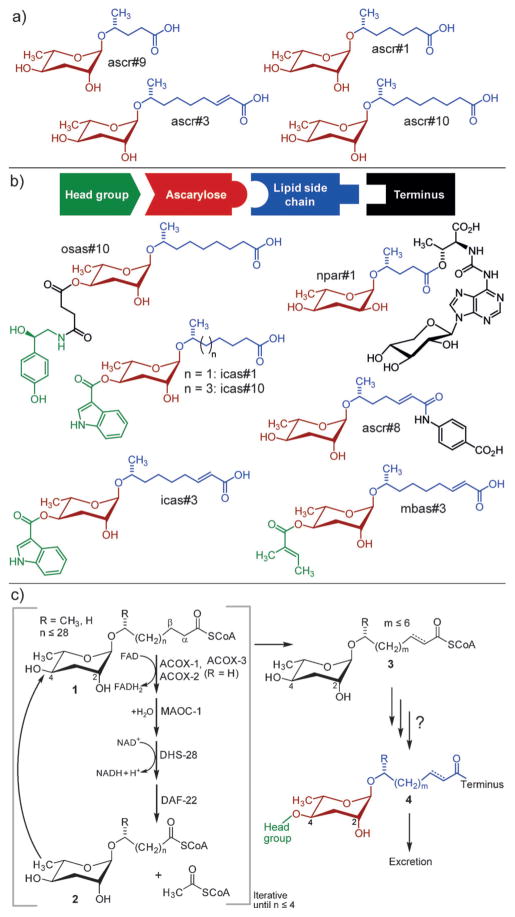
Nematodes, including the model organisms Caenorhabditis elegans and Pristionchus pacificus, produce a great diversity of ascaroside derivatives[1] (Figure 1). These signaling molecules are involved in virtually all aspects of nematode life history, including the regulation of aging,[2] dauer development,[3] morphology,[4] and behavior,[5] often acting through evolutionarily conserved signaling pathways.[1,6] The identification of the modular ascarosides demonstrated not only unexpected biosynthetic capabilities in an animal, but also provided an example of combinatorial generation of structural diversity that is reminiscent of synthetic combinatorial libraries in medicinal chemistry.[1]
Figure 1.
Structures and biosynthesis of the ascarosides. a) The simple ascarosides ascr#1, ascr#3, ascr#9, and ascr#10, which serve signaling functions in C. elegans and many other nematode species. b) Modular ascarosides incorporating building blocks from folate (ascr#8), neurotransmitter (osas#10), tryptophan (icas#3, icas#10), and short-chain fatty acid (mbas#3) metabolism from C. elegans, and npar#1 from the satellite model organism P. pacificus, which features a nucleoside-derived building block. c) A model for the biosynthesis of ascarosides. Iterative 4-step β-oxidation of long-chain ascarosides (1) produces short-chain precursors (3) of modular ascarosides (4).[19]
Assembly of the ascarosides identified from C. elegans and other nematodes proceeds with high selectivity. For example, the likely folate-derived p-aminobenzoic acid moiety in ascr#8 is selectively attached to an unsaturated 7-carbon sidechain, although the simple ascarosides based on saturated 7-carbon and unsaturated 9-carbon sidechains, ascr#1 and ascr#3, are much more abundant (Figure 1).[3c,7] Specific biological functions and highly selective assembly indicate that the modular ascarosides are products of dedicated biosynthetic pathways. Ultimately, the elucidation of these pathways will reveal how input from conserved primary metabolism is transduced to create signals that regulate development and behavior.
