Abstract
The clinical interpretation of genetic variants has come to rely heavily on reference population databases such as the Exome Aggregation Consortium (ExAC) database. Pathogenic variants in genes associated with severe, pediatric-onset, highly penetrant, autosomal dominant conditions are assumed to be absent or rare in these databases. Exome sequencing of a six-year-old female patient with seizures, developmental delay, dysmorphic features and failure to thrive identified an ASXL1 variant previously reported as causative of Bohring-Opitz syndrome (BOS). Surprisingly, the variant was observed seven times in the ExAC database, presumably in individuals without BOS. Although the BOS phenotype fit, the presence of the variant in reference population databases introduced ambiguity in result interpretation. Review of the literature revealed that acquired somatic mosaicism of ASXL1 variants (including pathogenic variants) during hematopoietic clonal expansion can occur with aging in healthy individuals. We examined all ASXL1 truncating variants in the ExAC database and determined most are likely somatic. Failure to consider somatic mosaicism may lead to the inaccurate assumption that conditions like Bohring-Opitz syndrome have reduced penetrance, or the misclassification of potentially pathogenic variants.
Keywords: Bohring-Opitz syndrome, ASXL1, Tatton-Brown-Rahman syndrome, DNMT3A, somatic mosaicism, variant interpretation, clonal hematopoiesis of indeterminate potential, Exome Aggregation Consortium
The pace of discovery of genes underlying inherited conditions has accelerated due to improved access to next-generation sequencing technology. Unfortunately, in some cases misattribution of disease-association to genetic variants occurred when a control set of unaffected individuals were undersampled (Andreasen et al. 2013). The advent of large, openly accessible population databases has helped address this issue (MacArthur et al. 2014). Guidelines for variant assessment incorporate population frequency data; absence from population controls is considered moderate evidence for pathogenicity. If the population frequency of a variant vastly exceeds the incidence of the associated condition, the variant would typically be considered benign, or at least only weakly penetrant (Richards et al. 2015).
Bohring-Opitz syndrome (BOS) (MIM# 605039) is a rare, autosomal dominant syndrome, characterized by severe intrauterine growth restriction, poor feeding, hypotonia, intellectual disability with associated MRI findings, hirsuitism, dysmorphism, and nevus flammeus of the face (Hoischen et al. 2011; Magini et al. 2012; Dangiolo et al. 2015; Russell et al. 2015). BOS is caused by de novo truncating variants in the additional sex combs-like 1 (ASXL1) (MIM #612990) gene, which encodes a putative member of the Polycomb Repressive Complex (Hoischen et al. 2011).
The molecular mechanism of ASXL1 pathogenic variants in BOS is unknown, but the C-terminal PHD (zinc finger domain), implicated in mediating interactions with other proteins, may be lost when ASXL1 protein is truncated. This has been proposed to ablate the auto-inhibition of ASXL1, which is consistent with observed over-activation of the mutant ASXL1-BAF1 complex (Balasubramani et al. 2015). The DNA-binding domain at the N-terminus of ASXL1 is predicted to remain intact. The same pathogenic variants can be found in myeloid malignancies and in BOS (Hoischen et al. 2011; Pratcorona et al. 2012), and a similar pathogenic mechanism may apply in both. Several BOS-associated variants occur in the last exon of ASXL1 (Hoischen et al. 2011; Magini et al. 2012) and would not be predicted to trigger nonsense mediated decay. Asxl1 heterozygous null mice exhibit craniofacial dysmorphism with reduced penetrance (Abdel-Wahab et al. 2013), though reduced penetrance has never been observed with Bohring-Opitz syndrome in humans. The following analysis will refer to putative protein truncating variants as pathogenic when they are known to cause BOS or truncating if there is no data on pathogenicity.
The proband was the fifth child born at term via normal spontaneous vaginal delivery after an uncomplicated pregnancy to non-consanguineous parents. Initial feeding difficulties, hypotonia, and an unusual cry led to Cri-du-chat syndrome testing that was negative. By 1 month of age, she had her first tonic seizure, which evolved into tonic-clonic seizures by 2 years of age. Also by 2 years of age, she was G-tube dependent and found to have a nearly absent corpus callosum by imaging. A multi-institution, extensive workup, including genetic (e.g., SNP-microarray; methylation studies; FOXG1, MECP2, UBE3A, PNPO, ALDH7A1 sequencing; and SMN1 dosage) and metabolic (e.g., lysosomal enzymes, very long-chain fatty acids, transferrin isoelectric focusing, and CSF neurotransmitters) studies, was inconclusive.
