Abstract
Wolf–Hirschhorn syndrome candidate 1 (WHSC1) is a histone 3 lysine 36 (H3K36) specific methyltransferase that is frequently deleted in Wolf–Hirschhorn syndrome (WHS). Whsc1 is also found mutated in a subgroup of B-cell derived malignant diseases by genomic translocation or point mutation, both of which resulted in hyperactivity of WHSC1 mediated H3K36 methylation and uncontrolled cell proliferation, suggesting that whsc1 functions as an oncogene. However, here we provided evidences to show that whsc1 also has tumor suppressor functions. We used zebrafish as an in vivo model and generated homozygous whsc1 mutant lines via clustered regularly interspaced short palindromic repeats-associated protein Cas9 (CRISPR/Cas9) technology. Then western-blot (WB) and immunofluorescence (IF) were performed to analysis the expression level of H3K36Me2 and H3K36Me3, and we identified the diseased tissue via hematoxylin–eosin (HE) staining, IF staining or immunohistochemistry (IHC). Whsc1 lose-of-function led to significant decrease in di- and tri-methylation of H3K36. A series of WHS related phenotypes were found in whsc1−/− zebrafish, including growth retardation, neural development defects and heart failure. In addition, loss of function of whsc1 led to defects in the development of swim bladder, possibly through the dis-regulation of key genes in swim bladder organogenesis and inhibition of progenitor cell differentiation, which was correlated with its expression in this organ during embryonic development. At later stage, these whsc1−/− zebrafishes are inclined to grow tumors in the swim bladder. Our work suggested that whsc1 may function as a tumor suppressor by governing progenitor cell differentiation.
Introduction
Epigenetics encompasses a wide range of heritable changes in gene expression without any alteration in DNA sequence, which includes DNA methylation, histone modification, nucleosome positioning etc [1], [2]. As the core components of nucleosome, histone proteins are subject to post-transcriptional covalent modifications which happen on specific residues at both their globular domains and unstructured N-terminal tails, including methylation, acetylation, ubiquitylation, sumoylation and phosphorylation. These histone modifications regulate a wide range of processes, including RNA transcription, DNA repair [3], [4], DNA replication [5] and chromosome condensation [6].
WHSC1, also known as multiple myeloma SET protein (MMSET) or nuclear receptor-binding SET domain-protein 2 (NSD2), is a SET domain histone methyltransferase, responsible for the methylation of H3K36. WHSC1 protein contains proline-tryptophan-tryptophan-proline (PWWP), high-mobility group (HMG box) and plant homeodomain (PHD) domains [7]. In human, WHS is characterized by a number of key features, including prominent forehead with widely spaced eyes, strabismus, mental retardation and growth retardation, and whsc1 is the only gene deleted in all the described cases and thus is believed to be responsible for major phenotypes of this syndrome [8], [9], [10]. In consistent with this notion, whsc1−/− mice showed growth retardation and congenital cardiovascular defects [7]. Whsc1 is also found mutated by genomic translocation or by point mutation in a subgroup of B-cell derived malignancies [11], [12], [13]. Through translocation, whsc1 gene is linked with strong B-cell specific enhancers from IgH locus which results in WHSC1 overexpression. Point mutation at amino acid 1099 (E1099K) in WHSC1 enhances its methyltransferase activity and leads to altered global H3K36 methylation [14].
Hyperactivated WHSC1 has been shown to promote cell-cycle progression, clonogenicity and tumorigenesis in B-cell melanoma and leukemia, suggesting that whsc1 functions as an oncogene [15]. Recently, WHSC1 has been found highly expressed and correlated with poor outcomes in a large variety of human solid tumors [16]. Like in malignant B-cells, overexpression of WHSC1 protein was demonstrated to be responsible for cell survival and proliferation in cells from solid tumors [17]. Thus these data suggested that whsc1 functions as an oncogene and thus is a potential target for cancer therapy.
In this study, we used zebrafish as an in vivo model to study the function of whsc1. We used CRISPR/Cas9 system to introduce indel mutations in whsc1 gene and disrupt its function. We identified two independent frame-shift lines, both of which exhibited several features of WHS. We found that a large number of whsc1−/− fishes failed to develop functional swim bladder, a homologue organ to mammalian lung, possibly due to the disability of progenitor cell differentiation, which is correlated with its high expression in this organ. At junior and adult stages, whsc1−/− zebrafishes are inclined to grow tumors from swim bladder. Thus, our work suggested that whsc1 may function as a tumor suppressor by governing cell differentiation.
Materials and Methods
Zebrafish Care
All the zebrafishes used in our experiment were AB line. Zebrafishes were raised and maintained according to Kimmel et al. [18]. Embryos were cultured in tank at 28.5 °C, and water was carefully changed every three days. Adult zebrafishes were maintained in the tank with an automatic fish housing system (ESEN). 3–5 months old healthy-female and male fishes, which were separately maintained, were chosen for mating once a week. The embryos used for observing were treated with PTU before 48 h (hour).
Cas9 Target Site Design and Cas9/sgRNA Synthesis
An online tool ZIFIT Targeter (http://zifit.partners.org/zifit/Introduction.aspx) was used to design the CRISPR/Cas9 targeting sites, and the restriction endonuclease cleavage sites were analyzed by an online tool NEBcutter 2.0 (http://nc2.neb.com/NEBcutter2/). sgRNA templates were generated from pT7-gRNA vector via PCR using primers NSD2-cas1F and gRNA-R(Table 1). Cas9 expression vector pCS2-Cas9 was linearized with NotI digestion. Cas9 mRNA and sgRNA were transcribed using the mMESSAGE mMACHINE T7 ULTRA kit and MEGAshortscript™ T7 Transcription Kit respectively (Ambion). RNAs were purified by LiCl precipitation and re-dissolved in RNase-free water. The RNA quality and concentration were analyzed by electrophoresis and nucleic acid spectrometer, respectively.
Table 1.
