Abstract
OBJECT: Preoperative knowledge of meningioma grade is essential for planning treatment and surgery. The purpose of this study was to investigate the diagnostic value of MRI texture and shape analysis in grading meningiomas. METHODS: A surgical database was reviewed to identify meningioma patients who had undergone tumor resection between January 2015 and December 2016. Preoperative MR images were retrieved and analyzed. Texture and shape analysis was conducted to quantitatively evaluate tumor heterogeneity and morphology. Three machine learning classifiers were trained with these features to build classification models. The performance of the features and classification models was assessed. RESULTS: A total of 131 patients were included in this study: 21 with high-grade meningiomas and 110 with low-grade meningiomas. Three texture features were selected: Horzl_RLNonUni, S(2,2)SumOfSqs, and WavEnHL_s-3; three shape features were selected: GeoFv, GeoW4, and GeoW5b. The Mann–Whitney test indicated that all six features were significantly different between high-grade and low-grade meningiomas. AUC values were generally greater than 0.50 (range, 0.73 to 0.88). Sensitivities and specificities ranged from 47.62% to 90.48% and 69.09% to 96.36%, respectively. Among the nine classification models obtained, the one built by training the SVM classifier with all six features achieved the best performance, with a sensitivity, specificity, diagnostic accuracy, and AUC of 0.86, 0.87, 0.87, and 0.87, respectively. CONCLUSIONS: Texture and shape analysis, especially when combined with a SVM classifier, can provide satisfactory performance in the preoperative determination of meningioma grade and is thus potentially useful for clinical application.
Introduction
Meningioma is the most common intracranial tumor in adults (36.6%) and has an annual incidence rate of 80.3 per million [1]. Although the majority of meningiomas are benign, a small subset exhibit aggressive behavior [2]. According to the latest CBTRUS report, high-grade meningiomas (grade 2 and 3) accounted for 18.6% of all newly diagnosed meningiomas between 2009 and 2013 [1]. High histopathological grade has an association with increased tumor recurrence and poor prognosis, and therefore, preoperative knowledge of meningioma grade is of considerable importance to aid in treatment and surgical planning [3], [4].
Traditional magnetic resonance imaging (MRI) remains the preferred modality for preoperative assessment of meningioma. Several studies have investigated imaging features capable of differentiating between high-grade and low-grade meningiomas, with the most frequently assessed features including tumor heterogeneity, shape, size, and location; tumor-brain interface (TBI); capsular enhancement (CapE); and peritumoral edema [5], [6], [7], [8]. The results indicated that some imaging features, such as tumor heterogeneity, tumor shape, and TBI, had different distributions between high-grade and low-grade meningiomas and hence may be of diagnostic value in grading these tumors. However, a major limitation of these studies is that the assessed features were measured subjectively, requiring considerable experience and expertise, and therefore being subject to intra-observer and inter-observer variability. This limitation restricts the use of the identified features in clinical practice. As such, objective quantitative methods may be better suited for clinical application.
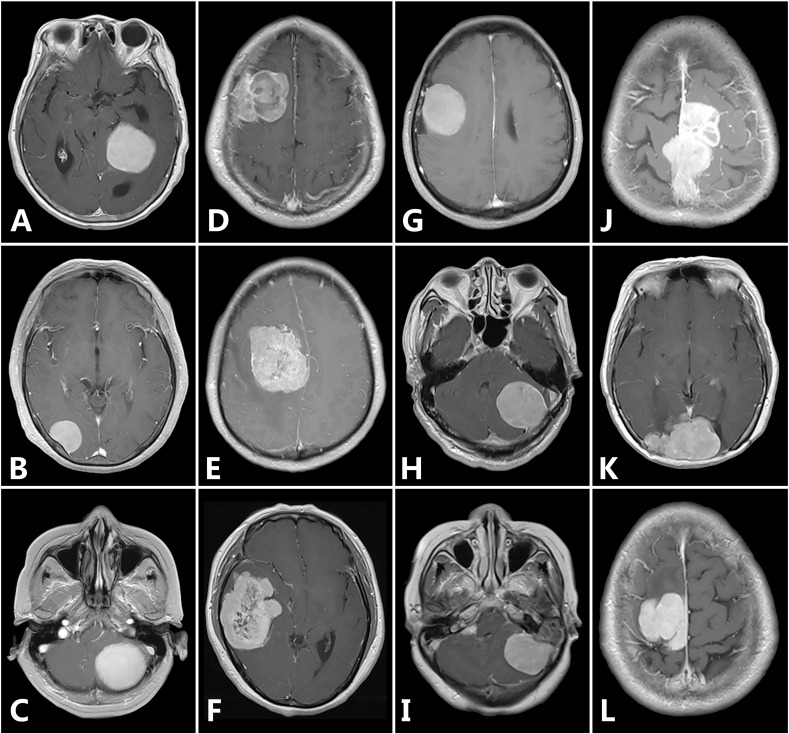
Medical images, which are digitally represented as a series of two-dimensional pixels, usually contain complex patterns (so-called image texture). While certain patterns (such as brightness and smoothness) can be observed qualitatively, many of these patterns remain imperceptible to the naked eye. Texture analysis is a well-established quantitative approach for image pattern recognition and works by extracting objective information through analysis of the spatial distribution of intensity variations in images [9], [10]. Texture and shape analysis has been widely used to evaluate medical images with promising results [11], [12], [13], [14], [15], [16]. However, to the best of our knowledge, no previous work has investigated the diagnostic value of preoperative MRI texture and shape analysis for grading meningiomas. Because heterogeneity and irregular shape are potential predictors for high-grade meningiomas (Figure 1), we hypothesized that texture and shape analysis, which can objectively evaluate the two features, could aid in meningioma grading. To test this hypothesis, we selected six texture and shape features and then built classification models using these features. Following this, the performance of the features and models in meningioma grading was assessed.
