Abstract
The current mandate for the drug discovery industry is to develop more efficient drugs faster while reducing the costs associated with their development. Incorporation of cell stimulation technologies during screening assays is expected to revolutionize the discovery of novel drugs as well as safety pharmacology. In this review, we highlight ‘classical’ and emerging cell stimulation technologies that provide the ability to evaluate the effects of drug candidates on cells in different functional states to assess clinically relevant phenotypes.
Keywords: drug discovery, drug screening, phenotypic screening, use-dependence, optical stimulation, electrical stimulation, optogenetic stimulation, optoelectrical stimulation, infrared stimulation, all-optical assays
Introduction
The pharmaceutical industry is facing immense pressure to develop better drugs in a less costly way. So far, drug discovery companies are losing this war. The current development pipeline is lengthy and inefficient, with the approval process alone taking on average 7–10 years. The cost of drug development has increased from US$1 billion to US$2.6 billion over the past 10 years [1], taking into account drugs that failed to gain the approval of the US Food and Drug Administration Agency (FDA) and, thus, to reach the market and patients. The cost threatens to increase geometrically in a trend termed ‘Eroom’s law’, which is the inverse of the well-known gains in efficiency enjoyed by the computer industry. Ultimately, developing tools for improved failure prediction of a drug during earlier stages of the development process is necessary to reverse current trends.
Part of the problem stems from the medical need for a diverse array of new drugs to treat a broad spectrum of disorders. Arguably, the ‘low-hanging fruit’ have been largely harvested, and the challenge now is to develop new drugs with more complex mechanisms of action (MOAs) and activity profiles. To identify such drugs, and to avoid costly failures, companies must be armed with a matching set of enabling methods and assays. Given that the common entry point in the discovery pipeline is a screen of 1 000 000+ compounds, special emphasis has been put on the automation, miniaturization, advanced robotics, ultra-high-density microplates, and computer hardware for data analysis. Although these advances in throughput have been impressive, drug discovery still remains time-consuming and costly because of the loss of critical information during high-throughput screening (HTS) assays. Their information content can be increased by incorporating cell stimulation technologies into earlier stages of drug discovery, which is expected to improve its efficiency, and lead to more promising drug candidates with a wider range of MOAs than was previously possible.
In in vivo settings, all living cells respond dynamically to environmental stimuli presented by changing their functional states. Effects of drugs on cells can vary as a function of their functional state. However, in in vitro settings, HTS assays acquire the data either from one functional state or an ensemble average of heterogeneous states, which is misleading or masks important dynamics of the response of a cell. Under these circumstances, drugs are being tested on randomly present rather than specifically defined functional states of targets of interest. As a result, some physiologically relevant drug effects can be missed. For instance, state dependence, an important property of many ion channel inhibitors, can be detected only if drugs are applied on ion channels in different states.
Phenotypic drug discovery is expected to benefit from cell stimulation technologies to an even higher degree than target-based drug discovery, because it uses fewer assumptions regarding underlying molecular mechanisms [2] and, thus, needs technologies to cycle cells through all possible functional states in the presence of drugs. Phenotypic screening already provides means for increasing assay predictiveness by targeting complex human cell behavior and exploring disease-relevant activities in live cells [2–4]. The leading edge of phenotypic drug discovery is the direct assessment of dynamic physiology of excitable cells, and especially human induced pluripotent stem cell (iPSC)-derived cells. Production of cardiomyocyte (CM), muscle, neuron, endocrine, and other cell types is now routine, and can yield cellular models of congenital disease [5–7]. Combining cell stimulation technologies with iPSC culture models of disease creates an unprecedented opportunity to probe new target space and novel MOAs applicable to the treatment of human disease.
An example of the challenges associated with detecting complex physiological effects is represented by the need to predict potentially lethal proarrhythmic effects of novel drugs as drug-induced long QT, and the potentially fatal polymorphic ventricular tachycardia ‘torsades de pointes’ (TdP) remains a major concern [8]. In 2013, the FDA convened a consortium of academics, governmental regulators, and industry practitioners that concluded that there is a significant need for more predictive pro-arrhythmia risk tests and suggested a new paradigm, the Comprehensive In Vitro Proarrhythmia Assay (CiPA) [9]. Given that the cell-based in vitro models of CMs are integrated into the CiPA testing, cell stimulation will be critical to effectively evaluate proarrhythmic proclivity of drug candidates in moderate throughput to augment single cardiac current (e.g., hERG potassium current) analyses. Therefore, enabling modulation of the cell membrane potential during HTS will enable researchers to develop better predictive and physiologically relevant assays that will be able to effectively identify novel drugs candidates with complex MOAs and good safety profiles.
Electrophysiological and optical assays that comprise functional drug screening [10,11] dramatically differ in their cell stimulation capabilities.
By design, electrophysiology already has technical capabilities to control the cell membrane potential and, thus, can stimulate cells with a high degree of precision. Given that electrophysiology also enables the simultaneous monitoring of cellular responses at a sub-millisecond timescale, it is no wonder that this method is considered the gold standard technology. To make this intricate technology screening friendly, several planar patch-clamp instruments have been developed over the last 15 years [11,12]. The highest throughput (simultaneous acquisition from 384 wells) was achieved in IonWorks Barracuda (Molecular Devices), SyncroPatch 384PE (Nanion), and Qube (Sophion). However, given the inherent limitations of miniaturization of electrical amplifiers, planar patch-clamp instruments are still not truly suitable for systematic tests or screening of 1 000 000-compound libraries. Moreover, several critical electrophysiology problems remain unresolved: (i) responses from only a small percentage of a cell population can be probed, and these tested cells might not accurately represent the whole population because of the inevitable selection of more resilient cells that can withstand patch clamping; (ii) automated electrophysiology does not work well on primary cells (e.g., neurons, or CMs)] because of their complex geometry, electrical connectivity, or contractions; (iii) there is a limited acquisition time because of the violation of the structural integrity of cells that is required for intracellular access during patch-clamping; and (iv) the cost of operation is high.
Optical assays have significant advantages over electrophysiology both in terms of throughput and extended time of data acquisition [13]. To evaluate cellular functional activity, these assays utilize a range of fluorescent sensors, either organic dyes or genetically encoded fluorescent indicators. In the HTS mode, an imaging instrument acquires one image from the whole plate (all wells at once). Such plate readers have significantly reduced optical sensitivity because only a few pixels are assigned to a single well. Information about individual cells is never collected, which works well for homogeneous cell populations. In the high-content screening (HCS) mode, an imaging instrument acquires data on individual cells within the field of view. The information gained at single-cell resolution can be useful for heterogeneous cell populations (e.g., neuronal cell cultures or stem cell-derived cells) or where the biological response is not uniform (e.g., in evaluating rare events, or electrical and mechanical conduction across a field of cells). Technological advances, including faster and more sensitive cameras as well as memory storage solutions, are speeding up HCS and allow its use during earlier stages of drug discovery.
The major challenge in optical assays is the limited options to change the cell membrane potential in a physiological, reversible, and temporarily precise manner. The list of cellular stimulation technologies compatible with optical HTS assays has significantly expanded over the past 10 years. Although ‘classical’ (e.g., chemical and electrical) stimulation methods are still being used, several novel [e.g., using optogenetics and infrared (IR) light], and emerging (e.g., nanotechnology-based) methods are poised to make a significant impact in enabling drug-screening assays that are both fast and informative. In this review, we discuss the instrumentation and technologies that have been developed to stimulate cells and capture the dynamic states and kinetics of their physiological responses. These advances promise to revolutionize the discovery of drugs for cardiac and neurological disorders as well as safety pharmacology.
Chemical stimulation
Changes in functional states of cells or targets of interest can be produced by chemical stimulation methods such as extracellular application of ‘high-potassium’ solutions or pharmacological agonists [14].
