Abstract
The investigation of pH-dependent membrane-associated folding has both fundamental interest and practical applications for targeting of acidic tumors and specific delivery of therapeutic molecules across membrane of cancer cells. We and others investigated molecular mechanism and medical uses of class of water soluble membrane peptides, pH (Low) Insertion Peptides (pHLIP® peptides). Here we employed optical spectroscopy methods to study interactions of the truncated pHLIP® peptide (Short pHLIP®) with lipid bilayer of membrane. Tryptophan fluorescence, CD and OCD data indicate on pH-triggered formation of transmembrane helical structure. Dual quenching and FRET assays demonstrated that Short pHLIP® peptide spans lipid bilayer of membrane similar to Long pHLIP® peptides. Truncated pHLIP® peptides with multiple charged and protonatable residues in their sequences potentially can make these peptides to be less hydrophobic compared to Long pHLIP® peptides, and might have utility in tumor imaging, and potentially, in pH-regulated cytoplasmic delivery of moderately hydrophobic drugs.
Abbreviations: 10DN, 10-doxylnonadecane; CD, circular dichroism; FRET, Förster resonance energy transfer; NBD-PE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-7-nitro-2-1,3-benzoxadiazol-4-yl ammonium salt; OCD, oriented circular dichroism; PET, positron emission tomography; pHLIP, pH Low Insertion Peptide; PMT, photomultiplier tube; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; TM, transmembrane; TMA-DPH, 1-(4-trimethylammoniumphenyl)−6-phenyl-1,3,5-hexatriene p-toluenesulfonate; UV, ultraviolet
Keywords: Fluorescence, CD, OCD, FRET, Dual quenching, Tumor targeting
Highlights
-
•
Protonation of single Asp triggers pH-dependent insertion into membrane.
-
•
Truncated pHLIP® peptide adopts TM helical orientation at low pH.
-
•
Truncated pHLIP® peptide spans lipid bilayer similar to full-length long peptide.
-
•
Potential applications in cytoplasmic delivery of small hydrophobic molecules.
1. Introduction
Study of polypeptide's pH-dependent insertion into the lipid bilayer of membrane and membrane-associated folding finds both fundamental interest and practical applications in targeting of acidic diseased tissues such as tumors. Here we have performed a comparative study of full-length and truncated versions of pH Low Insertion Peptide's (pHLIP® peptides) interaction with the lipid bilayer of a membrane. pHLIP® peptides are well investigated water-soluble membrane polypeptides, which insert into membrane and form a stable transmembrane (TM) alpha-helix in a result of pH drop [1], [2], [3], [4]. These peptides find wide applications in medicine, since they show excellent targeting of acidic diseased tissues and intracellular delivery of polar cell-impermeable cargo molecules [5]. Importantly, the mechanism of tumor targeting and intracellular delivery of cargo molecules is based on a pH-dependent membrane-associated folding of pHLIP® peptides. At neutral and high pHs, the peptides are in equilibrium between free in solution and membrane-bound forms [2]. Once the pH drops, protonatable Asp/Glu residues within the pHLIP® sequence become neutral, and the overall hydrophobicity of the peptide increases [6], [7], [8]. This triggers peptide partitioning into the membrane, which induces folding and formation of a TM helix [2]. The truncated pHLIP® peptides show lower affinity to the membrane at neutral pH [1]. As a result, it leads to the weaker interactions with the cellular membranes in blood and fast blood clearance [1], [9]. Fast blood circulation is essential in nuclear imaging for the delivery of fast decaying radioactive imaging probes to tumors. Also fast blood clearance of radioactive materials is needed to ensure safety. Recently, it was published several reports demonstrating high utility of truncated pHLIP® peptides in optical [1], [10], opto-acoustic [11] and PET [9] imaging.
2. Materials and methods
2.1. Peptides preparation
pHLIP® peptides were synthesized at W.M. KECK Biotechnology center at Yale. The synthesized peptides were dissolved in buffer containing 3 M urea and then passed through the fast spin G-10 column to remove urea. Concentrations of the peptides were calculated spectrophotometrically by measuring absorbance at 280 nm.
