Abstract
Interest in preclinical drug development for ovarian cancer has stimulated development of patient-derived xenograft (PDX) or tumorgraft models. However, the unintended formation of human lymphoma in severe combined immunodeficiency (SCID) mice from Epstein-Barr virus (EBV)–infected human lymphocytes can be problematic. In this study, we have characterized ovarian cancer PDXs which developed human lymphomas and explore methods to suppress lymphoproliferative growth. Fresh human ovarian tumors from 568 patients were transplanted intraperitoneally in SCID mice. A subset of PDX models demonstrated atypical patterns of dissemination with mediastinal masses, hepatosplenomegaly, and CD45-positive lymphoblastic atypia without ovarian tumor engraftment. Expression of human CD20 but not CD3 supported a B-cell lineage, and EBV genomes were detected in all lymphoproliferative tumors. Immunophenotyping confirmed monoclonal gene rearrangements consistent with B-cell lymphoma, and global gene expression patterns correlated well with other human lymphomas. The ability of rituximab, an anti-CD20 antibody, to suppress human lymphoproliferation from a patient's ovarian tumor in SCID mice and prevent growth of an established lymphoma led to a practice change with a goal to reduce the incidence of lymphomas. A single dose of rituximab during the primary tumor heterotransplantation process reduced the incidence of CD45-positive cells in subsequent PDX lines from 86.3% (n = 117 without rituximab) to 5.6% (n = 160 with rituximab), and the lymphoma rate declined from 11.1% to 1.88%. Taken together, investigators utilizing PDX models for research should routinely monitor for lymphoproliferative tumors and consider implementing methods to suppress their growth.
Abbreviations: PDX, patient-derived xenograft; TALs, tumor-associated lymphocytes
Introduction
Ovarian cancer is the most lethal gynecologic malignancy with heterogeneous biology and unpredictable clinical behavior [1]. The development of animal models to recapitulate human disease is critical for understanding its complex biology and to develop novel therapies. Classic cell lines derived from tumor tissue not only lack in vivo peritumoral components but contain significant genomic differences from high-grade serous ovarian cancer [2]. In contrast, ovarian cancer patient-derived xenograft (PDX) models closely recapitulate the histologic, molecular, and drug response characteristics of the matched primary patient tumor [3]. Accordingly, interest in PDXs for preclinical drug development has led to multi-institutional and international efforts to develop PDX models to better understand cancer biology and develop better therapies [4], [5].
A potential pitfall with creating and using PDX models is the potential for unintended co‐heterotransplantation of non‐carcinoma cell types. In ovarian cancer PDX models, tumor-associated plasma B cells can be detected by immunohistochemistry (IHC) of early-passage PDX tissue [6], [7], and the presence of circulating human immunoglobulin (Ig) G can be detected for up to 4 months after implantation [6]. Although this might be desirable for investigations of immunotherapies in these models, the potential for neoplastic transformation of lymphocytes or spontaneous transformation of murine lymphocytes [8] can confound PDX-based in vivo studies. Indeed, examples of human lymphocytes transformed in immunocompromised murine hosts after heterotransplantation with adenocarcinoma has been reported for PDXs derived from lung, colon, gastric, liver, breast, bladder, renal, and prostate cancer [9], [10], [11], [12], [13]. Although preliminary data with similar findings have been previously presented in ovarian PDX models [14], to our knowledge, this process has not been well characterized in ovarian PDX tissues, and strategies to manage lymphocyte transformation have not been reported.
Herein we describe our experience with 277 ovarian tumor transplants in severe combined immunodeficiency (SCID) mice. Although the majority of PDXs recapitulate the histologic, molecular, and therapeutic response of the source patient tumor [3], a subset of PDX tumors is CD45 positive and represents a clonal outgrowth of malignant B cells which are inhibited by rituximab without impacting ovarian tumor engraftment.
Materials and Methods
Tumor Engraftment and Cryopreservation
Fresh tissues from consenting, chemotherapy-naïve patients with ovarian, primary peritoneal, or fallopian tube cancer were collected at the time of primary debulking surgery at Mayo Clinic, Rochester. All biospecimens were coded with a patient heterotransplant (PH) number to protect patient identity in accordance with the Mayo Clinic Institutional Review Board and in accordance with the Health Insurance Portability and Accountability Act regulations through the Mayo Clinic Ovarian Tumor Repository. Tumorgrafts were developed as previously described by intraperitoneal injection into female SCID beige mice (C.B.-17/IcrHsd-Prkdcscid Lystbg; ENVIGO) [3], also in accordance with the Mayo Clinic Institutional Animal Care and Use Committee. Briefly, ~0.3 ml of minced fresh patient tumor was mixed 1:1 with McCoy's media in a 1-ml syringe and injected intraperitoneally through a 0.5-inch 16-gauge needle. No enzymatic or mechanical tumor dissociation was performed. Mice were monitored by routine palpation for engraftment, and tumors were harvested when moribund.
Tissue Processing and IHC
Tissues collected from mice or patients were fixed overnight in buffered formalin (Cat#23-011-120; Thermo Scientific, Wilmington, DE) and processed in the tissue core facility at Mayo Clinic, AZ. Deparaffinized and rehydrated 5- to 6-μm sections were unmasked for 15 minutes in EDTA buffer (1 mM EDTA, 0.05% Tween 20, pH 8.0) at 95°C to 99°C. Primary antibodies to determine epithelial and lymphocytic populations were purchased from Life Technologies (pan-cytokeratin clone AE1/AE3 at 1:300) and Dako North America (CD45 clone 2B11 + PD7/26 at 1:500, human specific) and incubated overnight at 4°C. Secondary antibody (Cat#8125S; Signal Stain Boost IHC detection system; Cell Signaling Technologies, Schwalbach, Germany) was applied for 30 to 60 minutes at room temperature. Chromogenic detection of protein expression was determined in the presence of DAB (Cat#DS900H, Betazoid DAB Chromogen Kit; Biocare Medical, Concord, CA) and visualized by light microscopy. A DakoCytomation autostainer was used for anti-CD20 (Cat# N1502; Dako North America, Carpinteria, CA) and anti-CD3 (Cat# A0452; DakoCytomation). The slides were removed from autostainer, rinsed, and counterstained with hematoxylin.