Our initial exam at 6 years of age identified a small child with length, weight, and head circumference below the first percentile. She had bilateral ptosis, optics for her myopia, a faint nevus flammeus of the face, protuberant cheeks, high palate with a narrow jaw and bruxism, tented lips and an open mouth, diffuse hirsutism including mild synophrys, and mild fat pads on her metacarpophalangeal joints (Figure 1A). She was non-verbal and her eyes did not track light but rather roamed continuously. She was able to reach and grab objects and bring them to her mouth, but was unable to sit independently. Flexion of the elbows and ulnar deviation and flexion of the wrists and metacarpophalangeal joints was not present.
Figure 1.
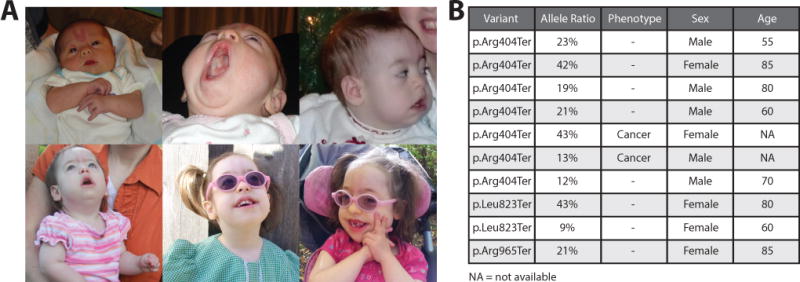
Bohring-Opitz syndrome (BOS) patient and BOS-associated variants in the Exome Aggregation Consortium (ExAC) browser. A) From top left: Patient as a newborn with a prominent nevus flammeus, at two months demonstrating her high palate, at one year with protuberant cheeks, at two years tented upper lip can be observed, at three and a half years wearing glasses for myopia, at age six when patient most recently presented in genetics clinic. Note the progressive fading of the nevus flammeus with age. B) Table with BOS-associated variants p.Arg404Ter (observed in our patient as well as Hoischen et al. 2011), p.Leu823Ter (Hoischen et al. 2011), and p.Arg965Ter (Magini et al. 2012). Allele ratio (allele balance or alternate allele frequency), phenotype (if the patient was part of a cancer cohort), sex, and age (rounded to the nearest five year mark) for each individual harboring the BOS-associated variants are listed.
Exome sequencing performed on the patient, both parents, and two unaffected siblings, identified a de novo variant, c.1210C>T; p.Arg404Ter, in the ASXL1 gene (NM_015338.5) (see Supp. Methods). When examined in IGV, the ASXL1 variant had high-quality reads inconsistent with a sequencing artifact, which was further supported by Sanger sequencing (Supp. Figure S1). This variant has been previously reported in BOS (Hoischen et al. 2011) and is listed in the dbSNP database (rs373145711) as pathogenic referencing the ClinVar entry (variant ID 30986). However, this variant was also reported in the Exome Aggregation Consortium (ExAC) browser (7/121378 chromosomes, with no homozygotes) and in the Exome Variant Server (2/13006 chromosomes, with no homozygotes) (Tennessen et al. 2012) (one of these samples is present in both datasets).
The presentation of the patient closely resembles BOS and the p.Arg404Ter variant had been reported in another BOS patient (Hoischen et al. 2011). Furthermore, all reported BOS patients (including this patient) that have de novo ASXL1 truncating variants are severely affected, and decreased penetrance or variable expressivity has not been described in BOS. ExAC individuals with the p.Arg404Ter variant are not part of a psychiatric subset of ExAC, so they are unlikely to have a mild BOS presentation. Therefore, the presence of this variant in these reference population databases was incongruous with the expectation of pathogenicity for this variant.