Figure 1.
Representative MR images to demonstrate differences in tumor heterogeneity and shape. Homogeneous enhancement (A-C), heterogeneous enhancement (D-F), regular tumor shape (G-I), and irregular tumor shape (J-L). Of the twelve meningiomas, five are high-grade (including D, E, F, J, and K), and the other seven are low-grade.
Material and Methods
Patient Population
Data were collected from our institution, a university-affiliated hospital. Approval from the institutional review board was obtained, and written informed consent was waived due to the retrospective nature of the study. We searched our departmental database for consecutive patients with histopathological confirmation of meningioma between January 2015 and December 2016. The exclusion criteria were previous intracranial surgery, radiotherapy, or injury; and incomplete preoperative MRI data.
Overall, 131 patients were included: 110 with low-grade (grade 1) meningioma (29 male, 81 female; mean age 53.00 ± 8.28 years, range 33–78 years) and 21 with high-grade meningioma (10 male, 11 female; mean age 52.38 ± 12.35 years, range 24–72 years). As part of routine clinical care, tumor samples were obtained by surgical resection, and histopathological diagnosis was made according to the 2007 World Health Organization (WHO) classification system for meningioma [2].
MRI Acquisition
All patients were examined using one of two scanners (Siemens Magnetom Avanto 1.5-T, and Siemens Magnetom Verio 3-T), each with an 8-channel radiofrequency coil. The MRI protocol used the following parameters: field of view, 230 × 230 mm; matrix size, 512 × 512; slice thickness, 5 mm; and flip angle, 90°. The repetition time (TR)/echo time (TE) for the T1-weighted sequence (T1WI), the T2-weighted sequence (T2WI), and the fluid-attenuated inversion recovery (FLAIR) sequence were 500/8.4 ms, 9000/89 ms, and 9000/105 ms, respectively. Contrast-enhanced T1-weighted images (CE-T1WI) were obtained in the sagittal and axial planes after intravenous administration of 0.2 ml/kg gadopentetate dimeglumine. All images were digitally stored in Picture Archiving and Communication Systems (PACS; Carestream Vue PACS, 11.3.4, Carestream Health, Rochester, NY, USA) and could be remotely accessed.
Image Analysis
Preoperative axial CE-T1WI images were retrieved and anonymized for image analysis. During image analysis, the investigators were blind to the patients' clinical information (sex, age, and tumor grade), and were allowed to zoom in/out the images and adjust the image levels/windows for better visualization. Two investigators (PFY and LY) independently reviewed each patient's CE-T1WI slices using MicroDicom (version 0.9.1, MicroDicom, Sofia, Bulgaria) to select the slice with the largest tumor cross-section; in case of discrepancy, consensus was achieved through discussion. Then, the corresponding slice was imported into ITK-SNAP (version 3.6.0, University of Pennsylvania) for tumor segmentation [17]. Each tumor was manually delineated using the “polygon mode” tool; contrast enhanced tissue and intratumoral cysts were included in the segmentation, whereas capsular enhancement and peritumoral edema were excluded. One investigator (PFY), experienced in the use of ITK-SNAP, participated in this process with the guidance of a neuroradiologist (ZZ, with 6 years of experience). As manual segmentation is a difficult and tedious task, to avoid potential errors, the investigators were asked to segment 10 images per day; they completed the whole patient population in 2 weeks. One month later, the two investigators together re-examined all the segmentations, and modifications were made when both agreed. The segmentations were then exported as BMP files, which would be used as region of interest (ROI) for feature calculation.
