Abstract
Microbes employ the thioredoxin system to defend against oxidative stress and ensure correct disulfide bonding to maintain protein function. Listeria monocytogenes has been shown to encode a putative thioredoxin, TrxA, but its biological roles and underlying mechanisms remain unknown. Here, we showed that expression of L. monocytogenes TrxA is significantly induced in bacteria treated with the thiol-specific oxidizing agent, diamide. Deletion of trxA markedly compromised tolerance of the pathogen to diamide, and mainly impaired early stages of infection in human intestinal epithelial Caco-2 cells. In addition, most trxA mutant bacteria were not associated with polymerized actin, and the rare bacteria that were associated with polymerized actin displayed very short tails or clouds during infection. Deletion or constitutive overexpression of TrxA, which was regulated by SigH, severely attenuated the virulence of the pathogen. Transcriptome analysis of L. monocytogenes revealed over 270 genes that were differentially transcribed in the ΔtrxA mutant compared to the wild-type, especially for the virulence-associated genes plcA, mpl, hly, actA, and plcB. Particularly, deletion of TrxA completely reduced LLO expression, and thereby led to a thoroughly impaired hemolytic activity. Expression of these virulence factors are positively regulated by the master regulator PrfA that was found here to use TrxA to maintain its reduced forms for activation. Interestingly, the trxA deletion mutant completely lacked flagella and was non-motile. We further confirmed that this deficiency is attributable to TrxA in maintaining the reduced intracellular monomer status of MogR, the key regulator for flagellar formation, to ensure correct dimerization. In summary, we demonstrated for the first time that L. monocytogenes thioredoxin A as a vital cellular reductase is essential for maintaining a highly reducing environment in the bacterial cytosol, which provides a favorable condition for protein folding and activation, and therefore contributes to bacterial virulence and motility.
Keywords: Listeria monocytogenes, thioredoxins, motility, host infection, redox interaction
Introduction
Listeria monocytogenes is a Gram-positive foodborne pathogen that causes listeriosis leading to high mortality, especially in the aging population, infants, and immunocompromised individuals (Vazquez-Boland et al., 2001; Corr and O'Neill, 2009). The organism is well-adapted to various physiological environments, employing different strategies to counteract hostile acidity, osmolality, oxygen tension, and other stress conditions (Cotter et al., 2001; Wemekamp-Kamphuis et al., 2004; Cheng et al., 2013) present in the environment and within the vacuolar compartment of phagocytic cells (Kim et al., 2007). Among these, oxidative stress is an imbalance in electrons that can damage DNA, iron-sulfur clusters, lipids, and proteins. Oxidative stress is both produced by the bacteria (endogenous) and encountered in the environment (exogenous; Imlay, 2003). Therefore, bacteria have evolved diverse detoxification mechanisms to survive in oxygen-rich environments, including production of antioxidants and enzymes that consume damaging reactive oxygen species (ROS; Whiteley et al., 2017). The intracellular life cycle of L. monocytogenes begins when the bacterium is phagocytosed by a host cell, where it transiently resides within the oxidizing environment of the phagosome. The bacteria then secrete the pore-forming toxin listeriolysin O to escape from the phagosome and enter into the cytosol that is a highly reducing environment (Schnupf and Portnoy, 2007). The rapid transit from the oxidizing phagosome to the reducing cytosol, making L. monocytogenes an ideal model for studying adaptive responses to redox changes during infection.
The thioredoxin (Trx) system comprising nicotinamide adenine dinucleotide phosphate (NADP), thioredoxin and the partner enzyme, thioredoxin reductase (TrxR), is widely distributed in several species, from archaea and bacteria to human. This antioxidant system transfers electrons from NADP through TrxR to Trx and then to a large range of proteins that play critical roles in DNA synthesis and oxidative stress (Holmgren, 1985; McCarver and Lessner, 2014). The oxidoreductase, thioredoxin, contains a common structural fold (Trx domain) and is involved in cellular defense against oxidative stress caused by reactive oxygen species (ROS), such as hydrogen peroxide, hydroxyl radicals, and superoxide anions (Boles and Singh, 2008; Yoshioka et al., 2012). Excessive amounts of these highly reactive compounds may damage DNA, proteins, and lipid membranes (Sampayo et al., 2003). In the microbial system, recovery from ROS stress is accomplished by Trx and TrxR (Yoshida et al., 2003). In several bacterial species, such as Staphylococcus aureus (Uziel et al., 2004), Escherichia coli (Ritz et al., 2000), Bacillus subtilis (Leichert et al., 2003), Rhodobacter sphaeroides (Li et al., 2003), and Brevibacillus choshinensis (Tanaka et al., 2004), thioredoxins play a major role in the protection of cells against ROS as well as maintaining the intracellular redox homeostasis. Importantly, the Trx system is the major cellular disulfide reductase in cells, which provides the electron donor for many critical enzymes, including ribonucleotide reductase, Prx, and methionine sulfoxide reductase, and is thus involved in a range of cellular functions (Lu and Holmgren, 2014b). The host cytosol is a highly reducing environment and upon entry into this compartment, all thiols are expected to be in the reduced form, which provides a favorable environment for protein folding and activation. A good example is for activation of PrfA, the master regulator controlling expression of most virulence genes in L. monocytogenes (Reniere et al., 2015). It was recently suggested that reduced glutathione (GSH) can function as an essential small molecule cofactor of PrfA through allosteric binding to the protein, thereby increasing the activity of PrfA at target genes(Hall et al., 2016). Moreover, PrfA thiols can be reversibly oxidized by the intracellular reactive oxygen and nitrogen species, temporarily inactivating the protein by inhibiting DNA binding, and leading to a downregulation of PrfA-regulated genes (Reniere et al., 2015). Therefore, we hypothesized that L. monocytogenes TrxA, as a reductase, might be highly involved in maintaining redox homeostasis in the bacterium for PrfA activation, and thus contribute to bacterial infection.
In addition, for oxidative protein folding in E. coli, Trx donates electrons to the disulfide bond (DSB) system and contributes to isomerization of incorrectly paired disulfide bonds (Rozhkova et al., 2004). Since many bacterial virulence factors require stable disulfide bonds for proper folding and function, studies on oxidoreductases are important for understanding the mechanisms underlying bacterial pathogenesis and the potential development of novel therapeutics. The disulfide bond formation (DSB) system in E. coli that catalyzes the oxidative folding step consists of several enzymes that form two distinct pathways: oxidative and isomerization (Ito and Inaba, 2008). The Trx domain-containing oxidase, DsbA, is responsible for the introduction of disulfide bonds into substrate proteins and subsequently obtaining the oxidizing equivalent from the membrane protein, DsbB, the second protein in the oxidative folding pathway (Missiakas et al., 1993). Due to DsbA misoxidation or external oxidative stress, isomerization of incorrectly paired disulfides in E. coli is performed by the DsbC-DsbD system. Mispaired disulfide bonds accept electrons from the homodimeric periplasmic protein, DsbC. DsbC accepts electrons from the membrane protein, DsbD, which, in turn, accepts electrons from the cytoplasmic Trx system (Missiakas et al., 1994, 1995). A DsbC homolog, DsbG, also located in the E. coli periplasm, is reported to function as a disulfide isomerase (Bessette et al., 1999).
Interestingly, many Gram-positive bacteria have different complements of DSB proteins. Searches for orthologs of B. subtilis DsbA and DsbB (known as BdbD and BdbC, respectively) in low GC Gram-positive bacteria revealed that many of these organisms contain either a DsbA or a DsbB protein, but not both (Kouwen et al., 2007). L. monocytogenes encodes two putative DSB proteins, Lmo0964 and Lmo1059, annotated as DsbA-like and DsbG in the GenBank database, respectively. Lmo0964 was recently designated as a putative thioredoxin similar to YjbH in Bacillus subtilis and shown to contribute to expression of the ActA protein, required for L. monocytogenes actin based motility (Reniere et al., 2016). Moreover, homologs of the Trx system-related genes have been identified in the sequenced genome of L. monocytogenes EGD-e via in silico analysis (Glaser et al., 2001). Based on a pattern search for the CXXC motif, the characteristic structure of Trx in the EGD-e genome, six thioredoxin family proteins (Lmo1233, 1609, 1903, 2152, 2424, 2830) and one TrxR (Lmo2478), were considered likely candidates (Gopal et al., 2009). Only the modeled structure of Lmo1233 (TrxA) coincided with the canonical conformation of thioredoxin containing the highly conserved CGPC motif. However, none of the components of the Trx system from L. monocytogenes have been characterized to date. In the current study, we aimed to fully elucidate the molecular functions and underlying mechanisms of the L. monocytogenes Trx system, with a view to determining whether it contributes to biological processes related to bacterial survival and infection. Our collective data indicate that L. monocytogenes thioredoxin A contributes to bacterial virulence and motility through redox interactions. The results from this study provide a valuable model for clarifying the pathways associated with the potential roles of thioredoxins from foodborne pathogens in improving survival in the external environment, and more importantly, successfully establishing infection within the host.
Results
Cys28 and Cys31 are required for the oxidoreductase activity of TrxA
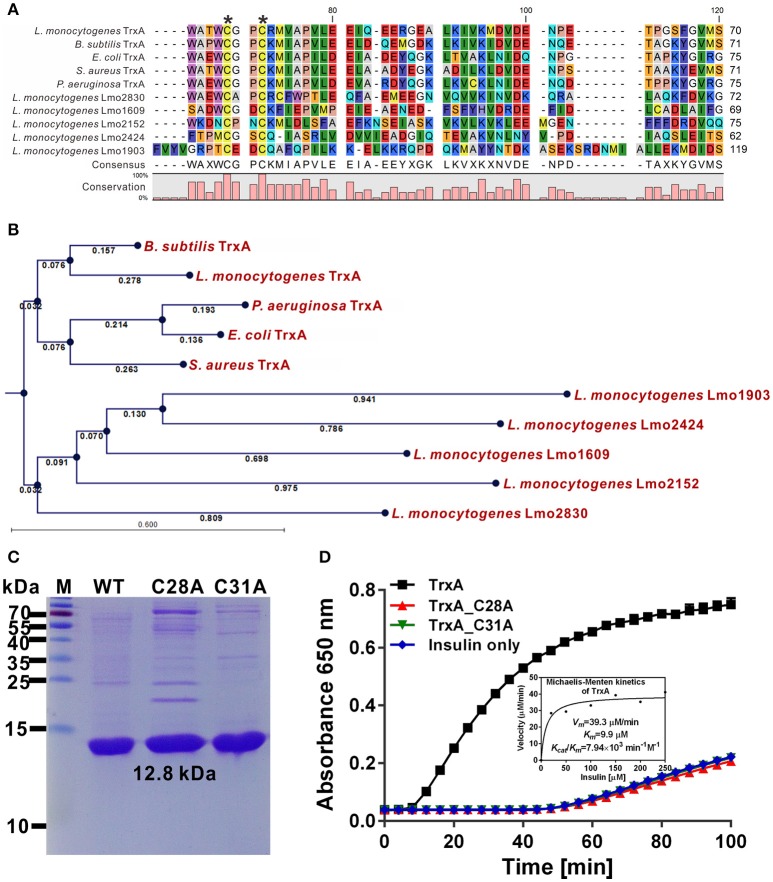
Six putative homologs, Lmo1233, 1609, 1903, 2152, 2424, and 2830, were annotated as thioredoxin-like proteins (Gopal et al., 2009). Among these, only TrxA contained the canonical CGPC active motif of the thioredoxins (Figure 1A). Moreover, L. monocytogenes TrxA shows 40.0–64.1% amino acid sequence homology to the Trxs from other microorganisms (Figure 1A). Phylogenetic analysis further revealed TrxA and its homolog from B. subtilis form a sister branch, while the other five thioredoxin homologures of L. monocytogenes form a separate clade (Figure 1B), indicating that these Trxs within this clade are distinct from the TrxA found in other bacteria species. Recombinant TrxA efficiently catalyzed the thiol-disulfide oxidoreduction of insulin in the presence of DTT as an electron donor (Figures 1C,D). Both C28A and C31A single mutations completely abolished the oxidoreductase ability of the protein to catalyze the reduction of insulin (Figure 1D), clearly indicating that L. monocytogenes trxA encodes a functional thioredoxin and the residues Cys28 and Cys31 are required for its catalytic activity.