2.2. Liposomes preparation
Large unilamellar and multilamellar vesicles were prepared by extrusion. POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids), or a mixture of POPC with 0.5% of 18:1 NBD-PE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-7-nitro-2-1,3-benzoxadiazol-4-yl ammonium salt (Avanti Polar Lipids) were dissolved in chloroform, desolvated in a rotary evaporator and dried under high vacuum for several hours. The phospholipid film was then rehydrated in 10 mM phosphate buffer pH 8, vortexed until the lipid bilayer was completely dissolved, and repeatedly extruded (15 times) through the membranes with 50 nm pore size.
2.3. Steady-state fluorescence, CD and OCD
Steady-state fluorescence, circular dichroism (CD) and oriented circular dichroism (OCD) measurements were carried out under a temperature control at 25 °C on a PC1 spectrofluorometer (ISS, Inc.) and MOS 450 spectropolarimeter (Bio-Logic, Inc.), respectively. The concentrations of the peptides and POPC lipids were varied from 3 to 7 µM and from 0.1 to 2.1 mM, respectively. The final measurements presented on the figures were performed with 7 µM of peptides and 2.1 mM of lipids (L/P ratio is 300). Tryptophan fluorescence of the peptides was excited at 295 nm. Emission was recorded with the excitation and emission slits set at 1 nm. The polarizers in the excitation and emission paths were set at the “magic” angle (54.7° from the vertical orientation) and vertically (0°), respectively. Peptide CD spectra were recorded from 195 to 255 nm (where no PMT saturation was observed) with 0.5 nm increment using a cuvette with an optical path length of 0.5 cm. Also, CD spectra were recorded using a stack of quartz slides with special polish for far UV measurements, with spacers of 0.2 mm thickness on one side of each slide (Starna). Oriented CD (OCD) was measured from the supported bilayers deposited on a stack of quartz slides. Quartz slides were cleaned by sonication for 10 min in cuvette cleaner solution (Decon Contrad 5% in water), 2-propanol, acetone, 2-propanol and rinsed with deionized water. Then the slides were immersed in a mixture of concentrated sulfuric acid and hydrogen peroxide (ratio 3:1) for 5–10 min to completely remove any remaining organic material. Slides were then thoroughly rinsed with and stored in deionized water (Milli-Q purified water kept at 25 °C). A POPC lipid monolayer was deposited on the clean quartz substrate by the Langmuir-Blodgett method using a KSV minitrough. For the Langmuir-Blodgett deposition, a POPC lipid solution in chloroform was spread on the subphase and allowed to evaporate solvent for about 30 min, followed by monolayer compression to 32 mN/m. An initial layer was deposited by retrieving the slide from the subphase at a rate of 15 mm/min. The second layer of the bilayer was created by fusion. For this step, the monolayer on the slide was incubated with a solution of POPC vesicles (50 nm in diameter obtained by extrusion) mixed with the peptide solution at pH 4 (0.5 mM POPC and 10 µM peptide). The fusion occurred during 6 h incubation at 100% humidity. Then, excess of vesicles was carefully removed and the slides were stacked to make a pile filled with the buffer solution at pH 4. Measurements were taken at 3 steps during the process: when the monolayers were incubated with the excess of liposomes, soon after spaces between slides were filled with the buffer solution and 6 h after the second measurement. 14 slides (28 bilayers) were assembled and OCD spectra were recorded on a MOS-450 spectrometer with 2 s sampling time. Control measurements were carried out of the peptide between slides with and without supported bilayers and in the presence of an excess of POPC liposomes in the range of 205–260 nm (where no PMT saturation was observed).
2.4. Dual quenching assay
POPC liposomes without and with 10% of the lipids replaced by 10-doxylnonadecane (10DN) (Avanti Polar Lipids) were prepared in 10 mM citrate-phosphate buffer at pH 8. Peptides and POPC liposomes were mixed to final concentrations of 7 µM peptide and 2.1 mM POPC without and with 10DN (L/P ratio is 300). In some of the samples the pH was lowered to pH 4 by addition of aliquot of 2 M citric acid and other samples were kept at pH 8. Acrylamide (Sigma-Aldrich) was added to the samples containing POPC liposomes without 10DN. The final concentration of acrylamide in the sample was 235 mM. The peptide concentration in all samples was kept constant by adding an appropriate amount of buffer at the required pH. To observe quenching of tryptophan fluorescence by 10DN or acrylamide the tryptophan fluorescence was recorded as described above. The appropriate POPC blanks were measured and subtracted from the measured spectra before analysis. The percentage of quenching was calculated by measuring area under the spectra in the presence of quencher and normalizing to the area under the spectra with no quencher added.