Primers Used in the Article
| Primer name | Seqeuence (5′-3′) |
|---|---|
| RT-NSD2-F | CCCACCAACCACGCAAATA |
| RT-NSD2-R | AACGCTTCCCTGATCAGACA |
| HyNSD2-F | TGCAGAAGCAGAATCAGACAC |
| HyNSD2-R | CAGTTCAAGAACGCACTACACAC |
| NSD2-cas1F | TAATACGACTCACTATAGGAAGCAAGTACCAGCAGACGTTTTAGAGCTAGAAATAGC |
| gRNA-R | GTGGCACCGAGTCGGTGCTTTTT |
| NSD2-F | CTGCAGAGATCCGTCATCCC |
| NSD2-R | TTCGTGAGCCTCTTTTGCCT |
| RT-tcf3a-F | TGACAGAAATGACTCCGGCT |
| RT-tcf3a-R | GCATCAGAAGCTGCGATGTT |
| RT-tcf3b-F | ACCAGGCCGTCAATGTCATA |
| RT-tcf3b-R | AGGTCACTTCTTCCAGGAGC |
| RT-fz2-F | CCTGCTGCTGTTTCAACTGT |
| RT-fz2-R | GAAAAGCAAGTCTGGGGAGC |
| RT-fz7b-F | ACTTTGCACCAATGACACCC |
| RT-fz7b-R | CGTTGTTTCCCCTTGGTTGT |
| RT-hoxc6a-F | CAGAAGAAGAGGTCGCCAGA |
| RT-hoxc6a-R | TTCCATTTCATGCGCCGATT |
| RT-hoxc8a-F | GGGCAATGAAGACGGTGAAG |
| RT-hoxc8a-R | ATTGGCGCGTCTTCTCTTTC |
| RT-lef1-F | TCCTCTGGGTTGGTTCTCAC |
| RT-lef1-R | CTCCTGCTCCTTTCTCTGCT |
| RT-Sox2-F | GCATGTCCTATTCGCAGCAA |
| RT-Sox2-R | CGCTCTGGTAATGTTGGGAC |
| RT-Wnt5b-F | CTCGTGCAGCTTGAAAACCT |
| RT-Wnt5b-R | ACCAGATCTTCACCTGTCGG |
| RT-Hoxc4a-F | GCACCGTCAACTCTAGTTACAATG |
| RT-Hoxc4a-R | GTTTGGCAGCCTATGGTCTTT |
| Hyfbp2-F | AGTGTCTGTCTTTGGAGCTGG |
| Hyfbp2-R | CAGAGACCATCGACCCAACA |
| Hyctgfa-F | GCCACTTGTGTGTTTGGTGG |
| Hyctgfa-R | ATCAGTGACATTGGGCGTCA |
Microinjection of Zebrafish Embryos
One-cell stage embryos were collected for microinjection by using an electronically regulated air-pressure micro-injector (Harvard Apparatus, NY, PL1–90). Cas9 mRNA (~50 pg) and sgRNA (~100 pg) were co-injected into embryos at one-cell stage.
DNA Isolation and Mutant Analysis
Genomic DNA was extracted by using the alkaline lysis method. In brief, 48 h zebrafish larvae (about 5) or tail fins cut from adult fishes were collected into PCR tubes, treated with 30 μl alkaline lysis buffer (50 mM NaOH) at 95 °C for 10 min, then 1/10 volume neutralization buffer (1 M Tris–HCl, pH 8.0) was added to neutralize the alkaline. The target site was amplified with NSD2-F and NSD2-R primers (Table 1), and then was analyzed with restriction enzyme digestion and Sanger sequencing.
Screening of Homozygous Mutant Zebrafish Lines
The F0 zebrafishes injected with Cas9/sgRNA were outcrossed with wild-type ones, presence of germ line transmitted indels was performed as described above. F1 fishes were raised to adult for indels screening. The heterozygous F1 zebrafishes with the same mutations were selected to mate, and the F2 embryos were raised to adult for screening homozygous mutant zebrafishes.
mRNA Isolation and Real-Time PCR(RT-PCR) Analysis
Total RNA was extracted from zebrafish embryos using TRIzol reagent (Invitrogen) and cDNA was synthesized using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara). RT-PCR was performed with SsoAdvanced SYBR Green Supermix (Bio-Rad) by using a Bio-Rad CFX96 Real-Time system(primers were listed in Table 1).
Western Blots
Approximately 30 embryos at 48 hpf (hour post fertilization) were lysed in RIPA (Beyotime Institute of Biotechnology, China) in the presence of cocktail inibitor (Invitrogen). The lysate was sonicated for 10 rounds of 3 seconds at 50% power, then the samples were mixed with 5× Laemmli sample buffer and boiled for 5 minutes. WB was carried out according to standard procedure using following antibodies: H3K36Me2 (Abcam), H3K36Me3 (Abcam), H3 (Protech) and β-actin (Protech).
Histology Analysis
Euthanized fishes were fixed with 4% paraformaldehyde overnight at room temperature. For younger zebrafishes (less than 1 month old), the fixed samples were washed in water for 2 h, followed by dehydration in gradient alcohol 70% (2× 20 minutes), 80% (30 minutes), 95% (2× 20 minutes), 100% (2× 30 minutes), and xylene (30 minutes), Paraffin I (30 minutes), Paraffin II (30 minutes), Paraffins III (15 minutes), then the samples were embedded in fresh paraffin. For zebrafishes older than 1 month, the fixed samples were decalcified by EDTA for at least two weeks and washed in water overnight. Embedded samples were sectioned (5 μm) transversally or sagittally. After deparaffinized and rehydrated, the slides were used for hematoxylin–eosin staining (HE) or immune-staining.
Immunofluorescence
Embryos or slides were washed with PBS for 3 times, each time 10 minutes, and blocked with blocking buffer (PBS containing 0.5% triton-100 and 10% goat serum) for 1 h at room temperature. Next, the samples were incubated with diluted primary antibody in PBST plus 1% goat serum overnight at 4 °C with gentle agitation. Then samples were washed with PBST for 3 times and were treated with diluted secondary antibodies for 2 h at room temperature. After washing, samples were treated with DAPI solution and were viewed under a fluorescence microscope (Leica Mikrosysteme Vertrieb GmbH, Bensheim, Germany).
Whole Mount in Situ Hybridization(WISH)
Antisense probes for whsc1, fbp2, ctgfa was in vitro transcribed with a DIG labeling kit in the presence of digoxigenin (Roche). Template for in vitro transcription was amplified with specific primers (HyNSD2-F, HyNSD2-R, Hyfbp2-F, Hyfbp2-R, Hyctgfa-F and Hyctgfa-R in Table 1). WISH were performed aspreviously described [19]. Hybridized transcripts were recognized in situ with antidigoxigenin antibodies and visualized with NBT/BCIP solutions. Then the embryos were imaged under a microscope (Leica) with an attached camera.