RNA Hybridization and Microarray Clustering
Purification of genomic RNA and DNA was simultaneously extracted following the manufacturer's protocol for Qiagen AllPrep DNA/RNA mini Kit (Cat#80204; Qiagen, Venlo, Netherlands). Nucleic acid concentration and purity were determined on a Thermo Scientific NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Scientific, Wilmington, DE). All samples met RNA integrity number and validated Agilent (Agilent Technologies, Santa Clara, CA) criteria. Total tumorgraft RNA was analyzed by Affymetrix HG U133 plus 2.0 arrays at the Mayo Medical Genome Facility according to the manufacturer's protocol.
Gene expression arrays were preprocessed and normalized by robust multichip analysis [15]. For differential gene expression studies, epithelial versus lymphoma tumorgrafts were compared using the linear models and empirical Bayes methods that share information across genes to estimate variance (LIMMA) [16], [17] and Java TreeView [18], [19]. Study samples and the Stanford NCI60 Cancer arrays underwent unsupervised, hierarchical clustering. The NCI 60 contains expression data for 64 cancers, including ovarian and leukocyte derived (lymphoid). Tree-structured dendrograms and heat maps were surveyed in an unsupervised fashion for clustering.
Epstein-Barr Virus Detection
Nucleic acid from each tissue specimen was extracted into elution buffer using the MagNA Pure (Roche Diagnostics, Indianapolis, IN) Total Nucleic Acid program. Quantitation for the presence of the Epstein-Barr virus (EBV) DNA was accomplished with the LightCycler 2.0 EBV Quant Kit (Roche Molecular Diagnostics, Pleasanton, CA) using standard real-time polymerase chain reaction (PCR) cycling conditions.
Lymphocyte Clonality Studies
Clonality assessment was used to discriminate between malignant and reactive lymphoproliferation. Final molecular clonality was interpreted using clinical, morphologic, and immunophenotypic features in collaboration with hematologists, pathologists, and immunologists [20]. Using a B-cell immunophenotyping PCR assay, 39 fluorescently tagged primers amplified microsatellite loci (InVivoScribe Clonality Assay, San Diego, CA). The primers were designed to amplify all theoretical rearrangements of the Ig heavy and Ig kappa light chain genes. Amplicon product fragments were separated by capillary gel electrophoresis using the Applied Biosystems machinery. Microsatellite analysis reported mean fluorescence intensity of base pair size fragments using Gene Mapper Software (Life Technologies, Carlsbad, CA). Immunoglobulin gene rearrangements producing clonal peaks were classified as monoclonal/oligoclonal or polyclonal when present in established base pair size ranges.
Detection of Human and Mouse Transcripts by Real-Time PCR
Snap-frozen tumor tissues were pulverized to a fine powder in liquid nitrogen by using the Cellcrusher Tissue Pulverizer (Cell Crusher Limited, Cork, Ireland) on dry ice, and total RNA was then extracted using the standard protocol for Qiagen RNeasy mini kit (Cat #74106; Qiagen, Venlo, Netherlands) with on-column DNA digestion. Total RNA concentration and purity were determined spectrophotometrically (OD260/280 > 1.8) (Nano Drop 2000 Spectrophotometer; ThermoFisher Scientific, Waltham MA). Between 300 and 500 ng of tissue RNA was reverse transcribed using ABI High Capacity RNA to cDNA kit (Cat#4387406; ThermoFisher Scientific, Waltham MA) as described in the manufacturer's instructions, and resulting cDNA was diluted 1:5 in molecular-grade RNase/DNase-free H2O. The quantitative real-time PCR was performed using the LightCycler 480-II with Dual Color Hydrolysis Universal Probe System (Roche Diagnostic Ltd., Basel, Switzerland).
Primers and probes were designed using Roche Universal Probe Library Assay Design Center (http://www.roche-applied-science.com) for human CD45 (NM_005608.2), human CK7 (NM_005556.3), and human PAX8 (NM_003466.2). Human CD45 primers (FW 5′-agtcaaagttattgttatgctgacaga-3′ and REV 5′-tgctttccttctccccagta-3′), human CK7 primers (FW 5′-caggctgagatcgacaacatc-3′ and REV 5′-cttggcacgagcatcctt-3′), and human PAX8 primers (FW 5′- tcgactcaccagacctaccc-3′ and REV 5′- tttcctgctttatggcgaag-3′) were synthesized at Mayo Foundation's Advanced Genomics Technology Center (Rochester, MN). The UPL probe #54 (for human CD45, Cat#04688511001), probe #21 (for human CK7; Cat#04686985001), and #87 (for human PAX8; Cat#04689127001) were obtained from the universal probe library database (Roche Diagnostic Ltd., Basel, Switzerland). The gene-specific probes and primers for housekeeping genes human glyceraldehyde-3-phophate dehydrogenase (GAPDH) (Cat#05190541001) and mouse ACT (β-actin) (Cat#05046190001) were obtained from Roche (Universal Probe Library Reference Gene Assay, Roche Diagnostic Ltd., Basel, Switzerland).
Each PCR was performed in triplicate using the Roche LightCycler 480-II (Roche Diagnostic Ltd., Basel, Switzerland) in the dual-color hydrolysis mode. After one cycle of 10 seconds of denaturation at 95°C, 40 cycles of PCR were performed: 95°C for 10 seconds, 60°C for 30 seconds, and 72°C for 1 second, followed by a cooling cycle of 40°C for 30 seconds. For in vivo rituximab studies, relative expression of target genes was determined by normalization to the GAPDH cycle threshold (CT) value and analyzed using the Roche LightCycler 480-II software (Roche Diagnostic Ltd., Basel, Switzerland).
Rituximab Monoclonal Antibody Treatment
Rituximab (Rituxan; Genentech, Inc., San Francisco, CA) 10 mg/kg was premixed with the tumor/McCoy's slurry and coinjected into mice at the time of heterotransplantation for the suppression studies. For the prospective lymphoma growth inhibition studies, rituximab was given as a single intraperitoneal injection at the same dose utilized for suppression studies at the time of tumor injection.
Statistical Analysis
Time to moribund (TTM) was defined in days as the duration between injection and necropsy. Two group outcomes compared medians using the Wilcoxon-Mann-Whitney two-sample rank-sum test, two tailed and reporting 10th to 90th percentile. Time to engraftment and engraftment rate were determined using a cumulative incidence approach to account for models still under observation for determination of engraftment. Comparisons between engraftment status and source patient characteristics were assessed using χ2 tests and Wilcoxon rank-sum tests. Spearman's correlation was used to assess association between continuous variables.