We explored the possibility that the observation of the ASXL1 p.Arg404Ter variant in a population database might be the result of somatic mosaicism. Hematopoietic clonal expansion of cells (clonal hematopoiesis of indeterminate potential or CHIP) with ASXL1 pathogenic variants has been reported as a frequent event acquired with aging and is present in roughly 10% of individuals over age 65 and nearly 20% of those over age 90 (Genovese et al. 2014; Jaiswal et al. 2014; Xie et al. 2014). The most commonly mutated genes in CHIP are DNMT3A (MIM# 602769), TET2 (MIM# 612839), and ASXL1; all three encode epigenetic modifiers.
Germline, de novo ASXL1 variants have been convincingly shown to cause BOS (Hoischen et al. 2011). It is important to distinguish between germline and somatic ASXL1 variants, since the later occurs with CHIP in healthy individuals. The gold standard to prove somatic mosaicism is to test DNA derived from other tissues such as buccal swabs or hair follicles, but due to the need to protect the anonymity of those who contribute their genetic information to reference databases further testing of these individuals is generally not possible. However, manual examination of the read support for seven individuals in ExAC who share our patient’s p.Arg404Ter ASXL1 variant revealed that five of the seven demonstrate considerable allelic imbalance (fewer than 35% of sequencing reads derived from the variant allele) (Figure 1B). Of the two individuals with a variant allele balance close to 50%, one was an individual with cancer (variant present in 43% of reads) and the other was an 85-year-old woman (variant present in 42% of reads), consistent with the known association of ASXL1 truncating variants with cancer risk and aging. Overall, two of the seven individuals carrying the p.Arg404Ter variant reportedly have cancer, none are from psychiatric cohorts, and the median rounded age of the five individuals from non-cancer cohorts is 70 (the youngest is 55 and the oldest 85). Though DNA from other tissues in these individuals is not available for testing, it is possible to infer that the incidence of the p.Arg404Ter ASXL1 variant in the ExAC database is likely due to somatic mosaicism.
The observation of the ASXL1 p.Arg404Ter variant in the ExAC database raised the possibility that other pathogenic ASXL1 variants might also be found in reference population databases. Another BOS-associated variant, p.Arg965Ter (Magini et al. 2012), was present in ExAC with an allele balance of 21% in an 85-year-old woman (Figure 1B). The BOS-associated p.Leu823Ter variant (Hoischen et al. 2011), though encoded by a different nucleotide change, was also observed in ExAC with an allele balance of 43% in an 80-year-old woman and 9% in a 60-year-old woman (Figure 1B). All putative truncating ASXL1 variants in the ExAC database were examined (see Supp. Methods and Supp. Table S1). A total of 342 individuals were carriers of 56 heterozygous, truncating ASXL1 variants. Four variants (observed in five individuals) were excluded because they were unlikely to cause ASXL1 truncation (p.Phe6fs and c.141-2A>G were associated with non-constitutive exons and two indels in close proximity (p.Ile1329fs and p.Pro1330fs) caused a Ile to Ser substitution rather than a frameshift (Supp. Figure S2)).
The two most common truncating variants, p.Gly646fs and p.Gly645fs, identified in 132 and 118 individuals respectively, were also excluded (Supp. Figure S2). These variants are located in an eight-nucleotide homopolymer tract of guanines (one variant is 7G and the other 9G) and potentially represent PCR artifacts. Intriguingly, deep sequencing of a large series of myeloid malignancies found the p.Gly646fs variant (confirmed in several cases by capillary electrophoresis or Sanger sequencing) to be the most common cancer-associated ASXL1 variant (identified in 47 out of 133 myeloid malignancies) (Van Ness et al. 2016). So far p.Gly645fs has not been observed even once in roughly 2000 myeloid malignancy panels and since polymerase slippage would be expected to create an equal number of 7G and 9G variants, Van Ness et al. conclude that the absence of p.Gly645fs variants suggests p.Gly646fs by comparison represents a genuine mutational hotspot. Our own analysis potentially supports this assessment, as p.Gly646fs (median allele balance 22%) was associated with TCGA samples in 66% of cases compared to 8% for p.Gly645fs (median allele balance 15%). However, despite acknowledging that a subset of the p.Gly646fs variants are likely genuine, without deeper coverage or additional confirmation (e.g. Sanger sequencing) it would be difficult to distinguish genuine variants from sequencing artifacts. For this reason we excluded all 250 individuals with either variant from further analysis.