Quantitative image analysis was performed with MaZda (version 4.6.0, Institute of Electronics, Technical University of Lodz, Lodz, Poland), which has been specifically designed for analyzing texture and shape features [18], [19]. Before analysis, image intensities were normalized between μ ± 3σ (μ: mean of image intensity inside the ROI; σ: standard deviation); the range obtained was then quantized to 6 bits/pixel. This procedure has been shown to reduce brightness and contrast variations and can minimize the influence of inter-scanner differences [20]. Then, 279 texture features (belonging to six categories) and 73 shape features were calculated. The six categories of texture feature included histogram-based parameters, gradient-based parameters, run-length matrix-based parameters, co-occurrence matrix-based parameters, wavelet parameters, and autoregressive model parameters (Table 1). A detailed description of these features could be found in previous literature [9], [10]. Attempting to analyze too many texture and shape features is clinically impractical and could influence classification performance; therefore, we selected three top-ranked texture features and three top-ranked shape features for further analysis using the “CfsSubsetEval” evaluator provided in the data mining software Weka (version 3.9.1, University of Waikato, New Zealand) [21], [22].
Table 1.
Summary of Different Texture Feature Categories
| Category | Texture Features | Number |
|---|---|---|
| Histogram | mean, variance, skewness, kurtosis, 1-% percentile, 10-% percentile, 50-% percentile, 90-% percentile, and 99-% percentile | 9 |
| Gradient | mean, variance, skewness, kurtosis, and percentage of pixels with nonzero gradient | 5 |
| Run-length matrix* | run length nonuniformity, gray level nonuniformity, long run emphasis, short run emphasis, and fraction of image in runs | 20 |
| Co-occurrence matrix† | angular second moment, contrast, correlation, sum of squares, inverse difference moment, sum average, sum variance, sum entropy, entropy, difference variance, and difference entropy | 220 |
| Wavelet analysis | teta1, teta2, teta3, teta4, and sigma | 5 |
| Autoregressive model‡ | WavEn | 20 |
Each feature in this category is computed for four directions for each tumor (d = 0°, 45°, 90°, and 135°).
Each feature in this category is computed for four directions and five between-pixel distances for each tumor (d = 0°, 45°, 90°, and 135°; θ = 1, 2, 3, 4, and 5).
This feature is computed at five scales within four frequency bands; thus, the total number of features calculated for this category is twenty.
Statistical Analysis
Statistical analysis was carried out using SPSS (version 22.0; SPSS Inc., Chicago, IL, USA) with a significance level of 0.05. Categorical values were expressed as numbers (with percentages), and continuous variables were expressed as the mean ± SD. The Mann–Whitney U test was used to examine whether each feature was significantly different between high-grade and low-grade meningiomas [23]. Receiver operating characteristic (ROC) curves were constructed for each feature, and the corresponding area under curve (AUC) values were calculated [24]. When calculating true-positive (TP), false-positive (FP), true negative (TN), and false negative (FN) rates, high-grade meningiomas were considered positive cases, whereas low-grade meningiomas were considered negative cases. Classification analysis was performed with Weka. Different classification models were built using three machine learning classifiers: logistic regression(LR), naive Bayes(NB), and support vector machine (SVM). Ten-fold cross-validation was used to assess classification performance [25]. Average accuracy, sensitivity, specificity, and AUC values were calculated for each model.
Results
In total, 131 patients were enrolled, of which 21 (16.03%) had high-grade meningiomas and 110 (83.97%) had low-grade meningiomas. The mean patient age was 52.89 ± 9.00 years (range, 24 to 78 years); 39 patients were male, and 92 were female (Table 2). The female to male sex ratio was 2.36. The mean period between preoperative MRI examination and surgery was 8.32 ± 3.55 days (range, 1 to 26 days).
Table 2.
Clinical Characteristics of Patients
| Characteristics | High-Grade Meningiomas (%; n = 21) | Low-Grade Meningiomas (%; n = 110) | P value* |
|---|---|---|---|
| Age | |||
| > = 65 years | 4 (19.05) | 11 (10.00) | = 0.41 |
| < 65 years | 17 (80.95) | 99 (90.00) | |
| Sex | |||
| male | 10 (47.62) | 29 (26.36) | = 0.09 |
| female | 11 (52.38) | 81 (73.64) | |
| Tumor-brain interface | |||
| clear | 7 (33.3) | 83 (75.45) | < 0.01 |
| unclear | 14 (66.7) | 27 (24.55) | |
| Peritumoral edema | |||
| present | 13 (61.90) | 42 (38.18) | = 0.08 |
| absent | 8 (38.10) | 68 (61.82) | |
| Capsular enhancement | |||
| present | 12 (57.14) | 86 (78.18) | = 0.08 |
| absent | 9 (42.86) | 24 (21.82) | |
| Tumor enhancement | |||
| homogenous | 4 (19.05) | 79 (71.82) | < 0.01 |
| heterogeneous | 17 (80.95) | 31 (28.18) | |
| Tumor shape | |||
| regular | 4 (19.05) | 92 (83.64) | < 0.01 |
| irregular | 17 (80.95) | 18 (16.36) |
Calculated using the chi-square test.