Figure 1.
Cys28 and Cys31 of TrxA are required for oxidoreductase activity. (A) Amino acid sequence alignment of L. monocytogenes putative thioredoxins (TrxA, Lmo1609, 1903, 2152, 2424, and 2830) against homologs from B. subtilis, E. coli, P. aeruginosa, and S. aureus. Key amino acid residues denoted with asterisks are involved in catalytic activity. (B) Phylogenetic tree of L. monocytogenes putative thioredoxins and homologs from other bacterial species. The tree was constructed with the Neighbor-Joining (NJ) program and a bootstrap test of 100 replicates used to estimate the confidence of branching patterns, where the numbers on internal nodes represent the support values. (C) SDS-PAGE (12% gel) analysis of recombinant TrxA and its mutants, C28A and C31A. (D) In vitro insulin reduction assay of recombinant TrxA and its mutants, C28A and C31A. The inset shows the Michaelis-Menten plot and kinetic parameters, Km, Vmax, and kcat, for TrxA. Data are representative of three independent experiments.
TrxA is responsible for bacterial resistance to thiol-specific oxidative stress
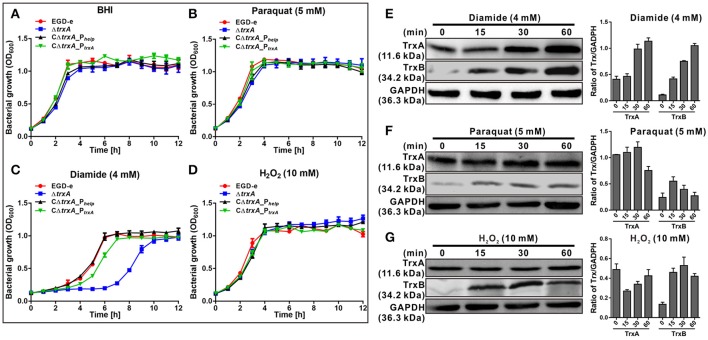
Previous work suggested a connection between the L. monocytogenes redox response and virulence (Reniere et al., 2016). These observations prompted us to investigate the role of the thioredoxin in L. monocytogenes. The trxA mutant strain exhibited similar growth to the parent strain in normal BHI medium (Figure 2A). To further determine whether the Trx system could contribute to oxidative tolerance, the bacteria were exposed to different oxidative agents. For oxidative stresses, paraquat was used as a superoxide generating compound, and H2O2 as a direct oxidant while diamide was used as a thiol-specific oxidizing agent. Notably, deletion of trxA led to a significantly increased lag phase in growth, starting from 4 h (P < 0.01), compared to its parental strain and two complement strains (CΔtrxA_PtrxA, expressing TrxA under its native promoter, and CΔtrxA_Phelp, carrying the constitutive Listeria promoter, Phelp) in BHI containing diamide, while H2O2 and paraquat did not exert marked effects on growth (Figures 2B–D). Immuno-blotting was used to determine expression of the Trx system under different oxidative stress conditions. As expected, diamide at a concentration of 4 mM induced expression of TrxA and TrxB (annotated as a thioredoxin reductase) to a significant extent, especially at the 30 and 60 min time-points (Figure 2E). However, no obvious induction was found in the presence of paraquat or H2O2 stress on TrxA expression, while TrxB was slightly induced by these two oxidants (Figures 2F,G). The results collectively suggest that diamide is a stronger inducer of TrxA expression than other oxidants, which enables L. monocytogenes to better survive in the thiol-specific oxidative conditions.
Figure 2.
TrxA is responsible for bacterial resistance to thiol-specific oxidative stress. (A–D) Growth of L. monocytogenes wild-type EGD-e, mutant ΔtrxA and its two complement strains, CΔtrxA_Phelp and CΔtrxA_PtrxA, in BHI broth supplemented with or without oxidative agents. Overnight bacteria were re-suspended in fresh BHI alone (A) or BHI supplemented with 5 mM paraquat (B), 4 mM diamide (C) or 10 mM H2O2 (D), and incubated at 37°C for 12 h. Kinetic growth at OD600nm was measured at 1 h intervals. Data are expressed as means ± SD of three independent experiments. (E–G) Western blot analysis of TrxA expression under oxidative conditions. Total bacterial cell-free proteins were isolated at the indicated times after treatment with 4 mM diamide (E), 4 mM paraquat (F) or 10 mM H2O2 (G), and protein levels of TrxA and TrxB determined via western blot. GAPDH was used as the internal control. Results are indicated as a grayscale ratio of TrxA to GADPH or TrxB to GADPH. Data are expressed as means ± SD of three independent experiments.
The L. monocytogenes trxA gene deletion mutant lacks flagella and is non-motile
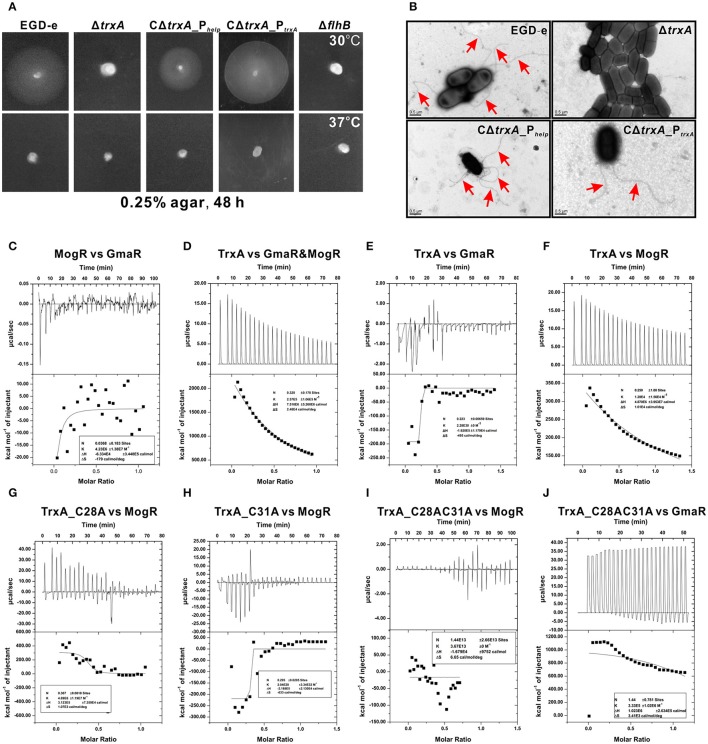
DsbA, a disulfide bond forming protein A comprising a Trx domain, catalyses the formation of disulfide bond that is involved in bacterial flagella synthesis (Heras et al., 2009). Thus, we examined whether TrxA is linked to motility in Listeria. Interestingly, motility of the ΔtrxA strain at 30°C was almost completely compromised, compared to its wild-type counterpart (Figure 3A). Impaired motility of the mutant was fully rescued in the complement strain, CΔtrxA_PtrxA, expressing TrxA under its native promoter, and partly restored in the other complement strain, CΔtrxA_Phelp, carrying the constitutive Listeria promoter, Phelp (Figure 3A), suggesting that L. monocytogenes TrxA contributes to bacterial motility. In contrast, no motility was observed at 37°C (Figure 3A), which inhibited flagellar formation in many Listeria strains (Grundling et al., 2004). Transmission electron microscopy further revealed that the trxA mutant is totally unable to produce visible flagella whereas morphologically normal-looking flagella were expressed by the wild-type and two trxA complement strains (Figure 3B). Our results demonstrate for the first time that L. monocytogenes TrxA participates in bacterial motility and flagellar production.
Figure 3.
TrxA contributes to L. monocytogenes motility and flagellar formation via interactions with MogR and GmaR (A,B). Motility assay (A) and transmission electron microscopy (TEM) (B) were performed on L. monocytogenes wild-type EGD-e, mutant ΔtrxA and its two complement strains, CΔtrxA_Phelp and CΔtrxA_PtrxA, grown on soft agar (0.25%) at 30° or 37°C. Mutant ΔflhB, which completely abolished bacterial motility, was taken as a reference control. (C–J) Isothermal titration calorimetry (ITC) measurement of protein-protein interactions. (C) ITC of interactions between MogR, the flagellar gene transcriptional repressor, and the anti-repressor, GmaR. (D) ITC of TrxA with MogR and GmaR. (E) ITC of TrxA binding to GmaR. (F) ITC of TrxA interactions with MogR. (G) ITC of interactions between the single mutant TrxA (C28A) and MogR. (H) ITC of interactions between the single mutant TrxA (C31A) and MogR. (I) ITC of interactions between the double Cys28/Cys31 mutant of TrxA (C28AC31A) and MogR. (J) ITC of interactions between the double Cys28/Cys31 mutant of TrxA (C28AC31A) and GmaR. For all titrations, raw data are presented in the upper panel and fitted binding curves in the lower panel.
TrxA is involved in the interactions between MogR and GmaR
Temperature-dependent expression of Listeria flagellar motility genes is mediated by the opposing activities of MogR, a DNA-binding transcriptional repressor, and GmaR, an anti-repressor that functions as a direct antagonist of MogR (Shen and Higgins, 2006; Shen et al., 2006; Kamp and Higgins, 2011). Given that MogR and GmaR contain 2 and 4 cysteines, respectively, the CGPC domain of TrxA is predicted to play a critical role in the interactions between MogR and GmaR, and therefore contribute to Listeria flagella formation. Calorimetric response data on MogR during reaction with GmaR showed significant scatter with no discernible trends, which was more evident in the corresponding binding isotherm plot (Figure 3C), indicating that in vitro interactions between MogR and GmaR are not strong. However, upon titration of purified TrxA into MogR and GmaR, the binding event exhibited endothermic heat of reaction. A good fit to the data for calorimetric titration was achieved using a single-site binding model (Figure 3D), suggesting that TrxA is required to increase the binding affinity of MogR to GmaR. TrxA displayed endothermic reactions for strong binding to MogR in a manner similar to calorimetric data obtained in Figure 3D, while showing weak binding affinity to GmaR (Figures 3E,F). Notably, the endothermic response clearly observed during the whole titration process between TrxA and MogR is believed to arise due to breakage of intermolecular bonds among MogR molecules as a consequence of titration of TrxA (Figure 3F), suggesting that this protein contributes to breaking the intermolecular bonds of MogR that bind DNA of the flagella-related gene promoter region as a dimer (Shen et al., 2009). More importantly, mutation of either C28A or C31A located within the critical motif of thioredoxin significantly weakened the binding affinity of TrxA to MogR and GmaR, as determined from calorimetric data (Figures 3G–J).
