Abstract
Lipid supplementation is a promising strategy for methane mitigation in cattle and has been evaluated using several different lipid sources. However, limited studies have assessed the effect of temperature on methane emissions from cattle and changes in incubation temperature have also not been extensively evaluated. The aim of this study was to evaluate the combined effect of pequi oil (high in unsaturated fatty acids) and incubation temperature on fermentation characteristics and microbial communities using the rumen simulation technique. A completely randomized experiment was conducted over a 28-day period using a Rusitec system. The experiment was divided into four periods of 7 days each, the first of which was a 7-day adaptation period followed by three experimental periods. The two treatments consisted of a control diet (no pequi oil inclusion) and a diet supplemented with pequi oil (1.5 mL/day) which increased the dietary fat content to 6% (dry matter, DM-basis). Three fermenter vessels (i.e., replicates) were allocated to each treatment. In the first experimental period, the incubation temperature was maintained at 39°C, decreased to 35°C in the second experimental period and then increased again to 39°C in the third. Pequi oil was continuously supplemented during the experiment. Microbial communities were assessed using high-throughput sequencing of the archaeal and bacterial 16S rRNA gene. Methane production was reduced by 57% following a 4°C decrease in incubation temperature. Supplementation with pequi oil increased the dietary fat content to 6% (DM-basis) but did not affect methane production. Analysis of the microbiota revealed that decreasing incubation temperature to 35°C affected the archaeal and bacterial diversity and richness of liquid-associated microbes, but lipid supplementation did not change microbial diversity.
Keywords: methanogens, fermentation process, greenhouse gas, ruminant nutrition, in vitro
Introduction
Methane (CH4) is produced in the rumen by a group of archaea known as methanogens. Methanogens have an unusual metabolism because they are capable of using substrates such as hydrogen, carbon dioxide (CO2), formate, methylated C1 compounds and acetate as energy and carbon sources for growth and produce CH4 as a major end product (Deppenmeier, 2002). The production of CH4 during fermentation in the bovine rumen also results in a loss of energy for the animal. Depending on feed intake as well as diet composition, around 2 to 12% of gross energy intake is converted into CH4 (Johnson and Johnson, 1995). Measures to mitigate enteric fermentation would not only reduce emissions, but may also raise animal productivity by increasing digestive efficiency.
A number of studies have reviewed methane mitigation strategies involving the modification of dietary composition or through dietary supplementation with lipids, organic acids, plant components, etc. (Hook et al., 2010). Among documented methane abatement strategies, dietary modification appears to be one of the promising technologies to reduce methane emissions from ruminants (Beauchemin et al., 2008; Martin et al., 2010; Patra, 2013). For example, the addition of fats to ruminant diets is one of the recognized means used to decrease enteric methane emissions (Beauchemin et al., 2008). Previous batch culture incubations in our research group indicated that pequi (Caryocar brasiliense) oil used at 1% v/v can reduce CH4 production by up to 74% (unpublished data).
Microbial activity in the rumen is known to be affected by dietary additives (Hristov et al., 2013) but information related to the effect of rumen temperature and/or incubation temperature on methane production is scarce. Graham et al. (1959) have shown that CH4 production in adult sheep decreased by 20% when the environmental temperature was reduced from 33 to 8°C. Others have also indicated a 30% CH4 reduction in cold-adapted sheep (Kennedy and Milligan, 1978) while Rogerson (1960) suggested that the effect of environmental temperatures changes between 20 and 40°C on CH4 production in cattle were variable. However, limited research has been conducted on the effect of incubation temperature changes on the ruminal microbiota and on fermentation characteristics. It has been suggested that microbial diversity in the bovine rumen decreases concurrently with the ambient temperature and that some bacteria adapt better to adverse environmental conditions (Romero-Perez et al., 2011), and this may also affect rumen function. Crater and Barboza (2007) demonstrated that the rumen of muskoxen (Ovibos moschatus) reached minimal temperatures of 31.4°C, 35.7°C, 32.5°C in early, middle and late winter, respectively. According to Crater and Barboza (2007) minimal rumen temperatures were much more variable than maxima. Ruminal cooling was often rapid (–6.24°C/min), dropping to 27.6°C within a few minutes.
We hypothesized that the inclusion of pequi oil and a 4°C decrease in rumen incubation temperature for 7 days would reduce CH4 emissions and alter the ruminal microbial community. As such, the objective of this study was to evaluate the effect of pequi oil supplementation and incubation temperature on CH4 production and the ruminal microbiota in a Rusitec (rumen simulation technique) system.
Materials and Methods
The donor cow was cared for in accordance with the guidelines of the German Animal Welfare Act approved by the Lower Saxony State Office for Consumer Protection and Food Safety (LAVES, approval number AZ 33.4-42505-04-13A373).
Experimental Design and Treatments
In vitro ruminal fermentation was conducted using a Rusitec unit to evaluate the effect of incubation temperature and dietary fatty acids from pequi (Caryocar brasiliense) on the rumen microbiota and methane production. The experiment was a completely randomized design with two treatments and three replicates per treatment. The two treatments consisted of a control diet (CON, no pequi oil inclusion) and a diet supplemented with pequi oil (1.5 mL/day) which increased the dietary fat content to 6% (DM-basis).
Plant Component
Pequi oil was selected to supplement the diets because of its high proportion of unsaturated fatty acids (Lima et al., 2007), which have been reported to be effective in reducing CH4 production without compromising diet digestibility and hence milk production in dairy cattle (Beauchemin et al., 2009).
Substrate and Rumen Inoculum
The substrate consisted of hay:concentrate (66.7:33.3 w/w; Table 1). Hay substrate was prepared using an electric wool clipper with a 76 mm blade. The particle size distribution of hay material [% of DM] (Soofi-Siawash and Mathison, 1994) indicated that 6.5, 46.7, and 46.8% of plant DM was retained on sieves with aperture sizes of ≥4-mm, 1-mm, and ≤0.63-mm, respectively. The prepared substrates were weighed into nylon bags (11.5 cm × 6.5 cm, pore size 150 μm) for a total mass of 11 g of substrate (DM basis).
Table 1.