Previous studies have demonstrated that the fatty acid like side chains in the ascarosides are derived from very long-chain precursors (1) through peroxisomal β-oxidation (Figure 1c).[5d,7,8] Other building blocks, for example, the indole carboxy moiety in icas#3 or the octopamine moiety in osas#10, are derived from canonical primary metabolites, for example, tryptophan in the case of icas#3[7] and tyrosine in the case of osas#10.[5c] Feeding experiments (see Methods in the Supporting Information and Ref.[7]) demonstrated that synthetic ascr#1 and ascr#3 are converted into 4′-substituted derivatives icas#1 and icas#3, thus suggesting that 4′-modification occurs subsequent to and independent of side-chain β-oxidation (Figure 1c). However, the enzymes and cellular compartments involved in the attachment of diverse moieties via ester and amide bonds remained unknown.
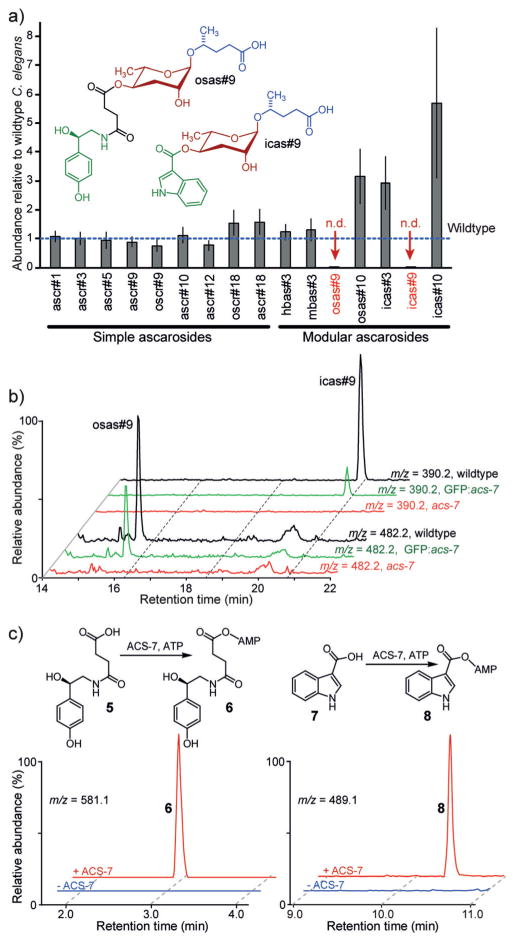
The 4′-acylated ascarosides include several important signaling molecules, for example, icas#3 (Figure 1) and icas#9, which are components of the C. elegans aggregation pheromone,[5b] and osas#9, a larval dispersal signal (Figure 2a).[5c] In search of enzymes that attach different acyl moieties to the 4′-position of ascarylose, we pursued a two-pronged approach, screening candidate genes based on either predicted function or subcellular localization. The C. elegans genome includes 61 genes annotated as O-acyltransferases, and for about 30 of these, likely loss-of-function mutants were available (Table S1 in the Supporting Information). Comparative metabolomic analysis of liquid cultures of these mutants and wildtype controls by LC-MS did not reveal any reduction in the amounts of 4′-modified ascarosides. Since shortening of the fatty acid like side chains of the ascarosides takes place in peroxisomes, we additionally screened several mutants of genes bearing a peroxisome targeting sequence (PTS),[9] including three putative acyl-CoA synthetases. In a pilot screen, we found that the production of two 4′-acylated derivatives of ascr#9, the aggregation pheromone icas#9 and the larval dispersal signal osas#9, was abolished in acs-7(tm6781) mutants (Figures 2a, b), whereas production of the structurally related icas#3, icas#10, and osas#10 was similar or elevated compared to the wildtype (Figures 1 and 2a, and Figure S2). The abundances of all other known ascarosides, including ascr#9, were close to those found in wildtype controls (Figure 2a). Similarly, the abundances of N-succinyl octopamine (5) and indole-3-carboxylic acid (7), which are plausible building blocks of osas#9 and icas#9, respectively, were not significantly changed in acs-7 mutants (Figure S1 in the Supporting Information). Ascaroside production was largely wildtype-like in the other mutants in our pilot screen (Figure S3a).
Figure 2.