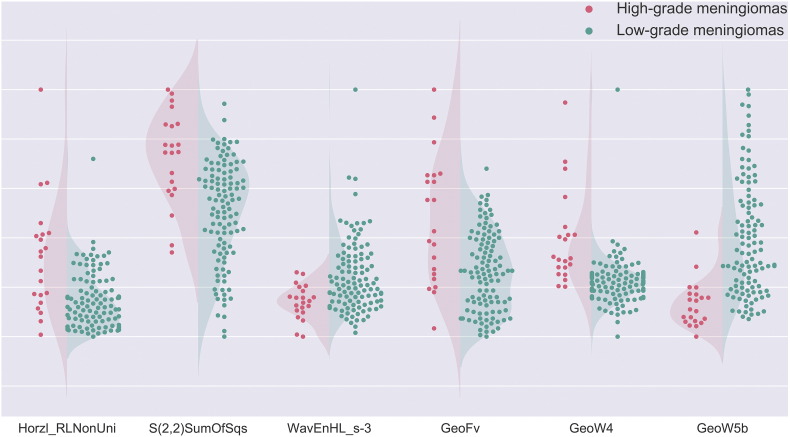
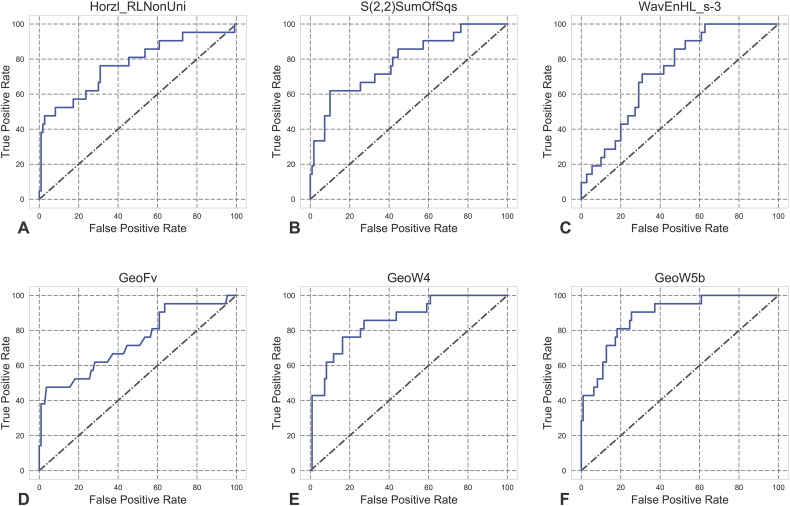
The selected top-ranked texture features were (1) Horzl_RLNonUni, (2) S(2,2)SumOfSqs, and (3) WavEnHL_s-3; the top-ranked shape features were (4) GeoFv, (5) GeoW4, and (6) GeoW5b. The Mann–Whitney test showed that features 1, 2, 4, and 5 were significantly greater in high-grade meningiomas (P values were =.0001, <.0001, =.0006, and <.0001, respectively), whereas features 3 and 6 were significantly greater in low-grade meningiomas (p values were =.0009 and <.0001, respectively; Figure 2). The ROC curves for the individual features are shown in Figure 3. The corresponding AUCs were 0.77, 0.79, 0.73, 0.74, 0.86, and 0.88 (Table 3); all of the values were significantly greater than 0.5, indicating their potential utility in meningioma grading.
Figure 2.
Patient distributions for each of the six texture/shape features. Red dots represent high-grade meningiomas, and green dots represent low-grade meningiomas. For features 1, 2, 4, and 5, the values for high-grade meningiomas are generally higher than those for low-grade meningiomas. In contrast, for features 3 and 6, the values of low-grade meningiomas are generally higher than those for high-grade meningiomas. This observation is consistent with Mann–Whitney test results, which indicated that all the features were significantly different between the two meningioma groups.
Figure 3.
ROC curves for each of the six texture/shape features for the prediction of high-grade meningiomas.
Table 3.