Results
Generation of whsc1 Null Zebrafish by Using CRISPR/Cas9
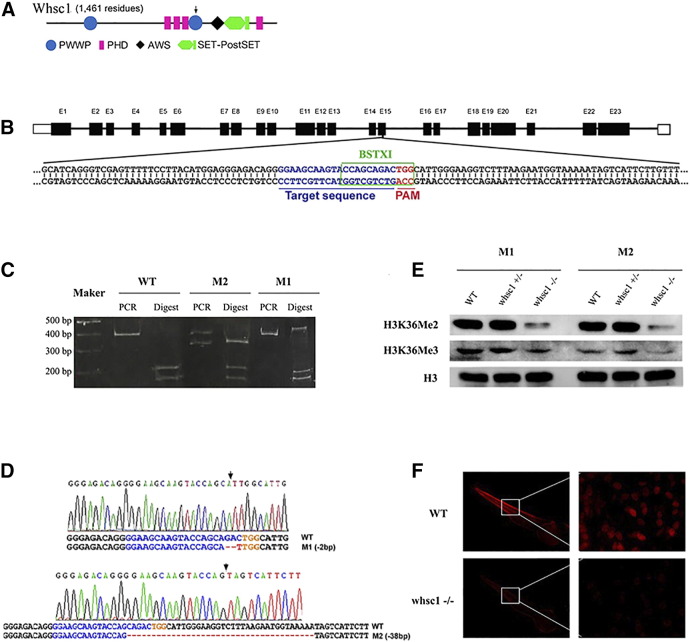
To study the function of whsc1 during the development of zebrafish, we used CRISPR/Cas9 technology to generate whsc1 knockout zebrafish. We first analyzed the genomic sequence of zebrafish whsc1, and designed a Cas9 sgRNA targeting the second PWWP domain of WHSC1, which is a conserved region and found in all vertebrate WHSC1 proteins (Figure 1, A and B). The whsc1 sgRNA and Cas9 mRNA were co-injected into the one-cell stage of zebrafish zygote, resulting in about 50% indels in its target site, as evidenced by restriction enzyme digestion analysis (data not shown). The injected embryos were raised to adulthood and screened for the presence of germ-line transmitted whsc1 mutation. Briefly, they were out-crossed with wild-type fishes and the offsprings were genotyped by PCR, restrict digestion and DNA sequencing. Three independent adult F0 zebrafishes were identified that harbor germ-line transmitted whsc1 mutation. Among these F1 fishes, two lines were kept for further research, with one harboring 2 bp deletion in exon 15 (referred as whsc1 M1) and the other harboring 38 bp deletion within the border of exon 15 and intron 15 (referred as whsc1 M2) (Figure 1, C and D).
Figure 1.
Generation of whsc1 mutant zebrafish with Crispr/Cas9 technology. (A) A diagram showing the structure of WHSC1 protein. (B) design of sgRNA against whsc1 gene. This sgRNA site targets the 15th exon. Target sequence was shown with blue letters, and PAM region was shown in red. Green box marked the reorganization site of BSTX1 enzyme that lies within sgRNA target site, loss of which indicated the presence of indels. (C) Restriction enzyme analysis of the presence of indel mutations in two independent mutant lines. (D) Sanger sequencing results of the whsc1 mutant lines. Arrows indicated the locations of indels. (E) WB analysis of the levels of H3K36Me2 and H3K36Me3 in whsc1 wild-type and mutant embryos. (F) Immunofluorescence analysis of H3K36Me2 level in whsc1 wild-type and mutant embryos. Photos of wildytpe and whsc1 mutant embryos were taken under same exposure time.
As WHSC1 is a H3K36-specific methyltransferase,we then performed WB to determine if the levels of H3K36Me2 and H3K36Me3 were reduced in the two mutation lines. As shown in Figure 1E, level of H3K36Me2 was significantly reduced in both whsc1 homozygous lines, and H3K36Me3 was slightly reduced. The reduced H3K36Me2 expression was further confirmed by immunofluorescence staining (Figure 1F). These results suggested that whsc1 is critical for bi-methylation of H3K36 during embryonic development, which is consistent with previous observations in mammals, and demonstrated that whsc1 has been knocked out successfully.
whsc1−/− Zebrafish Displayed Phenotypes Similar to WHS
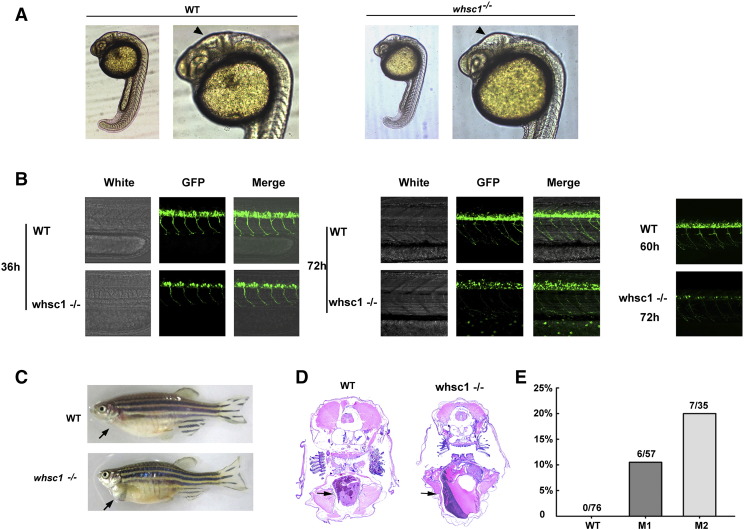
Whsc1−/− embryos did not show any obvious developmental defects before the 20-somites stage. However, at the stage of 25-somites to 30 h, the diencephalic ventricle in whsc1−/− zebrafish was apparently larger than that in wild-type embryos. Zebrafish diencephalic ventricle is a homologue of mammalian third ventricle, enlargement of which is frequently observed in WHS patients [20] (Figure 2A). Compared to age-matched wild-type embryos, whsc1−/− embryos showed reduced body length and less pigmentation (Figure 3A), suggesting that whsc1 mutation caused developmental delay, one of the key features of WHS. After hatching, whsc1−/− larvae showed obvious movement disorder. When stimulated by beating the fish tank, wild-type larvae responded quickly and swam away from the beating point, while most of whsc1−/− larvae preferred to stay at the bottom of the tank (data not shown). Therefore, we speculated that whsc1 loss-of-function might affect the development of neural system. We out-crossed whsc1−/− zebrafish with Hb9-GFP transgenic zebrafish that express GFP in motor neurons. We found that whsc1 mutation did not affect the differentiation of motor neurons because all whsc1−/− embryos express GFP signal in their spinal cord. However, the number of motor neurons was significantly reduced in whsc1−/− embryos. And the out-growth of motor neuron axons were slightly delayed (Figure 2B). At adult stage, a portion of (Figure 2E) whsc1−/− zebrafishes showed significantly enlarged pericardiums (Figure 2C), which was accompanied by ventricular dilatation and loss of cardiac muscle (Figure 2D). These phenotypes were comparable to WHS.
Figure 2.
WHS related phenotypes in whsc1−/− zebrafish. (A) Brain morphology in whsc1−/− and wild-type embryos. Arrow heads point to the diencephalic ventricle (homologue of mammalian third ventricle). (B) Motor neuron development in whsc1−/− and wild-type embryos. Note that the number of motor neurons in whsc1−/− mutant larva was significantly less than that in wild-type ones. (C) whsc1−/− adult zebrafish showed heart enlargement. (D) Tissue section analysis of the heart of wild-type or mutant adult fishes. Arrows point to the hearts. (E) Statistics of heart enlargement in these two mutation lines.