ACS-7 contributes to the biosynthesis of 4′-acylated ascarosides. a) Abundances of ascarosides in acs-7(tm6781) mutants relative to wildtype C. elegans. Error bars =standard error of three biological replicates. The variability in ascaroside biosynthesis in wildtype C. elegans is assessed in Figure S2. b) HPLC–MS (ESI−) ion chromatograms for icas#9 and osas#9 in wildtype, acs-7(tm6781) mutants, and acs-7(tm6781) mutants expressing gfp::acs-7. c) HPLC–MS (ESI−) ion chromatograms showing ACS-7-dependent formation of the adenylates of N-succinyl octopamine and indole-3-carboxylic acid (also see Figure S4).
These results suggested that the enzyme ACS-7 is involved in the biosynthesis of icas#9 and osas#9, specifically. acs-7 encodes a putative acyl-CoA synthetase with no known function in C. elegans and approximately 70% sequence homology to human acyl-CoA synthetase family member 2, a mitochondrial fatty acyl-CoA synthetase with a preference for medium-chain substrates. To confirm the role of ACS-7 in the biosynthesis of icas#9 and osas#9, we expressed green fluorescent protein (gfp)-tagged acs-7 (gfp::acs-7) under the native acs-7 promoter in an acs-7(tm6781) mutant background. LC-MS analysis of gfp::acs-7 cultures revealed that the biosynthesis of icas#9 and osas#9 was restored, whereas the amounts of other ascarosides remained unchanged (Figure 2b and Figure S3b). Additional untargeted metabolomics revealed no other significant differences between the metabolomes of acs-7 and wildtype worms. This observation confirmed that ACS-7 is involved specifically in the production of the 4′-modified ascarosides icas#9 and osas#9.
Based on sequence homology, we hypothesized that ACS-7 may activate indole-3-carboxylic acid (7) and N-succinyl octopamine (5), as building blocks of icas#9 and osas#9, respectively, for subsequent 4′-acylation of ascr#9. To test this, we purified recombinant his6–ACS-7 from E. coli. Incubation of the ACS-7 with indole-3-carboxylic acid, ATP, and coenzyme A rapidly produced indole-3-carboxy-AMP (8) (Km = 270 ± 90 μM, Figure 2c and Figure S4), whereas formation of indole carboxy-CoA was not observed. Similarly, N-succinyl octopamine (5) was converted into the corresponding adenylate 6. However, addition of ascr#9 or ascr#9-CoA into these assays did not lead to production of icas#9, osas#9, or the corresponding CoA-derivatives.
Similarly, ACS-7 did not convert ascr#9 or aliphatic fatty acids, which we included as control substrates, into either the corresponding adenylates or CoA derivatives (Figure S5). These results indicate that ACS-7 contributes to the biosynthesis of icas#9 and osas#9 by activating indole carboxylic acid and N-succinyl octopamine, respectively; however, 4′-attachment to ascr#9 or ascr#9-CoA may involve additional factors or require a specific cellular environment not mimicked in our assays.