High-potassium stimulation
The presence of inward rectifying potassium ion channels (Kir) in engineered cells ensures that, under physiological conditions, the resting membrane potential is set around −90 mV, because tonically active Kir channels keep the membrane potential close to the equilibrium potential for potassium. Changes in extracellular potassium concentrations lead to graded depolarization of the cell membrane in a concentration-dependent manner. As a result, voltage-gated transmembrane targets of interest co-expressed with Kir channels will transition from one functional state to another (e.g., closed–open–inactivated), which would enable the evaluation of the effects of drugs on different states [10]. The potassium stimulation protocol is fairly universal [15]. However, its applicability might be limited for voltage-gated ion channels that experience fast transient spontaneous conformational changes after being activated by membrane depolarization. To ensure the robust execution of the high-potassium stimulation protocol, several companies are offering dedicated heterologous channel-expressing cell lines that have ‘helper’ Kir channels co-expressed with other potential targets of interest (e.g., Chantest Cell Lines from Charles River Laboratories).
Pharmacological modifiers
For some targets, changes in their functional states can also be induced by pharmacological entities with a known MOA. For example, in engineered cells, sodium voltage-gated ion channels often reside in a nonconducting inactivated state and, therefore, drug application will not be able to induce any physiological changes that can be detected in screening assays. A depolarizing stimulus will not be helpful either: because of fast inactivation gating kinetics, sodium channels spontaneously exit an open state and do it faster than the temporal resolution of current optical detection methods. If inactivation is removed using channel openers, sodium channels stay in an open state, and the sodium influx via open channels will result in membrane depolarization. The most widely used sodium channel opener is veratridine [16,17], although a few alternatives are also available [18–20]. This approach might be effective only if MOAs of novel drug candidates are orthogonal to the MOA of veratridine. However, a caution should be exercised during such assays, because compound interactions might increase the false-positive or false-negative rates [21]. Overall, utilization of veratridine might affect the target pharmacology, can induce to veratridine-driven artifacts in cell physiology, and does not enable the detection of a use-dependent block.
In summary, chemical stimulation methods are nonphysiological, do not provide dynamic control of the cell membrane potential, cannot be used repetitively, have low temporal resolution, and might produce interactions between drug candidates and agonists or other depolarizing agents. Nevertheless, these methods are still currently being used for drug screening because they are robust and cost effective.
Electrical stimulation
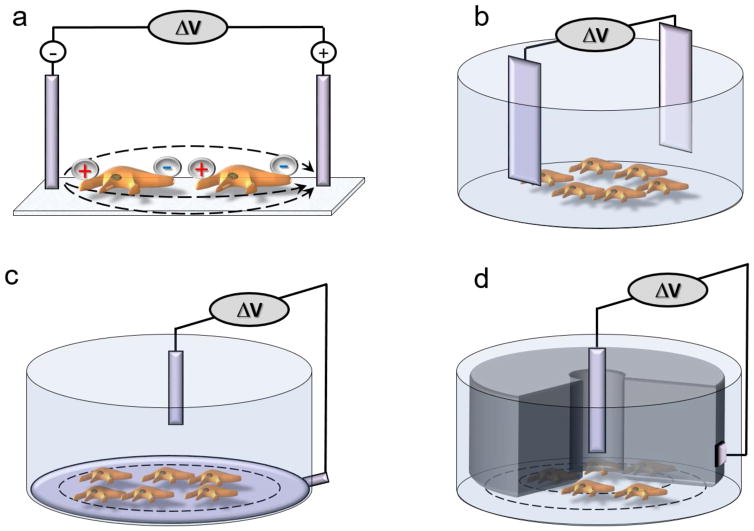
Changing the transmembrane electric gradient by applying external electric field (E-field) has been utilized for stimulation of cells for years [22–25]. When E-field is applied to a pair of metal electrodes immersed in the electrolytic physiological solution, it triggers electric currents that lead to changes in the membrane potential in cells located between the electrodes (Figure 1a). E-field stimulation (EFS) can stimulate cells repeatedly and with a desired temporal pattern.
Figure 1.
Electrical field stimulation (EFS) technology. (a) General E-field stimulation scheme. (b) Standard two-electrode EFS in a well as applied in the Electrical stimulation Voltage Ion Probe Reader (E-VIPR). (c) Two-electrode EFS as found in the Trans Cell Layer (TCL)-EFS VIPR. (d) Concentric EFS electrodes as found in the Cellaxess® Elektra Discovery.
EFS platforms have encountered significant challenges during the adaptation to a plate format. To create a spatially uniform E-field within each well of every plate with the same set of electrodes, there should be no variability in the well and plate geometry, which means that regular microtiter plates are not optimal for EFS [26]. Even in specialized geometrically correct plates, EFS produces E-field anisotropies near the edge of the well and creates non-uniform concomitant depolarization and hyperpolarization areas in a cell. Additionally, EFS produces dye bleaching, generates bubbles because of redox-triggered water hydrolysis, and might elicit cell damage [27–29]. These inherent technical limitations could negatively impact the assay outcome [13,29].
EFS was first enabled for drug screening over 15 years ago by Vertex Pharmaceuticals in a specialized imaging instrument with integrated stimulation and imaging components [Electrical stimulation Voltage Ion Probe Reader (E-VIPR)] [30]. Stimulation in E-VIPR (Figure 1b) was achieved by applying a voltage signal (e.g., 40 mV for 100 ms) to an array of eight pairs of platinum electrodes immersed into eight wells of a 96-well plate with cells. Optical detection of the activity of ion channels was performed using fluorescent voltage-sensitive probes. Vertex has been using E-VIRP only for in-house studies [31], and the instrument is not commercially available.
To increase the throughput, higher well density plates are necessary. However, adaption of two-electrode EFS to such plates was found to be challenging for several reasons: (i) to produce uniform E-fields in a well, the stimulation electrodes have to positioned far apart, which is challenging within the spatial confines of 384-and 1536-well plates; and (ii) the effect of carry-over of test compounds during repeated electrode insertions is significantly more pronounced in small wells.
To address the shortcomings of the two-electrode approach implemented in the E-VIPR, scientists at Merck developed the Trans Cell Layer Electric Field Stimulation (TCL-EFS) system for a Voltage Ion Probe Reader (VIPR™) [29]. In the TCL-EFS, a lower electrode was positioned beneath the cell layer along the perimeter of a filter well, while an upper electrode was placed above the cell layer at the top of the solution in each well (Figure 1c). Only the upper electrode is moved in and out of each well during drug screening, which could simplify the adoption of higher format microplates. The TCL-EFS system was customized for in-house screening, and currently is not operational.
Recognizing the importance of cellular stimulation of cells during drug screening assays, several companies have added an EFS module to their existing imaging instruments [32].
Cellectricon AB (Sweden) has modified its Cellaxess® Adherent Cell Electroporation (ACE) system to engineer an EFS module, and developed the Cellaxess® Elektra Discovery Platform that combines EFS with optical detection methods. The biophysics of electroporation and EFS is the same, with the only difference being that electroporation requires a greater magnitude of E-fields to efficiently disrupt the cell membranes. Voltage pulses are delivered through capillary electrodes that are arranged in a 96-well EFS head for stimulation of cells in 384-well plates. The electrodes each comprise two concentric titanium tubes separated by a polytrifluorochloroethylene (PTFCE) tip that serves as an electrical insulator and the contact with the well bottom (Figure 1d). Uniform E-fields are achieved within individual wells through specific contouring of the bottom of the PTFCE electrode tip. Advantages of this platform include: (i) low electrical currents that enable the minimization of electrochemical processes; and (ii) the tailored tip surface shape that can produce homogeneous E-field (over 3 mm, 100 V under the surface of capillary). Specialized optimized microplates are required for robust and uniform stimulation. The Cellaxess® Elektra is an HTS instrument, which means that whole-plate images are segmented for allocation to individual wells. Cellectricon recently became a service-based company and does not sell its products directly.
Interestingly, the idea to adapt a Cellectricon’s electroporation system for EFS-enabled imaging assays was first devised by Galenea Corp [26]. Specifically, Galenea integrated a customized CellaxessHT® system with the plate::vision plate reader (PerkinElmer) to develop synaptic vesicle-cycling assays in a 96-well format (MANTRA, or Multiwell Automated NeuroTRansmission Assay) for in-house studies.
