Abstract
Cadmium (Cd) is one of the most dangerous heavy metal pollutants in the environment and is toxic to animal and plant cells. On the other hand, iron (Fe) is an essential element for plant growth and development. The chlorosis of plant leaves under cadmium stress is similar to the typical symptom of iron deficiency. Recently, several Arabidopsis basic/helix-loop-helix (bHLH) transcription factors have been identified that are involved in the interactions between Cd and Fe. In the present study, over-expression the ORG3-like gene GmORG3, a bHLH transcription factor OBP3-responsive gene (ORG), enhanced Cd tolerance and stabilized Fe homeostasis. The domain analysis of GmORG3 showed that the protein contains a conserved 61-residue bHLH domain belonging to subfamily II. Moreover, subcellular localization experiments showed that GmORG3 is a nucleoprotein. GmORG3 was transcribed only in soybean roots and was significantly induced by external Cd stress in soybean plants. Heterologous expression of GmORG3 enhanced Cd tolerance in yeast. Furthermore, the overexpression of GmORG3 in soybean mosaic seedlings using a hairy root system showed that overexpressing plants increased the translocation ratio of Fe but reduced Cd translocation from the roots to shoots. In addition, the ectopic expression of GmORG3 in tobacco reduced phytotoxic effects induced by Cd stress and Fe deficiency, including the blockage of root elongation and decreased chlorophyll content. By integrating all these results, we found that GmORG3 plays an important role in response to Cd stress. The results provide new insights into the molecular mechanisms of Cd tolerance in soybean.
Keywords: soybean, bHLH transcription factor, GmORG3, cadmium tolerance, gene expression
Introduction
Soil heavy metal pollution is increasingly becoming a global environmental problem. Generally, heavy metals are naturally occurring in the soil, however, their amounts also increase due to human activities. Community development activities are causing the rapid expansion of non-agricultural construction and wastelands, and farming activities such as the increased use of agricultural fertilizers, especially phosphate fertilizer, as well as sewage sludge, wastewater, and pesticides are leading to soil heavy metal pollution. And these heavy metals may be absorbed by crops, thus enter the human body (Puga et al., 2015).
Direct or indirect human interference is continuously increasing levels of heavy metals (Larue et al., 2010; Huang et al., 2011), thus, the effects of this interference on soil occur worldwide. Cadmium (Cd) is a harmful heavy metal element and has become a serious problem for human health. Because of high water solubility, Cd is considered to be one of the most toxic heavy metal in the nature world. Even at very small concentrations, it is strong toxic to living organisms, and it is the fourth strength toxic heavy metal element to vascular plants, exists in soils as well (Huang et al., 2011; Xie et al., 2013; Pietrini et al., 2015). The concentrations of Cd as little as 8 mg/kg in the soil are toxic to plants (Pedro et al., 2013). Plants growing under cadmium stress exhibit leaf etiolation and arrested root elongation and can die (Kahle, 1993; Das et al., 1997). Cd gets into plant cells through Fe, zinc (Zn) and some other transporters/channels. There are several mechanisms are developed in plants for Cd detoxification, such as binding in cell wall, compartmentation in vacuole, chelation with phytochelatins, and enrichment in leaf epidermal hairs (Clemens, 2006).
Iron (Fe) is one of seven indispensable trace elements needed for plant growth. Iron deficiency is common in plants and animals (López-Millán et al., 2013; Boonyaves et al., 2017). Iron deficiency in plants is generally a problem of iron availability, rather than the supply of iron, because iron is the fourth highest rich element on the surface of the globe. Most of the plants can uptake and make use of soluble ferrous iron (Fe2+) that is ubiquitous in soils with the pH between 6.5 and 7.5 (Havlin et al., 2005). However, under alkaline conditions (pH > 8), Fe3+ in the soil is in an insoluble state that can’t dissolve in water, and this Fe3+ is difficult to be absorbed by most plant. Therefore, not enough Fe is introduced into food chain. In developing countries, most of the 0 to 10-year-old children and pregnant women (at least 30–40%) suffer from a lack of iron1. Anemia is used as an indirect indicator. Globally, anemia affects approximately 2 billion people or approximately 25% of the population (WHO, 2016). Iron deficiency chlorosis in plants caused a losses of about $120 million annually in the United States alone (Hansen et al., 2003). Therefore, there is a need to improve Fe uptake by plants to introduce Fe into the food chain.
The basic helix-loop-helix (bHLH) transcription factors compose a superfamily and have been studied comprehensively in eukaryotic organism cells, especially in mammalian cells, in which the genes expression, protein structure, function, and phylogenetic analyses have been thoroughly and comprehensively performed (Ledent and Vervoort, 2001; Heim et al., 2003; Toledo-Ortiz et al., 2003). The research data demonstrate that the bHLH transcription factors are a type of significant regulatory component in gene expressional networks in organisms, involved in a wide variety of processes, such as cell proliferation, differentiation and responses to abiotic stress (Grandori et al., 2000; Massari and Murre, 2000; Yuan et al., 2008; Wu et al., 2012). Wu et al. (2012) reported that AtbHLH38 and AtbHLH39 interact with AtbHLH29 to enhance the Cd tolerance of Arabidopsis seedlings via decreased cadmium transfer from roots to shoots and to improve the iron homeostasis and concentration of shoots. The data also showed that the overexpression of AtbHLH39 alone can improve the capability of Cd tolerance in the seedlings. The transcription factor AtbHLH29 is critical to the regulation of iron uptake (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). Under Fe deficiency, the expression of AtbHLH100 and AtbHLH101 is up-regulated in roots and leaves (Wang et al., 2007). The rice bHLH gene OsIRO2, which is orthologous to AtbHLH38 and AtbHLH39, is an essential regulator of the genes involved in Fe uptake under Fe deficiency conditions (Ogo et al., 2007).
In this study, we investigated the functions of a soybean bHLH transcription factor GmORG3 regarding Cd tolerance and Fe uptake. GmORG3 is an ortholog of AtbHLH39, which is involved in the absorption and translocation of Cd and Fe. The overexpression of GmORG3 in yeast enhanced yeast cell tolerance to Cd, and the overexpression of GmORG3 in soybean roots of mosaic seedlings reduced the translocation ratio of Cd from roots to shoots. GmORG3 transgenic tobacco plants were more tolerant than wild-type plants were under Fe deficiency.