2.5. NBD-FRET assay
First, POPC liposomes with NBD at the inner leaflet were prepared. To do so, POPC liposomes containing 0.5% PE lipids headgroup conjugated with NBD were prepared. Next, 1.2 mL of symmetrically NBD-labeled POPC liposomes at 6 mM concentration were incubated with 150 μL of 1 M freshly prepared membrane-impermeable dithionite in buffer at pH 8. The decrease of NBD fluorescence occurring as the result of quenching of NBD by dithionite was monitored at the excitation of 463 nm and emission of 530 nm. The dithionite quenching led to the reduction of about 60–65% of NBD fluorescence signal, which corresponds to the NBD on the outer leaflet of the bilayer. Next, the POPC solution was passed through a fast spin G-10 column to remove excess of dithionite. Asymmetrically labeled POPC liposomes (2.1 mM) were incubated with the peptides (7 µM), at L/P ratio of 300. FRET from tryptophan residues to NBD at inner leaflet of the bilayer was monitored at 295 nm excitation wavelength. The emission was recorded from 310 to 580 nm.
3. Results
3.1. Fluorescence, CD and OCD measurements
We have investigated full-length and truncated versions of pHLIP® peptide's (Table 1), and their interactions with a membrane. Full-length pHLIP® peptides, such as WT-pHLIP and Long pHLIP (called Var2 in Weerakkody et al. [1]), are well investigated water-soluble membrane polypeptides. They insert into the lipid bilayer of a membrane and form TM helix upon a drop of pH [4]. The truncated versions (Short WT-pHLIP and Short pHLIP) also demonstrate pH-dependent interaction with lipid bilayer of a membrane [1]. At the same time, the truncated version (Short pHLIP*), where Trp residue located at the beginning of putative TM part was replaced by Phe, lost it's pH-dependent ability to interact with membrane (Fig. 1a), which indicative of the lack of insertion into the lipid bilayer of membrane. Our further comparative investigation was performed with Long and Short pHLIP® sequences, where Trp residues were located at the beginning and end of putative TM region of the peptides. Such location of Trp residues, on one hand, is expected to stabilize TM orientation and, on other hand, is convenient for interpretation of the results of FRET and quenching assays.
Table 1.
Peptide sequences, the putative transmembrane part is underlined.
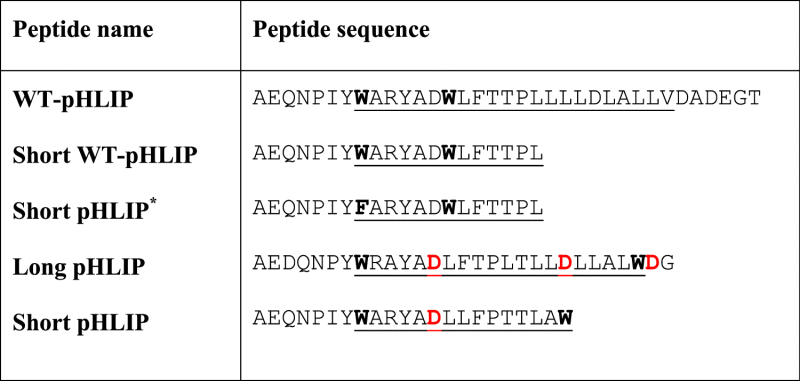 |
Fig. 1.
Three states of short pHLIP® peptide and OCD. The states of Short pHLIP*(a) and Short pHLIP® (b-c) were monitored by changes of the tryptophan fluorescence (a-b) and CD (c). The state I (black lines) represents peptides in solution at pH 8. The state II (blue lines) is a peptide in a solution in the presence of POPC liposomes at pH 8. The state III (red line) is a peptide in a solution in the presence of POPC liposomes at pH 4. The oriented circular dichroism (OCD) spectrum was measured for the Short pHLIP® peptide on the supported bilayers at pH 4 (d). The dashed line indicates zero level.