Results
Atypical Growth Characteristics in a Subset of Ovarian Cancer Heterotransplanted Tumors
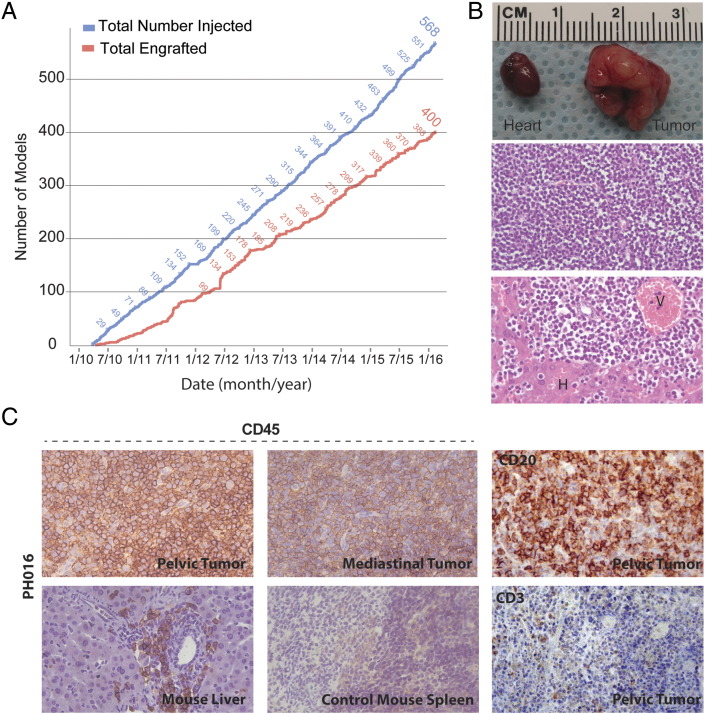
The ovarian cancer PDX resource has grown steadily with consistent engraftment of ovarian tumors (Figure 1A). However, a subset of eight early tumorgraft models demonstrated atypical patterns of dissemination. For instance, large mediastinal masses causing clinical signs of decompensation were found at necropsy (Figure 1B and Supplementary Figure 1), without the typical pattern of intraperitoneal tumor engraftment. By light microscopy of hematoxylin and eosin (H&E)–stained tissue sections, monotonous sheets of small blue cells and low cytoplasm bore no resemblance to the matched patient's primary tumor and did not resemble carcinoma in general. In the other atypical PDX tumors, pale hepatomegaly was common, and similar small blue cells can be seen in the intrahepatic perivascular space. When the mediastinal tumor, liver, and intraperitoneal tumors were stained for CD45, a nonspecific lymphocyte marker, strong expression was seen (Figure 1C). To ensure that the antibody did not cross-react with murine lymphocytes, staining of mouse spleen was shown to be negative. Following these initial observations, more extensive characterization of these tumors was performed.
Figure 1.
Injected and engrafted ovarian cancer PDX models.
(A) The number of injected and engrafted tumors is shown. (B) A representative mediastinal mass large enough to engulf the heart is shown (top panel). By light microscopy, H&E staining showed small blue cells without glandular features (middle panel). Similar cells were seen within the liver parenchyma clustered around perivascular spaces (V) and adjacent to hepatocytes (H) (lower panel). (C) IHC analysis of a representative model (PH016) revealed positive CD45 expression within the mediastinal tumor, pelvic tumor, and mouse liver (40×). Spleen from a control mouse, naïve to human tumor xenograft, showed no significant cross reactivity to murine CD45. PH016 was positive for CD20 B-cell marker but negative for CD3 T-cell marker, confirming a B-cell origin.
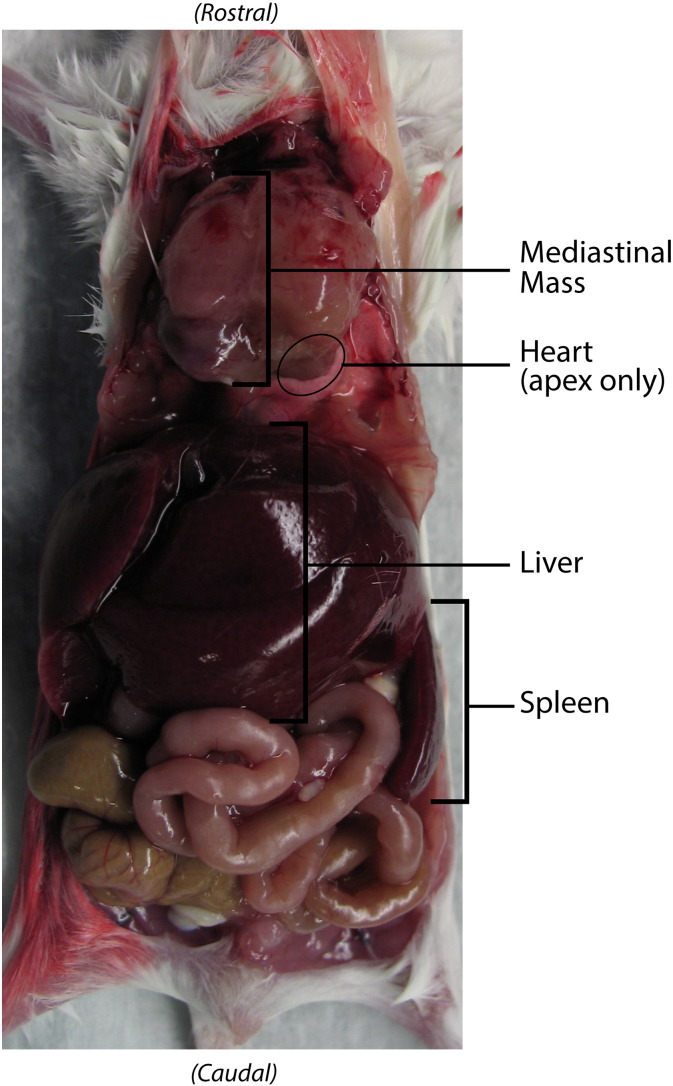
Supplementary Figure 1.
Mediastinal mass in situ.
Typical necropsy findings are shown with the abdominal wall and ventral thoracic cage resected. Within the abdomen and pelvis, no ovarian PDX tumor was observed, but hepatomegaly occupies roughly 50% of the abdomen. Splenomegaly is present but partially obscured by liver and bowel. Within the thoracic cavity, a large mediastinal mass encompasses the heart with only the cardiac apex visible.