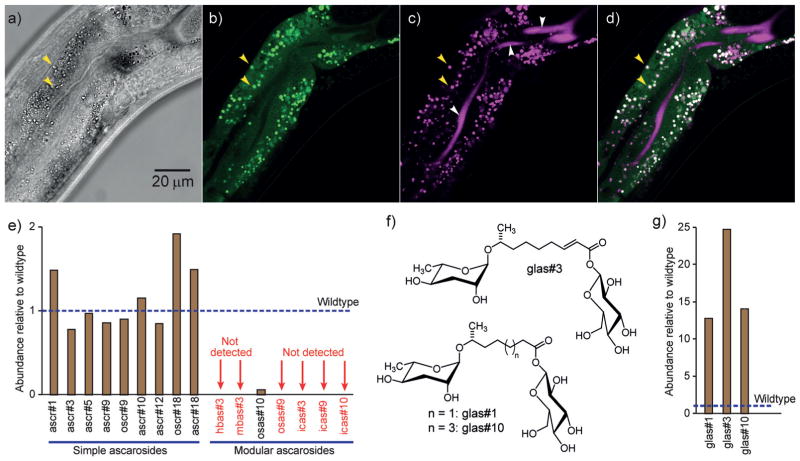
Next we asked whether ACS-7 localizes to peroxisomes, as predicted by its PTS. Surprisingly, we found that GFP::ACS-7 is primarily localized to punctate organelles (“gut granules”)[10] in intestinal cells, rather than peroxisomes (Figures 3a,b). C. elegans intestinal cells are known to contain several different types of gut granules, including acidic lysosome-related organelles (LROs).[10] LROs play a central role in the digestion of cellular waste and the regeneration of basic building blocks, but they are also suspected to contribute to the biosyntheses of diverse metabolites.[11] Staining with LysoTracker Red, a dye that selectively stains LROs in C. elegans,[10,12] revealed that GFP::ACS-7 localizes specifically to the LROs in the intestinal cells (Figures 3c,d). This suggests that the biosynthesis of the modular ascarosides icas#9 and osas#9 may proceed in the LROs and not in the peroxisomes.
Figure 3.
Identification of LROs as the site of the biosynthesis of modular ascarosides. a) A bright-field image of the midsection of a transgenic worm expressing gfp::acs-7 under the acs-7 promoter in acs-7 mutant background (young adult stage). b) GFP expression is localized to punctate organelles (e.g., yellow arrows). c) Staining of LROs with LysoTracker Red (white arrows: background staining of the intestinal lumen). d) Overlay of images in (b) and (c). e) Abundances of ascarosides in glo-1(zu437) mutants relative to wildtype C. elegans. f) Structures of ascaroside glucosyl conjugates. g) Abundances of the glucosyl conjugates glas#1, glas#3, and glas#10 are greatly increased in glo-1 knockout mutants. In (e) and (g), the values shown represent the averages of two replicates.
To test whether acidic LROs are required for the biosynthesis of icas#9, osas#9, and potentially other modular ascarosides, we investigated the ascaroside profiles of several mutants that are defective in specific classes of gut granules. glo-1 encodes a 24 kDa member of the Rab family of small GTPases, which is required for the biogenesis of acidic LROs.[10] Correspondingly, glo-1(zu437) mutants, which lack LROs, do not show any staining with LysoTracker Red.[10] glo-4 encodes a guanine nucleotide exchange factor that contributes to GLO-1 function.[10] Both glo-1 and glo-4 act downstream of apb-3, which encodes the worm orthologue of the β3 subunit of the mammalian adaptor protein complex AP-3.[13] We found that the biosynthesis of all known 4′-modified ascarosides is largely abolished in glo-1(zu437) mutants, which lack LROs (Figure 3e), and reduced in glo-4(ok623) and apb-3(ok429) mutants, in which LRO formation is reduced but not abolished (Figure S6). The amounts of other ascarosides in these mutants were similar to those in wildtype controls, except for the glycosylated derivatives glas#1, glas#3, and glas#10, which were starkly increased in glo-1(zu437) mutants relative to the wildtype (Figure 3f,g). Glycosylation generally serves as a detoxification mechanism in C. elegans,[14] and the buildup of glycosylated ascarosides in glo-1 worms may reflect the loss of alternative waste processing pathways owing to the lack of LROs in this mutant. Importantly, the abundances of indole-3-carboxylic acid and N-succinyl octopamine in glo-1 mutants were similar or only slightly reduced relative to the wildtype, suggesting that abolishment of 4′-modified ascarosides in this mutant is not due to depletion of any of the building blocks (Figure S1 in the Supporting Information). In contrast, the amounts of 4′-modified ascarosides in cup-5(ar465), haf-4(ok1042), and haf-9(gk23) mutants, which are defective in the formation of non-acidic gut granules but do have acidic LROs,[15] are similar or increased relative to the wildtype (Figure S6). These results indicate that acidic LROs are required for the biosynthesis of all 4′-modified ascarosides, including icas#9 and osas#9, the biosynthesis of which specifically requires ACS-7 (Figure 4).