Results
Phylogenetic Analysis of GmbHLH TFs and the Domain Analysis and Subcellular Localization of the GmORG3 Protein
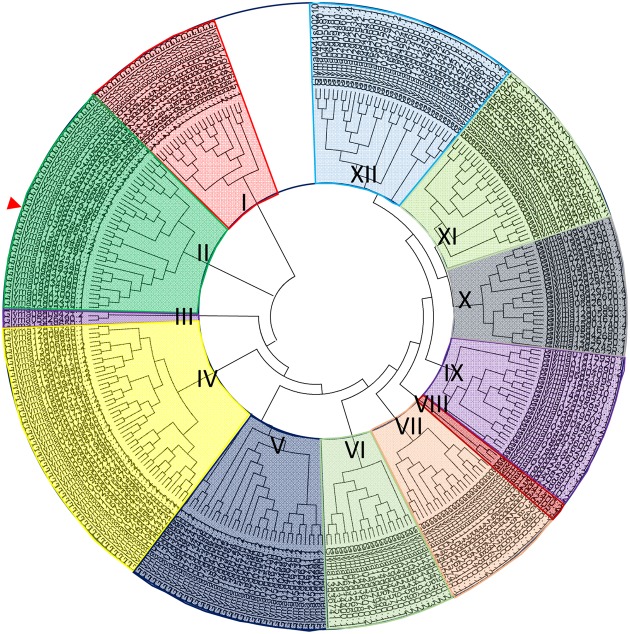
There are 308 bHLH loci in the soybean genome. Amino acid sequences of all GmbHLH genes were downloaded from the database of phytozome2. A comparative tree was constructed based on the amino acid sequence alignments of the 308 bHLH soybean genes (Supplementary Table S1). The clustering showed that there are 12 major subfamilies of related sequences that were strongly supported by bootstrap analysis; these subfamilies were titled I to XII (Figure 1). GmORG3 (Glyma03g28630), the red arrow-marked position, was classified as subfamily “II.” In a previous study, AtbHLH TFs were also divided into 12 subfamilies (Heim et al., 2003); the AtbHLH39, which is the orthologous gene of GmORG3, was classified as subfamily “Ib.” In Arabidopsis AtbHLH TFs were divided into 21 subfamilies in another study (Toledo-Ortiz et al., 2003); the AtbHLH39 was in the “2” subfamily.
FIGURE 1.
Phylogenetic analysis of GmbHLH TFs. Phylogenetic tree containing 308 full-length soybean bHLHs; GmORG3 is indicated by the arrow. Multiple sequence alignment containing the amino acid sequences of 308 bHLH proteins was performed using ClustalX2. The phylogenetic tree was constructed using the neighbor-joining method with MEGA software. The bHLH transcription factors were divided into 12 subfamilies titled I–XII. GmORG3 belonged to subfamily II.
The GmORG3 protein consists of 241 amino acids and contains one 61-residue-long conserved bHLH domain spanning amino acids 58–118 (Figure 2). The basic helix1-loop-helix2 domain sequence is PAMVKKLSHNASERDRR-KKVNDLVSSLRSLLP-GPDQTKKMS-IPATVSRVLKYIPELQH QVQ. Basic and helix domains are more conservative than are loop domains. There are conserved amino acids in the soybean bHLH transcription factors: the 4th residue in the basic part is L (Leucine)/V (Valine); the 3rd, 6th, and 13th residues in the helix1 part are V, L, and L, respectively; and 10th residue in the helix2 part is L. Those conserved amino acids should have important functions.
FIGURE 2.
Domain structure of the GmORG3 subfamily II. Multiple sequence alignment of the bHLH domains of the 32 GmbHLH members of subfamily II. The scheme at the top depicts the locations and boundaries of the basic, helix, and loop regions within the bHLH domain. The shading of the alignment presents identical residues in different color; conserved residues contain more bases. Dots denote gaps.
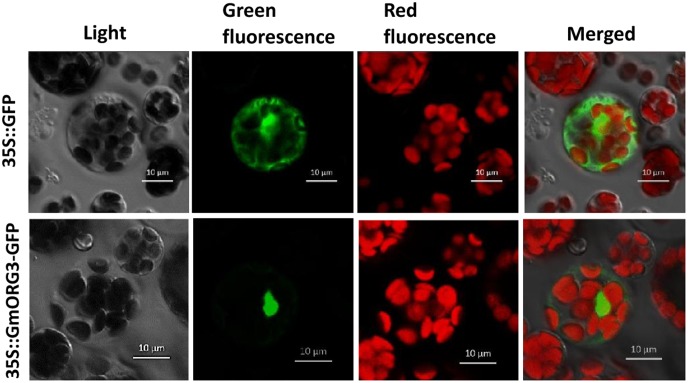
A GmORG3-green fluorescent protein (GFP) fusion construct and a control construct containing only GFP were created using the pJIT166-GFP vector. Transcription was driven by the CaMV 35S promoter in both constructs. Constructs were introduced into Arabidopsis protoplasts in accordance with the PEG4000-mediated method. The cells were inspected with a confocal laser-scanning microscope to visualize the subcellular localization of the proteins. The GmORG3-GFP protein was detected only in the nucleus; the control protein was observed throughout the cell (Figure 3). These results indicated that GmORG3 is a nuclear protein.
FIGURE 3.
Subcellular localization of the GmOGR3 protein in Arabidopsis protoplasts. A construct encoding a GmORG3-green fluorescent protein fusion (35S::GmORG3-GFP) was created using the pJIT166-GFP vector. The 35S::GmORG3-GFP and GFP control plasmid (35S::GFP) were transformed into Arabidopsis protoplasts using PEG4000 and observed using a confocal fluorescence microscope. Nucleic acids were detected using an excitation wavelength of 350 nm and an emission wavelength of 461 nm. The GFP and chloroplast autofluorescence were detected using an excitation wavelength of 480 nm and emission wavelengths of 461 and 685 nm, respectively. Scale bars = 10 μm.
Transcript Abundance of GmORG3
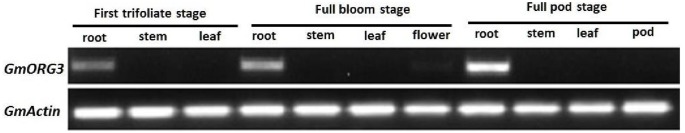
Transcript abundance of GmORG3 was determined in soybean plants. Total RNA was isolated from the roots, stems, leaves, flowers, and pods of soybean at the first-trifoliate, full-bloom, and full-pod stages. RNA was reverse-transcribed to obtain cDNA for semi-quantitative PCR analysis. The results showed that the transcripts of GmORG3 were detected only in the roots at all selected stages, and the highest transcript abundance in the root tissue was detected at the full-pod stage (Figure 4). GmActin was used as an internal control.
FIGURE 4.
Transcript abundance of GmORG3 in the roots, stems, leaves, flowers, and pods of soybean at the first-trifoliate, full-bloom and full-pod stages. The experiments were carried out using semi-quantitative RT-PCR; GmActin was used as an internal control for the normalization of cDNA templates. The PCR was repeated twice, each producing similar results.