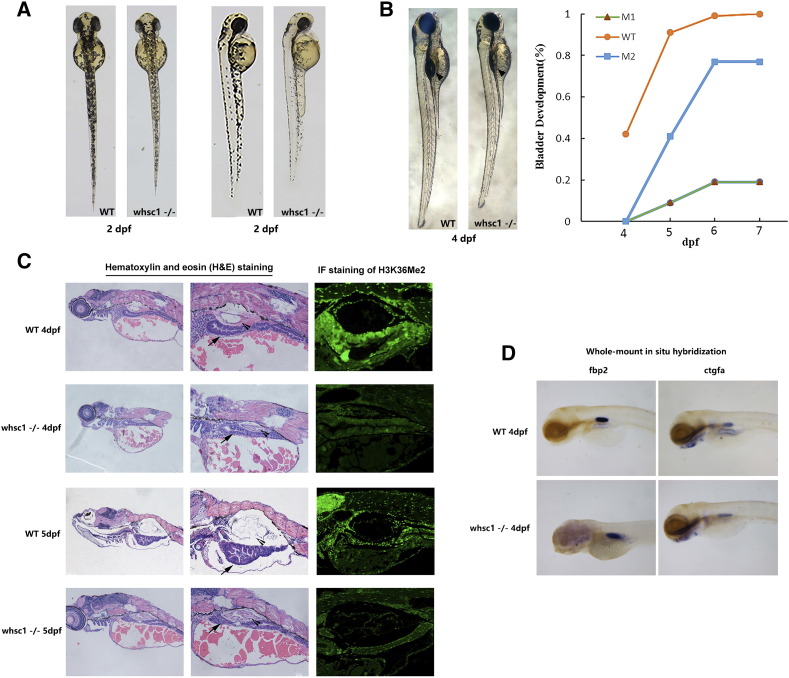
Figure 3.
whsc1−/− zebrafishes had developmental defects in swim bladder. (A) In the early development, compared to age-matched wild-type embryos, whsc1−/− embryos showed reduced body length and less pigmentation. (B) The left part shows that swim bladder inflation defects in whsc1−/− zebrafish. Arrow heads point to the swim bladders. Note that wild-type larva had already inflation, while whsc1−/− larva had not, suggesting their swim bladders were either developed abnormally or not functional. And the right part is the statistics of inflation of swim bladder in whsc1−/− zebrafish and wild-type zebrafish. (C) HE staining and immunofluorescence analysis of the swim bladder in whsc1−/− zebrafish and wild-type zebrafish at 4 dpf and 5 dpf. Arrow heads pointed to the bladder and arrows pointed to the gut. In the HE staining of zebrafish larvae at 4 and 5 dpf (left and middle panels), bladders in wild-type larvae had formed a three layer structure that was filled with air. However, bladders in whsc1 mutants were still cell masses and failed to develop three layers. Immunofluorescence staining of the H3K36me2 revealed that whsc1−/− larvae expressed significantly lower level of H3K36me2 than wild-type larvae did. (D) WISH analysis of swim bladder markers fbp2 and ctgfa in whsc1−/− zebrafish and wild-type zebrafish at 4 dpf.
whsc1−/− Affected the Development of Zebrafish Swim Bladder
In addition to these phenotypes, we also observed that a large portion of whsc1−/− embryos had defects in their swim bladder (Figure 3B). In wild-type zebrafishes, swim bladder began to inflate at about 4 dpf (day post fertilization), as shown in Figure 3B, 47.2% (58/123) wild-type larvae showed inflated swim bladder, while no obvious inflation in whsc1−/− larvae. At 6 and 7 dpf, 95.9% of wild-type zebrafishes showed inflated swim bladder, while only 20% M1 larvae (n = 125) and 74.2% M2 larvae (n = 132) showed apparent inflation, but much smaller than wild-type larvae. And the bladder of the mutant larvae without any inflation in the early 7 days couldn’t inflate anymore and most of them died out in the following two weeks.
To characterize the swim bladder of whsc1−/− larvae, we performed tissue section and whole-mount in situ hybridization (WISH) analyses. Tissue section analysis showed that the swim bladders of 4 dpf and 5 dpf wild-type larvae were formed with three layers, which were the outer mesothelium, the mesenchymal smooth muscle layer and inner epithelial layer (Figure 3C). However, the bladders of mutation larvae lost the normal three-layer structure and developed into a cell mass with increased cellularity. Cells within the bladder showed abnormal shape with increased cytoplasmic content, indicating that cell differentiation and arrangement were defective. WISH analysis with two early swim bladder markers, fbp2 and ctgfa, showed that whsc1 mutation did not affect both genes, suggesting that differentiation of endoderm into swim bladder progenitors is not affected in whsc1−/− larvae (Figure 3D). And the immunofluorescence analysis showed that the expression level of H3K36Me2 in mutant larvae was lower than that in wild-type ones. These results indicated that whsc1 is responsible for further differentiation of swim bladder progenitor cells into terminally differentiated functional cells.
Expression of whsc1 mRNA
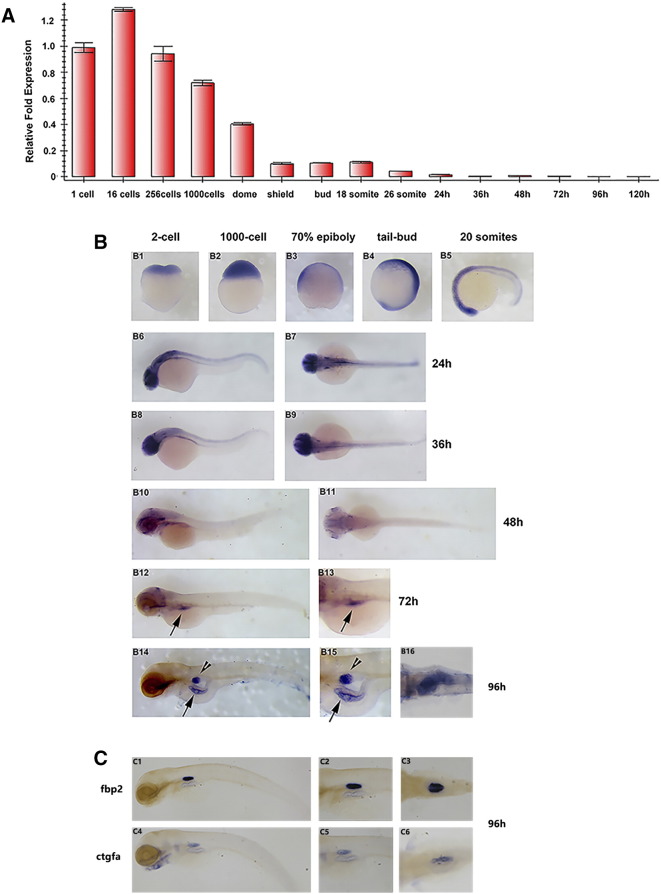
To investigate the whsc1 expression pattern during early embryonic development, embryos at different stages were collected for RT-PCR and WISH analysis. As shown in Figure 4A, whsc1 mRNA was detected by RT-PCR in all stages studied, suggesting that it is expressed both maternally and zygotically during zebrafish embryonic development. WISH analysis (Figure 4B) revealed that whsc1 mRNA was ubiquitously distributed at blastula and gastrula stages. At 20 somites stage to pharyngulation stage (36 hpf), whsc1 mRNA was mainly detected in neural tissue. Then, at 48 hpf, its level in neural tissue became very weak. At 72 and 96 hpf, whsc1 mRNA was mainly observed in endoderm derived tissues, such as the swim bladder and gut. To confirm the expression domain of whsc1 in swim bladder, we stained two additional swim bladder markers, fbp2 and ctgfa, in parallel (Figure 4C). These results suggested that the expression pattern of whsc1 was highly correlated with its developmental function.