Hamamatsu Photonics (Japan) recently introduced a 96-channel electrode array module that can be mounted on the FDSS/mCELL system. This commercially available EFS module can provide stimulation to all 96 wells in a microplate simultaneously while fluorescence or luminescence signals are monitored. The goal is to combine EFS with fluorescent measurements of intracellular calcium kinetics to perform in vitro assessment of cardiac toxicity of pharmacological compounds using CMs, in particular in toxicity screening during early stages of drug development, as well as in cardiovascular research. By varying the duration (5–50 ms) and magnitude of E-field (5–30 V), Hamamatsu optimized and tested various EFS protocols at frequencies from 0.5 Hz to 3 Hz for drug screening on different types of CMs, including rat primary CMs (voltage 5 V, duration 5 ms), mouse embryonic stem cell (ESC)-derived CMs, human iPSC-derived thin-layered and semi-clumped CMs, and human iPSC-derived CMs.
Vala Sciences (USA) also developed an optional EFS module for its IC200-KIC™ imaging instrument. In contrast to the HTS systems described above, IC200-KIC™ is an HCS instrument in which EFS is performed in one well at a time in a 384-well format while fluorescent signals are acquired at a single-cell resolution [32]. IC200-KIC™ has been usefully used for calcium, voltage, and contractility screening assays on CMs [33,34]. In general, HCS systems have lower throughput, but they can provide a wealth of information about drug effects on individual cells.
The highest density of electrode arrays on any EFS-capable imaging instrument does not exceed 96 pairs for parallel stimulation in 96 wells, suggesting the limited compatibility of the EFS method with HTS miniaturization requirements. Another potential issue for EFS in imaging assays is the limitations imposed by its IP status. For example, Hamamatsu Photonics warns that ‘The FDSS/mCELL EFS system should not be used for optically detecting change in transmembrane potential of the cells, and should not be used with the cells in which you/somebody expressed the target ion channels’. Such a ‘red label’ might present a significant limitation for not only target-based drug discovery programs, but also phenotypic voltage imaging assays that are essential for cardio- and neurotoxicity studies.
Optical stimulation
Light is a perfect trigger for the dynamic stimulation of cells during optical assays. Optical stimulation enables fast, reversible, remote stimulation of many cells simultaneously, and it can be performed in a miniaturized format. Several types of optical stimulation method are currently being pursued in labs, and have different degrees of readiness for transfer to drug screening. These methods fall into three major categories that are reviewed below: optogenetic, optoelectronic, and IR methods.
Optogenetic stimulation
Optogenetics is a revolutionary new technology that is utilizing light and genetics to manipulate the functional activity of biological systems and to visualize changes in such activity. Optogenetics offers genetically encoded tools for optical stimulation of cells (actuators) and for optical monitoring of cellular behavior (reporters or sensors). After being first introduced as a novel method for neuroscience research over 10 years ago, optogenetics underwent extensive multi-iteration development [35] that produced an extensive toolbox for many different applications, including drug discovery [36–40]. The situation with drug screening is less straightforward, because most HTS assays are performed in homogeneous cell populations. It means that the main advantages of optogenetics, such as selective incorporation of light-sensitive proteins into genetically distinct subpopulation of cells, have limited value in drug-screening assays.
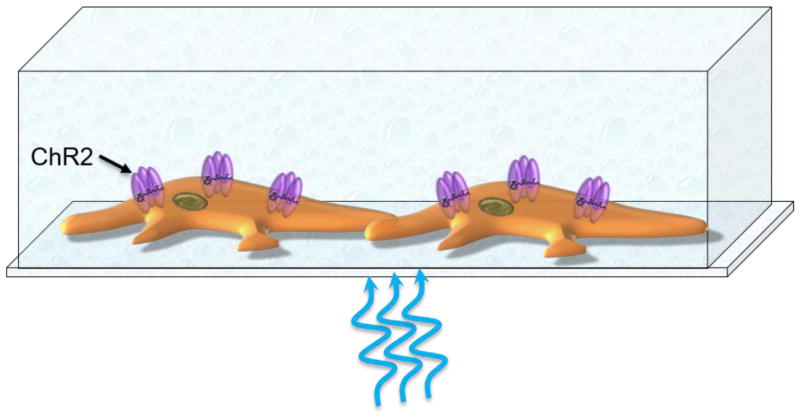
The cornerstone of optogenetic stimulation is a family of channelrhodopsins (ChRs), light-gated ion channels from microalgae [41–43]. Light absorption and subsequent isomerization of retinal inside ChRs leads to their opening (Figure 2). A flux of ions (i.e., ion currents) through now open ion channels trigger changes in the cell membrane potential. There are various mutants and variants of ChRs that differ in their spectral profile, amplitude, and response speed. As a general rule, ChRs with faster kinetics have weaker light sensitivity and, thus, smaller light-induced currents [44].
Figure 2.
Optogenetic stimulation based on a facility of light-sensitive ion channels (e.g., channelrhodopsin 2 (ChR2). Photoisomerizable 11-cis retinal is shown inside a ChR2 protein.
In all optical assays, optogenetic stimulation can be combined with either organic fluorescent dyes or optogenetic sensors for monitoring cellular responses during drug screening. Many excellent organic dyes [e.g., fluorescein-based calcium-sensitive dyes (e.g., Fluo4)) or voltage-sensitive dyes (e.g., FluoVolt)] are excited in the same spectral region as the most reliable optogenetic actuators (e.g., ChR2 or CheRiff). Therefore, optical assays with optogenetic stimulation are forced to choose spectrally compatible rather than the best-performing dyes. Hopefully, further development of optogenetic actuators and fluorescent dyes will improve this situation. Fluorescent dyes usually produce more homogeneous signals than do optogenetic sensors. However, dyes are only suitable for short-term monitoring, whereas optogenetic sensors are excellent for evaluating the cell activity at any time point [45]. By contrast, all-optogenetic assays have their own limitations. For example, the need to express exogenous sensors on top of actuators could place a greater strain on cells and decrease assay robustness because of variable expression levels of the actuators and sensors. Additionally, the chance for potential interference with tested drugs is increased, because sensors must stay continuously active during optical monitoring.
Optogenetic stimulation by itself is a complex phenomenon, which has to be taken into account while designing and analyzing optogenetic-enabled drug-screening assays. We discuss several important limitations below. Although some of these limitations could be mitigated by performing supplementary orthogonal screening assays, it will lead to an increase in costs and drug development time.
The high-level expression of exogenous light-sensitive proteins in cells and their functional integration into the intracellular machinery is necessary for the robust manipulation of the cell membrane potential. The mere presence of optogenetic actuators in the cell membrane and the properties of their light-induced activity (e.g., the gating kinetics and the activity of specific ions conducted by actuators) might lead to unexpected adverse effects on cellular functions [46]. Optogenetic actuators have intrinsic kinetics that can affect some endogenous processes in a cell (both voltage dependent and ion dependent). For example, slow depolarization triggered by some optogenetic actuators might result in calcium influx via voltage-gated ion channels [47], or prolonged light stimulation and subsequent depolarization might lead to inactivation of voltage-gated sodium channels and resultant block of action potential (AP) [48].
Intracellular ion homeostasis might be greatly altered by optogenetic actuators because of their permeability to multiple species of ions, mainly sodium and protons, but also calcium and potassium [43]. All these ions can, in principle, affect multiple aspects of cellular function and, thus, could distort the drug effects. Moreover, even if needed (e.g., for modeling of depolarization-induced tachycardia), prolonged optogenetic stimulation is not optimal for cells because of an even greater impact on ion homeostasis.
Given the complex gating kinetics, optogenetic stimulation does not immediately stop when light is turned off. Continuing post-light stimulation decays in a nonlinear fashion with toff of ~20 ms [e.g., ChR2(H134R) and CheRiff)] and, thus, can distort AP profiles [49] not only in neurons, but also in CMs, leading to errors in the pharmacological profiling of drug candidates.
Optogenetic actuators are essentially transmembrane ion channel proteins that have a pore and conduct different ions; therefore, drug candidates might directly affect actuators (closed and/or activated) instead or in addition to a target of interest. Such interference could produce false-positive and false-negative results in a drug-screening assay.