The remaining 88 individuals (with 50 different truncating ASXL1 variants) were investigated to determine the relative balance of variant alleles. The median variant allele balance was 25% (range 9–59%, interquartile range (IQR) 18–35%). ASXL1 variants may occur more frequently in individuals with cancer, either because they confer cancer susceptibility or arise in response to mutagenic cancer treatments such as chemotherapy or radiation. Therefore, we separated the 23 individuals from the TCGA cohort and the 65 individuals from non-cancer cohorts (across multiple ancestries in ExAC) and compared the variant allele balance in both groups. The aim of ExAC was to include germline samples, so individuals with hematological cancers were excluded; however, increased rates of hematopoietic mosaicism involving variants in ASXL1 is also seen in individuals with other cancer types (Artomov et al. 2016). The variant allele balance was not significantly different between the cancer (median 31%) and the non-cancer (median 22%) cohorts (Mann-Whitney U Test, p=0.18).
To ensure that the skewed variant allele balance was not related to sequencing of ASXL1, we assessed two other classes of variants predicted to be functionally neutral. First, individuals with 30 rare synonymous ASXL1 variants showed no evidence of allelic imbalance with a median allele balance of 47% (range 15–61%, IQR 44–50%). Only three out of 70 individuals had an allele balance <35%. The difference between the allele balances for non-cancer-associated, putative truncating variants (median 22%) and for the rare synonymous variants (median 47%) is significant (Mann-Whitney U Test, p=1.02E-14) (Figure 2C). Second, we were concerned whether difficulty in aligning indels, which constitute 40% of the observed truncating ASXL1 variants, could contribute to an apparent decreased allele balance. However, there was no evidence of variant allele imbalance in 20 ASXL1 non-frameshift indels (in-frame indels and noncoding indels) identified in 32 individuals from non-cancer cohorts, with a median variant allele balance of 48% (range 29–65%, IQR 41–50%). For the non-cancer cohorts, the allele balances were significantly different between frameshift indels versus non-frameshift indels (Mann-Whitney U Test, p=1.92E-4). Additionally, allelic imbalance of nonsense variants was also observed, which would not be affected by alignment issues. Overall, allelic imbalance was not identified for rare ASXL1 variants that are unlikely to impact protein function.
Figure 2.
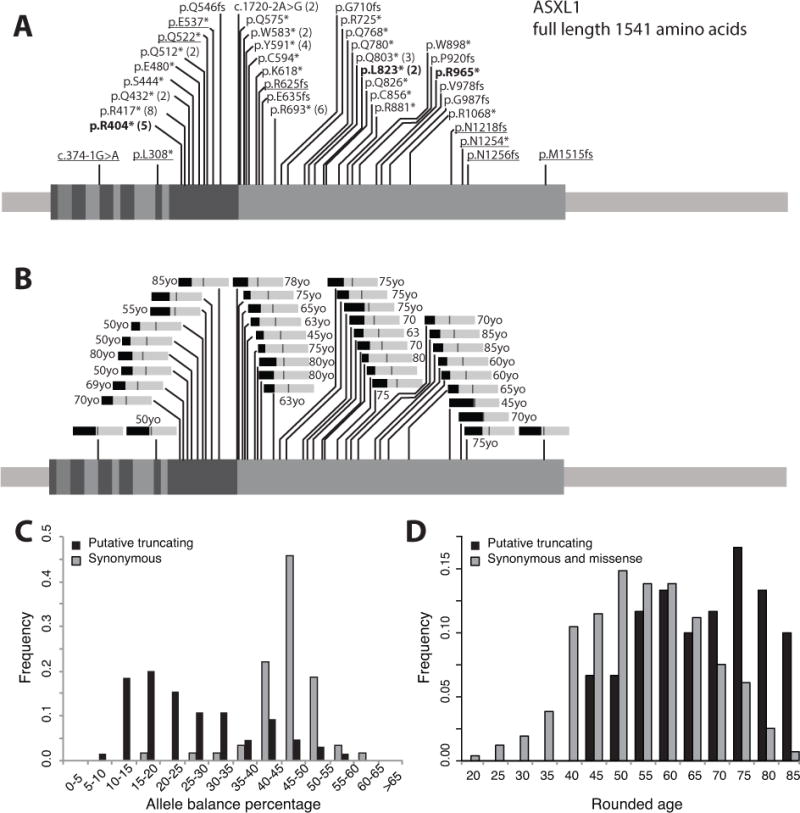
Likely somatic putative truncating variants in the Exome Aggregation Consortium (ExAC) browser excluding variants from cancer cohorts. A) 39 ASXL1 (exons indicated) putative truncating variants after filtering are observed in ExAC, with the number of individuals greater than one listed in parentheses (total = 65 individuals). The bolded variants have been observed in patients with Bohring-Opitz syndrome.Underlined variants are those with an average allele balance equal to or greater than 35%. B) Bar graphs of average allele balances for each variant (medium gray bar = 50%), with average rounded ages associated with each variant listed when available. C) Allele balances for filtered, rare ASXL1 variants. Black bars indicate individuals with putative truncating ASXL1 variants