DSB oxidative and isomerization pathways are required for protein folding
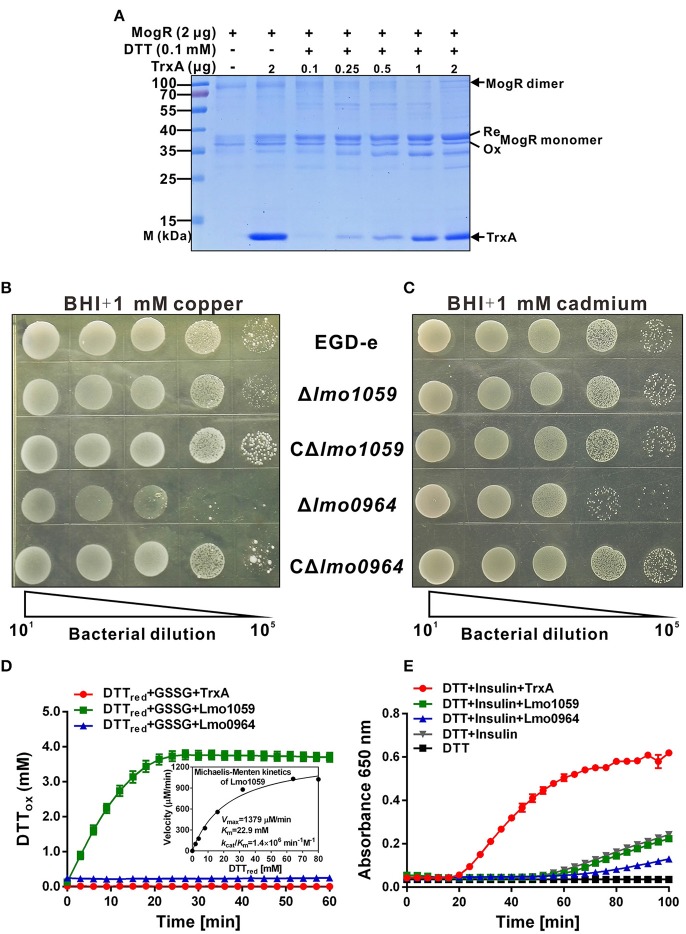
As indicated from isothermal titration calorimetry (ITC) data (Figure 3), TrxA could contribute to catalyzing the reduction of misfolded MogR disulfides and maintaining its intracellular monomer status. To further characterize the reduction properties of recombinant TrxA, we assayed the ability of the thioredoxin to reduce MogR in vitro. The thioredoxin assay directly measures the redox state of the substrate by using a gel-based assay. In the presence of SDS, a protein with free cysteine residues is less compact than the same protein in which these residues form disulfide bonds. The reduced form of a protein therefore migrates more slowly upon polyacrylamide gel electrophoresis (PAGE) in the presence of SDS than its oxidized counterpart. As indicated in the Figure 4A, the recombinant MogR monomer exists as a mixture of the reduced and oxidized forms. However, the reduced form amount of MogR monomers gradually increased when an increasing amount of the recombinant TrxA added in the presence of DTT as an electron donor, while the amount of MogR dimers decreased (Figure 4A), strongly confirming that TrxA could act to reduce disulfides in MogR and maintain the monomer status.
Figure 4.
The redox state of MogR and enzymatic analysis and metal sensitivity assay. (A) The in vitro redox state of MogR changes under the efficient reduction by TrxA. MogR is reduced following incubation with the increasing amount of TrxA in the presence of DTT, and the redox state of MogR was visualized by SDS-PAGE and Coomassie blue staining. The reduced and oxidized forms of MogR are indicated as Re and Ox, respectively. (B,C) L. monocytogenes WT EGD-e, Δlmo1059, Δlmo0964 and the complement strains, CΔlmo1059 and CΔlmo0964, were subjected to copper (A) and cadmium (B) sensitivity assays by spotting 107 cfu of each bacterial strain onto BHI plates containing 1 mM copper and 1 mM cadmium. (D) The disulfide interchange reaction was used to determine the isomerase activities of TrxA, Lmo1059, and Lmo0964. The inset depicts the Michaelis-Menten plot and kinetic parameters, Km, Vmax, and kcat, for Lmo1059. (E) In vitro insulin reductase activity was assayed for recombinant TrxA, Lmo1059, and Lmo0964 over a period of 100 min. Data in (D,E) are expressed as means ± SD of three independent experiments.
In addition, the thioredoxin system is the major cellular disulfide reductase in cells, which can facilitate correct oxidative protein folding together with protein disulfide isomerases (PDI) and DSB proteins (Lu and Holmgren, 2014b). Searches for orthologs of B. subtilis DsbA and DsbB in L. monocytogenes revealed that this pathogen only encodes two putative DSB proteins, DsbA (Lmo0964), and DsbG (Lmo1059), but does not encode DsbB or any other DSB proteins. Therefore, the roles of Lmo0964 and Lmo1059 in correct dimerization of MogR were further investigated. To investigate mutant sensitivity to oxidative stress, Listeria were grown in the presence of the redox-active metal, copper, which rapidly and randomly oxidizes unpaired cysteines through a superoxide mechanism (Hiniker et al., 2005). WT and Δlmo1059 had a similar resistance to copper while Δlmo0964 was extremely copper-sensitive (Figure 4B). Cadmium sensitivity was further tested as an indicator of oxidase capacity due to the high affinity of Cd2+ for protein thiol groups (Vallee and Ulmer, 1972). The lmo0964-null strain was especially Cd2+ sensitive, while wild-type and Δlmo1059 strains were resistant to a concentration of 1 mM Cd2+ (Figure 4C). Our results suggest that Lmo0964 acts not only as an oxidase but also an isomerase, whereas bacteria in the absence of lmo1059 remain cadmium- and copper-resistant. Interestingly, in vitro enzymatic reactions indicate that Lmo1059 exhibits high disulfide interchange activity (Figure 4D) but fails to catalyze reduction of insulin in the presence of DTT (Figure 4E), suggesting that this protein mainly functions as an isomerase but not an oxidoreductase in vitro. Intriguingly, distinct from E. coli DsbA or BdbD in B. subtilis, Lmo0964 is unable to catalyze reduction of insulin, at least under standard assay conditions, suggesting that this protein is more stable in its oxidized form and mainly functions as an oxidase in vitro.
TrxA alters global expression profiles of L. monocytogenes under oxidative conditions, especially expression of virulence-associated genes
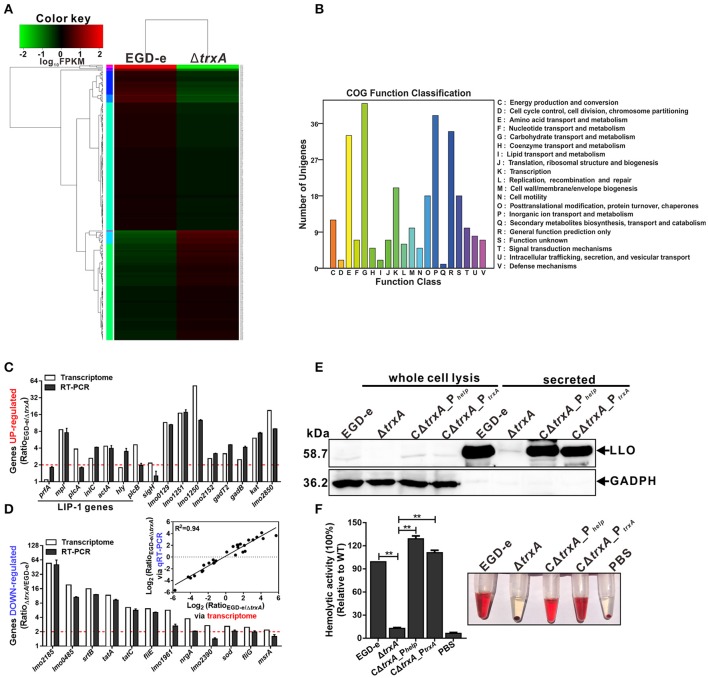
Whole-genome transcriptomic sequencing revealed that 160 genes show = 2.0-fold-higher transcript levels in parent EGD-e than the ΔtrxA mutant strain under oxidative conditions (expo0sure of stationary cells to 2 mM diamide for 1 h; Table S1, Figures 5A,B). Transcript levels of a total of 114 genes were =2.0-fold higher in the ΔtrxA mutant than the parent strain (Table S2, Figures 5A,B). Notably, the virulence genes, plcA, mpl, actA, and plcB, showed significantly higher expression in wild-type than the ΔtrxA mutant, with transcriptional ratios consistently >3.8 (Table S1). The internalin gene, inlC, also showed significant expression ratios of >2.0 (Table S1). In addition, three other virulence-associated genes, hly, inlA, and inlB, were slightly activated by TrxA (Table S1). Transcriptomic data were validated using quantitative RT-PCR, showing that the results of transcriptome were firmly reliable (Figures 5C,D). In addition, deletion of TrxA almost completely reduced cytoplasm LLO expression and significantly inhibited its secretion into culture supernatant, compared to the wild-type and complement strains (Figure 5E), which was further verified by the finding that loss of TrxA leads to thoroughly reduced LLO activity in lysis of sheep red blood cells (Figure 5F). These results collectively show for the first time that L. monocytogenes TrxA alters global expression profiles, especially those of genes encoding virulence-associated factors, and therefore most probably play a critical role in bacterial pathogenicity.
Figure 5.
L. monocytogenes TrxA alters global gene expression profiles, especially regulating genes encoding virulence-associated factors. (A) Cluster analysis of differentially expressed genes in the WT and ΔtrxA mutant strains via two-dimensional hierarchical clustering. The red color indicates upregulation, green indicates downregulation and black indicates no changes, compared to wild-type. The X-axis represents samples while the Y-axis represents genes. (B) Clusters of orthologous groups (COG) classification for differentially expressed unigenes in the WT and ΔtrxA mutant strains. A total of 274 proteins were aligned to the COG protein database and classified functionally into 25 classes. Each function class is denoted by different capital letters under the x-axis. The y-axis represents the number of unigenes in a corresponding function class. (C,D) qRT-PCR validation of the transcriptome. Twenty-eight genes were randomly selected for qRT-PCR validation, including 16 representative upregulated genes (C) and 12 representative downregulated genes (D). The inset shows normalized relationships between qRT-PCR and transcriptomes of randomly selected genes. Values are presented as the log2 ratio (EGD-e/ΔtrxA) for genes. The coefficient of determination (r2) is indicated. All qRT-PCR data are expressed as means ± SD of three independent experiments. (E,F) TrxA is essential for LLO expression and hemolytic activity. Bacterial overnight cultures of L. monocytogenes strains were diluted into 100 mL fresh BHI and grown to the stationary phase. Bacterial pellets and cultures were collected to obtain the different cell fractions, as described in Section Materials and Methods. (E) Proteins were separated via 12% SDS-PAGE and immunoblotted with α-LLO or α-GAPDH antisera. The MW of each protein is indicated on the left. GAPDH was used as an internal control. (F) LLO-associated hemolytic activity was assessed by lysis of sheep red blood cells from serial dilutions of culture supernatants of bacterial strains. Data are expressed as means ± SD of three independent experiments. **P < 0.01.