It is possible to address some of these issues by expressing optogenetic actuators in one cell subpopulation (‘sparks’ or ‘actuators’) while testing the effects of drugs on another subpopulation (naïve cells) [50]. A few different versions of this approach have been developed (e.g., ‘tandem-cell-unit’ or ‘spatial separation’) [40,50–52]. The proposed design relies on direct contact between two cell subpopulations and, thus, can decrease the assay reliability because of the intrinsic variability of an additional link in the stimulation path.
To enable optogenetics-based stimulation in immortalized cell lines, significantly higher depolarization is required compared with excitable cells, where minor changes in the cell membrane potential can lead to generation of APs in all-or-none manner. It would mean the higher expression level of optogenetic constructs and/or alternative optogenetic contracts with higher current amplitude (higher light sensitivity) but slower kinetics. Both these properties are not desirable in an HTS setting.
Currently, to introduce optogenetic actuators into CMs, viral gene transfer (transient transduction) has to be performed 24–48 h before experiments, which leads to extra steps in the assay protocol, the need to handle a virus, and variability in expression levels (if not at the maximal levels); more importantly, it also gives rise to heterogeneity that might negatively impact the assay robustness.
These considerations do not preclude the utilization of optogenetics-based stimulation in drug screening. However, it means that additional controls (which would result in a decrease in HT) and/or complex spatial arrangements will be required to ensure the reproducibility and reliability of such assays.
Several companies (e.g., Q-State Biosciences, AXXAM, and Vala Sciences) have been working on the adaption of optogenetic actuators for drug screening [53], which has resulted in a limited number of optogenetics-enabled assays. Currently, these assays are offered only on a contract basis.
AXXAM [53] was the first company to evaluate the feasibility of optogenetic stimulation in drug-screening assays in a miniaturized 384-well format using an industry-renowned HTS system, the FLIPR® (Fluorometric Imaging Plate Reader) Tetra (Molecular Devices). These optical assays tested the effects of benchmark blockers on human voltage-gated calcium channels (hCaV1.3; assay 1) or human hyperpolarization-activated cyclic nucleotide-gated channels (HCN2; assay 2) that were stably co-expressed in HEK293 cells with their respective optogenetic actuators [ChR2 D156A or Beggiatoa light-activated adenylyl cyclase (bPAC)]. Optogenetic actuators were activated by a short (msec) light pulse from a blue LED (470–495 nm), and cellular responses were monitored for many minutes using either fluorescent calcium-sensitive (assay 1) or voltage-sensitive (assay 2) probes.
The screening assay for state-dependent drugs acting on hCaV1.3 channels is of particular interest because of its unusual design. AXXAM realized that the wild-type ChR2 with its fast kinetics, but relatively small light-induced current, would not be able to trigger membrane depolarization sufficiently for full activation of hCaV1.3. Therefore, AXXAM switched to the ChR2(D156A) optogenetic actuator, which exhibits a larger photocurrent, but slower kinetics (toff = 10 min) [54]. During the assay, a brief light pulse activates ChR2(D156A), which triggers membrane depolarization and activation of hCaV1.3 channels. While the membrane potential is slowly returned to the baseline (30 min), the induced calcium influx is monitored with a calcium-sensitive probe. The timing of drug applications is chosen by: (i) taking into consideration the complex off-rate kinetics of ChR2-induced depolarization; and (ii) assuming the corresponding degree of inactivation of hCaV1.3 channels at any given level of depolarization. Although this assay can be considered as proof-of-concept for optogenetic stimulation, it can produce highly ambiguous results because of the need for assumptions regarding the activation state of both an actuator and a target of interest.
The development of all-optical drug screening assays is not limited to industry. An interesting design that combines optogenetic stimulation with optical monitoring using fluorescent organic indicators was recently tested in an academic setting [50,52]. Optogenetic actuation in CMs was first demonstrated 6 years ago [36,37]. The proposed screening platform is called OptoDyCe (for all-optical dynamic cardiac electrophysiology) and, as its name implies, it is designed exclusively for screening using CMs. Optical stimulation in OptoDyCe is enabled by the optogenetic actuator ChR2(H134R) [43], which is either virally introduced into CMs or transiently expressed in HEK293 cells. Optical monitoring is performed using fluorescent red-shifted organic dyes (e.g., di-4-ANBDQBS for voltage imaging and Rhod-4AM for calcium imaging assays). Although these dyes appear to be spectrally compatible with ChR2s, because of its wide absorption spectrum, di-4-ANBDQBS could be excited by the ChR2-designated blue light [55].
OptoDyCe assays are performed in an HCS mode on a conventional epifluorescent microscope in a single-well format using CMs cultured in 96-well plates. When fully automated, this assay configuration would only be able to test approximately 100 dose-responses per day, which falls below HTS requirements. The OptoDyCe platform was tested by using two benchmark compounds: nifedipine, a blocker of voltage-gated calcium channels, and dofetilide, a blocker of hERG channels. AP duration 80% below the peak (APD80) appeared to be an unreliable parameter for the evaluation of drug effects, probably because of the AP waveform distortion caused by lasting ChR2-mediated conductance. Additionally, nifedipine at extremely low concentrations (0.0001 MM) produced an unusually considerable blocking effect on both APs and CTs, which raises concerns about potential compound interference with the OptoDyCe platform.
A ‘tandem-cell-unit’ version of the OptoDyCe platform addressed these concerns by expressing optogenetic actuators in one cell subpopulation (e.g., ‘spark’ HEK293 cells) and testing drug effects on another cell subpopulation (e.g., CMs). ChR2-expressing HEK293 cells can either be co-cultured with CMs from the start or added (‘sprinkled’) to CMs 24–48 h before the experiment. While addressing potential interference between ChR2-driven processes and drug effects, this approach has created new challenges: (i) using an additional subpopulation of cells complicates the assay workflow and introduces an additional level of heterogeneity into an already complex system. For example, APs of CMs interfacing with ‘spark’ ChR2-expressing cells can vary as a function of the ‘spark’ cell concentration and distribution patterns. To enable controlled ‘spark’-cell delivery, localized and/or patterned pacing sites inside a well might be needed; (ii) after ‘spark’ HEK293 cells are ‘sprinkled’ onto CMs, they inevitably undergo the dye labeling procedure together with CMs and, thus, contribute to the fluorescent signal from a well. HTS would not be viable with this assay design. To separate the signals from two cell subpopulations, HCS assays at a single-cell resolution and image deconvolution software would be required. Future technological advances might help to overcome the limitations discussed above and make OptoDyCe assays more screening friendly.
Q-state Biosciences has developed a proprietary all-optogenetic Optopatch platform that combines optogenetic actuators with optogenetic sensors [56]. Optical stimulation in Optopatch is achieved with a channelrhodopsin variant, CheRiff [51]. Compared with ChR2(H134), CheRiff requires ninefold lower light intensity to trigger APs in neurons [56], which is more beneficial for cell health. Optical monitoring in Optopatch is performed using a fusion protein CaViar (‘calcium and voltage indicator’) comprising a red light-excited voltage-sensitive fluorescent protein, QuasAr2 [56,57] and a blue-light-excited calcium-sensitive protein, GCaMP6f [58]. Optopatch assays are currently performed in a single-well format using 10-mm glass-bottom dishes (Matek) because of the low quantum yield of QuasAr2 (4×10−3) and the need for the spatial separation of two cell subpopulations.