and grey bars indicate 30 randomly selected rare (<10 individuals) synonymous ASXL1 variants in ExAC. The median allele balance of putative truncating ASXL1 variants is 22% (n=65), while the median allele balance for those with rare synonymous ASXL1 variants is 47% (n=70). The difference is significant (Mann-Whitney U Test, p=1.02E-14). D) The median rounded age for individuals with putative truncating ASXL1 variants is 70 (n=60, no age data is available for five individuals), while the median rounded age for those with rare synonymous or missense ASXL1 variants is 55 (n=983). The difference is significant (Mann-Whitney U Test, p=3.98E-11). For the original data see Supp. Table S1.
Most ASXL1 putative truncating variants are found in the last exon (Figure 2A), which is also the largest exon and a location where truncating variants are likely to generate transcripts that escape nonsense mediated decay. Comparing the age distribution of the non-cancer cohort with putative truncating ASXL1 variants (median rounded age of 70 years old), to the age distribution of the non-cancer cohort with rare synonymous or missense ASXL1 variants (median rounded age of 55 years old) reveals a significant shift towards older individuals in the former group (Mann-Whitney U Test, p=3.98E-11) (Figure 2D).
In summary, by examining the data associated with 345 individuals cumulatively carrying 56 truncating ASXL1 variants in the ExAC database, we found most are unlikely to be germline variants. In a few cases, the variants themselves are suspect (i.e. dubious annotation or homopolymer variants), highlighting the importance of reviewing variant position on a genome browser. In other cases, allelic imbalance in a population that skews older (median age 70 compared to 55 for all ExAC individuals) indicated potential somatic mosaicism, emphasizing the need to examine variant read support on the ExAC browser. This may also yield direct evidence of somatic mosaicism, as is the case for p.Asn1256fs, a variant with an allele balance of 36% present in a single ExAC individual. This individual also has a common synonymous variant 5 nucleotides upstream of the rare frameshift variant. There were 109 reads covering both variants, 39% of which contained the frameshift deletion alone, 52% the synonymous variant alone, and 9% neither variant (Supp. Figure S2). The frameshift variant likely arose as a somatic mutation in trans to the synonymous variant.
Although ASXL1 provides an excellent illustration of how somatic mosaicism may confound the analysis of pathogenic variants, the issue is not unique to this gene. It is worth considering TET2 and DNMT3A, two other genes commonly mutated during clonal hematopoiesis (Genovese et al. 2014; Jaiswal et al. 2014; Xie et al. 2014). Germline TET2 variants are not associated with a known syndrome, however de novo DNMT3A variants cause autosomal dominant Tatton-Brown-Rahman syndrome (TBRS) (MIM# 615879), an overgrowth condition characterized by tall stature, macrocephaly, distinctive facial features, and intellectual disability (Tatton-Brown et al. 2014). Putative truncating variants in DNMT3A are present in ExAC and as with ASXL1 demonstrate signs of somatic mosaicism (data not shown). However, the majority of TBRS cases have been attributed to pathogenic missense variants (Tatton-Brown et al. 2014).