Lineage Determination of Lymphoproliferative Tumors
To determine if the CD45-positive cells were lineage specific, gene expression for B- and T-cell markers was performed. IHC analysis revealed that all eight atypical tumorgrafts stained CD20 positive and CD3 negative, consistent with a B-cell lineage (Figure 1C). In addition, tumors were negative for pan-cytokeratin expression (data not shown). Of note, none of the source patients had a hematologic malignancy or lymphoproliferative disorder prior to surgical debulking or at any time thereafter (median postsurgery follow-up time, 24.0 months).
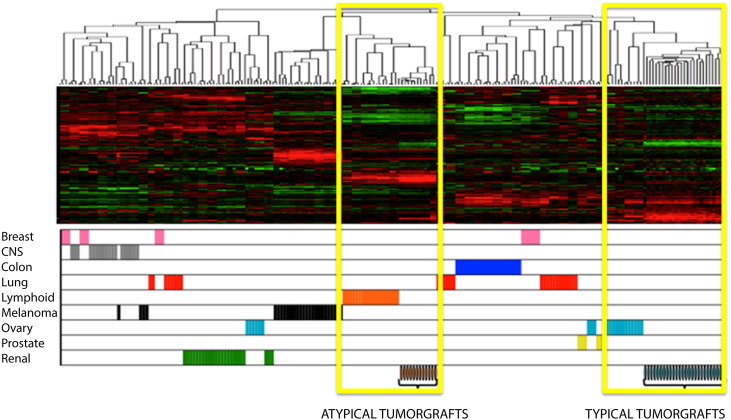
To evaluate the global gene expression profile, we conducted gene expression microarray studies of the eight atypical tumorgrafts compared to typical ovarian carcinoma PDX tumors (pan-cytokeratin positive, CD45 negative, histologic characteristics of carcinoma) [3]. Unsupervised clustering of the atypical tumorgrafts with the Stanford NCI60 data set containing expression from 60 cancer cell lines [21] revealed strong similarity between the PDX models but dissimilarity to the typical ovarian tumorgrafts (Supplementary Figure 2). The profiles of gene expression correlated well among atypical tumorgraft models and human lymphomas. Typical epithelial tumorgrafts clustered, unsupervised with human epithelial ovarian tumor arrays.
Supplementary Figure 2.
Gene expression microarray.
The first eight atypical tumorgraft models were compared to typical ovarian carcinoma tumorgrafts. These samples and the Stanford NCI60 Cancer arrays underwent unsupervised, hierarchical clustering. The atypical tumorgrafts revealed similarity among the models but dissimilarity to the typical ovarian tumors. Moreover, the gene expression profiles correlate among atypical tumorgraft and human lymphomas, corroborating the hypothesis of lymphoproliferation due to cells within the patient's tissues.
Clonality of B-Cell Lymphoproliferative Tumors
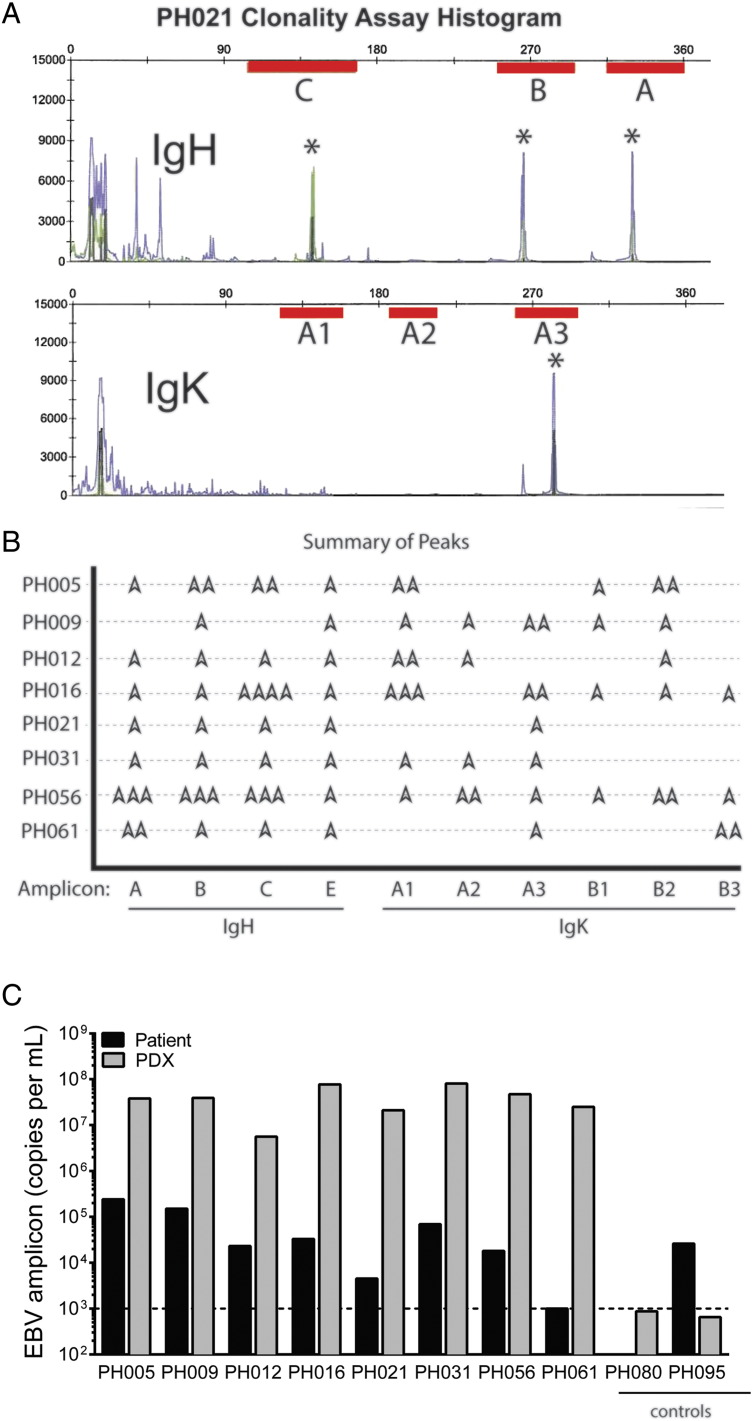
To determine if the lymphoproliferation is driven by a polyclonal reactive process or a monoclonal process indicative of malignant transformation, immunoglobulin heavy and light chain rearrangement studies were performed using a highly sensitive PCR-based differential immunofluorescence assay targeting the V-J region. Clonality was identified if one or two amplicons were detected within an assay-defined region (Figure 2A). Across all eight atypical PDX models, detection of clonal immunoglobulin heavy and kappa light chain gene rearrangement was consistent with the presence of a clonal cell population (Figure 2B). The negative control was a reactive lymph node and showed multiple peaks, as expected for a reactive or polyclonal process.