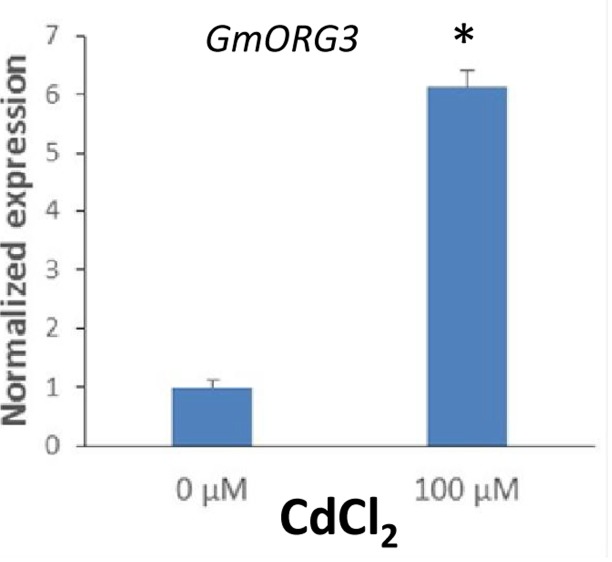
The transcript abundance response of GmORG3 was also detected in soybean plants growing under Cd stress. qRT-PCR analyses were carried out to determine the transcript abundance patterns of the GmORG3 gene in the roots of soybean plants. As shown in Figure 5, the expression of GmORG3 was clearly up-regulated in the roots of soybean seedlings after an overnight treatment with 100 μM CdCl2. The expression level of GmORG3 under CdCl2 was 6.13-fold higher than that under normal conditions.
FIGURE 5.
Transcript abundance of GmORG3 under Cd exposure. Three-week-old seedlings were transferred to 100-ml glass tubes containing Hoagland solution. The tubes were divided into two groups for Cd treatments: one with 0 μM and the other with 100 μM CdCl2. After overnight treatment, the normalized relative transcript abundance of the GmORG3 gene was detected in the roots using real-time quantitative RT-PCR. The y-axis shows RNA levels normalized to those of GmActin. N = 3, and values are the mean ± SD. ∗Significant difference in values at the level of P < 0.05 by Student’s t-test in comparison with those of 0 μM.
Overexpression Analysis of GmORG3 in Yeast under Cd Stress
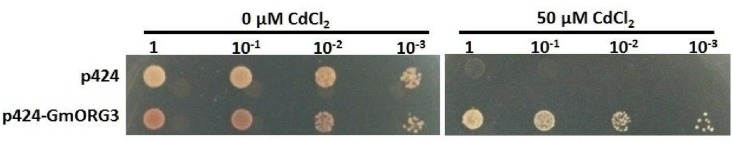
Heterologous expression is also an effective way to detect gene function. GmORG3 was fused to a p424 GAL vector and overexpressed in the yeast strain WB303B. In the control YPDA medium containing 0 μM CdCl2, the growth rate of yeast cells expressing GmORG3 was slightly slower than that of cells containing the empty plasmid (Figure 6). However, the expression of GmORG3 significantly improved the yeast tolerance to CdCl2, while yeast with empty plasmids did not survive on the YPDA medium with 50 μM CdCl2.
FIGURE 6.
Response of GmORG3 to cadmium tolerance in yeast. The GmORG3 gene was fused in frame to the GAL DNA-binding domain expression vector p424 GAL and then transformed into yeast strain W303B. An empty p424 vector was used as a control. The transformants with different diluted concentrations (1, 10-1, 10-2, 10-3) were added to plates containing YPDA medium with either 0 or 50 μM CdCl2.
Overexpression Analysis of GmORG3 in Soybean Mosaic Seedlings Grown under Cd Stress
The function of GmORG3 was investigated by its construction into the plant overexpression vector pCXSN and subsequent transformation to generate soybean mosaic seedlings via Agrobacterium rhizogenes mediation. Soybean mosaic seedlings were tested under Cd stress to determine Cd contents in roots and leaves.
Soybean mosaic seedlings having transgenic roots and non-transgenic shoots were generated in accordance with procedures explained elsewhere. PCR-validated transgenic seedlings of GmORG3 and those with empty vectors were grown in 100-ml glass tubes containing half-strength Hoagland solution for a few weeks to develop enough transgenic roots. Afterward, the mosaic seedlings were transferred to fresh ½-strength Hoagland solution that contained 20 μM CdCl2 and grown for 9 days. The leaves and roots of both seedlings with empty vector and GmORG3 mosaic seedlings were harvested separately at 0, 1, 2, 4, 6, and 9 days after treatment. The Cd content in the samples was determined using an atomic absorption spectrometer with a continuous light source following the manufacturer’s instructions/manual (ContrAA®300, Analytik Jena AG).
Soybean mosaic plants with GmORG3 transgenic hairy roots did not significantly differ from those with empty vector transgenic hairy roots after 9 days of growth under 20 μM CdCl2 stress (Figure 7A). However, the Cd content was lower in GmORG3 transgenic roots than in those of plants with empty vectors during the first 2 days, but this finding was opposite at 4 and 6 days. No Cd was detected immediately after treatment (0 days) (Figure 7Ba). It is interesting that the translocation ratio of Cd from roots to shoots (Cd amount in shoots/total Cd amount) was consistent at every sample collection time. The GmORG3 transgenic root seedlings had a lower ratio at all sample collection times (Figure 7Bc). Therefore, when roots absorb the same amount of Cd, GmORG3 transgenic roots will translocate less Cd to shoots.
FIGURE 7.