A DNMT3A missense variants associated with TBRS, p.Arg749Cys (Tatton-Brown et al. 2014), is reported in three individuals in ExAC. Based on allele balance (11%, 20%, and 24%), the presence of this pathogenic DNMT3A variant in ExAC can also likely be attributed to somatic mosaicism. The rounded ages of the two individuals with the 20% and 24% allele balance were 70, and the individual with the 11% allele balance was part of a cancer cohort for which age was not available. Another DNMT3A missense variant, p.Arg882His, has recently been associated with TBRS (Kosaki et al. 2017), with the germline nature of this variant confirmed via buccal samples. The arginine residue at position 882 represents the most prevalent hotspot for pathogenic DNMT3A variants in acute myeloid leukemia (Ley et al. 2010; Lu et al. 2016), and the p.Arg882His variant was detected in 66 individuals (55 from non-cancer cohorts), a frequency of more than 1 in 1,000 individuals in ExAC. The p.Arg882His variant also demonstrates allele imbalance (the median allele balance is 19% with an IQR of 8–30% for the 55 individuals from non-cancer cohorts), which is consistent with somatic mosaicism. Given the evidence required to consider a missense variant pathogenic (Richards et al. 2015), diagnoses of TBRS could be missed if less well-characterized DNMT3A missense variants are misclassified due to their presence in reference population databases.
Reference population databases such as ExAC are invaluable for characterizing the genetic diversity of human populations (MacArthur et al. 2014). These resources have enabled researchers and clinicians to better interpret variants as potentially pathogenic or benign based on allele frequency. However, these databases are known to contain individuals with pathogenic variants, including variants that cause recessive disorders, variants associated with incomplete penetrance, or variants associated with variable expressivity resulting in mildly affected individuals. Here, we present hematopoietic mosaicism as another caveat to consider in determining the pathogenicity of a variant based on its presence in a population database. When variants arise by somatic mutation, those that provide a growth advantage to hematopoietic cells can lead to clonal expansion resulting in these variants being present at a higher allele frequency than is typical for somatic variants. Such somatic variants with a relatively higher allele balance are more often mistakenly being called by variant calling algorithms that are designed to detect germline genetic variants.
The presence of any BOS-associated ASXL1 variant in the ExAC database is unexpected for a severe, pediatric-onset autosomal dominant condition that is assumed to be fully penetrant. Our analysis suggests that p.Arg404Ter is observed in individuals with allelic imbalance suggesting somatic mosaicism. Most of these individuals were elderly or from a cancer cohort (both associated with CHIP). Of course, without testing other tissues from the individuals in ExAC to prove somatic mosaicism it is not possible to rule out decreased penetrance or variable expressivity of BOS, and there have been reports of healthy individuals apparently resilient to highly penetrant, childhood-onset conditions (Chen et al. 2016). However, in light of our findings we believe the most parsimonious explanation for ASXL1 truncating variants in the ExAC database is hematopoietic somatic mosaicism.
Our results could extend to genes associated with other autosomal dominant disorders. A recent study questioned the penetrance of pathogenic variants for intellectual disability and related disorders, including ASXL1 (Ropers and Wienker 2015). The authors noted the difficulty of reconciling population database frequencies for pathogenic ASXL1 variants with inherited variants having never been observed in BOS. We propose that age-related somatic mosaicism, rather than incomplete penetrance, most likely accounts for the contradictory observations described by Ropers and Wieker. Awareness of potential somatic mosaicism is warranted when performing variant investigation for ASXL1, DNMT3A, and perhaps many other genes.
While our results suggest that somatic mosaicism is a likely reason for the observation of some pathogenic variants in non-syndromic individuals, we acknowledge the limitations of our study. Notably, we have presented evidence implying the presence of mosaicism but not proven it by testing other tissues. Also, we cannot discount the possibility that some of the variants excluded from our analysis (e.g. a subset of the p.Gly646fs variants) are genuine pathogenic variants. Similarly, we have not demonstrated that all the truncating variants included in our analysis are truly pathogenic variants, though the identification of two previously published BOS-associated variants suggests we are enriching for variants that impact ASXL1 function. Finally, we acknowledge that we could not explain all truncating ASXL1 variants observed in ExAC, particularly those with normal allele balance in younger individuals from non-cancer cohorts. However, by attributing many of the truncating ASXL1 variants to technological artifacts or somatic mosaicism, our analysis decreased the number of unexplained individuals with truncating ASXL1 variants in ExAC from 395 to approximately 16. We believe that this study helps address the issue of pathogenic variants observed in population databases (Ropers and Wienker 2015), and will aid variant assessment using the current American College of Medical Genetics guidelines (Richards et al. 2015).