Figure 2.
B-cell proliferation is clonal and associated with EBV.
(A) Representative clonality assay of PDX model PH021 shows the target amplification regions for PCR (red bars) in the heavy chain (upper panel) and light chain (lower panel) with peaks (*) depicting amplicons. (B) Eight individual PDX models are summarized (amplicon peaks =  ). (C) EBV DNA was detected in all of the eight PDX tissue samples analyzed, but no viral copies were detected above the assay threshold in two epithelial PDX models used as controls (PH080 and PH095).
). (C) EBV DNA was detected in all of the eight PDX tissue samples analyzed, but no viral copies were detected above the assay threshold in two epithelial PDX models used as controls (PH080 and PH095).
EBV Quantification
As latent EBV in lymphocytes can become activated in an immunocompromised background, we then investigated whether the clonal B-cell tumorgrafts were EBV positive. Indeed, all eight lines contained greater than 106 virus copies per ml (Figure 2C). In contrast, epithelial ovarian cancer PDX tissues, which are negative for CD45 expression, lacked detectable EBV DNA (</=1000 copies per ml). The source of these clonal B cells was presumably derived from physiologic tumor-associated lymphocytes (TALs). This was supported by the observation that viral copies were detected in the primary tumors but at lower levels. One patient's surgical specimen, PH061, did not contain detectible EBV DNA despite the high levels found in the corresponding PDX. In contrast, the presence of EBV was not sufficient to induce a clonal B-cell outgrowth in PH095.
Suppression of B-Cell Lymphoproliferation
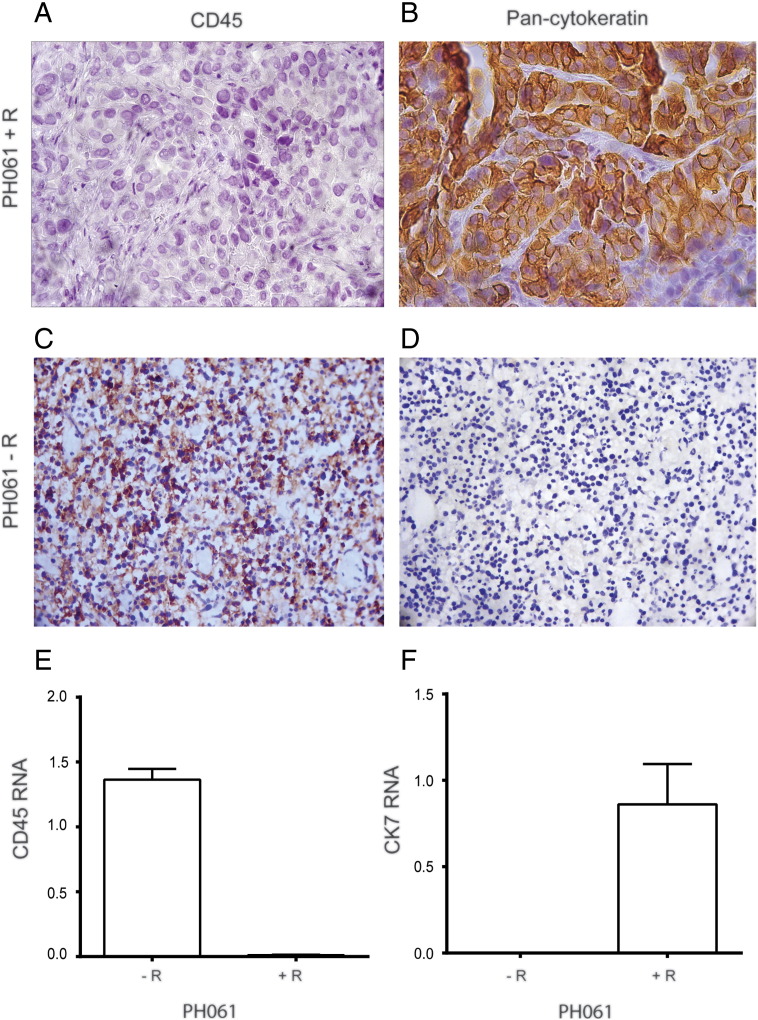
Because surplus patient tumor is cryogenically preserved at the time of initial surgery, a backup source of primary tumor is available to attempt a second engraftment of ovarian cancer if needed. To determine if rituximab treatment could be used to suppress B-cell proliferation in ovarian tumor heterotransplants, an aliquot of viable primary patient tumor PH061 (known to produce an EBV-associated B cell lymphoma PDX) was premixed with rituximab (10 mg/kg) and coinjected as a single treatment into a SCID mouse (PH061 + R). After engraftment and necropsy, the subsequent PDX was cytokeratin positive and CD45 negative by IHC (Figure 3, A and B). In contrast, the mouse injected with the same source patient tumor but without rituximab (PH061-R) engrafted a tumor lacking epithelial markers but strongly expressing the lymphocyte marker CD45 (Figure 3, C and D). Because real-time PCR allows for a more rapid assessment of gene expression, which is needed to make real-time decisions regarding the disposition of a newly established PDX model prior to expansion of tumor into additional mice, human-specific primers for CD45 and CK7 (epithelial marker) were developed, and the real-time PCR results were consistent with the protein expression (Figure 3, E and F).
Figure 3.
Expression of epithelial and lymphocyte markers with and without rituximab treatment at the time of engraftment.
PH061 + R primary patient tumor developed an intraperitoneal xenograft which was CD45 negative (A) and pan-cytokeratin positive (B). In contrast, the same primary patient tumor transplanted without rituximab (PH061-R) engrafted a tumor that was strongly CD45 positive (C) but lacking pan-cytokeratin (D). All images were captured with a 40× objective. After normalization to human GAPDH, human-specific real-time PCR results also showed that CD45 (E) and CK7 (F) gene expression was consistent with IHC data.