Results for ROC Analysis of Each Texture/Shape Feature
| Feature | Sensitivity | Specificity | AUC | Standard error | P value |
|---|---|---|---|---|---|
| Horzl_RLNonUni | 76.19 | 69.09 | 0.77 | 0.06 | <0.0001 |
| S(2,2)SumOfSqs | 61.9 | 90.00 | 0.79 | 0.06 | <0.0001 |
| WavEnHL_s-3 | 71.43 | 69.09 | 0.73 | 0.05 | <0.0001 |
| GeoFv | 47.62 | 96.36 | 0.74 | 0.07 | =0.0003 |
| GeoW4 | 76.19 | 83.64 | 0.86 | 0.04 | <0.0001 |
| GeoW5b | 90.48 | 74.55 | 0.88 | 0.04 | <0.0001 |
We subsequently built and assessed several classification models. The following feature subsets were assessed: (1) the three texture features (feature subset 1), (2) the three shape features (feature subset 2), and (3) all six texture and shape features (feature subset 3). Each subset was used to train the three classifiers. As a result, nine classification models were obtained. A detailed summary of the models' performance is presented in Table 4. The classification accuracy and AUCs of the models were generally satisfactory, ranging from 0.77–0.89 and 0.80–0.91, respectively. Among models built using LR and NB classifiers, those trained with feature subset 1 and 2 demonstrated similar performance. Models trained with feature subset 3 performed better and had a marked improvement in sensitivity, indicating that a greater number of high-grade meningiomas could be correctly classified using these models. Among models built using the SVM classifier, the one trained with feature subset 3 also demonstrated better performance with a marked improvement in specificity, indicating that a greater number of low-grade meningiomas could be correctly classified with this model. The NB and SVM classifiers performed better than the LR classifier in general, as they provided comparable specificities (0.918 and 0.873 vs. 0.936) and significantly higher sensitivities (0.762 and 0.857 vs. 0.667).
Table 4.
Details Regarding the Performance of the Classification Models
| Logistic Regression (LR) | Naive Bayes (NB) | Support Vector Machine (SVM) | |
|---|---|---|---|
| Feature subset 1* | |||
| True positive (TP) | 9 | 12 | 18 |
| False negative (FN) | 12 | 9 | 3 |
| True negative (TN) | 104 | 101 | 83 |
| False positive (FP) | 6 | 9 | 27 |
| Sensitivity | 0.43 | 0.57 | 0.86 |
| Specificity | 0.95 | 0.92 | 0.76 |
| Diagnostic accuracy | 0.86 | 0.86 | 0.77 |
| AUC | 0.84 | 0.88 | 0.81 |
| Feature subset 2 | |||
| True positive (TP) | 9 | 13 | 17 |
| False negative (FN) | 12 | 8 | 4 |
| True negative (TN) | 104 | 101 | 88 |
| False positive (FP) | 6 | 9 | 22 |
| Sensitivity | 0.43 | 0.62 | 0.81 |
| Specificity | 0.95 | 0.92 | 0.80 |
| Diagnostic accuracy | 0.86 | 0.87 | 0.80 |
| AUC | 0.86 | 0.88 | 0.81 |
| Feature subset 3 | |||
| True positive (TP) | 14 | 16 | 18 |
| False negative (FN) | 7 | 5 | 3 |
| True negative (TN) | 103 | 101 | 96 |
| False positive (FP) | 7 | 9 | 14 |
| Sensitivity | 0.67 | 0.76 | 0.86 |
| Specificity | 0.94 | 0.92 | 0.87 |
| Diagnostic accuracy | 0.89 | 0.89 | 0.87 |
| AUC | 0.85 | 0.91 | 0.87 |
Feature subset 1 contains the three texture features (Horzl_RLNonUni, S(2,2)SumOfSqs, and WavEnHL_s-3), feature subset 2 contains the three shape features (GeoFv, GeoW4, and GeoW5b), and feature subset 3 contains all six features.
Discussion
Although the majority of primary tumors theoretically originate from a single transformed cell and proliferate via clonal expansion, various degrees of intratumoral heterogeneity are often observed. Intratumoral heterogeneity is thought to reflect differences in gene expression, motility, metabolism, proliferation, angiogenesis and other biological characteristics [26], [27]. Modern imaging techniques, including traditional MRI, offer an ideal approach for detecting such heterogeneity in a quick, direct, and noninvasive manner. Several studies have sought to determine the correlation between meningioma heterogeneity and histopathological grade [6], [7], [8], [28]. Results of these studies are generally consistent with the view that heterogeneity is a characteristic feature of high-grade meningiomas.
The finding that tumor shape is an indicator of malignancy is controversial. Some studies have reported that irregular shape is significantly correlated with meningioma aggressiveness. For example, after reviewing the radiological features of seven malignant meningiomas, New et al. found that mushrooming shape was the most useful indicator of malignancy [29]. In another study by Nakasu et al., one hundred and one patients with meningioma were enrolled. These patients underwent surgery and were followed up for at least five years or until tumor recurrence. Fifteen meningiomas recurred during follow-up. Assessment of preoperative radiological examinations revealed that tumor shape was the only significant predictive factor for recurrence in univariate and multivariate analysis. Compared with regular-shaped meningiomas, lobulated and mushrooming ones were more likely to recur [30]. However, these findings are not supported by other studies. For instance, in a study by Kawahara et al., irregular shape increased the likelihood of high-grade meningioma in univariate analysis, but this correlation was not significant in multivariate analysis [7].