TrxA is required for early stages of L. monocytogenes infection and virulence within mice
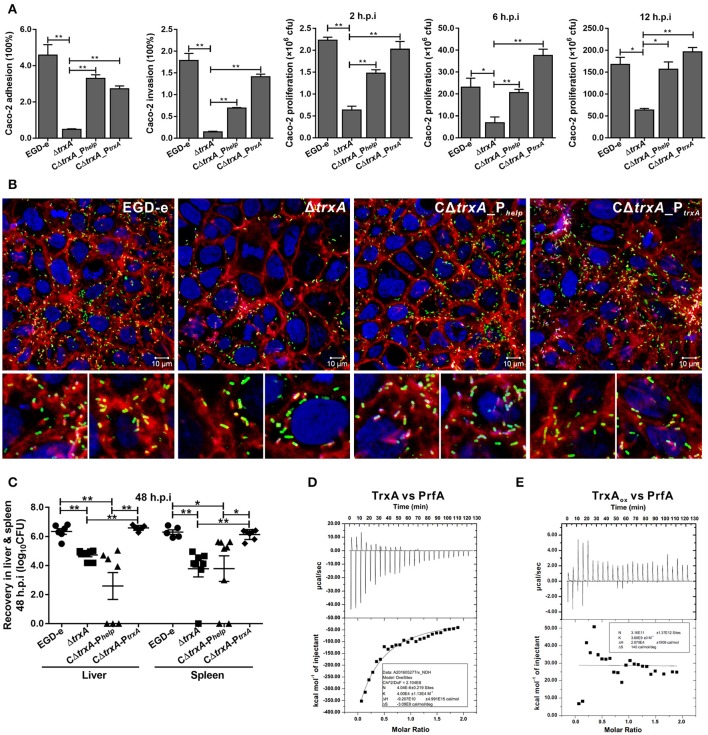
To investigate the involvement of TrxA in L. monocytogenes infection inside host cells, the epithelial cells Caco-2 was used to compare the wild-type and trxA mutant strains in terms of adhesion, invasion, and intracellular survival. The trxA mutant was significantly less adhesive and invasive, therefore resulting in the less efficient intracellular survival, compared to that of the wild-type strain (Figures 6A,B). Notably, EGD-e and the complement strains were able to associate with F-actin and formed long actin tails (Figure 6B), while a large proportion of the trxA mutant bacteria were not associated with polymerized actin, and the rare bacteria that were associated with polymerized actin displayed very short tails or clouds and were often located next to the plasma membrane (Figure 6B). However, unlike the circumstance in epithelial cells, the ΔtrxA mutant did not show a significantly decreased ability to grow intracellularly in RAW264.7 macrophages relative to the wild-type strain (data not shown), suggesting that TrxA mainly contributes to the early stages of L. monocytogenes infection within host. Moreover, the ΔtrxA mutant exhibited severely attenuated virulence, as infected mice exhibited bacterial burdens that were significantly lower relative to those infected with the wild-type strain at 48 h post infection (Figure 6C). Most importantly, the mutant strain containing the entire trxA CDS with its native promoter fully complemented virulence defects associated with loss of trxA, but not the Phelp promoter (Figure 6C), suggesting no expression or overexpression of TrxA in vivo would disturb the equilibrium of the intracellular redox potential and thus bring harm to this pathogen for its ability to establish infections. Previous work suggested that the sigma factor H (SigH) is important for the regulation of mycobacterial response to oxidative stress by regulating the expression of thioredoxins (trxB1 and trxC) and thioredoxin reductase (Bhat et al., 2012), which therefore prompted us to investigate the role of SigH in regulating on the Trx expression in L. monocytogenes. As determined by EMSA assay, the purified recombinant SigH bound to the fragment containing the promoter region of trxA or trxB. Besides, upon incubation of the promoter DNA fragment with increasing amounts of SigH, shifted, supershifted, and super-supershifted DNA complexes were observed (Figure S1), supporting the theory that the thioredoxin system of L. monocytogenes is strictly regulated by an alternative sigma factor. Taken together, these data demonstrate that TrxA-mediated regulation of virulence-associated factors constitutes an important aspect of L. monocytogenes infection within host. Importantly, the ITC experiment indicates that TrxA exhibits exothermic reactions for strong binding to the virulence regulator, PrfA, a member of the Crp/Fnr family of bacterial transcription factors (Figure 6D), while the oxidized forms of TrxA (TrxAox) almost showed no binding affinity to PrfA (Figure 6E). Given the fact that PrfA activation is highly dependent on the reducing environment and only the reduced PrfA dimers can bind to DNA and then activate transcription of virulence genes (Reniere et al., 2015), we therefore suggest that TrxA plays a critical role in reducing the intermolecular disulfide bonds of PrfA and subsequently maintaining the intracellular monomer status for further dimerization.
Figure 6.
TrxA contributes to L. monocytogenes infection and virulence within mice. (A) Adhesion, invasion, and proliferation of L. monocytogenes in human intestinal epithelial cells, Caco-2. Cells infected with L. monocytogenes strains at the indicated time points were lysed and viable bacteria serially plated on BHI agar plates. The number of bacteria able to invade cells and survive are expressed as means ± SD of the recovery rate for each strain. *P < 0.05; **P < 0.01. (B) Actin tail formation in Caco-2 cells infected with L. monocytogenes strains 6 h post inoculation. Bacteria were detected with anti-Lm (green), bacteria actin tails and host actin using phalloidine (red), and cell nucleus labeled with DAPI (blue). Scale bar is 10 μm. The high-magnification images displayed at the bottom of each image show F-actin (red), bacteria (green), and nuclei (blue). (C) Proliferation of L. monocytogenes in mouse organs. Mice inoculated i.p. with L. monocytogenes strains (1 × 106 CFU) were euthanized 48 h after infection, and organs (liver and spleen) recovered and homogenized. Homogenates were serially plated on BHI agar. Numbers of bacteria colonized in liver and spleen are expressed as means ± SD of the recovery rate per organ for each group. *P < 0.05; **P < 0.01; ns means no significance. (D,E) The interactions analysis of TrxA (D) or the oxidized forms of TrxA (TrxAox) (E) with PrfA, the central virulence regulator of Listeria, by using the ITC assay. For all titrations, raw data are presented in the upper panel and fitted binding curves in the lower panel.
Discussion
L. monocytogenes adapts to a range of environmental extremes, including high salt concentrations, low pH, and various oxidative stress conditions in nature and host cells (Begley et al., 2010). Thioredoxins play major roles in protection of cells against toxic oxygen species as well as maintaining the intracellular thio-disulfide balance in bacteria (Uziel et al., 2004). In our experiments, Listeria depleted of trxA displayed significant growth defects upon exposure to diamide. Diamide, a thiol-specific oxidizing agent, induces glutathione depletion by trapping the free-SH group to increase the redox potential, and is therefore used to mimic oxidative shift in the cellular thiol-disulfide redox state (disulfide stress; Kallifidas et al., 2010). Cysteine thiols in bacterial proteins fulfill an important and diverse set of cellular functions, and ROS and reactive nitrogen species, so-called “reactive electrophilic species” (RES), affect the thiol redox balance (Pother et al., 2009). The toxicity of diamide is based on the formation of nonnative inter- and intra-molecular disulfide bonds that result in damage of proteins (Pother et al., 2009). We here found that the Listeria Trx system is differentially induced by a variety of agents that generate oxidative stress, implying that different regulatory mechanisms are involved in controlling the expression of trxA and other genes that participate in the oxidative stress response.
The thioredoxin (Trx) system, which is composed of NADPH, thioredoxin reductase (TrxR), and thioredoxin, is a key antioxidant system in defense against oxidative stress through its disulfide reductase activity regulating protein dithiol/disulfide balance (Lu and Holmgren, 2014a; Shelar et al., 2015). Thioredoxins are proposed to participate directly in the oxidative stress response owing to their capability to reduce oxidized proteins. However, these proteins can also participate indirectly by affecting the expression of other genes involved in this response. Smits et al. (2005) previously reported the effects of thioredoxin depletion on global transcription levels in B. subtilis, whereby a total of 636 genes showed >2-fold changes in transcription levels. Effective depletion of thioredoxin A from B. subtilis induced genes involved in the oxidative stress response (but not those dependent on PerR), phage-related functions, and sulfur utilization. Moreover, several stationary phase processes, such as sporulation and competence, were affected. Since thioredoxins have not been reported to act as transcriptional regulators, the authors suggested that these transcriptional changes are likely to represent indirect effects of thioredoxin A (e.g., interactions with or influence on transcription factors or other proteins; Smits et al., 2005). Moreover, thioredoxins are involved in redox-dependent regulation of photosynthesis genes in Rhodobacter (Li et al., 2003). Importantly, in the current study, the known Listeria virulence genes, hly, plcA, mpl, actA, and plcB, showed significantly higher expression in wild-type than the ΔtrxA mutant strain. These data imply that downregulation of expression of virulence factors in the mutant strain contributes significantly to the virulence defects. Bacteria express a range of disulfide-bonded virulence factors, including secreted toxins, surface components, such as adhesins and pili, and secretion systems. To ensure functional activity, many of these proteins must be oxidatively folded (Heras et al., 2009). The thioredoxin system is the major cellular disulfide reductase in cells, which can provide a highly reducing environment and then function as an effector to facilitate correct oxidative protein folding, together with protein disulfide isomerases (PDI) and DSB proteins (Lu and Holmgren, 2014b). PrfA is a member of the cAMP receptor protein (Crp) family of transcription factors, which are characterized by their allosteric regulation via small-molecule activators. In L. monocytogenes, PrfA is exclusively activated in the cytosol of host cells, leading to the assumption that the activating cofactor for PrfA is specific to this compartment (Reniere et al., 2015). Recently, glutathione (GSH), either generated by bacteria or derived from host cells, was found to be the essential small molecule cofactor of PrfA through allosteric binding to the protein (Hall et al., 2016). The process of infection or intercellular spread requires that L. monocytogenes inhabit an oxidizing vacuole, which may contain both reactive oxygen and nitrogen species. Upon oxidation, glutathione dimerizes to GSSG, which does not bind PrfA, owning to the ternary complex structure of PrfA cannot accommodate the larger oxidized glutathione (GSSG; Wang et al., 2017). In addition, PrfA thiols may be reversibly oxidized, temporarily inactivating the protein by inhibiting DNA binding and leading to a downregulation of PrfA-regulated genes (Reniere et al., 2015). It has been proposed that PrfA activation constitutes a two-step process requiring reduced protein thiols for initial DNA binding and allosteric binding of glutathione to PrfA for fully transcriptional activation (Reniere et al., 2015), indicating that the GSH-activated PrfA is primed for DNA binding (Hall et al., 2016). Consistent with this, the glutathione synthase (gshF)-deficient L. monocytogenes was moderately more sensitive to oxidative stress in vitro, expressed lower levels of the PrfA dependent virulence factors (ActA, LLO, and Hpt) in cells, formed very small plaques in tissue culture assay, and thus was attenuated in vivo (Reniere et al., 2015). Similarly, it was recently reported that LLO can be posttranslationally modified by S-glutathionylation at the conserved cysteine residue (Cys484) and that either endogenously synthesized or exogenously added glutathione was sufficient to form this modification. When recapitulated with purified protein in vitro, this modification completely ablated the activity of LLO, and this inhibitory effect was fully reversible by treatment with reducing agents (Portman et al., 2017). Previously, it has been showed that, GILT, a soluble thiol reductase expressed constitutively within the lysosomes of antigen-presenting cells, is able to activate LLO within the phagosome by the thiol reductase mechanism shared by members of the thioredoxin family, and therefore acts a critical host factor that facilitates L. monocytogenes infection (Singh et al., 2008). Here, we showed that TrxA exhibits exothermic reactions for strong binding to PrfA, suggesting the thioredoxin could contribute to provide a highly reducing environment for reduced PrfA maintaining, and therefore acts as an indirectly-activating factor (like GshF) of PrfA, thereby providing a reasonable explanation for our finding that deletion of trxA resulted in a significant downregulation of PrfA-regulated virulence genes, and exhibited less virulent in mice. Based on the findings, we conclude that TrxA modulates the expression of Listeria virulence factors, possibly through participating in the correct folding mechanism and activating of PrfA. A similar mechanism has been illustrated in D. hafniense CprK (also belonging to the Crp/Fnr family), whose activation requires not only a structural change induced by o-chlorophenol binding, but also a redox switch. The reduced CprK specifically binds to DNA. In addition, CprK can be reversibly inactivated by oxygen through formation of intermolecular Cys11–Cys200 disulfide bridges (Levy et al., 2008). Given that PrfA contains four cysteine residues in each monomer, we thus propose that TrxA could maintain redox homeostasis in the bacterium to reduce the intermolecular disulfide bonds of PrfA for further dimerization (Figure 7).
Figure 7.