Q-State Biosciences validated the Optopatch platform for cardiotoxicity screening and successfully evaluated the effects of 12 pharmacologically diverse compounds on APs and CTs in CMs. CheRiff was found to provide a common electrophysiological background for all cells in a dish, thus decreasing the assay variability. To ensure that drug-screening results are free from potential artifacts from nonspecific ChR2 conductance, an optogenetic actuator CheRiff and an optogenetic reporter CaViar were expressed in two distinct subpopulations of CMs in a mono-layer cell culture. For monitoring of the cell membrane potential alone, CheRiff-expressing and CaViar-expressing cells can be intermixed. For simultaneous monitoring of both calcium signals and changes in the cell membrane potential, CaViar-expressing cells have to be spatially segregated from CheRiff-expressing cells to avoid optical crosstalk from the blue light needed to excite GCaMP6f. For these assays, CheRiff-expressing cells are plated on the periphery, whereas CaViar-expressing cells are placed in the center of a cell culture dish. Pulses of blue LED illumination (6 ms, 0.5 W/cm2, λ = 488 nm) are utilized to stimulate CheRiff-expressing CMs to pace the syncytium. Blue laser light (λ = 488 nm, 0.15 W/cm2) is utilized to excite GCaMP6f, whereas red laser light (λ = 640 nm, 50 W/cm2) is used to excite QuasAr2. Parallel acquisition of fluorescent signals from voltage- and calcium-sensitive reporters was performed using a dual-view imaging system that enable the simultaneous acquisition of emissions from GCaMP6f (525–575 nm) and QuasAr2 (660–760 nm).
The Optopatch platform was also utilized in screening assays for the evaluation of state-dependent drug effects on voltage-gated ion channels in a heterologous expression system [59]. To do so, four different transgenic constructs were expressed in electrophysiologically neutral cells (e.g., HEK293 cells): inward rectifying potassium ion channels Kir2.1 (to keep the membrane potential hyperpolarized); voltage-gated sodium channels Nav1.7 (the target of interest); CheRiff (optogenetic actuator); and QuasAr2 (voltage-sensitive reporter). The resultant ‘spiking’ HEK293 cells allowed the light-induced generation and monitoring of sodium channel-driven APs. Complex illumination patterns with varying light intensities and duration were developed to simulate voltage-clamp protocols that are routinely used for testing voltage-gated sodium channels. The outcome of an Optopatch-enabled screen of 320 FDA-approved compounds was in close agreement with patch-clamp measurements.
Optopatch assays are currently performed on a specialized custom-built imaging instrument in a 384-well plate format, one well at a time. Parallel measurements from all wells at once could be problematic partially because of a mismatch between the low quantum yield of QuasAr2 and the decreased sensitivity of plate readers. Additionally, water- or oil-immersion lenses are required for efficient fluorescent signal acquisition, which creates another limitation for performing Optopatch assays in an HTS mode. The high light intensity of 400 W/cm2 required for the continuous excitation QuasAr2 during an assay imposes the upper limit on the assay duration and/or the plate density because it can raise a solution temperature by approximately 10°C in 10 min in a single well of a 384-well plate. Regardless of these shortcomings, the Optopatch is currently the most advanced and well-developed platform for all-optical assays with an optogenetic stimulation capability.
Optoelectronic stimulation
Photovoltaic stimulation acts via a light-induced external electric field generated in the vicinity of a cell. Usually, a photovoltaic event has three steps: generation of charges (e.g., photogenerated excitons or electron-hole pairs), charge separation (e.g., exciton into free electrons and holes), and charge transport. In contrast to optogenetics-based optical stimulation, these optical stimulation platforms do not require the genetic modification of cells. Furthermore, they are more physiological, because they activate cells via a capacitive mechanism rather than via exogenous optogenetics-driven ion currents.
Several optical stimulation platforms based on photovoltaic effects have been introduced over the years. In these platforms, there are two possible mechanisms of charge transfer in the electrolytic solution: Faradaic and capacitive (also called nonFaradaic). The Faradaic mechanism involves the transfer of an electron between the electrode and electrolyte (i.e., redox reaction), whereas the capacitive mechanism involves only charge redistribution near the electrode–electrolyte and/or electrolyte–cell membrane interfaces [27]. Given the fundamental differences in physical properties of various photoactive materials, there is no ‘generally applicable model’ for the interaction of these materials with cells [60].
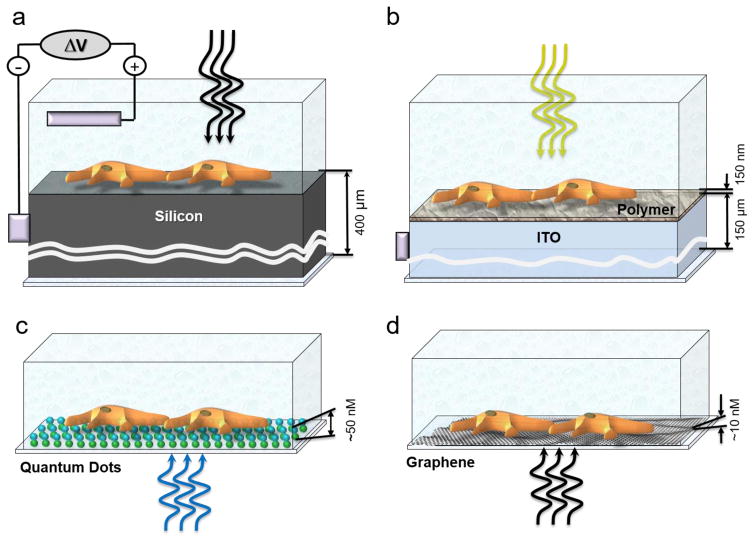
Silicon-based platforms (Figure 3a)
Figure 3.
Optoelectronic stimulation. (a) Silicon-based platform. (b) Conducting polymer-based platform. (c) Quantum dot (QD)-based platform. (d) Graphene-based platform. Dimensions of different components are shown to the left of corresponding schemes.
Given that silicon is an indirect band-gap semiconductor, generation of charges in silicon requires multiple particle interactions. A voltage bias has to be applied to the silicon–electrolyte interface to deplete the majority of charge carried from the silicon surface, and then generate a local photocurrent upon high-intensity light illumination [61]. This photocurrent flows from the silicon surface through cells and electrolyte solution to the counter ground electrode, resulting in depolarization of cells via Ohm’s law (V = I * R). The strong voltage bias regimen enables the generation of sustained DC photocurrents, but can lead to electrochemical reactions on the surface, including hydrolysis of water [62]. Some cells can be activated by transient light-triggered capacitive currents generated at a weaker voltage bias, probably when there is a tighter interaction between cells and silicon surface [64].
Conducting polymer-based platforms (Figure 3b)
These platforms do not require either voltage bias or an extracellular electrode [63]. The challenging step in this platform is charge separation, which is impeded by low dielectric permittivity and weak intermolecular forces in conducting organic polymers. Therefore, a layered architecture of electron-conducting and hole-conducting materials deposited on an indium tin oxide (ITO) layer serving as a cathode is required to enable efficient charge separation. Optical stimulation of neurons was demonstrated using the conducting polymers poly(3-hexylthiophene (P3HT) and phenyl-C61-butyric-acid-methyl ester (PCBM), with an absorption peak at 530 nm [64]. Studies suggested that no transport of photogenerated charges is involved in optical stimulation via polymer-based platform, but rather cells become depolarized as positive ions from the electrolytic solution move to neutralize photogenerated negative charges at the polymer–electrolyte interface.
Quantum dot (QD)-based platform (Figure 3c)
As a result of quantum confinement effects in semiconductor nanoparticles (also called QDs), this platform does not require either a voltage bias or a layered architecture. Instead, it relies on the QD geometrical parameters being larger than the Bohr radius of the electron wave function, which greatly facilitates the charge separation and subsequent charge transport. QDs have broadband absorption spectrum and, therefore, this platform can be easily combined with a range of fluorescent indicators for monitoring cellular activity. A QD-based platform can depolarize cells by changing the ion distribution near the cell membrane by either attracting positive ions to the QD layer (as in the case with a polymer-based platform) or by photogenerated electrons that diffused to the cell membrane [13,64].
So far, these optical stimulation platforms have not found their way into drug screening because they exhibited multiple shortcomings, including (i) low compatibility with optical detection methods because of low transparency; (ii) miniaturization problems because of their mechanical properties and complex architecture; and (iii) inadequate biocompatibility.