We also assessed several classical radiological features using the same data as in the present study. According to our results, three radiological features were significantly different between high-grade and low-grade meningiomas: tumor heterogeneity, tumor shape, and TBI. Heterogeneous enhancement, irregular shape, and unclear TBI were predictive factors for high-grade meningioma. The corresponding odds ratios (ORs) were 5.63 (95% CI 1.25–25.41, P = .0247), 6.32 (95% CI 1.47–27.14, P = .0132), and 3.82 (95% CI 0.97–15.12, P = .0559) (data not published).
One limitation of the above studies is that all radiological features were assessed subjectively. Thus, the analysis could have been influenced by factors such as intra-observer and inter-observer variability. Objective methods, in contrast, can provide quantitative evaluation and are more desirable for clinical application. We searched PubMed and Google scholar, and found only a few studies that had attempted to investigate the value of objective methods for the preoperative determination of meningioma grade. In a study by Schob et al., 66 meningioma patients were included, 10 with high-grade and 56 with low-grade tumors. The authors segmented the tumors and objectively measured their signal intensities in T1WI, FLAIR, and CE-T1WI, and no significant differences were found between the two groups [31]. In another study by Czyz et al., a method called fractal analysis was used for objective assessment. The average fractal dimension (FDa) and maximum fractal dimension (FDm) were found to be significantly higher in high-grade meningiomas, indicating that fractal analysis was potentially useful for meningioma grading [32].
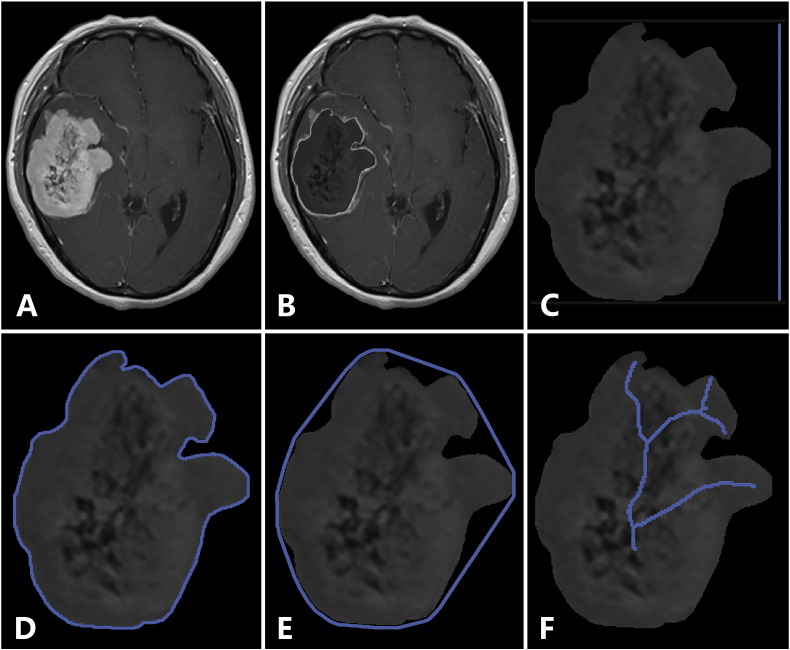
In the present study, we used texture and shape analysis to quantitatively evaluate tumor heterogeneity and shape. The three texture features obtained were Horzl_RLNonUni, S(2,2)SumOfSqs, and WavEnHL_s-3. Horzl_RLNonUni was a run-length matrix-based parameter, being one form of “run length nonuniformity” with θ being 0°; S(2,2)SumOfSqs was a co-occurrence matrix-based parameter, being one form of “sum of squares” with θ being 45°and d being 2; and WavEnHL_s-3 was a wavelet parameter. As the three features belonged to three different texture categories, they could be considered to reflect different aspects of tumor heterogeneity. The three shape features obtained were GeoFv, GeoW4, and GeoW5b. GeoFv was the vertical Feret's diameter; GeoW4 = GeoU1/GeoUw, where GeoU1 was the profile specific perimeter and GeoUw was the convex perimeter; and GeoW5b = GeoLsz/GeoF, where GeoLsz was the skeleton length and GeoF was the area (i.e., number of pixels inside the tumor; Figure 4). According to the definitions of these shape features, they were not closely correlated with each other and hence could be considered to reflect different aspects of tumor morphology.
Figure 4.
Illustration of the selected shape features. Original image (A), tumor segmentation (B), the vertical Feret's diameter (C), the profile specific perimeter (D), the convex perimeter (E), and the skeleton length (F).