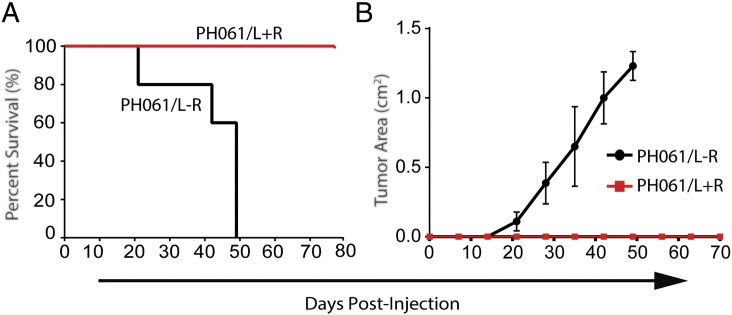
Although these data suggest that rituximab can suppress B-cell proliferation long enough to allow for epithelial ovarian carcinoma engraftment and growth before B-cell transformation occurs, the ability to prevent growth of an established B-cell lymphoma was also explored. In a prospective study, the PH061 PDX line, which was shown to have a clonal expansion of lymphocytes (PH061/L), was reconstituted into SCID mice and treated with rituximab (PH061/L + R) or saline (PH061/L-R), n = 5 mice each group, as a single dose at the time of tumor injection. The primary end point was TTM and change in tumor size by ultrasound. The secondary end point was tumor mass at necropsy. In the PH061/L-R cohort, the median TTM was 49 days, whereas the TTM was not reached in mice treated with a single dose of rituximab at the time of initial tumor injection (Figure 4A). Animal survival correlated with intraperitoneal tumor growth (Figure 4B). Three weeks after the last PH061/L-R mouse was sacrificed, all PH061/L + R mice underwent necropsy and no tumor was found, whereas the PH061/L-R animals had a mean tumor mass at necropsy of 1.17 g, confirmed as CD45 positive by PCR.
Figure 4.
Inhibition of xenograft B cell lymphoma growth with rituximab.
PDX line PH061 with a clonal expansion of malignant B-cell lymphocytes (PH061/L) was treated with rituximab (PH061/L + R) or saline (PH061/L-R) as a single dose at the time of tumor injection. (A) The median time to progression was 49 days for the PH061/L-R cohort but not reached for the PH061/L + R cohort. (B) The PH061/L + R group did not develop measurable tumor by ultrasound, whereas the PH061/L-R group exhibited persistent tumor growth.
Because rituximab suppressed the outgrowth of cotransplanted B cells from a patient's primary tumor, a laboratory practice change was instituted to determine the impact of premixing rituximab on the coengraftment of TALs. For this analysis, the first 277 of 400 sequentially engrafted PDX models were included. The prerituximab period (cohort 1) was comprised of 117 engrafted PDX models, and the postrituximab period (cohort 2) contained 160 PDX models. Real-time PCR was used to screen for tissue-specific markers and identify each tumor as carcinoma or lymphoproliferative tumor while ruling out a spontaneous mouse tumor (Table 1).
Table 1.
RT-PCR Interpretation
| Scenario | h-GAPDH | h-CD45 | h-CK7 or h-PAX8 | m-Actin | Interpretation |
|---|---|---|---|---|---|
| 1 | + | − | + | + | Ovarian/FT/PP PDX (carcinoma) |
| 2 | + | + | − | + | Human lymphoproliferation |
| 3 | + | + | + | + | Carcinoma + TAL |
| 4 | − | − | − | + | Spontaneous mouse tumor |
h, human; m, mouse; FT, fallopian tube; PP, primary peritoneal.
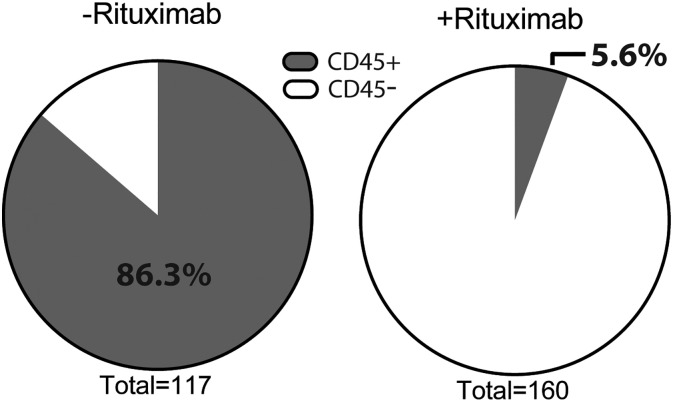
Irrespective of tumor dissemination patterns, at least one tumor from each PDX line in cohort 1 was tested. In cohort 2, only PDX models destined for imminent in vivo studies or those with atypical growth patterns were screened for CD45 expression. In addition, repeat testing was performed on any PDX tumor with atypical growth patterns in subsequent passages. Rituximab reduced the incidence of CD45-positive PDX tissue from 86.3% (101 of 117 in cohort 1) to 5.6% (9 of 160 in cohort 2) (Figure 5). Because the eight lymphomas described above (Figure 2) exhibited consistent IHC staining patterns (CD45+, CD20+, panCK−), IHC and H&E were used to validate any CD45-positive PDX by PCR in cohort 1 unless clonality studies were already done. In addition to the eight lymphomas confirmed by clonality studies, five other PDX models were determined to be lymphomas by IHC (CD45+, panCK−) and H&E (lymphoid cellularity without glandular features) similar to images in Figure 1. Taken together, the lymphoma rate in cohort 1 was 11.1% or 13 of 117. In contrast, IHC validation of the nine CD45-positive PDX tumors in cohort 2 revealed three lymphomas by histology and IHC, for a lymphoma rate of 1.88% or 3 of 160. Of note, one of these PDXs was derived from a splenic metastasis which presumably had an abundance of lymphocytes cotransplanted with the ovarian carcinoma.
Figure 5.
Rituximab reduces the incidence of CD45+ cells in ovarian cancer PDXs.
Real-time PCR was used to screen for expression of human CD45 in 277 early generation PDX models. The results shown in those pie charts are relative expression of human CD45 with or without rituximab treatment. The incidence of CD45-positive engrafted tumors decreased from 86.3% to 5.6% after the addition of rituximab.