Graphene-based platform (Figure 3d)
Graphene differs substantially from the above-mentioned photoactive materials and these properties make it a promising solution for optical stimulation. Graphene is a zero-band gap semiconductor (i.e., neither metal nor semiconductor), and the behavior of electrons in graphene is described by the equations of relativistic quantum physics that represent electrons as ‘massless’ quasiparticles. The crucial difference between graphene and conventional semiconductor materials is that light produces ‘hot’ ballistic electrons in graphene rather than electron-hole pairs [65]. The mean free path of these electrons can be up to 1 μm [66], which means that requirements for the proximity between cells and graphene surface are significantly more relaxed than for other materials. When photogenerated ‘hot’ ballistic electrons reach the cell membrane, they affect the charge distribution near the cell membrane by directly interacting with the Debye layer.
Graphene is an efficient light-to-electricity converter, because, as a result of zero-band gap and strong electron–electron interactions, photogenerated electrons in graphene preferentially distribute their energy to secondary electrons rather than produce lattice heating [67,68]. Therefore, light illumination of a graphene-based stimulation platform produces no light-induced heating effects [65].
The absorption of graphene materials is independent of light wavelength in the range 350–2500 nm, which allows greater flexibility in choosing fluorescent indicators for monitoring cellular activity. Given that the light intensity above a threshold level is required to enable optical stimulation via graphene surfaces, optical crosstalk between stimulation and monitoring processes can be easily avoided by combining a low-intensity (subthreshold) light of fluorophore-specific excitation wavelength and a high-intensity light of any wavelength outside fluorophore excitation spectrum. Light intensities required for optical stimulation of cells via a graphene-based platform and for optogenetic stimulation are in the same range. Although the optoelectronic properties of graphene suggest that light stimulation of graphene is an effective stimulation technology, substantial efforts would be required to fully validate it with different cell types and imaging instruments. Research is underway to develop and validate a graphene-based platform using chemically diverse reference compounds and iPSC-derived neurons [69] and CMs.
IR stimulation
Multiple reviews and research articles have provided a thorough discussion of the use of IR or near-IR (NIR) light for optical stimulation during drug screening [70–72]. Exposure to brief pulses of NIR or IR light (e.g., 790–850 nm, 1400–1600 nm, and 1840–2100 nm) effectively stimulates cells. IR stimulation has been shown to work in neurons [73–76], astrocytes [77], CMs [78–80], HeLa cells [81], and PC12 cells [82]. This method provides direct stimulation of cells, because it does not require any intermediaries, such as chemicals, electrodes, genetic modifications, or interfaces. However, the energy requirement for IR stimulation is relatively large (398–796 mJ cm−2), which is approximately 100 times higher than that for electrical stimulation, and 10 000 times higher than that for optogenetic stimulation.
Despite significant progress in this area, the mechanism behind IR stimulation of cells is not fully elucidated. Following the absorption of IR light by water in the biological tissue [83], subsequent thermal, mechanical, and/or chemical effects might lead to stimulation of cells via several downstream processes [74,84]. Light-induced thermal effects dominate during IR stimulation [84], because pulsed IR light induces a transient increase in temperature up to ~22.2°C for a 10-ms pulse (7.3 mJ, 5.8 J cm−2) [85]. The heating of tissue caused by water absorbing most of the incident energy could lead to potential thermal tissue damage and, therefore, this limits the rate and maximum radiant exposure of stimulation [74,84,85].
Light-induced transient temperature gradients can trigger changes in the plasma and intracellular cell membranes [85–87] and/or activate temperature-sensitive transient receptor potential cation ion channels [88]. It appears that changes in the electrical capacitance of the cell membrane are responsible for the primary effect, whereas direct temperature-dependent effects on ion channels have only a modulatory role [80], because the efficiency of IR stimulation is dependent on temperature gradients (which are, in turn, dependent on the radiant exposure), rather than on absolute temperature (Figure 4).
Figure 4.
Infrared (IR) stimulation. Red areas on cells highlight that both plasma and intracellular membranes are affected by temperature gradients triggered by IR illumination.
Furthermore, recent studies demonstrated that IR stimulation can modulate intracellular calcium cycling in CMs [80] and neurons [76] via IR-induced effects on mitochondria. Proposed mechanisms include the IR absorption by cytochrome C oxidase accelerating respiratory metabolism, or capacitive effects on mitochondrial membranes leading to changes in the mitochondrial membrane potential. These findings raise serious concerns about IR stimulation in the context of drug screening, because IR-induced changes in intracellular calcium concentrations also have widespread effects on almost all aspects of cellular physiology, from activation of calcium-dependent proteins (e.g., phosphatases, ion channels, etc.) to mitochondria signaling (e.g., redox processes) to neurotransmitter release. Thus, IR stimulation in screening assays might produce ambiguous results where drug effects are intermixed with stimulation artifacts.
Additional technical limitations of IR stimulation during drug-screening assays are associated with the fact that the main mechanism of IR stimulation is photothermal. Introducing temperature-dependent artifacts can influence: (i) dye fluorescence, photostability, and phototoxicity; (ii) temperature differences in wells across a microtiter plate; (iii) temperature-induced fluctuations in focus; 9iv) temperature-induced effects on the volume of wells, especially in high-density plates; (v) energy requirements and temperature distribution inside an imaging instrument; as well as (vi) temperature-dependent changes in the intrinsic cell physiology (e.g., arrhythmia proclivity). Thus, IR stimulation in screening assays might produce ambiguous results where drug effects are intermixed with stimulation artifacts.
Concluding remarks
The need for cellular stimulation methods compatible with optical recording methods is becoming appreciated and methods are advancing. The physiological context, properties of cells, and/or targets of interest have to be taken into consideration when a specific cellular stimulation protocol is chosen. Current stimulation methods each have significant limitations that can impair the predictiveness of primary screening assays. For instance, optogenetics, in its commonly implemented form, introduces ion channels that have the potential to alter cellular physiology. Similarly, IR stimulation introduces unwanted thermal effects. Emerging methods, in particular optoelectronic stimulation, have promising properties for resolving some of the most challenging issues, especially when aided by technological advances in imaging and computer hardware.
Optical stimulation methods described in this review are not capable of clamping the cell membrane potential and therefore cannot replace the patch-clamp method required for the thorough elucidation of MOA of novel drug candidates. However, all-optical assays have higher throughput and can evaluate the effects of drugs on thousands of cells in an unbiased manner. These assays have no cell type restrictions and, thus, are compatible with both target-based screening on heterologous expression systems and phenotypic screening on primary or iPSC-derived cells. Given the non-invasive nature of imaging, optical assays allow the evaluation of cellular behavior on extended timescales, and can offer insight into additional mechanisms.
Highlights.
Cell stimulation can increase the information content and predictiveness of HTS.
Chemical and electrical stimulation are robust methods that are still used in HTS.
Emerging optical stimulation technologies could revolutionize drug screening.
Teaser.
The dynamic stimulation of cells during drug screening helps to make the drug discovery process more efficient. Here, we review ‘classic’ and emerging cellular stimulation technologies that are compatible with optical screening assays.
Acknowledgments
This work is supported by grants from the NIH (1R01HL128072, 5R01HL113601), the California Institute for Regenerative Medicine (CIRM TR4-06857), and the Foundation Leducq ‘Shapeheart’ Transatlantic Alliance.
Biographies

Elena Molokanova
Elena Molokanova is the cofounder of Nanotools Biosciences. She has expertise in biophysics, neuroscience, biophotonics, electrophysiology, mathematical modeling, and advanced optical methods and materials. Her current research is focused on the development of nanotechnology-inspired innovative solutions for neuroscience drug discovery and drug-screening applications. After obtaining a MSc in physics, Dr Molokanova was awarded a PhD in biological sciences from the Bogomoletz Institute of Physiology (Kiev, Ukraine).

Mark Mercola
Mark Mercola is a professor of cardiovascular medicine and a member of the Cardiovascular Institute at Stanford University. His academic research is focused on developing and using quantitative assays of patient-specific cardiomyocyte function to discover druggable targets for preserving contractile function in heart failure and promoting regeneration following ischemic injury. Previously, Professor Mercola cofounded the Conrad Prebys Center for Chemical Genomics, one of the largest academic drug-screening centers. He was awarded a BSc and PhD in molecular biology from the University of California and subsequently trained at the Dana-Farber Cancer Institute in Boston. He has also held faculty positions at Harvard Medical School and University of California, San Diego before joining Stanford.