Short pHLIP® peptide demonstrated pH-dependent changes of the fluorescence and CD spectral signals (Figs. 1b and c). The increase of tryptophan emission and the short wavelength shift of the spectrum were observed. The overall strength of the CD signal was twice less than the strength of the full-length pHLIP® peptide's CD signals [1], [2], and the first minimum (at 208–210 nm) had higher amplitude compared to the second one at 222–225 nm. The observed CD might represent presence of a mixture of α-helical and random coil conformations of the peptide, and/or formation of a 310 helical segments, which are known to have stronger signal at 208 compared to 222 [12]. The ratio of CD signal at 222/208 nm for 310 helix was established previously to be in the range of 0.3–0.4 compared to the same ratio for an α-helix, which is close to 1 or higher [12]. According to our previous data the selected L/P ratio of 300 is expected to ensure shift of the equilibrium toward the membrane-bound form of the peptide [1], [3]. Thus, the population of the free peptide in solution (random coil) should be minimal.
To prove that Short pHLIP® peptide indeed adopts TM orientation we recorded OCD spectrum from the peptide on the supported bilayers (Fig. 1d). We observed a characteristic shift and increase of the positive band; and a disappearance of 208 negative band, which is indicative of a TM orientation of the peptide [13].
3.2. Dual-quenching assay
To identify location of Trp residues within a bilayer, we employed dual quenching and NBD-FRET fluorescent assays [14]. Both Long and Short pHLIP® peptides have Trp residues located at the beginning and end of the putative TM parts. The quenching of Trp fluorescence by acrylamide and 10DN was carried out to establish location of tryptophan residues within the lipid bilayer of membrane at high and low pHs. Effective quenching of Trp fluorescence by acrylamide occurs when tryptophan residues are exposed to the polar parts of outer or inner leaflets of a bilayer. At the same time tryptophan residues located in the middle of a membrane could be effectively quenched by 10DN. At pH 8 Short pHLIP® peptide just barely partitions into the bilayer and therefore tryptophan fluorescence is quenched by acrylamide very well (Fig. 2a and Table 2). Long pHLIP® peptide being more hydrophobic, is located much deeper into the bilayer, which correlates well with our previous data [1], [4], [15]. Lowering the pH reduces quenching of Trp fluorescence by acrylamide and increases quenching by 10DN. The overall trend of Short pHLIP® peptide's partition into the bilayer at low pH is similar to Long pHLIP® peptide. However, Trp residues in Short pHLIP® peptide are more exposed to the acrylamide compared to Trp residues of Long pHLIP® peptide.
Fig. 2.
Dual-quenching assay. The tryptophan fluorescence of Short (a, b) and Long (c, d) pHLIP® peptides in the presence of POPC liposomes at pH 8 (blue lines) and pH 4 (red lines) are shown. The emission of tryptophan residues of the peptides in the presence of POPC liposomes at both pHs is quenched by 10DN (magenta lines) or acrylamide (green lines). The amount of quenching is given in Table 2.
Table 2.
The percentage of quenching of Trp fluorescence of Long-pHLIP and Short-pHLIP in the presence of POPC liposomes at pH 8 and pH 4, by acrylamide and 10DN incorporated into liposomes. The data are calculated from the spectra shown on Fig. 2. The percentage of quenching was calculated by measuring area under the spectra in the presence of quencher and normalizing to the area under the spectra with no quencher added. The experiment was repeated three times for Short pHLIP peptide and mean and St.d. numbers are presented in the Table.