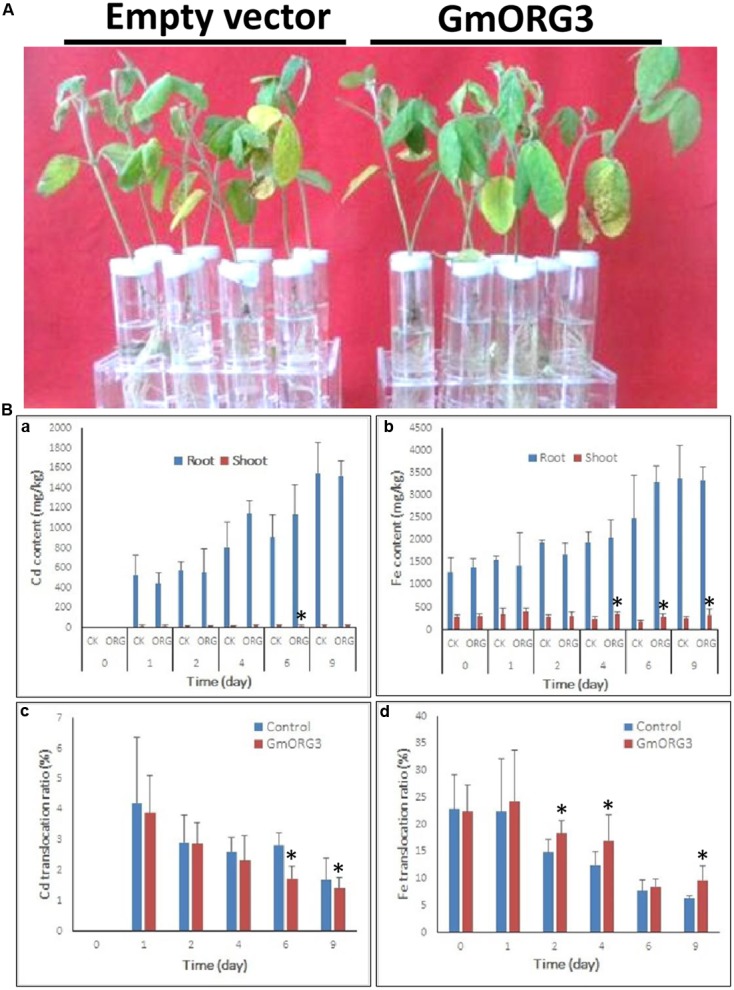
The response of mosaic soybean seedlings under Cd stress. GmORG3 overexpression mosaic soybean seedlings were transferred to one-half-strength Hoagland solution containing 20 μM CdCl2 and grown for 9 days. Cd and Fe contents were determined in root and shoot samples harvested at 0, 1, 2, 4, 6, and 9 days and analyzed for their translocation ratios from roots to shoots. (A) Response of soybean mosaic seedlings carrying an empty vector or a GmORG3 overexpression vector in their hairy roots grown under Cd stress. (Ba), Cd content in the roots or shoots of soybean mosaic seedlings carrying an empty vector of a control check (CK) or a GmORG3 overexpression vector. (Bb) Fe content in the roots or shoots of soybean mosaic seedlings carrying an empty vector of a control check (CK) or a GmORG3 overexpression vector. (Bc) Cd translocation ratio from roots to shoots in soybean mosaic seedlings under Cd stress. (Bd) Fe translocation ratio from roots to shoots in soybean mosaic seedlings under Cd stress. ∗Significant difference in values at the level of P < 0.05 by Student’s t-test in comparison with those of the empty vector of the control check (CK).
Considering that the absorption and transport of Fe is related to transporters that can also take up Cd and other metals, Fe accumulations in the roots and shoots of mosaic soybean seedlings were measured and analyzed (Figure 7Bb,d). The Fe content was higher in the roots than in the shoots, and a subtle increase in Fe uptake was evident with increasing time of Cd stress. GmORG3 showed slightly lower Fe content than did empty vector seedlings during the first 2 days, whereas at 4, 6, and 9 days of treatment, GmORG3 seedlings showed slightly higher or equal Fe content than did those with empty vectors (Figure 7Bc). Additionally, the Fe translocation ratio from roots to shoots (Cd amount in shoots/total Cd amount in plants) was also calculated. Interestingly, the GmORG3 transgenic mosaic seedlings had a higher ratio at all time points (Figure 7Bd).
GmORG3 Transgenic Tobacco Growth under Cd Stress and Fe Deficiency
Since GmORG3 significantly responded to cadmium stress and its overexpression conferred tolerance to Cd stress in yeast, further characterization of this gene was carried out. The heterologous ectopic over expression of GmORG3 in tobacco plants showed enhanced tolerance to cadmium stress. The GmORG3 CDS was used to prepare the construct of pCXSN-GmORG3, which have a 2X 35S promoter and a NOS terminator. In addition, the correct recombinant plasmid was transformed into tobacco by Agrobacterium-mediated leaf disk explants methods. The regenerated transformed seedlings were screened on medium containing hygromycin (100 mg/L). The putative tobacco transformants were identified using GmORG3 gene-specific PCR analysis. The T3 seeds of transgenic plant line 1 (L-1), line 2 (L-2), and line 3 (L-3) were used in subsequent experiments.
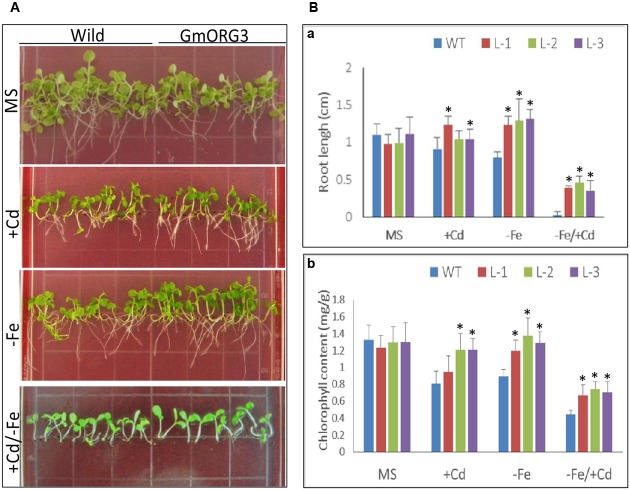
Wild-type (WT) and transgenic tobacco L-1, L-2, and L-3 seeds were sown on 1/2 MS medium (MS), 1/2 MS medium supplemented with 20 μM CdCl2 (MS/+Cd), 1/2 MS medium without Fe (MS/-Fe) and 1/2 MS medium with +Cd/-Fe (MS/+Cd-Fe) (Figure 8). The growth of transgenic seedlings on MS control medium was similar to that of WT seedlings (Figure 8A): the roots of transgenic plants were slightly shorter than those of WT. The root length assays indicated that the average root length of WT plants was 1.10 cm and that of transgenic plants L-1, L-2, and L-3 were 0.98, 0.99, and 1.11 cm, respectively (Figure 8Ba). The leaf color of both WT and transgenic plants was also similar. The content of chlorophyll was 1.30 mg/g of fresh weight (FW) in the leaves of WT and was 1.24, 1.30, and 1.30 mg/g (FW) in the leaves of transgenic lines L-1, L-2, and L-3, respectively (Figure 8Bb).
FIGURE 8.
The root length and chlorophyll content of GmORG3 transgenic tobacco plants under +Cd and/or –Fe stress. T3 GmORG3 transgenic tobacco L-1, L-2, and L-3 seeds were grown on square plastic Petri dishes filled with one-half-strength MS (Murashige and Skoog) medium for 3 weeks. Four media treatments were established: control (MS), MS medium with 20 μM Cd (MS/+Cd), MS medium without Fe (MS/–Fe), and MS medium with 20 μM Cd but without iron (MS/+Cd-Fe). (A,Ba), Root length of transgenic or wild-type tobacco seedlings on four different media treatments. (Bb), Chlorophyll content in fresh seedlings on four different media treatments. ∗Significant difference in values at the level of P < 0.05 by Student’s t-test in comparison with those of the wild-type (WT) seedlings.