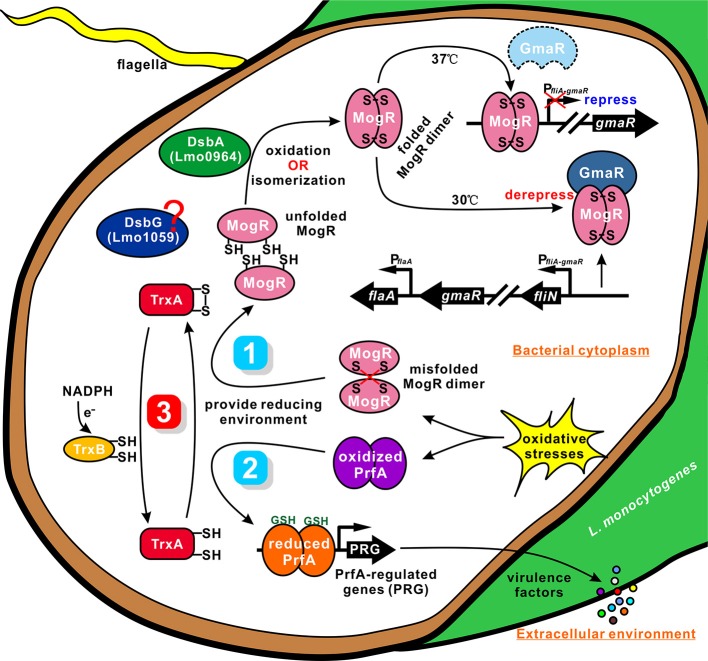
A proposed model for TrxA as a vital cellular reductase in L. monocytogenes. We propose that TrxA is essential for maintaining a highly reducing environment in the bacterial cytosol, which provides a favorable condition for protein folding and activation. Lmo0964 (DsbA) is responsible for direct oxidization of disulfide bonds in reduced MogR. Once MogR is correctly dimerized, the MogR:GmaR anti-repression complex is formed, which removes MogR from all flagellar motility gene promoters, enabling flagellar motility gene transcription at low temperatures. PrfA contains four cysteine residues in each monomer. TrxA contributes to maintain the reduced PrfA dimer that is exclusively activated in the cytosol in the presence of the co-factor GSH, and subsequently triggering expression of the virulence-associated factors.
Many bacteria with mutations in either dsbA or dsbB are nonmotile owing to their inability to produce functional flagella (Dailey and Berg, 1993; Hayashi et al., 2000; Hiniker and Bardwell, 2004; Coulthurst et al., 2008). This defect is best characterized in E. coli, in which DsbA catalyzes the formation of an intramolecular disulfide bond between two cysteine residues in the flagellar P-ring motor protein, FlgI (Dailey and Berg, 1993). The lack of correctly folded FlgI may prevent subsequent integration of the major FliC component in the flagella organelle (Hiniker and Bardwell, 2004). TrxA displays endothermic reactions for strong binding to MogR, indicating that the protein contributes to breakage of the intermolecular bonds of MogR (containing two cysteines) that binds DNA of the flagella-related gene promoter as a dimer in Listeria. Accordingly, TrxA is speculated to play a critical role as a strong reductase in reducing the mismatched intermolecular disulfide bonds of MogR and maintaining the intracellular monomer status. Based on the findings of TrxA in PrfA activation, we proposed a similar mechanism for TrxA-dependent modification of MogR to ensure correct dimerization and flagellar gene synthesis (Figure 7). As an oxidoreductase, TrxA plays a critical role in reducing the mismatched intermolecular disulfide bonds of MogR and maintaining the intracellular monomer status for further dimerization. Moreover, Lmo0964 is a bifunctional protein that oxidizes and isomerizes disulfide bonds required for direct oxidization of reduced MogR to form disulfide bonds. Deletion of Lmo0964 markedly impaired Listeria motility while no significant differences in motility were observed between the Lmo1059-null mutant and wild-type (Figure S2), we suggest that the isomerase Lmo1059 might act as a backup system. Correct dimerization of MogR facilitates the formation of a MogR:GmaR anti-repression complex, which removes MogR from all flagellar motility gene promoters, allowing transcription of genes associated with flagellar motility at low temperatures (Kamp and Higgins, 2011). In summary, we propose that TrxA as a vital cellular reductase is essential for maintaining a highly reducing environment in the bacterial cytosol, which provides a favorable condition for protein folding and activation, especially for the site-specific DNA-binding transcription regulators whose activation needs merization, and thus is involved in a range of biological functions.
In addition, TrxA expression might be regulated by the alternative sigma factor, SigH, through binding to the promoter region of thioredoxin system genes by EMSA assay. Actually, previous microarray assay has shown that L. monocytogenes trxA and trxB exhibited 1.5-fold higher transcript levels in the WT than in the ΔSigH strain (Chaturongakul et al., 2011). The role of SigH in oxidative stress was first demonstrated by Fernandes and co-workers using M. smegmatis sigH mutants. The group showed that SigH protects against oxidative stress by regulating the expression of thioredoxins (trxB1 and trxC) and thioredoxin reductase (Fernandes et al., 1999). Under reducing conditions, RshA, an anti-sigma factor, binds SigH, and inhibits the transcription of Trx and TrxR. However, under oxidative stress conditions, SigH is released from the complex and binds the promoters of Trx and TrxR to activate their transcription (Lu and Holmgren, 2014a). Consequently, we speculate that no expression or constitutive overexpression of TrxA in vivo would disturb the equilibrium of the intracellular redox potential and thus bring harm to L. monocytogenes for its ability to establish infections.
Generally, stress-tolerant Listeria strains are more invasive in vitro (Conte et al., 2000) and more virulent in vivo (Gahan et al., 1996; Cheng et al., 2015). Obviously, the capacity to tolerate oxidative stress is related to the virulence potential of L. monocytogenes and other bacterial species (Degnan et al., 2000; Chen et al., 2011). We demonstrated that the oxidative resistance ability is closely linked to L. monocytogenes pathogenicity, providing molecular insights into why the TrxA mutant is attenuated and nonmotile. Overall, our data should aid in clarifying the redox modification mechanisms of the thioredoxin system utilized by L. monocytogenes to adapt to niche environments outside and inside the host with a view to pathogen infection control from the public health perspective.
Materials and methods
Bacterial strains, plasmids, primers, and culture conditions
L. monocytogenes EGD-e was used as the wild-type strain. E. coli DH5α was employed for cloning experiments and as the host strain for plasmids pET30a(+) (Merck, Darmstadt, Germany), pIMK2 and pKSV7. E. coli Rosetta (DE3) was used for prokaryotic protein expression. Listeria strains were cultured in brain heart infusion (BHI) medium (Oxoid, Hampshire, England). E. coli strains were grown at 37°C in Luria-Bertani broth (LB) (Oxoid). Stock solutions of ampicillin (50 mg/ml), erythromycin (50 mg/ml), kanamycin (50 mg/ml), or chloramphenicol (50 mg/ml) were added to media when necessary. All chemicals were obtained from Sangon Biotech (Shanghai, China), Merck or Sigma-Aldrich (St. Louis, USA) and were of the highest available purity. All primers used in this study are listed in Table S3.
Bioinformatics analysis
The amino acid sequences of putative thioredoxin proteins from L. monocytogenes EGD-e and its homologs in other microbial species were obtained from the National Centre for Biotechnology Information database (NCBI). Trx amino acid sequences were aligned with MUSCLE using Geneious software (Edgar, 2004). The phylogenetic tree was constructed with the Neighbor-joining (NJ) method using 100 bootstrap replicates.
Prokaryotic expression and purification of recombinant proteins
The recombinant proteins used in this study were expressed as an N-terminal His tag fusion using pET30a(+) as the expression vector and Rosetta (DE3) as the expression host. The full-length ORF of the gene of interest from the EGD-e genome was amplified with the primer pair, inserted into the pET30a(+) vector, and finally transformed into Rosetta competent cells. E. coli cells harboring recombinant plasmids were grown in 500 mL LB supplemented with 50 μg/mL kanamycin at 37°C until cultures reached 0.8–1.0 at OD600nm. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.2 mM to induce expression of recombinant proteins for an additional 3 h under optimized conditions. His-tagged fusion proteins were purified using nickel-chelated affinity column chromatography.
Preparation of polyclonal antibodies against recombinant proteins
Rabbits were initially immunized via subcutaneous injection of 500 μg protein with an equal volume of Freund's complete adjuvant (Sigma). After 2 weeks, rabbits were boosted by subcutaneous injection of 250 μg protein each in incomplete Freund's adjuvant (Sigma) three times at 10 day intervals. Rabbits were bled ~10 days after the last injection.
Site-directed mutagenesis
To identify the predicted active sites of TrxA, single (C28A and C31A) and a double mutant (C28AC31A) were generated using the original vector template, pET30a-TrxA, and the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, Palo Alto, USA) with the primer pairs described in Table S3. Template DNA was removed via digestion with DpnI (Toyobo Co., Osaka, Japan) for 2 h at 37°C. All mutant constructs were sequenced to ensure that only the desired single mutations had been incorporated correctly. The corresponding mutant proteins were designated TrxA_C28A, TrxA_C31A, and TrxA_C28AC31A, and expressed and purified as described above.
In vitro oxidoreductase activity assays
The ability of TrxA, DsbA, and DsbC/G to catalyze reduction of human insulin (Sigma) in the presence of DTT was measured as described previously by Holmgren (1979). Briefly, reaction mixtures were prepared using 0.1 M potassium phosphate buffer (pH 7.0), 150 μM insulin, 2 mM EDTA, and 1.0 μM purified proteins in a final volume of 200 μL. Reactions were initiated by adding DTT to a final concentration of 1 mM, and monitored as the increase in absorbance at 650 nm every 3 min (100 min in total) at 25°C using the Micro-plate reader Synergy H1 (BioTek Solutions, Inc., Santa Barbara, USA). The calibration curve for quantitative analysis of reduced insulin was determined by incubating 0.5 μM purified TrxA in PBS containing 1 mM DTT with various concentrations of insulin (0, 40, 80, 120, 160, and 200 μM).
Construction of gene deletion mutants
Construction of L. monocytogenes gene deletion mutants were performed as described previously (Camilli et al., 1993). The temperature-sensitive pKSV7 shuttle vector was used for generating mutations in the L. monocytogenes strain EGD-e. A homologous recombination strategy with the splicing by overlap extension (SOE) PCR procedure was used for in-frame deletion to construct gene deletion mutants (Camilli et al., 1993; Monk et al., 2008). Specifically, DNA fragments containing homologous arms upstream and downstream of the gene of interest were obtained via amplification of EGD-e genomic DNA using SOE primer pairs. The obtained fragment was then cloned into the temperature-sensitive shuttle vector pKSV7 and transformed into E. coli DH5α. After confirmation by sequencing, the recombinant vector containing the target gene deletion cassette was then electroporated into the competent L. monocytogenes EGD-e cells. Transformants were selected on BHI agar plates containing chloramphenicol (10 μg/ml). A single transformant was serially passaged for ~30 generations at a non-permissive temperature (40°C) in BHI medium containing 10 μg/ml chloramphenicol to promote chromosomal integration. A single colony with chromosomal integration was successively passaged in BHI medium without antibiotic at a permissive temperature (30°C) for ~50 generations to enable plasmid excision and curing. The recombinants were identified as chloramphenicol-sensitive colonies, and the mutagenesis was further confirmed by PCR and DNA sequencing.
Complementation of gene deletion mutants
To complement the L. monocytogenes ΔtrxA strain, we constructed two complement strains using the integrative plasmid, pIMK2, which harbors a constitutive Listeria promoter Phelp (Monk et al., 2008). For the first complement strain, we amplified the complete open reading frame (ORF) of trxA from EGD-e genomic DNA using the primer pair CΔtrxA-a/CΔtrxA-b and inserted the fragment downstream of Phelp after digestion with the appropriate restriction enzymes. For the other complement strain, the complete ORF of trxA along with its promoter region was amplified using the primer pair CΔtrxA-c/CΔtrxA-d and cloned into pIMK2 following digestion with the appropriate restriction enzymes to remove the Phelp region. The resulting plasmids were electroporated into the L. monocytogenes ΔtrxA strain. Regenerated cells were plated on BHI agar containing kanamycin (50 μg/ml). The two complement strains were designated CΔtrxA_Phelp and CΔtrxA_PtrxA, respectively.