|
pH 8 |
pH 4 |
|||
|---|---|---|---|---|
| Acrylamide | 10-DN | Acrylamide | 10-DN | |
| Short-pHLIP | 84.1±4.6% | 7.7±0.2% | 48.9±1.5% | 34.4±0.8% |
| Long-pHLIP | 44% | 33% | 31% | 45% |
3.3. NBD-FRET assay
The dual-quenching assay provided information about the degree of partitioning of Trp residues into a bilayer. However it did not allow distinguishing between inner or outer leaflet locations of the acrylamide-accessible Trp residues. To further investigate location of tryptophan residues in membrane we performed NBD-FRET assay [16], [17]. Symmetrically-labeled (with NBD dye) POPC liposomes were prepared. Then, the membrane-impermeable dithionite was used to chemically modify and quench fluorescence of NBD in the outer leaflet of bilayer, followed by the removal of dithionite by gel filtration. As a result, asymmetrically-labeled liposomes with NBD at the inner leaflet were obtained. The absence of a flip-flopping of lipids was accessed by the absence of quenching of NBD fluorescence by addition of new portion of dithionite with time. FRET was monitored from the tryptophan residues of peptides to NBD at the inner leaflet of bilayer. Energy transfer occurs when both fluorophores (Trp and NBD) are in a close proximity to each other (the Förster distance for Trp-NBD donor-acceptor pair is about 10 Å [18]). Thus, when tryptophan residues are located at the outer leaflet of the bilayer, there is not any significant energy transfer to NBD at the inner leaflet of bilayer. This is the situation observed at pH 8 for both peptides, but was less pronounced for Long pHLIP® peptide, which partitions deeper into the membrane (Fig. 3). At the same time, at low pH the FRET signal was comparable for both peptides. We observed that the NBD fluorescence signal increased by 11.7 and 12.9 times for Short and Long pHLIP® peptides, respectively, in the presence of POPC at low pH compared to the baseline. It indicates that Trp residue in both Long and Short pHLIP® peptides is in close proximity to the headgroups of the inner leaflet of a bilayer. Previously it was shown that C-terminus of pHLIP® peptides propagates into membrane across lipid bilayer upon drop of pH [2], [4], [19].
Fig. 3.
NBD-FRET assay. The tryptophan fluorescence of Short (a, b) and Long (c, d) pHLIP® peptides in three states are shown. The state I (black lines) represents peptides in solution at pH 8. The state II (blue lines) is a peptide in a solution in the presence of asymmetrically-labeled (by NBD) POPC liposomes at pH 8. The state III (red line) is a peptide in a solution in the presence of asymmetrically-labeled (by NBD) POPC liposomes at pH 4. Energy transfer from tryptophan residues to NBD dye at the inner leaflet of bilayer was monitored (b, d). The numbers on panels b and d indicate the increase of NBD fluorescence (calculated by increase of area under the spectra in the range of 480–560 nm) in states III and II compared to the baseline (black lines).
4. Discussion
All obtained results allow us to make a conclusion that Short pHLIP® peptide inserts into the lipid bilayer of a membrane and spans the bilayer. Since Short pHLIP® peptide has truncated sequence a negative hydrophobic mismatch might occur. Usually, positive and/or negative hydrophobic mismatches lead to the energetic penalties, since the hydrophobic segments of the polypeptide sequence could be exposed to the polar environment or, vice versa, the polar segments of the polypeptide sequences could be exposed to the hydrophobic environment. As a result, it can lead to the structural perturbations in a polypeptide, alteration in a polypeptide's mobility and/or membrane thickness changes to compensate energetic penalties [20], [21], [22]. As it was proposed early, there are number of ways a system might reduce a negative energy of the hydrophobic mismatch, such as thinning of lipids, aggregation of peptides, anchoring aromatic residues to the lipid headgroups, and stretching from the alpha-helical to 310-helical conformations [23], [24], [25]. Our data does not point to the aggregation of the peptide in membrane; however we cannot exclude that as a possibility. Also, our data might point to the appearance of elements of stretched 310 helical conformation, or mixture of alpha- and 310-helices potentially with 310 components at the beginning and end of TM alpha-helix. Finally, our data indicate that anchoring of Trp residue at the lipid headgroups contributes to the stability of the inserted helical peptide at low pH.