Figure 4.
whsc1 expression pattern during early embryonic development. (A) Quantitative PCR analysis of whsc1 mRNA levels in embryos at various developmental stages. Relative ratio (fold changes) obtained from 1-cell stage embryos was set to 1. (B) Whole mount in situ hybridization analysis of whsc1 mRNA expression pattern in embryos at various developmental stages. B1-B2 were lateral view; B3-B4 were lateral view with dorsal to the right; B7, B9, B11 and B16 were dorsal view with head to the left; the rest were lateral view with head to the left. Note that whsc1 was expressed specifically in the swim bladder and gut (B14–15). Arrowheads point to the bladder and arrows point to the gut. (C) WISH analysis of swim bladder markers fbp2 and ctgfa. C1–2 and C4–5 were shown in lateral view with head to the left; C3 and C6 were shown in dorsal view with head to the left.
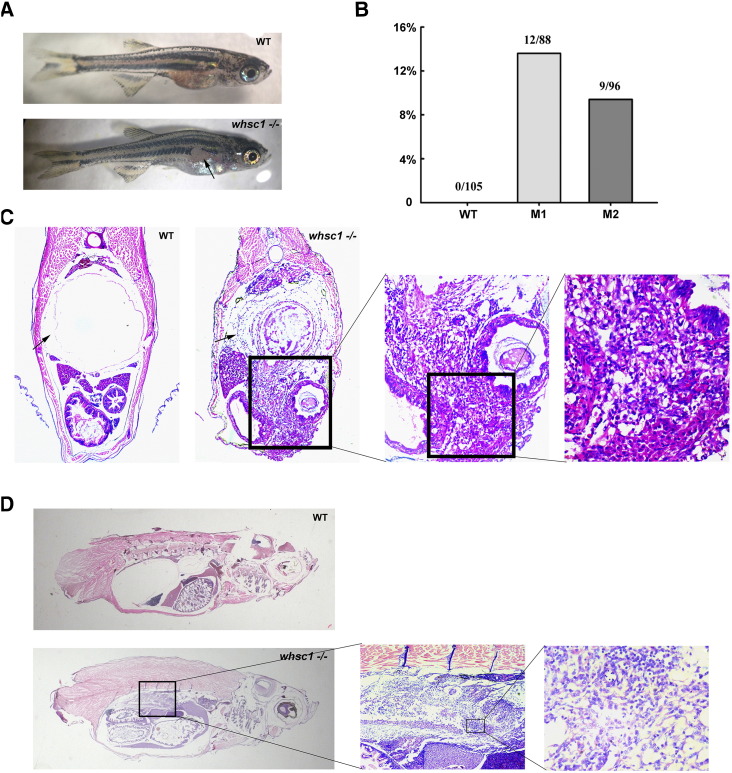
Swim Bladder of whsc1−/− Zebrafish Grew Tumors
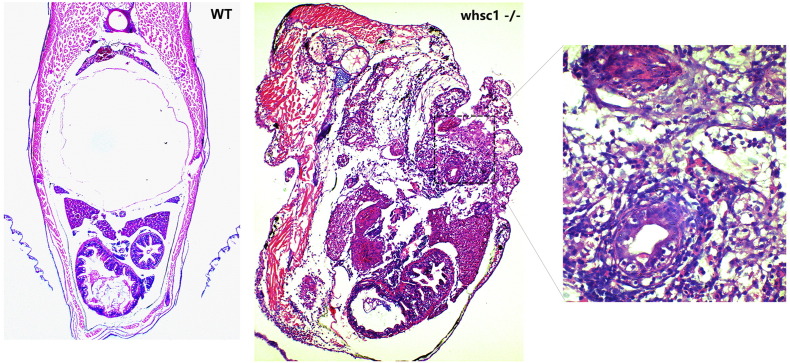
At 1 month old, a portion of whsc1−/− zebrafishes grew abnormal swells in their abdomen and eventually led to anabrosis in their skins, while no such swells were found in wild-type fishes (Figure 5, A and B). To determine if these swells were indeed tumors, we performed tissue section analysis on a panel of fishes with swells and anabrosis. Histologically, these swells were composed of cells that showed key features of tumor cells such as condensed chromatins, and prominent nucleoli. And these cells showed significant invasiveness (Figure 5C; Supplementary Figure S1). We further analyzed a panel of adult fishes by tissue section and found 18.2% of them (n = 11) grow tumors. Continuous sections revealed that those tumor cells were originated from swim bladder, which showed abnormal morphology, and were not occupied with air, but with tumor cells and extra-cellular matrix (Figure 5D).
Figure 5.
whsc1 mutant zebrafish developed swim bladder lesions. (A) One-month-old whsc1−/− zebrafishes developed skin lesions. Arrow pointed to the lesion. (B) The Statistics of wild-type and whsc1 mutant fishes with lesions. (C) HE staining of swim bladder in one-month-old wild-type and whsc1 mutant zebrafishes (cross-section). Tissues were serially sectioned from head to tail, and sections containing swim bladders (wild-type fishes) or skin lesions (mutant fishes) were HE stained for analysis. (D) HE analysis of swim bladder in three-month-old zebrafish (vertical section). Note that wild-type fish had normal and functional swim bladder, while whsc1 mutant fishes had no obvious swim bladder. The position of swim bladder in whsc1 mutant was occupied with cell masses that seemed to have no cell polarity.
Supplementary Figure S1.
HE analysis of pathological tissue in one-month-old zebrafish (cross-section).