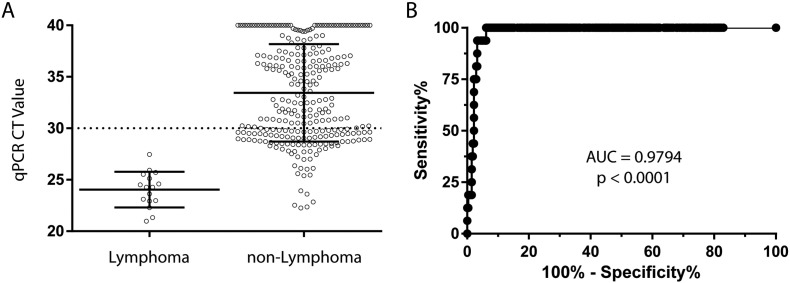
Given the sensitivity of real-time PCR, the detection of CD45 transcripts is not uncommon in early-passage ovarian cancer PDX tumors containing residual nontransformed TAL. To explore the correlation between CD45 CT values and engraftment of lymphomas, PDX models were stratified by tumor type (Supplementary Figure 3A). All lymphoma models exhibited CT values <30, and as a cohort, mean CT values were significantly lower than nonlymphoma tumors (two-tailed t test, P < .0001). Moreover, a receiver operator curve (ROC) showed that a CT value <27.49 gives a 100% sensitivity and 93.8% specificity in detecting lymphomas, supporting the utility of the assay as a screening procedure (Supplementary Figure 3B).
Supplementary Figure 3.
Distribution of CD45 expression by real-time quantitative PCR across engrafted PDX tumors. (A) The mean CT value of CD45 mRNA was lower in tumors confirmed to be lymphomas by either clonality studies or histology/IHC, indicating a greater abundance of human CD45 transcripts in lymphomas (two-tailed t test P < .0001). The dotted line shows the screening threshold use to flag PDX tumors as CD45 positive. Error bars show standard deviation. (B) An ROC of the same data showed a high area under the curve, indicative of high quantitative PCR accuracy for detecting lymphomas. Although the screening threshold for CD45 was CT <30, the ROC analysis showed that a CT value of <27.49 still gives 100% sensitivity for detecting lymphomas while maintaining a 93.8% specificity.
Discussion
In this study, we assessed 277 established PDX models from our experience transplanting 568 ovarian cancers into SCID mice. Although the unintended engraftment of TALs has been previously reported, to our knowledge, this effort represents the largest single-institution experience with PDX tumors in general and ovarian PDX tumors in particular. Similar to other tumors types, we have shown that ovarian tumors harbor TALs that can develop into B-cell lymphomas associated with latent EBV in an immunodeficient mouse host. The B-cell lineage was confirmed by H&E, IHC, and PCR-based clonality studies. Most importantly, treatment with an anti-CD20 monoclonal antibody, rituximab, not only inhibited the proliferation of established B-cell lymphoma in SCID mice but also prevented the occurrence of lymphomatous outgrowth in early-passage xenografts, allowing for the rescue of a PDX model known to develop a lymphoma. The routine use of rituximab at the time of engraftment has now become our standard practice.
The observation that lymphoproliferative lesions or lymphomas can develop from the cotransplantation of human epithelial tumors and TALs is consistent with data presented in PDX tumors derived from lung, colorectal, breast, prostate, hepatocellular, and gastric cancers. The data presented herein add to the hypothesis that this is not a phenomenon unique to any single tumor type but rather is a function of the cells (TALs and carcinoma) that constitute the primary tumor in the immunodeficient animal host. Moreover, the degree of immunodeficiency may also play a role, as demonstrated by the development of unintentional lymphoproliferative lesions or lymphomas in NSG, NOG, or NOD/SCID mice as frequent as 80% for prostate cancer [12] and as low as 22% for colorectal cancer [11], with multiple other tumors types in between: hepatocellular [13], gastric [10], breast [9], [22], lung [22], pancreas [9], bladder, and renal cancers [9]. For comparison, however, no lymphomas developed in nude mice [9], [10]. In contrast, SCID-Beige mice retain some NK cell activity [23] and complement [24], which may contribute to the relatively lower rate of lymphomas seen in our study with SCID-bg mice (11.1% or 13/117 engrafted tumors in the prerituximab period).
Prevention of unintentional B-cell lymphomas has important implications. B-cell–depleting anti-CD20 antibodies, such as rituximab, have been successfully used in the treatment of human B-cell malignancies. The presumed mechanisms of action include antibody-dependent cytotoxicity mediated by Fc receptor gamma, complement-mediated cell lysis, growth arrest, and B-cell apoptosis [25] to effectively reduce the persistence of lymphocytes in early PDX generations and prevent the outgrowth of B cell lymphomas. However, the benefits reported herein may not be as apparent for PDXs generated in mice with more substantial immunodeficiency. For instance, NSG, NOG, or NOD/SCID mice have absence of mature B and T cells, defective dendritic cells and macrophages, absence or defective natural killer cells, and absence of complement [26], all of which may limit the efficacy of rituximab. Indeed, in vivo studies with rituximab in rodents have demonstrated that a mechanism of cytotoxicity is mediated, at least in part, by complement [27], [28]. Although monitoring early generations for splenomegaly and/or hepatomegaly is advantageous to identify possible lymphoproliferative lesions, prevention would avoid the loss of valuable PDX lines due to lymphoma.
The use of PDX models in oncology has extended beyond the study of tumor biology and preclinical drug development [5], [29], with several clinical trials currently under way utilizing PDX models to personalize cancer care for patients with ovarian cancer (NCT02312245), sarcoma (NCT02720796), head and neck cancer (NCT02752932), and pancreatic cancer (NCT02795650). Screening all PDX tumors for the presence of lymphoid and epithelial, as well as murine, markers is imperative to identify unintentional lymphomas and spontaneous mouse tumors. More importantly, strategies to prevent the outgrowth of such tumors may help avoid the loss of valuable PDX models which may be difficult, if not impossible, to replace.
The following are the supplementary data related to this article.
Acknowledgements
The authors thank the following for their generous collaboration:
Clinical Parasitology and Virology Division of Clinical Microbiology, Mayo Clinic, Rochester, MN: Bobbie Pritt, MD, MSc, and Mark Espy.
Division of Hematology and Internal Medicine, Mayo Clinic, Rochester, MN: Stephen M. Ansell, MDm PhD; Phuong Nguyen, MD; and Steven Ziesmer.
Footnotes
Authors' disclosure of potential conflicts of interests: none.
Support: This research was supported by the Mayo Clinic Specialized Program in Research Excellence grant CA136393 and R01 CA184502 from the National Institutes of Health, Ovarian Cancer Research Fund Alliance, Minnesota Ovarian Cancer Alliance, and a private donation from the Petersen Family to support general lab efforts.
Contributor Information
Kristina A. Butler, Email: Butler.Kristina@mayo.edu.
Xiaonan Hou, Email: Hou.Xiaonan@mayo.edu.