The growth of transgenic seedlings on MS medium supplemented with 20 μM CaCl2 showed obvious differences with that of WT. The roots of transgenic seedlings were longer than those of WT; there were significant differences between root lengths of transgenic L-1 and WT seedlings but no significant differences between those of transgenic L-2 and L-3 with WT seedlings (Figure 8Ba). The chlorophyll contents varied significantly between all transgenic lines and WT (Figure 8Bb). On Fe-deficient medium, the root lengths of transgenic seedlings were significantly longer than those of WT, while chlorophyll contents in transgenic lines were higher than those of WT (Figure 8B). The responses of all transgenic and WT seedlings were worse on +Cd/-Fe medium than on other media; all seedlings grown on +Cd/-Fe medium were short. However, the chlorophyll content of all the transgenic lines was significantly (P < 0.05) higher than that of WT. Similarly, the root length of transgenic seedlings was also significantly (P < 0.01) longer than that of WT.
The above results demonstrated that overexpressing GmORG3 improves the tolerance of transgenic tobacco plants to cadmium stress.
Discussion
Basic/helix-loop-helix transcription factors, which are common types of transcription factors, is one of the largest gene families in plants with important biological functions (Pires and Dolan, 2010). In Arabidopsis, 162 bHLH-encoding genes have been identified, which indicate that bHLH transcription factors constitute the second largest transcription factor family after the MYB superfamily, which has 190 members (Riechmann et al., 2000; Dubos et al., 2010). Several studies have reported the existence of this superfamily in other species, such as rice (Oryza sativa L.), which has 180 bHLH transcription factors; tobacco (Nicotiana tabacum L.), which has 190; and grape (Vitis vinifera L.), which has more than 191 (Xiong et al., 2005; Jaillon et al., 2007; Rushton et al., 2008). We isolated 308 bHLH transcription factors in soybean (Figure 1), which may be the largest transcription factor family in the whole genome reported thus far. The members of this superfamily are involved in many biological processes such as stress responses (Osorio et al., 2012), nodule growth and NH4+ transport (Chiasson et al., 2014). In our study, the GmbHLHs were divided into 12 subfamilies (Figure 1); previous studies in other species reported six subfamilies in the animal genomes (Atchley and Fitch, 1997), 21 subfamilies in Arabidopsis (Toledo-Ortiz et al., 2003), and 22 subfamilies in rice (Li et al., 2006). This suggests that the classification of bHLH transcription factors in plants is more complex than in animals, and more research is needed to classify plant bHLH transcription factors.
GmORG3 belongs to the bHLH transcription factor superfamily. GmORG3 contains one 61-amino acid domain that comprises basic-helix1-loop-helix2 domains (Figure 2). It is interesting that the basic and helix domains were more conserved than the loop domain was. The basic region binds with the DNA E-box motif (5′-CANNTG-3′) (Toledo-Ortiz et al., 2003; Li et al., 2006). The helix domain needs stable hydrophobic amino acids such as the position 4 L (Leucine)/V (Valine) in helix1; the position 3 V, 6 L and 13 L in helix2; and the position 10 L in the bHLH structure to form homo- or heterodimers (Ferré-D’Amaré et al., 1993). Therefore, the results of the previous studies and our results suggest that GmORG3 may interact with itself or other GmbHLHs before binding with the E-box in the promoter, but this speculation needs more evidence.
BLASTing the Arabidopsis genome showed that the GmORG3 orthologous gene was the bHLH gene AtbHLH39 (AtORG3), which is expressed during both Fe deficiency stress (Yuan et al., 2008) and Cd stress (Wu et al., 2012). Studies have also shown that AtbHLH39 interacts with AtbHLH29 in the nucleus. The overexpression of AtbHLH39 with AtbHLH29 in Arabidopsis seedlings altered the expression pattern of the Fe uptake genes FRO2 (ferric chelate reductase 2) and IRT1 (iron-regulated transporter 1) from being induced to constitutive and enhanced seedling tolerance to Cd. The determination of the Fe and Cd content showed that co-overexpression of AtbHLH39 and AtbHLH29 increased Cd sequestration in roots and improved Fe homeostasis of shoots (Yuan et al., 2008; Wu et al., 2012). Based on our results, GmORG3 was also localized in the nucleus (Figure 3) and was significantly up-regulated by Cd stress (Figure 5). This suggests that the structure of similar genes may have a similar function. However, GmORG3 was only expressed in the roots during all growth stages (Figure 4). This may suggest that GmORG3 is important in the translocation of metals from the roots to shoots. The overexpression of GmORG3 in yeast W303B cells showed improved growth compared with that of the control on CdCl2-supplemented medium (Figure 6), which suggests a similar function of GmORG3 in plants. In the present study, we chose tobacco plants as the heterologous expression material for our phytoremediation study because of its fast growth, high biomass productivity without special care, and overall significance (Evangelou et al., 2007). GmORG3 transgenic tobacco lines were developed using Agrobacterium-mediated transformation. The T3 seeds of tobacco lines L-1, L-2, and L-3 were sown on MS medium with 20 μM CdCl2 and subjected to exogenous Cd stress. The transgenic tobacco plants showed enhanced Cd tolerance via significant increases in growth, root length, and chlorophyll content compared to those of wild-type controls (Figure 8). The Cd detoxification in plants is a very intricate proocess, involving a number of different physiological and molecular mechanisms, such as reduced uptaking from soil, binding in cell wall, enrichment in the epidermal hairs, chelation with phytochelatins and reduced translocation from roots to shoots. Plant phenotypes exhibiting tolerance usually cannot be explained simply by a single mechanism. Detoxification usually is a result of multiple mechanisms joint action. Several studies have indicated that Cd stress can hinder chlorophyll biosynthesis (Küpper et al., 2007; Redondo-Gómez et al., 2009; Rai et al., 2013; Çelekli et al., 2015; Li et al., 2015; Cheng et al., 2016). In the present study, GmORG3 reduced the damage of Cd stress in the transgenic tobacco plants and increased the chlorophyll content. This suggests that GmORG3 might have increased the stability of the photosynthesis system. However, the mechanism by which GmORG3 stabilizes the photosynthetic system is not clear; here, the mosaic soybean seedling experiments could provide the results (Figure 7). In conclusion, GmORG3 exhibited lower Cd translocation ratios from roots to shoots in mosaic soybean seedlings, and the results were in accordance with those of Wu et al. (2012). Recently, a major soybean QTL (quantitative trait locus) associated with low Cd concentrations in seeds was identified (Jegadeesan et al., 2010). This QTL accounted for 57.3% of the phenotypic variation. In this QTL, a gene for the plasma membrane H+-ATPase was found to be located at the locus, and this gene may be the key gene Cda1 (low Cd accumulation). Therefore, Cda1 could be the target gene of GmORG3. Future experiments should study the interaction between those two genes.