Chemical composition of the substrates (%; w/w).
| Hay | Concentrate | |
|---|---|---|
| Dry matter (DM) | 89.74 | 89.68 |
| Crude protein | 10.7 | 17.0 |
| Crude fat | 2.1 | 3.1 |
| Neutral detergent fiber | 53.8 | 21.2 |
| Ash | 5.81 | 6.51 |
Rumen inoculum from a fistulated cow (Soliva et al., 2011; Soliva et al., 2015) was collected 2 hours after morning feeding (e.g., bag exchange). The diet fed to the donor cow and the substrate proportion (forage:concentrate ratio) used in the experiment were identical. Extracted rumen content was strained through four layers of cheesecloth to separate the solid and liquid portion and used as the initial inoculum.
Chemical Composition
Feed was analyzed following AOAC (1995) methods for DM (Method 967.03), ash (Method 942), and ether extract (EE) content by extraction with petroleum ether using a Soxtherm Fat Extractor (Gerhardt Instruments, Königswinter, Germany; Method 920.39). Neutral detergent fibre (NDF) content was analyzed according to Van Soest et al. (1991) with the use of sodium sulfite and heat-stable a-amylase. Nitrogen (N) concentration was determined by the Kjeldahl method (Vapodest 20S, Gerhardt Instruments) with CP content calculated as N × 6.25.
Rusitec Fermentation
One Rusitec unit with six fermentation vessels (800 mL capacity each) submerged in a water bath was set up for the experiment following the incubation procedure described by Czerkawski and Breckenridge (1977). The experiment was conducted over 28 days, with the first 7 days used as an adaptation period at 39°C incubation, followed by an additional 7 days at 39°C, then 7 days at 35°C and finally 7 days at 39°C. On the first day of the experiment 800 mL of rumen fluid along with individual nylon bags containing 10 g of solid digesta and 11 g of the diet (DM basis) were placed in each fermentation vessel within the unit (water bath). The fermentation vessels were immediately infused with modified McDougall’s buffer pH 7.35 with an osmolality of 293 mosm/L at a dilution rate of 30 mL/h. Pequi oil was added (1.5 mL) each day to the nylon bags prior to the bag exchange, according to the experimental design. Following the initial 24 h incubation period, the bag with the rumen digesta (rumen solids) was replaced with a nylon bag containing the substrate and after 48 hours the bags were replaced each day. Therefore, each fermentation vessel always had two bags present during the experiment (Avila-Stagno et al., 2014).
Sample Collection and Analysis
Gas Production and Dry Matter (DM) Disappearance
Volumes of the effluent and total gas production were determined each morning before the nylon bags were exchanged. Accumulated effluent was collected in a 1 L glass flask and placed on ice to arrest microbial growth. Effluent output was measured daily during feed bag exchange. Total gas was collected in gas-tight bags (Plastigas, Linde AG, Munchen, Germany) and the volume of gas produced was measured using a drum-type meter (Ritter Apparatebau, Bochum, Germany) (Riede et al., 2013).
Throughout the experiment, pH and redox potential was measured daily during bag exchange using a Knick pH meter (digital pH meter 646, Knick, Berlin, Germany). DM disappearance was determined using the residue remaining in each nylon bag following 48 h of fermentation. Once removed, the nylon bags were washed with cold distilled water until the water ran clear. The clean bags were then dried at 55°C for 48 h (Narvaez et al., 2013; Li et al., 2014). The residue DM weight was then recorded and used for the calculation of DM disappearance.
Methane Production
Gas samples for methane analysis were taken using a 20 mL syringe and transferred into blood sample tubes (Venosafe type VF-109 SP, Terumo, Eschborn, Germany). The methane samples were analyzed using gas chromatography (GC 2014, Shimadzu Europa GmbH, Duisburg, Germany). Methane production was calculated by multiplying total gas volume by the percentage of CH4 in the gas sample and then corrected for standard temperature and pressure [0°C; 1013 hPa] (Riede et al., 2013).
Volatile Fatty Acids (VFA) and Ammonia Production
From d 1 to 28, before the replacement of substrate nylon bags, 5 mL of the effluent collected in the outflow flasks from each fermenter were taken and stored at –40°C. These samples were later used for volatile fatty acids (VFA) and ammonia (NH3) analyses. VFA concentration was analyzed by a gas chromatography system (model 5890 II, Hewlett Packard, Böblingen, Germany) equipped with a 1.8 m × 2 mm glass column packed with Chromosorb WAW (mesh 80/100) with 20% neopentyl glycol succinate and 2% orthophosphoric acid. Helium was used as a carrier gas with a flow rate of 25 mL/min. Injection port, detector and oven temperatures were 220, 250, and 130°C, respectively. VFA production was estimated by multiplying VFA concentration by the volume of effluent (Oeztuerk et al., 2005). For NH3 concentration analysis, 1 mL of sample was centrifuged at 4,600 × g for 10 min. From the supernatant, 50 μL were mixed with 5 mL phenol solution (106 mM phenol, 0.17 mM sodium nitroprusside dehydrate) and 5 mL sodium hypochlorite solution (1% v/v sodium hypochlorite; 125 mM NaOH) and kept at room temperature for 10 min. After an incubation step at 60°C for 10 min, NH3 concentration was determined photometrically at 546 nm in a spectrometer (DU 640, Beckman Coulter GmbH, Krefeld, Germany) using a NH4Cl standard solution (5 mM).
DNA Extraction and Illumina Sequencing of the Archaeal and Bacterial 16S rRNA Gene
16S rRNA gene sequencing was used to analyze the archaeal and bacterial communities of the original rumen fluid and digesta (rumen solids), as well as the LAM (liquid-associated microbes) and SAM (solid-associated microbes) samples taken at the end of every experimental period (day 14, 21, and 28). Total genomic DNA was extracted from each sample using a QIAamp Fast DNA stool mini kit (Qiagen), according to manufacturer’s instructions, with a lysis temperature of 80°C. DNA yield and purity was quantified using an Eppendorf BioPhotometer 6131 (Eppendorf, Hamburg, Germany). Extracted DNA was stored at –20°C until use.