Figure 4.
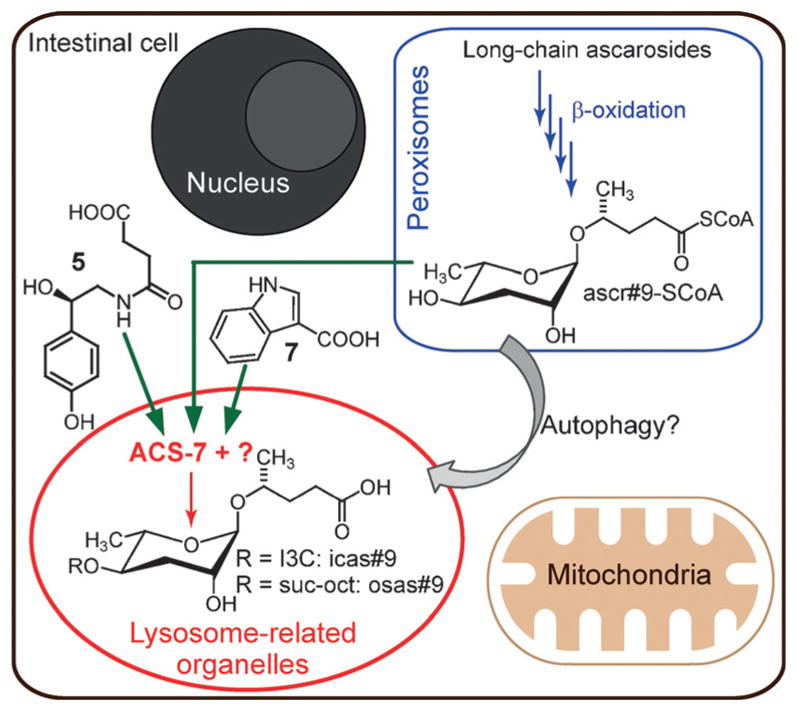
Simplified model for the biosynthesis of modular ascarosides in C. elegans. Long-chain ascaroside precursors undergo multiple rounds of peroxisomal β-oxidation to produce Coenzyme A esters of short-chain ascarosides such as ascr#9. Subsequently, short-chain ascarosides (or their CoA esters) may undergo 4′-modification in acidic lysosome-related organelles (LROs). I3C =indole-3-carboxy, suc-oct =N-succinyl octopamine.
The acidic LROs in C. elegans share many characteristics with lysosomes in higher animals.[10] The lysosome, classically known as the waste disposal system of the cell, is rapidly gaining attention as a central hub for intra- and intercellular chemical signaling.[11b,16] Lysosomal acid lipases play critical roles under starvation conditions to generate small-molecule signals such as polyunsaturated fatty acids[17] and oleoylethanolamide,[11c] which in turn activate diverse downstream responses, including autophagy, a starvation-activated process by which targeted components of the cell are transferred to the lysosome for recycling.[11b] We found that in atg-18(gk378) mutants,[18] which are deficient in autophagosome assembly, the abundances of most 4′-modified ascarosides are significantly reduced relative to wildtype (Figure S7), thus suggesting that autophagy contributes to the biosynthesis of modular ascarosides in the LROs.