As we outlined in the introduction, the molecular mechanism of pHLIP® peptide's pH-triggered insertion into membrane is based on protonation of Asp residues, with leads to the increase of peptide's hydrophobicity and peptide partitioning into bilayer and folding. The role of protonatable Asp residues in TM part and peptide's inserting end was investigated previously [1], [4], [7], [8]. However, all investigated peptides had multiple protonatable residues in the sequence. Here we investigated truncated peptide, which has single Asp residue. Our data indicate that protonation of a single Asp residue located in TM and free C-terminal group of the short pHLIP® peptide is enough to trigger insertion into the lipid bilayer of membrane at low pH. According to our previous kinetics data obtained on similar truncated pHLIP® peptide (Var12) the process of peptide insertion into membrane is completed within 100 ms [1].
The truncated pHLIP® peptides have less hydrophobic residues in their sequence and they still can insert across a membrane. So, the overall hydrophobicity of the truncated pHLIP® peptides is lower compared to the full-length pHLIP® peptides. Moreover, the truncated peptides might be designed with different number of protonatable Asp/Glu residues, which are charged at high pH and protonatable at low pH. Such short pHLIP® peptides might be attractive for tumor targeting and pH-dependent cellular delivery of moderately hydrophobic therapeutics cargo molecules. Small hydrophobic (drug like) molecules demonstrate fast blood clearance and lack of tumor targeting. pHLIP® peptides might alter pharmacokinetics and biodistribution and enhance pH-specific translation of these molecules across membrane of cancer cells, and potentially prevent translation across membrane of normal cells in healthy tissue. It would allow reducing therapeutic dose, enhancing therapeutic index and reducing side effects.
Funding
This work was supported by the National Institute of Health [grant number GM073857 to OAA and YKR].
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.10.001.
Appendix A. Transparency document
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Weerakkody D., Moshnikova A., Thakur M.S., Moshnikova V., Daniels J., Engelman D.M., Andreev O.A., Reshetnyak Y.K. Family of pH (low) insertion peptides for tumor targeting. Proc. Natl. Acad. Sci. USA. 2013;110:5834–5839. doi: 10.1073/pnas.1303708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reshetnyak Y.K., Segala M., Andreev O.A., Engelman D.M. A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers. Biophys. J. 2007;93:2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reshetnyak Y.K., Andreev O.A., Segala M., Markin V.S., Engelman D.M. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc. Natl. Acad. Sci. USA. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karabadzhak A.G., Weerakkody D., Wijesinghe D., Thakur M.S., Engelman D.M., Andreev O.A., Markin V.S., Reshetnyak Y.K. Modulation of the pHLIP transmembrane helix insertion pathway. Biophys. J. 2012;102:1846–1855. doi: 10.1016/j.bpj.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreev O.A., Engelman D.M., Reshetnyak Y.K. Targeting diseased tissues by pHLIP insertion at low cell surface pH. Front. Physiol. 2014;5:97. doi: 10.3389/fphys.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreev O.A., Dupuy A.D., Segala M., Sandugu S., Serra D.A., Chichester C.O., Engelman D.M., Reshetnyak Y.K. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc. Natl. Acad. Sci. USA. 2007;104:7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musial-Siwek M., Karabadzhak A., Andreev O.A., Reshetnyak Y.K., Engelman D.M. Tuning the insertion properties of pHLIP. Biochim. Biophys. Acta. 1798;2010:1041–1046. doi: 10.1016/j.bbamem.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrera F.N., Weerakkody D., Anderson M., Andreev O.A., Reshetnyak Y.K., Engelman D.M. Roles of carboxyl groups in the transmembrane insertion of peptides. J. Mol. Biol. 2011;413:359–371. doi: 10.1016/j.jmb.