The 16S rRNA gene sequence libraries were generated using a two-step PCR protocol. The first PCR step amplified the V4 region of the 16S rRNA gene using the universal bacterial and archaeal primers 515-F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806-R (5′-GGACTACVSGGGTATCTAAT-3′) (Caporaso et al., 2011). The second PCR step was used to add a unique 10-bp barcode at the 5′ end of the amplicon as well to add Illumina adapters. All PCR amplification and sequencing steps were carried out at Genome Quebec (Montreal, QC, Canada). Briefly, the 16S rRNA gene amplicons were quantified using a Quant-iT PicoGreen dsDNA assay kit (Invitrogen, Burlington, ON, Canada), pooled in equimolar ratios and then purified with AMPure XP beads (Beckman Coulter, Mississauga, ON, Canada). The 16S rRNA gene amplicons were then sequenced using an Illumina MiSeq (2 × 250) and the MiSeq Reagent Kit v2 (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions.
Analysis of Archaeal and Bacterial 16S rRNA Gene Sequences
All 16S rRNA gene sequences were processed and analyzed within the QIIME software package v. 1.9.1 (Caporaso et al., 2010b). Paired-end reads were joined using fastq-join with a minimum overlap of 35 bp and a maximum percent difference of 15 (Aronesty, 2013). These joined sequences were then quality filtered with sequences truncated following three consecutive base calls of a Phred score of less than 25. Only those sequences containing 75% or more of the original sequence following truncation were retained for further analysis. Chimeric sequences were removed using the UCHIME algorithm (Edgar et al., 2011) implemented in USEARCH v. 6.1.544 (Edgar, 2010). Sequences were then clustered into operational taxonomic units (OTUs) at 97% similarity using an open reference OTU picking method and the SILVA database [v. 111] (Quast et al., 2013). Sequences that did not match OTUs in the SILVA database were clustered into OTUs using the de novo approach and USEARCH. The UCLUST consensus taxonomy assigner (Edgar, 2010) was used to assign taxonomy to OTUs using the SILVA database, with a minimum similarity of 0.8 and max accepts of 3. Representative sequences for the OTUs were aligned using PyNast (Caporaso et al., 2010a) and a phylogenetic tree was created using FastTree (Price et al., 2010). All OTUs containing fewer than 10 sequences as well as those OTUs classified as chloroplasts were excluded from further analysis.
Each sample was randomly subsampled to 23,000 sequences per sample to account for uneven sequencing depth. The bacterial and archaeal diversity in each sample was calculated within QIIME using the Shannon index (Shannon, 1948) and phylogenetic diversity (PD whole tree) (Faith, 1992). Unweighted UniFrac distances (Lozupone and Knight, 2005) were used to assess the bacterial and archaeal community structure (beta-diversity) of each treatment, incubation temperature and sample type (solid vs. liquid). The subsequent distance matrices were visualized as principal coordinate analysis (PCoA) plots using Emperor (Vazquez-Baeza et al., 2013). All 16S rRNA gene sequences were submitted to the NCBI Sequence Read Archive1 under bioproject accession SRP066918.
Statistical Analysis
Fermentation data were analyzed using the MIXED procedure of SAS (SAS, Inc., 2015; SAS Online Doc 9.1.3). The model included the fixed effects of pequi oil, temperature and pequi oil × temperature interaction. Therefore, the individual fermenter was used as the experimental unit for statistical analysis. The minimum values of Akaike’s information criterion was used to select the covariance structure among compound symmetry, heterogeneous compound symmetry, autoregressive, heterogeneous autoregressive, Toeplitz, unstructured and banded for each parameter. Diversity metrics were analyzed using PROC MIXED in SAS 9.4 (SAS Inst., Inc., Cary, NC, United States) with fermenter as a random effect and pequi oil, temperature and interaction pequi oil × temperature as fixed effects. Treatment means were compared using the least squares mean linear hypothesis test (LSMEANS/DIFF). Significance was declared at P ≤ 0.05 and a trend was discussed when 0.05 < P ≤ 0.10.
The unweighted UniFrac distances were compared using ANOSIM (analysis of similarities) with 999 permutations within QIIME. Operational taxonomic units that were differentially abundant by sample type and treatment were identified using the G-test with a false discovery rate (FDR) < 0.05. Linear discriminant analysis effect size (LEfSe) was used to determine which phyla and genera were differentially abundant based on treatment, sample type and incubation temperature. The LEfSe uses the Kruskal–Wallis test to identify significantly different (P < 0.05) phyla and genera among sample groups and uses linear discriminant analysis (LDA) to estimate the effect size of each of these (Segata et al., 2011). A LDA score of 3.5 was used as the threshold for plotting differentially abundant genera.
Results
Effects of Incubation Temperature and Pequi Oil Supplementation on Fermentation Characteristics
Pequi oil supplementation had no effect on total gas, CH4 production or DM disappearance (P ≥ 0.58; Table 2). Decreasing the incubation temperature from 39°C to 35°C on d 15 had a negative effect on CH4 production reducing it from 80 mg/d to 34.3 mg/d. When incubation temperature was returned to 39°C on d 22, CH4 production was still lower (P < 0.01) than the initial 39°C incubation period (day 8–14). DM disappearance was greater (P < 0.01) during the 8–14 days incubation period and was diminished when the temperature was reduced to 35°C and through to the end of the experiment (Table 2).
Table 2.
Effect of incubation temperature and pequi oil on methane production and dry matter disappearance (DMD).
| 39°C | 35°C | 39°C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (Day 8–14) |
(Day 15–21) |
(Day 22–28) |
P-value |
|||||||
| Control | Pequi oil | Control | Pequi oil | Control | Pequi oil | SEM | Pequi oil | Temperature | Pequi oil × Temperature | |
| Total gas (mL/d) | 1851 | 1819 | 1505 | 1596 | 1597 | 1595 | 59.1 | 0.51 | <0.01 | 0.41 |
| CH4 (%) | 4.85 | 5.75 | 2.49 | 3.89 | 2.01 | 1.10 | 1.188 | 0.70 | 0.01 | 0.50 |
| CH4 (mL/d) | 109.5 | 114.8 | 44.0 | 52.1 | 44.5 | 37.5 | 10.65 | 0.81 | <0.01 | 0.75 |
| CH4 (mg/d) | 78.2 | 82.0 | 31.4 | 37.2 | 31.8 | 26.8 | 7.60 | 0.81 | <0.01 | 0.75 |
| DMD (%) | 68.9 | 71.5 | 61.6 | 63.0 | 63.6 | 64.3 | 2.52 | 0.58 | <0.01 | 0.59 |
Mean values with standard errors.