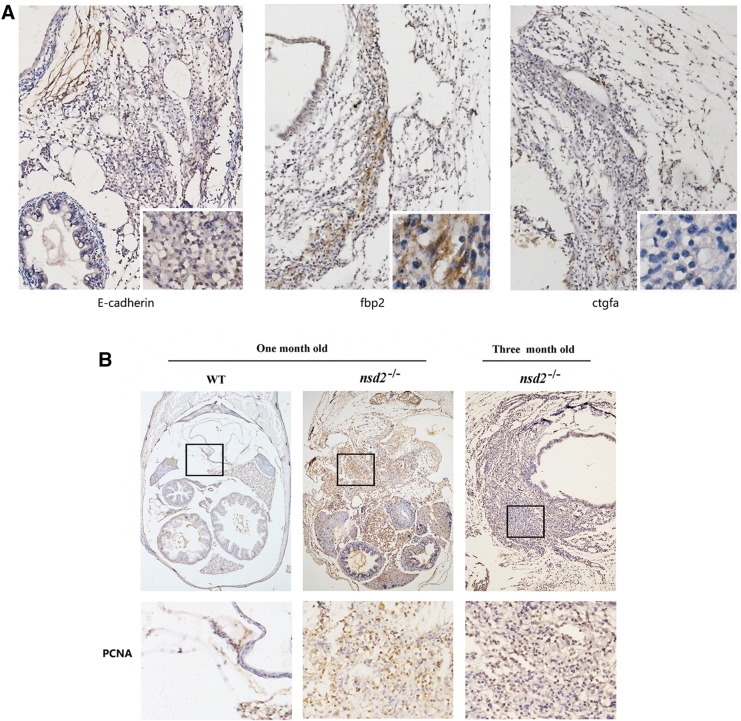
IHC staining revealed that these tumor cells were E-cadherin positive, suggesting that they were originated from epithelial cells (Figure 6A). We also stained the tumors with swim bladder specific markers, fbp2 and ctgfa, and found tumor cells were fbp2 positive but ctgfa negative (Figure 6A). Importantly, these tumor cells showed strong staining of proliferating cell nuclear antigen (PCNA), which is a specific marker of proliferating cells (Figure 6B). Taken together, these data demonstrated that the swells found in whsc1−/− zebrafishes were indeed tumors.
Figure 6.
IHC staining of the zebrafish swim bladder tumor. (A) IHC staining of E-cadherin, fbp2 and ctgfa in tumor tissues of whsc1 mutant zebrafishes. (B) IHC staining of the PCNA expression in normal zebrafish swim bladder and tumor tissues of whsc1 mutant fishes.
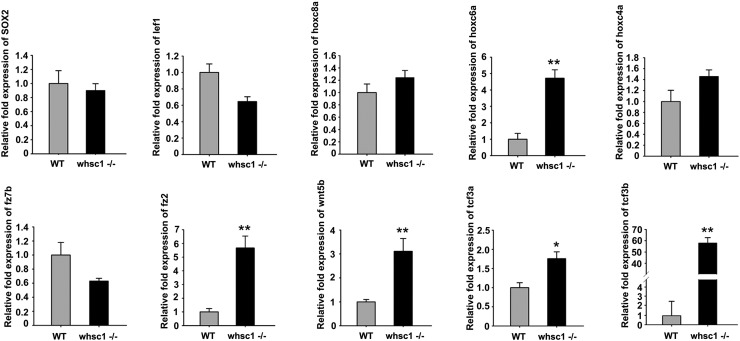
whsc1−/− Zebrafish Exhibited Abnormal Expression of Genes Involved in Swim Bladder Formation
To further investigate the molecular mechanism of swim bladder defects, we examined if the whsc1 loss-of-function could affect the expression of genes that are critical for swim bladder development. By using quantitative-PCR(qPCR), we analyzed the expression of a panel of genes that are essential for foregut differentiation and early swim bladder formation, including fz2, fz7b, lef1, sox2, tcf3a, tcf3b, wnt5b, hoxc4a, hoxc6a and hoxc8a [21], [22]. We found that the expression of tcf3b, a key suppressor of wnt signaling pathway, was dramatically increased in whsc1−/− zebrafishes at 48 hpf. The expression of fz2, hoxc6a, tcf3b and wnt5b was much higher in whsc1−/− zebrafishes than that in wild-type ones (Figure 7). Coincidently, abnormal expression of hoxc6a [23], [24], [25] and wnt5b [26], [27], [28] have been shown to be related with cancers.
Figure 7.
whsc1−/− zebrafish exhibited disordered expression of genes involved in swim bladder formation. mRNA levels of indicated genes were normalized to b-actin. Relative ratios (fold changes) obtained from wild-type embryos were set to 1. Note that expression of several key genes in swim bladder development and wnt signaling pathway, including tcf3b/3a, wnt5b and fz2, was significantly up-regulated.
Discussion
Histone methylation is one of the major epigenetic mechanisms that occurs at specific amino acids of the histone proteins [29], [30]. It plays fundamental roles in gene regulation and cell differentiation by forming synergistic signaling platforms within specific genomic loci, which are recognized by specific receptors, regulating transcription and chromatin remodeling [31]. Depending on which amino acids in the histone is modified and how many methyl groups are added, histone methylation can either increase or decrease the transcription of target genes [32], [33]. Several histone methyltranferases were found mutated in inherited diseases. For example, whsc1, encoding a protein in charge of H3K36 methylation, had been found deleted in all cases of WHS. Whsc1, also named nsd2, belongs to NSD family that includes nsd1, nsd2 and nsd3. Mutations in nsd1 are related to another inherited disease, Sotos syndrome. WHSC1 protein mainly catalyzes bi-methylation of H3K36, which is usually associated with actively transcribed regions and has been proposed to provide landmarks for continuing transcription. We showed that zebrafishes with whsc1 mutation developed several key features of WHS, including growth retardation, motor neuron proliferation and heart failure. Together with the observations in mice, our data confirmed that deletion of whsc1 gene is the major cause of WHS. Besides these phenotypes, we noticed that a large number of whsc1−/− fishes failed to develop functional swim bladder, a homologue organ to mammalian lung. In agreement with this, we detected high level of whsc1 expression in swim bladder, especially in the anterior part. Defective swim bladder development is also observed in other zebrafish mutants, which were concluded to be secondary to developmental delay or other phenotypes. As mentioned above, whsc1−/− embryos had obvious developmental delay characterized by less pigmentation and reduced body length, therefore developmental delay may in part responsible for the swim bladder phenotypes in whsc1 mutant.
At junior and adult stage, whsc1−/− fishes are inclined to develop tumors, which were demonstrated to be originated from their swim bladders. It is not surprising that mutations in epigenetic factors lead to tumorigenesis, because epigenetic mechanisms had long been recognized as fundamental driving force during genesis and progression of cancer cells. Accumulating evidences showed that the malignant transformation of normal cells requires an extensive epigenomic reconfiguration [34], [35], [36]. Being one of the major epigenetic mechanisms, histone modifications were often found dysregulated in cancer cells, which led to uncontrolled gene expression and cell proliferation. In prostate and ovarian tumors, a decreased expression of H3K9 acetylation (H3K9Ac) was correlated with high histological grading and poor clinical outcomes [37], [38], [39]. However, in hepatocellular carcinomas and gastric adenocarcinoma, an increase in H3K9Ac levels and decrease in H3K9Me3 were associated with poor prognosis, suggesting that uncontrolled levels of histone modifications can either promote or inhibit cancerous phenotypes depending on specific tissues [40].