This investigation provides important guidance for variant analysis. In certain cases, one cannot rely on the presence of a variant in reference population databases to indicate that the variant is unlikely to be pathogenic for a highly penetrant, pediatric-onset condition. Additional information regarding database individuals may need to be examined, such as evidence for potential allelic imbalance, age, and whether they belong to a disease cohort such as patients diagnosed with cancer. Special attention should be paid to genes susceptible to the accumulation of age-related somatic variants in hematopoietic lineages, particularly as this is the source of DNA in most clinical genetic testing. Although we identified this issue due to our understanding of the role of ASXL1 and DNMT3A in clonal hematopoiesis, this phenomenon might be overlooked in less well-characterized genes.
This work highlights the importance of certain features of the ExAC browser and suggests other areas where further annotation is needed. As truncating ASXL1 variants are associated with an increased risk of solid tumors (Artomov et al. 2016) the inclusion of exome data from TCGA may increase the prevalence of these variants. A non-TCGA ExAC data subset is available for download and may be more appropriate for some analyses. A reference database flag for CHIP-associated genes may also be worth considering. Additionally, in the last ExAC data release, aggregated age data is available for each variant. Examination of the raw sequence read data available on the ExAC browser is an important step of variant review, to identify any issues such as a complex indel or skewed allele balance. Further work is needed to identify systematic deviations in allele balance in reference population databases, and it is a future goal to flag such variants in the next version of ExAC. Clinical laboratories and clinicians tasked with variant interpretation should remain vigilant regarding somatic variants in all current databases.
The increasing volume of next-generation sequencing tests is already taxing a system that still largely relies on expert curation. Bioinformatics pipelines that filter out presumed benign variants are useful, but as our results have shown, over-dependence on variant frequencies in reference databases is dangerous, and establishing hard frequency thresholds for filtering will require careful consideration of complicating issues such as somatic mosaicism. As the trend toward automation continues it will be important not only to refine the rules of variant assessment, but also to recognize when the unique biology of certain genes may create important exceptions to those rules.
Supplementary Material
Acknowledgments
We are grateful to the family for their willingness to share their daughter’s case with the medical community. We also thank the clinical genomics laboratory at ARUP for performing exome sequencing. We would like to thank the members of the Exome Aggregation Consortium for creating this resource. We appreciate the efforts of MacArthur lab members, particularly Monkol Lek, Konrad Karczewski, and Kaitlin Samocha for the development of resources for analysis of the ExAC dataset, several of which were very helpful in this work. We would also like to acknowledge John Carey, Oliver Tam, Wei Shen, Philippe Szankasi, Michael Van Ness, Tim Tidwell, and Alex Chapin for helpful discussions in Utah. C.M.C. is supported by The University of Utah Department of Pathology and ARUP Laboratories and A.H.O.-L. is supported by a Pfizer/ACMG Foundation Translational Genomic Fellowship. ExAC analysis supported by NIH grants NIGMS R01 GM104371 and NIDDK U54 DK105566.