Taken together, our results indicate that in C. elegans, homologues of genes from canonical metazoan metabolism act in different cell compartments to produce modular specialized metabolites. The high specificity of ACS-7, which is required only for the biosynthesis of 4′-modified versions of the 5-carbon side-chain ascaroside ascr#9, that is, icas#9 and osas#9, but not the corresponding 7-carbon or 9-carbon side-chain versions, for example, icas#3 or osas#10 (Figures 1, 2a), indicates that other enzymes must exist that serve equivalent roles in the biosynthesis of 4′-modified ascarosides with longer fatty acid side chains. The specific localization of ACS-7 to the LROs, which serve as part of the cell waste disposal system, and the finding that LROs are required for the biosynthesis of all 4′-modified ascarosides will motivate organelle-specific proteomics and metabolomics studies to identify other components in the biosynthesis of modular ascarosides. Of particular interest will be the mechanisms that underlie the selection of different building blocks and govern the side-chain-length specificity of the modifications, thereby ultimately integrating metabolic state in a complex chemical message.
Supplementary Material
Acknowledgments
Strains used in this work were provided by NBRP (Japan) and CGC which is funded by NIH (P40 OD010440). This work was supported in part by the NIH (GM113692, GM088290 to F.C.S., T32-GM007616 to A.A., T32 GM008500 to J.C.J.), NSF (DBI-0618969 to BTI), and HHMI.
Footnotes
Dedicated to Professor Jerrold Meinwald on the occasion of his 90th birthday
Conflict of interest
The authors declare no conflict of interest.
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: http://dx.doi.org/10.1002/anie.201700103.
Contributor Information
Oishika Panda, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY (USA).
Allison E. Akagi, Howard Hughes Medical Institute and Division of Biology and Biological Engineering, California Institute of Technology Pasadena, CA (USA)
Dr. Alexander B. Artyukhin, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY (USA)
Dr. Joshua C. Judkins, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY (USA)
Henry H. Le, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY (USA)
Dr. Parag Mahanti, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY (USA)
Sarah M. Cohen, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY (USA). Howard Hughes Medical Institute and Division of Biology and Biological Engineering, California Institute of Technology Pasadena, CA (USA)
Prof. Paul W. Sternberg, Howard Hughes Medical Institute and Division of Biology and Biological Engineering, California Institute of Technology Pasadena, CA (USA)
Prof. Frank C. Schroeder, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY (USA), Homepage: http://bti.cornell.edu/schroeder
References
- 1.von Reuss SH, Schroeder FC. Nat Prod Rep. 2015;32:994–1006. doi: 10.1039/c5np00042d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ludewig AH, Izrayelit Y, Park D, Malik RU, Zimmermann A, Mahanti P, Fox BW, Bethke A, Doering F, Riddle DL, Schroeder FC. Proc Natl Acad Sci USA. 2013;110:5522– 5527. doi: 10.1073/pnas.1214467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Jeong PY, Jung M, Yim YH, Kim H, Park M, Hong E, Lee W, Kim YH, Kim K, Paik YK. Nature. 2005;433:541–545. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]; b) Butcher RA, Fujita M, Schroeder FC, Clardy J. Nat Chem Biol. 2007;3:420–422. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]; c) Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, Schroeder FC. Proc Natl Acad Sci USA. 2009;106:7708– 7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose N, Ogawa A, von Reuss SH, Yim JJ, Ragsdale EJ, Sommer RJ, Schroeder FC. Angew Chem Int Ed. 2012;51:12438–12443. doi: 10.1002/anie.201206797. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2012;124:12606–12611. [Google Scholar]