The main goal of our study was to grade meningiomas based on preoperative MRI. As each of the above six features contained specific information regarding tumor heterogeneity or morphology, it was reasonable to expect that combining these features would result in better classification performance. We built nine classification models by training three machine learning classifiers with these features. As expected, models built with all six features (feature subset 3) performed better than those built with fewer features. In addition, the SVM classifier seemed superior to the LR and NB classifiers, as it provided balanced performance in terms of sensitivity, specificity, diagnostic accuracy, and AUC. This finding is consistent with some previous reports demonstrating that SVM classifiers performed well on medical classification problems [33], [34], [35]. As there is readily available software to carry out texture/shape analysis and build classification models (such as MaZda, TexRad, Weka, Orange, and scikit-learn), it would not be difficult for clinicians to carry out such analyses in clinical settings.
This study has several limitations. First, compared with the low-grade cases, the number of high-grade cases was small (21 vs. 110). This data imbalance is similar to previous related studies, and the primary explanation appears to be the relatively low incidence of high-grade meningioma [6], [7], [8], [31]. Second, some degree of selection bias may exist. We excluded patients whose preoperative MR images were not available in PACS, and these excluded tumors could have unique heterogeneity and morphology properties that would have influenced the results of our analysis. Third, a manual approach was adopted to segment tumors in this study. Although manual segmentation generally works better than automatic methods and two investigators participated in the segmentation process, segmentation errors could still exist. Fourth, as mentioned before, the MR images in this study were obtained with two different machines. This may have influenced image quality and subsequent analysis. However, according to a previous study by Herlidou-Meme et al., texture analysis based on different MRI systems was highly reproducible. In addition, we applied a normalization technique before image analysis to reduce the effect of inter-scanner differences. Therefore, the influence of this factor should have been small.
Conclusions
In this study, we investigated the diagnostic value of preoperative MRI texture and shape analysis for meningioma grading. Three texture features (Horzl_RLNonUni, S(2,2)SumOfSqs, and WavEnHL_s-3) and three shape features (GeoFv, GeoW4, and GeoW5b) were shown to be helpful in the determination of meningioma grade. Classification models built with these features provided generally satisfactory results. Our findings suggest that texture and shape analysis is potentially a useful tool for preoperative differentiation of high-grade and low-grade meningiomas in clinical practice.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interest
None.
Acknowledgements
None.
Contributor Information
Hong-Yang Zhao, Email: zhaohyang@yahoo.com.
Jun Feng, Email: fengjun.doc@gmail.com.
References
- 1.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro Oncol. 2016;18(Suppl. 5):v1–75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marosi C, Hassler M, Roessler K, Reni M, Sant M, Mazza E, Vecht C. Meningioma. Crit Rev Oncol Hematol. 2008;67(2):153–171. doi: 10.1016/j.critrevonc.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 4.McCarthy BJ, Davis FG, Freels S, Surawicz TS, Damek DM, Grutsch J, Menck HR, Laws ER., Jr. Factors associated with survival in patients with meningioma. J Neurosurg. 1998;88(5):831–839. doi: 10.3171/jns.1998.88.5.0831. [DOI] [PubMed] [Google Scholar]
- 5.Hashiba T, Hashimoto N, Maruno M, Izumoto S, Suzuki T, Kagawa N, Yoshimine T. Scoring radiologic characteristics to predict proliferative potential in meningiomas. Brain Tumor Pathol. 2006;23(1):49–54. doi: 10.1007/s10014-006-0199-4. [DOI] [PubMed] [Google Scholar]
- 6.Hsu CC, Pai CY, Kao HW, Hsueh CJ, Hsu WL, Lo CP. Do aggressive imaging features correlate with advanced histopathological grade in meningiomas? J Clin Neurosci. 2010;17(5):584–587. doi: 10.1016/j.jocn.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Kawahara Y, Nakada M, Hayashi Y, Kai Y, Hayashi Y, Uchiyama N, Nakamura H, Kuratsu JI, Hamada JI. Prediction of high-grade meningioma by preoperative MRI assessment. J Neurooncol. 2012;108(1):147–152. doi: 10.1007/s11060-012-0809-4. [DOI] [PubMed] [Google Scholar]
- 8.Lin BJ, Chou KN, Kao HW, Lin C, Tsai WC, Feng SW, Lee MS, Hueng DY. Correlation between magnetic resonance imaging grading and pathological grading in meningioma: Clinical article. J Neurosurg. 2014;121(5):1201–1208. doi: 10.3171/2014.7.JNS132359. [DOI] [PubMed] [Google Scholar]