There was a tendency (P = 0.10) toward lower pH at 35°C compared to the last period at 39°C (6.76 vs. 6.78) and pequi oil supplementation decreased (P = 0.03) pH compared to the control (6.74 vs. 6.78). The redox potential was not affected by pequi oil addition but was lower (P < 0.01) during the 22–28 days period (39°C) compared to other time periods. An interaction between pequi oil supplementation and incubation temperature was observed for total VFA, acetate, propionate and iso-valerate production and the acetate:propionate ratio (P ≤ 0.05; Table 3). Supplementation with pequi oil only increased (P = 0.03) total VFA production during the 22–28 days fermentation period. Pequi oil increased (P ≤ 0.01) acetate and propionate production, following the initial adaptation period and throughout the experiment. The acetate:propionate ratio was decreased (P = 0.04) with the addition of pequi oil following the adaptation period (day 1–7). Ammonia production was also lower (P < 0.01) at the 35°C (day 15–22) incubation temperature compared to 39°C, but was not affected by pequi oil supplementation.
Table 3.
Effect of incubation temperature and pequi oil on pH, redox potential and fermentation characteristics.
| 39°C | 35°C | 39°C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (Day 8–14) |
(Day 15–21) |
(Day 22–28) |
P-value |
|||||||
| Control | Pequi oil | Control | Pequi oil | Control | Pequi oil | SEM | Pequi oil | Temperature | Pequi oil × Temperature | |
| pH | 6.8 | 6.7 | 6.8 | 6.7 | 6.8 | 6.8 | 0.02 | 0.03 | 0.10 | 0.51 |
| Redox potential (mV) | –177.0 | –185.3 | –190.4 | –197.0 | –235.0 | –235.6 | 11.27 | 0.64 | <0.01 | 0.97 |
| Total VFA (mmol/d) | 32.6 | 38.3 | 31.6 | 34.2 | 33.0b | 36.8a | 1.77 | 0.03 | <0.01 | 0.03 |
| Acetate (mmol/d) | 16.8b | 19.5a | 16.0 | 17.1 | 17.0b | 19.1a | 0.43 | 0.01 | <0.01 | 0.02 |
| Propionate (mmol/d) | 5.9b | 7.6a | 5.9b | 6.9a | 6.5b | 7.6a | 0.16 | <0.01 | <0.01 | <0.01 |
| Butyrate (mmol/d) | 5.5 | 6.5 | 5.4 | 5.6 | 6.1 | 6.2 | 0.25 | 0.47 | <0.01 | 0.13 |
| Valerate (mmol/d) | 2.4 | 2.6 | 2.0 | 2.1 | 2.2 | 2.3 | 0.15 | 0.57 | <0.01 | 0.76 |
| Iso-valerate (mmol/d) | 0.79b | 1.17a | 0.94 | 1.19 | 1.01b | 1.19a | 0.017 | 0.01 | <0.01 | 0.03 |
| Acetate:propionate | 2.85a | 2.57b | 2.71a | 2.48b | 2.62a | 2.51b | 0.080 | <0.01 | <0.01 | 0.04 |
| Ammonia (mmol/d) | 6.9 | 7.2 | 5.5 | 5.9 | 6.5 | 6.5 | 0.09 | 0.31 | <0.01 | 0.19 |
Different lowercase letters in rows indicate significantly different means (P < 0.05).
Rumen Microbiota in the Rusitec System
There was a total of 1,400,917 16S rRNA gene sequences with an average length of 262 bp following quality filtering. These sequences were clustered into 4,384 OTUs and classified into 20 different phyla and 89 distinct genera. The most relatively abundant phyla among all samples were: Bacteroidetes (45.8%), Firmicutes (25.9%), Fibrobacteres (8.8%), Spirochaetes (7.8%), Actinobacteria (5.9%) and Euryarchaeota (1.7%). The most relatively abundant genus was Prevotella (35.8%), followed by Fibrobacter (8.8%), Treponema (7.6%), Lactobacillus (7.2%) and Megasphaera (5.6%) (Figure 1). In terms of methanogens, overall there were three genera identified, all with a relative abundance of less than 0.1%: Methanobrevibacter (0.09%), Methanomicrobium (0.01%), and Methanosphaera (0.0008%). However, in the original rumen solid sample, Methanobrevibacter comprised 14.6% of the total microbial population.
FIGURE 1.
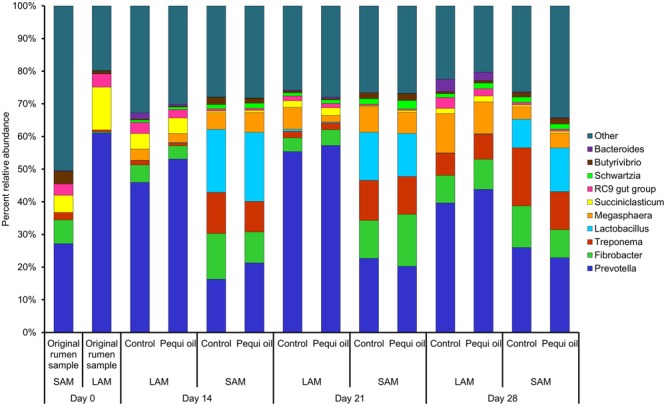
Classification of 16S rRNA gene sequences at the genus level for each individual sampling day, incubation temperature, sample type and treatment. Only the ten most relatively abundant genera are displayed (N = 3 for all samples). LAM = liquid-associated microbes, SAM = solid-associated microbes.