Materials and Methods
Plant Materials and Growth Conditions
Plants of the Chinese cultivar Dongnong 690 of soybean (Glycine max L. Merr.), the Columbia ecotype of Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum L.) were used in this study. Dongnong 690 plants was sown in 20-cm pots filled with a 1:1 mixture of vermiculite and nutrient-rich soil that were placed in the greenhouse. Arabidopsis seeds were sown in small pots containing vermiculite with sufficient water; the pots were covered with plastic wrap and placed into an incubator. Photoperiod and temperature were 12 h of light at 22°C and 12 h of darkness at 20°C. Seeds of tobacco and transgenic lines were sterilized with 10% NaClO for 10 min, then washed more than three times with double distilled water. After vernalization at 4°C in the dark for 2 days, the seeds were sown onto plates with 1/2 MS medium at pH 5.8. The plates were incubated at 22°C under a 12-h photoperiod. All the seedlings were then used for further analysis.
Sequence and Domain Analysis of GmORG3
Bioinformatics analyses were performed by various web-based software programs. The amino acid sequences of the soybean bHLH transcription factors including GmORG3 were downloaded from the Phytozome database2 and aligned using Clustal Omega web software3. Other bioinformatics software programs utilized included MEGA 5 and PowerPoint 10 for general sequence homology analysis, drawing of figures, domain analyses and the prediction of similarity and identity among different members of the subfamilies.
Semi-quantitative RT-PCR and Quantitative PCR
Total RNA was isolated from the tissues of roots, stems, leaves, flowers, and pods of Dongnong 690 at the first-trifoliate, full-bloom and full-pod stages using an SV total RNA kit (Promega, United States). RNA samples were quantified using a Nanodrop spectrophotometer (United States). cDNA was synthesized from 5 μg of total RNA with a SuperScript RT III first-strand cDNA synthesis kit (Invitrogen, San Diego, CA, United States) according to the manufacturer’s instructions. The synthesized cDNA was used as PCR template. GmActin was used as an internal control for semi-quantitative RT-PCR analysis. The GmORG3-specific primer pair, ORG3-F (5′-CGAGATCATCATAGCATTATCT-3′) and ORG3-R (5′-CTCCATTCTAATTTCCGAACT-3′), was utilized for expression analysis (semi-qPCR). The GmActin (accession number: NM_001250673) primer pair, GmActin-F (5′-CGGTGGTTCTATCTTGGCATC-3′) and GmActin-R (5′-GTCTTTCGCTTCAATAACCCTA-3′) was used as an internal control. PCR was carried out in 1x PCR buffer supplemented with dNTPs at 150 μM, 1.5 U of Taq DNA polymerase and 5 pmol of each gene-specific primer according to the following program: initial denaturation at 95°C for 5 min; 27 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s; and a final extension step at 72°C for 5 min. Semi-quantitative RT-PCR experiments were repeated three times. The amplification products were separated via agarose gel electrophoresis and analyzed using a fluorescence imaging system (Peiqing CN).
Total RNA from the roots of 3-week-old soybean seedlings grown under control and 50 μM CdCl2 stress conditions for 12 h was extracted, reverse-transcribed and used for real-time quantitative PCR analysis. Sample preparation and PCR analysis were performed using the SYBR Premix Ex Taq in a Roche Light Cycler 2.0 with Light Cycler software (build 4.1.1.21) (LightCycler Carousel-based system, F. Hoffmann-La Roche Ltd, Germany). A 10-μl reaction consisted of 5 μl of SYBR Premix Ex Taq, 0.8 μl of each primer (forward and reverse, 10 mM), 1 μl of template and 2.4 μl of ddH2O. The GmORG3-specific primer pair, qORG-F (5′-TCGTTCACTTCTTCCTGGGC-3′) and qORG-R (5′-ATAGTGGAGCCTTGTGAGCC-3′), was utilized for its expression study under control and Cd stress conditions. Forward (5′-CGGTGGTTCTATCTTGGCATC-3′) and reverse (5′-GTCTTTCGCTTCAATAACCCTA-3′) primers of the soybean GmActin gene were used as an internal control for the expression analysis of GmORG3. An equal amount of cDNA template was used for each sample reaction, including the internal control. qPCR amplification conditions included an initial denaturation for 30 s at 95°C followed by 35 quantification cycles consisting of denaturation for 5 s at 94°C, annealing for 10 s at 58°C, and extension for 30 s at 72°C. A melting curve analysis was then performed to confirm the specificity of the PCR products. The real-time quantitative PCR analysis was repeated in three independent experiments.
GmORG3 Mosaic Soybean Seedlings
An Agrobacterium rhizogenes-mediated soybean genetic transformation system was used to introduce the GmORG3 overexpression cassette into the hairy roots of soybean variety Dongnong 690. Soybean seeds were sown into pots containing vermiculite and nutrient-rich soil in a 1:1 ratio. Upon the emergence of cotyledons, healthy and robust seedlings were selected for transformation. Agrobacteria carrying the GmORG3 overexpression cassette/plant expression vector pCXSN-GmOGR3 were streaked onto an LB plate containing kanamycin and precultured at 28°C for 2 days. A single colony was used to inoculate liquid LB medium that contained kanamycin as a selectable marker. The culture was grown at 28°C for 12 h with shaking at 200 rpm. The culture was centrifuged at 4500 rpm for two min at room temperature to harvest the bacteria. The pellet was twice suspended gently in a 10 mM MgCl2 solution. The OD600 of the final bacterial suspension was adjusted to 0.4–0.6. Young seedlings with unfolded cotyledons were infected at the cotyledonary node and/or the hypocotyl with Agrobacterium rhizogenes carrying the pCXSN-GmORG3 construct. The injection point was covered immediately after inoculation with a wet vermiculite/soil mixture. The infection sites were kept in a high-humidity environment. Water was applied as required. The seedlings were grown in a greenhouse at 25°C with a 12-h photoperiod and approximately 60% relative humidity. Hairy roots were induced at the injection point after 1 week; the roots grew to 2–3 cm in length 1 week after inoculation. As such, the hairy roots developed 3 weeks after sowing. The main roots were removed when the emerged hairy roots could support the plants. Mosaic seedlings with one hairy root were transferred to 100-ml glass tubes containing ½-strength Hoagland solution. The glass tubes containing mosaic soybean seedlings were placed in a growth chamber with 25/22°C day/night temperature and a 12-h photoperiod for approximately 1 week. The Hoagland solution in the glass tubes was replaced every other day. Once the hairy roots reached bottom of the tube, the transformation of hairy roots was verified using PCR analysis. PCR was performed using a CaMV35S promoter-specific forward primer (5′-CAATCCCACTATCCTTCGCAAGACC-3′) and a GmORG3 gene-specific reverse primer. Genomic DNA from the hairy roots was used as a template. One set of soybean mosaic seedlings carrying a pCXSN empty vector was prepared for use as a control. Transgenic hairy roots of mosaic seedlings were immersed in ½-strength Hoagland solution containing 20 μM CdCl2 and grown for 9 days in a growth chamber. The Hoagland solution containing Cd was replaced every other day. At 0, 1, 2, 4, 6, and 9 days, roots and shoots tissues were harvested for analysis of Cd and Fe content. The experiment was repeated three times.