References
- Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, Lafave LM, Koche R, Shih AH, Guryanova OA, Kim E, Li S, Pandey S, Shin JY, Telis L, Liu J, Bhatt PK, Monette S, Zhao X, Mason CE, Park CY, Bernstein BE, Aifantis I, Levine RL. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen C, Nielsen JB, Refsgaard L, Holst AG, Christensen AH, Andreasen L, Sajadieh A, Haunsø S, Svendsen JH, Olesen MS. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet. 2013;21:918–928. doi: 10.1038/ejhg.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artomov M, Rivas MA, Genovese G, Daly MJ. Mosaic mutations in blood DNA sequence are associated with solid tumor cancers. bioRxiv. doi: 10.1101/065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramani A, Larjo A, Bassein JA, Chang X, Hastie RB, Togher SM, Lähdesmäki H, Rao A. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun. 2015;6:7307. doi: 10.1038/ncomms8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, Zhou H, Tian L, Prakash O, Lemire M, Sleiman P, Cheng W-Y, Chen W, Shah H, Shen Y, Fromer M, Omberg L, Deardorff MA, Zackai E, Bobe JR, Levin E, Hudson TJ, Groop L, Wang J, Hakonarson H, Wojcicki A, Diaz GA, Edelmann L, Schadt EE, Friend SH. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- Dangiolo SB, Wilson A, Jobanputra V, Anyane-Yeboa K. Bohring-Opitz syndrome (BOS) with a new ASXL1 pathogenic variant: Review of the most prevalent molecular and phenotypic features of the syndrome. Am J Med Genet A. 2015;167:3161–3166. doi: 10.1002/ajmg.a.37342. [DOI] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landén M, Höglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Grönberg H, Hultman CM, McCarroll SA. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, van Bon BWM, Rodríguez-Santiago B, Gilissen C, Vissers LELM, de Vries P, Janssen I, van Lier B, Hastings R, Smithson SF, Newbury-Ecob R, Kjaergaard S, Goodship J, McGowan R, Bartholdi D, Rauch A, Peippo M, Cobben JM, Wieczorek D, Gillessen-Kaesbach G, Veltman JA, Brunner HG, de Vries BBBA. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat Genet. 2011;43:729–731. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki R, Terashima H, Kubota M, Kosaki K. Acute myeloid leukemia-associated DNMT3A p.Arg882His mutation in a patient with Tatton-Brown-Rahman overgrowth syndrome as a constitutional mutation. Am J Med Genet A. 2017;173:250–253. doi: 10.1002/ajmg.a.37995. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, Dipersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang P, Parton T, Zhou Y, Chrysovergis K, Rockowitz S, Chen W-Y, Abdel-Wahab O, Wade PA, Zheng D, Wang GG. Epigenetic Perturbations by Arg882-Mutated DNMT3A Potentiate Aberrant Stem Cell Gene-Expression Program and Acute Leukemia Development. Cancer Cell. 2016;30:92–107. doi: 10.1016/j.ccell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magini P, Monica Della M, Uzielli MLG, Mongelli P, Scarselli G, Gambineri E, Scarano G, Seri M. Two novel patients with Bohring-Opitz syndrome caused by de novo ASXL1 mutations. Am J Med Genet A. 2012;158A:917–921. doi: 10.1002/ajmg.a.35265. [DOI] [PubMed] [Google Scholar]
- Pratcorona M, Abbas S, Sanders MA, Koenders JE, Kavelaars FG, Erpelinck-Verschueren CAJ, Zeilemakers A, Löwenberg B, Valk PJM. Acquired mutations in ASXL1 in acute myeloid leukemia: prevalence and prognostic value. Haematologica. 2012;97:388–392. doi: 10.3324/haematol.2011.051532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee . Genetics in medicine : official journal of the American College of Medical Genetics. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH, Wienker T. Penetrance of pathogenic mutations in haploinsufficient genes for intellectual disability and related disorders. Eur J Med Genet. 2015;58:715–718. doi: 10.1016/j.ejmg.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Russell B, Johnston JJ, Biesecker LG, Kramer N, Pickart A, Rhead W, Tan W-H, Brownstein CA, Kate Clarkson L, Dobson A, Rosenberg AZ, Vergano SAS, Helm BM, Harrison RE, Graham JM. Clinical management of patients with ASXL1 mutations and Bohring-Opitz syndrome, emphasizing the need for Wilms tumor surveillance. Am J Med Genet A. 2015;167A:2122–2131. doi: 10.1002/ajmg.a.37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, Zachariou A, Hanks S, O’Brien E, Aksglaede L, Baralle D, Dabir T, Gener B, Goudie D, Homfray T, Kumar A, Pilz DT, Selicorni A, Temple IK, Van Maldergem L, Yachelevich N, Childhood Overgrowth Consortium. van Montfort R, Rahman N. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO, NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness M, Szankasi P, Frizzell K, Shen W, Kelley TW. Analysis of ASXL1 Mutations in a Large Series of Myeloid Malignancies. Mod Pathol 29 Suppl. 2016;2:333–388. [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, Ozenberger BA, Welch JS, Link DC, Walter MJ, Mardis ER, Dipersio JF, Chen F, Wilson RK, Ley TJ, Ding L. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