Growth analysis of L. monocytogenes under oxidative conditions
L. monocytogenes wild-type strain EGD-e, mutant ΔtrxA and two complement strains, CΔtrxA_Phelp and CΔtrxA_PtrxA, were grown overnight at 37°C in BHI broth with shaking. Cultures were collected by centrifugation at 5,000 × g at 4°C, washed once in PBS (10 mM, pH 7.4) and initial OD600 nm adjusted to 1.0. Bacteria were diluted (1:100) in fresh BHI broth or BHI containing 5 mM paraquat, 4 mM diamide, or 10 mM H2O2, and incubated at 37°C for 12 h. Kinetic growth at OD600 nm was measured at 1 h intervals.
Changes in TrxA and TrxB expression levels under oxidative conditions
Bacteria were grown in BHI broth to the stationary growth phase and lysates prepared as described earlier (Ryan et al., 2009). Specifically, stationary bacteria were collected at the indicated times after treatment with 5 mM paraquat, 4 mM diamide, or 10 mM H2O2. Bacterial pellets were re-suspended in 1 mL of extraction solution (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0). One gram of glass beads (G8772, Sigma) was added and samples lysed using the homogenizer Precellys 24 (Bertin, Provence, France) at 6,000 rpm for 30 s with intermittent cooling for 30 s (2 cycles in total), followed by centrifugation at 12,000 g for 15 min. Supernatants were retained as cell-free extracts. Proteins were separated via 12% SDS-PAGE and blotted onto 0.22 μm PVDF membrane (Merck). Membranes were blocked for 1 h with 5% skimmed milk, and incubated for 1 h with polyclonal antisera against recombinant TrxA or TrxB (prepared in this study) in 0.5% skimmed milk. Next, membranes probed with anti-TrxA or anti-TrxB were developed using HRP-conjugated goat anti-rabbit IgG (Santa Cruz, California, USA) as the secondary antibody. Chemiluminescence was detected via an UVP EC3 imaging system (UVP Inc., Upland, USA).
Motility assay (swimming ability) and transmission electron microscopy (TEM)
Motility assay was performed essentially on soft LB agar (0.25%) as described (Paxman et al., 2009). L. monocytogenes were grown overnight in BHI and the cultures adjusted at OD600 nm to 0.40 (about 2 × 108 CFU/ml). Bacterial samples (5 μL) were dropped onto soft TSA agar and incubated at 30° or 37°C for 48 h to allow growth. Motility was assessed by examining migration of bacteria through agar from the center toward the periphery of the colony. TEM experiments were performed as described previously (Dingle et al., 2011) in the Institute of Agrobiology and Environmental Sciences of Zhejiang University. Briefly, L. monocytogenes colonies grown overnight at 30°C from BHI agar plates were suspended in 50 μL monoethanolamine buffer (pH 10.0), and 10 μL of the suspension applied to carbon-coated copper grids and allowed to stand for 2 min at room temperature. Excess liquid was subsequently removed using filter paper, and bacteria stained with 10 μL of 0.5% phosphotungstic acid (pH 7.0) placed on the grids for 10 s at room temperature. Excess stain was gently wicked away using filter paper, and the dried grids examined under a Hitachi H-7650 transmission electron microscope (Hitachi High-Technologies Corporation, Tokyo, Japan).
Determination of protein-ligand interactions via isothermal titration calorimetry (ITC)
The protein-protein interactions between TrxA (or its active site mutants), MogR, and GmaR were measured using isothermal titration calorimetry (ITC) with a VP-isothermal titration calorimeter from Microcal, Inc. (Northampton, USA). Calorimetric titrations of TrxA (80–200 μM in the syringe) and PrfA, MogR, or GmaR (8–20 μM in the cuvette) were carried out at 25°C in 25 mM Tris-HCl, pH 7.5, 100 mM NaCl. TrxA or its active site mutants (C28A, C31A, and C28AC31A) was titrated into PrfA, MogR, or GmaR in 15 μL injections with a spacing of 300 s between injections. Calorimetric data were analyzed by integrating heat effects normalized to the amount of injected protein and curve-fitting based on a 1:1 binding model using the Origin software package (Microcal). The dissociation constant was derived from data using standard procedures. ITC was carried out in the Life Sciences Institute of Zhejiang University.
In vitro redox status determination of MogR with TrxA
Reactions with recombinant TrxA were carried out in 50 mM Tris-HCl buffer, pH 7.5 with the increasing amount (0–2 μg) of reduced TrxA and 2 μg MogR with or without 0.1 mM DTT as an electron donor. After 1 h incubation, an equal amount of SDS–PAGE sample buffer was added to quench the reactions. Reactions were terminated with 10% (v/v) trichloroacetic acid (TCA) and then separated on 12% polyacrylamide gels, and stained with Coomassie blue to visualize the redox state of MogR.
Cadmium and copper sensitivity assays
Cadmium and copper sensitivity assays are usually performed to test oxidase and isomerase activities, respectively (Ren et al., 2014). L. monocytogenes wild-type strain EGD-e, isogenic lmo1059 and lmo0964 deletion mutants and the corresponding complementation strains were grown overnight in BHI broth. Bacteria were diluted to OD600nm of 1.0 (~109 cfu ml−1) in PBS. Bacteria were serially diluted 10-fold, and 10 μl of each dilution spotted onto BHI agar plates containing 1 mM cadmium chloride (Sangon) or 1 mM copper(II) chloride (Sangon). Following incubation at 37°C for 48–72 h, colony growth on each plate was assessed and imaged. All spot titer assays were performed in duplicate to verify the results.
Disulfide interchange reactions
Disulfide interchange reactions were used to investigate the ability of TrxA, DsbA, and DsbC/G to catalyze the reduction of oxidized glutathione (GSSG) by DTT. The DTT/glutathione equilibrium lies far on the side of oxidized DTT, and DTTox formation can be followed spectrophotometrically. GSSG (5 mM) was reduced by DTT (5 mM) at 25°C in 100 mM formic acid/NaOH (pH 4.0), 1 mM EDTA. Reactions were monitored based on the increase in absorbance at 287 nm.
Comparison of transcriptome profiles of wild-type and the isogenic trxA mutant upon exposure to oxidative stress
RNA preparation
Bacterial overnight cultures of wild-type EGD-e and its isogenic trxA mutant were diluted (1:100) in fresh BHI broth and grown to the early stationary phase. Pellets were centrifuged at 5,000 g for 10 min, re-suspended in equal volumes of fresh BHI containing 2 mM diamide, and incubated statically at 37°C for additional 1 h. Total RNA was extracted using the Column Bacterial total RNA Purification Kit (Sangon), according to the manufacturer's instructions, and genomic DNA removed using DNase I (TaKara, Japan). RNA purity was assessed using the NanoDrop (Thermo Fisher Scientific, Lafayette, USA) and RNA integrity determined via electrophoresis.
Transcriptome sequencing
RNA samples were sent to the commercial sequencing company, Majorbio, for library construction and transcriptome sequencing, based on the Illumina Hiseq2500 sequencing platform. The paired-end raw reads were trimmed and quality controlled with SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) using default parameters. Clean reads were separately aligned to the reference genome with the orientation mode using Bowtie 2 software (Langmead and Salzberg, 2012). Output data generated from sequencing were stored in the standard FASTQ format for use as inputs for subsequent analyses.
Differential expression analysis and functional annotation
RSEM (RNA-Seq by Expectation-Maximization; Li and Dewey, 2011) was used to quantify gene and isoform abundance. EdgeR (Empirical analysis of digital gene expression data in R) (Robinson et al., 2010) was utilized for differential expression analysis. The TMM (trimmed mean of M-values) method was selected to compute normalization factors and differentially expressed genes (DEGs) between two samples selected using the following criteria: (I) logarithmic of fold change >1.0 and (II) FDR (false discovery rate) < 0.05. To determine the functions of the differentially expressed genes, the unigenes were aligned by BLASTx against the NCBI non-redundant (nr), Swiss-Prot, Kyoto Encyclopedia of Genes and Genome (KEGG), and Cluster of Orthologous Groups (COG) protein databases. GO functional enrichment analyses were carried out using Goatools and KOBAS (Xie et al., 2011). DEGs were significantly enriched in GO terms and metabolic pathways at Bonferroni-corrected P-values of < 0.05.
qRT-PCR validation of transcriptome results
To validate of the reliability of transcriptome sequencing data, real-time quantification of random mRNAs was performed using qRT-PCR with the same total cellular RNAs used for transcriptome experiments. We randomly selected 28 genes for qRT-PCR validation, i.e., 16 representative downregulated genes and 12 representative upregulated genes in the ΔtrxA mutant. Specific primers for these genes were designed and purchased from Sangon Biotech. cDNA was synthesized with reverse transcriptase (Toyobo). Quantitative PCR was performed in 20 μL reaction mixtures containing SYBR quantitative PCR mix (Toyobo) to measure the transcriptional levels of genes of interest using the Mx3000P PCR detection system (Agilent) with specific primer pairs. Real-time quantitative PCR was performed in a 20 μL reaction volume containing 200 ng cDNA, 10 μL SYBR quantitative PCR mix and 1 μL gene-specific primers (200 nM). The housekeeping gene, rpoB, was used as an internal control for normalization in each sample. Triplicate assays were performed for each gene.
Changes in expression levels of LLO in the absence of trxA
To further establish, whether TrxA is involved in bacterial virulence, western blot was employed to analyze the changes in expression of the major virulence-associated factor, LLO. Bacterial overnight cultures of L. monocytogenes wild-type EGD-e, mutant ΔtrxA and two complement strains, CΔtrxA_Phelp and CΔtrxA_PtrxA, were diluted into 100 mL fresh BHI broth and grown to the stationary phase. For isolation of secreted proteins, the fractionation procedure was performed as described before (Lenz and Portnoy, 2002), with minor modifications. Briefly, bacterial cells were pelleted via centrifugation at 13,000 g for 20 min at 4°C, and the resulting culture supernatant collected and filtered through a 0.22 μm polyethersulfone membrane filter (Merck). Trichloroacetic acid (TCA) was added to the supernatant to obtain a final concentration of 10%. Proteins were TCA-precipitated on ice overnight and washed with ice-cold acetone. Washed precipitates of supernatant proteins were re-suspended in SDS-PAGE sample buffer (5% SDS, 10% glycerol, and 50 mM Tris-HCl, pH 6.8). Samples were boiled for 5 min and stored at -20°C before electrophoresis. Total cell protein isolation was performed as described above. Protein samples were separated using 12% SDS-PAGE and immunoblotted with α-LLO or α-GAPDH antisera (prepared in this study). GAPDH was employed as an internal control.
LLO-mediated hemolytic activity measurement
Measurement of LLO-associated hemolytic activity was done as described previously (Xayarath et al., 2015). L. monocytogenes wild-type and mutant strains were grown for 16 h with shaking in BHI broth at 37°C. All cultures were adjusted to OD600nm of 1.0 before supernatants were collected. Hemolytic activity was measured based on lysis of sheep red blood cells (SRBCs) using supernatants from cultures. The culture supernatant (50 μL) was diluted with an equal volume of PBS containing 6 mM cysteine (pH 5.6) and equilibrated to 37°C for 10 min. Next, 50 μL PBS-washed intact SRBCs were added to each sample and incubated at 37°C for 60 min. Samples were centrifuged and supernatants analyzed for hemoglobin absorption at 550 nm. PBS was employed as a negative control. The values corresponding to the reciprocal of the dilution of culture supernatant required to lyse 50% of HRBCs were used to compare the hemolytic activities in the different supernatants.