GmORG3-Transformed Tobacco
For the heterologous expression of the GmORG3 in tobacco, A. tumefaciens strain EHA105 containing the pCXSN:2xCaMV35S::GmORG3-NOS construct was used to get transgenic tobacco plant according to a reported protocol, with a little modification. The hygromycin were used as selection maker. The positive transformed tobacco seedlings were grown in a contained net house. When the plantlets were large enough, DNA was extracted from the leaves, and positive plantlets were screened using PCR. T1 seeds were then harvested, and selection continued until the T3-generation seeds were collected and cultured for further study.
Subcellular Localization
Arabidopsis protoplasts were isolated as previously described (Yoo et al., 2007). The CDS of GmORG3 was inserted into the pJIT166-GFP vector without a termination codon to create an in-frame fusion between GmORG3 and the green fluorescent protein. The fusion construct (p35S::GmORG3-GFP) and a control construct (p35S::GFP) were introduced into Arabidopsis protoplasts using the PEG4000 method as described by Abel and Theologis (1994). After the incubation of transformed Arabidopsis protoplasts in culture solution for 18–24 h without light at room temperature, imaging was performed using a confocal microscope (Zeiss, LSM510 Meta, Carl Zeiss AG) and a 40X lens. The fluorescence of GFP was measured at 495–545 nm, and chloroplast autofluorescence was measured at 480–685 nm.
Determination of Cd, Fe, and Chlorophyll Contents
Regarding the experiments to determine the function of GmORG3 under Cd toxicity stress, mosaic soybean seedlings were generated following the procedure described in the section “Development and treatment of GmORG3 mosaic soybean seedlings” above. Root and shoot tissue was harvested, and the Cd and Fe contents of the samples were then determined. Samples were kept at 105°C for 20 min and dried at 80°C to constant weight. Dry tissues were ground into a powder, and 1 ml of 100 mM acetic acid was added. Samples were incubated at 90°C for 6 h. Cadmium and iron contents were then determined using an atomic absorption spectrometer with a continuous light source (ContrAA®300, Analytik Jena AG). The analysis was repeated three times.
Tobacco seeds of the GmORG3 transgenic lines and wild-type plants were grown on square plastic Petri dishes filled with 1/2 MS medium with 20 μM CdCl2, without Fe, and 1/2 MS medium supplemented with 20 μM Cd but without Fe. The dishes were placed into a growth chamber. Three weeks later, the seedlings were imaged, chlorophyll contents were determined, root lengths were measured and samples were harvested. The estimation of chlorophyll from seedlings was performed in accordance with the method of Arnon (1949): 100 mg of tissue was homogenized thoroughly in 1 ml of 80% acetone and centrifuged at 3000 rpm for 2–3 min. The supernatant was retained, and absorbance was measured using a spectrophotometer at 663 and 645 nm. Total chlorophyll was estimated using the following equation: total chlorophyll = [(20.2 × A645 + 8.02 × A663)/(1000 × W)] × V. The experiment was repeated three times.
Author Contributions
ZX, XL, HM, and HS conceived, designed and conducted the experiments. XH, LX, YH, DZ, and BT analyzed the data and results. ZX wrote the manuscript. HS and HM monitored the experiments and critically commented on the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was sponsored by the project of the National Science Foundation of Jiangsu Province (BK20130727), the Project of Jiangsu Key Laboratory for Bioresources of Saline Soils (JKLBS2014002), the National Science Foundation of China (31101166), the One Hundred Talent Plan of Foreign Experts of Jiangsu Province (JSB2015005), the Autonomous Innovation Project of Jiangsu Agriculture Science & Technology [CX(15)1005] and the Shuangchuang Talent Plan of Jiangsu Province, China.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01098/full#supplementary-material
A comparative tree based on the amino acid sequence alignments of the 308 bHLH soybean genes.
References
- Abel S., Theologis A. (1994). Transient transformation of Arabidopsis leaf protoplasts, a versatile experimental system to study gene expression. Plant J. 5 421–427. 10.1111/j.1365-313X.1994.00421.x [DOI] [PubMed] [Google Scholar]
- Arnon D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley W. R., Fitch W. M. (1997). A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 94 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaves K., Wu T. Y., Gruissem W., Bhullar N. K. (2017). Enhanced grain iron levels in rice expressing an iron-regulated metal transporter, nicotianamine synthase and ferritin gene cassette. Front. Plant Sci. 8:130 10.3389/fpls.2017.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çelekli A., Gültekin E., Bozkurt H. (2015). Morphological and biochemical responses of spirogyra setiformis exposed to cadmium. CLEAN Soil Air Water 44 256–262. [Google Scholar]
- Cheng J., Qiu H., Chang Z., Jiang Z., Yin W. (2016). The effect of cadmium on the growth and antioxidant response for freshwater algae Chlorella vulgaris. Springerplus 5 1290 10.1186/s40064-016-2963-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiasson D. M., Loughlin P. C., Mazurkiewicz D., Mohammadidehcheshmeh M., Fedorova E. E., Okamoto M., et al. (2014). Soybean SAT1 (Symbiotic Ammonium Transporter 1) encodes a bHLH transcription factor involved in nodule growth and NH4+ transport. Proc. Natl. Acad. Sci. U.S.A. 111 4814–4819. 10.1073/pnas.1312801111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S. (2006). Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88 1707–1719. 10.1016/j.biochi.2006.07.003 [DOI] [PubMed] [Google Scholar]
- Colangelo E. P., Guerinot M. L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P., Samantaray S., Rout G. R. (1997). Studies on cadmium toxicity in plants: a review. Environ. Pollut. 98 29–36. 10.1016/S0269-7491(97)00110-3 [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Evangelou M. W., Ebel M., Schaeffer A. (2007). Chelate assisted phytoextraction of heavy metals from soil. Effect, mechanism, toxicity, and fate of chelating agents. Chemosphere 68 989–1003. [DOI] [PubMed] [Google Scholar]
- Ferré-D’Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. (1993). Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363 38–45. 10.1038/363038a0 [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley S. M., James L. P., Eisenman R. N. (2000). The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16 653–699. 10.1146/annurev.cellbio.16.1.653 [DOI] [PubMed] [Google Scholar]
- Hansen N. C., Schmitt M. A., Anderson J. E., Strock J. S. (2003). Iron deficiency of soybean in the upper Midwest and associated soil properties. Agron. J. 95 1595–1601. 10.2134/agronj2003.1595 [DOI] [Google Scholar]
- Havlin J. L., Beaton J. D., Tisdale S. L., Nelson W. L. (2005). Soil Fertility and Fertilizers: An Introduction to Nutrient Management, Vol. 515 Upper Saddle River, NJ: Pearson Prentice Hall [Google Scholar]