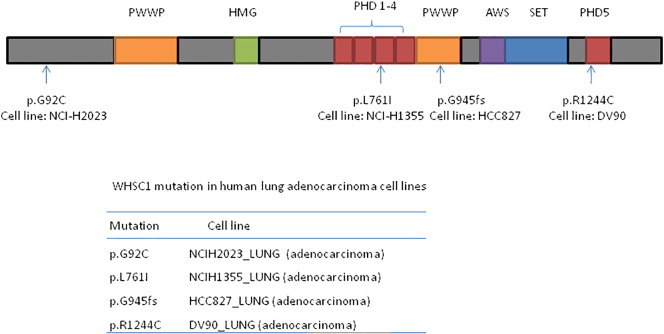
Abnormal histone modifications were resulted from either mutation or mis-expression of histone modifiers or their regulators. Whsc1 is frequently overexpressed in many solid tumors in comparison to the corresponding normal tissues. In addition, in several subtypes of B-cell derived lymphoma and leukemia, Whsc1 is mutated by genome translocation or point mutation, leading to WHSC1 hyper-activation [12], [41], [42]. Both of these conditions led to up-regulation of H3K36 methylation and uncontrolled expression of downstream target genes. In vivo experiments, artificial overexpression or knock down of Whsc1 could reduce malignant phenotypes. These data suggested that WHSC1 is an oncoprotein, at least in certain types of malignant cells. However, our work showed that fishes with whsc1 loss-of-function are inclined to grow tumor from swim bladder. We showed that whsc1 loss-of-function caused significant reduction in the levels of bi- and tri-methylation of H3K36 and led to developmental defects of swim bladder, which was possibly due to differentiation defects of progenitor cells. It is well known that disability of progenitor cell to differentiate is one of the most common driving forces in tumorigenesis [43], [44], [45], [46], [47], [48], [49], [50], [51]. A good example is the acute promyeloid leukemia, in which PML/RARα fusion protein generated by genome translocation (t15:17) significantly inhibits promyelocytes from differentiating past the promyeloid stage and leads to uncontrolled proliferation of leukemia cells [51]. Thus, based on our observations, it is reasonable to propose that whsc1 can also function as a tumor suppressor in certain conditions by permitting cell differentiations. In agreement with these results, we also found whsc1 is recurrently mutated in human lung adenocarcinomas through analyzing Cancer Cell Line Encyclopedia (CCLE) database (n = 67, Supplemental Figure S2).
Supplementary Figure S2.
Recurrent Whsc1 mutations identified in CCLE data base. (A) Scheme diagram showing the distribution of mutations in WHSC1 protein. (B) WHSC1 mutations in lung adenocarcinoma cell lines.
Conclusion
In summary, we created a whsc1 knockout zebrafish model and discovered an unexpected function of whsc1 in swim bladder development. We showed that whsc1 loss-of-function in zebrafish embryos led to significant down-regulation of bi-methylation of H3K36, dysregulation of gene expression and defects in differentiation of swim bladder progenitor cells, which eventually resulted in tumorigenesis. Thus, our work suggested that whsc1 may function as a tumor suppressor by governing progenitor cells differentiation.
The following are the supplementary data related to this article.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81372143, No. 31000646 and No. 81123003).
Contributor Information
Shaohua Yao, Email: shaohuayao@scu.edu.cn.
Yuquan Wei, Email: yuquanwei@scu.edu.cn.
References
- 1.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 2.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 3.Huertas D, Sendra R, Munoz P. Chromatin dynamics coupled to DNA repair. Epigenetics. 2009;4:31–42. doi: 10.4161/epi.4.1.7733. [DOI] [PubMed] [Google Scholar]
- 4.Hajdu I, Ciccia A, Lewis SM, Elledge SJ. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci U S A. 2011;108:13130–13134. doi: 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith O.M., Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 8.Hirschhorn K, Cooper HL. Chromosomal aberrations in human disease. A review of the status of cytogenetics in medicine. Am J Med. 1961;31:442–470. doi: 10.1016/0002-9343(61)90128-0. [DOI] [PubMed] [Google Scholar]
- 9.Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, Altherr MR, den Dunnen JT. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 10.Wright TJ, Ricke DO, Denison K, Abmayr S, Cotter PD, Hirschhorn K, Keinanen M, McDonald-McGinn D, Somer M, Spinner N. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet. 1997;6:317–324. doi: 10.1093/hmg/6.2.317. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Jima D, Moffitt AB, Liu Q, Czader M, Hsi ED, Fedoriw Y, Dunphy CH, Richards KL, Gill JI. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123:2988–2996. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyer JAHX, Zheng Y. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28:198–201. doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudlebusch HR, Theilgaard-Monch K, Lodahl M, Johnsen HE, Rasmussen T. Identification of ID-1 as a potential target gene of MMSET in multiple myeloma. Br J Haematol. 2005;130:700–708. doi: 10.1111/j.1365-2141.2005.05664.x. [DOI] [PubMed] [Google Scholar]
- 14.Oyer JA, Huang X, Zheng Y, Shim J, Ezponda T, Carpenter Z, Allegretta M, Okot-Kotber CI, Patel JP, Melnick A. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28:198–201. doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Z, Wu H, Chuai S, Xu F, Yan F, Englund N, Wang Z, Zhang H, Fang M, Wang Y. NSD2 is recruited through its PHD domain to oncogenic gene loci to drive multiple myeloma. Cancer Res. 2013;73:6277–6288. doi: 10.1158/0008-5472.CAN-13-1000. [DOI] [PubMed] [Google Scholar]
- 16.Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiaer E., Rossing HH, Johansen JV, Jorgensen M, Sauter G, Helin K. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17:2919–2933. doi: 10.1158/1078-0432.CCR-10-1302. [DOI] [PubMed] [Google Scholar]
- 17.Saloura V, Cho HS, Kiyotani K, Alachkar H, Zuo Z, Nakakido M, Tsunoda T, Seiwert T, Lingen M, Licht J. WHSC1 promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol Cancer Res. 2015;13:293–304. doi: 10.1158/1541-7786.MCR-14-0292-T. [DOI] [PubMed] [Google Scholar]
- 18.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 19.Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 20.Righini A, Ciosci R, Selicorni A, Bianchini E, Parazzini C, Zollino M, Lodi M, Triulzi F. Brain magnetic resonance imaging in Wolf-Hirschhorn syndrome. Neuropediatrics. 2007;38:25–28. doi: 10.1055/s-2007-981685. [DOI] [PubMed] [Google Scholar]