Effect of Pequi Oil, Sample Type, and Incubation Temperature on Archaeal and Bacterial Diversity and Microbiota Structure
There was greater archaeal and bacterial diversity and richness in the SAM samples compared with the LAM samples based on the alpha-diversity metrics listed in Table 4 (P < 0.05). Alpha-diversity analysis also suggested that a temperature effect was present only in the LAM samples as LAM samples incubated at 39°C had a higher Shannon diversity value and greater phylogenetic diversity (P < 0.01). The addition of pequi oil had no effect on any of the diversity or richness metrics for either LAM or SAM samples. Unweighted UniFrac distances were used to determine the effect that pequi oil and incubation temperature had on the archaeal and bacterial community structure. Similar to the within-sample diversity analysis, ANOSIM of unweighted UniFrac distances demonstrated that sample type (LAM vs. SAM) was the most important factor in determining the archaeal and bacterial community structure (Figure 2; R-value = 0.994; P < 0.01). Pequi oil did not have any significant effect on the archaeal and bacterial community structure among either the LAM (Figure 3A; R-value = –0.010; P = 0.44) or SAM (Figure 3B; R-value = –0.025; P = 0.59) samples. In contrast, there was a strong temperature and time effect present in both the LAM (Figure 4A, R-value = 0.395; P < 0.01) and SAM (Figure 4B; R-value = 0.410; P < 0.01) samples. However, it should be noted that the microbial community on day 28 did not return to a completely similar state as it was on day 14, when the incubation temperature was also 39°C.
Table 4.
Richness and diversity measures for each sampling day, incubation temperature, sample type and treatment (N = 3).
| 39°C | 35°C | 39°C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (Day 8–14) |
(Day 15–21) |
(Day 22–28) |
P-value |
|||||||
| Control | Pequi oil | Control | Pequi oil | Control | Pequi oil | SEM | Pequi oil | Temperature | Pequi oil × Temperature | |
| LAM | ||||||||||
| Shannon index | 4.44 | 4.38 | 4.12 | 4.19 | 4.51 | 4.47 | 0.084 | 0.87 | <0.01 | 0.75 |
| PD whole tree | 81.6 | 83.9 | 81.0 | 78.2 | 89.6 | 89.3 | 1.74 | 0.85 | <0.01 | 0.38 |
| Observed OTUs | 1292 | 1339 | 1323 | 1242 | 1465 | 1506 | 51.8 | 0.96 | <0.01 | 0.41 |
| SAM | ||||||||||
| Shannon index | 4.49 | 4.41 | 4.52 | 4.74 | 4.60 | 4.51 | 0.103 | 0.87 | 0.26 | 0.28 |
| PD whole tree | 85.7 | 85.4 | 83.1 | 87.4 | 88.1 | 86.3 | 1.38 | 0.53 | 0.35 | 0.11 |
| Observed OTUs | 1498 | 1488 | 1481 | 1624 | 1529 | 1473 | 38.0 | 0.42 | 0.27 | 0.05 |
LAM = liquid-associated microbes, SAM = solid-associated microbes.
FIGURE 2.
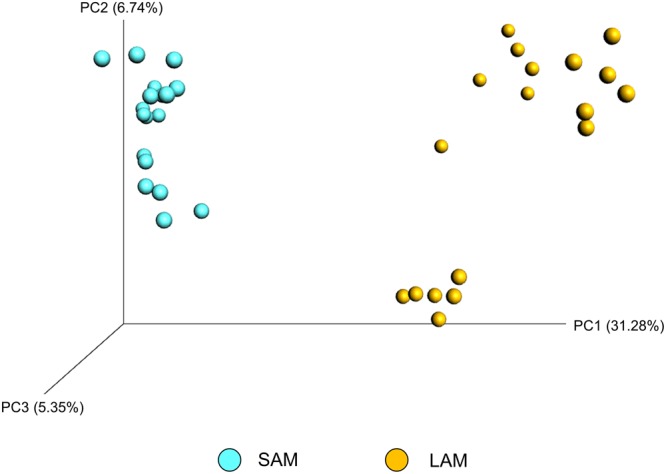
Principal coordinate analysis (PCoA) of the unweighted UniFrac distances for SAM and LAM samples. The percent variation explained by each principal coordinate is indicated on the axes. Original rumen samples are excluded. LAM, liquid-associated microbes; SAM, solid-associated microbes. N = 18 for both sample types.
FIGURE 3.

Principal coordinate analysis (PCoA) of the unweighted UniFrac distances for the effect of pequi oil supplementation on (A) LAM and (B) SAM samples. The percent variation explained by each principal coordinate is indicated on the axes. No rumen samples included. LAM, liquid-associated microbes; SAM, Solid-associated microbes.
FIGURE 4.
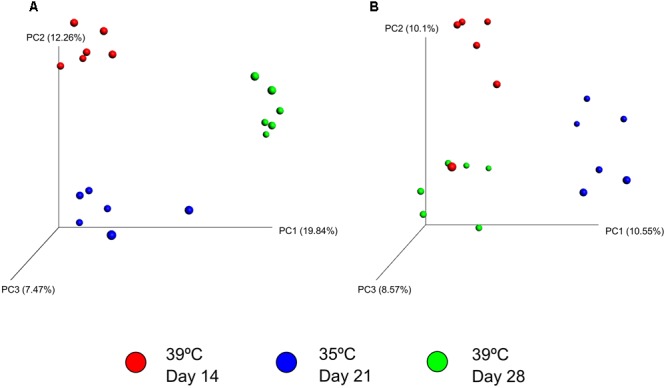
Principal coordinate analysis (PCoA) of the unweighted UniFrac distances for (A) LAM samples and (B) SAM samples for each sampling day and incubation temperature. The percent variation explained by each principal coordinate is indicated on the axes. Original rumen samples are excluded. LAM, liquid-associated microbes; SAM, solid-associated microbes. N = 6 for each sample type, sampling time and incubation time.
Differentially Abundant OTUs and Taxa between Sample Types, Incubation Temperatures, and Treatment Groups
Differentially abundant genera between pequi oil and control samples were identified using LEfSe. Anaerovibrio was found to be enriched in both the LAM and SAM samples under the pequi oil treatment compared with the control. The only other differentially abundant genus in either treatment group was Anaerovorax which was enriched in the control group of SAM samples (LDA score > 3.5; P < 0.05). When differences between LAM and SAM samples were assessed, a total of 11 genera were enriched in one of the two sample types (Figure 5; LDA score > 3.5). Of these genera, Prevotella was most notably enriched in the SAM samples and Lactobacillus in the LAM samples. Only three genera were enriched at one of the two incubation temperatures; Bacteroides and the RC9 gut group at 39°C and Streptococcus at 35°C (LDA score > 3.5; P < 0.05).