- Heim M. A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P. C. (2003). The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20 735–747. 10.1093/molbev/msg088 [DOI] [PubMed] [Google Scholar]
- Huang H., Yu N., Wang L., Gupta D. K., He Z., Wang K., et al. (2011). The phytoremediation potential of bioenergy crop Ricinus communis for DDTs and cadmium co-contaminated soil. Bioresour. Technol. 102 11034–11038. 10.1016/j.biortech.2011.09.067 [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J. M., Noel B., Policriti A., Clepet C., Casagrande A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–467. 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Wang H. Y., Reidt W., Weisshaar B., Bauer P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577 528–534. 10.1016/j.febslet.2004.10.062 [DOI] [PubMed] [Google Scholar]
- Jegadeesan S., Yu K., Poysa V., Gawalko E., Morrison M. J., Shi C., et al. (2010). Mapping and validation of simple sequence repeat markers linked to a major gene controlling seed cadmium accumulation in soybean [Glycine max (L.) Merr]. Theor. Appl. Genet. 121 283–294. 10.1007/s00122-010-1309-6 [DOI] [PubMed] [Google Scholar]
- Kahle H. (1993). Response of roots of trees to heavy metals. Environ. Exp. Bot. 33 99–119. 10.1016/0098-8472(93)90059-O [DOI] [Google Scholar]
- Küpper H., Parameswaran A., Leitenmaier B., Trtilek M., Šetlík I. (2007). Cadmium-induced inhibition of photosynthesis and long-term acclimation to cadmium stress in the hyperaccumulator Thlaspi caerulescens. New Phytol. 175 655–674. 10.1111/j.1469-8137.2007.02139.x [DOI] [PubMed] [Google Scholar]
- Larue C., Korboulewsky N., Wang R., Mévy J. P. (2010). Depollution potential of three macrophytes: exudated, wall-bound and intracellular peroxidase activities plus intracellular phenol concentrations. Bioresour. Technol. 101 7951–7957. 10.1016/j.biortech.2010.05.010 [DOI] [PubMed] [Google Scholar]
- Ledent V., Vervoort M. (2001). The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11 754–770. 10.1101/gr.177001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yang W., Yang T., Chen Y., Ni W. (2015). Effects of cadmium stress on leaf chlorophyll fluorescence and photosynthesis of Elsholtzia argyi—a cadmium accumulating plant. Int. J. Phytoremediation 17 85–92. 10.1080/15226514.2013.828020 [DOI] [PubMed] [Google Scholar]
- Li X., Duan X., Jiang H., Sun Y., Tang Y., Yuan Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141 1167–1184. 10.1104/pp.106.080580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán A. F., Grusak M. A., Abadía A., Abadía J. (2013). Iron deficiency in plants: an insight from proteomic approaches. Front. Plant Sci. 4:254 10.3389/fpls.2013.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M. E., Murre C. (2000). Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 20 429–440. 10.1128/MCB.20.2.429-440.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y., Nakanishi Itai R., Nakanishi H., Kobayashi T., Takahashi M., Mori S., et al. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51 366–377. 10.1111/j.1365-313X.2007.03149.x [DOI] [PubMed] [Google Scholar]
- Osorio M. B., Bücker-Neto L., Castilhos G., Turchetto-Zolet A. C., Wiebke-Strohm B., Bodanese-Zanettini M. H., et al. (2012). Identification and in silico characterization of soybean trihelix-GT and bHLH transcription factors involved in stress responses. Genet. Mol. Biol. 35 233–246. 10.1590/s1415-47572012000200005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro C. A., Santos M. S., Ferreira S. M., Gonçalves S. C. (2013). The influence of cadmium contamination and salinity on the survival, growth and phytoremediation capacity of the saltmarsh plant Salicornia ramosissima. Mar. Environ. Res. 92 197–205. 10.1016/j.marenvres.2013.09.018 [DOI] [PubMed] [Google Scholar]
- Pietrini F., Iori V., Bianconi D., Mughini G., Massacci A., Zacchini M. (2015). Assessment of physiological and biochemical responses, metal tolerance and accumulation in two eucalypt hybrid clones for phytoremediation of cadmium-contaminated waters. J. Environ. Manage. 162 221–231. 10.1016/j.jenvman.2015.07.053 [DOI] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27 862–874. 10.1093/molbev/msp288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga A. P., Abreu C. A., Melo L. C. A., Beesley L. (2015). Biochar application to a contaminated soil reduces the availability and plant uptake of zinc, lead and cadmium. J. Environ. Manage. 159 86–93. 10.1016/j.jenvman.2015.05.036 [DOI] [PubMed] [Google Scholar]
- Rai U. N., Singh N. K., Upadhyay A. K., Verma S. (2013). Chromate tolerance and accumulation in Chlorella vulgaris L.: role of antioxidant enzymes and biochemical changes in detoxification of metals. Bioresour. Technol. 136 604–609. 10.1016/j.biortech.2013.03.043 [DOI] [PubMed] [Google Scholar]
- Redondo-Gómez S., Cantos M., Mateos-Naranjo E., Figueroa M. E., Troncoso A. (2009). Heavy metals and trace element concentrations in intertidal soils of four estuaries of SW Iberian Peninsula. Soil Sediment Contam. 18 320–327. [Google Scholar]
- Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C. Z., Keddie J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110. 10.1126/science.290.5499.2105 [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Bokowiec M. T., Han S., Zhang H., Brannock J. F., Chen X., et al. (2008). Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 147 280–295. 10.1104/pp.107.114041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Y., Klatte M., Jakoby M., Bäumlein H., Weisshaar B., Bauer P. (2007). Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226 897–908. 10.1007/s00425-007-0535-x [DOI] [PubMed] [Google Scholar]
- WHO (2016). Iron Deficiency Anaemia. Available at: http://www.who.int/nutrition/topics/ida/en/ [accessed August 29 2016]. [Google Scholar]
- Wu H., Chen C., Du J., Liu H., Cui Y., Zhang Y., et al. (2012). Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol 158 790–800. 10.1104/pp.111.190983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W. Y., Huang Q., Li G., Rensing C., Zhu Y. G. (2013). Cadmium accumulation in the rootless macrophyte Wolffia globosa and its potential for phytoremediation. Int. J. Phytoremediation 15 385–397. 10.1080/15226514.2012.702809 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Liu T., Tian C., Sun S., Li J., Chen M. (2005). Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol. Biol. 59 191–203. 10.1007/s11103-005-6503-6 [DOI] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H., Sheen J. (2007). Arabidopsis mesophyll protoplasts, a versatile cell system for transient gene expression analysis. Nat Protocols 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Wu H., Wang N., Li J., Zhao W., Du J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18 385–397. 10.1038/cr.2008.26 [DOI] [PubMed] [Google Scholar]
- Yuan Y. X., Zhang J., Wang D. W., Ling H. Q. (2005). AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 15 613–621. 10.1038/sj.cr.7290331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A comparative tree based on the amino acid sequence alignments of the 308 bHLH soybean genes.