- 5.a) Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PEA, Malik RU, Edison AS, Sternberg PW, Schroeder FC. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho MC, O’Doherty OG, Edison AS, Sternberg PW, Schroeder FC. PLoS Biol. 2012;10:e1001237. doi: 10.1371/journal.pbio.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Artyukhin AB, Yim JJ, Srinivasan J, Izrayelit Y, Bose N, von Reuss SH, Jo Y, Jordan JM, Baugh LR, Cheong M, Sternberg PW, Avery L, Schroeder FC. J Biol Chem. 2013;288:18778–18783. doi: 10.1074/jbc.C113.477000. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Izrayelit Y, Srinivasan J, Campbell SL, Jo Y, von Reuss SH, Genoff MC, Sternberg PW, Schroeder FC. ACS Chem Biol. 2012;7:1321–1325. doi: 10.1021/cb300169c. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Kaplan F, Srinivasan J, Mahanti P, Ajredini R, Durak O, Nimalendran R, Sternberg PW, Teal PE, Schroeder FC, Edison AS, Alborn HT. PLoS ONE. 2011;6:e17804. doi: 10.1371/journal.pone.0017804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludewig AH, Schroeder FC. WormBook. 2013:1–22. doi: 10.1895/wormbook.1.155.1. [DOI] [PMC free article] [PubMed]
- 7.von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, Schroeder FC. J Am Chem Soc. 2012;134:1817– 1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY. Proc Natl Acad Sci USA. 2009;106:1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Izrayelit Y, Robinette SL, Bose N, von Reuss SH, Schroeder FC. ACS Chem Biol. 2013;8:314–319. doi: 10.1021/cb3004644. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang X, Feng L, Chinta S, Singh P, Wang Y, Nunnery JK, Butcher RA. Proc Natl Acad Sci USA. 2015;112:3955–3960. doi: 10.1073/pnas.1423951112. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Markov GV, Meyer JM, Panda O, Artyukhin AB, Claassen M, Witte H, Schroeder FC, Sommer RJ. Mol Biol Evol. 2016;33:2506– 2514. doi: 10.1093/molbev/msw090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Motley AM, Hettema EH, Ketting R, Plasterk R, Tabak HF. EMBO Rep. 2000;1:40– 46. doi: 10.1093/embo-reports/kvd010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR. Mol Biol Cell. 2005;16:3273– 3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Coburn C, Allman E, Mahanti P, Benedetto A, Cabreiro F, Pincus Z, Matthijssens F, Araiz C, Mandel A, Vlachos M, Edwards SA, Fischer G, Davidson A, Pryor RE, Stevens A, Slack FJ, Tavernarakis N, Braeckman BP, Schroeder FC, Nehrke K, Gems D. PLoS Biol. 2013;11:e1001613. doi: 10.1371/journal.pbio.1001613. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mony VK, Benjamin S, O’Rourke EJ. Autophagy. 2016;12:619–631. doi: 10.1080/15548627.2016.1147671. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, Wang MC. Science. 2015;347:83– 86. doi: 10.1126/science.1258857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soukas AA, Carr CE, Ruvkun G. PLoS Genet. 2013;9:e1003908. doi: 10.1371/journal.pgen.1003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boehm M, Bonifacino JS. Mol Biol Cell. 2001;12:2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Stupp GS, von Reuss SH, Izrayelit Y, Ajredini R, Schroeder FC, Edison AS. ACS Chem Biol. 2013;8:309–313. doi: 10.1021/cb300520u. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Soukup ST, Spanier B, Grunz G, Bunzel D, Daniel H, Kulling SE. PLoS One. 2012;7:e46914. doi: 10.1371/journal.pone.0046914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanji T, Nishikori K, Haga S, Kanno Y, Kobayashi Y, Takaya M, Gengyo-Ando K, Mitani S, Shiraishi H, Ohashi-Kobayashi A. BMC Cell Biol. 2016;17:4. doi: 10.1186/s12860-015-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber LA, Teis D. Curr Opin Cell Biol. 2016;39:8– 14. doi: 10.1016/j.ceb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 17.O’Rourke EJ, Kuballa P, Xavier R, Ruvkun G. Genes Dev. 2013;27:429– 440. doi: 10.1101/gad.205294.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Chang JT, Guo B, Hansen M, Jia K, Kovács AL, Kumsta C, Lapierre LR, Legouis R, Lin L, Lu Q, Meléndez A, O’Rourke EJ, Sato K, Sato M, Wang X, Wu F. Autophagy. 2015;11:9– 27. doi: 10.1080/15548627.2014.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.See www.smid-db.org for compound naming.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.