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viola-Villegas N.T., Carlin S.D., Ackerstaff E., Sevak K.K., Divilov V., Serganova I., Kruchevsky N., Anderson M., Blasberg R.G., Andreev O.A., Engelman D.M., Koutcher J.A., Reshetnyak Y.K., Lewis J.S. Understanding the pharmacological properties of a metabolic PET tracer in prostate cancer. Proc. Natl. Acad. Sci. USA. 2014;111:7254–7259. doi: 10.1073/pnas.1405240111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Monserrate Z., Roland C.L., Deng D., Arumugam T., Moshnikova A., Andreev O.A., Reshetnyak Y.K., Logsdon C.D. Targeting pancreatic ductal adenocarcinoma acidic microenvironment. Sci. Rep. 2014;4:4410. doi: 10.1038/srep04410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.C.W.KimbroughA.KhanalM.ZeidermanB.R.KhanalN.C.BurtonK.M.McMastersS.M.VickersW.E.GrizzleL.R.McNally, Targeting Acidity in Pancreatic Adenocarcinoma: Multispectral Optoacoustic Tomography Detects pH-low Insertion Peptide Probes in vivo, Clin Cancer Res, 2015. [DOI] [PMC free article] [PubMed]
- 12.Manning M.C., Woody R.W. Theoretical CD studies of polypeptide helices: examination of important electronic and geometric factors. Biopolymers. 1991;31:569–586. doi: 10.1002/bip.360310511. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y., Huang H.W., Olah G.A. Method of oriented circular dichroism. Biophys. J. 1990;57:797–806. doi: 10.1016/S0006-3495(90)82599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caputo G.A., London E. Using a novel dual fluorescence quenching assay for measurement of tryptophan depth within lipid bilayers to determine hydrophobic alpha-helix locations within membranes. Biochemistry. 2003;42:3265–3274. doi: 10.1021/bi026696l. [DOI] [PubMed] [Google Scholar]
- 15.Zoonens M., Reshetnyak Y.K., Engelman D.M. Bilayer interactions of pHLIP, a peptide that can deliver drugs and target tumors. Biophys. J. 2008;95:225–235. doi: 10.1529/biophysj.107.124156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIntyre J.C., Sleight R.G. Fluorescence assay for phospholipid membrane asymmetry. Biochemistry. 1991;30:11819–11827. doi: 10.1021/bi00115a012. [DOI] [PubMed] [Google Scholar]
- 17.Clausell A., Rabanal F., Garcia-Subirats M., Asuncion Alsina M., Cajal Y. Membrane association and contact formation by a synthetic analogue of polymyxin B and its fluorescent derivatives. J. Phys. Chem. B. 2006;110:4465–4471. doi: 10.1021/jp0551972. [DOI] [PubMed] [Google Scholar]
- 18.Galassi V., Nolan V., Villarreal M.A., Perduca M., Monaco H.L., Montich G.G. Kinetics of lipid-membrane binding and conformational change of L-BABP. Biochem Biophys. Res. Commun. 2009;382:771–775. doi: 10.1016/j.bbrc.2009.03.103. [DOI] [PubMed] [Google Scholar]
- 19.Reshetnyak Y.K., Andreev O.A., Lehnert U., Engelman D.M. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc. Natl. Acad. Sci. USA. 2006;103:6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondal S., Johnston J.M., Wang H., Khelashvili G., Filizola M., Weinstein H. Membrane driven spatial organization of GPCRs. Sci. Rep. 2013;3:2909. doi: 10.1038/srep02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomura T., Cranfield C.G., Deplazes E., Owen D.M., Macmillan A., Battle A.R., Constantine M., Sokabe M., Martinac B. Differential effects of lipids and lyso-lipids on the mechanosensitivity of the mechanosensitive channels MscL and MscS. Proc. Natl. Acad. Sci. USA. 2012;109:8770–8775. doi: 10.1073/pnas.1200051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gambin Y., Reffay M., Sierecki E., Homble F., Hodges R.S., Gov N.S., Taulier N., Urbach W. Variation of the lateral mobility of transmembrane peptides with hydrophobic mismatch. J. Phys. Chem. B. 2010;114:3559–3566. doi: 10.1021/jp911354y. [DOI] [PubMed] [Google Scholar]
- 23.Killian J.A. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta. 1998;1376:401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 24.de Planque M.R., Killian J.A. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 25.Salnikov E.S., Friedrich H., Li X., Bertani P., Reissmann S., Hertweck C., O’Neil J.D., Raap J., Bechinger B. Structure and alignment of the membrane-associated peptaibols ampullosporin A and alamethicin by oriented 15N and 31P solid-state NMR spectroscopy. Biophys. J. 2009;96:86–100. doi: 10.1529/biophysj.108.136242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material