FIGURE 5.

Differentially abundant genera between LAM and SAM samples as determined using linear discriminant analysis effect size (LEfSe) analysis. Only genera with a LDA score greater than 3.5 and an overall relative abundance of greater than 0.01% were included in the analysis.
Excluding the original rumen samples, there were 194 OTUs that were differentially abundant (FDR < 0.05) between solid (84 OTUs) and liquid (110 OTUs) samples (Supplementary Table S1). There were also 57 OTUs that were differentially abundant between the samples incubated at 35°C and 39°C, with 27 more abundant at 35°C and 30 at 35°C (Supplementary Table S2). The most relatively abundant of these OTUs for both temperatures were classified as Prevotella spp. Only 20 OTUs were differentially abundant between the pequi oil (13 OTUs) and control (7 OTUs) samples (Supplementary Table S3).
Discussion
One of the strategies for lowering enteric methane emissions in ruminants has been the supplementation of diets with fatty acids due to their ability to decrease available organic matter fermented in the rumen, reduce methanogen activity and protozoal populations (Johnson and Johnson, 1995). In the present study, we evaluated the combined effect of pequi oil supplementation and incubation temperature shifts in a Rusitec system. Pequi oil comprised 6% of dietary fat content (DM-basis), as it has been recommended that fat inclusion in diets for the purpose of mitigating methane production should not exceed 6-7% due to the depression in DM intake (Johnson and Johnson, 1995). Despite the high content of the unsaturated fatty acid C18:1 in pequi oil (Segall et al., 2006), we did not find an inhibitory effect on methane production. In contrast, Beauchemin et al. (2009) reported a daily reduction of 13% in CH4 production when diets were supplemented with crushed oilseeds (3.7% supplementation) that were high in fatty acids (C18:1).
Our findings on methane production are more in accordance with the results of Johnson et al. (2002), who found that dietary fat supplementation from oilseeds (cotton and canola seeds) did not affect CH4 emissions in vivo at inclusion concentrations of 2.3, 4.0, and 5.6%. However, information regarding methane reduction and monounsaturated fatty acids (e.g., C18:1) is more limited when compared with polyunsaturated fatty acids such as linoleic and linolenic acid (Martin et al., 2010). In the current study, we found that supplementation with pequi oil had a positive effect on propionate production and a negative effect on the acetate:propionate ratio (Table 3) following the adaptation period (day 1–7; data not shown). However, these effects were influenced by an interaction between supplementation and incubation temperature, with greater propionate production at 39°C and a lower acetate:propionate ratio at 35°C, although propionate production was greater compared with the control diet at 35°C.
The effect that lipids have on methane abatement is of interest due to their potential to decrease methanogenesis through inhibition of protozoa and increased production of propionic acid (Johnson and Johnson, 1995). However, pequi oil supplementation did not alter the archaeal community in the present study. The lack of effect of pequi oil could be related to the relatively small concentration of pequi oil used in relation to the total volume of the fermenter (1.5 mL pequi oil/820 mL fermenter). When expressed in terms of incubated substrate DM, pequi oil comprised 6% of dietary fat content. However, it represents only 0.18% of the total fermenter volume and therefore, this may account for the differences observed in the current study in relation to CH4 production compared to earlier work in our lab using batch culture and pequi oil (Unpublished data).
Environmental temperature has been associated with methane production and changes in feed digestibility (Shibata and Mukai, 1979; Nonaka et al., 2008; Shibata and Terada, 2010). In the current work, we observed that CH4 production was negatively affected by incubation temperature. Decreasing the fermentation temperature to 35°C from 39°C reduced CH4 production (mg/d) by 43%. Similarly, DM digestibility was also reduced at 35°C. To date, relatively few in vitro studies have evaluated the effect of temperature on fermentation parameters and methane production. Bhatta et al. (2006), for example, evaluated the effect of two different incubation temperatures in a Rusitec and concluded that a minor increase in temperature (39°C vs. 41°C) did not have any significant effect on methane production. These authors also reported that pH was the most important factor affecting microbial production of VFA and CH4, suggesting that altering pH might be a way to optimize VFA production and reduce CH4 production (Bhatta et al., 2006). Decreasing the incubation temperature to 35°C for 7 days and returning to normal rumen temperature of 39°C is a potentially novel way to affect CH4 production. In justification for this experiment, the reduction of the rumen temperature in vivo could be similarly achieved through the introduction of a specific device into the rumen which could adjust rumen temperature. This would allow for further evaluation of decreased rumen temperature on rumen fermentation and production parameters.
High-throughput sequencing of the archaeal and bacterial 16S rRNA gene revealed that the addition of pequi oil to the Rusitec system did not alter the ruminal microbiota. With the exception of the original rumen SAM sample, the relative abundance of methanogens remained low throughout the study (1.4% overall). Methanobrevibacter spp. were the most relatively abundant methanogens, a finding that is in accordance with Henderson et al. (2015). These authors reported that members of the Methanobrevibacter were widely distributed in the microbiota of several different ruminant species and accounted for 74% of all ruminal archaeal genera. Among other methanogens, only Methanosphaera and Methanomicrobium were identified, an expected finding given that the rumen archaeal microbiota is much less rich and diverse compared to the bacterial microbiota (Henderson et al., 2015).
We observed increased production of propionate and reduced acetate:propionate ratio in the present study. It is possible that this result was due to the high relative abundance of Prevotella in this experiment, as Prevotella spp. have been reported to be propionate producers (Russell and Hespell, 1981; McCabe et al., 2015) and members of this genus have shown clear dominance in the rumen (Stevenson and Weimer, 2007; Henderson et al., 2013). Although pequi oil supplementation did not have any effect on the structure of the archaeal and bacterial microbiota in the Rusitec, the genus Anaerovibrio was found to be enriched in the pequi oil-supplemented samples. Anaerovibrio has been reported to have an important role in the ruminal metabolism of lipids, producing substrates used for propionate synthesis (Jouany et al., 2007; Jenkins et al., 2008).