Alex Savtchenko
Alex Savtchenko is an Instructor in the Cardiovascular Institute at Stanford University. His expertise covers automated patch clamp electrophysiology, high-throughput screening, high content imaging, development of fluorescent biosensors, neuroscience drug discovery, and studies on the regeneration and development of new approaches to treat heart disease. Dr Savchenko was awarded an MSc in physics and chemistry by the Moscow Institute of Physics and Technology and a PhD in biological sciences from Bogomoletz Institute of Physiology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DiMasi JA, et al. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 3.Priest BT, Erdemli G. Phenotypic screening in the 21st century. Front Pharmacol. 2014;5:264–273. doi: 10.3389/fphar.2014.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng W, et al. Phenotypic screens as a renewed approach for drug discovery. Drug Discov Today. 2013;18:1067–1073. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avior Y, et al. Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- 6.Mercola M, et al. Induced pluripotent stem cells in cardiovascular drug discovery. Circ Res. 2013;112:534–548. doi: 10.1161/CIRCRESAHA.111.250266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grskovic M, et al. Induced pluripotent stem cells: opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 8.Waring MJ, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discov. 2015;14:475–486. doi: 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- 9.Sager PT, et al. Rechanneling the cardiac proarrhythmia safety paradigm: a meeting report from the Cardiac Safety Research Consortium. Am Heart J. 2014;167:292–300. doi: 10.1016/j.ahj.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 10.McManus OB. HTS assays for developing the molecular pharmacology of ion channels. Curr Opin Pharmacol. 2014;15:91–96. doi: 10.1016/j.coph.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Picones A, et al. Contribution of automated technologies to ion channel drug discovery. Adv Protein Chem Struct Biol. 2016;104:357–378. doi: 10.1016/bs.apcsb.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Obergrussberger A, et al. Automated patch clamp meets high-throughput screening: 384 cells recorded in parallel on a planar patch clamp module. J Lab Autom. 2015;21:779–793. doi: 10.1177/2211068215623209. [DOI] [PubMed] [Google Scholar]
- 13.Molokanova E, Savchenko A. Bright future of optical assays for ion channel drug discovery. Drug Discov Today. 2008;13:14–22. doi: 10.1016/j.drudis.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Felix JP, et al. Functional assay of voltage-gated sodium channels using membrane potential-sensitive dyes. Assay Drug Dev Technol. 2004;2:260–268. doi: 10.1089/1540658041410696. [DOI] [PubMed] [Google Scholar]
- 15.Yu HB, et al. High throughput screening technologies for ion channels. Acta Pharm Sin. 2016;37:34–43. doi: 10.1038/aps.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulbricht W. Effects of veratridine on sodium currents and fluxes. Rev Physiol Biochem Pharmacol. 1998;133:1–54. doi: 10.1007/BFb0000612. [DOI] [PubMed] [Google Scholar]
- 17.Benjamin ER, et al. State-dependent compound inhibition of Nav1. 2 sodium channels using the FLIPR Vm dye: on-target and off-target effects of diverse pharmacological agents. J Biomol Screen. 2006;11:29–39. doi: 10.1177/1087057105280918. [DOI] [PubMed] [Google Scholar]
- 18.Zhao F, et al. Development of a rapid throughput assay for identification of hNav1. 7 antagonist using unique efficacious sodium channel agonist, antillatoxin. Marine Drugs. 2016;14:36. doi: 10.3390/md14020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GS, et al. An integrated view of the molecular toxinology of sodium channel gating in excitable cells. Annu Rev Neurosci. 1987;10:237–267. doi: 10.1146/annurev.ne.10.030187.001321. [DOI] [PubMed] [Google Scholar]
- 20.Narahashi T, Herman MD. Overview of toxins and drugs as tools to study excitable membrane ion channels: I. Voltage-activated channels. Methods Enzymol. 1992;207:620–643. doi: 10.1016/0076-6879(92)07045-p. [DOI] [PubMed] [Google Scholar]
- 21.Kaczorowski GJ, et al. Ion channels as drug targets: the next GPCRs. J Gen Physiol. 2008;131:399–405. doi: 10.1085/jgp.200709946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross D, et al. Optical imaging of cell membrane potential changes induced by applied electric fields. Biophys J. 1986;50:339–348. doi: 10.1016/S0006-3495(86)83467-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tung L, Borderies JR. Analysis of electric field stimulation of single cardiac muscle cells. Biophys J. 1992;63:371–386. doi: 10.1016/S0006-3495(92)81632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krassowska W, Neu JC. Response of a single cell to an external electric field. Biophys J. 1994;66:1768–1776. doi: 10.1016/S0006-3495(94)80971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spira ME, Hai A. Multi-electrode array technologies for neuroscience and cardiology. Nat Nanotechnol. 2013;8:83–94. doi: 10.1038/nnano.2012.265. [DOI] [PubMed] [Google Scholar]
- 26.Hempel CM, et al. A system for performing high throughput assays of synaptic function. PLoS ONE. 2011;6:e25999. doi: 10.1371/journal.pone.0025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrill DR, et al. Electrical stimulation of excitable tissue: design of efficacious and safe protocols. J Neurosci Methods. 2005;141:171–198. doi: 10.1016/j.jneumeth.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Knollmann BC. Pacing lightly: optogenetics gets to the heart. Nat Methods. 2010;7:889–891. doi: 10.1038/nmeth1110-889. [DOI] [PubMed] [Google Scholar]
- 29.Bugianesi RM, et al. A cell-sparing electric field stimulation technique for high-throughput screening of voltage-gated ion channels. Assay Drug Dev Technol. 2006;4:21–35. doi: 10.1089/adt.2006.4.21. [DOI] [PubMed] [Google Scholar]
- 30.González J, Maher M. Cellular fluorescent indicators and voltage/ion probe reader (VIPR) tools for ion channel and receptor drug discovery. Receptors Channels. 2002;8:283–285. [PubMed] [Google Scholar]
- 31.Huang CJ, et al. Characterization of voltage-gated sodium-channel blockers by electrical stimulation and fluorescence detection of membrane potential. Nat Biotechnol. 2006;24:439–446. doi: 10.1038/nbt1194. [DOI] [PubMed] [Google Scholar]
- 32.Cerignoli F, et al. High throughput measurement of Ca2+ dynamics for drug risk assessment in human stem cell-derived cardiomyocytes by kinetic image cytometry. J Pharmacol Toxicol Methods. 2012;66:246–256. doi: 10.1016/j.vascn.2012.08.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu HR, et al. High throughput measurement of Ca++ dynamics in human stem cell-derived cardiomyocytes by kinetic image cytometery: a cardiac risk assessment characterization using a large panel of cardioactive and inactive compounds. Toxicol Sci. 2015;148:503–516. doi: 10.1093/toxsci/kfv201. [DOI] [PubMed] [Google Scholar]
- 34.Pfeiffer ER, et al. Specific prediction of clinical QT prolongation by kinetic image cytometry in human stem cell derived cardiomyocytes. J Pharmacol Toxicol Methods. 2016;81:263–273. doi: 10.1016/j.vascn.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–1225. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arrenberg AB, et al. Optogenetic control of cardiac function. Science. 2010;330:971–974. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- 37.Bruegmann T, et al. Optogenetic control of heart muscle in vitro and in vivo. Nat Methods. 2010;7:897–900. doi: 10.1038/nmeth.1512. [DOI] [PubMed] [Google Scholar]
- 38.Nussinovitch U, Gepstein L. Optogenetics for in vivo cardiac pacing and resynchronization therapies. Nat Biotechnol. 2015;33:750–754. doi: 10.1038/nbt.3268. [DOI] [PubMed] [Google Scholar]
- 39.Burton RA, et al. Optical control of excitation waves in cardiac tissue. Nat Photonics. 2015;9:813–816. doi: 10.1038/nphoton.2015.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song C, Knopfel T. Optogenetics enlightens neuroscience drug discovery. Nat Rev Drug Discov. 2016;15:97–109. doi: 10.1038/nrd.2015.15. [DOI] [PubMed] [Google Scholar]