- 9.Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;3(6):610–621. [Google Scholar]
- 10.Haralick RM. Statistical and structural approaches to texture. Proc IEEE. 1979;67(5):786–804. [Google Scholar]
- 11.Bayanati H, Thornhill RE, Souza CA, Sethi-Virmani V, Gupta A, Maziak D, Amjadi K, Dennie C. Quantitative CT texture and shape analysis: Can it differentiate benign and malignant mediastinal lymph nodes in patients with primary lung cancer? Eur Radiol. 2015;25(2):480–487. doi: 10.1007/s00330-014-3420-6. [DOI] [PubMed] [Google Scholar]
- 12.Hodgdon T, McInnes MD, Schieda N, Flood TA, Lamb L, Thornhill RE. Can quantitative CT texture analysis be used to differentiate fat-poor renal angiomyolipoma from renal cell carcinoma on unenhanced CT images? Radiology. 2015;276(3):787–796. doi: 10.1148/radiol.2015142215. [DOI] [PubMed] [Google Scholar]
- 13.Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol. 2016;85(4):824–829. doi: 10.1016/j.ejrad.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Sogawa K, Nodera H, Takamatsu N, Mori A, Yamazaki H, Shimatani Y, Izumi Y, Kaji R. Neurogenic and myogenic diseases: Quantitative texture analysis of muscle US data for differentiation. Radiology. 2017;160826 doi: 10.1148/radiol.2016160826. [DOI] [PubMed] [Google Scholar]
- 15.Waugh SA, Purdie CA, Jordan LB, Vinnicombe S, Lerski RA, Martin P, Thompson AM. Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol. 2016;26(2):322–330. doi: 10.1007/s00330-015-3845-6. [DOI] [PubMed] [Google Scholar]
- 16.Zacharaki EI, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem ER, Davatzikos C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med. 2009;62(6):1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Strzelecki M, Szczypinski P, Materka A, Klepaczko A. A software tool for automatic classification and segmentation of 2D/3D medical images. Nucl Instrum Methods Phys Res, Sect A. 2013;702:137–140. [Google Scholar]
- 19.Szczypiński PM, Strzelecki M, Materka A, Klepaczko A. MaZda—a software package for image texture analysis. Comput Methods Programs Biomed. 2009;94(1):66–76. doi: 10.1016/j.cmpb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging. 2004;22:81–91. doi: 10.1016/j.mri.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3(Mar):1157–1182. [Google Scholar]
- 22.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsl. 2009;11(1):10–18. [Google Scholar]
- 23.Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat. 1947;1:50–60. [Google Scholar]
- 24.Fawcett T. An introduction to ROC analysis. Pattern Recogn lett. 2006;27(8):861–874. [Google Scholar]
- 25.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. InIjcai. 1995;14(2):1137–1145. [Google Scholar]
- 26.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietemann JL, Heldt N, Burguet JL, Medjek L, Maitrot D, Wackenheim A. CT findings in malignant meningiomas. Neuroradiology. 1982;23(4):207–209. doi: 10.1007/BF00342542. [DOI] [PubMed] [Google Scholar]
- 29.New PF, Hesselink JR, O'Carroll CP, Kleinman GM. Malignant meningiomas: CT and histologic criteria, including a new CT sign. Am J Neuroradiol. 1982;3(3):267–276. [PMC free article] [PubMed] [Google Scholar]
- 30.Nakasu S, Nakasu Y, Nakajima M, Matsuda M, Handa J. Preoperative identification of meningiomas that are highly likely to recur. J Neurosurg. 1999;90(3):455–462. doi: 10.3171/jns.1999.90.3.0455. [DOI] [PubMed] [Google Scholar]
- 31.Schob S, Frydrychowicz C, Gawlitza M, Bure L, Preuß M, Hoffmann KT, Surov A. Signal Intensities in Preoperative MRI Do Not Reflect Proliferative Activity in Meningioma. Transl Oncol. 2016;9(4):274–279. doi: 10.1016/j.tranon.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czyz M, Radwan H, Li JY, Filippi CG, Tykocki T, Schulder M. Fractal Analysis May Improve the Preoperative Identification of Atypical Meningiomas. Neurosurgery. 2017;80(2):300–308. doi: 10.1093/neuros/nyw030. [DOI] [PubMed] [Google Scholar]
- 33.Emblem KE, Zoellner FG, Tennoe B, Nedregaard B, Nome T, Due-Tonnessen P, Hald JK, Scheie D, Bjornerud A. Predictive modeling in glioma grading from MR perfusion images using support vector machines. Magn Reson Med. 2008;60(4):945–952. doi: 10.1002/mrm.21736. [DOI] [PubMed] [Google Scholar]
- 34.Juntu J, Sijbers J, De Backer S, Rajan J, Van Dyck D. Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Imaging. 2010;31(3):680–689. doi: 10.1002/jmri.22095. [DOI] [PubMed] [Google Scholar]
- 35.Larroza A, Moratal D, Paredes-Sánchez A, Soria-Olivas E, Chust ML, Arribas LA, Arana E. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J Magn Reson Imaging. 2015;42(5):1362–1368. doi: 10.1002/jmri.24913. [DOI] [PubMed] [Google Scholar]