The archaeal and bacterial richness and diversity indices suggested a temperature effect, but only for the LAM samples, with lower diversity and richness observed in LAM samples incubated at 35°C (day 14 to 21). When the incubation temperature was returned to 39°C during days 21 to 28, the richness and phylogenetic diversity actually increased in comparison with the original 39°C incubation period. In addition, incubation temperature did not affect the diversity of the SAM samples, which may explain why methanogen populations were also not affected by incubation temperature. However, the reduction in methane production which continued when the temperature was returned to the original incubation temperature of 39°C (day 22–28), suggests that methanogen activity was affected but not the overall microbial community. In addition, it is possible that using a PCR primer pair that is more selective and sensitive for methanogens may provide better resolution of the ruminal methanogenic community.
Conclusion
We determined that a 4°C decrease in temperature resulted in a 57% reduction of methane production and also affected microbial diversity, but only for LAM samples. This could suggest that specific methanogens within the LAM samples have an important role in methane production. Although the supplementation with pequi oil in this study did not affect methane production or microbial diversity, propionate production was enhanced. The microbiota of both the LAM and SAM samples were also strongly affected by temperature/sampling time. Further in vivo studies manipulating the rumen temperature are advised to verify its effect on CH4 emissions and/or its effects on the microbiome.
Author Contributions
Study design: AC, GB, AD. Conducting RUSITEC study: AD, AC. Lab analysis: AD, KK, DH, TA, GB, AC. DNA extraction: AD, KK. Bioinformatics: DH, AD. Writing the manuscript: AD, DH, TA, GB, AC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to Alexander von Humboldt Foundation for supporting Alex V. Chaves during his fellowship. We acknowledge Marion Burmester for her unconditional lab assistance during this 6 months fellowship.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01076/full#supplementary-material
References
- Aronesty E. (2013). Comparison of sequencing utility programs. Open Bioinform. J. 7 1–8. 10.2174/1875036201307010001 [DOI] [Google Scholar]
- Avila-Stagno J., Chaves A. V., Ribeiro G. O., Jr., Ungerfeld E. M., McAllister T. A. (2014). Inclusion of glycerol in forage diets increases methane production in a rumen simulation technique system. Br. J. Nutr. 111 829–835. 10.1017/S0007114513003206 [DOI] [PubMed] [Google Scholar]
- Beauchemin K., Kreuzer M., O’mara F., McAllister T. (2008). Nutritional management for enteric methane abatement: a review. Anim. Prod. Sci. 48 21–27. 10.1071/EA07199 [DOI] [Google Scholar]
- Beauchemin K. A., McGinn S. M., Benchaar C., Holtshausen L. (2009). Crushed sunflower, flax, or canola seeds in lactating dairy cow diets: effects on methane production, rumen fermentation, and milk production. J. Dairy Sci. 92 2118–2127. 10.3168/jds.2008-1903 [DOI] [PubMed] [Google Scholar]
- Bhatta R., Tajima K., Kurihara M. (2006). Influence of temperature and pH on fermentation pattern and methane production in the rumen simulating fermenter (RUSITEC). Asian Australas J. Anim. Sci. 19 376 10.5713/ajas.2006.376 [DOI] [Google Scholar]
- Caporaso J. G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., Knight R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26 266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1) 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crater A. R., Barboza P. S. (2007). The rumen in winter: cold shocks in naturally feeding Muskoxen (Ovibos moschatus). J. Mammal. 88 625–631. 10.1644/06-MAMM-A-170R1.1 [DOI] [Google Scholar]
- Czerkawski J. W., Breckenridge G. (1977). Design and development of a long-term rumen simulation technique (Rusitec). Br. J. Nutr. 38 371–384. 10.1079/BJN19770102 [DOI] [PubMed] [Google Scholar]
- Deppenmeier U. (2002). The unique biochemistry of methanogenesis. Prog. Nucleic Acid Res. Mol. Biol. 71 223–283. 10.1016/S0079-6603(02)71045-3 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61 1–10. 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- Graham N. M., Wainman F., Blaxter K., Armstrong D. (1959). Environmental temperature, energy metabolism and heat regulation in sheep. I. Energy metabolism in closely clipped sheep. J. Agric. Sci. 52 13–24. 10.1017/S0021859600035632 [DOI] [Google Scholar]
- Henderson G., Cox F., Ganesh S., Jonker A., Young W. Global Rumen Census Collaborators et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567 10.1038/srep14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson G., Cox F., Kittelmann S., Miri V. H., Zethof M., Noel S. J., et al. (2013). Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS ONE 8:e74787 10.1371/journal.pone.0074787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook S. E., Wright A. D., McBride B. W. (2010). Methanogens: methane producers of the rumen and mitigation strategies. Archaea 2010:945785 10.1155/2010/945785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristov A., Oh J., Lee C., Meinen R., Montes F., Ott T., et al. (2013). “Mitigation of greenhouse gas emissions in livestock production: a review of technical options for non-CO2 emissions,” in Proceedings of the Food and Agriculture Organization of the United Nations Technical Workshop Report Rome: 1–255. [Google Scholar]
- Jenkins T. C., Wallace R. J., Moate P. J., Mosley E. E. (2008). Board-invited review: recent advances in biohydrogenation of unsaturated fatty acids within the rumen microbial ecosystem. J. Anim. Sci. 86 397–412. 10.2527/jas.2007-0588 [DOI] [PubMed] [Google Scholar]
- Johnson K. A., Johnson D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73 2483–2492. 10.2527/1995.7382483x [DOI] [PubMed] [Google Scholar]