- 41.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 42.Boyden ES, et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 43.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato HE, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leyton-Mange JS, et al. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Rep. 2014;2:163–170. doi: 10.1016/j.stemcr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen BD, et al. Principles of designing interpretable optogenetic behavior experiments. Learning Memory. 2015;22:232–238. doi: 10.1101/lm.038026.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang YP, Oertner TG. Optical induction of synaptic plasticity using a light-sensitive channel. Nat Methods. 2007;4:139–141. doi: 10.1038/nmeth988. [DOI] [PubMed] [Google Scholar]
- 48.Herman AM, et al. Cell type-specific and time-dependent light exposure contribute to silencing in neurons expressing Channelrhodopsin-2. Elife. 2014;3:e01481. doi: 10.7554/eLife.01481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park SA, et al. Optical mapping of optogenetically shaped cardiac action potentials. Sci Rep. 2014;4:6125. doi: 10.1038/srep06125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia Z, et al. Stimulating cardiac muscle by light: cardiac optogenetics by cell delivery. Circulation Arrhythmia Electrophysiol. 2011;4:753–760. doi: 10.1161/CIRCEP.111.964247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dempsey GT, et al. Cardiotoxicity screening with simultaneous optogenetic pacing, voltage imaging and calcium imaging. J Pharmacol Toxicol Methods. 2016;81:240–250. doi: 10.1016/j.vascn.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Klimas A, et al. OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat Commun. 2016;7:1–12. doi: 10.1038/ncomms11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agus V, et al. Bringing the light to high throughput screening: use of optogenetic tools for the development of recombinant cellular assays. SPIE. 2015;9305:93052T. [Google Scholar]
- 54.Bamann C, et al. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2009;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 55.Zhou WL, et al. Intracellular long-wavelength voltage-sensitive dyes for studying the dynamics of action potentials in axons and thin dendrites. J Neurosci Methods. 2007;164:225–239. doi: 10.1016/j.jneumeth.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zou P, et al. Bright and fast multi-colored voltage reporters via electrochromic FRET. Nature Communications. 2014;5:4625–4625. doi: 10.1038/ncomms5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, et al. Optical electrophysiology for probing function and pharmacology of voltage–gated ion channels. ELife. 2016;5:e15202. doi: 10.7554/eLife.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bareket-Keren L, Hanein Y. Novel interfaces for light directed neuronal stimulation: advances and challenges. Int J Nanomedicine. 2014;9(Suppl 1):65–83. doi: 10.2147/IJN.S51193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goda Y, Colicos MA. Photoconductive stimulation of neurons cultured on silicon wafers. Nat Protoc. 2006;1:461–467. doi: 10.1038/nprot.2006.67. [DOI] [PubMed] [Google Scholar]
- 62.Memming R, Bahnemann D. Semiconductor Electrochemistry. John Wiley & Sons; 2015. [Google Scholar]
- 63.Ghezzi D, et al. A hybrid bioorganic interface for neuronal photoactivation. Nat Commun. 2011;2:166. doi: 10.1038/ncomms1164. [DOI] [PubMed] [Google Scholar]
- 64.Molokanova E, et al. Quantum dots move beyond fluorescent imaging. Biophotonics Int. 2008;15:26–31. [Google Scholar]
- 65.Gabor NM, et al. Hot carrier-assisted intrinsic photoresponse in graphene. Science. 2011;334(6056):648–652. doi: 10.1126/science.1211384. [DOI] [PubMed] [Google Scholar]
- 66.Novoselov KS, et al. Electric field effect in atomically thin carbon films. Science. 2004;306:666–669. doi: 10.1126/science.1102896. [DOI] [PubMed] [Google Scholar]
- 67.Freitag M, et al. Photoconductivity of biased graphene. Nat Photonics. 2013;7:53–59. [Google Scholar]
- 68.Tielrooij K, et al. Photoexcitation cascade and multiple hot-carrier generation in graphene. Nat Physics. 2013;9:248–252. [Google Scholar]
- 69.Molokanova E, et al. Light-controlled activation of neurons via a graphene-based biocompatible optoelectronic interface. Annual Meeting of Society for Neuroscience. 2015;199(14) [Google Scholar]
- 70.Richter CP, Tan X. Photons and neurons. Hearing Res. 2014;311:72–88. doi: 10.1016/j.heares.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chernov M, Roe AW. Infrared neural stimulation: a new stimulation tool for central nervous system applications. Neurophotonics. 2014;1:011011. doi: 10.1117/1.NPh.1.1.011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson AC, et al. Optical stimulation of neurons. Curr Mol Imag. 2014;3:162–177. doi: 10.2174/2211555203666141117220611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wells J, et al. Optical stimulation of neural tissue in vivo. Optics Lett. 2005;30:504–506. doi: 10.1364/ol.30.000504. [DOI] [PubMed] [Google Scholar]
- 74.Richter CP, et al. Neural stimulation with optical radiation. Laser Photonics Rev. 2011;5:68–80. doi: 10.1002/lpor.200900044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bec JM, et al. Characteristics of laser stimulation by near infrared pulses of retinal and vestibular primary neurons. Lasers Surg Med. 2012;44:736–745. doi: 10.1002/lsm.22078. [DOI] [PubMed] [Google Scholar]
- 76.Lumbreras V, et al. Pulsed infrared radiation excites cultured neonatal spiral and vestibular ganglion neurons by modulating mitochondrial calcium cycling. J Neurophysiol. 2014;112:1246–1255. doi: 10.1152/jn.00253.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Y, et al. Photostimulation of astrocytes with femtosecond laser pulses. Optics Express. 2009;17:1291–1298. doi: 10.1364/oe.17.001291. [DOI] [PubMed] [Google Scholar]
- 78.Smith NI, et al. A femtosecond laser pacemaker for heart muscle cells. Optics Express. 2008;16:8604–8616. doi: 10.1364/oe.16.008604. [DOI] [PubMed] [Google Scholar]
- 79.Jenkins MW, et al. Optical pacing of the embryonic heart. Nat Photonics. 2010;4:623–626. doi: 10.1038/nphoton.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dittami GM, et al. Intracellular calcium transients evoked by pulsed infrared radiation in neonatal cardiomyocytes. J Physiol. 2011;589:1295–1306. doi: 10.1113/jphysiol.2010.198804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith NI, et al. Generation of calcium waves in living cells by pulsed-laser-induced photodisruption. Appl Phys Lett. 2001;79:1208. [Google Scholar]
- 82.Smith N, et al. Photostimulation of two types of Ca2+ waves in rat pheochromocytoma PC12 cells by ultrashort pulsed near-infrared laser irradiation. Laser Phys Lett. 2005;3:154. [Google Scholar]
- 83.Thompson AC, et al. Modeling of light absorption in tissue during infrared neural stimulation. J Biomed Optics. 2012;17:0750021–0750026. doi: 10.1117/1.JBO.17.7.075002. [DOI] [PubMed] [Google Scholar]
- 84.Wells J, et al. Biophysical mechanisms of transient optical stimulation of peripheral nerve. Biophys J. 2007;93:2567–2580. doi: 10.1529/biophysj.107.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shapiro MG, et al. Infrared light excites cells by changing their electrical capacitance. Nat Commun. 2012;3:736. doi: 10.1038/ncomms1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okunade O, Santos-Sacchi J. IR laser-induced perturbations of the voltage-dependent solute carrier protein SLC26a5. Biophys J. 2013;105:1822–1828. doi: 10.1016/j.bpj.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu CH, et al. Graphene photodetectors with ultra-broadband and high responsivity at room temperature. Nat Nanotechnol. 2014;9:273–278. doi: 10.1038/nnano.2014.31. [DOI] [PubMed] [Google Scholar]
- 88.Albert ES, et al. TRPV4 channels mediate the infrared laser-evoked response in sensory neurons. J Neurophysiol. 2012;107:3227–3234. doi: 10.1152/jn.00424.2011. [DOI] [PubMed] [Google Scholar]