Adhesion, invasion, and survival in Caco-2 cells
Bacterial survival in human intestinal epithelial Caco-2 cells was assessed as described previously (Reddy et al., 2016). Stationary L. monocytogenes wild-type strain EGD-e, mutant ΔtrxA and two complement strains, CΔtrxA_Phelp and CΔtrxA_PtrxA, at 37°C in BHI were washed and re-suspended in PBS (10 mM, pH 7.4). Caco-2 cells cultured to 80% confluence in Dulbecco's modified eagle medium (DMEM) containing 10% fetal calf serum were infected with the above strains for 60 min at a multiplicity of infection (MOI) of 10:1. For adhesion, cells were lysed after being washed three times with PBS. For estimation of invasion, cells were washed with PBS after 1 h infection and incubated for an additional hour in DMEM containing gentamicin at 200 μg/mL. Caco-2 cells were lysed by adding 1 mL of ice-cold sterile distilled water. Lysates were 10-fold diluted for enumeration of viable bacteria on BHI agar plates that were considered the 0 h numbers invading cells. For intracellular proliferation, cells were further incubated for 6 or 12 h in 5% CO2 at 37°C. Viable bacteria were enumerated as described above. Adhesion was expressed as the ratio of recovered colonies to colonies inoculated, while invasion was calculated as the ratio of colonies recovered after gentamicin treatment to colonies inoculated.
Actin-tail formation in Caco-2 cells
Cells were infected at MOI of 10:1 at 37°C with 5% CO2 for 1 h. Extracellular bacteria were then killed with 200 μg/mL gentamicin for 1 h and incubated for an additional 6 h. Cells were washed gently with PBS (10 mM, pH 7.4), fixed with 4% paraformaldehyde and then permeabilized with 0.5% Triton X-100. The bacterial cells were stained with polyclonal antibodies to L. monocytogenes for 1 h at 37°C, washed twice with PBS and probed with Alexa Fluor 488-conjugated donkey anti-rabbit antibody (Santa Cruz) for 1 h at 37°C. F-actin was then stained with phalloidin-Alexa Fluor 568 (Thermo Fisher Scientific). DAPI (4′,6-diamidino-2-phenylindole; Thermo Fisher Scientific) was used to stain the nuclei. Actin tails recruited by the bacteria were visualized under a ZEISS LSM510 confocal microscope (Zeiss Germany, Oberkochen, Germany).
Virulence in the mouse model
The L. monocytogenes wild-type strain EGD-e, mutant ΔtrxA and two complement strains, CΔtrxA_Phelp and CΔtrxA_PtrxA, were tested for recovery in liver and spleen sections of ICR mice (female, 18–22 g, purchased from Zhejiang Academy of Medical Sciences, Hangzhou, China). ICR mice (eight per group) were injected intraperitoneally with ~106 CFU of each strain. At 24 and 48 h post-infection, mice were sacrificed, and livers and spleens removed and individually homogenized in PBS (10 mM, pH 7.4). Surviving Listeria cells were enumerated by plating serial dilutions of homogenates on BHI agar plates. Results were expressed as means ± SD of the recovery rate per organ for each group.
Electrophoretic mobility shift assay (EMSA)
To further demonstrate, whether the L. monocytogenes Trx system is regulated by SigH, we determined the binding abilities between SigH and promoter regions of trxA and trxB in vitro using the electrophoretic mobility shift assay (EMSA). DNA fragments of the promoter regions of trxA and trxB were generated by PCR with the specific primer pairs, and purified with the PCR Purification Kit (Sangon) according to the manufacturer's instructions. Next, 200 ng DNA was incubated with varying concentrations of purified recombinant SigH and incubated in binding buffer (50 mM Tris-HCl, pH 8.0, 250 mM NaCl, 5.0 mM MgCl2, 2.5 mM DTT, 2.5 mM EDTA, and 20% glycerol) for 30 min at room temperature. Protein-DNA complexes were separated electrophoretically on a native 5% polyacrylamide gel at 80 V with 0.5 × Tris-acetate-EDTA (TAE) buffer and visualized using ethidium bromide staining.
Statistical analysis
All experiments were repeated at least three times. Data were analyzed using the two-tailed homoscedastic Student's t-test. Differences with P < 0.05 were considered statistically significant.
Ethics statements
All animal care and use protocols were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People's Republic of China. The protocol was approved by the Institutional Animal Care and Use Committee of Zhejiang A&F University (Permit Number: ZJAFU/IACUC_2011-10-25-02).
Author contributions
CC, WF, and HS conceived the study. CC, JS, XH, ZD, HW, LJ, CS, and TM carried out experiments. CC, YY, ZC, HH, and XW analyzed data. CC, WF, NF, and HS drafted the manuscript and all the authors contributed to preparing the final version of the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We specially thank Dr. Martin Wiedmann at Cornell University for kindly providing the shuttle plasmid pKSV7.
Footnotes
Funding. This work was supported by National Natural Science Foundation of China (Nos. 31470179, 31502083, and 31402215), Zhejiang Provincial Natural Science Foundation (Nos. LY17C180001 and LY15C010003), Initiative Design Project of Agricultural Research of Hangzhou (No. 20162012A07) and ZAFU talents starting program (No. 2014FR073 and 2015FR007). The funders had no role in design of the study or analysis and interpretation of the data.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00287/full#supplementary-material
The thioredoxin system is regulated by SigH. SigH bound to trxA and trxB promoters, as determined using the electrophoretic mobility shift assay. PtrxA and PtrxB fragments were obtained by PCR with the primers specified in Table S3, and incubated with recombinant SigH for 30 min at room temperature. Gel retardation of DNA-protein complexes was monitored after ethidium bromide staining. The promoter DNA fragment of rpoB (a house-keeping gene in L. monocytogenes, which is annotated as DNA-directed RNA polymerase subunit beta) was considered as a negative control.
Lmo0964 (DsbA) is responsible for bacterial motility. The motility assay was performed by growing L. monocytogenes WT EGD-e, Δlmo1059, Δlmo0964 and the complement strains, CΔlmo1059 and CΔlmo0964, on soft agar (0.25%) at 30°C.
Genes identified as significantly downregulated in the mutant strain of L. monocytogenes, ΔtrxA, via transcriptome analysis.
Genes identified as significantly upregulated in the mutant strain of L. monocytogenes, ΔtrxA, via transcriptome analysis.
PCR primers used in this study. The nucleotides introduced to create restriction enzyme sites are underlined. All primers were synthesized by Sangon Biotech, Inc., Shanghai, China.
References
- Begley M., Cotter P. D., Hill C., Ross R. P. (2010). Glutamate decarboxylase-mediated nisin resistance in Listeria monocytogenes. Appl. Environ. Microbiol. 76, 6541–6546. 10.1128/AEM.00203-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessette P. H., Cotto J. J., Gilbert H. F., Georgiou G. (1999). In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J. Biol. Chem. 274, 7784–7792. 10.1074/jbc.274.12.7784 [DOI] [PubMed] [Google Scholar]
- Bhat S. A., Singh N., Trivedi A., Kansal P., Gupta P., Kumar A. (2012). The mechanism of redox sensing in Mycobacterium tuberculosis. Free Radic. Biol. Med. 53, 1625–1641. 10.1016/j.freeradbiomed.2012.08.008 [DOI] [PubMed] [Google Scholar]
- Boles B. R., Singh P. K. (2008). Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. U.S.A. 105, 12503–12508. 10.1073/pnas.0801499105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A., Tilney L. G., Portnoy D. A. (1993). Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8, 143–157. 10.1111/j.1365-2958.1993.tb01211.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S., Raengpradub S., Palmer M. E., Bergholz T. M., Orsi R. H., Hu Y., et al. (2011). Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 187–200. 10.1128/AEM.00952-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Cheng C., Xia Y., Zhao H., Fang C., Shan Y., et al. (2011). Lmo0036, an ornithine and putrescine carbamoyltransferase in Listeria monocytogenes, participates in arginine deiminase and agmatine deiminase pathways and mediates acid tolerance. Microbiology 157(Pt 11), 3150–3161. 10.1099/mic.0.049619-0 [DOI] [PubMed] [Google Scholar]
- Cheng C., Chen J., Fang C., Xia Y., Shan Y., Liu Y., et al. (2013). Listeria monocytogenes aguA1, but not aguA2, encodes a functional agmatine deiminase: biochemical characterization of its catalytic properties and roles in acid tolerance. J. Biol. Chem. 288, 26606–26615. 10.1074/jbc.M113.477380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Yang Y., Dong Z., Wang X., Fang C., Yang M., et al. (2015). Listeria monocytogenes varies among strains to maintain intracellular pH homeostasis under stresses by different acids as analyzed by a high-throughput microplate-based fluorometry. Front. Microbiol. 6:15. 10.3389/fmicb.2015.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte M. P., Petrone G., Di Biase A. M., Ammendolia M. G., Superti F., Seganti L. (2000). Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macrophage-like cells. Microb. Pathog. 29, 137–144. 10.1006/mpat.2000.0379 [DOI] [PubMed] [Google Scholar]
- Corr S. C., O'Neill L. A. (2009). Listeria monocytogenes infection in the face of innate immunity. Cell. Microbiol. 11, 703–709. 10.1111/j.1462-5822.2009.01294.x [DOI] [PubMed] [Google Scholar]
- Cotter P. D., Gahan C. G., Hill C. (2001). A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40, 465–475. 10.1046/j.1365-2958.2001.02398.x [DOI] [PubMed] [Google Scholar]
- Coulthurst S. J., Lilley K. S., Hedley P. E., Liu H., Toth I. K., Salmond G. P. (2008). DsbA plays a critical and multifaceted role in the production of secreted virulence factors by the phytopathogen Erwinia carotovora subsp. atroseptica. J. Biol. Chem. 283, 23739–23753. 10.1074/jbc.M801829200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey F. E., Berg H. C. (1993). Mutants in Disulfide bond formation that disrupt flagellar assembly in Escherichia-Coli. Proc. Natl. Acad. Sci. U.S.A. 90, 1043–1047. 10.1073/pnas.90.3.1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan B. A., Fontaine M. C., Doebereiner A. H., Lee J. J., Mastroeni P., Dougan G., et al. (2000). Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68, 2441–2448. 10.1128/IAI.68.5.2441-2448.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle T. C., Mulvey G. L., Armstrong G. D. (2011). Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect. Immun. 79, 4061–4067. 10.1128/IAI.05305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes N. D., Wu Q. L., Kong D., Puyang X., Garg S., Husson R. N. (1999). A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181, 4266–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan C. G., O'Driscoll B., Hill C. (1996). Acid adaptation of Listeria monocytogenes can enhance survival in acidic foods and during milk fermentation. Appl. Environ. Microbiol. 62, 3128–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F., et al. (2001). Comparative genomics of Listeria species. Science 294, 849–852. 10.1126/science.1063447 [DOI] [PubMed] [Google Scholar]
- Gopal S., Srinivas V., Zameer F., Kreft J. (2009). Prediction of proteins putatively involved in the thiol: disulfide redox metabolism of a bacterium (Listeria): the CXXC motif as query sequence. In Silico Biol. 9, 407–414. 10.3233/ISB-2009-0409 [DOI] [PubMed] [Google Scholar]
- Grundling A., Burrack L. S., Bouwer H. G., Higgins D. E. (2004). Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. U.S.A. 101, 12318–12323. 10.1073/pnas.0404924101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M., Grundstrom C., Begum A., Lindberg M. J., Sauer U. H., Almqvist F., et al. (2016). Structural basis for glutathione-mediated activation of the virulence regulatory protein PrfA in Listeria. Proc. Natl. Acad. Sci. U.S.A. 113, 14733–14738. 10.1073/pnas.1614028114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Abe M., Kimoto M., Furukawa S., Nakazawa T. (2000). The DsbA-DsbB disulfide bond formation system of Burkholderia cepacia is involved in the production of protease and alkaline phosphatase, motility, metal resistance, and multi-drug resistance. Microbiol. Immunol. 44, 41–50. 10.1111/j.1348-0421.2000.tb01244.x [DOI] [PubMed] [Google Scholar]