- Johnson K. A., Kincaid R. L., Westberg H. H., Gaskins C. T., Lamb B. K., Cronrath J. D. (2002). The effect of oilseeds production in diets of lactating cows on milk emissions methane. J. Dairy Sci. 85 1509–1515. 10.3168/jds.S0022-0302(02)74220-3 [DOI] [PubMed] [Google Scholar]
- Jouany J.-P., Lassalas B., Doreau M., Glasser F. (2007). Dynamic features of the rumen metabolism of linoleic acid, linolenic acid and linseed oil measured in vitro. Lipids 42 351–360. 10.1007/s11745-007-3032-x [DOI] [PubMed] [Google Scholar]
- Kennedy P. M., Milligan L. P. (1978). Effects of cold exposure on digestion, microbial synthesis and nitrogen transformations in sheep. Br. J. Nutr. 39 105–117. 10.1079/BJN19780017 [DOI] [PubMed] [Google Scholar]
- Li X., Durmic Z., Liu S., McSweeney C. S., Vercoe P. E. (2014). Eremophila glabra reduces methane production and methanogen populations when fermented in a rusitec. Anaerobe 29 100–107. 10.1016/j.anaerobe.2013.10.008 [DOI] [PubMed] [Google Scholar]
- Lima A. D., Silva A. D. O., Trindade R. A., Torres R. P., Mancini-Filho J. (2007). Composição química e compostos bioativos presentes na polpa e na amêndoa do pequi (Caryocar brasiliense, Camb.). Rev. Bras. Frutic. 29 695–698. 10.1590/S0100-29452007000300052 [DOI] [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71 8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Morgavi D. P., Doreau M. (2010). Methane mitigation in ruminants: from microbe to the farm scale. Animal 4 351–365. 10.1017/S1751731109990620 [DOI] [PubMed] [Google Scholar]
- McCabe M. S., Cormican P., Keogh K., O’Connor A., O’Hara E., Palladino R. A., et al. (2015). Illumina MiSeq phylogenetic amplicon sequencing shows a large reduction of an uncharacterised Succinivibrionaceae and an increase of the Methanobrevibacter gottschalkii Clade in feed restricted cattle. PLoS ONE 10:e0133234 10.1371/journal.pone.0133234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaez N., Wang Y., Xu Z., Alexander T., Garden S., McAllister T. (2013). Effects of hop varieties on ruminal fermentation and bacterial community in an artificial rumen (rusitec). J. Sci. Food Agric. 93 45–52. 10.1002/jsfa.5725 [DOI] [PubMed] [Google Scholar]
- Nonaka I., Takusari N., Tajima K., Suzuki T., Higuchi K., Kurihara M. (2008). Effects of high environmental temperatures on physiological and nutritional status of prepubertal Holstein heifers. Livest. Sci. 113 14–23. 10.1016/j.livsci.2007.02.010 [DOI] [Google Scholar]
- Oeztuerk H., Schroeder B., Beyerbach M., Breves G. (2005). Influence of living and autoclaved yeasts of Saccharomyces boulardii on in vitro ruminal microbial metabolism. J. Dairy Sci. 88 2594–2600. 10.3168/jds.S0022-0302(05)72935-0 [DOI] [PubMed] [Google Scholar]
- Patra A. K. (2013). The effect of dietary fats on methane emissions, and its other effects on digestibility, rumen fermentation and lactation performance in cattle: a meta-analysis. Livest. Sci. 155 244–254. 10.1016/j.livsci.2013.05.023 [DOI] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riede S., Boguhn J., Breves G. (2013). Studies on potential effects of fumaric acid on rumen microbial fermentation, methane production and microbial community. Arch. Anim. Nutr. 67 368–380. 10.1080/1745039X.2013.830518 [DOI] [PubMed] [Google Scholar]
- Rogerson A. (1960). The effect of environmental temperature on the energy metabolism of cattle. J. Agric. Sci. 55 359–364. 10.1017/S0021859600023236 [DOI] [Google Scholar]
- Romero-Perez G. A., Ominski K. H., McAllister T. A., Krause D. O. (2011). Effect of environmental factors and influence of rumen and hindgut biogeography on bacterial communities in steers. Appl. Environ. Microbiol. 77 258–268. 10.1128/AEM.01289-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Hespell R. B. (1981). Microbial rumen fermentation. J. Dairy Sci. 64 1153–1169. 10.3168/jds.S0022-0302(81)82694-X [DOI] [PubMed] [Google Scholar]
- Segall S. D., Artz W. E., Raslan D. S., Ferraz V. P., Takahashi J. A. (2006). Triacylglycerol analysis of pequi (Caryocar brasiliensis Camb.) oil by electrospray and tandem mass spectrometry. J. Sci. Food Agric. 86 445–452. 10.1002/jsfa.2349 [DOI] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27 379–423. 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- Shibata M., Mukai A. (1979). Effect of heat stress and hay-concentrate ratios on milk production, heat production and some physiological responses of lactating cows. Jpn. J. Zoot. Sci. 50 630–644. [Google Scholar]
- Shibata M., Terada F. (2010). Factors affecting methane production and mitigation in ruminants. Anim. Sci. J. 81 2–10. 10.1111/j.1740-0929.2009.00687.x [DOI] [PubMed] [Google Scholar]
- Soliva C. R., Amelchanka S. L., Duval S. M., Kreuzer M. (2011). Ruminal methane inhibition potential of various pure compounds in comparison with garlic oil as determined with a rumen simulation technique (Rusitec). Br. J. Nutr. 106 114–122. 10.1017/S0007114510005684 [DOI] [PubMed] [Google Scholar]
- Soliva C. R., Amelchanka S. L., Kreuzer M. (2015). The requirements for rumen-degradable protein per unit of fermentable organic matter differ between fibrous feed sources. Front. Microbiol. 6:715 10.3389/fmicb.2015.00715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soofi-Siawash R., Mathison G. (1994). Factors influencing particle size distributions in finely ground forage. Can. J. Anim. Sci. 74 383–385. 10.4141/cjas94-053 [DOI] [Google Scholar]
- Stevenson D. M., Weimer P. J. (2007). Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75 165–174. 10.1007/s00253-006-0802-y [DOI] [PubMed] [Google Scholar]
- Van Soest P. J., Robertson J. B., Lewis B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74 3583–3597. 10.3168/jds.S0022-0302(91)78551-2 [DOI] [PubMed] [Google Scholar]
- Vazquez-Baeza Y., Pirrung M., Gonzalez A., Knight R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