Marc A. Becker, Email: becke344@gmail.com.
Valentina Zanfagnin, Email: Zanfagnin.Valentina@mayo.edu.
Sergio Enderica-Gonzalez, Email: endericagonzalez.sergio@gmail.com.
Daniel Visscher, Email: Visscher.Daniel@mayo.edu.
Kimberly R. Kalli, Email: Kalli.Kimberly@mayo.edu.
Piyawan Tienchaianada, Email: pt_kuk@hotmail.com.
Paul Haluska, Email: paul.haluska@merck.com.
S. John Weroha, Email: Weroha.Saravut@mayo.edu.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weroha SJ, Becker MA, Enderica-Gonzalez S, Harrington SC, Oberg AL, Maurer MJ, Perkins SE, AlHilli M, Butler KA, McKinstry S. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin Cancer Res. 2014;20:1288–1297. doi: 10.1158/1078-0432.CCR-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott CL, Becker MA, Haluska P, Samimi G. Patient-derived xenograft models to improve targeted therapy in epithelial ovarian cancer treatment. Front Oncol. 2013;3:295. doi: 10.3389/fonc.2013.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, Silver DF, Yang NP, Oflazoglu E, Hempling RE, Piver MS, Repasky EA. Characterization of human ovarian carcinomas in a SCID mouse model. Gynecol Oncol. 1999;72:161–170. doi: 10.1006/gyno.1998.5238. [DOI] [PubMed] [Google Scholar]
- 7.Bankert RB, Balu-Iyer SV, Odunsi K, Shultz LD, Kelleher RJ, Jr., Barnas JL, Simpson-Abelson M, Parsons R, Yokota SJ. Humanized mouse model of ovarian cancer recapitulates patient solid tumor progression, ascites formation, and metastasis. PLoS One. 2011;6:e24420. doi: 10.1371/journal.pone.0024420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Custer RP, Bosma GC, Bosma MJ. Severe combined immunodeficiency (SCID) in the mouse. Pathology, reconstitution, neoplasms. Am J Pathol. 1985;120:464–477. [PMC free article] [PubMed] [Google Scholar]
- 9.Bondarenko G, Ugolkov A, Rohan S, Kulesza P, Dubrovskyi O, Gursel D, Mathews J, O'Halloran TV, Wei JJ, Mazar AP. Patient-derived tumor xenografts are susceptible to formation of human lymphocytic tumors. Neoplasia. 2015;17:735–741. doi: 10.1016/j.neo.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi YY, Lee JE, Kim H, Sim MH, Kim KK, Lee G, Kim HI, An JY, Hyung WJ, Kim CB. Establishment and characterisation of patient-derived xenografts as paraclinical models for gastric cancer. Sci Rep. 2016;6:22172. doi: 10.1038/srep22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukohyama J, Iwakiri D, Zen Y, Mukohara T, Minami H, Kakeji Y, Shimono Y. Evaluation of the risk of lymphomagenesis in xenografts by the PCR-based detection of EBV BamHI W region in patient cancer specimens. Oncotarget. 2016;7:50150–50160. doi: 10.18632/oncotarget.10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wetterauer C, Vlajnic T, Schuler J, Gsponer JR, Thalmann GN, Cecchini M, Schneider J, Zellweger T, Pueschel H, Bachmann A. Early development of human lymphomas in a prostate cancer xenograft program using triple knock-out immunocompromised mice. Prostate. 2015;75:585–592. doi: 10.1002/pros.22939. [DOI] [PubMed] [Google Scholar]
- 13.Chen K, Ahmed S, Adeyi O, Dick JE, Ghanekar A. Human solid tumor xenografts in immunodeficient mice are vulnerable to lymphomagenesis associated with Epstein-Barr virus. PLoS One. 2012;7:e39294. doi: 10.1371/journal.pone.0039294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler K, Weroha S, Becker M, Enderica-Gonzalez S, Harrington S, Hou X, Visscher D, Haluska P. Ovarian cancer tumorgraft: viral latency propagates lymphoma. Gynecol Oncol. 2012;127:S16. [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 18.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 19.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 20.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 21.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 22.Fujii E, Kato A, Chen YJ, Matsubara K, Ohnishi Y, Suzuki M. Characterization of EBV-related lymphoproliferative lesions arising in donor lymphocytes of transplanted human tumor tissues in the NOG mouse. Exp Anim. 2014;63:289–296. doi: 10.1538/expanim.63.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDougall JR, Croy BA, Chapeau C, Clark DA. Demonstration of a splenic cytotoxic effector cell in mice of genotype SCID/SCID.BG/BG. Cell Immunol. 1990;130:106–117. doi: 10.1016/0008-8749(90)90165-n. [DOI] [PubMed] [Google Scholar]
- 24.Guhad FA, Jensen HE, Hau J. Complement activation in SCID and nude mice is related to severity of tissue inflammation in the Candida mastitis model. FEMS Microbiol Lett. 2000;192:27–31. doi: 10.1111/j.1574-6968.2000.tb09354.x. [DOI] [PubMed] [Google Scholar]
- 25.Weiner GJ. Rituximab: mechanism of action. Semin Hematol. 2010;47:115–123. doi: 10.1053/j.seminhematol.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shultz LD, Schweitzer PA, Christianson SW, Gott B, Schweitzer IB, Tennent B, McKenna S, Mobraaten L, Rajan TV, Greiner DL. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol. 1995;154:180–191. [PubMed] [Google Scholar]
- 27.Di Gaetano N, Cittera E, Nota R, Vecchi A, Grieco V, Scanziani E, Botto M, Introna M, Golay J. Complement activation determines the therapeutic activity of rituximab in vivo. J Immunol. 2003;171:1581–1587. doi: 10.4049/jimmunol.171.3.1581. [DOI] [PubMed] [Google Scholar]
- 28.Golay J, Cittera E, Di Gaetano N, Manganini M, Mosca M, Nebuloni M, van Rooijen N, Vago L, Introna M. The role of complement in the therapeutic activity of rituximab in a murine B lymphoma model homing in lymph nodes. Haematologica. 2006;91:176–183. [PubMed] [Google Scholar]
- 29.Scott CL, Mackay HJ, Haluska P., Jr. Patient-derived xenograft models in gynecologic malignancies. Am Soc Clin Oncol Educ Book. 2014:66. doi: 10.14694/EdBook_AM.2014.34.e258. [DOI] [PMC free article] [PubMed] [Google Scholar]