- Heras B., Shouldice S. R., Totsika M., Scanlon M. J., Schembri M. A., Martin J. L. (2009). DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7, 215–225. 10.1038/nrmicro2087 [DOI] [PubMed] [Google Scholar]
- Hiniker A., Bardwell J. C. (2004). In vivo substrate specificity of periplasmic disulfide oxidoreductases. J. Biol. Chem. 279, 12967–12973. 10.1074/jbc.M311391200 [DOI] [PubMed] [Google Scholar]
- Hiniker A., Collet J. F., Bardwell J. C. (2005). Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J. Biol. Chem. 280, 33785–33791. 10.1074/jbc.M505742200 [DOI] [PubMed] [Google Scholar]
- Holmgren A. (1979). Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254, 9627–9632. [PubMed] [Google Scholar]
- Holmgren A. (1985). Thioredoxin. Annu. Rev. Biochem. 54, 237–271. 10.1146/annurev.bi.54.070185.001321 [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418. 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- Ito K., Inaba K. (2008). The disulfide bond formation (Dsb) system. Curr. Opin. Struct. Biol. 18, 450–458. 10.1016/j.sbi.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Kallifidas D., Thomas D., Doughty P., Paget M. S. (2010). The sigmaR regulon of Streptomyces coelicolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology 156(Pt 6), 1661–1672. 10.1099/mic.0.037804-0 [DOI] [PubMed] [Google Scholar]
- Kamp H. D., Higgins D. E. (2011). A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog. 7:e1002153. 10.1371/journal.ppat.1002153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. P., Hahm B. K., Bhunia A. K. (2007). The 2-cys peroxiredoxin-deficient Listeria monocytogenes displays impaired growth and survival in the presence of hydrogen peroxide in vitro but not in mouse organs. Curr. Microbiol. 54, 382–387. 10.1007/s00284-006-0487-6 [DOI] [PubMed] [Google Scholar]
- Kouwen T. R., van der Goot A., Dorenbos R., Winter T., Antelmann H., Plaisier M. C., et al. (2007). Thiol-disulphide oxidoreductase modules in the low-GC Gram-positive bacteria. Mol. Microbiol. 64, 984–999. 10.1111/j.1365-2958.2007.05707.x [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichert L. I. O., Scharf C., Hecker M. (2003). Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185, 1967–1975. 10.1128/JB.185.6.1967-1975.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz L. L., Portnoy D. A. (2002). Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45, 1043–1056. 10.1046/j.1365-2958.2002.03072.x [DOI] [PubMed] [Google Scholar]
- Levy C., Pike K., Heyes D. J., Joyce M. G., Gabor K., Smidt H., et al. (2008). Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator. Mol. Microbiol. 70, 151–167. 10.1111/j.1365-2958.2008.06399.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Pasternak C., Klug G. (2003). Expression of the trxA gene for thioredoxin 1 in Rhodobacter sphaeroides during oxidative stress. Arch. Microbiol. 180, 484–489. 10.1007/s00203-003-0620-x [DOI] [PubMed] [Google Scholar]
- Lu J., Holmgren A. (2014a). The thioredoxin antioxidant system. Free Radic. Biol. Med. 66, 75–87. 10.1016/j.freeradbiomed.2013.07.036 [DOI] [PubMed] [Google Scholar]
- Lu J., Holmgren A. (2014b). The thioredoxin superfamily in oxidative protein folding. Antioxid. Redox Signal. 21, 457–470. 10.1089/ars.2014.5849 [DOI] [PubMed] [Google Scholar]
- McCarver A. C., Lessner D. J. (2014). Molecular characterization of the thioredoxin system from Methanosarcina acetivorans. FEBS J. 281, 4598–4611. 10.1111/febs.12964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D., Georgopoulos C., Raina S. (1993). Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc. Natl. Acad. Sci. U.S.A. 90, 7084–7088. 10.1073/pnas.90.15.7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D., Georgopoulos C., Raina S. (1994). The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 13, 2013–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D., Schwager F., Raina S. (1995). Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J. 14, 3415–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk I. R., Gahan C. G., Hill C. (2008). Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 74, 3921–3934. 10.1128/AEM.00314-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxman J. J., Borg N. A., Horne J., Thompson P. E., Chin Y., Sharma P., et al. (2009). The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes. J. Biol. Chem. 284, 17835–17845. 10.1074/jbc.M109.011502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman J. L., Huang Q., Reniere M. L., Iavarone A. T., Portnoy D. A. (2017). Activity of the pore-forming virulence factor listeriolysin O is reversibly inhibited by naturally occurring S-glutathionylation. Infect. Immun. 85:e00959-16. 10.1128/IAI.00959-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pother D. C., Liebeke M., Hochgrafe F., Antelmann H., Becher D., Lalk M., et al. (2009). Diamide triggers mainly S thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 191, 7520–7530. 10.1128/JB.00937-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S., Akgul A., Karsi A., Abdelhamed H., Wills R. W., Lawrence M. L. (2016). The role of Listeria monocytogenes cell wall surface anchor protein LapB in virulence, adherence, and intracellular replication. Microb. Pathog. 92, 19–25. 10.1016/j.micpath.2015.12.012 [DOI] [PubMed] [Google Scholar]
- Ren G., Champion M. M., Huntley J. F. (2014). Identification of disulfide bond isomerase substrates reveals bacterial virulence factors. Mol. Microbiol. 94, 926–944. 10.1111/mmi.12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere M. L., Whiteley A. T., Portnoy D. A. (2016). An In vivo selection identifies Listeria monocytogenes genes required to sense the intracellular environment and activate virulence factor expression. PLoS Pathog. 12:e1005741. 10.1371/journal.ppat.1005741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere M. L., Whiteley A. T., Hamilton K. L., John S. M., Lauer P., Brennan R. G., et al. (2015). Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517, 170–173. 10.1038/nature14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz D., Patel H., Doan B., Zheng M., Aslund F., Storz G., et al. (2000). Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J. Biol. Chem. 275, 2505–2512. 10.1074/jbc.275.4.2505 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkova A., Stirnimann C. U., Frei P., Grauschopf U., Brunisholz R., Grutter M. G., et al. (2004). Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. EMBO J. 23, 1709–1719. 10.1038/sj.emboj.7600178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S., Begley M., Gahan C. G., Hill C. (2009). Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11, 432–445. 10.1111/j.1462-2920.2008.01782.x [DOI] [PubMed] [Google Scholar]
- Sampayo J. N., Gill M. S., Lithgow G. J. (2003). Oxidative stress and aging - the use of superoxide dismutase/catalase mimetics to extend lifespan. Biochem. Soc. Trans. 31, 1305–1307. 10.1042/bst0311305 [DOI] [PubMed] [Google Scholar]
- Schnupf P., Portnoy D. A. (2007). Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 9, 1176–1187. 10.1016/j.micinf.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Shelar S. B., Kaminska K. K., Reddy S. A., Kumar D., Tan C. T., Yu V. C., et al. (2015). Thioredoxin-dependent regulation of AIF-mediated DNA damage. Free Radic. Biol. Med. 87, 125–136. 10.1016/j.freeradbiomed.2015.06.029 [DOI] [PubMed] [Google Scholar]
- Shen A., Higgins D. E. (2006). The MogR transcriptional repressor regulates nonhierarchal expression of flagellar motility genes and virulence in Listeria monocytogenes. PLoS Pathog. 2:e30. 10.1371/journal.ppat.0020030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A., Higgins D. E., Panne D. (2009). Recognition of AT-rich DNA binding sites by the MogR repressor. Structure 17, 769–777. 10.1016/j.str.2009.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A., Kamp H. D., Grundling A., Higgins D. E. (2006). A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 20, 3283–3295. 10.1101/gad.1492606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Jamieson A., Cresswell P. (2008). GILT is a critical host factor for Listeria monocytogenes infection. Nature 455, 1244–1247. 10.1038/nature07344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits W. K., Dubois J. Y. F., Bron S., van Dijl J. M., Kuipers O. P. (2005). Tricksy business: transcriptome analysis reveals the involvement of thioredoxin a in redox homeostasis, oxidative stress, sulfur metabolism, and cellular differentiation in Bacillus subtilis. J. Bacteriol. 187, 3921–3930. 10.1128/JB.187.12.3921-3930.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Kosugi K., Mizukami M., Ishibashi M., Tokunaga H., Tokunaga M. (2004). Expression and purification of thioredoxin (TrxA) and thioredoxin reductase (TrxB) from Brevibacillus choshinensis. Protein Expr. Purif. 37, 385–391. 10.1016/j.pep.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Uziel O., Borovok I., Schreiber R., Cohen G., Aharonowitz Y. (2004). Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186, 326–334. 10.1128/JB.186.2.326-334.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L., Ulmer D. D. (1972). Biochemical effects of mercury, cadmium, and lead. Annu. Rev. Biochem. 41, 91–128. 10.1146/annurev.bi.41.070172.000515 [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland J. A., Kuhn M., Berche P., Chakraborty T., Dominguez-Bernal G., Goebel W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640. 10.1128/CMR.14.3.584-640.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Feng H., Zhu Y., Gao P. (2017). Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes. Protein Cell 8, 308–312. 10.1007/s13238-017-0390-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemekamp-Kamphuis H. H., Wouters J. A., de Leeuw P. P., Hain T., Chakraborty T., Abee T. (2004). Identification of sigma factor sigma B-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70, 3457–3466. 10.1128/AEM.70.6.3457-3466.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley A. T., Ruhland B. R., Edrozo M. B., Reniere M. L. (2017). A redox-responsive transcription factor is critical for pathogenesis and aerobic growth of Listeria monocytogenes. Infect. Immun. 10.1128/IAI.00978-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xayarath B., Alonzo F., III., Freitag N. E. (2015). Identification of a peptide-pheromone that enhances Listeria monocytogenes escape from host cell vacuoles. PLoS Pathog. 11:e1004707. 10.1371/journal.ppat.1004707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Oka S., Masutani H., Nakamura H., Yodoi J. (2003). The role of thioredoxin in the aging process: involvement of oxidative stress. Antioxid. Redox Signal. 5, 563–570. 10.1089/152308603770310211 [DOI] [PubMed] [Google Scholar]
- Yoshioka J., Chutkow W. A., Lee S., Kim J. B., Yan J., Tian R., et al. (2012). Deletion of thioredoxin-interacting protein in mice impairs mitochondrial function but protects the myocardium from ischemia-reperfusion injury. J. Clin. Invest. 122, 267–279. 10.1172/JCI44927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The thioredoxin system is regulated by SigH. SigH bound to trxA and trxB promoters, as determined using the electrophoretic mobility shift assay. PtrxA and PtrxB fragments were obtained by PCR with the primers specified in Table S3, and incubated with recombinant SigH for 30 min at room temperature. Gel retardation of DNA-protein complexes was monitored after ethidium bromide staining. The promoter DNA fragment of rpoB (a house-keeping gene in L. monocytogenes, which is annotated as DNA-directed RNA polymerase subunit beta) was considered as a negative control.
Lmo0964 (DsbA) is responsible for bacterial motility. The motility assay was performed by growing L. monocytogenes WT EGD-e, Δlmo1059, Δlmo0964 and the complement strains, CΔlmo1059 and CΔlmo0964, on soft agar (0.25%) at 30°C.
Genes identified as significantly downregulated in the mutant strain of L. monocytogenes, ΔtrxA, via transcriptome analysis.
Genes identified as significantly upregulated in the mutant strain of L. monocytogenes, ΔtrxA, via transcriptome analysis.
PCR primers used in this study. The nucleotides introduced to create restriction enzyme sites are underlined. All primers were synthesized by Sangon Biotech, Inc., Shanghai, China.