- 21.Yin A, Korzh S, Winata CL, Korzh V, Gong Z. Wnt signaling is required for early development of zebrafish swimbladder. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng W, Wang Z, Collins JE, Andrews RM, Stemple D, Gong Z. Comparative transcriptome analyses indicate molecular homology of zebrafish swimbladder and mammalian lung. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujiki K, Duerr EM, Kikuchi H, Ng A, Xavier RJ, Mizukami Y, Imamura T, Kulke MH, Chung DC. Hoxc6 is overexpressed in gastrointestinal carcinoids and interacts with JunD to regulate tumor growth. Gastroenterology. 2008;135:907–916. doi: 10.1053/j.gastro.2008.06.034. [916 e901-902] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramachandran S, Liu P, Young AN, Yin-Goen Q, Lim SD, Laycock N, Amin MB, Carney JK, Marshall FF, Petros JA. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q., Jin XS, Yang ZY, Wei M, Liu BY, Gu QL. Upregulated Hoxc6 expression is associated with poor survival in gastric cancer patients. Neoplasma. 2013;60:439–445. doi: 10.4149/neo_2013_057. [DOI] [PubMed] [Google Scholar]
- 26.Takeshita A, Iwai S., Morita Y, Niki-Yonekawa A, Hamada M, Yura Y. Wnt5b promotes the cell motility essential for metastasis of oral squamous cell carcinoma through active Cdc42 and RhoA. Int J Oncol. 2014;44:59–68. doi: 10.3892/ijo.2013.2172. [DOI] [PubMed] [Google Scholar]
- 27.Sun D, Qin L., Xu Y, Liu JX, Tian LP, Qian HX. Influence of adriamycin on changes in Nanog, Oct-4, Sox2, ARID1 and Wnt5b expression in liver cancer stem cells. World J Gastroenterol. 2014;20:6974–6980. doi: 10.3748/wjg.v20.i22.6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato S, Hayakawa Y, Sakurai H, Saiki I, Yokoyama S. Mesenchymal-transitioned cancer cells instigate the invasion of epithelial cancer cells through secretion of WNT3 and WNT5B. Cancer Sci. 2014;105:281–289. doi: 10.1111/cas.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prusov AN, Smirnova TA, Kolomijtseva GY. Influence of chromatin structure, antibiotics, and endogenous histone methylation on phosphorylation of histones H1 and H3 in the presence of protein kinase A in rat liver nuclei in vitro Biochemistry. Biokhimiia. 2013;78:176–184. doi: 10.1134/S0006297913020065. [DOI] [PubMed] [Google Scholar]
- 30.Ginsburg DS, Anlembom TE, Wang J, Patel SR, Li B, Hinnebusch AG. NuA4 links methylation of histone H3 lysines 4 and 36 to acetylation of histones H4 and H3. J Biol Chem. 2014;289:32656–32670. doi: 10.1074/jbc.M114.585588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler JS, Koutelou E, Schibler AC, Dent SY. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4:163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.du Preez LL, Patterton HG. Secondary structures of the core histone N-terminal tails: their role in regulating chromatin structure. Subcell Biochem. 2013;61:37–55. doi: 10.1007/978-94-007-4525-4_2. [DOI] [PubMed] [Google Scholar]
- 33.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 34.Kanwal R, Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81:303–311. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ushijima T. Cancer epigenetics: now harvesting fruit and seeding for common diseases. Biochem Biophys Res Commun. 2014;455:1–2. doi: 10.1016/j.bbrc.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Vogel T, Lassmann S. Epigenetics: development, dynamics and disease. Cell Tissue Res. 2014;356:451–455. doi: 10.1007/s00441-014-1916-7. [DOI] [PubMed] [Google Scholar]
- 37.Zhen L, Gui-lan L, Ping Y, Jin H, Ya-li W. The expression of H3K9Ac, H3K14Ac, and H4K20TriMe in epithelial ovarian tumors and the clinical significance. Int J Gynecol Cancer. 2010;20:82–86. doi: 10.1111/IGC.0b013e3181ae3efa. [DOI] [PubMed] [Google Scholar]
- 38.Mohamed MA, Greif PA, Diamond J, Sharaf O, Maxwell P, Montironi R, Young RA, Hamilton PW. Epigenetic events, remodelling enzymes and their relationship to chromatin organization in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. BJU Int. 2007;99:908–915. doi: 10.1111/j.1464-410X.2006.06704.x. [DOI] [PubMed] [Google Scholar]
- 39.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 40.Park YS, Jin MY, Kim YJ, Yook JH, Kim BS, Jang SJ. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann Surg Oncol. 2008;15:1968–1976. doi: 10.1245/s10434-008-9927-9. [DOI] [PubMed] [Google Scholar]
- 41.Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, Bhang HE, Taylor JE, Hu M, Englund NP. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45:1386–1391. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huether R, Dong L, Chen X, Wu G, Parker M, Wei L, Ma J, Edmonson MN, Hedlund EK, Rusch MC. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simons JW. Genetic, epigenetic, dysgenetic and non-genetic mechanisms in tumorigenesis. II. Further delineation of the rate limiting step. Anticancer Res. 1999;19:4781–4789. [PubMed] [Google Scholar]
- 44.Piao YJ. Dedifferentiation and regeneration of damaged cells and tissues. Di Yi Jun Yi Da Xue Xue Bao. 2004;24:736–737. [PubMed] [Google Scholar]
- 45.Hewitt Z, Priddle H, Thomson AJ, Wojtacha D, McWhir J. Ablation of undifferentiated human embryonic stem cells: exploiting innate immunity against the Gal alpha1-3Galbeta1-4GlcNAc-R (alpha-Gal) epitope. Stem Cells. 2007;25:10–18. doi: 10.1634/stemcells.2005-0481. [DOI] [PubMed] [Google Scholar]
- 46.Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harb Symp Quant Biol. 2008;73:171–174. doi: 10.1101/sqb.2008.73.041. [DOI] [PubMed] [Google Scholar]
- 47.Moritani S, Ichihara S, Hasegawa M, Takada S, Takahashi T, Kato E, Mii S, Iwakoshi A. Dedifferentiation and progression of an intracranial solitary fibrous tumor: autopsy case of a Japanese woman with a history of radiation therapy of the head during infancy. Pathol Int. 2011;61:143–149. doi: 10.1111/j.1440-1827.2010.02627.x. [DOI] [PubMed] [Google Scholar]
- 48.Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Bai F, Chan HL, Scott A, Smith MD, Fan C, Herschkowitz JI, Perou CM, Livingstone AS, Robbins DJ, Capobianco AJ. BRCA1 suppresses epithelial-to-mesenchymal transition and stem cell dedifferentiation during mammary and tumor development. Cancer Res. 2014;74:6161–6172. doi: 10.1158/0008-5472.CAN-14-1119. [DOI] [PubMed] [Google Scholar]
- 50.Zaragosi LE, Dadone B, Michiels JF, Marty M, Pedeutour F, Dani C, Bianchini L. Syndecan-1 regulates adipogenesis: new insights in dedifferentiated liposarcoma tumorigenesis. Carcinogenesis. 2015;36:32–40. doi: 10.1093/carcin/bgu222. [DOI] [PubMed] [Google Scholar]
- 51.Kakizuka A, Miller WH, Jr., Umesono K, Warrell RP, Jr., Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]